A. kawachii has been traditionally used for production of the distilled spirit shochu in Japan. Citric acid produced by A. kawachii plays an important role in preventing microbial contamination during the shochu fermentation process. This study characterized homologous laeA genes; using CAGE, complementation tests, and ChIP-qPCR, it was found that laeA is required for citric acid production through the regulation of cexA in A. kawachii. The epigenetic regulation of citric acid production elucidated in this study will be useful for controlling the fermentation processes of shochu.
KEYWORDS: Aspergillus luchuensis mut. kawachii, citric acid, putative methyltransferase, laeA, citrate exporter, cexA
ABSTRACT
The putative methyltransferase LaeA is a global regulator of metabolic and development processes in filamentous fungi. We characterized the homologous laeA genes of the white koji fungus Aspergillus luchuensis mut. kawachii (A. kawachii) to determine their role in citric acid hyperproduction. The ΔlaeA strain exhibited a significant reduction in citric acid production. Cap analysis gene expression (CAGE) revealed that laeA is required for the expression of a putative citrate exporter-encoding cexA gene, which is critical for citric acid production. Deficient citric acid production by a ΔlaeA strain was rescued by the overexpression of cexA to a level comparable with that of a cexA-overexpressing ΔcexA strain. In addition, chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR) analysis indicated that LaeA regulates the expression of cexA via methylation levels of the histones H3K4 and H3K9. These results indicate that LaeA is involved in citric acid production through epigenetic regulation of cexA in A. kawachii.
IMPORTANCE A. kawachii has been traditionally used for production of the distilled spirit shochu in Japan. Citric acid produced by A. kawachii plays an important role in preventing microbial contamination during the shochu fermentation process. This study characterized homologous laeA genes; using CAGE, complementation tests, and ChIP-qPCR, it was found that laeA is required for citric acid production through the regulation of cexA in A. kawachii. The epigenetic regulation of citric acid production elucidated in this study will be useful for controlling the fermentation processes of shochu.
INTRODUCTION
Shochu is a traditional Japanese distilled spirit (1). The black koji fungus Aspergillus luchuensis and its albino mutant, white koji fungus Aspergillus luchuensis mut. kawachii (A. kawachii), are primarily used for the production of shochu. A. luchuensis and A. kawachii produce enzymes that decompose the starch contained in ingredients such as rice, barley, buckwheat, and sweet potato (2). In addition, they also produce a large amount of citric acid during the fermentation process, which prevents microbial contamination.
A. luchuensis is phylogenetically related to Aspergillus niger, which has been used for industrial citric acid fermentation (3–5). Studies investigating citric acid production have been performed for A. niger with respect to various aspects (6–8). In previous studies, nonacidifying mutant strains of A. niger were analyzed and laeA was found to play a significant role in the production of citric acid and secondary metabolites (9). In addition, laeA disruption also caused a significant decrease in citric acid production by Aspergillus carbonarius, a species closely related to A. niger (10). LaeA was initially identified as a regulator of secondary metabolism in Aspergillus spp. (11). Subsequently, LaeA has been primarily studied as a regulator of secondary metabolic and development processes in filamentous fungi (12, 13). A transcriptomic study also supported the idea that laeA overexpression and disruption caused the production of secondary metabolites to dramatically change in A. niger (14). However, why LaeA is required for citric acid production in A. niger and A. carbonarius remains to be determined.
In this study, we characterized three laeA homologous genes, namely, laeA, laeA2, and laeA3, to determine the regulatory mechanism underlying citric acid production in A. kawachii. Study of gene disruption indicated that laeA significantly reduced citric acid production; therefore, we further analyzed LaeA-dependent gene expression by cap analysis gene expression (CAGE) and found that laeA is required for expression of a putative citrate exporter-encoding cexA gene, which plays a crucial role in citric acid production (15, 16). Further analysis via complementation test and chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR) indicated that LaeA is required for citric acid production through epigenetic regulation of cexA in A. kawachii.
RESULTS
LaeA-like putative methyltransferases in A. kawachii.
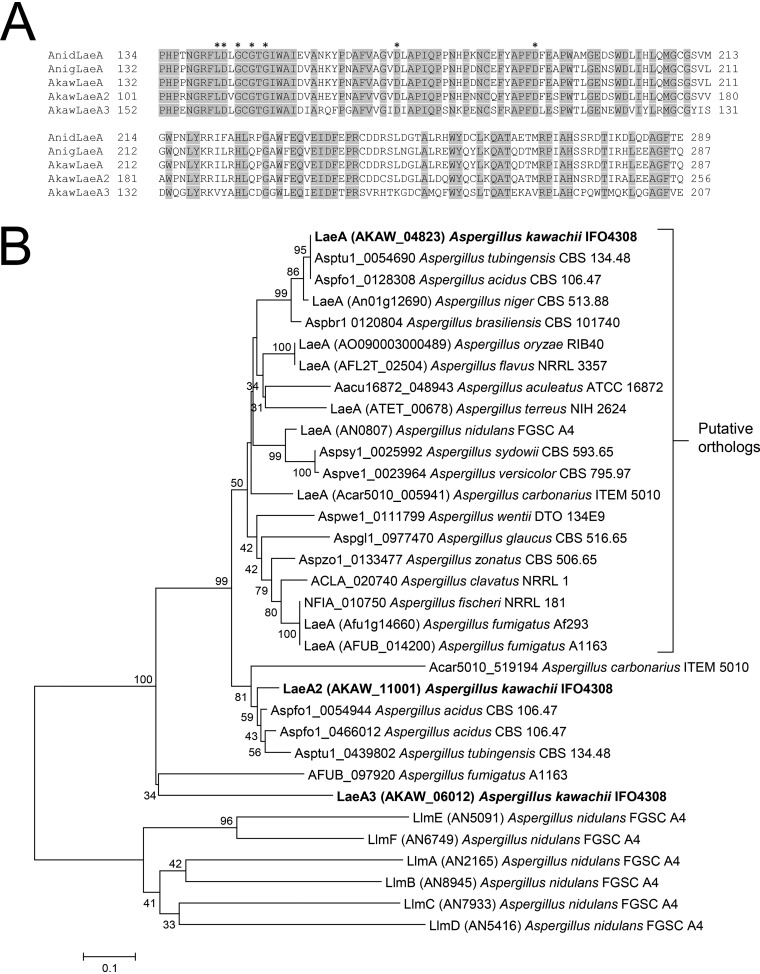
Protein BLAST analysis (https://www.ncbi.nlm.nih.gov/) using the amino acid sequences of Aspergillus nidulans LaeA (AN0807) and A. niger LaeA (An01g12690) as search queries identified three LaeA-like putative methyltransferases in A. kawachii using criteria of >50% identity and E values of <1.00E−100: AKAW_04823, AKAW_11001, and AKAW_06012. AKAW_04823 is extremely similar to A. niger LaeA, with 94% identity over 375 amino acid residues, likely owing to the close phylogenetic relationship between A. kawachii and A. niger. In addition, all three A. kawachii proteins showed a reciprocal best BLAST hit to A. nidulans LaeA, and alignments of these proteins demonstrated that structural homology with the S-adenosylmethionine (SAM) binding motif (11) is highly conserved (Fig. 1A). Thus, AKAW_04823, AKAW_11001, and AKAW_06012 as LaeA-like putative methyltransferases were termed LaeA, LaeA2, and LaeA3, respectively.
FIG 1.
(A) Putative methyltransferase domain sequences of A. nidulans (Anid) LaeA, A. niger (Anig) LaeA, and predicted A. kawachii (Akaw) LaeA-like proteins. The domains were identified by Pfam (https://pfam.xfam.org/). Sequence alignment was performed using ClustalW program in the BioEdit Sequence Alignment Editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Putative intron sequences of A. kawachii laeA and laeA2 were confirmed by transcriptome sequencing (RNA-seq; data not shown) and translated to the amino acid sequences. Asterisks indicate putative SAM binding sites (11). (B) Phylogenetic tree of amino acid sequences of LaeA-like methyltransferases identified from the Aspergillus genome database (http://www.aspgd.org/). The tree was constructed via the neighbor-joining method with complete gap deletion on MEGA version 6.0 (53). Bootstrap values (1,000 replicates) are indicated at the branches. Low bootstrap values (<25) were removed.
To further clarify the relationship between LaeA, LaeA2, and LaeA3, we performed a phylogenetic analysis of LaeA-like methyltransferases conserved in Aspergillus spp. (Fig. 1B). The data set for analysis was obtained by protein BLAST of the Aspergillus Genome Database (AspGD; http://www.aspgd.org/) using the amino acid sequences of A. kawachii LaeA, LaeA2, and LaeA3 as search queries. A. kawachii LaeA is classified into the putative LaeA ortholog group. Conversely, LaeA2 and LaeA3 were assigned to different positions; however, they remain closer to LaeA than to A. nidulans Llm (LaeA-like-methyltransferase) proteins (17). This result supports the hypothesis that LaeA2 and LaeA3 are also LaeA-like methyltransferases.
Colony formation and citric acid production by control, ΔsC ΔlaeA, ΔsC ΔlaeA2, and ΔsC ΔlaeA3 strains.
To investigate the role of LaeA-like methyltransferases in citric acid production by A. kawachii, we constructed strains with each gene disruption and observed their colony formation (Fig. 2A). For comparison with the respective disruption, the A. kawachii control strain was defined to show the same auxotrophic background.
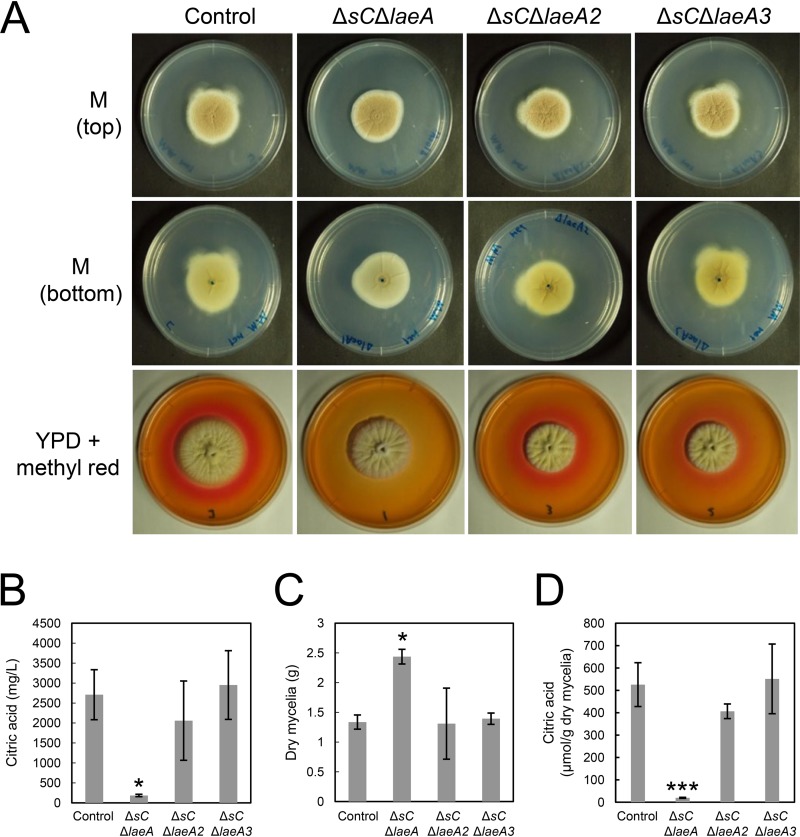
FIG 2.
(A) Colony formation of A. kawachii control, ΔsC ΔlaeA, ΔsC ΔlaeA2, and ΔsC ΔlaeA3 strains. Strains were cultured at 30°C for 5 days on M medium or YPD medium containing methyl red. Agar medium was inoculated with 1 × 104 conidiospores. Methyl red was added as a pH indicator; it turned red at pH values of ≤4.4. (B to D) Citric acid level in culture supernatant (B), mycelial biomass production (C), and extracellular citric acid production per mycelial weight (D) by A. kawachii strains. The control, ΔsC ΔlaeA, ΔsC ΔlaeA2, and ΔsC ΔlaeA3 strains were precultured in M medium for 36 h and then transferred to CAP medium for 48 h. Means and SDs were determined from the results of 3 independent cultivations. Asterisks indicate significant difference (*, P < 0.05; ***, P < 0.001; Student’s t test) from the results for the control strain.
The ΔsC ΔlaeA strain exhibited colony morphology similar to that of the control strain, but its color was found to be paler than that of control, ΔsC ΔlaeA2, and ΔsC ΔlaeA3 strains upon observation of the bottom of the minimal (M) medium agar plate (Fig. 2A). This result indicates that LaeA positively regulates the production of the hyphal yellow pigment in A. kawachii in addition to being involved in secondary metabolism in A. niger (9, 14). However, a difference was observed in the A. kawachii ΔsC ΔlaeA strain in that the A. niger ΔlaeA strain showed yellowish mycelia compared with those of the A. niger control strain on the minimal medium agar plate (9).
In addition, red color surrounding the colonies disappeared only with the ΔsC ΔlaeA strain in yeast extract-peptone-dextrose (YPD) medium containing methyl red as a pH indicator, indicating that the ΔsC ΔlaeA strain shows a nonacidifying phenotype (Fig. 2A). Therefore, we compared the organic acid productivities of control, ΔsC ΔlaeA, ΔsC ΔlaeA2, and ΔsC ΔlaeA3 strains to further investigate the role of LaeA-like methyltransferases in citric acid production. The strains were precultured in M medium at 30°C for 36 h and then transferred to citric acid production (CAP) medium and further cultured at 30°C for 48 h. Next, the measurement of citric acid levels in culture supernatant (Fig. 2B) and mycelial biomass (Fig. 2C) allowed for the determination of extracellular citric acid production per mycelial weight (Fig. 2D). CAP medium was used for evaluating organic acid production because it contains a high concentration of a carbon source (10% [wt/vol] glucose) as well as appropriate trace elements (6–8).
The citric acid level in culture supernatant for the ΔsC ΔlaeA strain was 14.3-fold lower than that for the control strain (Fig. 2B). In contrast, the production of mycelial biomass by the ΔsC ΔlaeA strain was 1.82-fold higher than that by the control strain (Fig. 2C). Based on citric acid level in culture supernatant and amount of mycelial biomass produced, the ΔsC ΔlaeA strain exhibited a 25-fold-lower citric acid production than the control strain, whereas the ΔsC ΔlaeA2 and ΔsC ΔlaeA3 strains exhibited similar citric acid levels (Fig. 2D). This result indicates that LaeA plays a significant role in citric acid production and hyphal growth of A. kawachii.
Test for complementation of the ΔlaeA strain with wild-type laeA.
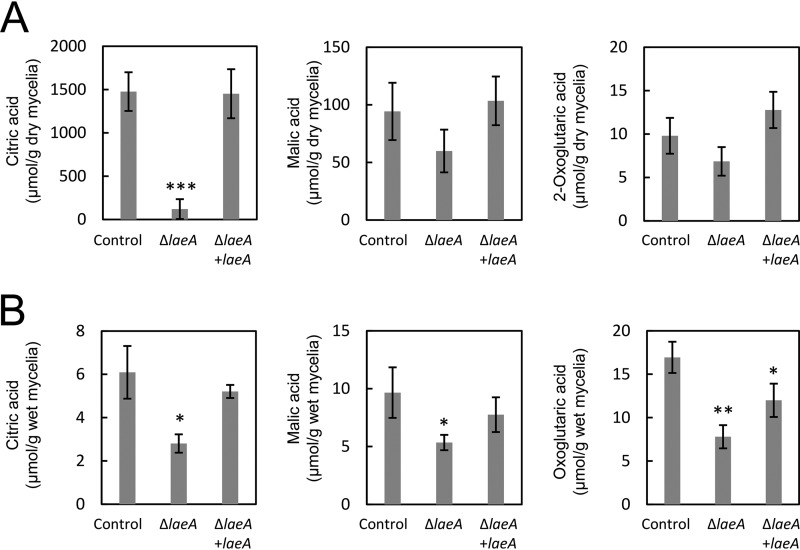
Because laeA disruption caused a significant decline in citric acid production by A. kawachii, we examined the complementation of wild-type laeA in the ΔsC ΔlaeA strain using an sC marker. Control, ΔlaeA, and ΔlaeA plus laeA strains were cultured as described above and culture supernatant and mycelia were separated as the extracellular and intracellular fractions, respectively. In the extracellular fraction, the ΔlaeA strain exhibited 12.5-fold-lower citric acid levels than the control strain (Fig. 3A). Conversely, decreases in the production of malic acid and oxoglutaric acid (1.6-fold and 1.4-fold, respectively) were not statistically significant. Decreases in the production of citric acid, malic acid, and oxoglutaric acid were rescued by the complementation of wild-type laeA.
FIG 3.
Extracellular (A) and intracellular (B) organic acid production by A. kawachii strains. The control, ΔlaeA, and ΔlaeA plus laeA strains were precultured in M medium for 36 h and then transferred to CAP medium for 48 h. Means and SDs were determined from the results of 3 independent cultivations. Asterisks indicate significant difference (*, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test) from the results for the control strain.
Additionally, in the intracellular fraction, the ΔlaeA strain exhibited productivities of 2.2-fold-lower citric acid, 1.8-fold-lower malic acid, and 2.2-fold-lower oxoglutaric acid (Fig. 3B). These decreases in organic acid production were rescued by the complementation of wild-type laeA, although the ΔlaeA plus laeA strain still exhibited 1.4-fold-lower oxoglutaric acid production than that of the control strain.
These results indicate that LaeA plays a significant role in both extracellular and intracellular organic acid production in A. kawachii. In addition, extracellular citric acid accumulation was most significantly affected by laeA disruption.
Colony formation of control, ΔlaeA, and ΔlaeA plus laeA strains.
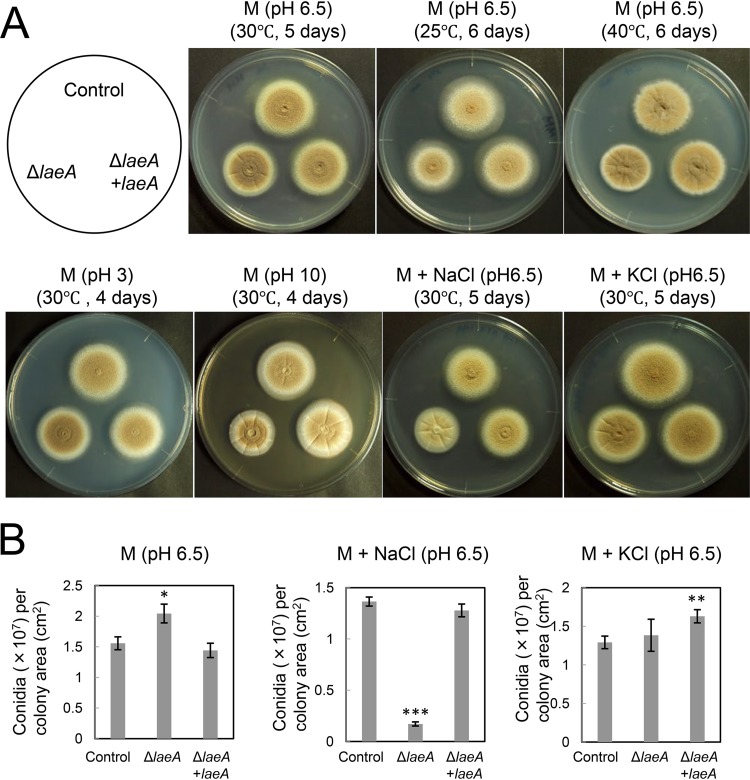
To explore the physiologic roles of LaeA, we characterized the colony morphology of A. kawachii control, ΔlaeA, and ΔlaeA plus laeA strains (Fig. 4A). The ΔlaeA strain exhibited a slightly smaller average colony diameter than those of the control and ΔlaeA plus laeA strains on M medium at all tested temperatures at pH 6.5. This growth deficiency was restored at pH 3, whereas it worsened at pH 10.
FIG 4.
Strains were cultured on M medium with or without stress conditions, and agar medium was inoculated with 1 × 104 conidia. (A) Colony morphology of A. kawachii control, ΔlaeA, and ΔlaeA plus laeA strains. (B) Conidiation of A. kawachii control, ΔlaeA, and ΔlaeA plus laeA strains. All results are expressed as means and SDs. Asterisks indicate significant difference (*, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test) from the results for the control strain.
In addition, colonies of the ΔlaeA strain were paler than those of the control and ΔlaeA plus laeA strains in M medium with 0.8 M sodium chloride but not in that with 0.6 M potassium chloride (Fig. 4A). Thus, we next assessed conidium formation (Fig. 4B). Strains were cultured on M medium with or without 0.8 M sodium chloride or 0.6 M potassium chloride at 30°C for 5 days, at which time the number of conidia formed was determined. The number of conidia per square centimeter of the ΔlaeA strain was significantly reduced, to approximately 13% of the number formed by the control strain, in M medium containing 0.8 M sodium chloride. Alternatively, the number of conidia per square centimeter for the ΔlaeA strain was 1.32-fold higher and similar to that of the control strain in M medium and M medium with 0.6 M potassium chloride, respectively. Complementation of laeA rescued the deficient conidium formation of the ΔlaeA strain, although the ΔlaeA plus laeA strain exhibited 1.26-fold-higher conidium formation in M medium containing 0.6 M potassium chloride.
These results indicate that LaeA is required for asexual development, particularly in the presence of sodium-specific stress versus high osmotic stress. The deficiency in conidium formation was consistent with previous reports that the laeA disruption caused a significant defect in conidium formation in A. oryzae (18) and Penicillium chrysogenum (19). Although LaeA plays a significant role in the sexual development of A. nidulans (20), sexual development has not been observed in A. kawachii.
Gene expression related to citric acid production.
To identify LaeA-regulated genes related to citric acid production, the expression profiles of A. kawachii control and ΔlaeA strains during citric acid production were compared. These strains were precultured in M medium at 30°C for 36 h and then transferred to CAP medium and further cultured at 30°C for 12 h, at which time the A. kawachii control strain vigorously produced citric acid. Next, gene expression profiles were compared using CAGE.
Gene expression change of a total of 9,647 genes was evaluated by CAGE (see Data Set S1 in the supplemental material). The change in gene expression was considered to be significant if the false-discovery rate (FDR) was <0.05 and the log2 fold change was less than −0.5 or greater than 0.5. Using these criteria, a total of 1,248 differentially expressed genes were identified, including 590 upregulated and 658 downregulated genes. Gene ontology (GO) term enrichment analysis of these gene data sets revealed the enrichment of GO terms related to transport and metabolic processes (data not shown); however, it was difficult to interpret the results to understand the reason as to why laeA disruption reduced citric acid production.
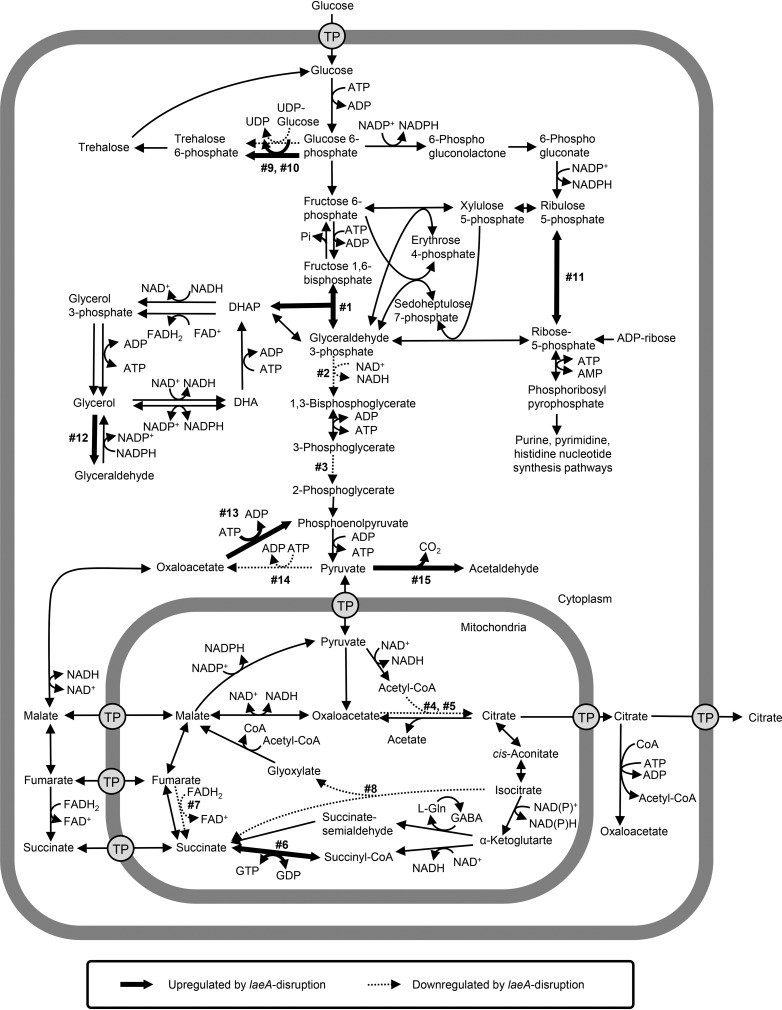
Next, we mapped the differentially expressed genes, 15 of which were mapped to metabolic pathways related to citric acid production (Fig. 5; Table 1). For example, genes related to glycerol synthesis (reactions 1 [AKAW_07170] and 12 [AKAW_08691] in Fig. 5) and pentose phosphate (reaction 11 [AKAW_00489]) pathways were upregulated by laeA disruption, whereas the Embden-Meyerhof-Parnas (EMP) pathway (reactions 2 [AKAW_04737] and 3 [AKAW_03026]) and citrate synthase in the tricarboxylic acid (TCA) and glyoxylate cycles (reactions 4 [AKAW_00170] and 5 [AKAW_06223]) were downregulated by laeA disruption. The citrate synthase-encoding gene (AKAW_06223) showed most significant reduction in gene expression (−11.35 log2 fold); however, it was not the most highly expressed citrate synthase gene, indicating that the reduced expression level of AKAW_06223 might not explain the deficient citric acid production.
FIG 5.
Differentially expressed genes mapped on the proposed metabolic pathways of A. kawachii. The pathway was constructed based on the metabolic model of A. niger and A. nidulans (51, 54, 55). The glyoxylate shunt was placed in the mitochondria. Bold and dotted arrows indicate downregulated and upregulated reactions due to laeA disruption, respectively. Numbers (1 to 15) next to the arrows correspond to the numbering of locus tags in Table 1. Abbreviations: CoA, coenzyme A; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; FAD, flavin adenine dinucleotide; Gln, glutamine; TP, transporter.
TABLE 1.
Citric acid production-related genes among up- and downregulated genes following laeA disruption
|
Reaction no. in Fig. 5 |
Locus tag | Putative function |
Log2 fold change |
FDR |
|---|---|---|---|---|
| 1 | AKAW_07170 | Fructose-bisphosphate aldolase | 4.01 | 0.0011 |
| 2 | AKAW_04737 | Glyceraldehyde 3-phosphate dehydrogenase | −1.05 | 0.0005 |
| 3 | AKAW_03026 | Phosphoglycerate mutase | −0.78 | 0.0335 |
| 4 | AKAW_00170 | Citrate synthase | −0.92 | 0.0014 |
| 5 | AKAW_06223 | Citrate synthase | −11.35 | 1.10E−98 |
| 6 | AKAW_07594 | Succinate-CoA ligase (GDP-forming) alpha chain | 1.27 | 0.0052 |
| 7 | AKAW_07352 | Succinate dehydrogenase | −0.71 | 0.0352 |
| 8 | AKAW_00119 | Isocitrate lyase | −1.40 | 1.063E−05 |
| 9 | AKAW_05189 | Alpha,alpha-trehalosephosphate synthase (UDP forming) 1 (TpsA, TpsB) | −0.76 | 0.0108 |
| 10 | AKAW_03597 | Alpha,alpha-trehalosephosphate synthase (UDP forming) 1 (TpsA, TpsB) | 0.87 | 0.0024 |
| 11 | AKAW_00489 | Ribose-5-phosphate isomerase (B) | 1.03 | 0.0035 |
| 12 | AKAW_08691 | Alcohol oxidase | 1.65 | 3.18E−09 |
| 13 | AKAW_02095 | Phosphoenolpyruvate carboxykinase (ATP) | 0.80 | 0.0139 |
| 14 | AKAW_00804 | Pyruvate carboxylase (cytosol) | −1.15 | 0.0168 |
| 15 | AKAW_06947 | Pyruvate decarboxylase | 0.83 | 0.0205 |
Next, we focused on the citric acid export process, as it is considered to play a significant role in the high citric acid production by A. niger (21–23) (Table 2). Our group previously reported that the two mitochondrial citrate transporters CtpA (ctpA [AKAW_03754]) and YhmA (yhmA [AKAW_06280]) (24) are involved in the transport of citric acid from mitochondria to cytosol: however, their gene expression levels did not significantly change. Conversely, a significant reduction in gene expression (−6.09 log2 fold) of the putative citrate exporter-encoding cexA (AKAW_07989). CexA was identified as the main citrate exporter of the plasma membrane in A. niger (15, 16). AKAW_07989 showed the best reciprocal BLAST hit to A. niger CexA, with 96% identity over 502 amino acid residues, indicating that AKAW_07989 represents CexA in A. kawachii.
TABLE 2.
Gene expression change of citric acid excretion-related genes following laeA disruption
| Gene | Locus tag | Putative function | Log2 fold change | FDR |
|---|---|---|---|---|
| ctpA | AKAW_03754 | Mitochondrial citrate transporter | 0.08 | 0.9432 |
| yhmA | AKAW_06280 | Mitochondrial citrate transporter | 0.09 | 0.9313 |
| cexA | AKAW_07989 | Plasma membrane localized citrate exporter | −6.09 | 3.72E−35 |
Citric acid was the most significantly decreased organic acid in the extracellular fraction resulting from the disruption of laeA (Fig. 3A); however, citric acid, malic acid, and oxoglutaric acid productions were reduced at similar levels within the intracellular fraction of the ΔlaeA strain (Fig. 3B). Thus, we focused on the significantly reduced expression level of cexA, which could explain the reduced citric acid accumulation in the extracellular fraction (Table 2).
Test of complementation of laeA disruption by cexA overexpression.
To examine whether the downregulation of cexA was crucial for the decrease in citric acid production caused by laeA disruption, we compared citric acid production by control and ΔlaeA strains with that by ΔcexA, ΔcexA plus OEcexA (cexA overexpression with deletion of cexA), and ΔlaeA plus OEcexA (cexA overexpression with deletion of laeA) strains (Fig. 6A). gpdA encodes glyceraldehyde-3-phosphate dehydrogenase (25, 26) and was used for overexpression of cexA because there was no significant change in gpdA expression between control and ΔlaeA strains in the CAGE data set (Data Set S1), indicating that the gpdA promoter is not controlled by LaeA.
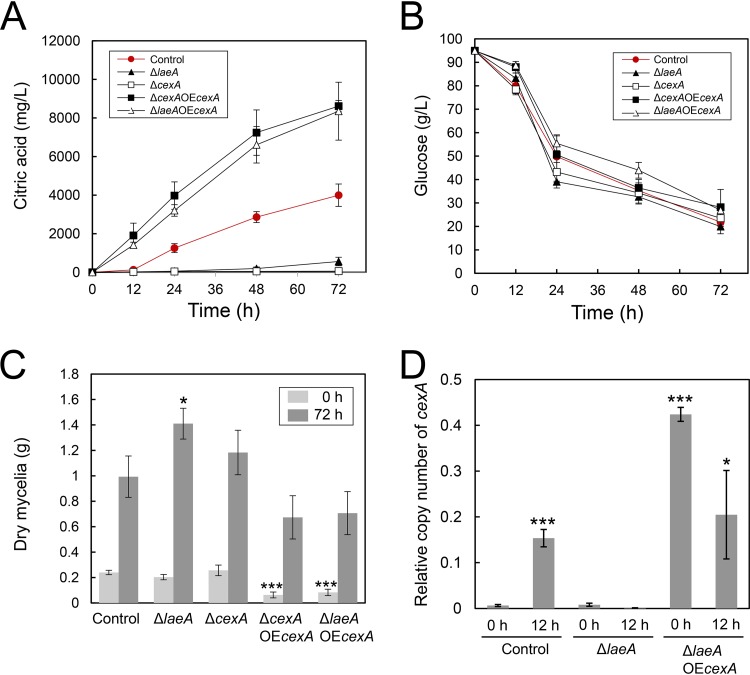
FIG 6.
Citric acid level (A) and glucose level (B) in culture supernatant and mycelial biomass production (C) by A. kawachii strains. Control, ΔlaeA, ΔcexA, ΔcexA plus OEcexA, and ΔlaeA plus OEcexA strains were precultured in M medium for 36 h, then transferred to CAP medium, and further cultured for 0, 12, 24, 48, and 72 h. Means and SDs were determined from the results of 3 independent cultivations. Asterisks indicate significant difference (*, P < 0.05; ***, P < 0.001; Student’s t test) from the results for the control strain. (D) Transcriptional levels of cexA. The control, ΔlaeA, and ΔlaeA plus OEcexA strains were precultured in M medium for 36 h, then transferred to CAP medium, and further cultured for 0 or 12 h. Means and SDs were determined from the results of 3 independent cultivations. Asterisks indicate significant difference (*, P < 0.05; ***, P < 0.001; Student’s t test) from the results for the control strain cultured for 0 h.
A. kawachii strains were precultured with M medium at 30°C for 36 h and were then transferred to CAP medium and further cultured at 30°C for 72 h. Then we measured the citric acid (Fig. 6A) and glucose (Fig. 6B) levels in culture supernatant. In addition, we measured the amount of mycelial biomass produced after precultivation in M medium for 36 h (defined as 0 h), which is the time point just before strain transfer to CAP medium, and after further cultivation in CAP medium for 72 h (Fig. 6C).
The ΔcexA strain exhibited lower extracellular citric acid production than the control strain, similar to that observed for the ΔlaeA strain, whereas the ΔcexA plus OEcexA and ΔlaeA plus OEcexA strains exhibited higher extracellular citric acid production than the control strain throughout the cultivation period of 72 h (Fig. 6A). There was no significant difference in citric acid production between the ΔcexA plus OEcexA and ΔlaeA plus OEcexA strains, indicating that the overexpression of cexA alone was sufficient to rescue deficient citric acid production in the ΔlaeA strain.
The amount of mycelial biomass produced by the ΔlaeA strain was not considerably different from that produced by the control strain at 0 h; however, this amount became 1.42-fold higher than that produced by the control strain after further cultivation in CAP medium for 72 h (Fig. 6C). This result indicates that the hyphal growth of the ΔlaeA strain was enhanced in CAP medium compared with that of the control strain. In addition, the amounts of mycelial biomass produced by the ΔlaeA plus OEcexA and ΔcexA plus OEcexA strains were 3.8-fold and 2.9-fold lower than those produced by the control strain, respectively, at 0 h (Fig. 6C). This result indicates that the overexpression of cexA negatively affects the hyphal growth of A. kawachii in M medium.
The glucose level in culture supernatant was 1.3-fold lower in the ΔlaeA strain than that of the control strain at 24 h (Fig. 6B). Therefore, the ΔlaeA strain exhibited a higher glucose consumption rate than the control strain in CAP medium. Because the initial amounts of mycelial biomass for the control and ΔlaeA strains were at similar levels at 0 h, the higher glucose consumption rate led to a higher mycelial biomass yield by the ΔlaeA strain at 72 h (Fig. 6C). In contrast, glucose levels were 1.1-fold higher for the ΔcexA plus OEcexA strain than for the control strain at 12 h (Fig. 6B). In addition, glucose levels were 1.1-fold, 1.2-fold, and 1.2-fold higher for the ΔlaeA plus OEcexA strain than for the control strain at 12, 48, and 72 h, respectively. This may be due to the smaller initial amounts of mycelial biomass for the ΔcexA plus OEcexA and ΔlaeA plus OEcexA strains than that for the control strain at 0 h (Fig. 6C).
Transcriptional levels of cexA in the control, ΔlaeA, and ΔlaeA plus OEcexA strains.
Because the ΔlaeA plus OEcexA strain exhibited higher citric acid production than the control strain (Fig. 6A), we determined gene expression levels of cexA in the control, ΔlaeA, and ΔlaeA plus OEcexA strains. Strains were precultured in M medium at 30°C for 36 h and then transferred to CAP medium. The time point of 36 h of precultivation just before transfer was defined as 0 h (i.e., starting time). We compared the gene expression levels of cexA at 0 and 12 h of cultivation in CAP medium via quantitative reverse transcription-PCR (RT-PCR) analysis (Fig. 6D). The control strain showed a 24-fold-greater expression level of cexA at 12 h than that at 0 h, whereas the ΔlaeA strain showed similar expression levels of cexA at 0 and 12 h. These results indicate that cexA expression is induced under the condition of citric acid production via LaeA.
Control and ΔlaeA plus OEcexA strains showed similar expression levels of cexA at 12 h; however, the ΔlaeA plus OEcexA strain exhibited a 66-fold-higher expression level of cexA than the control strain at 0 h, likely because the gpdA promoter is active in M as well as CAP medium. Thus, the higher citric acid production by the ΔlaeA plus OEcexA strain (Fig. 6A) could be attributable to a higher gene expression level of cexA when beginning culture in CAP medium.
Histone trimethylation level in the cexA promoter region.
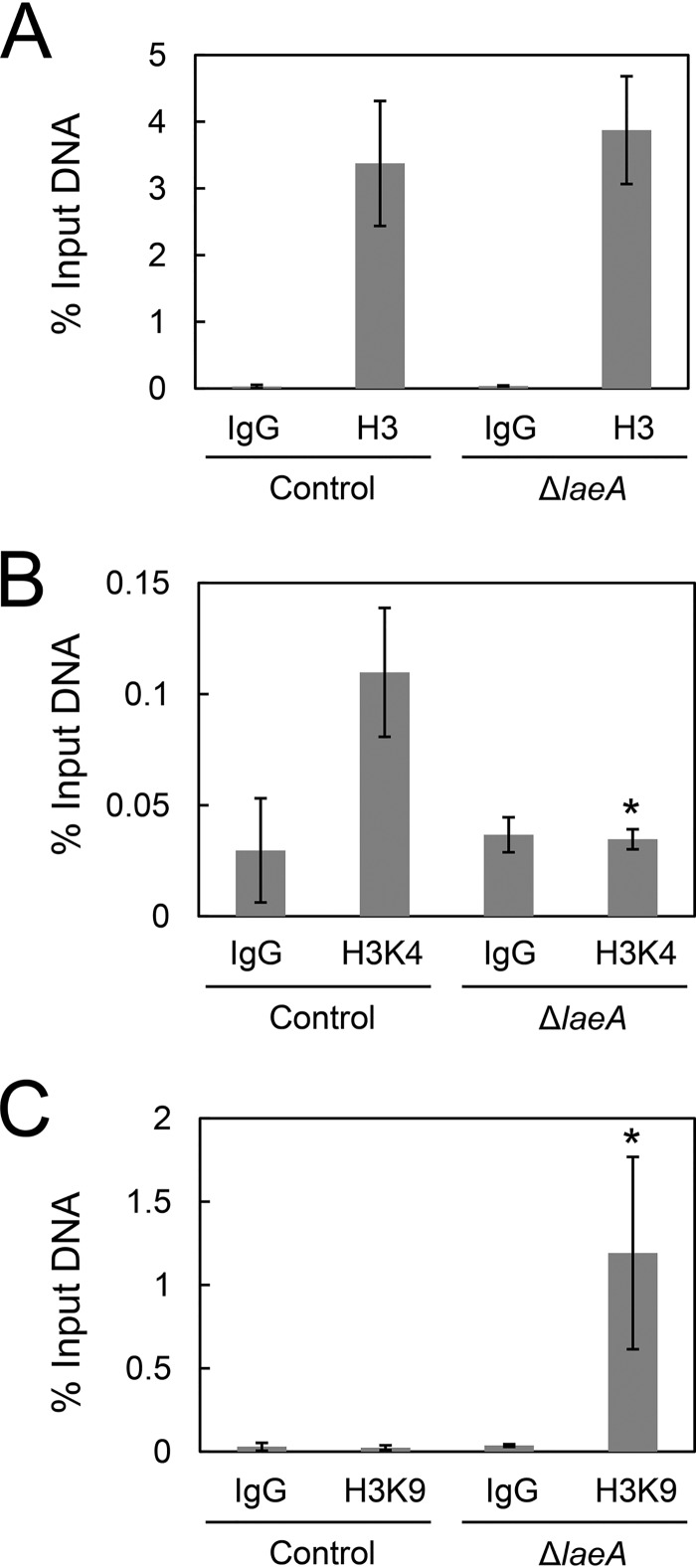
LaeA is believed to widely regulate gene expression via controlling the methylation level of histones (11–13). To determine the mechanism underlying LaeA-dependent cexA expression through histone methylation, we performed ChIP-qPCR analysis of histone H3, histone H3 trimethyl K4 (H3K4me3), and histone H3 trimethyl K9 (H3K9me3) in control and ΔlaeA strains. H3K4me3 and H3K9me3 are known to be euchromatin and heterochromatin markers, respectively.
The histone H3 occupancy at the cexA promoter did not change between the control and ΔlaeA strains (Fig. 7A). However, the euchromatin marker H3K4me3 at the cexA promoter was decreased to a level similar to that of the negative control (i.e., normal anti-mouse IgG) in the ΔlaeA strain (Fig. 7B). In addition, the heterochromatin marker H3K9me3 was greatly enriched in the ΔlaeA strain compared with that in the control strain (Fig. 7C). These results indicated that LaeA controls cexA expression via modulation of euchromatin/heterochromatin ratios at the cexA promoter region in A. kawachii.
FIG 7.
ChIP-qPCR of cexA promoter region. Control and ΔlaeA strains were cultured as described in the text. Histone H3 (A), histone H3K4me3 (B), and histone H3K9me3 (C) occupancies of the cexA promoter region were investigated. The means and SDs were determined from the results of 3 independent cultivations. Asterisks indicate significant difference (*, P < 0.05; Student’s t test) from the results for the control strain.
DISCUSSION
In this study, we characterized three LaeA-like methyltransferases, LaeA, LaeA2, and LaeA3, in A. kawachii to reveal their role in citric acid production. LaeA2 and LaeA3 are not conserved in all Aspergillus spp. (Fig. 1B), implying that these two LaeA-like methyltransferases are paralogs. For example, LaeA-like methyltransferases, similar to A. kawachii LaeA2, were only conserved in the Aspergillus section Nigri (A. carbonarius ITEM 5010, Aspergillus acidus CBS 106.47, and Aspergillus tubingensis CBS 134.48), with the exception of A. niger CBS 513.88. In addition, LaeA-like methyltransferases similar to A. kawachii LaeA3 were identified only in A. fumigatus A1163.
Among the three LaeA-like methyltransferase genes, only laeA disruption caused significant reduction in citric acid production by the A. kawachii strain (Fig. 2D). This result is consistent with the results of previous studies showing that LaeA is required for citric acid accumulation in culture supernatants of A. niger and A. carbonarius (9, 10).
As LaeA is known to control secondary metabolism and development in filamentous fungi (11–13), the ΔlaeA strain showed deficient production of yellow mycelium pigment (Fig. 2A) and formation of conidia (Fig. 4B). The ΔlaeA strain exhibited deficient formation of conidia in the presence of sodium chloride but not in the presence of potassium chloride (Fig. 4B), thereby indicating that LaeA is required for the formation of conidia, particularly in the presence of sodium-specific stress. Sodium is known to demonstrate toxic effects above a certain Na+/K+ ratio and has to be excreted from the cytosol to prevent the toxic effects caused by sodium transporters (27–29). However, there is no significant change in the expression of genes encoding putative sodium transporters Nha1 (AKAW_09133 and AKAW_03679) and Nhx1 (AKAW_07982) in the CAGE data set (Data Set S1). In addition, CAGE could not link the transcription start sites with predicted coding sequences of Ena1 (AKAW_08368 and AKAW_08719), which are also the homologs of sodium transporters.
CAGE identified 1,248 genes differentially expressed between the control and ΔlaeA strains (Data Set S1). These genes included approximately equal numbers of upregulated and downregulated genes (590 and 658, respectively). LaeA should regulate these genes not only directly but also indirectly because they include at least 9 transcription factor gene homologs. For example, the upregulation of lreB (AKAW_00032) (30), tamA (AKAW_02564) (31), and dbaA (AKAW_01762) (32) and the downregulation of amyR (AKAW_09854) (33), zipA (AKAW_05495) (34), oefC (AKAW_03045) (35), farB (AKAW_07498) (36), cpcA (AKAW_00007) (37), and devR (AKAW_08419) (38) were identified in this process.
CAGE identified considerable reduction in the gene expression of the citrate exporter-encoding cexA gene by laeA disruption (Table 2). A further study showed that cexA overexpression remedied deficient citric acid production by ΔlaeA and ΔcexA strains at similar levels (Fig. 6A). These results indicated that the deficient gene expression of cexA led to the deficient citric acid production by the ΔlaeA strain. For the evaluation of citric acid production, A. kawachii strains were cultivated in CAP medium after precultivation in M medium. CAP medium was used for enhancing citric acid production concomitant with reduced hyphal growth (24). Fungal pellet morphology is known to affect the production of organic acids, proteins, and secondary metabolites (39), and elevated citric acid production occurs with shorter hyphae or hyperbranched phenotypes in A. niger (40, 41). The production of mycelial biomass by the ΔlaeA strain did not considerably change during precultivation in M medium compared with that by the control strain (Fig. 6C); however, it considerably increased after cultivation in CAP medium (Fig. 2C, Fig. 6C), thereby indicating that the hyphal growth of the ΔlaeA strain was enhanced compared with that of the control strain under the condition of citric acid production. On the other hand, the production of mycelial biomass by the ΔcexA plus OEcexA and ΔlaeA plus OEcexA strains reduced during precultivation in M medium compared with that by the control strain (Fig. 6C). In addition, we observed that there were no considerable changes in the pellet sizes of the control, ΔlaeA, ΔcexA, ΔcexA plus OEcexA, and ΔlaeA plus OEcexA strains after cultivation in CAP medium; however, the surface roughness of the pellets of the ΔcexA plus OEcexA and ΔlaeA plus OEcexA strains was smoother than those of the control strains (data not shown). Unlike in the control strain, these growth phenotypes and morphological changes due to laeA deletion and cexA overexpression might be related to citric acid production (Fig. 6A).
Subcultivation of the control strain from M medium to CAP medium led to elevated transcript levels of cexA, whereas the ΔlaeA strain lost its transcriptional response, thereby indicating that LaeA is involved in the transcriptional regulation of cexA (Fig. 6D). Although details concerning the molecular function of LaeA remain unclear (11–13), LaeA is known to counteract the heterochromatinization of the promoter region of secondary metabolite gene clusters by histone H3K9 methylation via heterochromatin protein 1 (HepA) and a H3K9 methyltransferase (ClrD) in A. nidulans (42). Our findings indicated that the euchromatin structure of the cexA promoter nearly disappeared with laeA disruption (Fig. 7B), and therefore, the heterochromatin level of the cexA promoter might be enriched by HepA (AKAW_02119) and ClrD (AKAW_07568) orthologs in A. kawachii (Fig. 7C). Gene expression levels of AKAW_02119 and AKAW_07568 were not significantly altered in the ΔlaeA strain (Data Set S1), but loss of LaeA might affect histone modification balance and euchromatin/heterochromatin ratios. The molecular mechanism of LaeA-dependent histone modification should be confirmed through additional experiments because the methyl-accepting substrate of LaeA still remains elusive (43). In addition, whether gene expression of cexA requires a specific DNA-binding transcriptional factor remains unclear and should be further studied.
In conclusion, LaeA plays a significant role in citric acid production in A. kawachii by controlling cexA expression via histone modification at the cexA promoter region. Because A. kawachii is widely used in the production of shochu and elsewhere in the fermentation industry, our findings are expected to enhance the understanding of citric acid production mechanism(s) and facilitate optimization of strategies for controlling A. kawachii activity.
MATERIALS AND METHODS
Strains and growth conditions.
The Aspergillus kawachii strains used in this study are listed in Table 3, and strain SO2 (44) was used as the parental strain.
TABLE 3.
Aspergillus kawachii strains used in this study
| Strain name or description | Genotype | Reference |
|---|---|---|
| SO2 (ΔsC ΔargB) | ligD− argB::hph sC− | 44 |
| ΔsC | ligD− argB::argB sC− | This study |
| ΔsC ΔlaeA | ligD− argB::hph sC− laeA::argB | This study |
| ΔsC ΔlaeA2 | ligD− argB::hph sC− laeA2::argB | This study |
| ΔsC ΔlaeA3 | ligD− argB::hph sC− laeA3::argB | This study |
| CK2 | ligD− argB::argB sC::sC | 24 |
| ΔlaeA | ligD− argB::hph sC::sC laeA::argB | This study |
| ΔlaeA plus laeA | ligD− argB::hph sC− laeA::laeA-sC-argB | This study |
| ΔcexA | ligD− argB::hph sC::sC cexA::argB | This study |
| ΔcexA plus OEcexA | ligD− argB::hph sC− cexA::argB pGS-PgpdA-cexA | This study |
| ΔlaeA plus OEcexA | ligD− argB::hph sC− laeA::argB pGS-PgpdA-laeA | This study |
For construction and characterization, the strains were grown in minimal medium (1% [wt/vol] glucose, 0.6% [wt/vol] NaNO3, 0.052% [wt/vol] KCl, 0.052% [wt/vol] MgSO4·7H2O, and 0.152% [wt/vol] KH2PO4, plus Hutner’s trace elements [pH 6.5]). The medium was adjusted to the required pH using HCl or NaOH. For the cultivation of sC− and argB− strains, 0.02% (wt/vol) methionine and/or 0.211% (wt/vol) arginine was added to M medium, respectively.
To evaluate acidification occurring on agar medium, strains were grown in YPD with methyl red (2% [wt/vol] glucose, 1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% methyl red solution) prepared as follows: 100 mg of methyl red (Nakalai Tesque, Kyoto, Japan) was dissolved in 100 ml of ethanol and titrated by 0.1% (wt/vol) NaOH solution until observation of an obvious color change from red to yellow; the solution was then sterile filtered (0.2-μm pore size; Toyo Roshi Kaisha, Japan).
To investigate citric acid production, A. kawachii strains were also grown in CAP medium (10% [wt/vol] glucose, 0.3% [wt/vol] (NH4)2SO4, 0.001% [wt/vol] KH2PO4, 0.05% [wt/vol] MgSO4·7H2O, 0.000005% [wt/vol] FeSO4·7H2O, 0.00025% [wt/vol] ZnSO4·5H2O, 0.00006% [wt/vol] CuSO4·5H2O [pH 4.0]). CAP medium was adjusted to the required pH with HCl.
Construction of putative methyltransferase gene disruption strain.
laeA, laeA2, and laeA3 were disrupted in A. kawachii SO2 (44) by insertion of argB. A gene replacement cassette encompassing the homology arm at the 5′ end of the putative methyltransferase genes, an argB selection marker, and the homology arm at the 3′ end of the putative methyltransferase genes was constructed by recombinant PCR using the primer pairs AKlaeX-FC/AKlaeX-R1, argB-F2/argB-R2, and AKlaeX-F3/AKlaeX-RC, respectively (where “X” indicates A, A2, or A3) (Table 4). For amplification of the argB gene, the pDC1 plasmid was used as the template DNA (45). For amplification of other DNA fragments, A. kawachii IFO 4308 wild-type genomic DNA was used as the template DNA. The resultant DNA fragment amplified with primers AKlaeX-F1 and AKlaeX-R3 was used to transform A. kawachii SO2, yielding ΔsC ΔlaeA, ΔsC ΔlaeA2, and ΔsC ΔlaeA3 strains. M agar plates lacking arginine were used for the selection of transformants. Introduction of argB into each methyltransferase gene locus was confirmed by PCR using the primer pair AKlaeX-FC and AKlaeX-RC (Fig. S1).
TABLE 4.
Primers used in this study
| Primer name | Sequence (5′–3′) | Reference |
|---|---|---|
| AKlaeA-FC | TGATGCTCGGAAGGCACCAG | This study |
| AKlaeA-F1 | AGCCAGGCGCATTTCCCACC | This study |
| AKlaeA-R1 | GCATGCAAGCTTTCGCGAGCCGTCGCCATGGTGATTGGTGGGTGC | This study |
| AKlaeA-F3 | CCGGGTACCGAGCTCGAATTCGCGGGGGCGGCCCGTCCCTTTG | This study |
| AKlaeA-R3 | CGGCGCGGATGAGGATATTG | This study |
| AKlaeA-RC | CCCGACGTTCTCACAATCG | This study |
| AKlaeA2-FC | CTGATGAGGATGACTCAGC | This study |
| AKlaeA2-F1 | GCAGTTGAAGACGTCAAAGG | This study |
| AKlaeA2-R1 | GCATGCAAGCTTTCGCGAGCCGTCGGCTCCTCTTCATCACACGG | This study |
| AKlaeA2-F3 | CCGGGTACCGAGCTCGAATTCGCGGCACGTATACCAAGCACGG | This study |
| AKlaeA2-R3 | CAGCCTGGTCATTGACCATG | This study |
| AKlaeA2-RC | GGACTGGGATACCGACGAG | This study |
| AKlaeA3-FC | CTCAACGTGATTGCGGCAC | This study |
| AKlaeA3-F1 | CCACTTGGAGTGTCGACGAG | This study |
| AKlaeA3-R1 | GCATGCAAGCTTTCGCGAGCCGTCGCGTCAATCTGCGTCTTGG | This study |
| AKlaeA3-F3 | CCGGGTACCGAGCTCGAATTCGCGGCGCGTCGCCCCATGTAG | This study |
| AKlaeA3-R3 | TGCAGCCGAAACAGGCAC | This study |
| AKlaeA3-RC | CCAGCCTCAATATAGCGCGC | This study |
| argB-F2 | CGACGGCTCGCGAAAGCTTGCATGC | This study |
| argB-R2 | CCGCGAATTCGAGCTCGGTACCCGG | This study |
| sC-comp-F | CAATCACGCAAGCCGAGCTG | 24 |
| sC-comp-R | CTCACCGATGTAGGTCATG | 24 |
| AKlaeAcomp-R1 | CTGGGACACCATGACAACGGCAGCATTTTCAAAGGGACGGGC | This study |
| AKlaeAcomp-F2 | GCCCGTCCCTTTGAAAATGCTGCCGTTGTCATGGTGTCCCAG | This study |
| sC-argB-F2 | CCGTTGTCATGGTGTCCCAGCAGCA | 24 |
| sC-argB-R2 | AATTCGAGCTCGGTACCCGG | 24 |
| pGS-PgpdA-cexA-inf-F | CCGCCGAACAGTCGAACACATCTACACAATGTCTTCAACCACATCTTCATC | This study |
| pGS-PgpdA-cexA-inf-R | CTCCCATATGGTCGAcctccCTAATTTCCGTTGGC | This study |
| cexA-ChIP-F | GTCTTCAACCACATCTTCATCAAG | This study |
| cexA-ChIP-R | GAGACATCATCCAGGGCAGG | This study |
| cexA-RT-F | GGTCCCTGTACCACAGGTCA | This study |
| cexA-RT-R | GTGGCTTCTCGGACGACTGA | This study |
| actA-RT-F | GGTATGGGTCAGAAGGACTC | 51 |
| actA-RT-R | CTCCATGTCATCCCAGTTCG | 51 |
The SO2 strain was transformed using the argB cassette to employ the same auxotrophic genetic background strains for comparative study. This argB gene cassette was generated with PCR using A. kawachii genomic DNA and pDC1 as the template DNA and was used to transform the SO2 strain, yielding the ΔsC strain (Table 3). Transformants were selected on M agar medium lacking arginine.
Complementation of the laeA disruption strain.
For complementation analysis of laeA disruption using wild-type laeA, a gene replacement cassette encompassing a homology arm at the 5′ end of laeA, wild-type laeA, an sC selection marker, and a homology arm at the argB locus was constructed with recombinant PCR using the primer pairs AKlaeA-FC/AKlaeAcomp-R1 and AKlaeAcomp-F2/argB-R2 (Table 4). For amplification of DNA fragments, A. kawachii IFO 4308 wild-type genomic DNA and a plasmid carrying tandemly connected sC and argB were used as the template DNA. The resultant DNA fragment amplified with primers AKlaeA-F1/argB-R2 was used to transform laeA disruption, yielding the ΔlaeA plus laeA strain. Transformants were selected on M agar medium lacking methionine. Introduction of laeA and sC into the target locus was confirmed by PCR using primers AKlaeA-FC and argB-R2 (Fig. S2).
The ΔsC ΔlaeA strain was transformed using the sC cassette for use of the same auxotrophic genetic background strains for the comparative study. The sC cassette was generated by PCR using A. kawachii genomic DNA as the template and primers sC-comp-F and sC-comp-R (Table 4) and was used to transform the ΔsC ΔlaeA strain and yield the ΔlaeA strain (Table 3). Transformants were selected on M agar medium lacking methionine.
Construction of putative citrate exporter gene disruption strain.
cexA was disrupted in A. kawachii SO2 (44) by insertion of argB. A gene replacement cassette encompassing the homology arm at the 5′ end of the cexA, argB selection marker, and homology arm at the 3′ end of cexA was constructed using recombinant PCR with the primer pairs AKcexA-FC/AKcexA-R1, AKcexA-F2/AKcexA-R2, and AKcexA-F3/AKcexA-RC, respectively (Table 4). For amplification of argB, plasmid pDC1 was used as the template DNA (45). For amplification of the other DNA fragment, A. kawachii IFO 4308 wild-type genomic DNA was used as a template. The resultant DNA fragment was amplified with primers AKcexA-F1 and AKcexA-R3 and was used to transform A. kawachii SO2 and yield the ΔsCΔcexA strain. M agar plates lacking arginine were used for selection of transformants. Introduction of argB into the cexA locus was confirmed by PCR using the primer pair AKcexA-FC and AKcexA-RC (Fig. S3). After confirmation of gene disruption, the ΔsC ΔcexA strain was transformed with an sC cassette to use the same auxotrophic genetic background strains for comparative study. This cassette was synthesized by PCR using primers sC-comp-F and sC-comp-R and A. kawachii genomic DNA as the template (Table 4). Transformants were selected on M agar medium lacking methionine.
Construction of the putative citrate exporter overexpression strain.
Plasmid pGS-PgpdA (46), which carries A. kawachii sC (28), was used for overexpression of cexA. The cexA gene was amplified by PCR with primers pGSG-cexA-inf-F/pGSG-cexA-inf-R using A. kawahcii genomic DNA as the template (Table 4). The amplicon was inserted into the SalI site of pGS-PgpdA, thereby yielding pGS-PgpdA-cexA, which was used to transform the ΔsC ΔcexA and ΔsC ΔlaeA strains, yielding the ΔcexA plus OEcexA and ΔlaeA plus OEcexA strains, respectively (Table 3). Transformants were selected on M agar medium lacking methionine. Single-copy integration of pGS-PgpdA-cexA was confirmed by real-time RT-PCR (data not shown). In addition, the transformants were transferred to YPD with methyl red; then the appearance of a red border around the colonies was confirmed.
Measurement of extracellular and intracellular organic acids and extracellular glucose.
Levels of extracellular and intracellular organic acids were measured as described previously (24). Briefly, 2 × 107 conidial cells of A. kawachii were inoculated into 100 ml of M medium, precultured with shaking (180 rpm) at 30°C for 36 h, and then transferred into 50 ml of CAP medium and further cultured with shaking (163 rpm) at 30°C for 12, 24, 48, or 72 h. Culture supernatant was harvested as the extracellular fraction. Mycelia were used for preparation of the intracellular fraction using a hot-water extraction method (47) with modifications. The freeze-dried and wet mycelial weights were measured as extracellular and intracellular fractions, respectively. Wet mycelia were ground into a powder using mortar and pestle in the presence of liquid nitrogen and then dissolved in 10 ml of hot water (80°C) per 1 g of mycelial powder, vortexed, and centrifuged at 18,800 × g at 4°C for 30 min. The supernatant was taken as the intracellular fraction.
To measure organic acid level, extracellular and intracellular fractions were filtered through a polytetrafluoroethylene (PTFE) filter (0.2-μm pore size; Toyo Roshi Kaisha) and analyzed with HPLC on a Prominence HPLC system (Shimadzu, Kyoto, Japan) equipped with a CDD-10AVP conductivity detector (Shimadzu). The organic acids were separated using two tandem Shimadzu Shim-pack SCR-102H columns (internal diameter, 8 by 300 mm; Shimadzu) at 50°C using 4 mM p-toluenesulfonic acid monohydrate as the mobile phase at a flow rate of 0.8 ml/min. The flow rate of the postcolumn reaction solution (4 mM p-toluenesulfonic acid monohydrate, 16 mM bis-Tris, and 80 μM EDTA) was 0.8 ml/min.
To measure the glucose level, the filtrated extracellular fraction was analyzed with the Prominence HPLC system equipped with a RID-10A refractive index detector (Shimadzu). Glucose was separated using a COSMOSIL Sugar-D column (internal diameter, 4.6 by 250 mm; Nacalai Tesque, Kyoto, Japan) at 40°C using acetonitrile and water (3:1) as the mobile phase at a flow rate of 1.0 ml/min.
CAGE analysis.
Total RNA was extracted from mycelia. Briefly, 2 × 107 conidial cells of the A. kawachii strains were inoculated into 100 ml of M medium, precultured with shaking (180 rpm) at 30°C for 36 h, and then transferred to 50 ml of CAP medium and further cultured with shaking (163 rpm) at 30°C for 12 h. The mycelia were ground to a powder as described above. Then, RNA was extracted using RNAiso Plus reagent (TaKaRa Bio, Shiga, Japan). RNA samples were treated with the SV total RNA isolation system (Promega, Madison, WI) according to the manufacturer’s protocol.
Library preparation, sequencing, and data analysis for CAGE (48) were performed by Kabushiki Kaisha DNAFORM (Kanagawa, Japan). All CAGE experiments were performed three times with RNA samples obtained from independently prepared mycelia. First-strand cDNAs were transcribed to the 5′ end of capped RNAs and attached to CAGE barcode tags, and these tags were sequenced using the NextSeq 500 system (Illumina, San Diego, CA) and mapped to the A. kawachii IFO 4308 genome (49) using BWA software (v0.5.9) after discarding ribosomal or non-A/C/G/T-base-containing RNAs. For tag clustering, CAGE tag 5′ coordinates were input for RECLU clustering (50). The criteria for linking transcriptional start sites and predicted coding sequences were within 600 bp upstream or downstream of the predicted start codon. Triplicate data were analyzed and the expression ratio was also calculated as the log (base 2) ratio through the RECLU pipeline.
Transcriptional analysis.
For RNA extraction from mycelia, conidia (2 × 107 cells) of the A. kawachii control, ΔlaeA, and ΔlaeA plus OEcexA strains were inoculated into 100 ml of M medium and cultured with shaking (180 rpm) for 36 h at 30°C. After incubation, mycelia were collected and divided into two equal portions, transferred individually to CAP medium, and cultured with shaking (163 rpm) for 12 h at 30°C. Mycelia were ground as described above and RNA was extracted using RNAiso Plus (TaKaRa Bio), and then cDNA was synthesized from total RNA using a PrimeScript Perfect real-time reagent kit (TaKaRa Bio) according to manufacturer’s protocols. Real-time RT-PCR was performed using a Thermal Cycler Dice real-time system MRQ (TaKaRa Bio) with SYBR Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa Bio). The following primer sets were used: AKcexA-RT-F and AKcexA-RT-R for cexA and AKactA-RT-F and AKactA-RT-R for actA (Table 4) (51).
ChIP-qPCR.
A. kawachii conidia were cultured as described above. ChIP was performed as previously described (52) using normal anti-mouse IgG as a negative control (Cosmo Bio, Tokyo, Japan), as well as anti-histone H3 (Medical and Biological Laboratories, Nagoya, Japan), anti-H3K4me3 (Medical and Biological Laboratories), and anti-H3K9me3 (Medical and Biological Laboratories) antibodies. Two micrograms of antibody was used with 200 mg of total protein in each ChIP experiment. DNA quantification was performed with real-time qPCR using SYBR Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa Bio) and the primer set cexA-ChIP-F and cexA-ChIP-R (Table 4). Positions of the primers relative to the ATG site of cexA were +2 to +26 for cexA-ChIP-F and +238 to +257 for cexA-ChIP-R. Relative amounts of DNA (i.e., percent input DNA) were calculated by dividing immunoprecipitated DNA by input DNA.
Data availability.
CAGE data were deposited in the Gene Expression Omnibus under accession number GSE135849 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135849).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kenta Hamada and Aoi Miyamoto for helpful discussion and technical support.
This work was supported by JSPS KAKENHI grants (18K05394 and 19K05773) and a grant from the Noda Institute for Scientific Research. C.K. was supported by a grant-in-aid for JSPS Research Fellows (17J02753).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Akiyama H. 2010. Sake: the essence of 2000 years of Japanese wisdom gained from brewing alcoholic beverages from rice. Brewing Society of Japan, Tokyo, Japan. [Google Scholar]
- 2.Suganuma T, Fujita K, Kitahara K. 2007. Some distinguishable properties between acid-stable and neutral types of alpha-amylases from acid-producing koji. J Biosci Bioeng 104:353–362. doi: 10.1263/jbb.104.353. [DOI] [PubMed] [Google Scholar]
- 3.Yamada O, Takara R, Hamada R, Hayashi R, Tsukahara M, Mikami S. 2011. Molecular biological researches of Kuro-Koji molds, their classification and safety. J Biosci Bioeng 112:233–237. doi: 10.1016/j.jbiosc.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Hong SB, Lee M, Kim DH, Varga J, Frisvad JC, Perrone G, Gomi K, Yamada O, Machida M, Houbraken J, Samson RA. 2013. Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS One 8:e63769. doi: 10.1371/journal.pone.0063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong SB, Yamada O, Samson RA. 2014. Taxonomic re-evaluation of black koji molds. Appl Microbiol Biotechnol 98:555–561. doi: 10.1007/s00253-013-5332-9. [DOI] [PubMed] [Google Scholar]
- 6.Karaffa L, Kubicek CP. 2003. Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol 61:189–196. doi: 10.1007/s00253-002-1201-7. [DOI] [PubMed] [Google Scholar]
- 7.Magnuson JK, Lasure LL. 2004. Organic acid production by filamentous fungi, p 307–340. In Tkacz JS, Lange L (ed), Advances in fungal biotechnology for industry, agriculture, and medicine. Springer, Boston, MA. [Google Scholar]
- 8.Legisa M, Mattey M. 2007. Changes in primary metabolism leading to citric acid overflow in Aspergillus niger. Biotechnol Lett 29:181–190. doi: 10.1007/s10529-006-9235-z. [DOI] [PubMed] [Google Scholar]
- 9.Niu J, Arentshorst M, Nair PD, Dai Z, Baker SE, Frisvad JC, Nielsen KF, Punt PJ, Ram AF. 2015. Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 (Bethesda) 6:193–204. doi: 10.1534/g3.115.024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linde T, Zoglowek M, Lübeck M, Frisvad JC, Lübeck PS. 2016. The global regulator LaeA controls production of citric acid and endoglucanases in Aspergillus carbonarius. J Ind Microbiol Biotechnol 43:1139–1147. doi: 10.1007/s10295-016-1781-3. [DOI] [PubMed] [Google Scholar]
- 11.Bok JW, Keller NP. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3:527–535. doi: 10.1128/ec.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss J, Reyes-Dominguez Y. 2011. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol 48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarikaya-Bayram Ö, Palmer JM, Keller N, Braus GH, Bayram Ö. 2015. One Juliet and four Romeos: VeA and its methyltransferases. Front Microbiol 6:1. doi: 10.3389/fmicb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Lv Y, Li X, Lin Y, Deng H, Pan L. 2018. Profiling of secondary metabolite gene clusters regulated by LaeA in Aspergillus niger FGSC A1279 based on genome sequencing and transcriptome analysis. Res Microbiol 169:67–77. doi: 10.1016/j.resmic.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Odoni DI, Vazquez-Vilar M, van Gaal MP, Schonewille T, Martins Dos Santos VAP, Tamayo-Ramos JA, Suarez-Diez M, Schaap PJ. 2019. Aspergillus niger citrate exporter revealed by comparison of two alternative citrate producing conditions. FEMS Microbiol Lett 366:fnz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiger MG, Rassinger A, Mattanovich D, Sauer M. 2019. Engineering of the citrate exporter protein enables high citric acid production in Aspergillus niger. Metab Eng 52:224–231. doi: 10.1016/j.ymben.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Palmer JM, Theisen JM, Duran RM, Grayburn WS, Calvo AM, Keller NP. 2013. Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet 9:e1003193. doi: 10.1371/journal.pgen.1003193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawauchi M, Nishiura M, Iwashita K. 2013. Fungus-specific sirtuin HstD coordinates secondary metabolism and development through control of LaeA. Eukaryot Cell 12:1087–1096. doi: 10.1128/EC.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff B, Kamerewerd J, Sigl C, Mitterbauer R, Zadra I, Kürnsteiner H, Kück U. 2010. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryot Cell 9:1236–1250. doi: 10.1128/EC.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayram O, Braus GH. 2012. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 21.Torres N. 1994. Modelling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus niger. I. Model definition and stability of the steady state. Biotechnol Bioeng 44:104–111. doi: 10.1002/bit.260440115. [DOI] [PubMed] [Google Scholar]
- 22.Torres N. 1994. Modelling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus niger. II. Sensitivity analysis. Biotechnol Bioeng 44:112–118. doi: 10.1002/bit.260440116. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Vasquez F, González-Alcón C, Torres NV. 2000. Metabolism of citric acid production by Aspergillus niger: model definition, steady-state analysis and constrained optimization of citric acid production rate. Biotechnol Bioeng 70:82–108. doi:. [DOI] [PubMed] [Google Scholar]
- 24.Kadooka C, Izumitsu K, Onoue M, Okutsu K, Yoshizaki Y, Takamine K, Goto M, Tamaki H, Futagami T, Kadooka C, Izumitsu K, Onoue M, Okutsu K, Yoshizaki Y, Takamine K, Goto M, Tamaki H, Futagami T. 2019. Mitochondrial citrate transporters CtpA and YhmA are required for extracellular citric acid accumulation and contribute to cytosolic acetyl coenzyme A generation in Aspergillus luchuensis mut. kawachii. Appl Environ Microbiol 85:e03136-18. doi: 10.1128/AEM.03136-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punt PJ, Dingemanse MA, Kuyvenhoven A, Soede RD, Pouwels PH, van den Hondel CA. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109. doi: 10.1016/0378-1119(90)90142-e. [DOI] [PubMed] [Google Scholar]
- 26.Punt PJ, Zegers ND, Busscher M, Pouwels PH, van den Hondel CA. 1991. Intracellular and extracellular production of proteins in Aspergillus under the control of expression signals of the highly expressed Aspergillus nidulans gpdA gene. J Biotechnol 17:19–34. doi: 10.1016/0168-1656(91)90024-P. [DOI] [PubMed] [Google Scholar]
- 27.Benito B, Garciadeblás B, Pérez-Martín J, Rodríguez-Navarro A. 2009. Growth at high pH and sodium and potassium tolerance in media above the cytoplasmic pH depend on ENA ATPases in Ustilago maydis. Eukaryot Cell 8:821–829. doi: 10.1128/EC.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinclova-Zimmermannova O, Gaskova D, Sychrova H. 2006. The Na+, K+/H+-antiporter Nha1 influences the plasma membrane potential of Saccharomyces cerevisiae. FEMS Yeast Res 6:792–800. doi: 10.1111/j.1567-1364.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Navarro A. 2000. Potassium transport in fungi and plants. Biochim Biophys Acta 1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 30.Purschwitz J, Müller S, Kastner C, Schöser M, Haas H, Espeso EA, Atoui A, Calvo AM, Fischer R. 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol 18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 31.Downes DJ, Davis MA, Wong KH, Kreutzberger SD, Hynes MJ, Todd RB. 2014. Dual DNA binding and coactivator functions of Aspergillus nidulans TamA, a Zn(II)2Cys6 transcription factor. Mol Microbiol 92:1198–1211. doi: 10.1111/mmi.12620. [DOI] [PubMed] [Google Scholar]
- 32.Gerke J, Bayram O, Feussner K, Landesfeind M, Shelest E, Feussner I, Braus GH. 2012. Breaking the silence: protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans. Appl Environ Microbiol 78:8234–8244. doi: 10.1128/AEM.01808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomi K. 2019. Regulatory mechanisms for amylolytic gene expression in the koji mold Aspergillus oryzae. Biosci Biotechnol Biochem 83:1385–1401. doi: 10.1080/09168451.2019.1625265. [DOI] [PubMed] [Google Scholar]
- 34.Yin WB, Reinke AW, Szilágyi M, Emri T, Chiang YM, Keating AE, Pócsi I, Wang CC, Keller NP. 2013. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 159:77–88. doi: 10.1099/mic.0.063370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee BY, Han SY, Choi HG, Kim JH, Han KH, Han DM. 2005. Screening of growth- or development-related genes by using genomic library with inducible promoter in Aspergillus nidulans. J Microbiol 43:523–528. [PubMed] [Google Scholar]
- 36.Hynes MJ, Murray SL, Duncan A, Khew GS, Davis MA. 2006. Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans. Eukaryot Cell 5:794–805. doi: 10.1128/EC.5.5.794-805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann B, Valerius O, Andermann M, Braus GH. 2001. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol Biol Cell 12:2846–2857. doi: 10.1091/mbc.12.9.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tüncher A, Reinke H, Martic G, Caruso ML, Brakhage AA. 2004. A basic-region helix-loop-helix protein-encoding gene (devR) involved in the development of Aspergillus nidulans. Mol Microbiol 52:227–241. doi: 10.1111/j.1365-2958.2003.03961.x. [DOI] [PubMed] [Google Scholar]
- 39.Cairns TC, Zheng X, Zheng P, Sun J, Meyer V. 2019. Moulding the mould: understanding and reprogramming filamentous fungal growth and morphogenesis for next generation cell factories. Biotechnol Biofuels 12:77. doi: 10.1186/s13068-019-1400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papagianni M, Mattey M, Kristiansen B. 1994. Morphology and citric acid production of Aspergillus niger PM1. Biotechnol Lett 16:929–934. doi: 10.1007/BF00128627. [DOI] [Google Scholar]
- 41.Yin X, Shin HD, Li J, Du G, Liu L, Chen J. 2017. Comparative genomics and transcriptome analysis of Aspergillus niger and metabolic engineering for citrate production. Sci Rep 7:41040. doi: 10.1038/srep41040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. 2010. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol 76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patananan AN, Palmer JM, Garvey GS, Keller NP, Clarke SG. 2013. A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine. J Biol Chem 288:14032–14045. doi: 10.1074/jbc.M113.465765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadooka C, Onitsuka S, Uzawa M, Tashiro S, Kajiwara Y, Takashita H, Okutsu K, Yoshizaki Y, Takamine K, Goto M, Tamaki H, Futagami T. 2016. Marker recycling system using the sC gene in the white koji mold, Aspergillus luchuensis mut. kawachii. J Gen Appl Microbiol 62:160–163. doi: 10.2323/jgam.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Aramayo R, Adams TH, Timberlake WE. 1989. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics 122:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimoto D, Kadooka C, Saenrungrot P, Okutsu K, Yoshizaki Y, Takamine K, Goto M, Tamaki H, Futagami T. 2019. Pex16 is involved in peroxisome and Woronin body formation in the white koji fungus, Aspergillus luchuensis mut. kawachii. J Biosci Bioeng 127:85–92. doi: 10.1016/j.jbiosc.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Canelas AB, ten Pierick A, Ras C, Seifar RM, van Dam JC, van Gulik WM, Heijnen JJ. 2009. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal Chem 81:7379–7389. doi: 10.1021/ac900999t. [DOI] [PubMed] [Google Scholar]
- 48.Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, Hayashizaki Y, Carninci P. 2006. CAGE: cap analysis of gene expression. Nat Methods 3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 49.Futagami T, Mori K, Yamashita A, Wada S, Kajiwara Y, Takashita H, Omori T, Takegawa K, Tashiro K, Kuhara S, Goto M. 2011. Genome sequence of the white koji mold Aspergillus kawachii IFO 4308, used for brewing the Japanese distilled spirit shochu. Eukaryot Cell 10:1586–1587. doi: 10.1128/EC.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohmiya H, Vitezic M, Frith MC, Itoh M, Carninci P, Forrest AR, Hayashizaki Y, Lassmann T, FANTOM Consortium. 2014. RECLU: a pipeline to discover reproducible transcriptional start sites and their alternative regulation using capped analysis of gene expression (CAGE). BMC Genomics 15:269. doi: 10.1186/1471-2164-15-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Futagami T, Mori K, Wada S, Ida H, Kajiwara Y, Takashita H, Tashiro K, Yamada O, Omori T, Kuhara S, Goto M. 2015. Transcriptomic analysis of temperature responses of Aspergillus kawachii during barley koji production. Appl Environ Microbiol 81:1353–1363. doi: 10.1128/AEM.03483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernreiter A, Ramon A, Fernández-Martínez J, Berger H, Araújo-Bazan L, Espeso EA, Pachlinger R, Gallmetzer A, Anderl I, Scazzocchio C, Strauss J. 2007. Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol Cell Biol 27:791–802. doi: 10.1128/MCB.00761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen MR, Nielsen ML, Nielsen J. 2008. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol 4:178. doi: 10.1038/msb.2008.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flipphi M, Sun J, Robellet X, Karaffa L, Fekete E, Zeng AP, Kubicek CP. 2009. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol 46:S19–S44. doi: 10.1016/j.fgb.2008.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CAGE data were deposited in the Gene Expression Omnibus under accession number GSE135849 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135849).