Abstract
The multispecific organic anion transporters, OAT1 (SLC22A6) and OAT3 (SLC22A8), the main kidney elimination pathways for many common drugs, are often considered to have largely-redundant roles. However, whereas examination of metabolomics data from Oat-knockout mice (Oat1 and Oat3KO) revealed considerable overlap, over a hundred metabolites were increased in the plasma of one or the other of these knockout mice. Many of these relatively unique metabolites are components of distinct biochemical and signaling pathways, including those involving amino acids, lipids, bile acids, and uremic toxins. Cheminformatics, together with a “logical” statistical and machine learning-based approach, identified a number of molecular features distinguishing these unique endogenous substrates. Compared with OAT1, OAT3 tends to interact with more complex substrates possessing more rings and chiral centers. An independent “brute force” approach, analyzing all possible combinations of molecular features, supported the logical approach. Together, the results suggest the potential molecular basis by which OAT1 and OAT3 modulate distinct metabolic and signaling pathways in vivo. As suggested by the Remote Sensing and Signaling Theory, the analysis provides a potential mechanism by which “multispecific” kidney proximal tubule transporters exert distinct physiological effects. Furthermore, a strong metabolite-based machine-learning classifier was able to successfully predict unique OAT1 versus OAT3 drugs; this suggests the feasibility of drug design based on knockout metabolomics of drug transporters. The approach can be applied to other SLC and ATP-binding cassette drug transporters to define their nonredundant physiological roles and for analyzing the potential impact of drug–metabolite interactions.
Keywords: kidney, transporter, multidrug transporter, mouse, metabolomics, cheminformatics, machine-learning, organic anion transporter, SLC22A6, SLC22A8
Introduction
Organic anion transporter 1 (OAT1,3 SLC22A6 (solute carrier family 22 member 6) originally described as NKT (1)) and OAT3 (SLC22A8 (solute carrier family 22 member 8), also known as Roct (2)) are multispecific members of the solute carrier 22 (SLC22) family (3–7). SLC22 transporters are highly conserved across organisms (including humans) and, along with other multi-, oligo-, and mono-specific SLC and ATP-binding cassette (ABC) transporters, function collectively as an integrated network of influx and efflux transporters involved in metabolism, signaling, and other aspects of physiological homeostasis (8–12). Consistent with this notion, SLC22 transporters are expressed in many tissues and have the ability to interact with a diverse range of endogenous and exogenous molecules (13–16).
OAT1 and OAT3, which are both expressed on the basolateral aspect of renal proximal tubule epithelial cells, represent the key rate-limiting step for the transport and removal of protein-bound organic anion molecules from the blood and, as such, are essential for the elimination of a wide array of small organic molecule drugs, toxins, metabolites, signaling molecules, uremic toxins, and natural products (5, 17). Although OAT1 and OAT3 are generally considered the main organic anion “drug” transporters in the kidney, as well as other tissues (6, 7, 18–20), their physiological role in transporting endogenous metabolites (21–25) and in mediating inter-organ cross-talk (the “Remote Sensing and Signaling Theory”) is rapidly becoming apparent (5, 9, 26–31).
Nevertheless, much remains to be elucidated about the mechanisms driving the interaction of SLC22 transporters, like OAT1 and OAT3, with their endogenous (nonxenobiotic) substrates or ligands, particularly with regard to the specificity of the transporters. With respect to pharmaceutical drugs, it is often assumed, based on overlapping lists of drug ligands (3, 4, 6, 7), that OAT1 and OAT3 have similar substrate specificity. In the absence of protein crystal structures in multiple functional states, much research has been performed using ligand-based analyses of the substrates (13, 32–36). However, it has generally not been easy, using earlier data and methods, to clearly distinguish drugs that bind OAT1 from those that bind OAT3—with the caveat that some OAT3 ligands appear to have a cationic character (33, 37).
The drug–transporter interaction datasets, apart from being based on in vitro assays done by different labs (6, 7), are unavoidably limited in scope. For instance, certain classes of FDA-approved drugs transported by OATs (e.g. β-lactam antibiotics and NSAIDs) are over-represented. In addition, the drug-interaction dataset relies heavily on data derived from transport inhibition assays rather than actual substrate transport data, which tend to be more limited (33, 38). Thus, the usefulness of the drug dataset is likely to be less than ideal if one is interested in identifying the molecular properties that might uniquely target a ligand for interaction with a particular transporter (e.g. OAT1 or OAT3). An experimental evaluation of an unbiased selection of transporter substrates is a preferred input for a training set.
Recent metabolomics analyses of the plasma of OAT1 and OAT3 knockout mice (Oat1KO and Oat3KO) have considerably expanded the list of potential endogenous OAT ligands to over 200 (28, 34, 36, 39). Such studies are based on the idea that the absence (deletion) of an uptake/influx transporter, such as OAT1 or OAT3, from the basolateral membrane of the proximal tubule cell in the knockout kidney should result in the inability to remove endogenous substrates that are exclusively dependent upon the absent transporter for clearance from the blood. Consequently, this should lead to the accumulation of endogenous metabolite ligands of the deleted transporter in the plasma of each knockout animal (28, 34, 36, 39). Comparison of such metabolomics data between the two knockout animals should theoretically allow for the elucidation of potentially unique endogenous ligands for each of these two SLC transporters. This is, in fact, the case with almost 100 endogenous metabolites being found that “uniquely” accumulate in the plasma of the Oat1KO (but not in the Oat3KO), whereas another nearly 50 metabolites uniquely accumulate in the plasma of the Oat3KO mice (but not the Oat1KO) (28, 34, 36, 39).
Although some comparisons of the physicochemical features of endogenous metabolites interacting with these two multispecific transporters have been performed (28) and identified a few general features, a thorough computational and systematic analysis based on specific molecular properties/features that predispose endogenous metabolites to interact with OAT1 or OAT3 has yet to be carried out. This is important from both a physiological as well as a pharmaceutical perspective. Physiologically, OAT1 and OAT3 are major pathways through which the proximal tubule of the kidney interfaces with the rest of the body through the transport of numerous small organic molecules that function in normal metabolism, signaling, regulation of redox state, and the uremic metabolism of chronic kidney disease. If OAT1 and OAT3 have different structural preferences for metabolites and signaling molecules, this implies different effects on systemic metabolism, which has been implied by genome-scale metabolic reconstructions (36, 40, 41). Moreover, the possibility of differentially regulating OATs and other multispecific transporters that transport different sets of metabolites in various organs would also suggest, as described in the Remote Sensing and Signaling Theory, a mechanism for a relatively small number of transporters to exert major effects on metabolism in normal and disease settings (42).
In this study, cheminformatics and machine-learning techniques were combined to analyze the quantitative molecular properties of unique metabolites accumulating in vivo in the Oat1KO mice versus unique metabolites in the Oat3KO, with the goal of identifying a set of molecular properties (features) that enables clear and concise distinction of the physiologically-relevant molecules resulting from loss of either of the two transporters.
The analyses performed here provide a deeper understanding of the important physicochemical features driving endogenous ligand selectivity for OAT1 or OAT3 in vivo. Importantly, this analysis also connects OAT1 versus OAT3 unique endogenous metabolite physicochemical features to unique pathways fundamental to fatty acid, bile acid, amino acid, and peptide metabolism and signaling. The approaches employed here, combining multispecific drug transporter knockout metabolomics, cheminformatics, and machine learning to define molecular properties of endogenous ligands, can be applied to many other SLC and ABC drug transporters. This will not only help define the physiological roles of drug transporters but should also provide a new basis for metabolite-based drug design, tissue targeting of drugs, and analyzing drug–metabolite interactions. This is supported by our evaluation of the ability of the metabolite-based machine-learning model to predict OAT1- or OAT3-selective drugs.
Results
Overview of strategy (Fig. 1)
Figure 1.
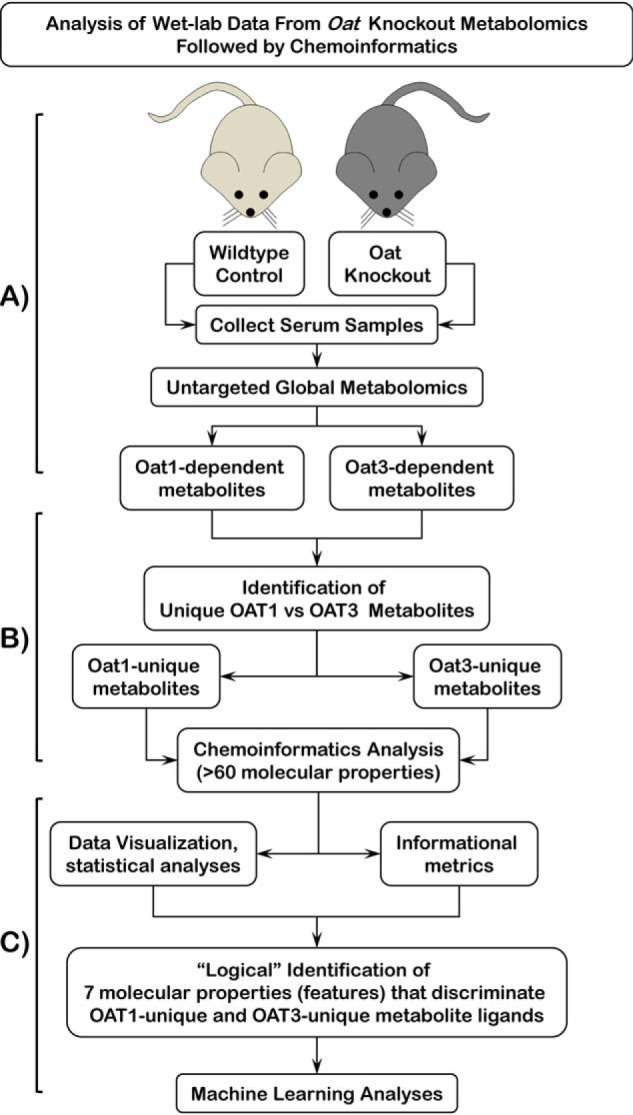
Schematic of workflow for “logical” identification of molecular properties for discrimination of unique OAT1 and OAT3 metabolites. A, OAs ultimately excreted by the kidney are cleared from the blood by the SLC transporters, OAT1 and OAT3, located in the basolateral membranes of proximal tubule cells of the kidney. Deletion of either of these transporters results in the plasma accumulation of OAs. Serum, obtained from WT and OatKOs, was subjected to untargeted, global LC-MS metabolomics analyses. B, resulting metabolomics data were used to identify Oat1 and Oat3 metabolites uniquely accumulating in each knockout mouse. Cheminformatics methods were used to identify over 60 molecular properties/features of the metabolites. C, data visualization and statistical analysis in Orange and Python libraries (Pandas, Matplotlib, Seaborn, and SQLAlchemy) of over 60 molecular properties for Oat1 and Oat3 unique metabolites were used to logically narrow down to a set of seven molecular properties to be used for machine-learning approaches to identify a set of features that classifies metabolites as uniquely Oat1 or uniquely Oat3.
The goal of this study was to identify the molecular features/characteristics determining ligand–substrate interaction with either OAT1 or OAT3; thus, we focused our attention on those metabolites that accumulate in the plasma of one of the knockouts but not the other (i.e. increased in the plasma of the Oat1KO but not the Oat3KO, or vice versa). In this way, 138 metabolites that appear to be unique for the Oat1KO (90 metabolites) or unique for the Oat3KO (48 metabolites) were identified (Figs. 1 and 2; Table S1). Although some overlap exists, OAT1-unique and OAT3-unique metabolites were unexpectedly found to have relatively unique effects on some biochemical pathways (Fig. S1). For example, ∼8% of the unique OAT1 metabolites were found to be components of the pathway involved in the metabolism of γ-glutamyl amino acids (although none of the unique OAT3 metabolites were involved in this pathway), whereas the metabolism of the primary and secondary bile acids comprises a substantial fraction of the OAT3-unique metabolites (although none of the unique OAT1 metabolites were involved in primary or secondary bile acid metabolism) (Fig. S1). Other OAT1-unique pathways included those for the degradation of valine, leucine, and isoleucine, as well as the metabolism of acylcarnitine and acylglycine fatty acids, which were found to be exclusive to OAT1-unique metabolites (Fig. S1). Other OAT3-unique pathways included the metabolism of arginine and proline (Fig. S1).
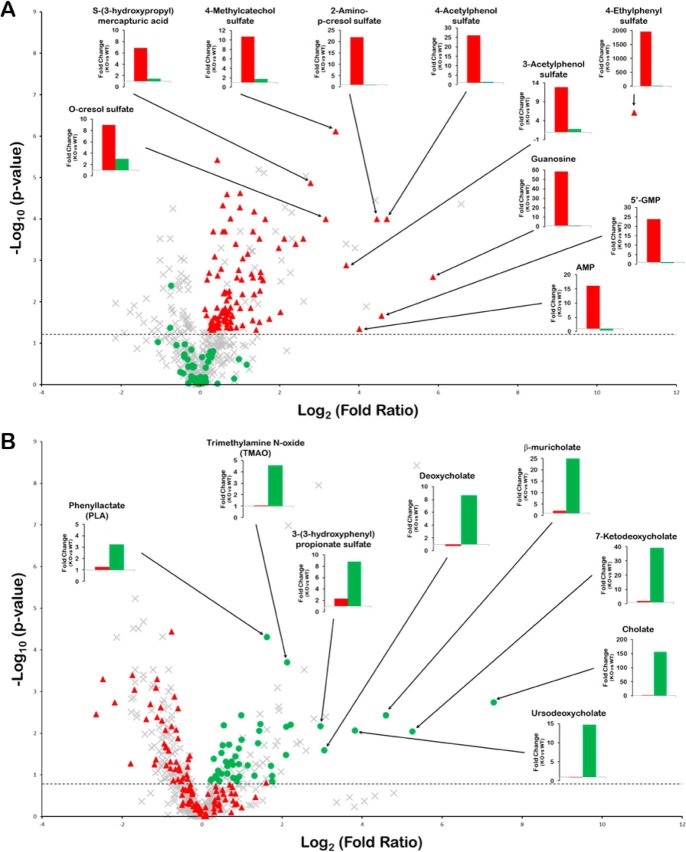
Figure 2.
Unique metabolites for the Oat1KO and Oat3KO. Volcano plots displaying metabolomics data derived from the analysis of the serum obtained from Oat1KO (KO versus WT) (A) and Oat3KO (KO versus WT) (B). The negative logarithm of the p value for each metabolite is plotted against the logarithm to the base 2 of the fold-change (KO versus WT). Each point in the plot represents a metabolite; the red triangles indicate those metabolites accumulating in the serum of the Oat1KO (p ≤0.05; dotted horizontal line), but not the Oat3KO; the green circles indicate metabolites accumulating in the Oat3KO (generally p ≤0.1 (dotted horizontal line) and see under “Experimental procedures”), but not the Oat1KO; the gray crosses indicate those metabolites either unchanged or that show similar changes in both knockout animals. The bar graphs in each volcano plot show the fold-change for the indicated metabolites in either the Oat1KO (red bars) or Oat3KO (green bars).
Cheminformatics approach to defining molecular properties of Oat1KO-unique metabolites and Oat3KO-unique metabolites
To define the molecular properties of the metabolites that would help distinguish unique metabolites of Oat1KO from those of Oat3KO, more than 60 physicochemical molecular properties (Fig. 1; Table S2) were collected or calculated and then analyzed with a focus on their potential utility in interpreting the results from a biochemical and physiological perspective, for example, in developing a biologically interpretable Decision Tree or other type of classification of the relevant molecular properties.
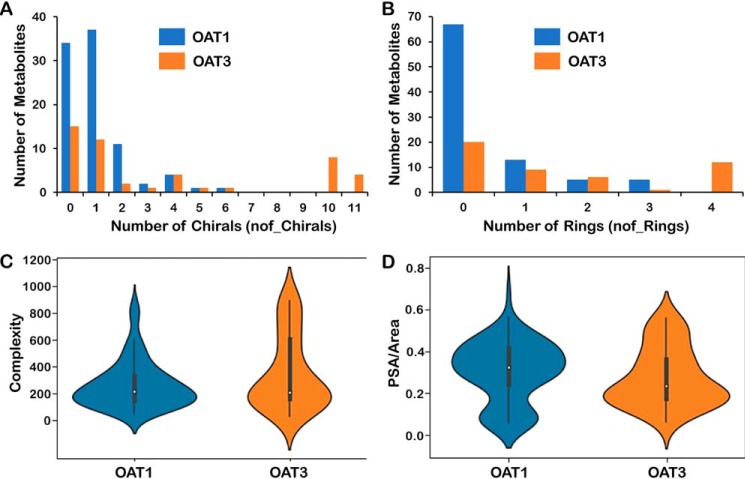
The goal of these initial analyses was to narrow down this extensive list of molecular properties to a more manageable number that could be: 1) readily understood in the context of metabolite structures, and 2) interpreted in the context of known transporter biology, and which were, individually or in combination, capable of helping to distinguish Oat1KO unique metabolites from those unique to the Oat3KO. Molecular properties that seemed of particular interest from this viewpoint were visualized in multiple ways (e.g. scatterplot, histograms, distribution plots, and violin plots) (Fig. 3). For example, the distribution plot for the number of rings indicated that the Oat3KO-specific metabolites were greatly over-represented among the molecules with more than two rings (Fig. 3 and Table 1). Although taurocholate is one of the classical transported substrates of OAT3 (43), and bile acids had among the greatest fold-changes among all metabolites in the Oat3KO, it is possible they may be overemphasized here due to over-representation on the targeted metabolomics platform and also due to the criteria used to identify unique metabolites (see “Experimental procedures”).
Figure 3.
Comparison of unique Oat1 and unique Oat3 molecular properties. A, distribution plot of a number of chirals (OAT1, blue; OAT3, red). B, distribution plot of a number of rings (OAT1, blue; OAT3, red). C, violin plot of complexity. D, violin plot of polar surface area over molecular area. From these plots, one can see that OAT1-unique metabolites are generally smaller molecules with less complexity and fewer chiral centers and less ringed structures, whereas the OAT3-unique metabolites generally are larger and more complex molecules with more chiral centers and more ringed structures. See Table S1 for the full list of metabolites and Table S2 for the full list of molecular properties.
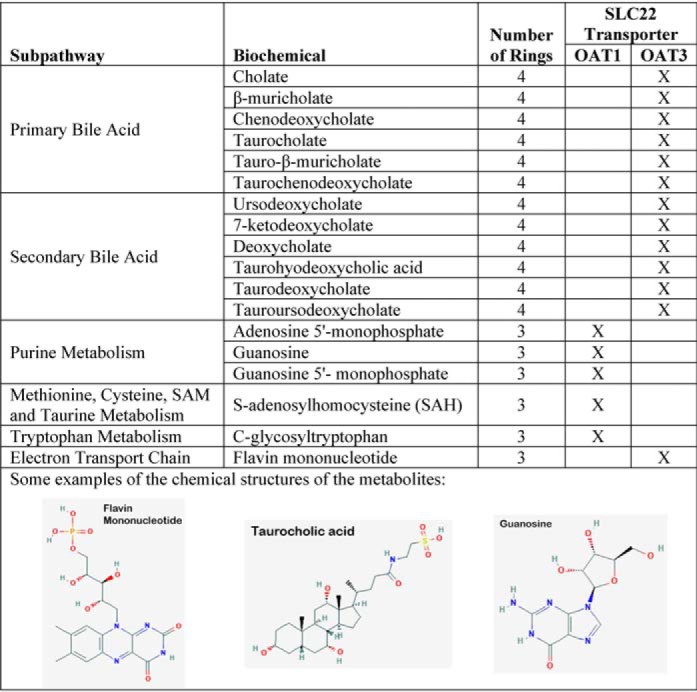
Table 1.
List of metabolites with 3 or more rings (indicating a preponderance of unique Oat3 metabolites)
Importance of a subset of molecular properties based on statistical and information metrics
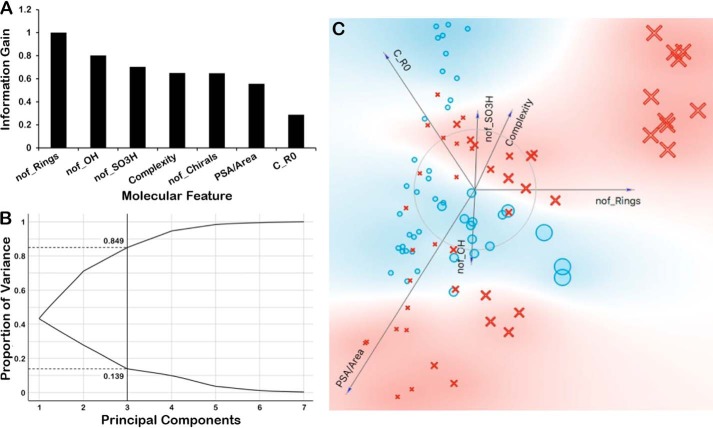
Molecular properties were ranked according to information gain and contribution to model performance. The results indicated the likely high importance of such molecular properties as the number of rings (nof_Rings), the number of chiral centers (nof_Chirals), and the complexity in separating Oat1KO unique metabolites from Oat3KO unique metabolites present in this dataset (Fig. 4). Nevertheless, in different measures (e.g. FreeViz) (44), other molecular properties such as polar surface area (PSA) over the molecular area (PSA/area) appeared important (Fig. 4). The inclusion of such features turned out to be important for machine-learning methods like random forests and decision trees (see below).
Figure 4.
Ranking of molecular properties according to information gain and other metrics indicate their likely high importance. A, bar graph of the ranking of molecular properties according to information gain in order to tease out importance of some molecular properties. The information gain for each molecular feature was normalized to the feature displaying the greatest gain (nof_Rings). B, principal component analysis reveals that first three components account for ∼90% of the variance. C, FreeViz visualization of various molecular properties and their importance in separating out OAT1 versus OAT3. In the FreeViz graphical representation, the magnitude of each vector indicates the relative importance of each molecular feature as determined by the FreeViz algorithm, whereas the direction of each vector indicates the relative preference of that feature for OAT1 (blue background) or OAT3 (red background). In the representation, the blue circles depict the OAT1-unique metabolites, and the red crosses depict the OAT3-unique metabolites. The size of each symbol corresponds to the number of rings. For example, the large red crosses in the upper right-hand portion of the representation are OAT3-unique metabolites that have a large number of rings.
Although in the iterative machine-learning process that followed (described below), the relative importance changed, at this point in the analyses the key molecular properties included the following: 1) the number of chirals (nof_Chirals); 2) the number of rings (nof_Rings); 3) the polar surface area relative to the total area (PSA/area); 4) an index of molecular complexity (Complexity); 5) a measure of molecular size (e.g. molArea, molVolume); 6) a measure of charge; and 7) the number of carbons with sp3 hybridization (c_sp3) (which is sometimes considered indicative of a molecule's three-dimensionality). By principal components analysis, three principal components were found to contribute 80% or more of the variance (Fig. 4). Later, other molecular properties not included in this original set of seven features, many of which are associated with modifications catalyzed by drug-metabolizing enzymes, were also found to be important for classification and substituted for several of the aforementioned molecular properties. These are discussed below.
Machine-learning analysis
Supervised machine-learning methods can be used to identify features that are important for classification. In this case, the cheminformatics analysis of metabolites uniquely accumulating in the Oat1KO and the Oat3KO initially resulted in >60 molecular properties (features) for each metabolite. Through the various visualizations and metrics already described, this set of features was “logically” narrowed down to <10 features, sets of which were then analyzed using the machine-learning methods described below to arrive at a set of seven features that were capable of classifying a metabolite as either OAT1 or OAT3 with about 75–80% accuracy.
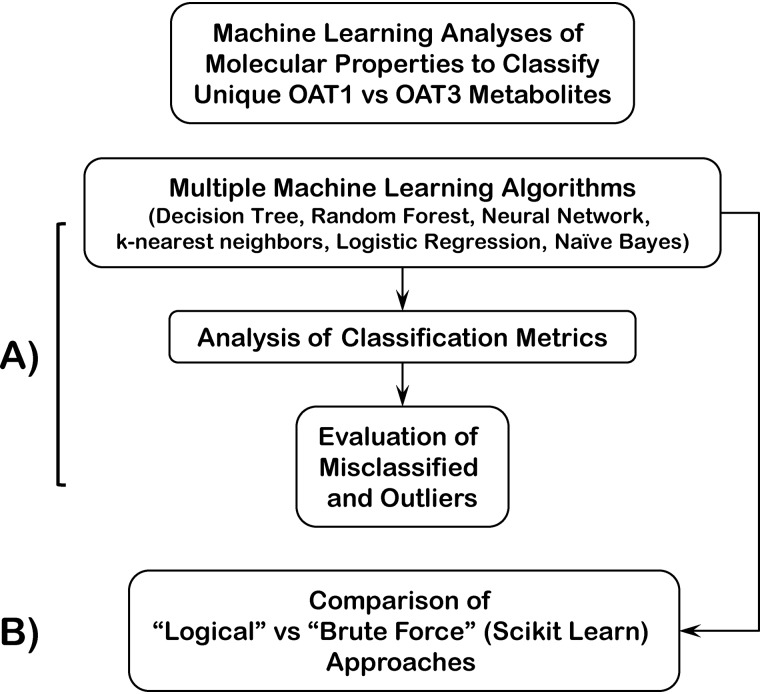
Within the Orange environment, a number of machine learning approaches were applied, including Decision Tree, Random Forest, nearest neighbor classifiers, Naive Bayes, as well as Logistic Regression and Neural Network models (Fig. 5; Table 2). Confusion Matrices were created (Fig. S2), and mis-categorized instances were evaluated. Some of the machine-learning results were also “double-checked” by direct coding using the Python Machine Learning Library SciKit-Learn (44–48).
Figure 5.
Schematic of workflow for machine-learning analyses of molecular properties to classify unique OAT1 versus OAT3 metabolites. A, machine learning in Orange and Python SciKit-Learn library using seven chosen molecular properties (features) ultimately focusing on random forests and decision trees. Analysis of accuracy score, AUC, confusion matrix, misclassified instances, and outliers was performed. B, brute force approach was used in parallel to identify seven molecular properties using Python SciKit-Learn library.
Table 2.
Output classification metrics of classification accuracy, AUC of the receiver operating curve, precision, and recall for the various machine-learning approaches
| Method | AUC | CA | F1 | Precision | Recall |
|---|---|---|---|---|---|
| Random Forest | 0.927 | 0.875 | 0.875 | 0.878 | 0.875 |
| k-Nearest Neighbor | 0.914 | 0.812 | 0.812 | 0.813 | 0.812 |
| Decision Tree | 0.886 | 0.812 | 0.812 | 0.815 | 0.812 |
| Neural Network | 0.798 | 0.76 | 0.757 | 0.775 | 0.76 |
| Naive Bayes | 0.756 | 0.667 | 0.659 | 0.682 | 0.667 |
| Logistic regression | 0.778 | 0.656 | 0.652 | 0.665 | 0.656 |
In Table 2, the various Orange machine-learning classification accuracy, area under the curve (AUC) and other scores (leave-one out method) are shown for one of the highest-scoring sets of seven molecular properties: nof_Rings; nof_Chirals; PSA/Area; nof_Sulfates; nof_OH; C_RO; and Complexity. Among the 11 metabolites misclassified by one Random Forest classification run (using the seven final features), two had been flagged in exploratory data analysis with the larger set of features as outliers by the Orange SVM-based Outlier widgets (Table S3). The confusion matrices for several machine-learning models also revealed different sets of misclassified metabolites, despite often having similar scores on classification metrics, and those misclassified sets of metabolites had a somewhat different overlap with outliers. Thus, although we have mainly focused on Random Forest classification in what follows, these considerations emphasize the need to examine the Confusion Matrix for each model.
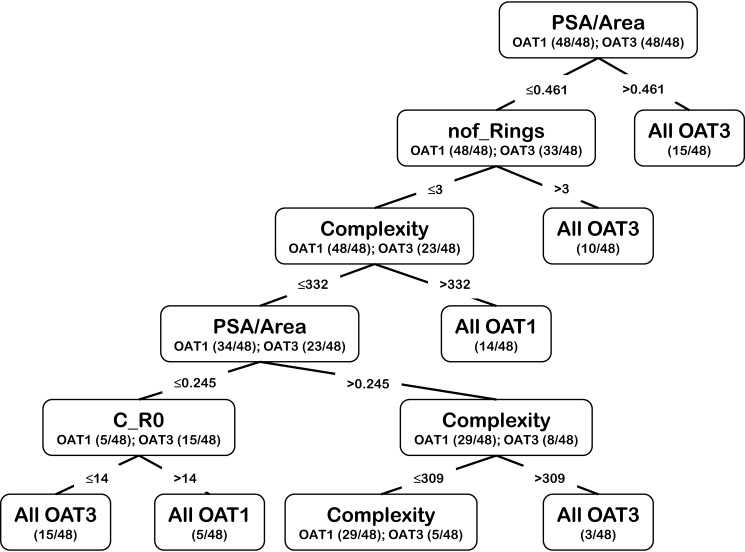
Although Random Forest classification generally gave the best score (>0.8 classification accuracy), Decision Tree classification often came close (Table 2; Fig. S3), and it was generally the easiest to interpret in terms of those molecular properties favoring classification of unique Oat1KO metabolites versus unique Oat3KO metabolites. As expected, with sampling (i.e. different sets of 48 out 90 OAT1 instances) different trees were generated, some of which were complicated. One of the more straight-forward yet representative trees is shown in Fig. 6.
Figure 6.
Example of a decision tree using the seven chosen molecular properties to classify unique OAT1 versus unique OAT3 metabolites. An example of a decision tree generated using the seven molecular features used to classify the metabolites.
Importance of including phase I and phase II modifications by drug-metabolizing enzymes as molecular properties
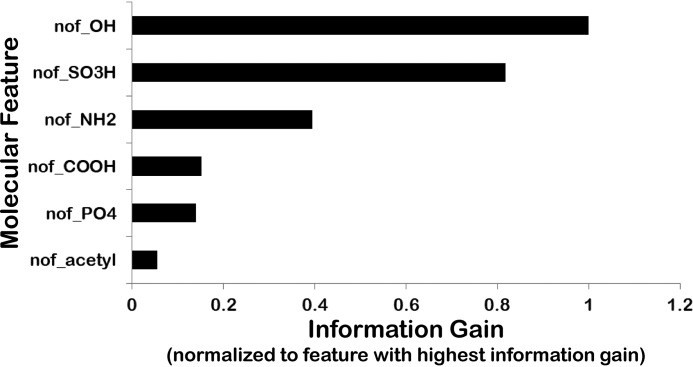
Many of the metabolites accumulating in the two knockouts included presumptive modifications by phase I and phase II drug-metabolizing enzymes (DMEs), as well as other enzymes involved in sulfation, hydroxylation, acetylation, GSH conjugation, and phosphorylation (49, 50). Phase I DMEs, which include members of the cytochrome P450 family, are important for redox modifications and hydroxylation. Phase II DMEs include acetyltransferases, glucuronosyltransferases, sulfotransferases, and other enzymes. Although these phase I and phase II DMEs are generally viewed as important in drug inactivation and/or making drugs more soluble (by adding polar groups) and thus more easily eliminated into the bile or urine, they also create many active endogenous metabolites and signaling molecules (49, 50). For example, the phase II sulfation of indoxyl creates indoxyl sulfate, which can bind nuclear receptors and modulate kinases as well as functioning as an endogenous toxin (9, 29, 51). Hence, evaluating these DME modifications has the potential to yield additional biological relevance to the analysis. Although considering these molecular properties only slightly improved the classification accuracy over what is described above, the numbers of hydroxyl groups (nof_OH) and sulfates (nof_SO3H) were important for some models; this was consistent with their importance in measures of information gain and in FreeViz analysis (Fig. 7). Other modifications appeared less important based on data visualizations, as well as statistical and information metrics, and were not considered further.
Figure 7.
Importance of phase I and phase II modifications by drug-metabolizing enzymes as molecular properties. Bar graph showing various DME modifications ranked according to information gain (normalized to the DME modification displaying the greatest information gain (nof_OH)). The number of hydroxyls (nof_OH) and number of sulfates (nof_SO3H) are clearly the two highest-ranked attributes contributing to the classification.
Comparison with a “brute force” Random Forest approach to identifying key molecular properties (features) for OAT1 versus OAT3 classification
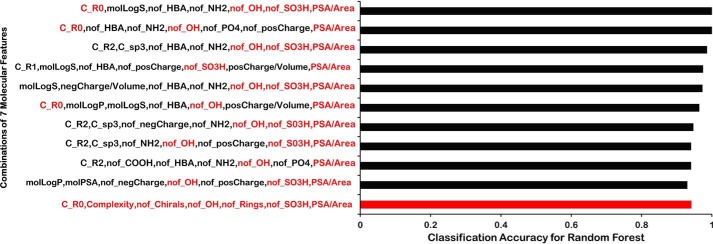
Using the Python machine-learning package SciKit-Learn (Fig. 5) (47), a brute force approach was used to identify seven molecular properties that, as features in the machine-learning analysis for classification by Random Forests, gave the highest classification accuracies (Fig. 8). As with the more “logical approach” using measures of feature importance and other techniques already described, multiple combinations of seven features gave comparably high classification accuracies (Fig. 8). (Using the Decision Tree algorithm in SciKit-Learn, the classification accuracy was generally slightly lower than Random Forests for the same features (data not shown).)
Figure 8.
Brute force random forest analysis. Bar graph shows the CA for a random forest classification using a brute-force approach. The CA is shown for each of the top 10 instances for each combination of seven molecular features in the brute-force approach. The bottom bar (red) shows the CA for the set of logically identified seven molecular features used in the machine-learning approach described in the text. These seven molecular features are also highlighted in each of the 10 sets (red text) to indicate their presence in the various combinations of molecular features showing the greatest classification accuracy.
Based on this brute force analysis, two points are to be emphasized. 1) Nearly all the features chosen using the aforementioned logical approach (nof_Rings, nof_Chirals, PSA/Area, nof_SO3H, nof_OH, C_R0, and Complexity) could be found consistently, with Complexity appearing least often in the brute-force approach. 2) The aforementioned set of logically-chosen features (nof_Rings, nof_Chirals, PSA/Area, nof_SO3H, nof_OH, C_R0, and Complexity) scored nearly as well (within ∼2% classification accuracy) as the highest-scoring random combinations of seven features (Fig. 8). This lends high confidence that the set of seven features finally settled on logically–and that captures multiple molecular properties known to be functionally important for OAT1 versus OAT3 interaction in experimental in vivo and in vitro biological systems and is thus more biologically interpretable–are about as good in terms of classification accuracy as the best-scoring set of seven features when a very large number of combinations are tested.
High-performing OAT1 versus OAT3 Random Forest classification model predicts OAT1- versus OAT3-interacting drugs with high accuracy
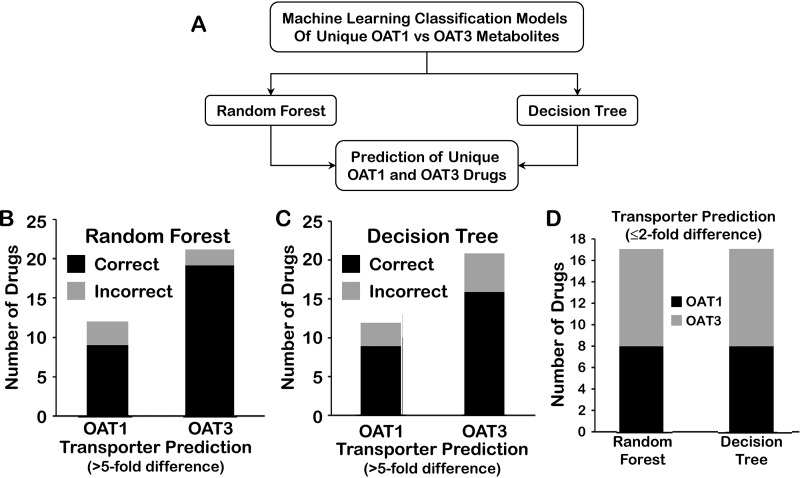
There has been much discussion in the literature about the relationship of small organic molecule drugs and metabolites (9, 32, 52, 53). Thus, we sought to determine the effectiveness of the Random Forest model (using the set of seven molecular properties chosen using the logical approach), which classified unique OAT1 versus OAT3 metabolites with >80% accuracy, for the classification of drugs known to preferentially interact with OAT1 or OAT3 (Fig. 9). We therefore initially analyzed a set of 55 such drugs, 21 of which are known to preferentially interact by >2-fold with OAT1 in cell-based in vitro assays and 34 of which are known to interact with OAT3.
Figure 9.
Prediction of unique OAT1 and OAT3 drugs. A, ability of a strong machine-learning model for classification of unique Oat1 and unique Oat3 metabolites to predict unique Oat1 and unique Oat3 small molecule drugs was analyzed. B–D, seven molecular features that were found to perform well in the classification of metabolites as either OAT1-unique or OAT3-unique were tested for their ability to correctly classify drugs known to interact with either OAT1 or OAT3 with reasonable specificity in random forest and decision tree approaches. Random forest (B) and decision tree (C) classification models were used to predict OAT1 and OAT3 drugs with ≥5-fold (B and C) or ≤2-fold (D) preferential affinities for OAT1 or OAT3. Consideration of drugs with a ≥5-fold preferential affinity for either OAT1 or OAT3 resulted in predictions of the unique metabolite-based random forest or decision tree models with ≥75% accuracy for unique OAT1 and OAT3 drugs, whereas ≤2-fold preferential affinity had around 50% accuracy.
We hypothesized that the Random Forest model, trained using the unique metabolite dataset, would predict OAT1- versus OAT3-interacting drugs significantly better than random. The ability of the (metabolite-based) model to correctly classify OAT1 drugs was only around 55–60%, whereas the ability of the model to correctly classify OAT3 drugs as such was ∼70%.
However, when we examined which drugs were misclassified, we found that they were mainly ones that had a >2-fold but less than 5-fold preferential affinity for either OAT1 or OAT3 (Table S4). When we considered only drugs with a ≥5-fold preferential affinity for either OAT1 or OAT3, the data set was reduced to 33 instances, but importantly, the predictions of the unique metabolite-based Random Forest or Decision Tree models improved considerably; they were 75% accurate for unique OAT1 drugs and >80% accurate for unique OAT3 drugs (Fig. 9).
Discussion
Our results are of both physiological and pharmaceutical interest. OAT1 and OAT3 are well-established as in vitro drug and toxin transporters (5, 42). Some of these in vitro findings have been supported in vivo in the Oat1KO and the Oat3KO mice or in ex vivo assays utilizing knockout tissues. For example, the role of one or both of the OATs has been demonstrated in the handling of diuretics, β-lactam antibiotics, and antivirals, as well as environmental toxins such as organic mercury conjugates and aristolochic acid (23, 54–62). In addition, a growing number of studies have pointed to the physiological roles of OAT1 and OAT3, including urate homeostasis, creatinine, endogenous signaling, and metabolism, uremic toxin, and uremic solute handling, and blood pressure regulation (14, 27, 28, 34, 36, 39–41, 54, 58, 63–69).
It is often held that OAT1 and OAT3 have largely overlapping substrates, and thus the two transporters have been sometimes suggested to be “redundant” when expressed in the same cell type, such as the kidney proximal tubule cell or the choroid plexus cell (5, 9). This idea is mostly based on in vitro assays of binding or transport in cells overexpressing OAT1 and OAT3; furthermore, most of the substrates tested have been drugs (7). However, our extensive metabolomics analysis of the Oat1KO and Oat3KO mice have revealed very different sets of metabolites, i.e. aerobic metabolism intermediates, signaling molecules, vitamins, gut microbiome products, fatty acids, bile acids, and odorants, accumulating in the plasma of the two knockout mice due to the lack of renal OAT-mediated transport from the blood to the urine (28, 34, 36, 39, 41, 43, 63).
In our study of metabolites uniquely accumulating in vivo due to the inability to produce functional OAT1 or OAT3 transporter, differences were initially analyzed by statistical and informational metrics and then using machine-learning tools. A previous study that included a few of the approaches used here (although with machine-learning algorithms not based in the Python SciKit-Learn package) compared the unique FDA-approved drug substrates of OAT1 and OAT3 (33). Consistent with prevailing “textbook” views, this previous study revealed considerable similarity in the molecular features (properties) of pharmaceutical drugs interacting with OAT1 and OAT3, although one molecular property was found to separate OAT1 from OAT3-interacting drugs–a greater propensity for OAT3 to interact with cationic drugs over OAT1, a fact supported by in vitro analysis of certain drugs (33).
However, it is possible that in vitro studies do not fully reflect in vivo reality. It is currently not feasible to analyze the behaviors of hundreds of drugs in the Oat1KO and Oat3KO animals, and in many cases, the behavior of FDA-approved drugs may not reflect the in vivo behaviors of metabolites, signaling molecules, antioxidants, and other known endogenous ligands of OAT1 and OAT3. Moreover, the OAT-transported FDA-approved drug dataset may over-represent certain classes of medically-effective molecules (e.g. NSAIDs) and/or be nonrepresentative of the relevant chemical space because of factors related to drug design and patenting, the approval process, and commercial issues.
In contrast, the use of metabolomics data offers a comparatively-efficient approach to the problem without the aforementioned biases that might accompany drug datasets. Because analysis of the endogenous substrates/ligands should allow one to identify the “true” molecular features driving ligand interaction for OAT1 or OAT3, and metabolites that accumulate in the plasma of mice bearing deletions in either OAT1 or OAT3 likely represent endogenous substrates/ligands of the transporters (unlike molecules in common drug datasets), the utilization and analysis of in vivo metabolomics data appear to be a reasonable approach to characterize the unique molecular properties/features driving in vivo interaction of the metabolites with the two transporters. Nevertheless, to obtain a large number of relatively unique metabolites to perform a machine-learning analysis, we had to combine results from several studies; these targeted and untargeted studies were done at different times, using different platforms or methods, and calls were sometimes made based on different criteria. Although this is not ideal, some of these issues are unavoidable with drug transport data as well, where the pooled data come from multiple labs using somewhat different methods.
Nevertheless, we note that among the Oat1KO and Oat3KO unique metabolites, many molecules of similar structure to these in vivo molecules have been tested in in vitro binding assays or transport assays (7, 42). For example, taurocholate is considered a prototypical in vitro–transported substrate of OAT3.
Machine-learning considerations and potential pharmaceutical relevance of findings
As we have shown, our combined cheminformatics and machine-learning approach gave very good classification accuracy for unique Oat1KO metabolites versus unique Oat3KO metabolites. We were able to ultimately identify a limited set of seven critical molecular properties/features that proved quite useful in classifying in vivo endogenous metabolites as substrates/ligands of OAT1 or OAT3 with high accuracy.
Statistical and informational metrics, as well as data visualizations, allowed us to reduce the >60 molecular properties to <10 molecular properties that seem to be key in determining the transporter specificity of the ligand. For example, it was possible to reduce the number of molecular properties because of high correlations (e.g. r >0.9) with other molecular properties. Our goal was to reduce the set of features to 10% or less of the number of total instances. Thus, from a set of several highly-correlated molecular properties, generally only one was chosen.
In the process of defining the key features, we also found that simple feature engineering—such as the use of PSA/molecular area instead of PSA, often led to improved classification accuracy using the machine-learning tools. Database queries were sometimes helpful for examining the biochemical nature of metabolites that were being captured by one or more molecular properties. For example, a search for molecules with higher numbers of rings yielded primarily Oat3KO metabolites; among these were flavonoids, bile acids, or conjugated sex steroids (Table 1).
The combined data analysis indicated that Oat3KO metabolites had more rings, more chiral centers, and greater complexity than Oat1KO metabolites (Fig. 3; Table 1). Similar analysis of the Oat1KO metabolites supports the view that Oat1 substrates have a higher PSA/area and more carbons outside of ring structures. Furthermore, by evaluating the importance of phase I and phase II DME modifications of metabolites as separate features (e.g. hydroxyl groups and sulfate groups) for machine-learning classification and finding that, at least in certain machine-learning models, one or both of these molecular properties affect classification accuracy, we provide additional biological context. Thus, at least for Oat1 and Oat3, the endogenous substrates should not be considered only from the viewpoint of their interaction with transporters but also in the context of modifications by DMEs.
Decision Tree classification and k-nearest neighbor classification were often nearly as good as Random Forest classification. Moreover, an independent brute-force approach was also used to randomly identify sets of seven molecular properties that could be used to classify the metabolites according to their preferred transporter. The classification accuracies of the best sets (out of >10∧5 analyzed) was similar to that obtained with the logically selected molecular properties as described above. Together, the results support the view that metabolites uniquely accumulating in vivo in the Oat1 and Oat3 knockouts can, indeed, be separated using a limited set of relatively independent molecular properties that seems to capture the diversity of unique metabolites accumulating in the two Oat knockouts in vivo.
We also asked whether the Random Forest classification model predicting unique OAT1 and OAT3 metabolites would be useful for classification of drugs known to preferentially interact with OAT1 and OAT3. There is a body of literature supporting the view that drugs might act like metabolites with similar structural features during transport (9, 32–34, 52, 53). In the case of the OATs, which, to our knowledge, are the drug transporters for which the most in vivo metabolite data are currently available, this appears to be the case, at least for relatively unique drugs known to interact with either OAT1 or OAT3 with a 5-fold or greater difference in affinity measures. For this set of drugs, ≥75% were correctly classified as either OAT1 or OAT3. Nevertheless, the number of drugs that fulfilled this criterion was only 33, or roughly one-third the size of the unique metabolite data set used to generate the Random Forest classification model. Thus, although the ratio of features to instances in the unique metabolite-based model was 1:14, the ratio of features to instances for the drug predictions was 1:5. Thus, it is possible that the predictions are somewhat overdetermined by the seven feature-based metabolite model. That said, with a set of drugs that had a less than 2-fold difference in affinity between OAT1 and OAT3, indicating a similar selectivity for OAT1 and OAT3, the model was unable to classify them, essentially making random calls. These results are what would be expected for a good model. Additional confidence comes from the fact that the Random Forest classification model that was successful in making drug calls was built from unique in vivo metabolites accumulating in knockout mice while the unique OAT1 or unique OAT3 drugs were identified based on in vitro transport assays. Of course, the metabolites are endogenous molecules, whereas the drugs are generally FDA-approved chemical compounds produced by pharmaceutical companies. So, the metabolite and drug datasets are independent from more than one viewpoint.
Identification of the similarities and differences in molecular properties between unique Oat1KO metabolites and Oat3KO metabolites can be helpful, particularly in light of tissue expression patterns, for predicting the likely route of distribution or elimination for a new compound, especially if it is similar in structure to metabolites. Furthermore, phase I and/or phase II DME modifications (i.e. hydroxylation and sulfation) appeared important in determining whether a particular endogenous molecule is handled by OAT1 or OAT3; this kind of information may be helpful in determining the fate of drugs (OAT1 versus OAT3 elimination) that undergo DME modifications. It follows that this type of analysis should help with the design of drugs that, because they have similar properties to particular sets of endogenous metabolites, can be targeted to various tissues through the drug transporters expressed and/or targeting drugs for elimination via a specific transporter.
Physiological relevance
Although more optimal drug design, tissue targeting of drugs, and a better understanding of drug–metabolite interactions are obvious practical applications of our results, the preceding extensive discussion on the machine-learning approach and the pharmaceutical relevance of the results does not take away from the potential physiological importance of our work.
The most significant biological result of our study, in the context of previously published knockout metabolomics and metabolic reconstructions of OAT1- or OAT3-regulated biochemical pathways (9, 42), is the general debunking of the view that OAT1 and OAT3 are redundant when expressed in the same tissue. This is clearly not true from a physiological standpoint and suggests that differential expression of OAT1 and OAT3 in tissues like the pancreas, retina, blood–brain barrier, and also during kidney development and regeneration means different regulation of local and/or systemic metabolism and signaling. Our results connect physiochemical features of endogenous metabolites uniquely changed in vivo in the OAT1 versus OAT3 knockouts to unique metabolic and signaling pathways regulating specific sets of fatty acids, bile acids, amino acids, and peptides.
The data presented here, based on in vivo metabolomics data, provide clear evidence to support the view that while there is overlap in the types of (bio)chemical structures interacting with OAT1 and OAT3, that overlap is not as great as believed previously; OAT1 and OAT3 handle distinct sets of metabolites involved in different biochemical and signaling pathways. These results, taken together with the other aforementioned studies, challenge existing summaries of the roles of OAT1 and OAT3 presented in numerous pharmacology and toxicology textbooks and review articles. We have been able to leverage the fact that hundreds of metabolites have been found to accumulate in the two Oat knockouts, presumably reflecting both unique, as well as common, physiological roles of OAT1 and OAT3. Assuming similar evidence regarding the true endogenous physiological substrates is found for other multispecific ABC and SLC transporters still generally thought to primarily function as transporters of drugs, it could cumulatively call into question the very notion of drug transporters (9).
Experimental procedures
Animals
All experimental protocols were approved by the University of California San Diego Institutional Animal Care and Use Committee (IACUC), and the animals were handled in accordance with the Institutional Guidelines on the Use of Live Animals for Research. Adult (n = 5) WT, Oat1KO male mice were housed separately under a 12-h light/dark cycle and were provided access to food (standard diet) and water ad libitum.
Metabolomic analysis, compound identification, quantification, data curation, and statistics for the Oat1KO
Individual, unpooled serum samples obtained from adult male wildtype (WT; control) and Oat1KO mice were stored at −80 °C and shipped on dry-ice to Metabolon (Durham, NC) for preparation and metabolomic profiling analysis as described previously (28, 63).
In this analysis of the Oat1KO, a total of 731 metabolites of known identity were detected and identified utilizing a reference library of chemical standard entries using software developed at Metabolon, as described previously. Quantification and statistical analyses were performed by Metabolon as described previously (36, 63).
Data collection from previous metabolomics analyses
Considerable targeted and untargeted metabolomics data from the Oat1KO and Oat3KO mice have been previously published by us (28, 34, 36, 39, 54, 63, 70). Although these older previous studies utilized different platforms, different reference libraries, and slightly different protocols (e.g. the Oat3KO metabolomics analyses included data from both male and female knockout mice), for the machine-learning analyses performed here, the available data from previous and the current metabolomics studies were combined to enable relative quantification of over 500 metabolites with known identities (28, 36, 63, 71).
In this case, each metabolite was compared across the various platforms employed, and only those metabolites present on all of the various platforms were considered. From this list of common metabolites, those unique to a particular Oat knockout were initially defined as those displaying significant (p < 0.05) increases in plasma concentration in one or the other of the knockout mice (e.g. significantly increased in the plasma of the Oat1KO, but not in the Oat3KO). In this way, a much larger number of unique metabolites (∼90) were present in the Oat1KO at a significance of p < 0.05, whereas a lower number were uniquely present in the Oat3KO at this level of significance. Therefore, to augment the number of unique Oat3KO-interacting metabolites, those compounds displaying accumulation in the serum of the Oat3KO (compared with the WT), but not the Oat1KO, with a significance of 0.05 > p < 0.1 (trending toward significance) or a particularly high fold-change with the inability to meet significance criteria apparently due to an anomalous value were included; this resulted in a set of 48 unique OAT3-interacting metabolites.
Although there were an unequal number of these unique metabolites between the two knockouts, the numbers of unique Oat1KO metabolites and Oat3KO metabolites were kept balanced in training sets for machine-learning application. The Data Sampler widget in Orange (44) was used for randomly sampling 48 metabolites from the 90 total Oat1KO metabolites; thus, the total number of metabolites used in the machine-learning classification analyses was 96 (48 unique Oat1KO and 48 unique Oat3KO).
Generation of molecular properties
A set of 67 structural and physiochemical properties of identified OAT1 and OAT3 metabolites were calculated with tools found in the computational environment of the commercially available computational chemistry software, ICM (version 3.8–6) (Molsoft LLC, San Diego, CA). Complexity values were obtained from PubChem, which describes “complexity” as follows. “The complexity rating of a compound is a rough estimate of how complicated a structure is, seen from both the point of view of the elements contained and the displayed structural features, including symmetry. This complexity rating is computed using the Bertz/Hendrickson/Ihlenfeldt formula (58).”
Elimination of highly-correlated molecular properties to reduce a number of features for machine-learning analyses
An initial evaluation of the molecular properties revealed highly-correlated descriptors. For example, a number of molecular properties, such as molWeight, molVolume, molArea, and number of atoms, are highly correlated with each other, essentially providing different but related measures of correlated properties. Pairwise correlation coefficients for the molecular features were determined, and a number of highly cross-correlated features were eliminated. In this way, the list of more than 60 molecular properties was reduced to a smaller number (∼25) for the size of the number of molecules in the training set and from the viewpoint of visualization and data analysis.
Machine learning
Orange, versions 3.13 and 3.16, was used. The Orange software package uses Python libraries, including Scikit-Learn, numpy, and scipy (45). A series of analyses, including PCA, ranking based on information gain, FreeViz (44), outlier analysis, and machine-learning analyses, i.e. k-means classification, decision trees, random forests, neural networks, Naive Bayes, and logistic regression, were performed. Other, mostly confirmatory and complementary, machine-learning analyses was performed using the Python-based package SciKit-Learn (47). Default parameters were generally kept.
Data visualization and SQL queries
Data visualizations were mostly done in Orange (distributions, scatterplots, and FreeViz). Additional data visualizations were done in Python using a Jupyter notebook with the pandas, matplolib, and seaborn libraries imported. For SQL queries, Excel or csv files were imported into SQL Studio for data analysis, or the queries were coded in Python using SQLAlchemy.
Identification of optimal seven properties with highest random forest classification accuracies by a systematic enumeration of property combinations
First, the set of 138 metabolites was divided into a training set of 48 each (OAT1, OAT3) and resampled test set of 42 each. All possible combinations of features were run, and the top 2500 AUC were recorded. Then, we took each set in this 2500 and used k-fold cross-validation as our metric for accuracy from the SciKit-Learn library. We could not simply run k-fold on the entire set of combinations due to computational limitations. To find the ten best sets of seven features, the classification accuracy along with its standard deviation was found by re-sampling k-fold-cross-validation 200 times to address variance from the random sampling. (By testing over 50 to 2000 iterations, the standard deviation remained at about 1.5%, thus providing confidence that these were consistent scores.) The highest average Random Forest scores were then chosen as the best feature sets.
Brute-force approach with SciKit-learn Random Forest classifier
Of the many numeric molecular properties previously described (Table S2), 25 properties were selected by selecting single representatives from several highly correlated properties. These 25 were then used as the “pool” of features from which sets of seven features were randomly chosen for a brute force analysis. In this way, all possible combinations of seven features from the list of 25 attributes (out of 480,700 possible combinations) were tested using the Random Forest classification algorithm in SciKit-Learn, and the classification accuracies were determined.
Predictions of unique OAT1 and unique OAT3 drugs by the unique OAT1 versus unique in vivo metabolite-based classification model
The metabolite-based Random Forest classifier model built, using the seven features described in the text that were able to correctly classify unique OAT1 versus unique OAT3 metabolites >80% of the time, was used to test whether unique OAT1 and unique OAT3 drugs could be predicted. The model was applied using the Predictions widget in Orange. As described under “Results,” the final drug dataset used consisted of drugs known to have ≥5-fold differences in affinity for OAT1 versus OAT3 when analyzed in vitro by cell-based assays. As a control, we used a drug dataset where there was no difference or minimal (<2-fold) difference in affinity between OAT1 and OAT3.
Author contributions
R. A. and S. K. N. conceptualization; G. L., D. S., and K. T. B. data curation; V. B., R. A., and S. K. N. supervision. A. K. N., K. L., and S. K. N. investigation; A. K. N., and S. K. N. writing-original draft; A. K. N., J. G. L., D. S., K. T. B., V. B., R. A., and S. K. N. writing-review and editing.
Supplementary Material
This work was supported by NIDDK Grant DK109392 and Eunice Kennedy Shriver NICHD Grant HD090259 (to S. K. N.). R. A. is a founder of Molsoft LLC. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1–S4.
- OAT
- organic anion transporter
- OA
- organic anion
- Oat1KO
- Oat1 knockout
- Oat3KO
- Oat3 knockout
- nof_Rings
- number of rings
- nof_Chirals
- number of chirals
- PSA
- polar surface area
- molArea
- molecular area
- molVol
- molecular volume
- nof_hydroxyls
- number of hydroxyls
- nof_SO3H
- number of sulfates
- CA
- classification accuracy
- ABC
- ATP-binding cassette
- NSAID
- nonsteroidal anti-inflammatory drug
- DME
- drug-metabolizing enzyme
- FDA
- Food and Drug Administration
- AUC
- area under the curve.
References
- 1. Lopez-Nieto C. E., You G., Bush K. T., Barros E. J., Beier D. R., and Nigam S. K. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J. Biol. Chem. 272, 6471–6478 10.1074/jbc.272.10.6471 [DOI] [PubMed] [Google Scholar]
- 2. Brady K. P., Dushkin H., Förnzler D., Koike T., Magner F., Her H., Gullans S., Segre G. V., Green R. M., and Beier D. R. (1999) A novel putative transporter map to the osteosclerosis (oc) mutation and is not expressed in the oc mutant mouse. Genomics 56, 254–261 10.1006/geno.1998.5722 [DOI] [PubMed] [Google Scholar]
- 3. Emami Riedmaier A., Nies A. T., Schaeffeler E., and Schwab M. (2012) Organic anion transporters and their implications in pharmacotherapy. Pharmacol. Rev. 64, 421–449 10.1124/pr.111.004614 [DOI] [PubMed] [Google Scholar]
- 4. Morrissey K. M., Stocker S. L., Wittwer M. B., Xu L., and Giacomini K. M. (2013) Renal transporters in drug development. Annu. Rev. Pharmacol. Toxicol. 53, 503–529 10.1146/annurev-pharmtox-011112-140317 [DOI] [PubMed] [Google Scholar]
- 5. Nigam S. K. (2018) The SLC22 transporter family: a paradigm for the impact of drug transporters on metabolic pathways, signaling, and disease. Annu. Rev. Pharmacol. Toxicol. 58, 663–687 10.1146/annurev-pharmtox-010617-052713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roth M., Obaidat A., and Hagenbuch B. (2012) OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. Br. J. Pharmacol. 165, 1260–1287 10.1111/j.1476-5381.2011.01724.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. VanWert A. L., Gionfriddo M. R., and Sweet D. H. (2010) Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm. Drug Dispos. 31, 1–71 10.1002/bdd.693 [DOI] [PubMed] [Google Scholar]
- 8. Saito H. (2010) Pathophysiological regulation of renal SLC22A organic ion transporters in acute kidney injury: pharmacological and toxicological implications. Pharmacol. Ther. 125, 79–91 10.1016/j.pharmthera.2009.09.008 [DOI] [PubMed] [Google Scholar]
- 9. Nigam S. K. (2015) What do drug transporters really do? Nat. Rev. Drug Discov. 14, 29–44 10.1038/nrd4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu C., Nigam K. B., Date R. C., Bush K. T., Springer S. A., Saier M. H. Jr., Wu W., and Nigam S. K. (2015) Evolutionary analysis and classification of OATs, OCTs, OCTNs, and other SLC22 transporters: structure-function implications and analysis of sequence motifs. PLoS ONE 10, e0140569 10.1371/journal.pone.0140569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. You G. F., and Morris M. E. (eds) (2014) Drug Transporters: Molecular Characterization and Role in Drug Disposition, John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 12. Rosenthal S. B., Bush K. T., and Nigam S. K. (2019) A network of SLC and ABC transporter and DME genes involved in remote sensing and signaling in the gut-liver-kidney axis. Sci. Rep. 9, 11879 10.1038/s41598-019-47798-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaler G., Truong D. M., Khandelwal A., Nagle M., Eraly S. A., Swaan P. W., and Nigam S. K. (2007) Structural variation governs substrate specificity for organic anion transporter (OAT) homologs. Potential remote sensing by OAT family members. J. Biol. Chem. 282, 23841–23853 10.1074/jbc.M703467200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lepist E. I., and Ray A. S. (2017) Beyond drug–drug interactions: effects of transporter inhibition on endobiotics, nutrients and toxins. Expert Opin. Drug Metab. Toxicol. 13, 1075–1087 10.1080/17425255.2017.1372425 [DOI] [PubMed] [Google Scholar]
- 15. Nigam S. K., Wu W., Bush K. T., Hoenig M. P., Blantz R. C., and Bhatnagar V. (2015) Handling of drugs, metabolites, and uremic toxins by kidney proximal tubule drug transporters. Clin. J. Am. Soc. Nephrol. 10, 2039–2049 10.2215/CJN.02440314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ullrich K. J. (1997) Renal transporters for organic anions and organic cations. Structural requirements for substrates. J. Membr. Biol. 158, 95–107 10.1007/s002329900247 [DOI] [PubMed] [Google Scholar]
- 17. Ahn S. Y., and Bhatnagar V. (2008) Update on the molecular physiology of organic anion transporters. Curr. Opin. Nephrol. Hypertens. 17, 499–505 10.1097/MNH.0b013e32830b5d5d [DOI] [PubMed] [Google Scholar]
- 18. Koepsell H. (2013) The SLC22 family with transporters of organic cations, anions and zwitterions. Mol. Aspects Med. 34, 413–435 10.1016/j.mam.2012.10.010 [DOI] [PubMed] [Google Scholar]
- 19. Robertson E. E., and Rankin G. O. (2006) Human renal organic anion transporters: characteristics and contributions to drug and drug metabolite excretion. Pharmacol. Ther. 109, 399–412 10.1016/j.pharmthera.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 20. Ivanyuk A., Livio F., Biollaz J., and Buclin T. (2017) Renal drug transporters and drug interactions. Clin. Pharmacokinet. 56, 825–892 10.1007/s40262-017-0506-8 [DOI] [PubMed] [Google Scholar]
- 21. Masereeuw R., and Russel F. G. (2010) Therapeutic implications of renal anionic drug transporters. Pharmacol. Ther. 126, 200–216 10.1016/j.pharmthera.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 22. Rizwan A. N., and Burckhardt G. (2007) Organic anion transporters of the SLC22 family: biopharmaceutical, physiological, and pathological roles. Pharm. Res. 24, 450–470 10.1007/s11095-006-9181-4 [DOI] [PubMed] [Google Scholar]
- 23. Prentice K. J., Luu L., Allister E. M., Liu Y., Jun L. S., Sloop K. W., Hardy A. B., Wei L., Jia W., Fantus I. G., Sweet D. H., Sweeney G., Retnakaran R., Dai F. F., and Wheeler M. B. (2014) The furan fatty acid metabolite CMPF is elevated in diabetes and induces beta cell dysfunction. Cell Metab. 19, 653–666 10.1016/j.cmet.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 24. Wang X., Han L., Li G., Peng W., Gao X., Klaassen C. D., Fan G., and Zhang Y. (2018) From the cover: identification of natural products as inhibitors of human organic anion transporters (OAT1 and OAT3) and their protective effect on mercury-induced toxicity. Toxicol. Sci. 161, 321–334 10.1093/toxsci/kfx216 [DOI] [PubMed] [Google Scholar]
- 25. An G., Wang X., and Morris M. E. (2014) Flavonoids are inhibitors of human organic anion transporter 1 (OAT1)-mediated transport. Drug Metab. Dispos. 42, 1357–1366 10.1124/dmd.114.059337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahn S. Y., and Nigam S. K. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol. Pharmacol. 76, 481–490 10.1124/mol.109.056564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhatnagar V., Richard E. L., Wu W., Nievergelt C. M., Lipkowitz M. S., Jeff J., Maihofer A. X., and Nigam S. K. (2016) Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin. Kidney J 9, 444–453 10.1093/ckj/sfw010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bush K. T., Wu W., Lun C., and Nigam S. K. (2017) The drug transporter OAT3 (SLC22A8) and endogenous metabolite communication via the gut–liver–kidney axis. J. Biol. Chem. 292, 15789–15803 10.1074/jbc.M117.796516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nigam S. K., and Bush K. T. (2019) Uraemic syndrome of chronic kidney disease: altered remote sensing and signalling. Nat. Rev. Nephrol. 15, 301–316 10.1038/s41581-019-0111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu W., Dnyanmote A. V., and Nigam S. K. (2011) Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol. Pharmacol. 79, 795–805 10.1124/mol.110.070607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martinez D., Muhrez K., Woillard J. B., Berthelot A., Gyan E., Choquet S., Andrès C. R., Marquet P., and Barin-Le Guellec C. (2018) Endogenous metabolite-mediated communication between OAT1/OAT3 and OATP1B1 may explain the association between SLCO1B1 SNPs and methotrexate toxicity. Clin. Pharmacol. Ther. 104, 687–698 10.1002/cpt.1008 [DOI] [PubMed] [Google Scholar]
- 32. Kouznetsova V. L., Tsigelny I. F., Nagle M. A., and Nigam S. K. (2011) Elucidation of common pharmacophores from analysis of targeted metabolites transported by the multispecific drug transporter-organic anion transporter1 (Oat1). Bioorg. Med. Chem. 19, 3320–3340 10.1016/j.bmc.2011.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu H. C., Goldenberg A., Chen Y., Lun C., Wu W., Bush K. T., Balac N., Rodriguez P., Abagyan R., and Nigam S. K. (2016) Molecular properties of drugs interacting with SLC22 transporters OAT1, OAT3, OCT1, and OCT2: a machine-learning approach. J. Pharmacol. Exp. Ther. 359, 215–229 10.1124/jpet.116.232660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wikoff W. R., Nagle M. A., Kouznetsova V. L., Tsigelny I. F., and Nigam S. K. (2011) Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J. Proteome Res. 10, 2842–2851 10.1021/pr200093w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu W., Bush K. T., Liu H. C., Zhu C., Abagyan R., and Nigam S. K. (2015) Shared ligands between organic anion transporters (OAT1 and OAT6) and odorant receptors. Drug Metab. Dispos. 43, 1855–1863 10.1124/dmd.115.065250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu W., Jamshidi N., Eraly S. A., Liu H. C., Bush K. T., Palsson B. O., and Nigam S. K. (2013) Multispecific drug transporter Slc22a8 (Oat3) regulates multiple metabolic and signaling pathways. Drug Metab. Dispos. 41, 1825–1834 10.1124/dmd.113.052647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahn S. Y., Eraly S. A., Tsigelny I., and Nigam S. K. (2009) Interaction of organic cations with organic anion transporters. J. Biol. Chem. 284, 31422–31430 10.1074/jbc.M109.024489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsson P., and Bergström C. A. (2015) Computational modeling to predict the functions and impact of drug transporters. In Silico Pharmacol. 3, 8 10.1186/s40203-015-0012-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eraly S. A., Vallon V., Vaughn D. A., Gangoiti J. A., Richter K., Nagle M., Monte J. C., Rieg T., Truong D. M., Long J. M., Barshop B. A., Kaler G., and Nigam S. K. (2006) Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem. 281, 5072–5083 10.1074/jbc.M508050200 [DOI] [PubMed] [Google Scholar]
- 40. Ahn S. Y., Jamshidi N., Mo M. L., Wu W., Eraly S. A., Dnyanmote A., Bush K. T., Gallegos T. F., Sweet D. H., Palsson B. Ø., and Nigam S. K. (2011) Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J. Biol. Chem. 286, 31522–31531 10.1074/jbc.M111.272534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu H. C., Jamshidi N., Chen Y., Eraly S. A., Cho S. Y., Bhatnagar V., Wu W., Bush K. T., Abagyan R., Palsson B. O., and Nigam S. K. (2016) An organic anion transporter 1 (OAT1)-centered metabolic network. J. Biol. Chem. 291, 19474–19486 10.1074/jbc.M116.745216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nigam S. K., Bush K. T., Martovetsky G., Ahn S. Y., Liu H. C., Richard E., Bhatnagar V., and Wu W. (2015) The organic anion transporter (OAT) family: a systems biology perspective. Physiol. Rev. 95, 83–123 10.1152/physrev.00025.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sweet D. H., Miller D. S., Pritchard J. B., Fujiwara Y., Beier D. R., and Nigam S. K. (2002) Impaired organic anion transport in kidney and choroid plexus of organic anion transporter 3 (Oat3 [Slc22a8]) knockout mice. J. Biol. Chem. 277, 26934–26943 10.1074/jbc.M203803200 [DOI] [PubMed] [Google Scholar]
- 44. Demsar J., Leban G., and Zupan B. (2007) FreeViz–an intelligent multivariate visualization approach to explorative analysis of biomedical data. J. Biomed. Inform. 40, 661–671 10.1016/j.jbi.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 45. Demsar J., Curk T., Erjavec A., Gorup C., Hocevar T., Milutinovic M., Mozina M., Polajnar M., Toplak M., Staric A., Stajdohar M., Umek L., Zagar L., Zbontar J., Zitnik M., and Zupan B. (2013) Orange: data mining toolbox in python. J. Mach. Learn. Res. 14, 2349–2353 [Google Scholar]
- 46. Mierswa I., Wurst M., Klinkenberg R., Scholz M., and Euler T. (2006) in Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Philadelphia, PA, August 20–23, 2006 (Ungar L., Craven M., Gunopulos D., and Eliassi-Rad T.) pp. 935–940, Association for Computing Machinery, New York [Google Scholar]
- 47. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., VanderPlas J., Passos A., Cournapeau D., Brucher M., Perrot M., and Duchesnay E. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 [Google Scholar]
- 48. Ritthoff O., Klinkenberg R., Fisher S., Mierswa I., and Felske S. (2001) YALE: Yet Another Learning Environment. in LLWA'01–Tagungsband der GI-WorkshopWoche Lernen–Lehren–Wissen Adaptivitat, University of Dortmund, Dortmund, Germany [Google Scholar]
- 49. Almazroo O. A., Miah M. K., and Venkataramanan R. (2017) Drug metabolism in the liver. Clin. Liver Dis. 21, 1–20 10.1016/j.cld.2016.08.001 [DOI] [PubMed] [Google Scholar]
- 50. Stanley L. A. (2017) in Pharmacognosy (Delgoda R., ed) pp. 527–545, Academic Press, Boston, MA [Google Scholar]
- 51. Jansen J., Jansen K., Neven E., Poesen R., Othman A., van Mil A., Sluijter J., Sastre Torano J., Zaal E. A., Berkers C. R., Esser D., Wichers H. J., van Ede K., van Duursen M., Burtey S., et al. (2019) Remote sensing and signaling in kidney proximal tubules stimulates gut microbiome-derived organic anion secretion. Proc. Natl. Acad. Sci. U.S.A. 116, 16105–16110 10.1073/pnas.1821809116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Hagan S., and Kell D. B. (2015) Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front. Pharmacol. 6, 105 10.3389/fphar.2015.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. O'Hagan S., and Kell D. B. (2017) Analysis of drug-endogenous human metabolite similarities in terms of their maximum common substructures. J. Cheminform 9, 18 10.1186/s13321-017-0198-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eraly S. A., Vallon V., Rieg T., Gangoiti J. A., Wikoff W. R., Siuzdak G., Barshop B. A., and Nigam S. K. (2008) Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol. Genomics 33, 180–192 10.1152/physiolgenomics.00207.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nagle M. A., Wu W., Eraly S. A., and Nigam S. K. (2013) Organic anion transport pathways in antiviral handling in choroid plexus in Oat1 (Slc22a6) and Oat3 (Slc22a8) deficient tissue. Neurosci. Lett. 534, 133–138 10.1016/j.neulet.2012.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Torres A. M., Dnyanmote A. V., Bush K. T., Wu W., and Nigam S. K. (2011) Deletion of multispecific organic anion transporter Oat1/Slc22a6 protects against mercury-induced kidney injury. J. Biol. Chem. 286, 26391–26395 10.1074/jbc.M111.249292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Truong D. M., Kaler G., Khandelwal A., Swaan P. W., and Nigam S. K. (2008) Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J. Biol. Chem. 283, 8654–8663 10.1074/jbc.M708615200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vallon V., Eraly S. A., Rao S. R., Gerasimova M., Rose M., Nagle M., Anzai N., Smith T., Sharma K., Nigam S. K., and Rieg T. (2012) A role for the organic anion transporter OAT3 in renal creatinine secretion in mice. Am. J. Physiol. Renal Physiol. 302, F1293–F1299 10.1152/ajprenal.00013.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vallon V., Rieg T., Ahn S. Y., Wu W., Eraly S. A., and Nigam S. K. (2008) Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am. J. Physiol. Renal Physiol. 294, F867–F873 10.1152/ajprenal.00528.2007 [DOI] [PubMed] [Google Scholar]
- 60. Vanwert A. L., Bailey R. M., and Sweet D. H. (2007) Organic anion transporter 3 (Oat3/Slc22a8) knockout mice exhibit altered clearance and distribution of penicillin G. Am. J. Physiol. Renal Physiol. 293, F1332–F1341 10.1152/ajprenal.00319.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vanwert A. L., Srimaroeng C., and Sweet D. H. (2008) Organic anion transporter 3 (oat3/slc22a8) interacts with carboxyfluoroquinolones, and deletion increases systemic exposure to ciprofloxacin. Mol. Pharmacol. 74, 122–131 10.1124/mol.107.042853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. VanWert A. L., and Sweet D. H. (2008) Impaired clearance of methotrexate in organic anion transporter 3 (Slc22a8) knockout mice: a gender specific impact of reduced folates. Pharm. Res. 25, 453–462 10.1007/s11095-007-9407-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu W., Bush K. T., and Nigam S. K. (2017) Key role for the organic anion transporters, OAT1 and OAT3, in the in vivo handling of uremic toxins and solutes. Sci. Rep. 7, 4939 10.1038/s41598-017-04949-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Masereeuw R., Mutsaers H. A., Toyohara T., Abe T., Jhawar S., Sweet D. H., and Lowenstein J. (2014) The kidney and uremic toxin removal: glomerulus or tubule? Semin. Nephrol. 34, 191–208 10.1016/j.semnephrol.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 65. Vriend J., Hoogstraten C. A., Venrooij K. R., van den Berge B. T., Govers L. P., van Rooij A., Huigen M. C. D. G., Schirris T. J. J., Russel F. G. M., Masereeuw R., and Wilmer M. J. (2019) Organic anion transporters 1 and 3 influence cellular energy metabolism in renal proximal tubule cells. Biol. Chem. 400, 1347–1358 10.1515/hsz-2018-0446 [DOI] [PubMed] [Google Scholar]
- 66. Dragojević J., Mihaljević I., Popović M., and Smital T. (2019) Zebrafish (Danio rerio) Oat1 and Oat3 transporters and their interaction with physiological compounds. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 236, 110309 10.1016/j.cbpb.2019.110309 [DOI] [PubMed] [Google Scholar]
- 67. Murray M., and Zhou F. (2017) Trafficking and other regulatory mechanisms for organic anion transporting polypeptides and organic anion transporters that modulate cellular drug and xenobiotic influx and that are dysregulated in disease. Br. J. Pharmacol. 174, 1908–1924 10.1111/bph.13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lozano E., Briz O., Macias R. I. R., Serrano M. A., Marin J. J. G., and Herraez E. (2018) Genetic heterogeneity of SLC22 family of transporters in drug disposition. J. Pers. Med. 8, E14 10.3390/jpm8020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nagle M. A., Truong D. M., Dnyanmote A. V., Ahn S. Y., Eraly S. A., Wu W., and Nigam S. K. (2011) Analysis of three-dimensional systems for developing and mature kidneys clarifies the role of OAT1 and OAT3 in antiviral handling. J. Biol. Chem. 286, 243–251 10.1074/jbc.M110.139949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vallon V., Eraly S. A., Wikoff W. R., Rieg T., Kaler G., Truong D. M., Ahn S. Y., Mahapatra N. R., Mahata S. K., Gangoiti J. A., Wu W., Barshop B. A., Siuzdak G., and Nigam S. K. (2008) Organic anion transporter 3 contributes to the regulation of blood pressure. J. Am. Soc. Nephrol. 19, 1732–1740 10.1681/ASN.2008020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Evans A. M., DeHaven C. D., Barrett T., Mitchell M., and Milgram E. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81, 6656–6667 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.