Abstract
The environmental stress response (ESR) is critical for cell survival. Yeast cells unable to synthesize inositol pyrophosphates (PP-InsPs) are unable to induce the ESR. We recently discovered a diphosphoinositol pentakisphosphate (PP-InsP5) phosphatase in Saccharomyces cerevisiae encoded by SIW14. Yeast strains deleted for SIW14 have increased levels of PP-InsPs. We hypothesized that strains with high inositol pyrophosphate levels will have an increased stress response. We examined the response of the siw14Δ mutant to heat shock, nutrient limitation, osmotic stress, and oxidative treatment using cell growth assays and found increased resistance to each. Transcriptional responses to oxidative and osmotic stresses were assessed using microarray and reverse transcriptase quantitative PCR. The ESR was partially induced in the siw14Δ mutant strain, consistent with the increased stress resistance, and the mutant strain further induced the ESR in response to oxidative and osmotic stresses. The levels of PP-InsPs increased in WT cells under oxidative stress but not under hyperosmotic stress, and they were high and unchanging in the mutant. Phosphatase activity of Siw14 was inhibited by oxidation that was reversible. To determine how altered PP-InsP levels affect the ESR, we performed epistasis experiments with mutations in rpd3 and msn2/4 combined with siw14Δ. We show that mutations in msn2Δ and msn4Δ, but not rpd3, are epistatic to siw14Δ by assessing growth under oxidative stress conditions and expression of CTT1. Msn2-GFP nuclear localization was increased in the siw14Δ. These data support a model in which the modulation of PP-InsPs influence the ESR through general stress response transcription factors Msn2/4.
Keywords: Saccharomyces cerevisiae, stress response, transcription regulation, gene expression, yeast genetics, inositol pyrophosphate, MSN2, SIW14
Introduction
The cumulative response to unfavorable conditions is known as the environmental stress response (ESR)5 and it is necessary for cells to adapt and survive external changes (1). Environmental stresses include conditions such as temperature extremes, nutrient limitation, acidic or basic conditions, and osmotic differences (2, 3). In the yeast Saccharomyces cerevisiae, the ESR is activated in general, and additional distinct stress responses are activated to varying degrees depending on the type of stress and its duration (4). The ESR includes the total transcriptional responses that cells have during stressful conditions, ∼300 genes are induced and ∼600 genes are repressed (1). Repressed genes are involved in promoting growth and include ribosome biogenesis and protein synthesis (1, 4). The induced genes in the ESR are involved in handling cellular damage (e.g. defense against reactive oxygen species (ROS), DNA repair, protein refolding), altering carbohydrate and protein metabolism, and generating intracellular signals (1, 5).
Transcriptional responses in the ESR are dependent upon the partially redundant transcription factors Msn2 and Msn4 (Msn2/4) that promote transcription of the general stress response genes (3, 6–8). The ability of these factors to activate transcription is regulated by multiple pathways, including TOR (target of rapamycin) and ras-cAMP-PKA, which affect subcellular localization, protein interactions, and protein half-life (3, 6, 9, 10). Under log-phase growth conditions, Msn2/4 proteins are differentially phosphorylated and sequestered in the cytoplasm (11, 12). Upon stress, Msn2/4 are dephosphorylated and move into the nucleus where Msn2/4 bind stress response elements to promote the transcription of stress response genes (reviewed in Refs. 3 and 5).
Transcriptional responses in the ESR are also regulated by the histone deacetylase (HDAC) complex Rpd3L. The canonical role for HDACs is to form repressive chromatin structures to inhibit transcription. Interestingly, the Rpd3L complex also functions to promote transcription during heat and oxidative stresses (13, 14). Rpd3L is required for the binding of Msn2 to promoters (14). Unexpectedly, inositol pyrophosphates were discovered to be critical for transcriptional responses during oxidative and osmotic stresses (15); transcriptional responses were lost in mutants unable to synthesize inositol pyrophosphates as well as in an rpd3-mutant with substitutions in the putative inositol phosphate-binding site (15).
Inositol pyrophosphates are high energy signaling molecules found ubiquitously across eukaryotes and are involved in diverse pathways such as DNA repair, yeast virulence, human immune response, glycolysis, energy homeostasis, and the general stress response (15–20). Inositol pyrophosphates are fully phosphorylated myo-inositol rings with an additional β-phosphate at the 1- or 5-position, or at both positions. In S. cerevisiae, the most abundant inositol pyrophosphate is 5-diphosphoinositol pentakisphosphate (5PP-InsP5, a specific isomer of InsP7) (21). This molecule is synthesized by the kinase Kcs1, which adds the β-phosphate to InsP6 at the 5-position (22). The kinase Vip1 pyrophosphorylates the 1-position of 5PP-InsP5, resulting in 1,5-bisdiphosphoinositol tetrakisphosphate (1,5PP-InsP4, also known as InsP8), as well as on InsP6, resulting in 1PP-InsP5, an isomer of InsP7 (23, 24). Deletion of both KCS1 and VIP1 prevents cells from producing inositol pyrophosphates; importantly, these cells are unable to induce the environmental stress response with either osmotic or oxidative stresses based on transcriptional profiling assays (15).
We identified the novel inositol pyrophosphate phosphatase Siw14 that specifically cleaves the β-phosphate from the 5-position of InsP7 (25). When SIW14 is deleted, the levels of InsP7 increase 6.5-fold and the levels of InsP8 increase 1.6-fold (25). The impact of the increased inositol pyrophosphate levels on the ESR in the siw14Δ mutant is unknown, and was investigated here by assessing growth phenotypes, transcriptional responses, and inositol pyrophosphate levels. We also sought to determine how inositol pyrophosphates may influence the stress response by using epistasis to examine roles for Msn2/4 and Rpd3L. These data demonstrate that increased intracellular levels of inositol pyrophosphates partially induce the ESR through Msn2/4 and support the role for the SIW14-encoded InsP7 phosphatase in regulating levels of inositol pyrophosphates.
Results
The siw14Δ mutant is resistant to a range of environmental stresses
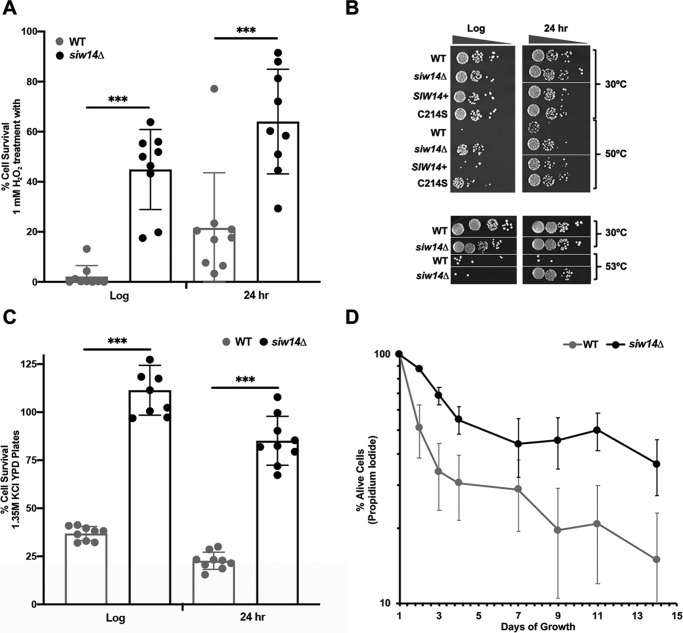
The siw14Δ mutant has elevated inositol pyrophosphates (25), leading us to hypothesize that it would be resistant to environmental stresses. To test this, we examined the response of the mutant strain to oxidative and osmotic stresses, heat shock, and nutrient deprivation. To test for oxidative stress, we assessed resistance to hydrogen peroxide. Cells were grown either to mid-log phase or after the diauxic shift (∼24 h) in SC medium, and were treated with 1 mm H2O2 for 3 h before plating onto solid YPD medium to determine survival. As shown in Fig. 1A, ∼40% of the siw14Δ mutant cells in log-phase survived H2O2 treatment as compared with only 2% of the treated WT cells; this level of resistance from strain BY4741 is in agreement with previous reports (26, 27). We found that post-diauxic shift cells responded similarly: ∼60% of the siw14Δ mutant survived H2O2 treatment compared with only 20% of WT cells (Fig. 1A).
Figure 1.
The siw14Δ mutant is resistant to environmental stresses. A, WT and siw14Δ mutant strains were grown to log phase or for 24 h (post-diauxic shift phase) and treated with 1 mm H2O2. Cells were plated, incubated at 30 °C for 2 days, cfu were determined, and the percent survival was calculated (cfu treated/cfu untreated, ×100). Bars represent the average of 12 biological replicates over 4 assays. B, representative growth of the WT + pRS316 vector and siw14Δ transformed with vector, plasmid-borne SIW14+, or SIW14-C214S (top) or untransformed (bottom). Strains were exposed to 50 (top) or 53 °C (bottom) heat stress for 10 min. Cells were normalized to the same OD600, serially diluted in buffered saline with glucose, and 2.5 μl was spotted on SC-ura medium (top) or YPD (bottom). The experiments were performed in triplicate on independent transformants; log-phase cultures were spotted on the same plate for each experiment, although not necessarily adjacent to each other; the same occurred for the 24-h cultures. C, WT and siw14Δ mutant strains were grown as described under A, and were spread onto YPD or YPD containing 1.35 m KCl medium for osmotic stress. Percent survival was quantified as in A; and bars represent the average of 6 replicates over two separate assays. D, WT and siw14Δ mutant strains were grown for 14 days to measure chronological aging. Aliquots of cells were removed every 24 h; cells were stained with propidium iodide and assayed by flow cytometry. Points represent the average of three biological replicates, and error bars are mean ± S.E. *, p values ≤ 0.05; **, p values ≤ 0.01; ***, p values ≤ 0.001.
To examine heat tolerance, the siw14Δ mutant and isogenic WT cells were grown in YPD medium to mid-log phase or post-diauxic shift, subjected to heat stress at 50 °C (for mid-log cells) or 53 °C (for post-diauxic cells) for 10 min and plated onto solid YPD medium (Fig. 1B). Virtually none of the WT cells survived the heat shock, whereas the siw14Δ mutant survived well (Fig. 1B). Complementation of the siw14Δ mutant with SIW14 on a plasmid restored the WT-sensitive phenotype to cells, whereas complementation with the catalytically dead allele, in which the active-site cysteine was mutated to serine (siw14-C214S), failed to restore the normal phenotype (Fig. 1B).
To test for resistance to osmotic stress, WT and siw14Δ mutant strains were grown to mid-log or post-diauxic phases in YPD medium, and were spread onto YPD medium or YPD medium containing 1.35 m KCl for high osmotic conditions. The siw14Δ mutant showed a 3-fold increase in survival in log-phase cultures and a 1.5-fold increase in post-diauxic phase cultures as compared with the WT strain (Fig. 1C).
In response to nutrient deprivation, yeast cells transition to stationary phase and induce many genes involved in the stress response (28). The chronological aging assay measures the survival of cells in stationary phase for a prolonged period of time (29). WT and siw14Δ mutant strains were inoculated into minimal medium and cultured at 30 °C for 14 days; aliquots were removed daily. Cells were stained with propidium iodide, which is excluded by living cells, and the percentage of living cells was determined relative to the total number of cells. We found that the siw14Δ mutant strain had fewer propidium iodide-stained cells compared with the WT strain each day during the 14-day period. The WT cells showed a much greater variation in survival than the mutant during the time course; even so, differences were significant at days 2 and 3 (Fig. 1D). These results showed that the siw14Δ mutant survived nutrient depletion better than the WT strain.
The stress response is partially induced in unstressed siw14Δ cells
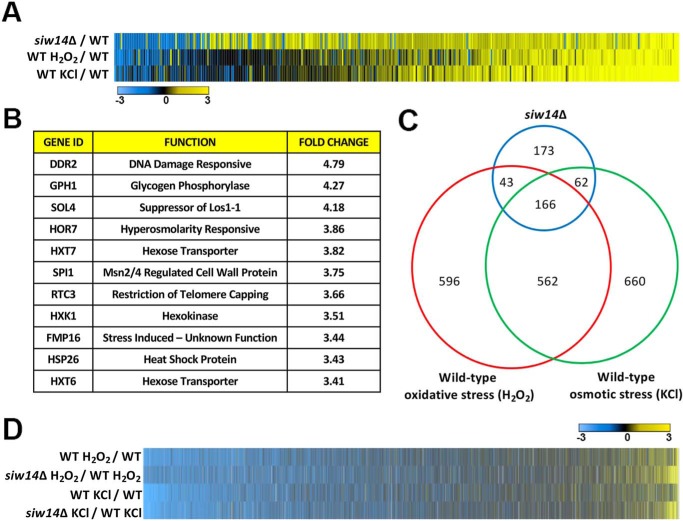
To determine whether siw14Δ mutant cells are stress-resistant due to increased expression of the ESR genes, we measured gene expression in WT (BY4741) and siw14Δ mutant cells during log-phase growth, oxidative stress, and osmotic stress using two-color DNA microarrays (15). In the WT, we found that 1,450 genes were affected for osmotic stress and 1,367 genes in the profile for oxidative stress using a 2-fold or greater cut-off (Table S1). Using the definition of the ESR as the overlap between these stresses (1), we found 728 genes were affected under both stress conditions.
When the siw14Δ mutant was compared with the WT under log-phase growth conditions (SC medium), 354 genes showed increased expression and 90 genes showed decreased expression of at least 2-fold. The genes differentially regulated in the siw14Δ mutant (271 of 444 genes) partially overlapped with the set of genes with altered expression in stressed WT cells (Fig. 2, A and C). Indeed, the genes with the greatest up-regulation in the siw14Δ mutant are ones that are typically induced under stress (Fig. 2B). Furthermore, we found minimal effects on expression of ribosomal biogenesis and ribosomal protein genes, consistent with the normal growth rate of the siw14Δ mutant (data not shown). Genes that were differentially regulated in the siw14Δ (173 genes, Fig. 2C) and did not overlap with the stress-response genes are involved in diverse processes such as glycolysis, gluconeogenesis, ATP generation/electron transport chain function, and oxidation-reduction reactions.
Figure 2.
The transcriptional stress response is partially induced in the unstressed siw14Δ mutant. A, heat map displaying the results of microarray analysis. Genes indicated in blue are down-regulated and those in yellow are up-regulated. The scale is log2 with the bluest shade as a log2 value of ≤−3 (8-fold) difference and the yellowest shade as a log2 value of ≥3; black indicates no difference in expression. The heat map shows the 444 genes that are misregulated in the siw14Δ mutant; these genes are aligned with the corresponding genes in the WT strain stressed with either 1.3 m KCl (osmotic) or 1 mm H2O2 (oxidative). B, the top 10 induced genes in the siw14Δ mutant are genes previously discovered to be induced in WT cells placed under stress (1). C, the Euler plot shows the overlap in the number of genes that are differentially expressed in the siw14Δ mutant and WT cells stressed with hydrogen peroxide or potassium chloride. D, heat map of the complete genome comparing the WT response to hydrogen peroxide and potassium chloride aligned with the siw14Δ mutant stressed with hydrogen peroxide and potassium chloride normalized to the corresponding WT stress response.
When placed under osmotic or oxidative stress conditions, the siw14Δ mutant is able to mount a stress response. Under stress conditions, the siw14Δ mutant induces 1328 genes under osmotic and 1247 genes under oxidative stress conditions when compared with log-phase growth conditions (Fig. 2D). The induced ESR genes were expressed at higher levels in the siw14Δ strain when compared with the WT strain (Fig. 2D). Thus, the siw14Δ mutant has a partially induced stress response under normal growth conditions (i.e. the 444 genes), and it is able to further mount a strong ESR in response to external stresses.
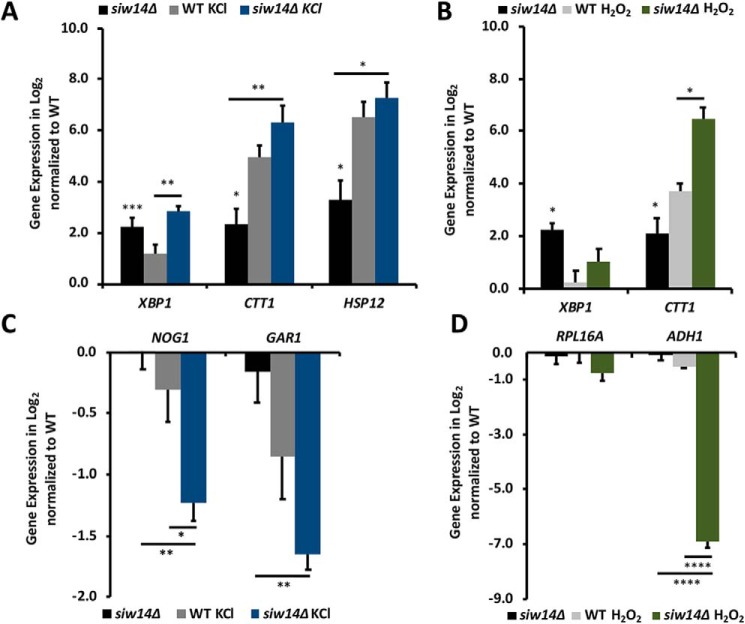
To confirm the results of the DNA microarray analysis, we examined gene expression using RT-qPCR, following the mRNA levels of several genes that had previously been shown to be induced and repressed during the ESR (Fig. 3, A–D). The induced genes selected for further expression analysis were: CTT1, which encodes catalase, the enzyme that converts H2O2 to water (30), and is under the control of the general stress response transcription factors Msn2/4 during oxidative stress (31–33); HSP12, which encodes a plasma-membrane–associated protein important for membrane integrity and is also a downstream target of Msn2/4 (34, 35); XBP1, a transcriptional repressor that down-regulates 15% of all genes when yeast transition to stationary phase and is known to be up-regulated in the ESR (36). The repressed genes selected for further expression analysis were: ADH1, which encodes alcohol dehydrogenase (37); NOG1, which is one of the most down-regulated genes in a WT strain under low nutrient stress conditions and is important for 60S ribosomal subunit biogenesis (38); GAR1, which is involved in ribosomal biogenesis (39); and RPL16A, which encodes the large ribosomal subunit 16A (40).
Figure 3.
The siw14Δ mutant cells have partially induced the ESR in unstressed conditions and are able to mount a stress response. A–D, RT-qPCR fold-changes represented as log2 values normalized to both the reference gene UBC6 and unstressed WT cells. A and C, cells were stressed with 0.4 m KCl, or B and D, 1 mm H2O2. Bars represent the average expression of triplicate samples and error bars are the mean ± S.E. Statistical differences were determined comparing the WT (which would be 0 on the graphs) and siw14Δ mutant (black bar), between treated WT and mutant (indicated by the short line above the gray and blue or green bars), and between untreated and treated siw14Δ mutant (long line above the black and blue or green bars); *, p value ≤ 0.05; **, p value ≤ 0.01; ***, p value ≤ 0.001; and ****, p value ≤ 0.0001.
Based on the microarray data, we expected that CTT1, HSP12, and XBP1 would show increased expression in the unstressed siw14Δ mutant relative to the WT and that they would be further induced upon stress treatment. Furthermore, we expected the ribosomal biogenesis genes to show no expression differences between the WT and siw14Δ mutant, and that they would be down-regulated in response to stress in both strains. Indeed, the unstressed siw14Δ mutant significantly up-regulates XBP1, CTT1, and HSP12 by 4.8-, 5-, and 10-fold, respectively (the black bars in Fig. 3, A and B). As expected, expression of NOG1, GAR1, RPL16A, and ADH1 in the siw14Δ showed no significant difference from the WT (the black bars in Fig. 3, C and D). HSP12 and CTT1 expression increased in the siw14Δ mutant under osmotic stress, by 81- and 152-fold, respectively (Fig. 3A, blue bars). This increase was higher than the 31- and 91-fold induction of the same genes during osmotic stress in WT cells (Fig. 3A, gray bars). NOG1 and GAR1 were further down-regulated in the siw14Δ mutant compared with the WT strain under osmotic stress (Fig. 3C, blue versus gray bars). These results together are consistent with the ability of the siw14Δ mutant to mount an enhanced stress response.
We also evaluated gene expression patterns for these genes under oxidative stress. There was a different expression response in the WT and mutant cells relative to the osmotic stress. CTT1 was induced 81-fold in stressed siw14Δ mutant cells and 13-fold in stressed WT (Fig. 3B, green versus gray bars). The expression of XBP1 was induced 2-fold by H2O2 in the siw14Δ and 1.2-fold in the WT (Fig. 3B, green versus gray bars). Hydrogen peroxide treatment of the siw14Δ mutant led to the down-regulation of RPL16A to 60% expression of the WT and expression of ADH1 decreased to 1% of the WT (Fig. 3D) .
Inositol pyrophosphate levels increase in WT cells during oxidative stress
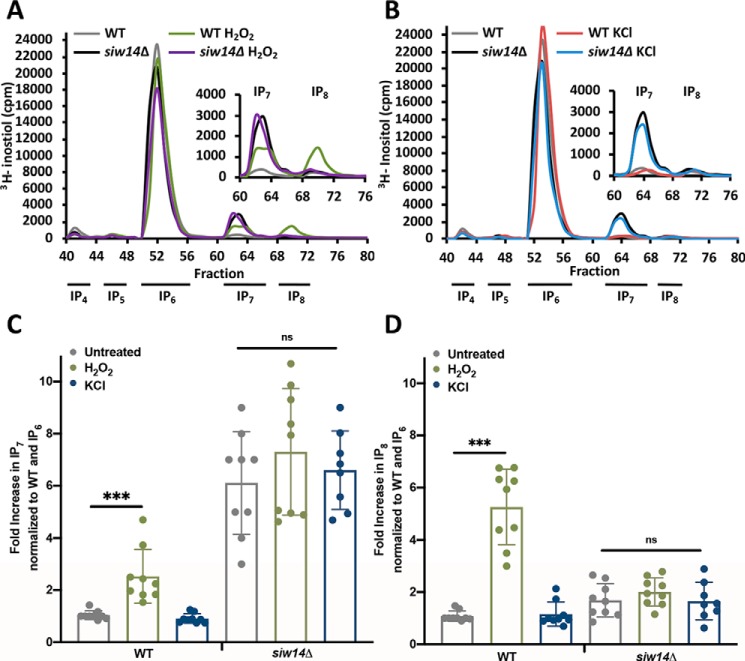
As inositol pyrophosphates are necessary for induction of the environmental stress response (15), we wondered whether inositol pyrophosphate levels might increase when yeast strains are exposed to environmental stresses. Strains were radiolabeled with myo-[3H]inositol, grown to mid-log phase and treated with hydrogen peroxide or potassium chloride for 20 min to induce oxidative or osmotic stress responses, respectively. After extracts were prepared, labeled inositol pyrophosphates were separated by HPLC and scintillation counting was performed. InsP7 and InsP8 levels were determined relative to InsP6, and then normalized to the unstressed WT strain (Fig. 4). Treatment with H2O2 led to a significant increase in both InsP7 (2.1 ± 0.3-fold) and InsP8 (5.3 ± 0.8-fold) in WT cells. Interestingly, we detected no increase in inositol pyrophosphates with KCl treatment (Fig. 4). The siw14Δ mutant exhibited high levels of InsP7 and InsP8, as we previously reported (25), and these levels did not change further under either oxidative or osmotic stresses (Fig. 4).
Figure 4.
Inositol pyrophosphate levels increase in WT cells under oxidative stress. Representative inositol phosphate profiles for WT and siw14Δ mutant stressed with (A) 1 mm H2O2 or (B) 0.4 m KCl for 20 min. Quantified levels of (C) InsP7 or (D) InsP8 relative to InsP6 in unstressed cells or cells stressed with 0.4 m KCl or 1 mm H2O2 in both WT or siw14Δ mutant strains. All fold-changes were normalized to unstressed WT levels. Bars represent the average of 9 biological replicates in 3 separate experiments and error bars represent mean ± S.D.; ***, p values ≤ 0.001.
The Siw14 phosphatase is reversibly inhibited by hydrogen peroxide
One mechanism to link oxidative stress with an increase in inositol pyrophosphate levels is through inhibition of Siw14. The active site of Siw14 contains a cysteine (HCX5R) required for catalysis. Reversible oxidation of the active site cysteine is a known mechanism for this family of phosphatases (41, 42). If oxidation inactivates Siw14 in WT cells, there would be an increase in inositol pyrophosphate pools.
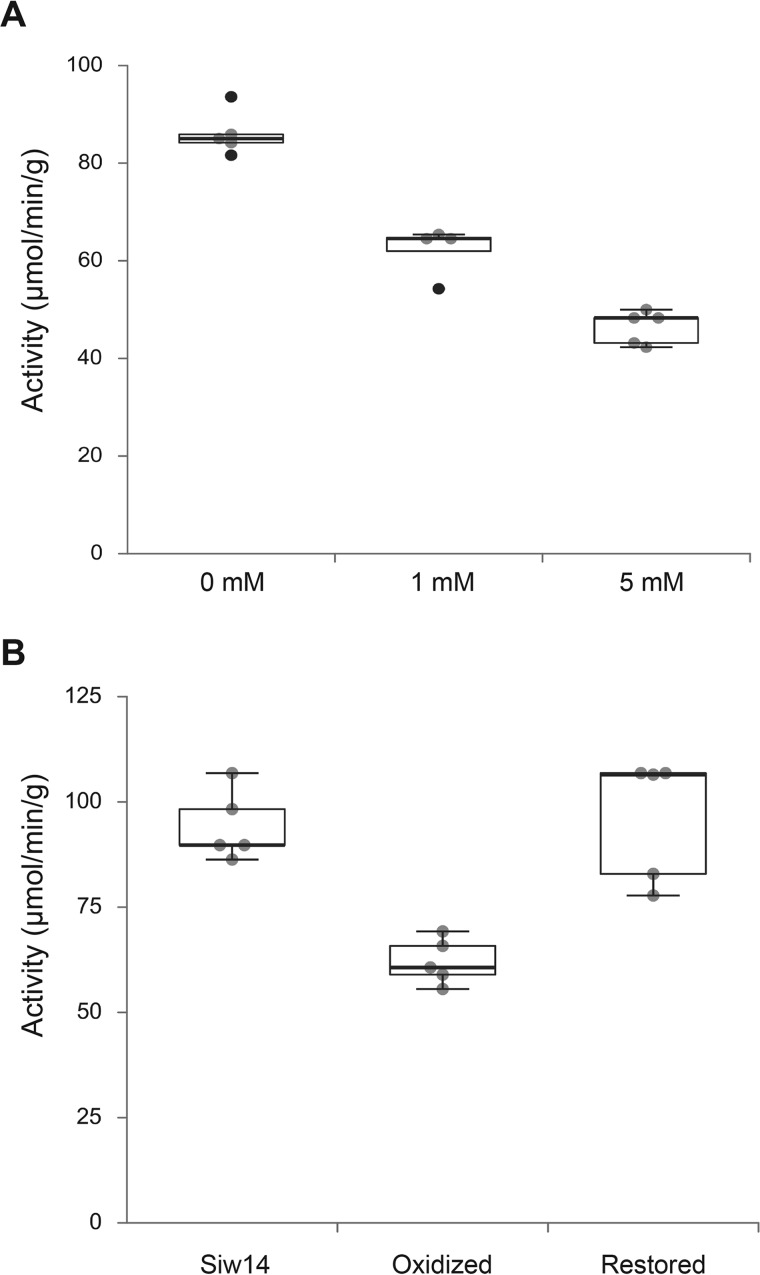
To test this model for inhibition of Siw14 by hydrogen peroxide, we purified recombinant His6-MBP-Siw14, and treated the model with 1 and 5 mm hydrogen peroxide for 30 min on ice. Enzyme activity was assayed using p-nitrophenyl phosphate as the phosphatase substrate (25). As shown in Fig. 5A, hydrogen peroxide treatment led to a decrease in the phosphatase activity of Siw14.
Figure 5.
Reversible inhibition of Siw14 phosphatase activity in vitro. A, recombinant His6-MBP-Siw14 protein was purified and treated with 0, 1, and 5 mm hydrogen peroxide for 30 min on ice, and assayed for phosphatase activity using p-nitrophenol phosphate as the substrate. Absorbance was measured at OD405 and activity was determined relative to a standard curve with p-nitrophenol. Activity was defined as 1 unit = 1 μmol/min/g of protein. B, recombinant, purified His6-MBP-Siw14 protein was treated with 1 mm hydrogen peroxide as in panel A, and then treated with 1 unit of catalase for 30 min on ice followed by the addition of 100 μm DTT. Phosphatase activity was measured as described above. The oxidized enzyme has enzyme activity that is different from the untreated enzyme (p = 0.005, Student's t test) and the restored enzyme is not different from the untreated enzyme (p = 0.367).
To determine whether the inhibition was reversible, we performed reversible oxidation assays as described (41, 42). We treated the purified recombinant enzyme with 1 mm hydrogen peroxide as above (30 min on ice), and then added catalase to a parallel sample of the treated enzyme to degrade any remaining hydrogen peroxide (30 min on ice) followed by addition of 0.1 mm DTT. Hydrogen peroxide treatment significantly decreased the activity to 62.2 ± 5.3 units (p = 0.005, Student's t test), a 28% reduction from the untreated activity of 86.1 ± 4.5 units (Fig. 5B). Removal of hydrogen peroxide with catalase restored enzyme activity to 80.3 ± 10.6 units, which was not statistically significantly different from the untreated sample (p = 0.367). These findings indicate that Siw14 is reversibly oxidized, that oxidation decreases enzyme activity, and the decreased activity could account for the increase in 5PP-InsP5 detected upon hydrogen peroxide treatment (Fig. 4).
Inositol pyrophosphates affect the environmental stress response through Msn2/4 signaling
We considered possible mechanisms to link the transcriptional changes to inositol pyrophosphates. It was previously reported that transcriptional profiles are similar in the rpd3Δ strain, in the kcs1Δ vip1Δ strain that cannot produce inositol pyrophosphates, and in the rpd3ibs mutant that has substitutions in the amino acids that contact the inositol pyrophosphate (15). These results suggest that the histone deacetylase activity of Rpd3L is regulated by inositol pyrophosphates, and therefore we hypothesized that increased inositol pyrophosphate levels would affect Rpd3L activity leading to increased gene expression and stress resistance. To test this hypothesis, we performed an epistasis experiment combining the rpd3ibs mutant and the siw14Δ mutant. Using an oxidative stress assay, we expected the rpd3ibs mutant to be sensitive to H2O2 (this expectation was based on the gene expression profile, although this phenotype had not been previously reported (15)); the siw14Δ mutant is resistant to H2O2 (Fig. 1A). If increased levels of inositol pyrophosphates regulate the activity of HDAC, then the phenotype of the rpd3ibs siw14Δ double mutant would be sensitive to H2O2 and rpd3ibs would be epistatic to the siw14Δ mutant. As shown in Fig. 6, A and B, the WT and rpd3ibs mutant have the same sensitivity to H2O2 (blue and red bars) and the double mutant was as resistant to oxidative stress as the siw14Δ mutant (orange and red-hatched bars). Thus, siw14Δ acted epistatic to rpd3ibs; this result is not consistent with a model in which varying levels of inositol pyrophosphates would regulate the activity of the histone deacetylase Rpd3L. However, this result is consistent with the role for inositol pyrophosphates to act in a structural role within the Rpd3L complex (see “Discussion”).
Figure 6.
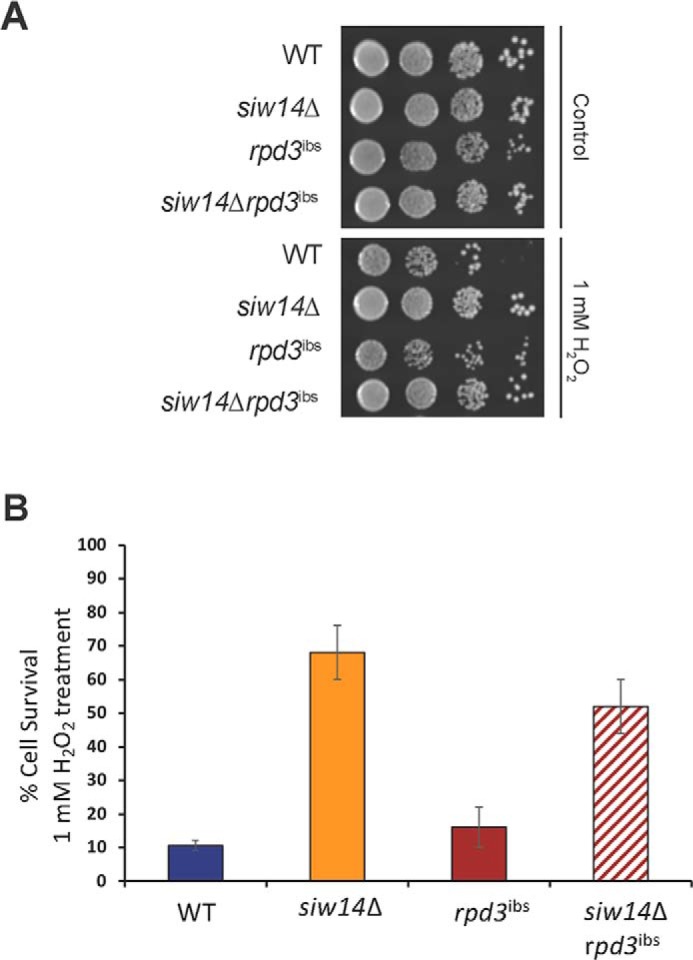
The siw14Δ mutation is epistatic to the rpd3ibs mutation. A, WT, siw14Δ, rpd3ibs, and siw14Δ rpd3ibs strains were grown to mid-log phase and treated with 1 mm H2O2 for 3 h, as described in the legend to Fig. 1. Cells were plated and incubated at 30 °C for 2 days. Colony-forming units were determined, and the percent survival was calculated (cfu treated/cfu untreated, ×100). B, bars represent the average of 9 biological replicates over 3 assays. Error bars represent mean ± S.D. **, p value ≤ 0.001.
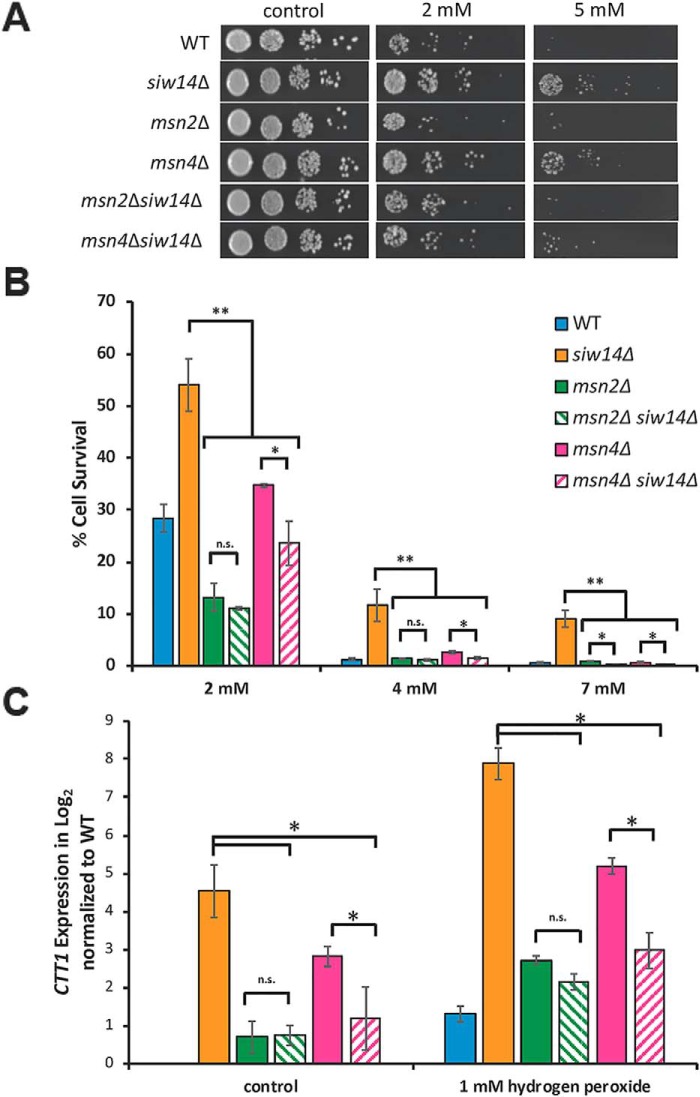
We next addressed the hypothesis that inositol pyrophosphates affect transcription through the Msn2/4 transcription factors. Following a similar approach, we examined the epistatic relationship between siw14Δ and strains carrying msn2Δ or msn4Δ mutations. If oxidative stress resistance, which is due to increased inositol pyrophosphate levels in the siw14Δ mutant, occurs through Msn2/4, we would expect that the msn2Δ and msn4Δ mutations would be epistatic to the siw14Δ mutation. As shown in Fig. 7, A and B, the siw14Δ mutant is resistant to oxidative stress (solid orange bars) and the msn2Δ and msn4Δ single mutants (green and pink bars, respectively) are as or more sensitive to H2O2 as the WT strain (blue bar). Importantly, both the msn2Δ siw14Δ and msn4Δ siw14Δ double mutants are as sensitive to H2O2 as the msn2Δ and msn4Δ single mutants (hatched green and pink bars, respectively). This result indicated that the msn2Δ and msn4Δ mutations are epistatic to siw14Δ.
Figure 7.
The msn2Δ and msn4Δ mutations are epistatic to the siw14Δ mutation. A, WT, siw14Δ, msn2Δ, and msn4Δ single mutant, and the msn2Δ siw14Δ and msn4Δ siw14Δ double mutant strains were grown overnight in YPD and normalized to OD600 of 0.2 in YPD medium. Cells were grown for 30 min and treated with 2 and 5 mm hydrogen peroxide for 3 h (61). Cells were serially diluted 10-fold, plated on YPD medium, and incubated at 30 °C for 2 days. B, strains were grown and prepared as described above; however, cells were treated with 2, 4, and 7 mm hydrogen peroxide. Instead of spotting, 20 μl of appropriately diluted sample were spread onto YPD plates. Colony-forming units were determined, and the percent survival was calculated (cfu treated/cfu untreated, ×100). Bars represent the average of 9 biological replicates over 3 assays. Error bars represent mean ± S.D. C, expression of CTT1 measured by RT-qPCR. Cells were grown as described in the legend to Fig. 3. Values were normalized to UBC6 and to the WT grown in YPD. The graph shows the mean from triplicate samples (biological replicates) and error bars represent mean ± S.D. Significance, **, p ≤ 0.01; *, p ≤ 0.05; n.s., not significant.
To examine this epistasis relationship further, we examined the expression of CTT1 that depends on Msn2/4 (31–33), using RT-qPCR analysis in strains with and without hydrogen peroxide treatment. As shown in Fig. 7C, the expression of CTT1 in the msn2Δ siw14Δ double mutant was virtually the same as in the msn2Δ single mutant, indicating that msn2Δ is epistatic to siw14Δ. The msn4Δ mutant exhibited expression higher than WT under normal conditions and further increased expression upon hydrogen peroxide treatment, and this level of expression was unexpected given that the strain carried the normal allele of MSN2. Expression in the double msn4Δ siw14Δ mutant was lower than that of the siw14Δ single mutant, indicating that msn4Δ acts epistatic to siw14Δ. Together, these data are consistent with the hydrogen peroxide resistance phenotype of the siw14Δ mutant occurring through the Msn2/4 transcription factors.
Msn2 shows increased nuclear localization in the siw14Δ mutant
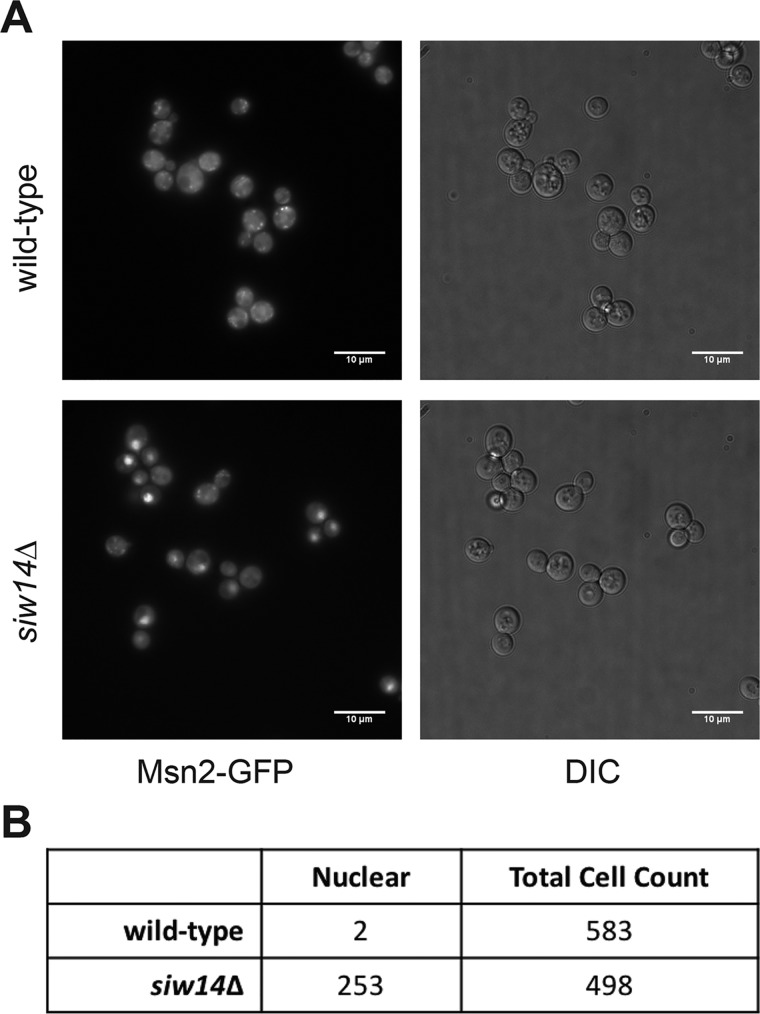
These epistatic data, coupled with the transcriptional analyses (Figs. 2 and 3), support a model in which inositol pyrophosphate levels affect the activity of Msn2/4. One mechanism to account for the increased expression of CTT1 is that Msn2 localizes to the nucleus more frequently in the siw14Δ mutant. To test this, we examined the localization of an Msn2-GFP fusion protein using live-cell fluorescence microscopy for each WT and siw14Δ strains (Fig. 8A). Nuclear localization of Msn2-GFP was scored in cells that had a bright concentrated GFP fluorescence (12). We found that Msn2-GFP localized to the nucleus in 2 of 583 WT cells (0.3%) and in 253 of 498 siw14Δ cells (50.8%) (Fig. 8B). The increased nuclear localization is consistent with the increased expression of ESR genes as well as the stress resistance phenotypes seen in the siw14Δ mutant.
Figure 8.
Msn2-GFP shows increased nuclear localization in the siw14Δ mutant. A, images were taken from cells prepared from multiple cultures grown on different days. Overnight cultures were grown in SC medium at room temperature, diluted 1:50 into fresh medium, and grown to mid-log phase at room temperature. Each sample (1 ml) was placed into borosilicate chamber coverslip slides and allowed to settle for 30 min at room temperature. Images were taken using an inverted microscope as described under “Experimental procedures.” Representative images are shown. B, cells were scored for the localization of Msn2-GFP: intense nuclear localization was scored as positive and diffuse cytosolic fluorescence was scored as negative. Localization was determined in WT (RR694) and siw14Δ mutant (RR695) strains.
Discussion
Using growth assays and gene expression analysis, we demonstrated that SIW14 is a negative regulator of the stress response at least partially through Msn2/4. The siw14Δ mutant is resistant to heat, osmotic, oxidative, and nutritional stresses, extending and confirming previously reported findings (27, 43–46). Transcription of the environmental stress response is partially induced under nonstress conditions in the siw14Δ mutant, consistent with the increased stress resistance. The siw14Δ mutant strain is capable of inducing a transcriptional ESR, indicating that the stress response signaling pathway is intact and that the steady-state levels of inositol pyrophosphates are implicated in setting a baseline. The results from the epistasis and localization experiments indicate that high levels of inositol pyrophosphates affect the nuclear localization of the transcription factor Msn2.
SIW14 encodes a phosphatase specific for the β-phosphate on 5PP-InsP5 and the mutant has 6-fold elevated levels of inositol pyrophosphates (25). Inositol pyrophosphates, synthesized by the kinases Kcs1 and Vip1, are required for induction of the environmental stress response (15) and for resistance to environmental stresses (47). Our results here demonstrate higher endogenous levels of inositol pyrophosphates in the mutant led to a partially-induced transcriptional ESR. These observations suggest that cells might modulate inositol pyrophosphate levels as an intracellular signal to affect the ESR under stress conditions.
Consistent with a role for inositol pyrophosphates as signaling molecules, hydrogen peroxide treatment increased the levels of inositol pyrophosphates in WT cells, and was particularly high for InsP8 (a 5.3 ± 0.8-fold increase). Conversely, the kcs1Δ vip1Δ double mutant, which does not produce inositol pyrophosphates, exhibited a severe transcriptional defect by completely failing to induce an ESR during H2O2 stress (15). Our results here linking inositol pyrophosphate levels to transcriptional responses are consistent with previous gene expression studies (15), and with the increased levels of InsP7 and H2O2 resistance of the vip1Δ mutant (21, 25). An intriguing possibility is that the active site cysteine of Siw14 directly senses cellular ROS levels, and the phosphatase is catalytically inactive when oxidized. Reversible oxidation of the active site cysteine could regulate the enzyme activity of Siw14, as has been described for other members of this phosphatase family (41, 42). If oxidative stress conditions inhibit Siw14 in WT cells, there would be an increase in inositol pyrophosphate pools; these could in turn partially induce the ESR. This model is also consistent with the absence of an increase in inositol pyrophosphate levels in the siw14Δ mutant upon oxidative stress (Fig. 4).
We found that neither InsP7 nor InsP8 levels changed during osmotic stress in either the WT or siw14Δ strains; our results with the WT are consistent with a previous study (48). Results from the kcs1Δ vip1Δ double mutant showed that a transcriptional stress response was partially mounted during osmotic stress (15), indicating a divergence in the role of inositol pyrophosphates in the osmotic and oxidative stress responses, and demonstrating that the transcriptional response to oxidative stress is more dependent on inositol pyrophosphate levels. It is possible that osmotic stress affects only a small pool of inositol pyrophosphates that is below our detection limits, whereas oxidative stress affects multiple localized pools (e.g. cytosolic, membrane-associated, and nuclear) such that the global changes are detectable by HPLC analysis.
One possible model linking inositol pyrophosphates to the induction of the ESR in yeast could be through the HDAC Rpd3L. Rpd3L is recruited to the promoters of many ESR genes (13, 14) and is proposed to have an inositol phosphate-binding pocket (15) based on amino acid conservation with an inositol polyphosphate-binding pocket found in the human HDAC3 (49, 50). This model suggests that inositol pyrophosphates directly bind Rpd3L to influence HDAC activity.
To test the model that inositol pyrophosphates activate the ESR through Rpd3L, we performed epistasis experiments with the siw14Δ mutant and the Rpd3L inositol-binding site (rpd3ibs) mutant. Unexpectedly, we found the stress-resistant phenotype of the siw14Δ mutant is inconsistent with regulation of HDAC activity by modulating PP-InsP levels. Although our experiments indicate an alternative mechanism for ESR activation, they do not rule out a structural role for inositol pyrophosphates in the Rpd3L complex. Worley and colleagues (15) showed that the transcription profiles of the rpd3Δ, rpd3ibs, and kcs1Δvip1Δ mutant strains are the same in nonstress and stress conditions, which demonstrated that PP-InsPs, the inositol phosphate-binding site, and catalytic activity by Rpd3L are all required for the appropriate transcriptional responses.
Our results led us to pursue an alternative model by which inositol pyrophosphates function through the stress response transcription factors Msn2/4. These partially redundant transcription factors regulate a large portion of ESR genes by binding to upstream stress response elements (32). Using known Msn2/4 target genes, we analyzed our microarray data to identify target genes that were differentially expressed in the siw14Δ mutant. Indeed, 122 of the 444 genes differentially expressed in the siw14Δ mutant are transcriptionally regulated by Msn2/4. The epistasis and localization results support a model in which modulation of inositol pyrophosphate levels affect signaling of cellular stress to Msn2/4.
This study has strengthened the connection between inositol pyrophosphates and the environmental stress response. Interestingly, a role for inositol pyrophosphates in stress responses have been found in other eukaryotes, but has remained understudied (51–53). For example, Cryptococcus neoformans requires the IP6 kinase Kcs1 and production of 5PP-InsP5 for adaptation to host cell environments and pathogenesis (54, 55). In Saccharomyces, inositol pyrophosphates are required for pseudohyphal growth, a response to nutrient limitation (56), and for prion propagation (57). Inositol pyrophosphates are important for signaling of heat and osmotic stress in mammalian cells (48, 58) and are critical for jasmonate-dependent defenses against herbivorous insects and necrotrophic fungi in plants (59). These examples highlight the broad role for inositol pyrophosphates in stress responses. New insights into the metabolism of inositol pyrophosphates may lead to novel therapeutics and treatments, as well as a deeper understanding of their cellular roles.
Experimental procedures
Strains and plasmids
Strains used in this study are listed in Table 1. Parental WT strain BY4741 and siw14Δ::KANMX, msn2Δ::KANMX, and msn4Δ::KANMX mutant strains were purchased from OpenBioSystems, and strains W3O3 and ACY614 were obtained from A. Capaldi (7). The msn2Δ siw14Δ and msn4Δ siw14Δ mutant strains were constructed by PCR amplification of the siw14Δ::URA3 allele, and homologous recombination at SIW14 with selection on SC-Ura medium, as described (25), in the BY4741 msn2Δ or msn4Δ mutant strains. The MSN2-GFP strain (60) was kindly provided by Mark Rose; to introduce MSN2-GFP into the siw14Δ::KANMX strain RR643, the MSN2-GFP::HIS3 allele was amplified by PCR, integrated by homologous recombination and selection on SC-His medium. The plasmids carrying the SIW14 gene or the phosphatase-dead allele siw14-C214S were previously reported (25).
Table 1.
Strains used in this article
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MATa his3Δ met15Δ leu2Δ ura3Δ | 71 |
| RR642 | MATa his3Δ met15Δ leu2Δ ura3Δ siw14::KANMX4 | 71 |
| RR650 | MATa his3Δ met15Δ leu2Δ ura3Δ msn2Δ::KANMX4 | 71 |
| RR651 | MATa his3Δ met15Δ leu2Δ ura3Δ msn4Δ::KANMX4 | 1 |
| RR667 | MATa his3Δ met15Δ leu2Δ ura3Δ msn2Δ::KANMX4 siw14Δ::URA3 [parent strain: RR650] | This study |
| RR668 | MATa his3Δ met15Δ leu2Δ ura3Δ msn4Δ::KANMX4 siw14Δ::URA3 [parent strain: RR651] | This study |
| W303 (ACY044) | MATa trp1–1 leu2–3,112 ura3–1 his3–11,15 can1–100 GAL1 + psi + ADE2 + | 72 |
| ACY614 | MATa trp1–1 leu2–3,112 ura3–1 his3–11,15 can1–100 GAL1 + psi + ADE2 + rpd3ibs | 15 |
| RR657 | MATa trp1–1 leu2–3,112 ura3–1 his3–11,15 can1–100 GAL1 + psi + ADE2 + rpd3ibs siw14Δ::KANMX4 rpd3ibs | This study |
| RR694 | MATa his3Δ met15Δ leu2Δ ura3Δ MSN2-GFP::HIS3 | 60 |
| RR695 | MATa his3Δ met15Δ leu2Δ ura3Δ MSN2-GFP::HIS3 siw14Δ::KANMX4 | This study |
Growth conditions
Cells were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) and plasmid-bearing cells were grown in SC-Ura (0.17% YNB (Sunrise Science Products), 0.5% ammonium sulfate, 0.069% CSM-Ura amino acid mix (MP Biomedicals), 2% dextrose, 0.75 μm adenine); strains without plasmids were grow on SC-Ura medium supplemented with 0.9 mm uracil. Overnight cultures were inoculated into fresh medium at an OD600 of 0.1–0.15, and were allowed to grow to log phase (OD600 of ∼0.6) or for 24 h for post-diauxic shift. Cultures were normalized to the same OD600 and serially diluted 1:10 four times in buffered saline with glucose (10 mm Tris-HCl, pH 7.5, 85 mm NaCl, 10 mm glucose) unless otherwise noted. Post-diauxic shift cultures were serially diluted 1:10 in 10 mm Tris-HCl, pH 7.5, 85 mm NaCl without glucose unless otherwise noted. For semi-quantitative measurement of yeast growth, 2.5 μl of each dilution was spotted onto solid medium. For quantitative measurement of yeast growth, 20–25 μl of the 10−3 or 10−4 dilutions were spread onto solid medium such that the cfu were in the range of 30–300 colonies and cell counts were made. All plates were allowed to grow for 2 days at 30 °C unless otherwise noted.
For osmotic stress, cultures were grown to early log phase (OD600 of 0.3) or post-diauxic shift (24 h), and plated on YPD medium containing 1.35 m KCl. For oxidative stress, cultures were grown to early log phase (OD600 of 0.3) or post-diauxic shift in SC medium, and 1 mm H2O2 was added for 3 h (modified from (73). Cells were serially diluted 1:10 in sterile deionized H2O and plated onto solid YPD medium. For cultures with mutations in msn2Δ or msn4Δ, cells were grown overnight and diluted to an OD600 of 0.2 in YPD and allowed to grow for 30 min and then treated with H2O2 (61). Cells were then serially diluted as described above. For heat shock, overnight cultures were diluted to an OD600 of 0.1 in YPD or SC-ura medium (for plasmid containing strains) and grown to mid-log phase or for 24 h. Cells were subjected to 50 or 53 °C for 10 min, serially diluted, and plated on SC-Ura solid medium (for plasmid containing strains) or YPD (modified from Ref. 43).
Chronological aging assays
Cells were grown overnight in YPD and then normalized in SC media at an OD600 of ∼0.1. Cultures were allowed to grow for 14 days at 30 °C with shaking. Samples were removed every 24 h and normalized to an OD600 of 0.3, which equaled about 106 cells. They were stained with ∼6 μg/ml of propidium iodide to test for dead cells (29). Using flow cytometry (Becton Dickinson FACSort), stained cells were counted and the percent alive were calculated for each strain at each time point in triplicate.
Microarray analysis
BY4741 and the siw14Δ mutant were grown to 0.6 OD600 in S.D. medium (0.67% YNB (BD Biosciences), 0.1% amino acid mixture (U. S. Biological Corp.), 2% dextrose). Half of the cells were immediately harvested, and the remaining half were osmotically stressed by the addition of KCl (0.4 m) or oxidatively stressed with 1 mm H2O2 for 20 min and harvested (15). W303 cells were grown to 0.6 OD600 in S.D. medium and were stressed in medium lacking dextrose for 20 min. RNA was isolated by hot acid phenol followed by purification using the RiboPureTM Yeast RNA Purification Kit (Ambion). RNA was converted into cDNA using oligo(dT) before being labeled with Cy3 or Cy5 for transcript level measurement on Agilent G4813A DNA microarrays and an Axon 4000B scanner, as described previously (15). The microarray data have been deposited in NCBI's Gene Expression Omnibus (62, 63) and are accessible through GEO Series accession number GSE135546. Heat maps were generated using the online source Morpheus (https://software.broadinstitute.org/Morpheus)6 (64). The Euler Plot in Fig. 2C was generated as described (65) using eulerAPE. GO term analysis, performed for the data in supporting Table S1, was generated using the Saccharomyces Genome Database GO Term Finder tool (66).
RT-qPCR
Overnight cultures were inoculated into fresh YPD medium and grown to log phase. Cultures were split and each set of cultures were treated with 1 mm H2O2, 0.4 m KCl, or were untreated for 20 min at 30 °C (15). Cells were immediately placed on ice and centrifuged at 2500 rpm for 2 min, and pellets were frozen at −80 °C (67). RNA was extracted using the RiboPureTM RNA Kit following the manufacturer's instructions with slight modifications: 3 μg of RNA was treated with 1 μl of DNase I in a 50-μl reaction volume for 1 h at 37 °C. RNA (200 ng) was used to synthesize the cDNAs using the SensiFAST cDNA synthesis kit (Bioline), following the manufacturer's instructions. The synthesized cDNA was diluted 5-fold into the diethyl pyrocarbonate water. The RT-qPCR was performed using the SensiFAST SYBR No-ROX Kit (Bioline), following the manufacturer's instructions with two technical replicates for each cDNA sample. Three μl of 5-fold diluted cDNA was added into the reaction with 15 μl final volume and the primers used for each gene are as follows: CTT1, forward 5′-AGAGAGTTACGCAATACTTTGG-3′ and reverse 5′-CCTTCAAGGTCAACAGGTTC-3′; HSP12, forward 5′-GCAGACCAAGCTAGAGATTAC-3′ and reverse 5′-TTCTTGGTTGGGTCTTCTTC-3′; XBP1, forward 5′-CCACTTTCCCTCAACCTTATG-3′ and reverse 5′-GTATTATGAGCTGGTCGTTGG-3′; UBC6, forward 5′-GATACTTGGAATCCTGGCTGGTCTGTCTC-3′ and reverse 5′-AAAGGGTCTTCTGTTTCATCACCTGTATTTGC-3′; GAR1, forward 5′-GCTGACAAACTATTGCCTATTG-3′ and reverse 5′-GGCACCACTTCTCTTCTTC-3′; NOG1, forward 5′-GGAGAAAGCTGCATGGATTAG-3′ and reverse 5′-AGTTTAGAACGTGGCATGATAG-3′; ADH1, forward 5′-CAAGTCGTCAAGTCCATCTC-3′ and reverse 5′-CAAGCCGACAACCTTGAT-3′; and RPL16A, forward 5′-GCCAAATTGGAAGCAAAGAG-3′ and reverse 5′-TTCAGCAGCAGTAGCATTAG-3′.
For CTT1 expression (Fig. 7), RNA was prepared using the Qiagen RNeasy Plus Mini kit, following the manufacturer's instructions, and the Luna Universal One Step RT-qPCR Kit (from New England Biolabs) was used for detection. The relative gene expression was calculated by the ΔCT method using UBC6 as the reference gene as described (68) and were normalized to the WT.
Extraction of [3H]inositol phosphates and HPLC analysis
Overnight cultures were grown in YPD medium and normalized to an OD600 of 0.005 in SC-inositol medium. Cells were radiolabeled with 75 μCi of myo-[3H]inositol for ∼20 h, stressed with either 0.4 m KCl or 1 mm H2O2 for 20 min, and harvested. Extracts were prepared and inositol pyrophosphates separated and assessed as described (25).
Purification of Siw14 and enzyme assay
Recombinant His6-MBP-Siw14 was expressed in Escherichia coli BL21(DE3) and purified following the protocol described (69), with modification. Briefly, E. coli containing the pGro7 chaperone plasmid and pDest-566-Siw14 plasmid were grown overnight in nutrient-rich 2× YT medium, inoculated 1:100 into fresh 2× YT containing 0.07% l-arabinose, pH 7.5, and grown at 37 °C to mid-log phase. Isopropyl β-d-thiogalactopyranoside (IPTG) was added to 100 μm and cultures were grown at 4 °C for 2 days. Cells were pelleted by centrifugation and lysed by sonication. Protein was purified in batch using nickel-nitrilotriacetic acid-Sepharose beads (GE Healthcare); beads were washed twice with Buffer 1 (20 mm Tris-HCl, pH 7.5, 20 mm imidazole, 300 mm NaCl) and once with Buffer 2 (20 mm Tris-HCl, pH 7.5, 20 mm imidazole, 50 mm NaCl). Protein was eluted in Buffer 3 (20 mm Tris-HCl, pH 7.5, 400 mm imidazole, 50 mm NaCl). To remove the inhibitory imidazole, buffer was exchanged using centrifugal filter units (Amicon Ultra-15, Ultracel-30K) to a buffer containing 20 mm Tris-HCl, pH 7.5, and 50 mm NaCl. Purification was assessed by PAGE and staining with Coommassie Brilliant Blue.
For oxidation reactions, 10 μg of purified Siw14 was incubated with hydrogen peroxide (0, 1, or 5 mm) for 30 min on ice. A phosphatase assay was performed in quintuplicate, as described (25), using p-nitrophenyl phosphate as the phosphatase substrate. To test reversible oxidation, the purified Siw14 was incubated with 1 mm hydrogen peroxide for 30 min on ice, and then 1 unit of catalase (Sigma C30-100MG) was added for 30 min to degrade residual hydrogen peroxide. Immediately following the catalase reaction, samples were treated with 100 μm DTT for 30 min on ice, followed by phosphatase assay (25). Absorbance values were recorded at OD405 and converted to activity; units reported as 1 unit = 1 μmol/min/g of protein and calculated using a standard curve. The dot plots were generated using an online tool (70).
Microscopy
Strains RR694 and RR695 were grown overnight in SC medium at room temperature, and then diluted to an OD600 of 0.1, and cultured for 6 h at room temperature. One ml of cells was transferred to a chamber slide (2-well borosilicate chamber coverglass slide) and allowed to settle for 30 min before imaging. Images were acquired using a deconvolution microscopy system (DeltaVision; Applied Precision, LLC) equipped with an inverted microscope (TE200; Nikon) and a ×100 objective with numerical aperture of 1.4. Image analysis were performed using Precision softWoRx and ImageJ. For each strain, three biological replicates and at least 450 cells were counted.
Author contributions
E. A. S., V. A. M., and R. J. R. conceptualization; E. A. S., V. A. M., K. F., L. C., A. C. R., and A. P. C. investigation; E. A. S. and V. A. M. writing-original draft; E. A. S., V. A. M., K. F., L. C., A. C. R., A. P. C., and R. J. R. writing-review and editing; A. C. R. and A. P. C. resources; A. C. R., A. P. C., and R. J. R. supervision; R. J. R. funding acquisition; R. J. R. project administration.
Supplementary Material
Acknowledgments
We thank Tanaporn Wangsanut for help with RNA extractions, Erica Raphael for help with strain construction, and Chamel Khoury for initial observations that started this project. We thank Audrey Gasch and Mark Rose for providing yeast strains, Jingwen Hu and Jeff Huang for providing catalase, and Mark Rose for use of the fluorescent microscope and for advice.
This work was supported by a Georgetown Pilot Grant (to R. J. R.), a GradGov Research Project Award (to V. A. M.), National Institutes of Health Grant R01GM097329 (to A. P. C.), and National Science Foundation CAREER Grant MCB-1253809 (to A. C. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
The microarray data reported in this paper have been submitted to the Gene Expression Omnibus (GEO) database under GEO accession no. GSE135546.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ESR
- environmental stress response
- InsP6
- inositol hexakisphosphate
- 5PP-InsP5
- 5-diphosphoinositol pentakisphosphate
- InsP7
- unspecified isomer of diphosphoinositol pentakisphosphate
- ROS
- reactive oxygen species
- HDAC
- histone deacetylase complex
- SC
- synthetic complete (a defined growth medium)
- PKA
- protein kinase A
- ibs
- inositol-binding site
- cfu
- colony-forming unit
- qPCR
- quantitative PCR.
References
- 1. Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., and Brown P. O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kültz D. (2003) Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J. Exp. Biol. 206, 3119–3124 10.1242/jeb.00549 [DOI] [PubMed] [Google Scholar]
- 3. Morano K. A., Grant C. M., and Moye-Rowley W. S. (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 10.1534/genetics.111.128033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gasch A. P. (2007) Comparative genomics of the environmental stress response in ascomycete fungi. Yeast 24, 961–976 10.1002/yea.1512 [DOI] [PubMed] [Google Scholar]
- 5. Gasch A. P. (2003) The environmental stress response: a common yeast response to diverse environmental stresses. in Yeast Stress Responses: Topics in Current Genetics (Hohmann S. and Mager W. H., eds) pp. 11–70, Springer, Berlin, Heidelberg [Google Scholar]
- 6. Lee P., Kim M. S., Paik S.-M., Choi S.-H., Cho B.-R., and Hahn J.-S. (2013) Rim15-dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae. FEBS Lett. 587, 3648–3655 10.1016/j.febslet.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 7. Hughes Hallett J. E., Luo X., and Capaldi A. P. (2014) State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198, 773–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. AkhavanAghdam Z., Sinha J., Tabbaa O. P., and Hao N. (2016) Dynamic control of gene regulatory logic by seemingly redundant transcription factors. Elife 5, e18458 10.7554/eLife.18458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadeh A., Movshovich N., Volokh M., Gheber L., and Aharoni A. (2011) Fine tuning of the Msn2/4-mediated yeast stress response as revealed by systematic deletion of Msn2/4 partners. Mol. Biol. Cell 22, 3127–3138 10.1091/mbc.e10-12-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inoki I., Ouyang H., Li Y., and Guan K. L. (2005) Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 69, 79–100 10.1128/MMBR.69.1.79-100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beck T., and Hall M. N. (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692 10.1038/45287 [DOI] [PubMed] [Google Scholar]
- 12. Görner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., Hamilton B., Ruis H., and Schüller C. (1998) Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protien kinase A activity. Genes Dev. 12, 586–597 10.1101/gad.12.4.586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruiz-Roig C., Viéitez C., Posas F., and de Nadal E. (2010) The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 76, 1049–1062 10.1111/j.1365-2958.2010.07167.x [DOI] [PubMed] [Google Scholar]
- 14. Alejandro-Osorio A. L., Huebert D. J., Porcaro D. T., Sonntag M. E., Nillasithanukroh S., Will J. L., and Gasch A. P. (2009) The histone deacetylatse Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 10, R57 10.1186/gb-2009-10-5-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Worley J., Luo X., and Capaldi A. P. (2013) Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell. Rep. 3, 1476–1482 10.1016/j.celrep.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fleischer B., Xie J., Mayrleitner M., Shears S. B., Palmer D. J., and Fleischer S. (1994) Golgi coatomer binds, and forms K+-selective channels gated by, inositol polyphosphates. J. Biol. Chem. 269, 17826–17832 [PubMed] [Google Scholar]
- 17. Ali N., Duden R., Bembenek M. E., and Shears S. B. (1995) The interaction of coatomer with inositol polyphosphates is conserved in Saccharomyces cerevisiae. Biochem. J. 310, 279–284 10.1042/bj3100279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saiardi A., Resnick A. C., Snowman A. M., Wendland B., and Snyder S. H. (2005) Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 1911–1914 10.1073/pnas.0409322102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. York S. J., Armbruster B. N., Greenwell P., Petes T. D., and York J. D. (2005) Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 280, 4264–4269 10.1074/jbc.M412070200 [DOI] [PubMed] [Google Scholar]
- 20. Szijgyarto Z., Garedew A., Azevedo C., and Saiardi A. (2011) Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 10.1126/science.1211908 [DOI] [PubMed] [Google Scholar]
- 21. Onnebo S. M., and Saiardi A. (2009) Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 423, 109–118 10.1042/BJ20090241 [DOI] [PubMed] [Google Scholar]
- 22. Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P., and Snyder S. H. (1999) Synthesis of diphophoinositol pentakisphosphate by a newly identified family of higher inositol pholyphosphate kinases. Curr. Biol. 9, 1323–1326 10.1016/S0960-9822(00)80055-X [DOI] [PubMed] [Google Scholar]
- 23. Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., and York J. D. (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 10.1126/science.1139099 [DOI] [PubMed] [Google Scholar]
- 24. Lin H., Fridy P. C., Ribeiro A. A., Choi J. H., Barma D. K., Vogel G., Falck J. R., Shears S. B., York J. D., and Mayr G. W. (2009) Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 284, 1863–1872 10.1074/jbc.M805686200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steidle E. A., Chong L. S., Wu M., Crooke E., Fiedler D., Resnick A. C., and Rolfes R. J. (2016) A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 protein selectively cleaves the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J. Biol. Chem. 291, 6772–6783 10.1074/jbc.M116.714907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martins D., and English A. M. (2014) Catalase activity is stimulated by H2O2 in rich culture medium and is required for H2O2 resistance and adaptation in yeast. Redox Biol. 2, 308–313 10.1016/j.redox.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Altinta A., Martini J., Mortensen U. H., and Workman C. T. (2016) Quantification of oxidative stress phenotypes based on high-throughput growth profiling of protein kinase and phosphatase knockouts. FEMS Yeast Res. 16, fov101 10.1093/femsyr/fov101 [DOI] [PubMed] [Google Scholar]
- 28. Werner-Washburne M., Braun E., Johnston G. C., and Singer R. A. (1993) Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57, 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pereira C., and Saraiva L. (2013) Interference of aging media on the assessment of yeast chronological life span by propidium iodide staining. Folia Microbiol. (Praha) 58, 81–84 10.1007/s12223-012-0186-6 [DOI] [PubMed] [Google Scholar]
- 30. Grant C. M., Perrone G., and Dawes I. W. (1998) Glutathione and catalase provide overlapping defenses for protection against hydrogen peroide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253, 893–898 10.1006/bbrc.1998.9864 [DOI] [PubMed] [Google Scholar]
- 31. Martínez-Pastor M. T., Marchler G., Schüller C., Marchler-Bauer A., Ruis H., and Estruch F. (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235 10.1002/j.1460-2075.1996.tb00576.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmitt A. P., and McEntee K. (1996) Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 93, 5777–5782 10.1073/pnas.93.12.5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boy-Marcotte E., Perrot M., Bussereau F., Boucherie H., and Jacquet M. (1998) Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Praekelt U. M., and Meacock P. (1990) HSP12, a new small heat schock gene of Saccharomyces cerevisiae: analysis of structure, regulation and function. Mol. Gen. Genet. 223, 97–106 10.1007/BF00315801 [DOI] [PubMed] [Google Scholar]
- 35. Welker S., Rudolph B., Frenzel E., Hagn F., Liebisch G., Schmitz G., Scheuring J., Kerth A., Blume A., Weinkauf S., Haslbeck M., Kessler H., and Buchner J. (2010) Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 39, 507–520 10.1016/j.molcel.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 36. Miles S., Li L., Davison J., and Breeden L. L. (2013) Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state. PLoS Genet. 9, e1003854 10.1371/journal.pgen.1003854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bennetzen J. L., and Hall B. D. (1982) The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J. Biol. Chem. 257, 3018–3025 [PubMed] [Google Scholar]
- 38. Kallstrom G., Hedges J., and Johnson A. (2003) The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Biol. Cell 23, 4344–4355 10.1128/MCB.23.12.4344-4355.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tremblay A., Lamontagne B., Catala M., Yam Y., Larose S., Good L., and Elela S. A. (2002) A physical interaction between Gar1p and Rntp is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol. Cell. Biol. 22, 4792–4802 10.1128/MCB.22.13.4792-4802.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Planta R. J., and Mager W. H. (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14, 471–477 [DOI] [PubMed] [Google Scholar]
- 41. Ostman A, Frijhoff J., Sandin A., and Böhmer F.-D. (2011) Regulation of protein tyrosine phosphatases by reversible oxidation. J. Biochem. 150, 345–356 10.1093/jb/mvr104 [DOI] [PubMed] [Google Scholar]
- 42. Denu J. M., and Tanner K. G. (1998) Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry 37, 5633–5642 10.1021/bi973035t [DOI] [PubMed] [Google Scholar]
- 43. Care A., Vousden K. A., Binley K. M., Radcliffe P., Trevethick J., Mannazzu I., and Sudbery P. E. (2004) A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics 166, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown J. A., Sherlock G., Myers C. L., Burrows N. M., Deng C., Wu H. I., McCann K. E., Troyanskaya O. G., and Brown J. M. (2006) Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol. Syst. Biol. 2, 2006.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jarolim S., Ayer A., Pillay B., Gee A. C., Phrakaysone A., Perrone G. G., Breitenbach M., and Dawes I. W. (2013) Saccharomyces cerevisiae genes involved in survival of heat shock. G3 3, 2321–2333 10.1534/g3.113.007971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garay E., Campos S. E., González de la Cruz J., Gaspar A. P., Jinich A., and Deluna A. (2014) High-resolution profiling of stationary-phase survival reveals yeast longevity factors and their genetic interactions. PLoS Genet. 10, e100468 10.1371/journal.pgen.1004168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dubois E., Scherens B., Vierendeels F., Ho M. M., Messenguy F., and Shears S. B. (2002) In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 277, 23755–23763 10.1074/jbc.M202206200 [DOI] [PubMed] [Google Scholar]
- 48. Choi K., Mollapour E., and Shears S. B. (2005) Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 17, 1533–1541 10.1016/j.cellsig.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 49. Watson P. J., Fairall L., Santos G. M., and Schwabe J. W. (2012) Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340 10.1038/nature10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Watson P. J., Millard C. J., Riley A. M., Robertson N. S., Wright L. C., Godage H. Y., Cowley S. M., Jamieson A. G., Potter B. V., and Schwabe J. W. (2016) Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 7, 11262 10.1038/ncomms11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilson M. S., Livermore T. M., and Saiardi A. (2013) Inositol pyrophosphates: between signaling and metabolism. Biochem. J. 452, 369–379 10.1042/BJ20130118 [DOI] [PubMed] [Google Scholar]
- 52. Shears S. B. (2015) Inositol pyrophosphates: why so many phosphates? Adv. Biol. Regul. 57, 203–216 10.1016/j.jbior.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thota S. G., and Bhandari R. (2015) The emerging roles of inositol pyrophoshates in eukaryotic cell physiology. J. Biosci. 40, 593–605 10.1007/s12038-015-9549-x [DOI] [PubMed] [Google Scholar]
- 54. Li C., Lev S., Saiardi A., Desmarini D., Sorrell T. C., and Djordjevic J. T. (2016) Identification of a major IP5 kinase in Cryptococcus neoformans confirms that PP-IP5/IP7, not IP6, is essential for virulence. Sci. Rep. 6, 23927 10.1038/srep23927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lev S., Li C., Desmarini D., Saiardi A., Fewings N. L., Schibeci S. D., Sharma R., Sorrell T. C., and Djordjevic J. T. (2015) Fungal inositol pyrophosphate IP7 is crucial for metabolic adaptation to the host environment and pathogenicity. MBio 6, e00531–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Norman K. L., Shively C. A., De La Rocha A. J., Mutlu N., Basu S., Cullen P. J., and Kumar A. (2018) Inositol polyphosphates regulate and predict yeast pseudohyphal growth phenotypes. PLos Genet. 14, e1007493 10.1371/journal.pgen.1007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wickner R. B., Kelly A. C., Bezsonov E. E., and Edskes H. K. (2017) [PSI+] prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. U.S.A. 114, E8402–E8410 10.1073/pnas.1714361114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pesesse X., Choi K., Zhang T., and Shears S. B. (2004) Signaling by higher inositol polyphosphates. Synthesis of bisdiphophoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J. Biol. Chem. 279, 43378–43381 10.1074/jbc.C400286200 [DOI] [PubMed] [Google Scholar]
- 59. Laha D., Johnen P., Azevedo C., Dynowski M., Weiß M., Capolicchio S., Mao H., Iven T., Steenbergen M., Freyer M., Gaugler P., de Campos M. K., Zheng N., Feussner I., Jessen H. J., Van Wees S. C., Saiardi A., and Schaaf G. (2015) VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27, 1082–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huh W.-K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 61. Hasan R., Leroy C., Isnard A. D., Labarre J., Boy-Marcotte E., and Toledano M. B. (2002) The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol. Microbiol 45, 233–241 10.1046/j.1365-2958.2002.03011.x [DOI] [PubMed] [Google Scholar]
- 62. Edgar R., Domrachev M., and Lash A. E. (2002) Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barrett T., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., Marshall K. A., Phillippy K. H., Sherman P. M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C. L., Serova N., Davis S., and Soboleva A. (2013) NCBI GEO: archive for functional genomics data sets–pdate. Nucleic Acids Res. 41, D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morpheus (2019) On-line Heat Map Generator, Broad Institute, Cambridge, MA [Google Scholar]
- 65. Micallef L., and Rodgers P. (2014) eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS ONE 9, e101717 10.1371/journal.pone.0101717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cherry J. M., Hong E. L., Amundsen C., Balakrishnan R., Binkley G., Chan E. T., Christie K. R., Costanzo M. C., Dwight S. S., Engel S. R., Fisk D. G., Hirschman J. E., Hitz B. C., Karra K., Krieger C. J., et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 10.1093/nar/gkr1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ghosh A. K., Wangsanut T., Fonzi W. A., and Rolfes R. J. (2015) The GRF10 homeobox gene regulates filamentous growth in the human fungal pathogen Candida albicans. FEMS Yeast Res. 15, fov093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Teste M. A., Duquenne M., François J. M., and Parrou J. L. (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 10, 99 10.1186/1471-2199-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang H., Gu C., Rolfes R. J., Jessen H. J., and Shears S. B. (2018) Structural and biochemical characterization of Siw14: a protein-tyrosine phosphatase fold that metabolizes inositol pyrophosphates. J. Biol. Chem. 293, 6905–6914 10.1074/jbc.RA117.001670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weissgerber T. L., Savic M., Winham S. J., Stanisavljevic D., Garovic V. D., and Milic N. M. (2017) Data visualization, bar naked: A free tool for creating interactive graphics. J. Biol. Chem. 292, 20592–20598 10.1074/jbc.RA117.000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 72. Capaldi A. P., Kaplan T., Liu Y., Habib N., Regev A., Friedman N., and O'Shea E. K. (2008) Structure and function of a transcriptional network activated by the MAPK Hog1. Nat. Genet. 40, 1300–1306 10.1038/ng.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vernis L., Facca C., Delagoutte E., Soler N., Chanet R., Guiard B., Faye G., and Baldacci G. (2009) A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS One 4, e4376 10.1371/journal.pone.0004376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.