Abstract
Colorectal cancer (CRC) is the most common type cancers in the world. CRC occurs sporadically in the majority of cases, indicating the predominant cause of the disease are environmental factors. Diet-induced changes in gut-microbiome are recently supposed to contribute on epidemics of CRC. This study was aimed to investigate the association of metagenomics and metabolomics in gut extracellular vesicles (EVs) of CRC and healthy subjects. A total of 40 healthy volunteers and 32 patients with CRC were enrolled in this study. Metagenomic profiling by sequencing 16 S rDNA was performed for assessing microbial codiversity. We explored the small molecule metabolites using gas chromatography-time-of-flight mass spectrometry. In total, stool EVs were prepared from 40 healthy volunteers and 32 patients with CRC. Metagenomic profiling demonstrated that bacterial phyla, particularly of Firmicutes and Proteobacteria, were significantly altered in patients with colorectal cancer. Through metabolomics profiling, we determined seven amino acids, four carboxylic acids, and four fatty acids; including short-chain to long chain fatty acids that altered in the disease group. Binary logistic regression was further tested to evaluate the diagnostic performance. In summary, the present findings suggest that gut flora dysbiosis may result in alternation of amino acid metabolism, which may be correlated with the pathogenesis of CRC.
Subject terms: Metabolomics, Microbiology techniques, Colon cancer
Introduction
Colorectal cancer (CRC) is the most common type cancers in the world1. The majority of CRC occurs sporadically, indicating that environmental influences are the predominant cause of the disease2. Dietary pattern has long been considered as the most important lifestyle risk factor for CRC. In vivo and in vitro studies have investigated the effect of protein intake on CRC risk and suggest that consumption of excessive protein could lead to DNA damage and influence on the maintenance of colonocyte intergrity3,4. Diet-induced changes in gut-microbiome are recently supposed to contribute on epidemics of CRC. Accordingly, studies have suggested that the intestinal microbiome might be important for CRC initiation and progression, since tumors preferentially develop in the distal colon and rectum, which are colonized by approximately 70% of host microbiomes2,5. The microbiome has the potential to generate a microenvironment that favors the development of CRC, presumably by recruiting mediators such as interleukins, tumor necrosis factor-alpha, and reactive oxygen species6,7. Furthermore, metabolic products of the gut microbiota might increase the risk of developing colorectal cancer. For example, high levels of acetaldehyde produced by the gut microbiota can break down colonial folate, thereby increasing CRC risk7.
Microbe-derived extracellular vesicles (EVs) are emerging as an important new research subject in understanding the intersection of the gut-microbial communities and human health. Gut microbiota can secrete different types of EVs, including outer membrane vesicles (OMVs), shedding vesicles, and apoptotic bodies8,9. EVs are mainly composed of lipids, proteins, nucleic acids, and metabolites10–12. Although the underlying mechanisms are still unclear, their primary role is to transport active biomolecules into cells over long distances, providing drug delivery to target sites or regulating host cellular responses11,13.
Recent studies have provided mechanistic evidences for the participation of the gut flora in CRC development. An in vivo study demonstrated that genetically engineered animal model of CRC develop fewer tumors under germ-free conditions compared to those with a conventional microbiota12. Further, Enterococcus faecalis and Escherichia coli produce extracellular genotoxins and free radicals targeting DNA that can contribute to CRC development14. However, it is not yet clear which disease-causing signals are produced by bacteria in the gut. In this study, we profiled the microbiome and metabolites within EVs from CRC patients and healthy controls using 16 S ribosomal DNA (rDNA) amplicon sequencing and global metabolomics, respectively, to develop diagnostic models to assess the risk of CRC.
Materials and Methods
Research subjects
A total of 32 patients with colorectal cancer from Seoul National University Bundang Hospital and Chung-Ang University Hospital and 40 healthy control individuals from Haewoondae Baek Hospital participated in the present study between April 2016 and April 2018. All patients with colorectal cancer were diagnosed for the first time according to the diagnostic criteria proposed by the International Union Against Cancer and the American Joint Committee on Cancer in 201315. The patients characteristics, such as age, sex, stage, tumor location, and carcinoembryonic antigen (CEA) test, were examined. Healthy subjects recruited for this study visited the hospital for a regular health screening. After the checkup, we selected healthy controls who were confirmed to have no known diseases and normal laboratory test results. The exclusion criteria for healthy controls included gut disease diagnosis, taking medication for gut disease, and previous CRC diagnosis. For healthy control individuals, general characteristics were recorded, including age, sex, and medical history. Patient and healthy subject exclusion criteria included colorectal cancer recurrence post-surgery, chemotherapy, complication of colorectal cancer with any other cancers or metabolic diseases, medication, or antibiotic treatment within 1 month of sample collection. Characteristics of subjects are shown in Table S1. The present study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (IRB No. B-1708/412-301) and Haewoondae Baek Hospital (IRB No. 129792-2015-064), and was conducted in accordance with the principles of the Declaration of Helsinki. Informed consent was obtained from all subjects.
Sample collection and EV isolation
Stool samples were collected prior to surgery or bowel preparation. All participants consumed a bland diet and did not smoke or consume alcohol 1 day prior to sample collection. A stool sample was collected from the center of the stool using a sterilized cotton swab and stored at −20 °C. Detailed procedure of sample collection was followed a previous study16. Prior to separation of bacterial EVs from stool, a stool sample (1 g) was mixed with 10 mL of phosphate-buffered saline (PBS) followed by vibration for 24 h. The samples were then incubated to separate the EVs from human stool; EVs from the stool samples were then isolated using centrifugation at 10,000 × g for 10 min at 4 °C. Bacteria and foreign particles contained in the supernatant were thoroughly eliminated by filtration using a 0.22-µm pore size17.
Gas chromatography time-of-flight mass spectrometry analysis
Frozen EV samples were thawed and prepared to analyze using gas chromatography time-of-flight mass spectrometry. Detailed experimental procedure is described in our previous studies18,19.
DNA extraction and sequencing
Bacterial EVs were boiled using a heat block for 40 min at 100 °C and then the remaining particles and waste were removed by centrifugation at 13,000 rpm for 30 min at 4 °C. The DNA was extracted from supernatants using a DNeasy PowerSoil kit (QIAGEN, Germany). The DNA of bacterial EVs in each sample was quantified by QIAxpert (QIAGEN, Germany). V3-V4 regions of the 16 S rDNA gene was amplified with primers; 16S_V3_F (5′ - TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG -3′) and 16S_V4_R (5′ - GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC -3′). The library preparation was performed using PCR products and each amplicon was sequenced by MiSeq (Illumina, USA).
Bioinformatics
Paired-end reads that matched the adapter sequences were trimmed by cutadapt (version 1.1.6)20. The resulting FASTQ files containing paired-end reads were merged with CASPER and then quality filtered with Phred (Q) score based criteria described by Bokulich21,22. Any read shorter than 350 bp or longer than 550 bp after merging was also discarded. To identify the chimeric sequences, a reference-based chimera was detected by VSEARCH against the SILVA gold database23. And then the clustering into Operational Taxonomic Units (OTUs) was conducted using VSEARCH with the de novo clustering algorithm under a 97% sequence similarity. Exclusion criteria of OTUs was a containing one read sequence in only a sample. The representative sequences of the OTUs were finally classified using the SILVA 128 database with UCLUST (script on QIIME version 1.9.1)24. We applied normalization to the data sets using the total count method as described by Previous study25. Chao indices, estimators of taxa richness per individual, were estimated to measure the alpha diversity of each sample. The metagenome biomarkers selection in the diagnostic model was based on the relative abundances at the genus level. The criteria of false discovery rate (FDR)-adjusted p-values as determined by Wilcoxon test, fold-changes and average relative abundances were less than 0.05, greater than 2-fold and greater than 0.5% in any group, respectively. In addition, we selected metabolome biomarkers with adjusted P-values less than 0.05 and changes greater than 2-fold. All diagnostic models were calculated by logistic regression based on Akaike information criteria using stepwise selection method with training and test sets selected randomly at an 80:20 ratio. The performance values, such as AUC, sensitivity, specificity, and accuracy, were reported using validataion set. A logistic regression model was built using individual omics data from metagenomic and metabolic biomarkers; its accuracy was then compared to a combined model of metagenomics and metabolic biomarkers to discriminate cancer from healthy controls.
Statistics for metabolomics data
Multivariate and univariate analyses were conducted using Metaboanalyst 4.0. Normalized data sets using log transformation and pareto scaling were analyzed and principal component analysis (PCA) was used to examine differentiation in overall metabolic profiles between the groups. Univariate analysis using false discovery rate (FDR)-adjusted P-value was used for the selection of metabolic candidates. Significant differences between the healthy control group and CRC patient group were determined using the Wilcoxon test for continuous variables. Findings were considered significant if the p-value was less than 0.05.
Statistics for metagenomic data
Alpha diversity of microbial composition for richness and evenness was analyzed using the Chao1 index and Shannon’s index to compare diversity between the healthy control and CRC patient groups. Principal coordinate analysis (PCoA) based on Bray-Curtis similarity for beta diversity was used to visualize relationships between samples. R (version 3.5.1) was used for all statistical analyses.
Results
Microbiome analysis of microbe-derived EVs in stool samples from CRC patients and healthy subjects
To investigate the microbial compositions of stool EVs from the CRC patients and healthy controls, metagenome analysis was performed based on 16 S rDNA amplicon sequencing. Comparison of alpha diversity in the CRC patients and healthy controls revealed no significant differences based on the Chao1 and Shannon indexes, as shown in Fig. 1(A). Beta diversity at the phylum and genus levels was represented through principal coordinate analysis (PCoA) (Fig. 1(B)). A comparison of beta diversity at the genus level demonstrated a clear separation between the groups compared to the phylum-level clusters.
Figure 1.
Alpha and beta diversity comparisons of microbiomes collected from CRC patients and healthy controls. Analysis was performed using sequencing data for the 16S rDNA V3 and V4 regions, with a rarefaction depth of 10,000 reads per sample. (A) Chao1 and Shannon indexes indicate alpha diversity. Whiskers in the boxplots represent the range of the minimum and maximum alpha diversity values within a population, excluding outliers. Principal coordinate analysis (PCoA) plots represent beta diversity at the (B) phylum and (C) genus levels. Red circles represent healthy control individuals and blue circles represent CRC patients. CRC, colorectal cancer.
Heat maps further allowed for the visualization of relative changes in microbial abundance at the phylum and genus levels (Fig. 2(A,C)). In the comparison of phylum levels, three individual phyla were found; Firmicutes was significantly increased in CRC patients, whereas the level of Proteobacteria and Tenericutes was decreased (Fig. 2(B)). The bar graphs in Fig. 2(D) demonstrate that the microbial compositions changed at the genus level in CRC patients compared to those in healthy controls. Detailed records of those data are listed in Table 1. There was a significant difference observed in 34 bacterial genera between the CRC group and the healthy control group. As presented in Table 1, the proportions of Actinomyces, Rothia, Propionibacterium, Bacteroidiales S24-7 group, Chloroplast, Lachnospiraceae NK4A136 group, Ruminococcaceae UCG-014, Staphylococcus,Methylobacterium, Solanum melongena, Sphingomonas, Escherichia-shigella, Proteus, Pseudomonas, Saccaribacteria, and Mollicutes were decreased in the CRC patients compared to those in the healthy controls (P < 0.05), whereas the proportions of Bifidobacterium, Collinsella, Blautia, Lachnoclostridium, Lachnospiraceae UCG-008, Dorea, Eubacterium coprostanoligenes group, Ruminococcus 2, Faecalibacterium, Ruminococcaceae NK4A214, Ruminococcaceae UCG-002, Ruminococcus, Subdoligranulum, Ruminococcaceae, Catenibacterium, Parvimonas, Ruminiclostridium 5, Enterobacter, and Diaphorobacter were significantly enriched (P < 0.05). The predominant observation regarding these changes was that microbial compositions of Proteobacteria were larger while the compositions of Firmicutes were reduced, except for those of Lachnospiraceae UCG-008, Ruminococcaceae UCG-014, and Staphylococcus. Moreover, among Proteobacteria, Proteus spp. were dramatically altered in the CRC patients and was absent in healthy controls. Taxonomic profiles are presented in Fig. 3.
Figure 2.
Comparison of bacterial composition of EVs from CRC patients and healthy controls. Heat maps represent the relative abundance of microbes (A) at the phylum level and (C) at the genus level in CRC patients and healthy controls. Cells with an abundance value close to zero are represented in light blue, and those with an abundance value larger than 0.5 are indicated in dark blue. The total abundance is 1. Bar graphs represent microbial populations that were significantly different in abundance between CRC patients and healthy controls (B) at the phylum level and (D) at the genus level. The gray-colored bar indicates the relative abundance in CRC patients, and the black-colored bar indicates the relative abundance in healthy controls. **P < 0.05, ***P < 0.01 between CRC patients and healthy controls. EV, extracellular vesicle; CRC, colorectal cancer.
Table 1.
List of microbiomes significantly changed in CRC.
| Phylum | Genus | MAV of healthy controls (%) | MAV of CRC patients (%) | Log2 | p-value | FDR adjusted p-value | Regulation |
|---|---|---|---|---|---|---|---|
| (Fold change) | (Wilcoxon) | (Wilcoxon) | |||||
| Actinobacteria | Actinomyces | 0.646 | 0.053 | −3.613 | 0.000 | 0.001 | Down |
| Bifidobacterium | 0.871 | 1.832 | 1.073 | 0.001 | 0.014 | Up | |
| Rothia | 0.533 | 0.016 | −5.099 | 0.000 | 0.000 | Down | |
| Propionibacterium | 0.685 | 0.164 | −2.061 | 0.000 | 0.000 | Down | |
| Collinsella | 0.065 | 0.953 | 3.880 | 0.000 | 0.000 | Up | |
| Bacteroidetes | Bacteroidales S24-7 group (f) | 0.610 | 0.491 | −0.313 | 0.003 | 0.031 | Down |
| Cyanobacteria | Chloroplast (o) | 0.507 | 0.059 | −3.104 | 0.000 | 0.000 | Down |
| Firmicutes | Blautia | 0.187 | 0.599 | 1.677 | 0.000 | 0.000 | Up |
| Lachnoclostridium | 0.096 | 0.740 | 2.943 | 0.000 | 0.000 | Up | |
| Lachnospiraceae NK4A136 group | 0.561 | 0.170 | −1.718 | 0.002 | 0.019 | Down | |
| Lachnospiraceae UCG-008 | 0.591 | 1.069 | 0.855 | 0.000 | 0.000 | Up | |
| Dorea | 0.450 | 0.510 | 0.181 | 0.000 | 0.008 | Up | |
| [Eubacterium] coprostanoligenes group | 1.332 | 5.696 | 2.096 | 0.000 | 0.000 | Up | |
| Ruminococcaceae UCG-002 | 0.484 | 2.180 | 2.171 | 0.000 | 0.002 | Up | |
| Ruminococcus 2 | 0.530 | 2.329 | 2.136 | 0.000 | 0.002 | Up | |
| Subdoligranulum | 0.562 | 2.408 | 2.099 | 0.000 | 0.000 | Up | |
| Ruminococcaceae (f) | 0.317 | 1.187 | 1.905 | 0.000 | 0.000 | Up | |
| Ruminococcaceae UCG-014 | 1.098 | 0.706 | −0.638 | 0.000 | 0.004 | Down | |
| Faecalibacterium | 4.624 | 11.974 | 1.373 | 0.000 | 0.003 | Up | |
| Ruminococcaceae NK4A214 group | 0.209 | 1.045 | 2.319 | 0.001 | 0.009 | Up | |
| Staphylococcus | 0.819 | 0.410 | −0.998 | 0.003 | 0.033 | Down | |
| Catenibacterium | 0.041 | 0.729 | 4.161 | 0.000 | 0.000 | Up | |
| Parvimonas | 0.028 | 0.812 | 4.840 | 0.001 | 0.013 | Up | |
| Ruminiclostridium 5 | 0.083 | 0.527 | 2.672 | 0.001 | 0.010 | Up | |
| Proteobacteria | Methylobacterium | 2.846 | 0.027 | −6.743 | 0.000 | 0.007 | Down |
| Solanum melongena (eggplant) | 1.046 | 0.141 | −2.887 | 0.000 | 0.000 | Down | |
| Sphingomonas | 0.548 | 0.172 | −1.674 | 0.000 | 0.000 | Down | |
| Diaphorobacter | 0.000 | 0.974 | 12.397 | 0.000 | 0.001 | Up | |
| Escherichia-Shigella | 3.427 | 0.871 | −1.975 | 0.000 | 0.000 | Down | |
| Proteus | 1.169 | 0.000 | « | 0.000 | 0.000 | Down | |
| Pseudomonas | 2.913 | 0.242 | −3.589 | 0.000 | 0.000 | Down | |
| Enterobacter | 0.158 | 0.816 | 2.370 | 0.004 | 0.035 | Up | |
| Saccharibacteria | Saccaribacteria (p) | 0.540 | 0.132 | −2.029 | 0.001 | 0.017 | Down |
| Tenericutes | Mollicutes RF9 (o) | 2.053 | 0.208 | −3.304 | 0.001 | 0.011 | Down |
MAV indicates mean abuandance value; Fold-change indicates the difference in the mean abundance values between CRC patients and healthy controls; for comparison of the relative abundance of the two groups, false discovery rate-adjusted p-values were calculated.
Figure 3.
Taxonomic profile of microbe-derived EVs in CRC patients and healthy controls. Analyses were performed for 16S rDNA V3 and V4 regions data, with a rarefaction depth of 10,000 reads per sample. Relative taxon abundance plots for CRC patients and healthy controls at the (A) phylum and (B) genus levels. Individuals are represented along the horizontal axis and relative taxon frequency is denoted on the vertical axis. EV, extracellular vesicle; CRC, colorectal cancer.
Metabolic profiling of stool EVs from CRC patients and healthy subjects
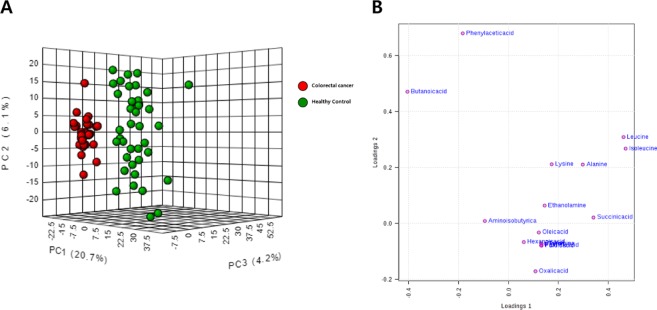
To assess the profile of small-molecule metabolites in EVs, we conducted a global metabolomics analysis using GC-TOF-MS. In three-dimensional PCA score plots, as shown in Fig. 4(A), three PCs (PC1–3) clearly separated the metabolomics profiles of healthy controls and CRC patients. The metabolites identified by multivariate analysis were selected according to their Q-values, which are P-values adjusted for the FDR. The metabolites that showed statistical significance (Q < 0.05) are listed in Table 2 and Table S2. The loading plot in Fig. 4(B) shows the metabolites that effectively differentiated CRC patients from healthy controls. The most frequent small-molecule metabolites were classified as amino acids that were more abundant in CRC patients. Furthermore, metabolites with alcohol forms (ethanolamine and phenol), carboxylic acids (furoic acid, succinic acid, and oxalic acid), and fatty acids (hexanoic acid, palmitic acid, and oleic acid) were enhanced in CRC patients compared to those in healthy controls. Notably, bacterial metabolites such as aminoisobutyric acid and butanoic acid were reduced.
Figure 4.
Distinct metabolic profiling between CRC patients and healthy controls. Metabolic profiling was performed using GC-TOF-MS. (A) Score plot of the three-dimensional principal component analysis (PCA) shows metabolic patterns of stool EV samples. Red circles indicate colorectal cancer and green circles indicate healthy controls. (B) Loading plots of PC1 and PC2 from the PCA results of differentially accumulating metabolites from CRC patients versus healthy controls. EV, extracellular vesicle; CRC, colorectal cancer; PC, principal component.
Table 2.
List of metabolites significantly changed in CRC.
| Metabolite | Class | Fold-change | p-value | Regulation |
|---|---|---|---|---|
| Leucine | Amino acid | 2.433 | 5.75E-03 | Up |
| Isoleucine | 2.192 | 5.23E-03 | Up | |
| Alanine | 1.667 | 4.31E-02 | Up | |
| Lysine | 1.441 | 1.48E-02 | Up | |
| Tyramine | 1.423 | 1.57E-22 | Up | |
| Aminoisobutyric acid | −1.116 | 6.66E-03 | Down | |
| Ethanolamine | Amino alcohol | 1.384 | 4.56E-04 | Up |
| Phenol | Aromatic alcohol | 1.589 | 1.99E-12 | Up |
| Furoic acid | Carboxylic acid | 1.417 | 1.31E-29 | Up |
| Succinic acid | 3.154 | 4.46E-09 | Up | |
| Oxalic acid | 1.396 | 1.66E-03 | Up | |
| Butanoic acid | Fatty acid | −1.300 | 2.26E-04 | Down |
| Hexanoic acid | 1.302 | 9.38E-06 | Up | |
| Palmitic acid | 1.436 | 4.37E-25 | Up | |
| Oleic acid | 1.317 | 3.09E-04 | Up |
Fold-change indicates the difference in the mean abundance values between CRC patients and healthy controls; for comparison of the relative intensities of the two groups, false discovery rate-adjusted p-values were calculated.
Correlation between microbiome and metabolic profiles in stool EVs
A Pearson rank correlation analysis demonstrated a close correlation between the gut microbiota and certain metabolic products (Fig. 5(A)). The relative abundance of most metabolic markers was highly positively correlated with the Firmicutes genera. Specifically, several amino acids were enriched according to the consistent regulation of gut flora in CRC patients. These bacteria shared a significant relationship with tyramine, phenol, and hexanoic acid (r > |0.5|, P < 0.05). Observations for Proteobacteria were opposite to those for Firmicutes, wherein the Proteobacteria family was negatively correlated with these metabolic biomarkers (r < |0.5|, P < 0.05). Among the metabolic biomarkers, carboxylic acids (such as furoic acid, succinic acid, and oxalic acid) and long chain fatty acids (such as palmitic acid and oleic acid) moderately correlated with levels of the entire gut flora.
Figure 5.
Pearson correlation analysis and the power of the relative abundance of microbiomes and metabolites to discriminate CRC from healthy controls. (A) Pearson correlation analysis was performed to investigate the association between metagenomic and metabolomic analysis data. The y-axis represents the result of the metagenomic analysis, and the x-axis represents metabolic biomarkers, both obtained through statistical comparisons. Each square shows the correlation coefficient value. Red squares indicate a positive correlation and blue squares indicate a negative correlation between microbial and metabolite abundances. (B) Receiver operating characteristic (ROC) curves for the markers leucine, butanoic acid, isoleucine, succinic acid, Bifidobacterium, Lactobacillus, Faecalibacterium, Pseudomonas, and Methylobacterium based on their ability to discriminate CRC patients from healthy controls. The red line indicates the model based on metagenomics analysis, the blue line indicates the model based on metabolomics analysis, and the green line indicates the model based on combination of metabolomics and metagenomics data. CRC, colorectal cancer.
CRC diagnostic models based on microbiome and metabolic profiles in stool EVs
To further define the useful biomarkers from the metagenomic and metabolomic biomarkers, a binary logistic regression analysis and an optimized algorithm of the forward stepwise method were employed to construct the best model using these retained biomarkers to distinguish CRC-positive individuals from healthy controls. Ultimately, two metabolites (leucine and oxalic acid) and two bacterial genera (Collinsella and Solanum melongena) were selected. Figure 5(B) shows the receiver operation curve of the logistic regression model to discriminate CRC-positive samples from healthy controls. Using the two metabolic biomarkers, the predictability of CRC was 92.0%with 80.0% sensitivity and 100% specificity. The two selected metagenomics biomarkers resulted AUC value (95.0%) with 90.0% sensitivity and 100% specificity. Each AUC values were slightly lower in test set compared to training set. Integration of these two panels of omics data led to an AUC of 100% with relevant accuracy in discriminating between CRC-positive samples and healthy controls (Figure 5(B) and Table S3). A permutation test of the logistic regression model was conducted for assessment and to exclude over-fitting (Table S4). Although the patients were diagnosed more accurately in the combined model, metagenomic biomarkers were found to fit on this model more efficiently compared to metabolic biomarkers (Table S4). These data suggest that these potential representative markers of CRC, a combination of metagenomic and metabolomic biomarkers, might diagnose CRC more accurately than a single omics biomarker.
Discussion
In the present study, we performed metabolic analysis and microbiome profiling of EVs obtained from stools of CRC patients and healthy volunteers to identify metabolites that change with pathophysiology and to suggest possible correlations with gut microbes, respectively. Through 16 S rDNA sequencing, we found compositional changes in bacteria belonging to the Firmicutes and Proteobacteria phyla in CRC patients compared to that in healthy controls. Based on global metabolomics profiling, several amino acids and carboxylic acids were more abundant in the presence of cancer, whereas some microbe-associated metabolites such as aminoisobutyric acid and butanoic acid were less abundant. To the best of our knowledge, this is the first study to report the association of CRC development with the microbiome and metabolomics using stool EVs.
Accumulating studies show that several bacterial species seem to be involved in the pathogenesis of CRC. For instance, Streptococcus bovis is predominant in patients with colon cancer, which colonize approximately 20–50% of the gut but less than 5% in healthy individuals26. Elevation of the Bacteroides and Prevotella population is also an indicative marker of CRC based on metagenome analysis27. In the present study, we observed dynamic changes in the Firmicutes and Proteobacteria phyla from the EVs of CRC patients compared to those in healthy controls. A higher abundance of Firmicutes and Fusobacteria has been primarily reported, whereas Proteobacteria were less abundant in individuals with CRC27. Firmicutes including taxa such as Eubacterium, Clostridium, Lactobacillus, and Peptostreptococcaceae, have been shown to be involved in energy resorption28. This might depend on the bacterium’s ability to rapidly exploit unabsorbed, labile amino acids and peptides from the diet. Most of these organisms have proteolytic activity, thereby degrading recalcitrant proteins that have relatively long transit times in the gut28,29. In this study, we demonstrated that several genera of Firmicutes were increased in CRC patients, such as Eubacterium, Faecalibacterium, those of the Ruminococcaceae family, and Catenibacterium. However, there were some exceptional cases wherein Lactobacillus and Clostridium were reduced compared to levels in healthy controls.
Firmicutes and Bacteroidetes are the dominant phyla in the gut microbial community, whereas other phyla such as Proteobacteria, Actinobacteria, and Verrucomicrobia, are generally less abundant. Compositional changes in the phyla, such as increased prevalence of Proteobacteria, can be easily influenced by inflammation of the gastrointestinal tract. An increased proportion of Proteobacteria including the families Enterobacteriaceae, Pasteurellaceae, and Neisseriaceae distinguishes a Crohn’s disease-related bacterial community from that of healthy subjects30. One possible explanation for the richness of Proteobacteria is that whereas the mucosal immune system is obligated to clear pathogens, an inappropriate immune response abolishes the homeostasis of the gut flora, leading to dysbiosis and triggering local and systemic inflammation and malfunction of the endogenous metabolism of the host7,29,31. The potential distinct functions of Proteobacteria in colon tumors are still unclear, although they are known as commensal bacteria that possess potential pathogenic features. Here, we suggest possible influential factors that might contribute to their functional repertoire, including toxic byproducts, virulence factors, and other parameters that propagate interactions between the bacteria and their gut environment, rather than acute and chronic inflammation.
Enhanced amino acid levels are closely related to CRC risk. Possible reasons for this might include the following: changes in dietary habits, as high protein intake has long been regarded the most important lifestyle risk factor for colorectal cancer32; inflammation, which diminishes the absorption of nutrients in patients with cancer5,27; degradation of dietary protein by fermenting bacteria in the distal colon of patients with CRC, which elevates the levels of amino acid metabolites in stool16. One study on amino acid utilization and catabolism in bacteria from the human intestine identified bacteria belonging to the Clostridium clusters (Bacillus, Lactobacillus and some Proteobacteria) as those mostly responsible for the fermentation of amino acids29. These organisms can utilize amino acids in the gut, such as lysine, proline, phenylalanine, and tryptophan, to produce small molecules including ammonia, hydrogen sulfide, nitric oxide, polyamines, and alcoholic compounds33,34. As mentioned earlier, Firmicutes can catalyze amino acids for energy recycling, whereas Proteobacteria can degrade amino acids including undigested proteins. This dysbiosis preferentially affects amino acid metabolism. Short-chain fatty acids (SCFAs) including butanoic acid and aminoisobutyric acid are a well-established energy source in the human intestine. These microbial metabolites play a role in modulating host metabolic and immune responses35. The current study demonstrated that diet-related SCFAs prevent disease and provide therapeutic implications for CRC36, as these bacterial metabolites are restricted to CRC patients and not present in healthy controls.
In the present study, we observed EVs with metagenomic profiles similar to those from previous studies utilizing stool-based metagenome analyses of CRC patients. However, alterations in microbial compositions based on EVs do not directly reflect the proportional changes of gut microbes. Bacteria secrete small vesicles of various forms, such as OMVs, EVs, and ectosomes, to transfer cellular components and modulate signaling pathways. These vesicles have become promising research tools to discover therapeutic targets, develop drug delivery systems, and quantify microbial compositions by utilizing their properties8,37. Thus, systematic and comprehensive studies integrating multiple sources are required to understand the complexity of EV-triggered intercellular and interkingdom communication. Global metabolomic analyses, based on the technique of ultra-high performance gas chromatography, have been established to profile a broad range of metabolites existing in the EVs, enabling the identification of cooperation between microbiomes and cancer development. These findings based on stool-based analysis require confirmation, and further functional studies are needed to determine whether the bacteria influence cancer development.
This study is the first to demonstrate the correlation between microbial changes and metabolic alternations within EV samples from patients with CRC. There was a strong association between the abundance of gut flora (Firmicutes and Proteobacteria) and relevant candidate metabolites (predominantly amino acids). This suggests that the altered composition of macronutrient-fermenting and degrading bacteria in CRC might result in the accumulation of amino acids and the depletion of energy sources. Moreover, our findings indicate that EVs secreted by gut microbes carry a dynamic range of metabolic information reflecting the host’s nutritional state, metabolism, and immune responses in the presence of disease.
Supplementary information
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) under grant number NRF 2017M3A9F3047498.
Author contributions
Y.-K.K. and J.-Y.C. conceptualized the study; D.H.L., S.M.K., Y.S.P and J.G.K. helped with recruitment; J.Y., H.S. and W.H.L. were involved in metagenome analysis; D.J.K. was involved in metabolomics analysis and drafting; I.-J.J., Y.-K.K. and J.-Y.C. were involved in revision of manuscript and critical appraisal.
Data availability
The raw sequence data and processed data of metagenome analysis are available through the Sequence Read Archive under BioProject ID: PRJNA601555. The metabolomics data are available in the electronic Supplementary Material and at the NIH Common Fund's National Metabolomics Data Repository (NMDR) website [Project ID: PR000888].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Da Jung Kim, Jinho Yang, Yoon-Keun Kim and Joo-Youn Cho.
Contributor Information
Yoon-Keun Kim, Email: ykkim@mdhc.kr.
Joo-Youn Cho, Email: joocho@snu.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-020-59529-8.
References
- 1.Maisonneuve C, Irrazabal T, Martin A, Girardin SE, Philpott DJ. The Impact of the Gut Microbiome on Colorectal Cancer. Annu. Rev. Canc Biol. 2018;2:229–249. doi: 10.1146/annurev-cancerbio-030617-050240. [DOI] [Google Scholar]
- 2.Turner ND, Ritchie LE, Bresalier RS, Chapkin RS. The microbiome and colorectal neoplasia: environmental modifiers of dysbiosis. Curr. Gastroenterol. Rep. 2013;15:346. doi: 10.1007/s11894-013-0346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toden S, Bird AR, Topping DL, Conlon MA. Resistant starch prevents colonic DNA damage induced by high dietary cooked red meat or casein in rats. Cancer Biol. Ther. 2006;5:267–272. doi: 10.4161/cbt.5.3.2382. [DOI] [PubMed] [Google Scholar]
- 4.Andriamihaja M, et al. Colon luminal content and epithelial cell morphology are markedly modified in rats fed with a high-protein diet. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G1030–1037. doi: 10.1152/ajpgi.00149.2010. [DOI] [PubMed] [Google Scholar]
- 5.Gagniere J, et al. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp. Biol. Med. 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 7.Homann N, Tillonen J, Salaspuro M. Microbially produced acetaldehyde from ethanol may increase the risk of colon cancer via folate deficiency. Int. J. Cancer. 2000;86:169–173. doi: 10.1002/(SICI)1097-0215(20000415)86:2<169::AID-IJC4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Badi, S. A. et al. Microbiota-Derived Extracellular Vesicles as New Systemic Regulators. Front Microbiol8, 10.3389/fmicb.2017.01610 (2017). [DOI] [PMC free article] [PubMed]
- 9.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012;80:1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandu, R., Oh, J. W. & Kim, K. P. Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Exp Mol Med51, 10.1038/s12276-019-0218-2 (2019). [DOI] [PMC free article] [PubMed]
- 11.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Biosci. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uronis JM, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M, H. R. et al. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int J Mol Sci18, 10.3390/ijms18030538 (2017). [DOI] [PMC free article] [PubMed]
- 14.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 15.Hari DM, et al. AJCC Cancer Staging Manual 7th edition criteria for colon cancer: do the complex modifications improve prognostic assessment? J. Am. Coll. Surg. 2013;217:181–190. doi: 10.1016/j.jamcollsurg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Wang J, Rao B, Deng L. Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp. Ther. Med. 2017;13:2848–2854. doi: 10.3892/etm.2017.4367. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Yang, J. et al. Consumption of a Leuconostoc holzapfelii-enriched synbiotic beverage alters the composition of the microbiota and microbial extracellular vesicles. Exp Mol Med51, 10.1038/s12276-019-0288-1 (2019). [DOI] [PMC free article] [PubMed]
- 18.Kim DJ, et al. Ursodeoxycholic acid exerts hepatoprotective effects by regulating amino acid, flavonoid, and fatty acid metabolic pathways. Metabolomics. 2019;15:30. doi: 10.1007/s11306-019-1494-5. [DOI] [PubMed] [Google Scholar]
- 19.Kim, D. J. et al. Comprehensive Metabolomic Search for Biomarkers to Differentiate Early Stage Hepatocellular Carcinoma from Cirrhosis. Cancers (Basel)11, 10.3390/cancers11101497 (2019). [DOI] [PMC free article] [PubMed]
- 20.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17:3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 21.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang XQ, et al. Gut microbiota as important modulator of metabolism in health and disease. Rsc Adv. 2018;8:42380–42389. doi: 10.1039/c8ra08094a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira MB, Wallroth M, Jonsson V, Kristiansson E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genomics. 2018;19:274. doi: 10.1186/s12864-018-4637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srikanth CV, McCormick BA. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: a three-way crosstalk. Interdiscip. Perspect. Infect. Dis. 2008;2008:626827. doi: 10.1155/2008/626827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, Z. G., Guo, B. M., Gao, R. Y., Zhu, Q. C. & Qin, H. L. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol6, 10.3389/fmicb.2015.00020 (2015). [DOI] [PMC free article] [PubMed]
- 28.Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai ZL, Wu ZL, Hang SQ, Zhu WY, Wu GY. Amino acid metabolism in intestinal bacteria and its potential implications for mammalian reproduction. Mol. Hum. Reprod. 2015;21:389–409. doi: 10.1093/molehr/gav003. [DOI] [PubMed] [Google Scholar]
- 30.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Lai, R. X. et al. The association between dietary protein intake and colorectal cancer risk: a metaanalysis. World J Surg Oncol15, 10.1186/s12957-017-1241-1 (2017). [DOI] [PMC free article] [PubMed]
- 33.Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front. Biosci. 2011;16:1768–1786. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- 34.Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 35.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 36.Gomes, S. D. et al. The Role of Diet Related Short-Chain Fatty Acids in Colorectal Cancer Metabolism and Survival: Prevention and Therapeutic Implications. Curr Med Chem, 10.2174/0929867325666180530102050 (2018). [DOI] [PubMed]
- 37.Yang J, Kim EK, McDowell A, Kim Y-K. Microbe-derived extracellular vesicles as a smart drug delivery system. Transl. Clin. Pharmacol. 2018;26:103–110. doi: 10.12793/tcp.2018.26.3.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data and processed data of metagenome analysis are available through the Sequence Read Archive under BioProject ID: PRJNA601555. The metabolomics data are available in the electronic Supplementary Material and at the NIH Common Fund's National Metabolomics Data Repository (NMDR) website [Project ID: PR000888].