Abstract
SMYD3 (SET and MYND domain-containing protein 3) is involved in histone modification, which initiates oncogenesis by activating transcription of multiple downstream genes. To investigate associations of variable numbers of tandem repeats (VNTR) variants in the SMYD3 gene promoter, SMYD3 serum levels and SMYD3 mRNA expression in hepatitis B virus (HBV) infection and clinical progression of related liver disease. SMYD3 VNTRs were genotyped in 756 HBV patients and 297 healthy controls. SMYD3 serum levels were measured in 293 patients and SMYD3 mRNA expression was quantified in 48 pairs of hepatocellular tumor and adjacent non-tumor liver tissues. Genotype SYMD3 VNTR 3/3 was more frequent among HCC patients than in controls (Padjusted = 0.037). SMYD3 serum levels increased according to clinical progression of liver diseases (P = 0.01); HCC patients had higher levels than non-HCC patients (P = 0.04). Among patients with SMYD3 VNTR 3/3, HCC patients had higher SMYD3 levels than others (P < 0.05). SMYD3 mRNA expression was up-regulated in HCC tumor tissues compared to other tissues (P = 0.008). In conclusion, upregulation of SMYD3 correlates with the occurrence of HCC and SMYD3 VNTR 3/3 appears to increase the risk of HCC through increasing SMYD3 levels. SMYD3 may be an indicator for HCC development in HBV patients.
Subject terms: Hepatocellular carcinoma, Hepatitis B
Introduction
Although effective hepatitis B virus (HBV) vaccines are in use worldwide, HBV-related liver diseases are still a major health concern with approximately 257 million chronic infections and 887,000 deaths in 20151. HBV is the main cause of primary hepatocellular carcinoma (HCC)2. The risk of HCC development is approximately 40 times higher in chronic HBV carriers (CHB) than in non-carriers3. Vietnam has a high prevalence of HBV infections, ranging from 10–20% in the general population and 20–40% among high-risk groups2,4. As a result, Vietnam is one of the countries with a high incidence of HCC with >25,000 new cases reported in 20185,6.
Further to viral factors contributing to cancer development, methylation of histone proteins is an important mechanism involved in multiple types of cancers including HCC7,8. Methylation of histone proteins at lysine residues can lead to chromatin remodelling, transcription, and signal transduction9. The SET and MYND domain containing proteins (SMYDs) belong to a family of the multi-domain SET-containing histone lysine methyltransferases and play a crucial role in histone methylation. To date, five SMYD family members have been recognized (SMYD1-SMYD5). SMYD3 is the most important member, as several findings have demonstrated its role in tumor cell growth and its increased expression in various cancers.
SYMD3 promotes dimethylation and trimethylation of histone H3 lysine 4 (H3K4), which initiates oncogenesis by activating transcription of multiple downstream target genes10,11. SYMD3 overexpression causes cell proliferation, migration and adhesion, whereas suppression by RNAi inhibits cell proliferation and migration, indicating that SMYD3 plays an important role in carcinogenesis12–14. SYMD3 was found upregulated in HCC, colorectal and in breast and lung cancers12,15,16. In vitro interaction of SMYD3 with HBV has been demonstrated, showing that SMYD3 expression was upregulated by hepatitis B x protein (HBx) in HepG2 cells, promoting HCC development and clinical progression17. Moreover, SMYD3 is a HBx-interacting protein, and this interaction induces activation of the activator protein 1 (AP-1), which increases the risk of HCC formation18,19.
The common variable number of tandem repeat (VNTR) CCGCC sequence in the SMYD3 promoter region is the binding site for the E2F transcription factor 1 (E2F-1) and shown to be a susceptibility factor for human malignancies20. It has been suggested that, compared to the genotype containing two copies, the genotype involving three copies of the CCGCC motif might enhance the binding affinity to E2F-1 and, as a result, promote cancer progression by activating transcription of multiple oncogenes such as myc gene (myc), signal transducer and activator of transcription 3 (STAT3) and β-catenin (β-cat)14,20,21. Moreover, the VNTR genotype 3/3 was associated with a higher risk of colorectal cancer, HCC, breast cancer, ovarian cancer and esophageal squamous cell carcinoma20,22,23. Particularly in liver diseases, VNTR genotype 3/3 contributed an over 3-fold increased risk of HCC in a Japanese and a Chinese population20,24. The present study aimed to investigate the association of VNTR polymorphisms in the SMYD3 promoter and SMYD expression with HBV infection and clinical progression of HBV-related liver diseases, in particular progression to HCC.
Patients and Methods
Patients
Seven hundred and fifty-six Vietnamese chronic HBV-infected patients enrolled between 2013 and 2015 at the 108 Military Central Hospital, Hanoi, Vietnam, were recruited. All patients were negative for antibodies for hepatitis C virus (anti-HCV) and antibodies for human immunodeficiency virus (anti-HIV) nor had a history of alcohol or drug abuse. Patients were categorized into the subgroups of chronic hepatitis B (CHB) without liver cirrhosis (LC) or HCC (n = 246), HBV-related LC patients (n = 174) and HBV-related HCC patients (n = 336). The clinical and diagnostic characteristics of the study group have been described previously25. LC and HCC patients were further classified according to Child-Pugh scores A, B, and C26 and HCC patients were categorized according to the Barcelona Clinic Liver Cancer (BCLC) staging system27. The control group (healthy controls; HC) consisted of 297 healthy blood donors. All HCs were HBV surface antigen (HBsAg), anti-HCV and anti-HIV negative. Five mls of venous blood were collected from each participant. Serum was stored at −80 °C until further use.
In order to determine SMYD3 mRNA expression, pairs of HCC tumor and adjacent non-tumor tissue specimens were obtained from 48 HCC patients who underwent surgery at the 108 Military Central Hospital. The tumor stage was scored following the BCLC system28.
Ethics statement
Informed written consent was obtained from all participants after detailed explanation of the study at the time of blood sampling. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the institution’s human research committee. The Institutional Review Board of the 108 Military Central Hospital, Hanoi, Vietnam (108MCH/Res/Epi-HBV-HDV-HEV-D2-14-03-2014) and the Ethics committee of University of Tübingen (206/2012B02) approved the study.
SMYD3 VNTR genotyping
Genomic DNA of all participants was isolated from whole blood (DNA purification kit; Qiagen, Hilden, Germany). The promoter region of the SMYD3 gene was amplified using primers SMYD3_F (5′-CGC CTG TCT TCT GCG CAG TCG-3′) and SMYD3_R (5′-CCC GAG AAG GCA GCG GTC G-3′). Amplicons underwent DNA sequencing.
Quantification of SMYD3 serum levels by ELISA
Of the 756 patients, 293 individuals were tested for SMYD3 serum levels, measured by a commercially available human SMYD3 sandwich ELISA kit (Wuhan Fine Biological Technology Co. Ltd, Wuhan City, China). In order to determine SMYD3 concentrations, a standard curve was plotted (https://www.curveexpert.net/) based on mean of OD values and the known concentration of the standards. Finally, SMYD3 concentrations were interpolated based on the standard curve. The detectable range of the kit was 31.25–2000 pg/ml.
Quantification of SMYD3 mRNA by RT-PCR
Total RNA of the liver tissues was extracted with Trizol reagent (Life Technologies, Carlsbad, CA, USA), followed by reverse transcription into cDNA (QuantiTect Reverse Transcription Kit; Qiagen, Hilden, Germany). SMYD3 mRNA levels were assessed through quantification of SMYD3 cDNA by qRT-PCR using SYBR Green PCR master mix (Bioline, Luckenwalde, Germany). The GAPDH gene (glyceraldehyde-3-phosphate dehydrogenase) was used as reference. Primer sequences were SMYD3_F: 5′-GTT GGC CTA TAT CCC AGT ATC TCT TTG CTC -3′, and SMYD3_R: 5′-ACC AGT TAG CAT ATC AGC ATC CTT GTC CTG -3′, GAPDH_F: 5′-CCA CCC ATG GCA AAT TCC ATG GCA-3′ and GAPDH_R: 5′-TCT AGA CGG CAG GTC AGG TCC ACC-3′. All qRT-PCR reactions were performed in duplicate and repeated twice (LightCycler® 480 real-time PCR system; Roche, Basel, Switzerland). The fold change of SMYD3 mRNA was normalized based upon the ΔΔCt method against expression of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Statistical analysis
Chi-square and Fisher´s exact tests were used to test for differences of categorical variables. Kruskal-Wallis and Mann-Whitney-Wilcoxon tests were applied to compare quantitative variables. Hardy-Weinberg equilibrium was assessed. Binary logistic regression models adjusted for age and gender were applied to determine SMYD3 VNTR associations with HBV-related liver diseases. Adjusted odds ratios (OR) with 95% confidence intervals (CI) were calculated. Paired-samples t-test was used to compare SMYD3 mRNA levels in tumor and adjacent non-tumor tissues. A linear regression model was applied to analyze the relationship of clinical parameters of patients and SMYD3 levels. Pearson’s correlation coefficient test was used to analyze correlations between SMYD3 levels and clinical parameters. Statistical analyses were performed using SPSS version 22 (SPSS Statistics, IBM, Armonk, NY, USA) and GraphPad Prism 7 (http://www.graphpad.com). Significance was set at P < 0.05.
Results
Patient characteristics
Demographic, laboratory and clinical parameters of the 1053 study participants are summarized in Table 1. In the HC group, the mean age was 43 years (range: 18–69), and majority of HCs were male (66.7%). Of the 756 patients, 630 (83.3%) were male; the mean age was 52 years (12–91). The median age of patients and the proportion of males increased according to the degree of progression of liver diseases (P < 0.0001). As expected, albumin, prothrombin levels and platelet counts were higher among CHB patients compared to the other patient subgroups (P < 0.0001). HBV DNA levels did not differ among patient subgroups (P = 0.4). Higher total bilirubin and direct bilirubin were observed in LC patients compared to the other groups (P < 0.0001). Alpha-fetoprotein (AFP) levels were significantly higher in HCC patients compared to CHB and LC patients (P < 0.0001) (Table 1).
Table 1.
Demographic and clinical characteristics of healthy controls and HBV patients.
| Characteristics | HC (n = 297) | HBV patients (n = 756) | CHB (n = 246) | LC (n = 174) | HCC (n = 336) | P value |
|---|---|---|---|---|---|---|
| Age (years) | 43 [16–69] | 52 [12–91] | 41 [12–85] | 56 [20–86] | 57 [15–91] | <0.0001# |
| Male (%) | 66.7 | 83.3 | 75.2 | 82.8 | 93.7 | <0.0001β |
| Child-Pugh | NA | |||||
| Child A | NA | 53/169 | 249/335 | |||
| Child B | NA | 75/169 | 65/300 | |||
| Child C | NA | 41/169 | 21/335 | |||
| Missing | NA | 5 | 1 | |||
| Clinical parameters | ||||||
| AST (IU/L) | NR | 132 [14–6206] | 187 [14–6206] | 119 [15–1221] | 101 [17–983] | <0.0001# |
| ALT (IU/L) | NR | 132 [8–3390] | 222 [9–3390] | 82 [8–1426] | 72 [11–1095] | 0.04# |
| Total bilirubin (µmol/L) | NR | 39.1 [4.1–571] | 34 [5.5–551] | 65.2 [4.1–571] | 29.4 [4.3–392] | <0.0001# |
| Direct bilirubin (µmol/L) | NR | 17.2 [0.4–349] | 16.1 [0.7–349] | 29.5 [0.4–291] | 11.6 [0.4–247.3] | <0.0001# |
| Albumin (g/L) | NR | 37 [15–48] | 42 [25–48] | 31.8 [15–47] | 37 [15–48] | <0.0001# |
| Prothrombin (% of standard) | NR | 82 [13–269] | 94 [17–267] | 60 [13–101] | 84 [20–269] | <0.0001# |
| <0.0001# | ||||||
| WBC (×103/mL) | NR | 6.6 [1.7–20.5] | 6.7 [4.1–13.44] | 6.2 [1.7–20.5] | 6.8 [2.7–17.9] | <0.0001# |
| RBC(×106/mL) | NR | 4.5 [1.7–6.8] | 4.9 [3.1–6.8] | 3.9 [1.9–6.7] | 4.5 [1.7–6.8] | <0.0001# |
| PLT (×103/ml) | NR | 174 [17–441] | 218 [66–379] | 106 [17–441] | 177 [34–432] | <0.0001# |
| HBV DNA (log10 copies/ml) | NR | 5.1 [1–10] | 5.2 [2–10] | 5 [1–10] | 5.1 [1–9] | 0.4# |
| AFP (IU/L) | NR | 142 [1–4029] | 7.4 [1–250] | 40 [1.18–707] | 280 [1–4029] | <0.0001# |
CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HC, healthy control; RBC, red blood cells; WBC, white blood cells; PLT, platelets. AST and ALT, aspartate and alanine aminotransferase; AFP, alpha-fetoprotein; NR, normal range, NA, not applicable. Values given are medians and ranges. (#) Kruskal-Wallis test (β): chi-square test.
SMYD3 VNTRs and HBV-related liver diseases
The allele frequencies and genotype distributions of SMYD3 VNTR polymorphisms are given in Table 2. Allele frequencies of VNTR 2 and 3 were 21.7% and 78.3% among controls and 12% and 88% in HBV patients, respectively. The three tandem repeat allele (VNTR 3) was more prevalent among HBV patients than HCs (OR = 1.4, 95% CI = 1.07–1.86, adjusted P = 0.017,). Genotype VNTR 3/3 was observed more frequently in patients compared to HCs (OR = 1.4, 95% CI = 1.02–1.97, adjusted P = 0.036). Although the frequencies of VNTRs 2/2 and 2/3 were higher among HCs than in the patient group, the differences were not significant.
Table 2.
Association of the variable number of tandem repeats of SMYD3 with HBV-related liver diseases.
| SMYD3 tandem repeat | HC n = 297 (%) | CHB n = 246 (%) | LC n = 174 (%) | HCC n = 336 (%) | CHB + LC n = 420 (%) | HBV total n = 756 (%) | CHB vs HC | LC vs HC | HCC vs HC | HBV vs HC | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P value | OR(95%CI) | P value | OR(95%CI) | P value | OR(95%CI) | P value | |||||||
| Genotypes | ||||||||||||||
| VNTR 2/2 | 21 (7.1) | 14 (5.7) | 6 (3.4) | 7 (2.1) | 20 (4.8) | 27 (3.6) | Reference | Reference | Reference | Reference | ||||
| VNTR 2/3 | 64 (21.5) | 37 (15) | 29 (16.7) | 61 (18.2) | 66 (15.7) | 127 (16.8) | 0.75 (0.32–1.76) | 0.5 | 1.8 (0.59–5.6) | 0.3 | 2.7 (0.89–7.92) | 0.079 | 1.2 (0.58–2.44) | 0.6 |
| VNTR 3/3 | 212 (71.4) | 195 (79.3) | 139 (79.9) | 268 (79.8) | 334 (79.5) | 602 (79.6) | 1.2 (0.6–2.6) | 0.5 | 2.2 (0.81–6.12) | 0.1 | 2.9(1.07–8) | 0.037 | 1.7 (0.88–3.15) | 0.1 |
| Allele | ||||||||||||||
| VNTR 2 | 106 (21.7) | 65 (13.2) | 41 (11.8) | 75 (11.2) | 106 (12.6) | 181 (12) | Reference | Reference | Reference | Reference | ||||
| VNTR 3 | 488 (78.3) | 427 (86.8) | 307 (88.2) | 597 (88.8) | 734 (87.4) | 1331 (88) | 1.4 (0.97–1.92) | 0.078 | 1.5 (1–2.38) | 0.05 | 1.5 (1–2.14) | 0.05 | 1.4 (1.07–1.86) | 0.017 |
| Dominant | ||||||||||||||
| VNTR 2/2&2/3 | 85 (28.6) | 51 (20.7) | 35 (20.1) | 68 (20.2) | 86 (20.5) | 154 (20.4) | Reference | Reference | Reference | Reference | ||||
| VNTR 3/3 | 212 (71.4) | 195 (79.3 | 139 (79.9) | 268 979.8) | 334 (79.5) | 602 (79.6) | 1.5 (0.99–2.23) | 0.057 | 1.4 (0.88–2.4) | 0.1 | 1.3 (0.86–2.05) | 0.2 | 1.4 (1.02–1.97) | 0.036 |
| Recessive | ||||||||||||||
| VNTR 2/2 | 21 (7.1) | 14 (5.7) | 6 (3.4) | 7 (2.1) | 20 (4.8) | 27 (3.6) | Reference | Reference | Reference | Reference | ||||
| VNTR 2/3&3/3 | 276 (92.9) | 232 (94.3) | 168 (96.6) | 329 (97.9) | 400 (95.2) | 729 (96.4 | 1.1 (0.55–2.35) | 0.7 | 2.2 (0.8–6.02) | 0.1 | 2.9 (1.07–8.03) | 0.036 | 1.6 (0.82–2.94) | 0.2 |
CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HC, healthy controls; n, numbers individuals; OR, Odd ratio. P values were calculated using binary logistic regression model adjusted for age and gender. Bold values reflect statistical significance.
We compared the distribution of allele and genotype frequencies of the SMYD3 VNTRs in the patient subgroups (CHB, LC, HCC) with those in HCs. Genotype VNTR 3/3 occurred more frequently in HCC patients than in HCs (OR = 2.9, 95%CI = 1.09–8, adjusted P = 0.037,), while genotype VNTR 2/2 was nearly three times more frequent in HCs compared to HCC patients (HCC vs HC, OR = 0.34, 95% CI = 0.13–0.93, adjusted P = 0.036). However, no differences were observed in pairwise comparisons between the CHB, LC and HC groups.
We also analyzed associations of SMYD3 VNTR polymorphisms with laboratory parameters of HBV infection (Alpha-fetoprotein (AFP), aspartate transaminase (AST), alanine aminotransferase (ALT), total and direct bilirubin, prothrombin, albumin, viral loads). The levels of these parameters were not different in all patients with different SMYD3 VNTR genotypes as well as in patient subgroups (CHB, LC and HCC) (P > 0.05).
SMYD3 serum levels in HBV-related liver diseases
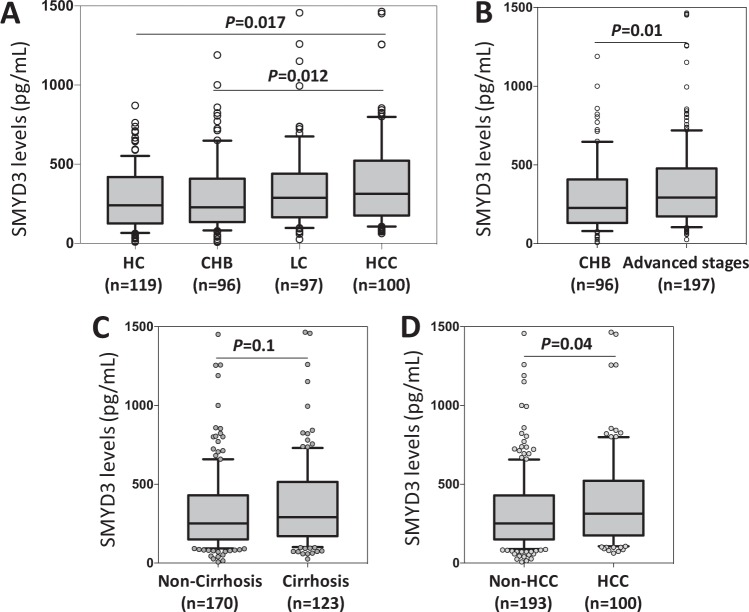
Levels of SMYD3 increased significantly according to the progression of liver diseases (Fig. 1A,B). The SMYD3 median was 227.1 pg/mL in CHB patients, 287.3 pg/mL in LC patients and 311.8 in HCC patients and 292.6 pg/mL in patients with both HCC plus LC (P < 0.05). The post-hoc analysis showed a significant difference between the HCC and CHB groups (P = 0.012). A corresponding result was observed when comparing the median of SMYD3 levels in non-HCC and in HCC patients (251.1 pg/mL and 311.8 pg/mL, respectively, P = 0.04; Fig. 1D), whereas the median of SMYD3 levels did not differ between non-LC and LC individuals (Fig. 1C). These results indicate that SMYD3 plays a role in HCC development. When analyzing the correlation of SMYD3 serum levels with the laboratory parameters in all HBV patients and in patient subgroups, SMYD3 serum levels did not correlate with these parameters in all HBV patients and in the subgroups.
Figure 1.
Association of the SMYD3 serum levels with HBV related liver diseases. The comparison of SMYD3 levels (A) among healthy controls and groups of patients according to HBV-related diseases, CHB, chronic hepatitis B; LC, liver cirrhosis without HCC; HCC, hepatocellular carcinoma; (B) between CHB patients with advanced stages patients (including LC and HCC), (C) between patients with and without cirrhosis, (D) between patients with and without HCC. Box-plots illustrate medians with 25 and 75 percentiles with whiskers to 10 and 90 percentiles; P value were calculated by using Kruskal-Wallis test or Mann-Whitney-Wilcoxon test.
SMYD3 VNTRs and SMYD3 serum levels and progression of HBV-related liver diseases
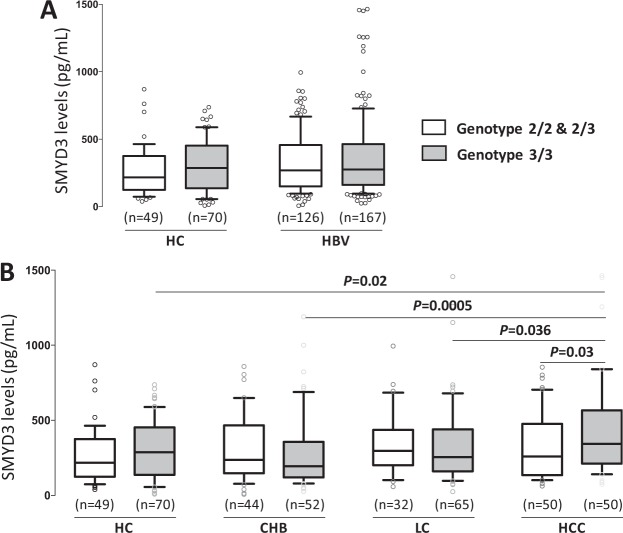
We analyzed the relationship between SMYD3 VNTR genotypes and SMYD3 serum levels in HBV patients (Fig. 2). SMYD3 levels in patients with SMYD3 genotype 3/3 were similar to that in patients with genotypes 2/2 or 2/3 (P > 0.05, Fig. 2A). In the HCC group, patients with genotype VNTR 3/3 had higher SMYD3 levels than those with genotypes VNTR 2/2 and 2/3 (P = 0.03, Fig. 2B). This was not observed in CHB and LC patients (Fig. 2B). In addition, in patients with genotypes 2/2 and 2/3 the levels of SYMD3 did not differ between the CHB, LC and HCC groups. However, patients with genotype VNTR 3/3 had increased SMYD3 serum levels according to the progression of liver diseases (HCC vs. CHB: P = 0.0005, LC vs. CHB: P = 0.036, Fig. 2B). These results indicate that genotype VNTR 3/3 may increase SMYD3 levels and, thus, increase the risk of HCC development.
Figure 2.
Association of the VNTRs of SMYD3 with SMYD3 serum levels in HBV-related liver diseases. Comparison of the SMYD3 serum levels among healthy controls and patients with HBV-related liver diseases who carry different SMYD3 VNTR genotypes. CHB, chronic hepatitis B, LC, liver cirrhosis without HCC, HCC, hepatocellular carcinoma. (A) Among healthy controls and HBV patients, (B) among healthy controls and patient subgroups including CHB, LC and HCC patients, Box-plots illustrate medians with 25 and 75 percentiles with whiskers to 10 and 90 percentiles; P values were calculated by using Mann-Whitney-Wilcoxon test or Kruskal-Wallis test.
SMYD3 mRNA expression in HCC
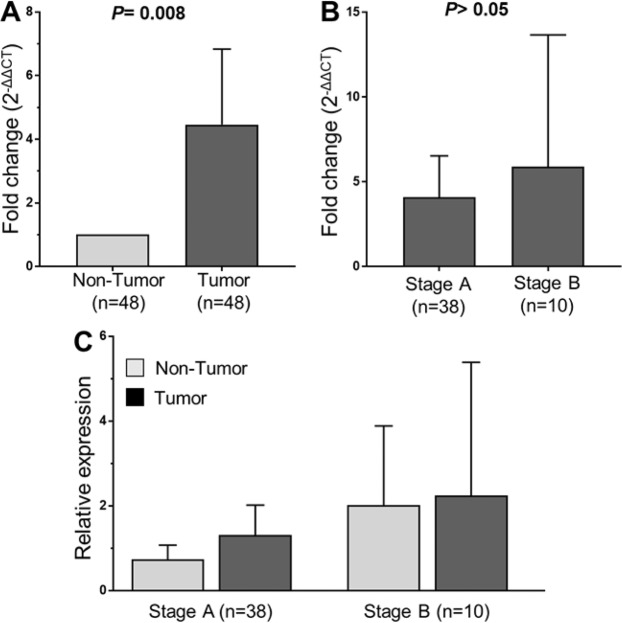
In order to examine whether SMYD3 mRNA is upregulated in cancer stages as seen in other cancers14,15, we analyzed the expression of SMYD3 mRNA in HCC tissue specimens and in adjacent non-tumor liver tissues obtained from 48 HBV-related HCC patients. Expression of SMYD3 mRNA in tumor tissues was significantly higher than that in adjacent non-tumor tissues (P = 0.008, Fig. 3A). We then examined whether SMYD3 mRNA expression was associated with development of HCC by correlating SMYD3 mRNA expression with BCLC stages. SMYD3 mRNA expression was higher in stage-B tumor tissues compared to that in stage-A tissues (Fig. 3B). A similar trend was seen when SMYD3 mRNA expression was compared between non-tumor tissues obtained from stage-A and stage-B HCC patients; however, the difference did not reach significance (Fig. 3C). These results indicate that SMYD3 expression is associated with HBV-related HCC.
Figure 3.
SMYD3 mRNA expression in liver specimens from 48 HCC patients. Relative quantitative real-time PCR analysis of SMYD3 mRNA levels. (A) Relative SMYD3 mRNA expression in tumor tissues and in adjacent non-tumor tissues. (B) The fold changed SMYD3 mRNA levels between the group of stage A-HCC patients and the group of stage B-HCC patients, (C) SMYD3 mRNA expression in stage-A and stage-B tumor tissues and in adjacent non-tumor tissues. P values were calculated by using Mann-Whitney-Wilcoxon test. Data are shown as mean values with 95% confidence intervals.
Discussion
HBV infection is a major cause of HCC. In Vietnam, the prevalence of HBV infection is over 10% in the general population1,5. Further to HBV, other factors contributing to the development of HCC include epigenetic changes such as methylation and histone modifications of regulatory genes7,8,25. SMYD3, a histone H3-K4 specific methyltransferase, is an example of histone modification which is considered a crucial epigenetic factor contributing to the development of various human cancers, including liver cancer12,15. So far, the role of SYMD3 and its encoding gene SYMD3 in HBV-related liver diseases is not clear. We studied the association of genetic variation and expression of variant SYMD3 VNTRs with susceptibility to HBV infection and with liver disease progression. We show that both SMYD3 VNTRs in the promoter region and SYMD3 overexpression are associated with HBV-related HCC.
SMYD3 VNTR variability has been reported to be a significant factor in HCC20,24. An earlier study has reported that VNTR 3/3 homozygosity conferred an over 3-fold increased risk of HCC in a Japanese population20. Accordingly, a study in a Chinese population found that the frequency of the SMYD3 VNTR 3/3 genotype was higher in HCC patients than in controls24. Consistent with these studies, we observed that the frequencies of the SMYD3 VNTR three repeat allele and the 3/3 genotype was higher in HBV patients than in controls (Table 2). Further analysis showed that in the HCC group the SMYD3 repeat 3/3 genotype was more frequent than in HCs (OR = 2.9, P < 0.05, Table 2). Notably, Wang et al.29 did not find an association between the risk of HCC and the three tandem repeat allele.
The SMYD3 VNTRs have been shown to be a susceptibility factor for human cancers, especially for colorectal cancer, breast cancer and HCC20. We observed that the SMYD3 VNTR 3/3 genotype increases the risk of HBV-related HCC. A possible explanation is that SMYD3 VNTR 3/3 can effectively promote its affinity with E2F-1, which is considered an important transcription factor stimulating cellular proliferation and cell cycle progression20,30,31. In HCC development, overexpression of SMYD3 was previously found in distinct cell lines17,32, and in HCC tumors33. However, an association between SMYD3 serum levels and HCC has not been shown so far. In our study, SMYD3 serum levels were significantly increased according to the various HBV-related liver disease stages. SMYD3 levels were higher in advanced liver disease (LC, HCC) compared to CHB, and HCC patients had higher SMYD3 levels than non-HCC patients. These findings indicate that increased SMYD3 levels may be associated with the occurrence of HCC, and SMYD3 serum levels may be considered a potential marker for HCC. Moreover, SMYD3 serum levels in HCC patients with the SMYD3 VNTR genotype 3/3 were higher than in those with genotypes 2/2 and 2/3; this difference was not observed in CHB and LC patients.
Finally, we assessed SMYD3 mRNA expression in liver tissues. In vitro evidence indicates that SMYD3 expression is up-regulated in HCC cell lines12,32. SMYD3 expression was analyzed in HCC tissues, and upregulation was significantly associated with an unfavourable prognosis of HCC33,34. SMYD3 mRNA expression was upregulated in HCC tumor compared to adjacent non-tumor tissues, providing further evidence of a role of SMYD3 in HCC development. Several studies have shown that SMYD3 can interact with the HBV-HBx protein, which can induce upregulation of SMYD3 in HCC, these interactions promote the development of HCC by regulating ERK- and AKT/GSK-3β signaling pathways17–19,35. The SMYD3 VNTR polymorphisms might regulate HCC development possibly by increased SMYD3 expression in the liver. A recent study has shown that SMYD3 may interact with cyclin-dependent kinase 2 (CDK2) and matrix metallopeptidase 2 (MMP2), which are involved in controlling cell cycle, proliferation and invasion of hepatocytes, and thus contributing to the tumorigenicity of HCC33. SMYD3 was also associated with ankyrin repeat and KH domain-containing protein 1 (ANKHD1) that regulate the initiation, progression and metastasis in HCC36 Although our data underline the significance of the SMYD3 VNTR 3/3 genotype and SMYD3 overexpression in HBV-related HCC development, the interaction between HBV and SYMD3 needs to be investigated further.
In conclusion, our study shows the association of the SMYD3 VNTR 3/3 as well as upregulation of SMYD3 serum levels and mRNA expression with HCC development, suggesting that SYMD3 has some potential for the prediction of HCC in HBV patients.
Acknowledgements
We thank all patients and healthy individuals for their participation. T.P.V. acknowledges the financial support by DAAD-PAGEL (57140033) for fellowships of Vietnamese students as well as support from the Federal Ministry of Education and Research, Germany (BMBF01DP19006A, BMBF01DP17047). The funder had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
T.P.V. designed, supervised the study, contributed materials and reagents, and wrote the manuscript. P.G.K. contributed to materials and reagents. M.T.B. recruited patients and collected samples, performed the experiments, carried out the statistical analyses, interpreted data and wrote the manuscript. N.X.H. recruited patients and collected samples, carried out the statistical analyses. H.V.T., C.-T.B. and H.W. contributed to the analysis, interpreted data and the manuscript. D.P.G. contributed to the experimental design. L.H.S., N.L.T. and M.H.B. recruited patients. C.G.M. revised the draft and edited the final version of the manuscript. All authors agreed with the results and conclusions.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. Hepatitis B. (2017).
- 2.Nguyen VT. Hepatitis B infection in Vietnam: current issues and future challenges. Asia Pac. J. Public. Health. 2012;24:361–373. doi: 10.1177/1010539510385220. [DOI] [PubMed] [Google Scholar]
- 3.Lee MH, et al. Prediction models of long-term cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: risk scores integrating host and virus profiles. Hepatology. 2013;58:546–554. doi: 10.1002/hep.26385. [DOI] [PubMed] [Google Scholar]
- 4.WHO. Hepatitis B in Vietnam. (2018).
- 5.GLOBOCAN. The cancer rates in Vietnam. (2018).
- 6.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J. Gastroenterol. 2014;20:333–345. doi: 10.3748/wjg.v20.i2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCabe MT, Mohammad HP, Barbash O, Kruger RG. Targeting Histone Methylation in Cancer. Cancer J. 2017;23:292–301. doi: 10.1097/PPO.0000000000000283. [DOI] [PubMed] [Google Scholar]
- 9.Spellmon N, Holcomb J, Trescott L, Sirinupong N, Yang Z. Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunizaki M, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 11.Zou JN, et al. Knockdown of SMYD3 by RNA interference down-regulates c-Met expression and inhibits cells migration and invasion induced by HGF. Cancer Lett. 2009;280:78–85. doi: 10.1016/j.canlet.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Hamamoto R, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 13.Luo XG, et al. Effects of SMYD3 overexpression on transformation, serum dependence, and apoptosis sensitivity in NIH3T3 cells. IUBMB Life. 2009;61:679–684. doi: 10.1002/iub.216. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Overexpression of SMYD3 was associated with increased STAT3 activation in gastric cancer. Med. Oncol. 2015;32:404. doi: 10.1007/s12032-014-0404-y. [DOI] [PubMed] [Google Scholar]
- 15.Hamamoto R, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazur PK, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nat. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, He J, Chen L, Wang G. Hepatitis B virus X protein upregulates expression of SMYD3 and C-MYC in HepG2 cells. Med. Oncol. 2009;26:445–451. doi: 10.1007/s12032-008-9144-1. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi M, et al. Interaction of the hepatitis B virus X protein with the lysine methyltransferase SET and MYND domain-containing 3 induces activator protein 1 activation. Microbiol. Immunol. 2016;60:17–25. doi: 10.1111/1348-0421.12345. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Guo Y, Xiao S, Shi X. Protein kinase p-JNK is correlated with the activation of AP-1 and its associated Jun family proteins in hepatocellular carcinoma. Life Sci. 2005;77:1869–1878. doi: 10.1016/j.lfs.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Tsuge M, et al. A variable number of tandem repeats polymorphism in an E2F-1 binding element in the 5′ flanking region of SMYD3 is a risk factor for human cancers. Nat. Genet. 2005;37:1104–1107. doi: 10.1038/ng1638. [DOI] [PubMed] [Google Scholar]
- 21.Sarris ME, Moulos P, Haroniti A, Giakountis A, Talianidis I. Smyd3 Is a Transcriptional Potentiator of Multiple Cancer-Promoting Genes and Required for Liver and Colon Cancer Development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Liu TT, et al. SET and MYND Domain-Containing Protein 3 (SMYD3) Polymorphism as a Risk Factor for Susceptibility and Poor Prognosis in Ovarian Cancer. Med. Sci. Monit. 2016;22:5131–5140. doi: 10.12659/MSM.898095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, et al. Association of the variable number of tandem repeats polymorphism in the promoter region of the SMYD3 gene with risk of esophageal squamous cell carcinoma in relation to tobacco smoking. Cancer Sci. 2008;99:787–791. doi: 10.1111/j.1349-7006.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li RD, et al. The SMYD3 VNTR 3/3 polymorphism confers an increased risk and poor prognosis of hepatocellular carcinoma in a Chinese population. Pathol. Res. Pract. 2018;214:625–630. doi: 10.1016/j.prp.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Hoan NX, et al. SOCS3 genetic variants and promoter hypermethylation in patients with chronic hepatitis B. Oncotarget. 2017;8:17127–17139. doi: 10.18632/oncotarget.15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cholongitas E, et al. Systematic review: The model for end-stage liver disease–should it replace Child-Pugh’s classification for assessing prognosis in cirrhosis? Aliment. Pharmacol. Ther. 2005;22:1079–1089. doi: 10.1111/j.1365-2036.2005.02691.x. [DOI] [PubMed] [Google Scholar]
- 27.Diaz-Gonzalez A, Reig M, Bruix J. Treatment of Hepatocellular Carcinoma. Dig. Dis. 2016;34:597–602. doi: 10.1159/000445275. [DOI] [PubMed] [Google Scholar]
- 28.Cillo U, et al. Prospective validation of the Barcelona Clinic Liver Cancer staging system. J. Hepatol. 2006;44:723–731. doi: 10.1016/j.jhep.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 29.Wang XQ, Miao X, Cai Q, Garcia-Barcelo MM, Fan ST. SMYD3 tandem repeats polymorphism is not associated with the occurrence and metastasis of hepatocellular carcinoma in a Chinese population. Exp. Oncol. 2007;29:71–73. [PubMed] [Google Scholar]
- 30.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes. Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 31.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002;14:684–691. doi: 10.1016/S0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 32.Chen LB, Xu JY, Yang Z, Wang GB. Silencing SMYD3 in hepatoma demethylates RIZI promoter induces apoptosis and inhibits cell proliferation and migration. World J. Gastroenterol. 2007;13:5718–5724. doi: 10.3748/wjg.v13.i43.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Amplification of SMYD3 promotes tumorigenicity and intrahepatic metastasis of hepatocellular carcinoma via upregulation of CDK2 and MMP2. Oncogene. 2019;38:4948–4961. doi: 10.1038/s41388-019-0766-x. [DOI] [PubMed] [Google Scholar]
- 34.Fei X, Ma Y, Liu X, Meng Z. Overexpression of SMYD3 Is Predictive of Unfavorable Prognosis in Hepatocellular Carcinoma. Tohoku J. Exp. Med. 2017;243:219–226. doi: 10.1620/tjem.243.219. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, et al. A Novel lncRNA IHS Promotes Tumor Proliferation and Metastasis in HCC by Regulating the ERK- and AKT/GSK-3beta-Signaling Pathways. Mol. Ther. Nucleic Acids. 2019;16:707–720. doi: 10.1016/j.omtn.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, et al. ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:18. doi: 10.1186/s13046-018-1011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]