This study provides a unique insight into the effects of food safety interventions implemented in one sector of the food industry on the transmission routes of a major foodborne agent. Following the implementation of food safety interventions by the poultry industry, shifts in the molecular epidemiology of Campylobacter jejuni infections in a sentinel region of New Zealand were observed. Targeted interventions to reduce disease incidence are effective but require continued surveillance and analysis to indicate where further interventions may be beneficial.
KEYWORDS: Campylobacter, chicken meat, foodborne diseases, ruminants
ABSTRACT
In 2006, New Zealand had the highest notification rate of campylobacteriosis in the world, and poultry was considered the leading source of campylobacteriosis. Implementation of food safety interventions by the poultry industry led to a decrease in the campylobacteriosis notification rate. The aim is to examine the impact of targeted food safety interventions implemented by the New Zealand poultry industry on the source attribution of Campylobacter jejuni infections in a sentinel region. Campylobacter jejuni isolates collected from the Manawatu region of New Zealand between 2005 and 2007 (“before intervention”) and 2008 and 2015 (“after intervention”) from human clinical cases, chicken meat, ruminant feces, environmental water, and wild bird sources were subtyped by multilocus sequence typing. Viable counts of Campylobacter spp. from carcasses were analyzed using a zero-inflated Poisson regression model. In the period before intervention, sequence type 474 (ST-474) was the most common sequence type (ST) recovered from human cases, accounting for 28.2% of the isolates. After intervention, the proportion of human cases positive for ST-474 reduced to 9.3%. Modeling indicated that chicken meat, primarily from one supplier, was the main source of C. jejuni infection in the Manawatu region before intervention. However, after intervention poultry collectively had a similar attribution to ruminants, but more human cases were attributed to ruminants than any single chicken supplier. Viable counts on carcasses were lower in all poultry suppliers after intervention. This study provides evidence of changes in the source attribution of campylobacteriosis following targeted food safety interventions in one sector of the food supply chain.
IMPORTANCE This study provides a unique insight into the effects of food safety interventions implemented in one sector of the food industry on the transmission routes of a major foodborne agent. Following the implementation of food safety interventions by the poultry industry, shifts in the molecular epidemiology of Campylobacter jejuni infections in a sentinel region of New Zealand were observed. Targeted interventions to reduce disease incidence are effective but require continued surveillance and analysis to indicate where further interventions may be beneficial.
INTRODUCTION
Farm-to-fork monitoring of foodborne pathogens helps to improve and maintain the safety of food supply chains. Campylobacteriosis is a significant burden on health care systems due to its high incidence. Campylobacter jejuni is the predominant species isolated from human campylobacteriosis cases in New Zealand and worldwide (1–3). Most research on campylobacteriosis has focused on C. jejuni not only due to its high prevalence in cases of acute diarrhea but also for its association with serious sequelae, such as Guillain-Barré and Miller Fisher syndromes (4–6).
Contaminated chicken and red meat, environmental water, milk, and contact with pets and farm animals are the most studied sources of human campylobacteriosis (7, 8). To assess the contribution of different infection sources to the burden of C. jejuni infection, DNA subtyping by multilocus sequence typing (MLST) is widely applied (9). Several studies, worldwide (10–13), indicated the consumption of chicken meat as the main risk factor for C. jejuni infections. This finding is supported by the high level of contamination with C. jejuni often found in chicken meat (14, 15). However, other infection sources cannot be discounted. For instance, in Finland, bovines and poultry are identified as equally important sources of C. jejuni infections (16).
A case-control study performed in the 1990s in New Zealand indicated the odds of campylobacteriosis were directly and strongly correlated with recent consumption of raw or undercooked chicken and with chicken eaten in restaurants (17). In 2006, New Zealand had the highest rate of campylobacteriosis notification in the world, with >380 cases per 100,000 population (18). Poultry was estimated to be the leading source of campylobacteriosis, associated with 58% to 76% of cases, followed by the ruminant source (20% to 30%) (19). This triggered the New Zealand Food Safety Authority to announce the implementation of a Campylobacter Risk Management Strategy in collaboration with the poultry industry, starting in April 2008 (20). These interventions (21) eventually led to a 60% decrease in the campylobacteriosis notification rate (22). Preliminary data in the Manawatu region of New Zealand, the sentinel site for campylobacteriosis (23), showed that the decline in cases in the first 2 years of the postintervention period was associated with a decline in poultry-associated cases, which was accompanied by a relative increase in the contribution of other sources (24).
The aim of the current study was to compare the source attributions of campylobacteriosis caused by C. jejuni during two time periods, namely, before the implementation of the Campylobacter Risk Management Strategy (“before intervention” [2005 to 2007]) and an extended period “after intervention” (2008 to 2015). The Campylobacter microbial loads on chicken carcasses in the two time periods were also compared.
RESULTS
Description of the samples.
A total of 7,951 samples were collected from humans, chickens, ruminants, environmental water, and wild bird sources between 2005 and 2015 and were analyzed for Campylobacter sp. by culture. A comparison between samples collected before intervention and after intervention is shown in Table 1. The predominant species identified in both periods, among all sources, was C. jejuni, which accounted for >95% of human clinical isolates, followed by Campylobacter coli, accounting for a maximum of 12% in chicken samples.
TABLE 1.
Characteristics of samples collected before intervention and after interventiona
| Source | No. of collected samples | No. of Campylobacter sp.-positive samples | No. of C. jejuni isolates | No. of C. coli isolates | No. of isolates not further processedb | No. of C. jejuni isolates typed by MLST |
|---|---|---|---|---|---|---|
| Human | 774/1,525 | 670/1,185 | 652/1,128 | 17/38 | 1/19 | 652/1,040 |
| Chicken | 480/963 | 376/750 | 331/597 | 37/90 | 8/63 | 330/507 |
| Ruminant | 1,058/1,135 | 347/503 | 236/319 | 26/41 | 85/143 | 221/244 |
| Environmental water | 308/270 | 140/77 | 77/40 | 6/6 | 57/31 | 76/29 |
| Wild bird | 77/1,361 | 24/682 | 21/354 | 0/1 | 3/327 | 21/193 |
| Total | 2,697/5,254 | 1,557/3,197 | 1,317/2,438 | 86/176 | 154/583 | 1,300/2,013 |
Before intervention and after intervention are separated by a slash throughout the table.
Campylobacter species culture-positive isolates were not further identified if the isolates were PCR negative for C. jejuni and C. coli.
C. jejuni genetic relatedness.
A total of 3,313 C. jejuni isolates from all sources were typed by MLST (see Tables S1 and S2 in the supplemental material). Before intervention, sequence type 474 (ST-474) was the predominant ST, accounting for 17.2% of the 112 STs, followed by ST-45, ST-50, and ST-48, accounting for 11.9%, 8.0%, and 6.5% of the STs, respectively. After intervention, the proportions of the major STs (ST-45, ST-50, and ST-48) did not change significantly, except for ST-474, which dropped from 17.2% to 6.6%. ST-474 declined among the human clinical isolates from 28.2% (184/652) to 9.3% (97/1,040) and among the chicken meat isolates from 9.7% (32/330) to 5.3% (27/507). There was no strong variation in the frequency of most of the clonal complexes (CCs) between the two periods, except for CC ST-48, which decreased from 27.0% to 14.9%, and CC ST-45, which increased from 15.0% to 18.3%. The decrease in frequency of CC ST-48 and the increase of CC ST-45 were significant (Fisher’s two-tailed exact test, P < 0.05). Tables S1 and S2 in the supplemental material show the frequencies of all the C. jejuni CCs and STs in the different sources.
Figure 1 shows the rarefaction curves of C. jejuni STs from different sources before intervention (Fig. 1a) and after intervention (Fig. 1b). In both periods, human C. jejuni had the greatest number of STs. Rarefaction curves for the environmental water source before intervention and wild bird STs after intervention are steep, but the curves for chicken and ruminant STs appear to approach the asymptote in both periods, indicating a more exhaustive sampling of STs than that in the environmental water and wild bird populations.
FIG 1.
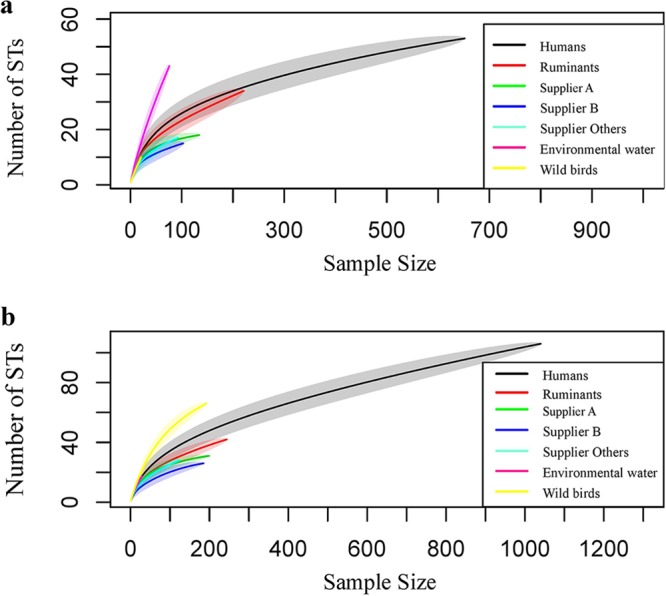
Rarefaction curves before intervention (a) and after intervention (b) of the humans, chicken suppliers (A, B, and “others”), ruminants, environmental water, and wild bird C. jejuni STs. The shaded area represents the 95% CrI. In panel a, only ruminants’ upper boundary of the 95% CrI reach the point estimate of the human curve at maximum sample size. The wild bird curve overlaps the human curve, and the environmental water curve is steep. In panel b, none of the sources’ upper boundaries of the 95% CrI reach the point estimate of the human curve at maximum sample size.
C. jejuni population genetic structure.
Minimum spanning trees visualizing the STs before intervention and after intervention are shown in Fig. S1 and S2, respectively, in the supplemental material. Most of the highly abundant STs in both periods occurred in at least three sources (humans, chickens, and ruminants), except for ST-48, which occurred only in human clinical cases and chicken. STs from environmental water isolates (Fig. S1) and wild bird isolates (Fig. S2) generally appeared to be dissociated from the other sources.
Campylobacteriosis source attribution.
Before intervention, the proportional similarity index (PSI) between supplier A and human C. jejuni was significantly higher than that between C. jejuni from other sources and humans (Table 2). Supplier B, supplier “others,” and ruminant sources shared almost similar PSI results, and their PSIs were significantly greater than the PSI between isolates from environmental water or wild bird sources and human isolates. However, after intervention, all isolates from chicken suppliers and ruminant sources had very similar PSIs with human clinical isolates, but these remained significantly greater than the PSI between isolates from human and environmental water or wild bird sources. There was a significant increase, from the before intervention to the after intervention period, in the PSI between isolates from human and supplier B, supplier “others,” and ruminant sources, with a decrease in the PSI between human and supplier A isolates from 0.61 to 0.49.
TABLE 2.
Multilocus sequence type proportional similarity indices between different C. jejuni sources and human clinical isolates before intervention and after intervention
| Source | PSI (95% CrI) |
|
|---|---|---|
| Before intervention | After intervention | |
| Supplier A | 0.61 (0.51–0.66) | 0.49 (0.42–0.52) |
| Supplier B | 0.34 (0.27–0.38) | 0.48 (0.40–0.51) |
| Supplier “others” | 0.35 (0.27–0.39) | 0.49 (0.39–0.52) |
| Ruminants | 0.37 (0.32–0.41) | 0.52 (0.46–0.56) |
| Environmental water | 0.16 (0.07–0.21) | 0.18 (0.06–0.19) |
| Wild birds | 0.10 (0.07–0.12) | 0.22 (0.17–0.24) |
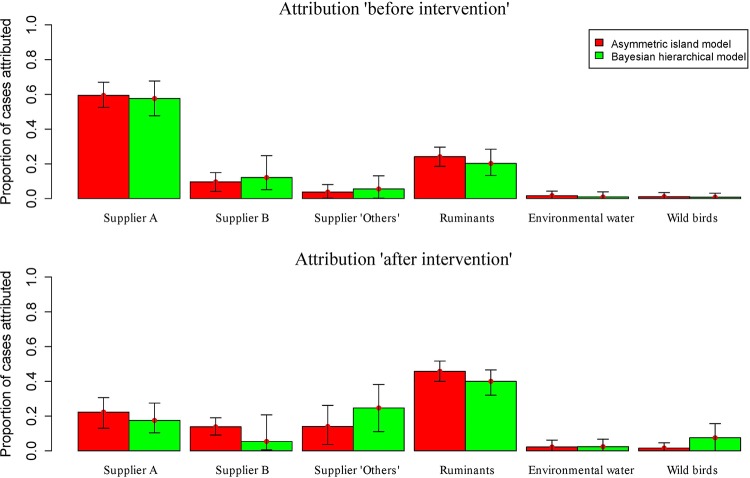
The asymmetric island and Bayesian hierarchical models showed similar attribution estimates (Fig. 2), attributing 59% (95% credible interval [CrI], 52.5% to 67%) and 57% (95% CrI, 47.7% to 67.7%) of human infections, respectively, to supplier A, before intervention. In this period, ruminant sources were the second most common source of infection attributed by the asymmetric island and Bayesian hierarchical models, with attributable values of 24.1% (95% CrI, 18.5% to 29.6%) and 20.2% (95% CrI, 13.3% to 28.4%), respectively. However, both models defined ruminants as the main source for human infection compared with any single chicken supplier after intervention (the asymmetric island model attributed 45.8% [95% CrI, 40.0% to 51.7%] and the Bayesian hierarchical model 40.0% [95% CrI, 32.0% to 46.6%]). Both models showed a significant decrease in the infections attributable to supplier A in the second period.
FIG 2.
Proportion of human campylobacteriosis cases, caused by C. jejuni, attributable to each source based on the asymmetric island model and the Bayesian hierarchical model in both periods. Error bars represent the 95% credible interval.
Altogether, the above models attributed the majority of human campylobacteriosis infections to ruminant and chicken sources, whereas exposure to environmental water and wild bird sources appeared to contribute significantly less to the burden of disease.
Bacterial count results.
A total of 1,126/1,443 (78%) chicken meat samples were Campylobacter sp. positive by culture. The zero-inflated Poisson (ZIP) model (Table 3) showed that all suppliers had a drop in mean log10 counts after the intervention. The estimated proportions of negatives (i.e., probability of zero) before and after intervention were similar among suppliers A and B; the point estimate increased for supplier A and decreased for supplier B, but the confidence intervals overlapped for both suppliers. However, supplier “others” had a higher probability of zero after intervention in addition to lower mean counts. Therefore, considering both the probability of zero counts and the mean counts, all suppliers showed reduced contamination after intervention.
TABLE 3.
ZIP model predictionsa
| Supplier | Probability of negative sample (CIb ) | Mean log10/ml (CI) | Data range (CFU/ml) |
|---|---|---|---|
| Before intervention | |||
| A | 0.14 (0.08–0.23) | 2.08 (2.08–2.08) | 0–29,300 |
| B | 0.43 (0.35–0.54) | 2.00 (2.00–2.00) | 0–101,000 |
| Others | 0.26 (0.15–0.41) | 1.52 (1.51–1.53) | 0–37,500 |
| After intervention | |||
| A | 0.20 (0.15–0.25) | 1.59 (1.59–1.59) | 0–12,880 |
| B | 0.33 (0.27–0.38) | 1.15 (1.15–1.16) | 0–4,500 |
| Others | 0.45 (0.40–0.51) | 1.16 (1.16–1.17) | 0–3,160 |
The data range is from the raw data, while the other two values (probability of negative sample and mean log10/ml) are from the ZIP model.
CI, confidence interval.
DISCUSSION
We compared the molecular epidemiology and source attribution of human C. jejuni infection in the Manawatu region of New Zealand before and after food safety interventions implemented by the poultry industry. C. jejuni was the dominant species among all sources in both periods, accounting for more than 95% of the clinical isolates, which is consistent with reports from other countries (1, 2, 25, 26). Both periods combined, the estimated percentage of Campylobacter sp.-positive chicken carcasses by culture remained high (∼78%) and was also similar to the percentage observed in other countries (14, 27, 28). However, our model results (Table 3) suggest that the implemented food safety interventions may have led to a significant decrease in the Campylobacter sp. CFU counts on chicken carcasses, which is important to reduce disease incidence.
Our data show a marked drop in the prevalence of the major C. jejuni clonal complex (CC) ST-48, from 27% before intervention, to 13.6% after intervention. This change in CC ST-48 prevalence was attributable to a large decrease in the isolation of the (internationally rare but common in New Zealand) genotype ST-474 among all sources, from 17.2% (224/1,300) to 6.6% (132/2,013). Moreover, ST-474 was a dominant ST isolated from clinical cases before intervention but dropped from 28.2% to 9.3% after intervention and dropped from 9.7% to 5.3% after intervention in all chicken sources, which was also shown in a 2013 study (29). The significant decrease in ST-474 prevalence observed in the sentinel Manawatu region might explain the national 60% decrease in campylobacteriosis notification rate (21). Higher resolution genotyping, such as whole-genome MLST or single nucleotide polymorphism (SNP)-based analyses of ST-474, may help elucidate whether this decline was associated with a change in the population structure of ST-474. Another significant change was the increase in the prevalence of CC ST-45 from 15% to 18.3%. This was associated with a significant increase in the proportion of ST-583 and the detection of 16 different STs belonging to CC ST-45 that were not detected between 2005 and 2007.
The overlap between human- and chicken-associated genotypes is consistent with poultry being important in disease transmission in New Zealand (15, 17, 24, 30), albeit at a lower level after intervention. The PSIs were calculated before intervention and after intervention to evaluate the similarity between ST frequency distributions. Before intervention, the largest similarity was between STs from isolates from humans and supplier A, mainly due to the high prevalence of ST-474, both among clinical cases and supplier A, and also to the low prevalence of ST-474 in samples from nonpoultry sources in New Zealand (31). On the other hand, the similarity between isolates from clinical cases and supplier B, supplier “others,” and ruminant sources increased significantly after intervention and dropped between clinical cases and those from supplier A (the 95% CrIs overlapped among chicken and ruminant sourced isolates). Before intervention, there was a good agreement between the results of the models, where supplier A was the largest significant contributor to human infections (Fig. 2). After intervention, the contribution of the ruminant source increased significantly to become the major contributor to human infections, unlike the supplier A source, which decreased significantly according to the Bayesian hierarchical and asymmetric island models. However, if we combine all the suppliers as one poultry source, the contributions to human infections by poultry and ruminant sources are similar (see Table S3 in the supplemental material), which was not the case before intervention, where chicken contributed between 58% and 76% and ruminants contributed between ∼20% and ∼30% (19).
In summary, our results indicate that >95% of human campylobacteriosis cases are due to C. jejuni in the Manawatu region sentinel site study in New Zealand. The majority of human campylobacteriosis infections could be attributed to ruminant and chicken sources, whereas exposure to environmental water and wild bird sources appeared to contribute significantly less to the burden of disease. Our source attribution comparative analysis after intervention estimated ruminant and chicken sources as equal contributors to the human disease burden, unlike other countries where chickens remain the dominant risk factor, followed by ruminants (32, 33). This apparent change in the source of C. jejuni infections following food safety interventions implemented by the poultry industry provides valuable information to inform decision making by industry and food safety regulators.
MATERIALS AND METHODS
C. jejuni isolates.
(i) Clinical human isolates. We used C. jejuni isolated from human feces that was submitted to clinical microbiology laboratories before intervention (February 2005 to December 2007) and after intervention (January 2008 to December 2015) in the Manawatu region (23). Officially, the implementation of the Campylobacter Risk Management Strategy started in April 2008, but the poultry companies started trial interventions months earlier (24).
(ii) Chicken meat isolates. Over the same period, fresh chicken carcasses from different commercial chicken suppliers were sampled each month from supermarkets in Palmerston North City (suppliers are designated supplier A, supplier B, and five smaller suppliers collectively designated “others”). Culture for Campylobacter sp. was performed as previously described (3).
(iii) Isolates from environmental water, wild birds, and farmed ruminants. (a) Environmental water isolates. C. jejuni bacteria isolated from recreational waterways and pretreatment drinking water samples from the Manawatu area between 2006 and 2014 were used.
(b) Isolates from wild birds and ruminants. A total of 214 C. jejuni isolates from feces of wild birds were obtained between 2005 and 2013. A total of 465 isolates from cattle and sheep feces from 30 different farms obtained between 2005 and 2015 as part of a monitoring program implemented in the area were also used.
Isolates from pigs were not included due to the low prevalence of Campylobacter sp. from this source in New Zealand (we obtained 5 C. jejuni isolates from 650 pig meat samples; data not shown).
C. jejuni isolation, identification, and subtyping.
Methods for Campylobacter sp. isolation from water, feces, and meat samples and for identification and subtyping by MLST have been previously described (3), except that between August 2014 and December 2015, human clinical Campylobacter spp. were identified by multiplex PCR to detect the ceuE gene associated with C. coli and the hipO gene associated with C. jejuni (34).
Campylobacter sp. viable bacterial counts from chicken meat samples.
Enumeration of Campylobacter colonies from the chicken samples was done using manual spread plating and a spiral plater (Wasp; Don Whitley, West Yorkshire, UK). During the rinsing of chicken carcasses, duplicate modified charcoal-cefoperazone-deoxycholate agar (mCCDA) (Fort Richard Laboratories, Auckland, New Zealand) plates were inoculated with 1 ml (manual spread plate) and 50 μl (spiral plater) of rinse and 100-μl (spiral plater) aliquots of resuspended rinse pellet. Plates were incubated microaerobically at 42°C for 48 h and Campylobacter sp.-like colonies were counted using a plate reader (aCOLyte; Synbiosis, England) or manually. The number of CFUs in 1 ml of 200 ml chicken rinse was then extrapolated.
Analysis of C. jejuni genetic relatedness.
The genetic diversities of C. jejuni from different sources were compared by rarefaction analysis using the package vegan in R version 3.1.3 (35).
Minimum spanning trees were used to visualize allelic differences between the MLST of C. jejuni isolates from different sources, using pairwise Hamming distances. Two trees were generated to compare before intervention and after intervention, using the Prim’s algorithm (36), as implemented in the Bionumerics software (Applied Maths).
C. jejuni source attribution.
The relative contribution of various C. jejuni sources to human disease was estimated over the two sampling periods. The similarity between the frequency distribution of human C. jejuni STs and those of the different sources was estimated using proportional similarity indices (PSIs) and their bootstrap credible intervals (CrIs), as previously described (37). The PSI measures the area of intersection between two frequency distributions (38) and ranges between 0 and 1, where 0 indicates no similarity and 1 indicates identical frequency distributions (39). Calculations were performed using R version 3.1.3 (35).
The asymmetric island model was used to probabilistically assign each human isolate to one of four sources (chickens, ruminants, environmental water, and wild birds) (13).
A Bayesian hierarchical model, based on the modified Hald model (40), was used to implement a nonparametric source attribution model to attribute human campylobacteriosis cases to sources in a Bayesian framework with source and sequence type effects (41).
Analysis of Campylobacter colony counts.
A zero-inflated Poisson (ZIP) model was used to assess differences in the Campylobacter viable counts between chicken meat samples collected in the two sampling periods. This model was used because the variance of the viable counts was higher than the mean due to the presence of samples with zero counts, which led to overdispersion. The R Studio version 3.1.3 software (“pscl” package) (35) was used to apply the following equation:
where is the total number of colonies across all replicates in isolate i from supplier j, is the probability of a zero count, is the total volume of rinse that is plated across replicates, and is the average colony count per ml, where
These equations allow both the probability of a zero count (i.e., probability of a negative) and the expected colony count, given that it is positive, to differ between the two sampling periods and between suppliers. The use of total counts and volumes across replicates for each isolate removes potential clustering by isolate, such that each observation in the model may be regarded as independent given supplier and time period. This does not result in a loss of information, as the Poisson model is invariant to aggregation across data with a constant rate.
Supplementary Material
ACKNOWLEDGMENTS
The sentinel site, as well as microbiological and molecular analyses performed therein, was funded by the New Zealand Ministry for Primary Industries (formerly the New Zealand Food Safety Authority). N.P.F. is supported by the New Zealand Food Safety Science and Research Centre.
We thank Rukhshana Akhter and Lynn Rogers from the mEpiLab team for their laboratory work contribution, Ahmed Fayaz for the extraction of the data, and Phil Carter from the Institute of Environmental Science and Research for help with sequence typing.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Horrocks SM, Anderson RC, Nisbet DJ, Ricke SC. 2009. Incidence and ecology of Campylobacter jejuni and coli in animals. Anaerobe 15:18–25. doi: 10.1016/j.anaerobe.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Moore JE, Corcoran D, Dooley JSG, Fanning S, Lucey B, Matsuda M, McDowell DA, Mégraud F, Cherie Millar B, O’Mahony R, O’Riordan L, O’Rourke M, Rao JR, Rooney PJ, Sails A, Whyte P. 2005. Campylobacter. Vet Res 36:351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- 3.Nohra A, Grinberg A, Midwinter AC, Marshall JC, Collins-Emerson JM, French NP. 2016. Molecular epidemiology of Campylobacter coli isolated from different sources in New Zealand between 2005 and 2014. Appl Environ Microbiol 82:4363–4370. doi: 10.1128/AEM.00934-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ang CW, De Klerk MA, Endtz HP, Jacobs BC, Laman JD, van der Meché FG, van Doorn PA. 2001. Guillain-Barré syndrome- and Miller Fisher syndrome-associated Campylobacter jejuni lipopolysaccharides induce anti-GM1 and anti-GQ1b antibodies in rabbits. Infect Immun 69:2462–2469. doi: 10.1128/IAI.69.4.2462-2469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs BC, Rothbarth PH, van der Meché FGA, Herbrink P, Schmitz PIM, de Klerk MA, van Doorn PA. 1998. The spectrum of antecedent infections in Guillain-Barré syndrome: a case-control study. Neurology 51:1110–1115. doi: 10.1212/wnl.51.4.1110. [DOI] [PubMed] [Google Scholar]
- 6.Rees JH, Soudain SE, Gregson NA, Hughes RAC. 1995. Campylobacter jejuni infection and Guillain-Barré syndrome. N Engl J Med 333:1374–1379. doi: 10.1056/NEJM199511233332102. [DOI] [PubMed] [Google Scholar]
- 7.Friedman CJ, Neiman J, Wegener HC, Tauxe RV. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialised nations. ASM Press, Washington, DC. [Google Scholar]
- 8.Kapperud G, Espeland G, Wahl E, Walde A, Herikstad H, Gustavsen S, Tveit I, Natås O, Bevanger L, Digranes A. 2003. Factors associated with increased and decreased risk of Campylobacter infection: a prospective case-control study in Norway. Am J Epidemiol 158:234–242. doi: 10.1093/aje/kwg139. [DOI] [PubMed] [Google Scholar]
- 9.Dingle KE, Colles FM, Falush D, Maiden MCJ. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol 43:340–347. doi: 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kittl S, Heckel G, Korczak BM, Kuhnert P. 2013. Source attribution of human Campylobacter isolates by MLST and Fla-typing and association of genotypes with quinolone resistance. PLoS One 8:e81796. doi: 10.1371/journal.pone.0081796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doorduyn Y, Van Den Brandhof WE, Van Duynhoven YTHP, Breukink BJ, Wagenaar JA, Van Pelt W. 2010. Risk factors for indigenous Campylobacter jejuni and Campylobacter coli infections in the Netherlands: a case-control study. Epidemiol Infect 138:1391–1404. doi: 10.1017/S095026881000052X. [DOI] [PubMed] [Google Scholar]
- 12.Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MC, Forbes KJ. 2009. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis 48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet 4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer JM, Frost JA, Bolton FJ, Wareing DRA. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J Food Prot 63:1654–1659. doi: 10.4315/0362-028x-63.12.1654. [DOI] [PubMed] [Google Scholar]
- 15.Müllner P, Collins-Emerson JM, Midwinter AC, Carter P, Spencer SEF, van der Logt P, Hathaway S, French NP. 2010. Molecular epidemiology of Campylobacter jejuni in a geographically isolated country with a uniquely structured poultry industry. Appl Environ Microbiol 76:2145–2154. doi: 10.1128/AEM.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Haan CPA, Kivistö RI, Hakkinen M, Corander J, Hänninen M-L. 2010. MLST of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol 10:200. doi: 10.1186/1471-2180-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberhart-Phillips J, Walker N, Garrett N, Bell D, Sinclair D, Rainger W, Bates M. 1997. Campylobacteriosis in New Zealand: results of a case-control study. J Epidemiol Community Health 51:686–691. doi: 10.1136/jech.51.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Institute of Environmental Science and Research Ltd. 2006. Notifiable and other diseases in New Zealand: annual report 2006. Institute of Environmental Science and Research Ltd, Porirua, New Zealand. [Google Scholar]
- 19.Mullner P, Spencer SEF, Wilson DJ, Jones G, Noble AD, Midwinter AC, Collins-Emerson JM, Carter P, Hathaway S, French NP. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Genet Evol 9:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Institute of Environmental Science and Research Ltd. 2014. Notifiable diseases in New Zealand: annual report 2014. Institute of Environmental Science and Research Ltd, Porirua, New Zealand. [Google Scholar]
- 21.Sears A, Baker MG, Wilson N, Marshall JC, Muellner P, Campbell DM, Lake RJ, French NP. 2011. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg Infect Dis 17:1007–1015. doi: 10.3201/eid/1706.101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.New Zealand Food Safety Authority. 2008. Campylobacter risk management strategy 2008–2011. New Zealand Food Safety Authority, Wellington, New Zealand: http://gdsindexnz.org/wp-content/uploads/2019/04/60.-Campylobacter_Risk_Management_Strategy_2008-2011-NZFSA.pdf. [Google Scholar]
- 23.Bolwell CF, Gilpin BJ, Campbell D, French NP. 2015. Evaluation of the representativeness of a sentinel surveillance site for campylobacteriosis. Epidemiol Infect 143:1990–2002. doi: 10.1017/S0950268814003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muellner P, Marshall JC, Spencer SE, Noble AD, Shadbolt T, Collins-Emerson JM, Midwinter AC, Carter PE, Pirie R, Wilson DJ, Campbell DM, Stevenson MA, French NP. 2011. Utilizing a combination of molecular and spatial tools to assess the effect of a public health intervention. Prev Vet Med 102:242–253. doi: 10.1016/j.prevetmed.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 25.WHO. 2011. Campylobacter. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs255/en/. [Google Scholar]
- 26.Schielke A, Rosner BM, Stark K. 2014. Epidemiology of campylobacteriosis in Germany—insights from 10 years of surveillance. BMC Infect Dis 14:30. doi: 10.1186/1471-2334-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, White DG, Wagner D, Meng J. 2001. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, D.C., area. Appl Environ Microbiol 67:5431–5436. doi: 10.1128/AEM.67.12.5431-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern NJ, Line JE. 1992. Comparison of three methods for recovery of Campylobacter spp. from broiler carcasses. J Food Prot 55:663–666. doi: 10.4315/0362-028X-55.9.663. [DOI] [PubMed] [Google Scholar]
- 29.Muellner P, Pleydell E, Pirie R, Baker MG, Campbell D, Carter PE, French NP. 2013. Molecular-based surveillance of campylobacteriosis in New Zealand—from source attribution to genomic epidemiology. Euro Surveill 18(3):pii=20365 https://www.eurosurveillance.org/content/10.2807/ese.18.03.20365-en. [PubMed] [Google Scholar]
- 30.Baker M, Wilson N, Ikram R, Chambers S, Shoemack P, Cook G. 2006. Regulation of chicken contamination is urgently needed to control New Zealand’s serious campylobacteriosis epidemic. N Z Med J 119:U2264. [PubMed] [Google Scholar]
- 31.French N. 2008. Enhancing surveillance of potentially foodborne enteric diseases in New Zealand: human campylobacteriosis in the Manawatu. Ministry for Primary Industries, Wellington, New Zealand: https://www.mpi.govt.nz/dmsdocument/23128-enhancing-surveillance-of-potentially-foodborne-enteric-diseases-in-new-zealand-human-campylobacteriosis-in-the-manawatu-final-report. [Google Scholar]
- 32.Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, Cauchie HM, van Pelt W, Ragimbeau C. 2016. Human campylobacteriosis in Luxembourg, 2010–2013: a case-control study combined with multilocus sequence typing for source attribution and risk factor analysis. Sci Rep 6:20939. doi: 10.1038/srep20939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boysen L, Rosenquist H, Larsson JT, Nielsen EM, Sorensen G, Nordentoft S, Hald T. 2014. Source attribution of human campylobacteriosis in Denmark. Epidemiol Infect 142:1599–1608. doi: 10.1017/S0950268813002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, Woodward DL, Rodgers FG. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol 40:4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Development Core Team. 2005. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 36.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrett N, Devane ML, Hudson JA, Nicol C, Ball A, Klena JD, Scholes P, Baker MG, Gilpin BJ, Savill MG. 2007. Statistical comparison of Campylobacter jejuni subtypes from human cases and environmental sources. J Appl Microbiol 103:2113–2121. doi: 10.1111/j.1365-2672.2007.03437.x. [DOI] [PubMed] [Google Scholar]
- 38.Rosef O, Kapperud G, Lauwers S, Gondrosen B. 1985. Serotyping of Campylobacter jejuni, Campylobacter coli, and Campylobacter laridis from domestic and wild animals. Appl Environ Microbiol 49:1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feinsinger P, Spears EE, Poole RW. 1981. A simple measure of niche breadth. Ecology 62:27–32. doi: 10.2307/1936664. [DOI] [Google Scholar]
- 40.Mullner P, Jones G, Noble A, Spencer SEF, Hathaway S, French NP. 2009. Source attribution of food-borne zoonoses in New Zealand: a modified Hald model. Risk Anal 29:970–984. doi: 10.1111/j.1539-6924.2009.01224.x. [DOI] [PubMed] [Google Scholar]
- 41.Miller P, Marshall J, French N, Jewell C. 2017. sourceR: classification and source attribution of infectious agents among heterogeneous populations. PLoS Comput Biol 13:e1005564. doi: 10.1371/journal.pcbi.1005564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.