Abstract
The widely expressed bromodomain and extraterminal motif (BET) proteins bromodomain-containing protein 2 (BRD2), BRD3, and BRD4 are multifunctional transcriptional regulators that bind acetylated chromatin via their conserved tandem bromodomains. Small molecules that target BET bromodomains are being tested for various diseases but typically do not discern between BET family members. Genomic distributions and protein partners of BET proteins have been described, but the basis for differences in BET protein function within a given lineage remains unclear. By establishing a gene knockout-rescue system in a Brd2-null erythroblast cell line, here we compared a series of mutant and chimeric BET proteins for their ability to modulate cell growth, differentiation, and gene expression. We found that the BET N-terminal halves bearing the bromodomains convey marked differences in protein stability but do not account for specificity in BET protein function. Instead, when BET proteins were expressed at comparable levels, their specificity was largely determined by the C-terminal half. Remarkably, a chimeric BET protein comprising the N-terminal half of the structurally similar short BRD4 isoform (BRD4S) and the C-terminal half of BRD2 functioned similarly to intact BRD2. We traced part of the BRD2-specific activity to a previously uncharacterized short segment predicted to harbor a coiled-coil (CC) domain. Deleting the CC segment impaired BRD2's ability to restore growth and differentiation, and the CC region functioned in conjunction with the adjacent ET domain to impart BRD2-like activity onto BRD4S. In summary, our results identify distinct BET protein domains that regulate protein turnover and biological activities.
Keywords: epigenetics, transcription regulation, chromatin, protein structure, bromodomain-containing protein 4 (BRD4), bromodomain and extraterminal motif (BET) proteins, bromodomain-containing protein 2 (BRD2), bromodomain-containing protein 3 (BRD3), RNA polymerase II-associated factor (PAF)
Introduction
Bromodomain and extraterminal motif (BET)3 proteins mechanistically link chromatin acetylation to gene transcription (1, 2). The BET family consists of BRD2, BRD3, and BRD4, which are widely expressed across tissue types, and the testis-specific BRDT (2). BET proteins share a general domain structure consisting of tandem N-terminal bromodomains and a C-terminal ET domain conserved across species (3). The bromodomains bind to acetylated lysine residues on histones and transcription factors (1). The ET domain is a protein-protein interaction motif that recruits additional co-activators (4–9). BET proteins thereby function as molecular scaffolds for transcriptional regulators. Small molecules that target the bromodomains partially displace BET proteins along with associated co-factors from chromatin (10, 11). BET inhibitors are in clinical trials for the treatment of several cancers and may have additional use in inflammatory, metabolic, and cardiac disorders (2, 12). Because the bromodomains are similar between BET proteins, molecules targeting them do not strongly discriminate between the individual family members (10, 11, 13). The lack of selectivity might limit their use and underscores the need for more targeted BET inhibitors. Moreover, clinical exploitation of BET inhibition requires a better understanding of selective BET protein functions.
Structurally, the long isoform of BRD4 (BRD4L) is the most distinct BET protein in that it possesses a unique extended C-terminal tail that recruits factors required for the transition of RNA polymerase II from transcriptional initiation into productive elongation (14–16). The short isoform of BRD4 (BRD4S) lacks this domain and is thus structurally more similar to BRD2 and BRD3, but its expression and function in most cell types remains poorly characterized (17, 18). Depletion of BRD2 or BRD3 results in transcriptional changes that are distinct from those caused by BRD4 depletion (19, 20), suggesting nonoverlapping functions among BET proteins. Additionally, BRD2- and BRD4-deficient mice exhibit different phenotypes (21–24). BRD3-null mice have not been reported. It has been proposed that BET-specific functions are conveyed in part via selectivity of the bromodomains for acetylated substrates (25–30). Specificity might also be mediated by additional domains that influence chromatin occupancy (31, 32) or by differences in protein interactions (9, 33). However, the ET protein-interaction domain is highly conserved across BET proteins, making its contribution to BET-selective interactions unclear (4–8). Thus, whereas all BET proteins are enriched at acetylated chromatin, the mechanisms underlying BET-specific functions remain unresolved.
GATA1-mediated erythroid cell differentiation has provided a useful model for the dissection of BET protein biology (34). GATA1 is a master erythroid transcription factor that drives the terminal maturation of erythroid precursor cells (35). BRD3 and to a lesser extent BRD4 bind to GATA1 in an acetylation-dependent manner and co-localize with GATA1 on chromatin (36, 37). Point mutations in the BET-binding motif of GATA1, or pan-BET inhibition, impair GATA1 occupancy on chromatin (36–38). GATA1 transcriptional activity has been extensively studied in G1E-ER4 cells, an erythroid progenitor line that stably expresses a conditional form of GATA1 fused to the ligand binding domain of the estrogen receptor (GATA1-ER) (39). Using this system, GATA1-ER target genes can be activated upon treatment with estradiol, which faithfully recapitulates terminal erythroid maturation (39). BRD2 and BRD4 are both necessary for GATA1-ER–mediated gene expression (36). In contrast, BRD3 appears to be dispensable, but BRD3 loss exacerbates defects associated with BRD2 deficiency, suggesting overlapping functions of these two proteins (36). Accordingly, overexpression of BRD3 can partially compensate for BRD2 loss (36). BRD3 is expressed ∼4-fold less than BRD2 based on RNA-Seq in G1E-ER4 cells, potentially explaining its lack of compensatory activity when not overexpressed (40). It is unclear whether BRD2 and BRD3 possess structural features that could facilitate functions that are distinct from BRD4.
To better understand functional differences between BRD2, BRD3, and BRD4, we took advantage of Brd2-null G1E-ER4 erythroblasts. These cells are viable but fail to undergo GATA1-mediated terminal maturation. We asked whether reconstitution of these cells with exogenous BET proteins rescues their growth rate, maturation, and gene expression. We found that the BRD4 isoforms are functionally different from BRD2 and BRD3. Moreover, the bromodomains contribute surprisingly little in terms of BET-specific functions. We define a small region adjacent to the ET domain that contains a predicted coiled-coil structure in BRD2 and BRD3 but not BRD4 that we show to contribute to BRD2-selective activity and to bind in vitro to the RNA polymerase II-associated factor (PAF) and casein kinase II (CK2) complexes.
Results
BRD2 is essential for GATA1-dependent erythroid differentiation and gene expression
A mechanistic explanation for specific functions of individual BET proteins remains unresolved. We therefore took advantage of a cellular system in which BET proteins can be directly compared. We generated four Brd2-null G1E-ER4 cell lines (BRD2 KO1–KO4) using CRISPR-Cas9 (see “Experimental procedures”). The absence of BRD2 was confirmed by Western blotting with antibodies against BRD2 N- and C-terminal epitopes (Fig. S1A). We measured BRD3 and BRD4L protein levels and found them to be unchanged in the BRD2 KO cells (Fig. S1A), indicating that BRD2 is not necessary for BRD3 and BRD4L expression and that any defects in the BRD2 KO cells cannot be attributed to their loss. We were unable to detect endogenous BRD4S in whole-cell lysates using commercially available antibodies (Fig. S1B). To assess BRD4S expression by different means, we extracted RNA-Seq reads mapping to a BRD4S-specific exon-exon junction. This revealed that BRD4S is the minor isoform, with approximately ∼33% of BRD4 transcripts corresponding to it in parental G1E-ER4 cells, ∼24% in BRD2 KO1 cells, and ∼19% in KO2 cells. It is unclear why this isoform cannot be detected at the protein level, but it may be due to insufficient sensitivity of the antibody.
To test whether GATA1-driven erythroid maturation depends on BRD2, we exposed BRD2 KO clonal lines to estradiol for 24 h, a time point at which GATA1-ER target genes are fully expressed (39). Consistent with our previous studies (36), all BRD2 KO cells failed to induce the expression of major erythroid GATA1 target genes, including Hbb-b1, Alas2, Slc4a1, and Spta1 (Fig. S1C). Importantly, BRD2 depletion did not impact RNA levels of the Gata1-ER transgene or housekeeping genes Actb and Gapdh. Consistent with the Western blots, Brd3 and Brd4 RNA levels were unchanged in BRD2 KO cells.
Identification of a BRD2-specific transcriptional signature
To measure the global transcriptional changes associated with BRD2 loss, we performed RNA-Seq on parental G1E-ER4 and two BRD2 KO clones (KO1 and KO2) after 24 h of GATA1-ER induction. Pearson correlation coefficients demonstrated strong concordance among the three replicates for each cell line and higher similarity of KO1 and KO2 (Fig. S2A). Dimensionality reduction using principle component analysis showed separation of the two KO clones from the parental G1E-ER4 cells on PC1 (37.3% of variance), revealing the transcriptional impact of BRD2 depletion. Separation of the three cell lines on PC2 (18.9% of variance) suggested clonal variation between the cell lines (Fig. S2B). We therefore classified differentially expressed genes as those that exhibited a ≥2-fold change (false discovery rate (FDR) < 0.01) in both KO clones compared with parental G1E-ER4 cells and that were unchanged between the two KO clones (Table S1 and Fig. S2C). This identified 158 down-regulated genes and 261 up-regulated genes. A less stringent cutoff of ≥1.5-fold change (FDR < 0.05) resulted in 433 and 626 genes that were down- and up-regulated, respectively (Fig. S2C). Expression of several highly expressed maturation-associated genes (Hbb-b1, Hbb-b2, Hba-a1, Hba-a2, Alas2, Slc4a1, and Spta1) was decreased in the BRD2 KO cells compared with the parental cells, whereas many genes whose expression is high in immature cells and down-regulated during erythroid maturation (Gata2, Myc, Myb, Kit, Il2rg, Lyl1, and Vim) were elevated in at least one of the BRD2 KO cell lines (Fig. S2D). This indicates a general failure of BRD2 KO cells to fully mature upon GATA1-ER activation.
To determine what biological pathways could be affected by the observed transcriptional changes, we queried the differentially expressed genes for enrichment of hallmark gene sets in the Molecular Signatures Database. The gene sets most associated with the down-regulated genes were HEME_METABOLISM, MTORC1_SIGNALING, and CHOLESTEROL_HOMEOSTASIS (Fig. S2E). The gene sets most associated with the up-regulated genes include IL2_STAT5_SIGNALING, TNFA_SIGNALING_VIA_NFKB, and P53_PATHWAY. The relevance of these pathways is not immediately clear, but they could reflect a failure to silence genes important for cell proliferation. An analysis using the less stringent list of differentially expressed genes (-fold change ≥1.5, FDR <0.05) yielded similar gene set enrichments (not shown).
The pharmacologic BET inhibitor JQ1 targets all BET proteins with comparable efficacy (10, 11, 13). We assessed to what extent the cellular response to JQ1 can be attributed to BRD2 inhibition by comparing the transcriptome analyses of BRD2 KO cells to our previously generated data sets in which these cells were exposed to JQ1. Because the prior experiments employed a microarray platform (36), we first intersected genes detected by both methods, resulting in 9132 genes (see “Experimental procedures”). Comparison of the gene expression changes in the two data sets revealed a positive correlation between genes impacted by BRD2 loss and JQ1 exposure (Pearson correlation coefficient of r = 0.47 (p < 2.2e−16)) (Fig. S2F). Of the 248 genes up-regulated in BRD2 KO cells, 104 were also up-regulated (-fold change >2, FDR <0.01) upon JQ1 treatment (p < 2.2e−16), and of the 137 down-regulated genes, 54 were also down-regulated upon JQ1 treatment (p < 2.2e−16) (Fig. S2G). A limitation to this comparison is that alterations in gene expression upon chronic BRD2 loss are a composite of direct and indirect effects. In contrast, JQ1 treatment enables detection of an immediate response to BET inhibition. However, JQ1 does not fully displace BET proteins from chromatin (13, 41), which might limit effect size. Despite these limitations, these results suggest that impaired BRD2 function accounts for at least part of the effects of JQ1 treatment. In sum, BRD2 is critical for the regulation of a wide array of erythroid genes involved in multiple aspects of GATA1-mediated differentiation, and loss of BRD2 activity accounts for at least part of the JQ1 effects.
BET-specific functions during erythroid cell growth and differentiation
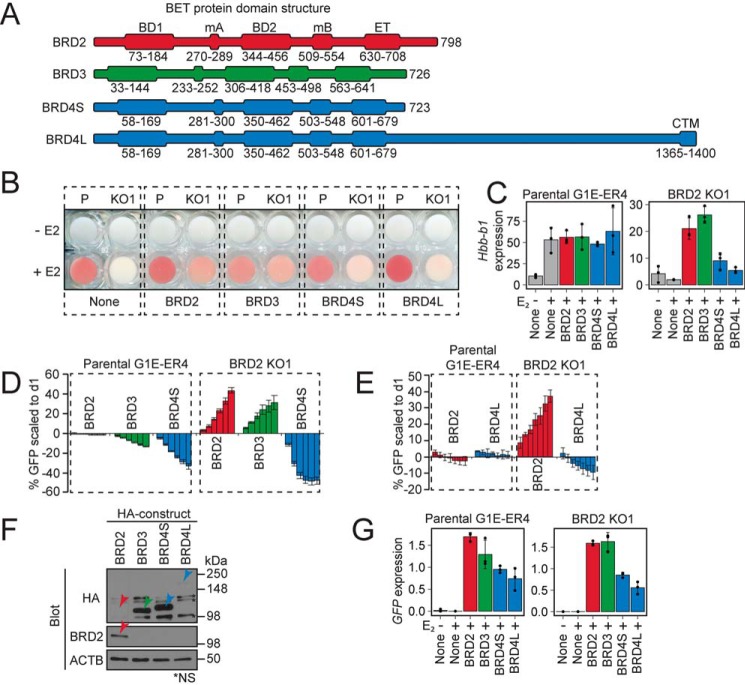
BET proteins are structurally similar, but little is known about whether the conserved domains of BET proteins contribute to any cell- or gene-specific functions (Table S2 and Fig. 1A). We previously reported that BRD2 and BRD3 can partially compensate for each other in G1E-ER4 cells (36). Here, we expanded our analysis to test whether a similar functional overlap exists between BRD2/3 and BRD4S/4L. Although endogenous BRD4S protein levels remain uncertain (see above), we included it in this functional comparison due to its structural resemblance to BRD2 and BRD3. We used a murine retroviral expression vector to transduce HA-tagged forms of BRD2, BRD3, BRD4S, or BRD4L cDNA into parental G1E-ER4 cells or BRD2 KO1 cells. We used three assays to study the relative activities of these proteins: 1) a reddening of the cells indicating activation of genes involved in hemoglobin synthesis, 2) expression of erythroid maturation–associated genes, and 3) cell growth. To visualize hemoglobinization, we induced GATA1-ER activity for 48 h, at which time the cells normally turn red. As expected, BRD2 was required for hemoglobin production (Fig. 1B). Expression of BRD2 restored the ability of BRD2 KO1 cells to produce hemoglobin. BRD3 was also able to rescue hemoglobinization, but both BRD4S and BRD4L displayed much lower activity in this assay. We next measured mRNA levels of Hbb-b1, which was one of the most differentially regulated genes in the BRD2 KO cells based on the previous analysis. Activation of GATA1-ER failed to induce expression of Hbb-b1 in the BRD2 KO1 cells. However, expression of BRD2 and BRD3 restored Hbb-b1 expression in BRD2 KO1 cells to similar levels, whereas BRD4S and BRD4L showed little activity (Fig. 1C). Note that overexpression of BET proteins in parental cells did not overtly perturb hemoglobin production or Hbb-b1 expression (Fig. 1, A and B), suggesting that excess levels of a given BET protein are compatible with cell maturation.
Figure 1.
Differential function and expression of BRD2, BRD3, and short and long isoforms of BRD4 in G1E-ER4 cells. A, schematic of BET protein domains in mouse BRD2, BRD3, BRD4S, and BRD4L. BD1 and BD2, bromodomains 1 and 2; mA and mB, motifs A and B; ET, extraterminal domain/motif; CTM, C-terminal motif. B, visualization of hemoglobin production (redness) with or without 48 h of GATA1-ER activation (+E2) in parental (P) or Brd2-null (KO1) G1E-ER4 cells stably expressing the indicated HA-tagged BET proteins. C, Hbb-b1 mRNA levels normalized to Pabpc1 after 24 h of GATA1-ER activation. D and E, competitive growth assay in which %GFP is tracked after retroviral transduction with bicistronic vector encoding the indicated BET protein and GFP selection marker. Bars, consecutive days of GFP percentage measurement from left to right. F, Western blots of HA-tagged BET protein expression in BRD2 KO1 cells, representative of similar expression patterns in parental G1E-ER4 cells. ACTB is a loading control. *NS, nonspecific bands associated with HA antibody. G, GFP mRNA levels (indicative of transgene expression) normalized to Pabpc1. Averages and S.D. (error bars) are derived from three independent replicates.
To further compare the activities of BET proteins, we assessed the impact of BET expression on cell growth. The retroviral vector encoding the BET cDNAs contained an IRES-GFP module to mark cells with stably integrated constructs. We designed a competitive growth assay which monitors the fraction of GFP+ cells in a mixture of infected and uninfected cells over time using flow cytometry. Restoration of BRD2 expression in BRD2 KO1 cells increased the fraction of GFP+ cells by ∼40% over the time course (Fig. 1D). BRD3 expression augmented growth almost to the same extent as BRD2. In contrast, BRD4S expression failed to rescue BRD2 KO1 cell growth and even lowered cell numbers. We also measured growth rates in cells expressing BRD4L and found BRD4L to be incapable of restoring BRD2 KO1 cell growth (Fig. 1E). In parental cells, BRD2 overexpression had no impact on cell growth, whereas BRD3 resulted in a ∼20% decrease, BRD4S in a ∼40% decrease, and BRD4L in a 10% decrease (Fig. 1, D and E), suggesting that excess levels of these molecules can impair proliferation or viability. In sum, both differentiation and growth assays point at overlapping functions between BRD2 and BRD3, whereas BRD4S, which is overall structurally similar, and BRD4L exert distinct functions in these cells.
The abundance of BRD2 and BRD4S is strongly influenced by their N-terminal halves
Although the BET constructs were expressed by the same vector, the resulting protein amounts were markedly variable as determined by Western blotting with anti-HA antibodies (Fig. 1F). Specifically, HA-BRD3 and HA-BRD4S were readily detected, whereas HA-BRD2 could only be detected by using a BRD2-specific antibody. Differences in exogenous BET expression potentially confound interpretation of the results. However, comparisons between the effects of BRD3 and BRD4S might still be instructive because these proteins are expressed at similar levels. For example, BRD3 and BRD4S exert opposite effects on BRD2 KO1 cell growth, with BRD3 restoring their expansion and BRD4S lowering cell numbers (Fig. 1D). Similarly, whereas BRD3 rescued Hbb-b1 expression to levels comparable with that achieved by BRD2, BRD4S failed to do so (Fig. 1C).
Variable production of BET proteins could be due to differences in mRNA stability, translation, and/or protein turnover. To address this, we measured mRNA levels of the HA-Brd2, Brd3, Brd4S, and Brd4L transgenes in parental G1E-ER4 and BRD2 KO1 cells using primers common to all constructs (specific to the GFP-coding portion of the mRNAs). We found that mRNA levels varied substantially less than protein levels. In fact, HA-Brd2 transcripts were higher than those of HA-Brd4S (Fig. 1G), indicating that the decreased BRD2 protein was due to a post-transcriptional mechanism.
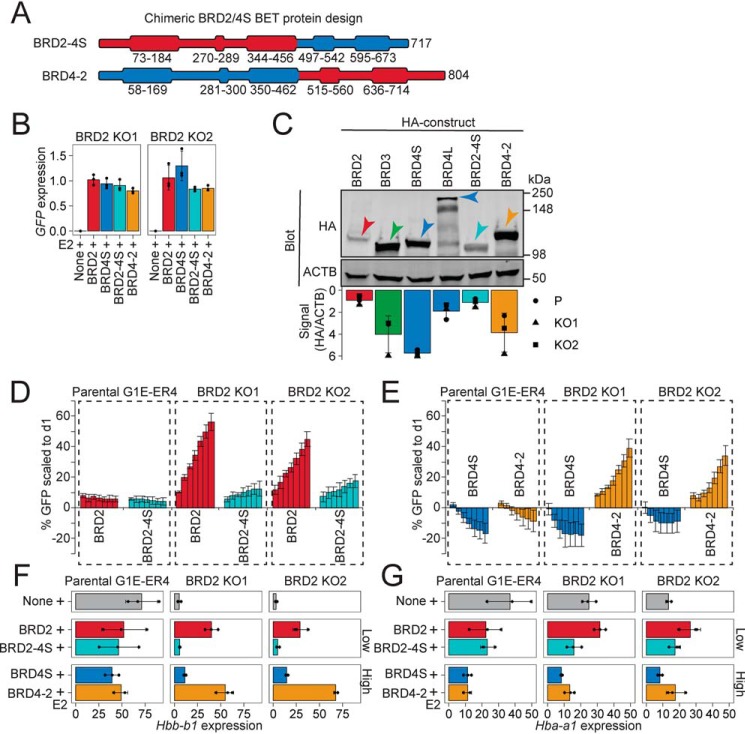
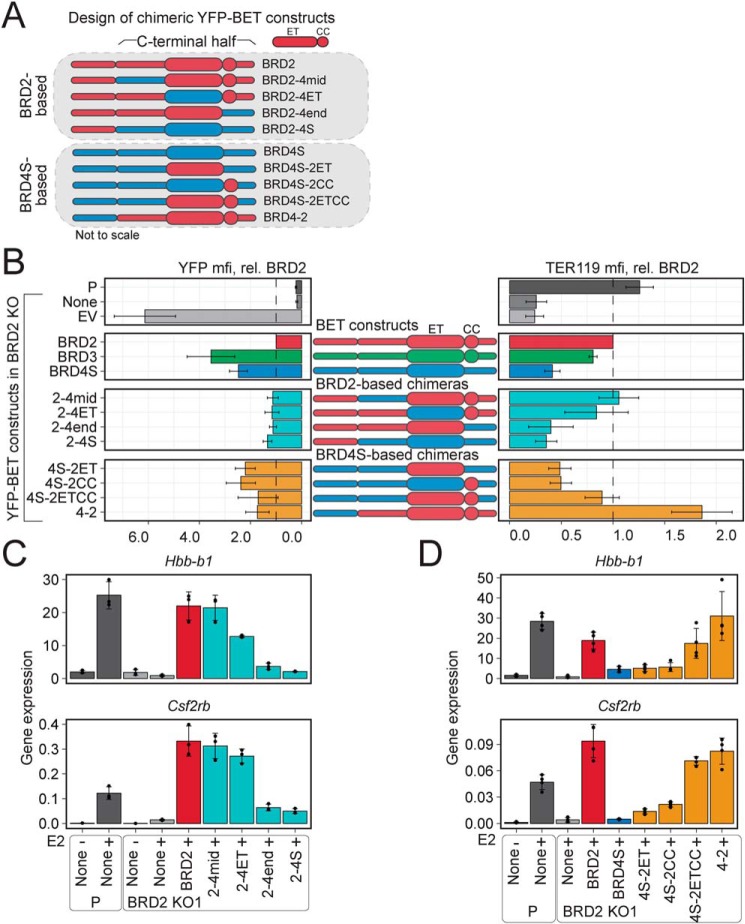
To identify which features may be dictating the contrasting nature of BRD2 and BRD4S function and stability, we began to dissect their differences by designing chimeric BET constructs consisting of the N-terminal and C-terminal halves of BRD2 and BRD4S fused immediately downstream of the second bromodomain. These chimeric BET proteins were termed BRD2-4S and BRD4-2, respectively (Fig. 2A). This strategy preserved their overall modular domain structure, including the N- and C-terminal nuclear localization signals (42, 43). These constructs were introduced into the same retroviral vector as described before. We measured mRNA levels produced by each construct and found them to be similar (Fig. 2B). Given the dramatic differences in expected protein levels (Fig. 1F), we employed the LI-COR quantification system to enable better comparison of protein expression among all BET constructs. Strikingly, the protein levels of BRD2 and the chimeric BRD2-4S were dramatically lower than those of BRD4S and chimeric BRD4-2 (Fig. 2C). BRD3 levels were similarly to those of BRD4S and BRD4-2, whereas BRD4L levels were intermediate. These data indicate that BET proteins vary in their steady-state levels, which are determined at least in part by sequences within the N-terminal halves of the protein.
Figure 2.
Structure, expression and function of chimeric BET proteins composed of the N- and C-terminal halves of BRD2 and BRD4S. A, design of chimeric BET proteins. B, GFP mRNA levels (indicative of transgene expression) normalized to Pabpc1 after 24 h of GATA1-ER activation (+E2). C, quantitative Western blotting of HA-tagged BET protein expression in parental (P) and BRD2 KO lysates. ACTB is a loading control. ACTB normalized signal is plotted below. D and E, competitive growth assay in which %GFP is tracked after retroviral transduction with bicistronic vector encoding the indicated BET protein and GFP selection marker. Bars, consecutive days of GFP percentage measurement from left to right. F and G, Hbb-b1 and Hba-a1 mRNA levels normalized to Pabpc1 after 24 h of GATA1-ER activation. Constructs grouped by expression levels. Averages and S.D. (error bars) are derived from three independent replicates.
Selective functions of BET proteins are determined by their C-terminal halves
To determine whether there are BET-selective functions that are conveyed by domains that do not affect protein levels, we compared the abilities of BRD2 and BRD2-4S and of BRD4S and BRD4-2 proteins to restore BRD2 KO cell growth and differentiation-associated gene expression. BRD2-4S, which was expressed at similar levels as BRD2, was impaired in its ability to restore cell growth compared with BRD2 (Fig. 2D), whereas BRD4-2, which was expressed similarly to BRD4S, had a positive effect on BRD2 KO cell growth (Fig. 2E). Given that BRD2 but not BRD4S rescued the expression of Hbb-b1 in BRD2 KO cells (Fig. 1C), we asked whether the C-terminal halves of BRD2 and BRD4S were responsible for this differential activity. BRD4-2 rescued Hbb-b1 expression at a level comparable with BRD2, whereas BRD2-4S had no measurable effect (Fig. 2F). The same trends held true for the expression of the other major globin chain gene Hba-a1, although compared with Hbb-b1, Hba-a1 expression was less affected by BRD2 depletion (Fig. 2G). Together, these observations suggest that the C-terminal half of BRD2 encodes its specific activity, which can be grafted onto the N-terminal half of BRD4S to substantially convert it into a BRD2-like molecule. This further suggests that the specific functions of BET proteins are not critically determined by differences in the bromodomains, but rather by differences in the C-terminal halves of the proteins.
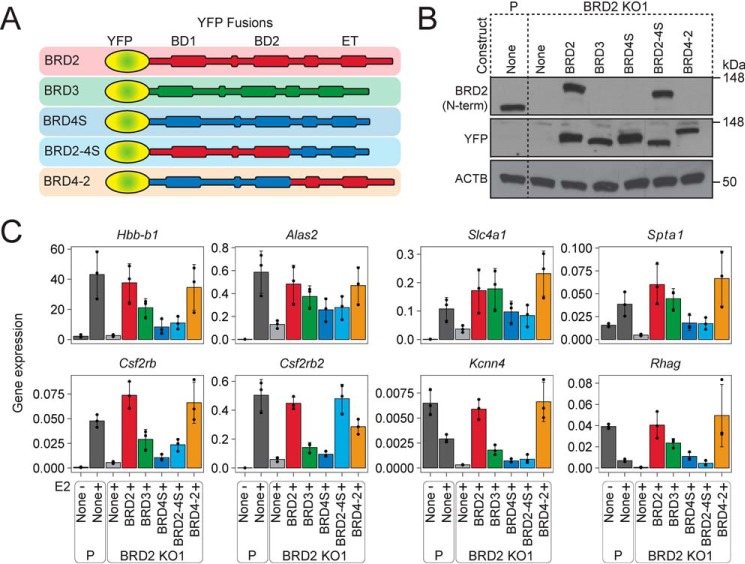
So far, comparisons among proteins were largely limited to those that were expressed at comparable levels, but the drastic differences in protein abundance conferred by the N-terminal halves of BRD2, BRD3, and BRD4S confounded direct functional comparison across all of the constructs. To overcome these limitations, all BET constructs were fused to YFP and stably introduced into BRD2 KO1 cells via retroviral transduction (Fig. 3A). Generation of cell lines with comparable expression of each construct required careful titering of viruses and multiple rounds of FACS using stringent gating on the YFP signal. In that manner, we eventually succeeded in isolating subpopulations of cells in which all proteins were produced at nearly equal levels that in turn were similar to the amount of endogenous BRD2 in parental G1E-ER4 cells (Fig. 3B). YFP-BET transgene mRNAs were measured with qPCR primers that target the YFP sequence and the 5′ and 3′ end of Brd2 (Fig. S3A). These measurements indicate that the RNA levels corresponding to the YFP-BRD2 and YFP-BRD2-4S proteins were slightly higher than the RNA levels of YFP-BRD3, YFP-BRD4S, and YFP-BRD4-2. This is consistent with the lower protein yields for BRD2 and BRD2-4S, requiring selection of cells with higher RNA expression. However, the discrepancy between relative mRNA and protein levels was not as drastic as that observed with versions lacking YFP. It is possible that the YFP moiety affects BRD2 mRNA or protein in a manner resulting in a stronger correlation between its RNA and protein levels. Of note, levels of Gata1-ER, Actb, and Gapdh mRNA were similar in the cell lines (Fig. S3A).
Figure 3.
The C-terminal halves provide distinct functions to BRD2 and BRD4S. A, schematic of YFP-fusion constructs utilized in this experiment. B, Western blotting of exogenous YFP-fused BET proteins in BRD2 KO1 lysates and endogenous BRD2 for reference. ACTB was used as a loading control. C, gene expression of BRD2-dependent and control genes normalized to Pabpc1 after 24 h of GATA1-ER activity (+E2). Averages and S.D. (error bars) are derived from three independent replicates.
We next assessed whether the reconstituted BRD2 KO1 cells could activate eight BRD2-dependent genes during erythroid maturation. As expected, expression of Hbb-b1, Alas2, Slc4a1, Spta1, Csf2rb, Csf2rb2, Kcnn4, and Rhag were all reduced in the unreconstituted BRD2 KO1 cells compared with the parental G1E-ER4 cells (Fig. 3C). Expression of all of these genes was restored by YFP-BRD2 and to a lesser extent by YFP-BRD3, indicating that the increased bulk added by the YFP fusion onto the N terminus of BET proteins did not impact their overall function. YFP-BRD4S had a marginal impact on the expression of these BRD2-dependent genes and was clearly less potent than YFP-BRD3. We next assessed the functions of the chimeric YFP-BRD2-4S and YFP-BRD4-2 BET proteins. YFP-BRD2-4S was unable to restore expression of seven of the eight genes. In contrast, YFP-BRD4-2 was able to restore expression of all eight genes to levels comparable with YFP-BRD2 and parental G1E-ER4 cells. We also examined the ability of the YFP-BRD2, YFP-BRD4S, and the two chimeric YFP-BRD2-4S and YFP-BRD4-2 proteins to rescue hemoglobinization in BRD2 KO1 and KO2 cells after sorting for equal YFP-BET levels (Fig. S3B). As observed previously, parental G1E-ER4 cells fully hemoglobinized, whereas the BRD2 KO1 and KO2 cells failed to turn red (Fig. S3C). YFP-BRD2 expression in both BRD2 KO cell lines rescued hemoglobin production to levels comparable with the parental G1E-ER4 cells, whereas YFP-BRD4S–expressing BRD2 KO cells failed to turn red. Chimeric YFP-BRD2-4S failed to rescue BRD2 KO cells, whereas YFP-BRD4-2 restored hemoglobinization comparably with YFP-BRD2. These data confirm that the C-terminal but not the N-terminal half of BRD2 conveys functional specificity.
BRD2 and BRD3 contain a putative coiled-coil domain that contributes to BRD2 activity
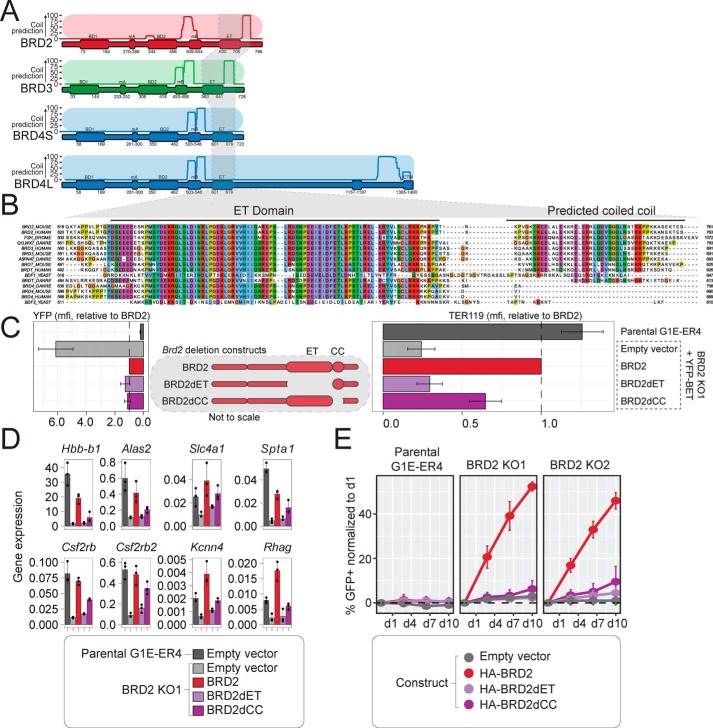
The previous experiments reveal that BRD2-like activity shared by BRD2 and BRD3 maps to the C-terminal half of BRD2. We therefore examined the amino acid sequences of the C-terminal halves of both BRD2 and BRD3 for shared features that may distinguish them from BRD4S. Coiled-coil prediction of the mouse BET proteins using MARCOIL (44) revealed multiple putative coiled-coils (Fig. 4A). We identified a region immediately downstream of the ET domain that is predicted to harbor a coiled-coil (that we refer to as the CC domain) in BRD2 and BRD3 but not in BRD4. The CC domain is conserved across the BRD2, BRD3, and BRDT paralogues in humans, mice, and zebrafish (Fig. 4B). It is also conserved in the Drosophila BET paralog FSH as well as one of the yeast paralogs, BDF1, but not BDF2. We therefore asked whether this region could account for some of the observed differences between BRD2/BRD3 and BRD4S.
Figure 4.
A predicted CC domain adjacent to the ET domain contributes to BRD2 function. A, MARCOIL coiled-coil prediction across the mouse BRD2, BRD3, BRD4S, and BRD4L amino acid sequences. B, conservation of the ET and predicted coiled-coil region between BET proteins and across species. Blue, hydrophobic; red, positive charge; magenta, negative charge; green, polar; pink, cysteines; orange, glycines; yellow, prolines; cyan, aromatic; white, not conserved. C, schematic of BRD2 constructs with the ET and CC domains deleted (middle). Protein expression (left) and TER119 surface levels (right) in BRD2 KO cells are after 24 h of GATA1-ER activation. Values are normalized to YFP-BRD2. mfi, mean fluorescence intensity. D, gene expression levels normalized to Pabpc1 after 24 h of GATA1-ER activation. E, competitive growth assay in which %GFP is tracked after retroviral transduction with bicistronic vector encoding the indicated BET protein and GFP selection marker. Averages and S.D. (error bars) are derived from three independent replicates.
To expedite analyses of several constructs in multiple independent cell lines, we developed a flow cytometry–based version of the erythroid maturation gene complementation assay in G1E-ER4 cells. This approach quantifies the fluorescent tag on each construct as a proxy for protein abundance and the erythroid-specific surface antigen TER119 as a measure of GATA1-mediated differentiation (Fig. S4A). TER119 was only expressed on G1E-ER4 cells upon GATA1-induced maturation (Fig. S4B). We validated the use of the TER119-based erythroid maturation assay by demonstrating that YFP-BRD2, but not a YFP-only vector, restored GATA1-mediated TER119 expression in BRD2 KO cells to levels observed in the parental G1E-ER4 cells (Fig. S4, B and C). We also compared erythroid gene expression levels with TER119 surface staining after 24 h of GATA1-ER activation in parental G1E-ER4 cells or BRD2 KO1 cells expressing YFP-fused BRD2, BRD3, BRD4S, or chimeric BRD2-4S or BRD4-2 (Fig. S4D). There was a strong correlation between the amount of TER119 expression on the surface of these cells and the mRNA levels of the erythroid genes Alas2 (r = 0.81, p = 0.026), Hbb-b1 (r = 0.84, p = 0.017), Slc4a1 (r = 0.82, p = 0.024), and Spta1 (r = 0.88, p = 0.009). For example, whereas the parental G1E-ER4 cells exhibited high levels of mRNA and TER119 staining, the BRD2 KO1 cells without YFP-BET contained low mRNA levels of these genes and exhibited minimal TER119 staining. However, the YFP-BRD2–rescued BRD2 KO1 cells expressed mRNA and TER119 at levels comparable with the parental G1E-ER4 cells. YFP-BRD3 and YFP-BRD4-2 likewise rescued the BRD2 KO1 cells, albeit to varying degrees, whereas YFP-BRD4S and YFP-BRD2-4S failed to rescue mRNA and TER119 expression. These data indicate that the gene encoding the TER119 antigen is BRD2-dependent and is representative of erythroid maturation.
The ET region is a protein-protein interaction domain that recruits transcriptional co-activators to chromatin (4–8). Given the proximity between the CC and ET domains, it is possible that the CC contributes to the ET-mediated interactions; alternatively, it might function independently. To test whether either domain contributes to BRD2 function, we deleted both the ET (BRD2dET) and CC (BRD2dCC) regions individually and expressed these constructs in BRD2 KO cells (Fig. 4C, middle). Both genes were expressed at levels similar to full-length BRD2 based on YFP fluorescence (Fig. 4C, left). Deletion of the ET domain completely abrogated TER119 expression compared with full-length BRD2 (Fig. 4C, right). Deletion of the CC domain also reduced TER119 expression, albeit to a lesser extent. To measure effects on gene expression, we isolated BRD2 KO1 cell lines stably expressing HA-tagged versions of these constructs or an empty vector (encoding only GFP) and confirmed by Western blotting that BRD2, BRD2dET, and BRD2dCC protein and RNA levels (based on Brd2 and GFP in bicistronic transgene) were similar (Fig. S5, A and B). Levels of Gata1-ER, Brd3, Brd4, Actb, and Gapdh were likewise similar across the cell lines. Restoration of BRD2 rescued the expression of multiple BRD2-dependent genes (Hbb-b1, Alas2, Slc4a1, Spta1, Csf2rb2, Kcnn4, and Rhag) (Fig. 4D). Deletion of the ET domain completely abrogated the ability of BRD2 to rescue these genes, whereas deletion of the CC domain had a partial effect. These data indicate that whereas the ET domain is absolutely critical for proper BRD2 functioning during erythroid maturation, the CC region on its own is only partially contributory.
In addition to restoring erythroid gene expression, BRD2 also restores the cell growth rate of BRD2 KO cells. We therefore tested whether the ET and CC regions contributed to BRD2-mediated cell growth. As seen previously, reconstitution of BRD2 in BRD2 KO1 and KO2 cells led to a ∼45% increase over the time course (Fig. 4E). Deletion of the ET domain drastically decreased BRD2 function in this assay. Interestingly, although the CC domain contributed only partially to erythroid gene expression as seen above, its deletion had a significant impact on BRD2 function in cell growth, indicating that the erythroid genes assayed are not completely reflective of the overall gene expression changes associated with loss of the BRD2 CC region. In summary, both the ET and CC regions contribute to BRD2 activity, but the CC region may play a role for only a subset of BRD2-dependent genes.
The combined ETCC region functionally distinguishes BRD2 and BRD4S
Like the bromodomains, the ET region is highly conserved between BET proteins. The adjacent CC region, however, is present only in BRD2 and BRD3. We therefore asked whether either the ET, the CC, or the combined ETCC module distinguishes BRD2 from BRD4S. We devised additional chimeric BET proteins using BRD2 and BRD4S as the backbones in which the sequences surrounding or containing the ET and CC domains were exchanged (Fig. 5A). For the BRD2-based chimeric constructs, we tested which, if any, of the C-terminal segments (the middle region between BD2 and ET, the ET, or the end segment) resulted in a loss of BRD2 function when the corresponding region from BRD4S was substituted. For the BRD4S-based chimeric constructs, we tested whether the BRD2 ET domain, the CC region, or a segment containing both could impart BRD2 function to BRD4S. As controls, we compared these new chimeric constructs with the original BRD2-4S and BRD4-2 versions, as well as with BRD2 and BRD3, which possess the CC region, and with BRD4S, which lacks the CC region. We used a combination of TER119 marker expression and erythroid gene activation to uncover any functional differences between these BET constructs.
Figure 5.
The ETCC region functionally distinguishes BRD2 and BRD4S). A, schematic of the YFP-BET fusion constructs in which C-terminal regions have been interchanged. B, protein expression levels (left) and TER119 surface levels (right) of YFP-BET constructs are after 24 h of GATA1-ER activation. Values are normalized to YFP-BRD2. mfi, mean fluorescence intensity. C and D, expression levels of mRNA normalized to Pabpc1 after 24 h of GATA1-ER activity (+E2). Averages and S.D. (error bars) are derived from 3–4 independent replicates.
We first examined TER119 expression by flow cytometry. Of note, we used low-titer viruses that statistically guarantee single-copy gene integration into the cells. Therefore, any differences in protein expression detected by YFP fluorescence were not due to gene copy number. YFP fluorescence revealed that BRD3 and BRD4S were expressed ∼2–3-fold higher than BRD2. This difference is less than that observed without the use of YFP fusion (Fig. 1F), confirming that the YFP sequence stabilizes BRD2 mRNA or protein to some extent. Next, we examined erythroid maturation based on TER119 surface phenotyping. As expected, a YFP-only vector did not alter TER119 expression in BRD2 KO cells, whereas virus encoding BRD2 restored TER119 close to parental G1E-ER4 levels (Fig. 5B). BRD3 also restored TER119 expression, although to a lesser extent than BRD2. BRD4S had minimal impact on TER119 expression.
We next examined the relative abilities of the BRD2-based chimeric BET proteins to restore TER119 and erythroid gene expression. All of the BRD2-based constructs are expressed at levels similar to BRD2 based on YFP fluorescence (Fig. 5B). Replacing the middle region (BRD2–4mid) or the ET domain (BRD2–4ET) from BRD2 with the corresponding regions from BRD4S had no appreciable impact on TER119 expression. However, replacing the end segment of BRD2 (BRD2–4end), which contains the CC region, completely abrogated its activity to the same extent as replacing the entire C-terminal half (BRD2-4S). To examine gene expression, we established stable BRD2 KO1 cells expressing the above constructs at similar levels (Fig. S6A). Expression of the Brd2-based transgenes and control genes (Actb and Gapdh) was similar across the cell lines. Examination of erythroid gene expression revealed differences in the ability of these constructs to rescue expression of Hbb-b1, Csf2rb, Csf2rb2, and Rhag and, to a lesser extent, Alas2, Spta1, and Kcnn4 that largely reflect the differences in TER119 expression (Fig. 5C and Fig. S6A). For some of these genes, all of the chimeric constructs exhibited reduced activity. However, BRD2–4end had the least activity and most resembled BRD2-4S for seven of the eight genes examined. In summary, the activity of the BRD2-based chimeras indicates that the region that discriminates between BRD2 and BRD4S is the C-terminal end segment containing the CC domain. The middle segment and the ET domain appear interchangeable with minimal disruption of BRD2 activity.
We next assessed the activities of the BRD4S-based chimeras. These variants were expressed at similar levels, and about 2-fold higher than BRD2 based on YFP fluorescence (Fig. 5B). Replacing the BRD4 ET domain with that of BRD2 (BRD4S-2ET) did not increase TER119 expression. Adding the BRD2 CC domain adjacent to the BRD4 ET domain (BRD4S-2CC) likewise did not impact TER119 expression. However, when both the ET and CC domains from BRD2 were inserted into BRD4S (BRD4S-2ETCC), TER119 expression was restored to levels close to that achieved by BRD2. The BRD4-2 protein containing the entire C-terminal half of BRD2 promoted TER119 expression even more strongly, indicating that additional C-terminal domain(s) may contribute to BRD2 function. To examine gene expression, we established BRD2 KO1 cell lines stably expressing the above constructs. Protein levels of the BRD4S-based constructs were about 2-fold higher than BRD2 protein levels but were similar across the cell lines (Fig. S6B). Despite the lower protein level of YFP-BRD2, its mRNA was slightly higher than that of the YFP-BRD4S–based BET proteins as measured by qPCR primers targeting YFP or the 3′ end of Brd2, consistent with the decreased protein/RNA ratio previously observed for BRD2 (Fig. S6B). Levels of mRNA for Gata1-ER, Actb, and Gapdh were similar. As with TER119 staining, the ET and CC domains were insufficient to confer BRD2-like activity to BRD4S based on the inability of the BRD4S-2ET and BRD4S-2CC proteins to restore erythroid gene activation (Fig. 5D and Fig. S6B). However, the combined ETCC domain (BRD4S-2ETCC) clearly imparted BRD2-like activity to BRD4S, as Hbb-b1, Csf2rb, Csf2rb2, and Kcnn4 expression were at least partially increased above the levels achieved by BRD4S. As was the case for TER119 staining, the BRD4-2 construct had the most BRD2-like activity at these genes.
To test whether the ETCC region likewise rescued hemoglobin synthesis and cell growth, we expressed HA-tagged versions of BRD2, BRD4S, or the BRD4S-based chimeric constructs in BRD2 KO1 cells. The addition of the ET, CC, combined ETCC, or entire BRD2 C-terminal half progressively increased hemoglobinization and cell growth (Fig. S6, C and D).
In summary, these results support the idea that neither the ET domain nor the CC region alone suffices to convert BRD4S into a functional BRD2 molecule. Instead, the combined ETCC domain can impart BRD2-like activity onto BRD4S.
The BRD2 and BRD3 CC domains are helical modules that do not dimerize
To assess the structure of human BRD2-CC(715–757) and human BRD3-CC(641–688), we expressed and purified these regions and recorded far-UV CD spectra of each protein (Fig. S7A). The spectra show minima at 222 nm, indicative of the presence of some helical structure. Raussens' method estimates that each of the two peptides contains ∼33% helical structure (http://perry.freeshell.org/raussens.html)4 (69). Because coiled-coils often form dimers (45), we assessed whether either the BRD2-CC or BRD3-CC domains can form a homomeric coiled-coil using size-exclusion chromatography (SEC) coupled to multiangle laser light scattering (MALLS). As shown in Fig. S7B, both peptides run with molecular masses that are close to the monomeric masses (5.3 and 7.3 kDa, respectively), indicating that they do not significantly self-associate in solution at a concentration of ∼100 μm (accounting for dilution on the column). We also assessed the ability of the peptides to associate with each other using the same approach, but again no significant self-association was observed. We conclude that if these sequences form coiled-coils, then they must do this by partnering with other proteins.
The ETCC region binds to the PAF and CK2 complexes
To identify partner proteins, we generated GST-fusion proteins and exposed them to nuclear extracts from MEL cells (see “Experimental procedures”; Fig. S8A). As a control, we included the mB region, which is also a putative coiled-coil. We also included two different sets of mutations in the CC predicted to abrogate coiled structure (CCp and CCd). Each sample was compared with a GST-only control. Mass spectrometry (see “Experimental procedures”) revealed enrichment of a total of 392 proteins (log2 ratio >1 over GST alone at FDR <0.0001, Table S3). Several known BET binding partners were among these, validating our approach. For example, ATAD5, CHD8, SMARCA4/BRG1, WHSC1/NSD2, WHSC1L1/NSD3, and CDK9 (5, 7, 8, 16, 46) were enriched significantly by both the ET and ETCC domains (Fig. S8B). Likewise, NIPBL, which has recently been shown to interact with the ET domain of BRD4 (47, 48), was also enriched in these samples. Two other previously reported ET-mediated interactions, CHD4 (5, 8) and CCNT1 (16), did not meet the thresholds set in our analysis but were nonetheless enriched in the ET and ETCC samples. Likewise, mB interaction partner LYAR (49, 50) was elevated specifically in the mB sample. The 392 enriched proteins were distributed in the samples as follows: ET (165), ETCC (179), mB (232), CC (0), CCp (2), and CCd (8). These data suggest that the CC does not engage in protein interactions on its own (Table S3 and Fig. S8B).
To identify the proteins that associate most strongly with the GST-fusion proteins, we increased the threshold to log2 ratio >1.5. This identified interactions that had not been previously reported. For example, the ET domain associated most strongly with AHNAK, AHNAK2, CBX4, ZMYND8, and ZNF592. Interestingly, when combined with the CC domain, ETCC associated with several additional proteins, including all five members of the PAF complex (PAF1, LEO1, CTR9, CDC73, and WDR61) as well as the PAF-associated protein IWS1 (Fig. S8, C and D), which are involved in transcriptional elongation (51–53). The ETCC domain also enriched all three members of CK2, which, like PAF, has been shown to occupy active chromatin (54) and associate with elongation factors (55). Western blots with antibodies against components of the PAF and CK2 complexes confirmed association with GST-ETCC but less so or not at all with GST-ET or GST-CC (Fig. S8E). The ETCC-mediated interactions with PAF and CK2 were also observed when using the ETCC regions from BRD3 and BRDT, but not their ET domains alone (Fig. S8F). These results suggest that the BRD2 and BRD3 CC domains augment the ability of the ET domains to engage in specific protein contacts.
Discussion
We used BET-dependent G1E-ER4 cell growth, maturation, and gene expression to define functional similarities and differences among BET proteins. We exploited the fact that BRD2 is required for G1E-ER4 growth and GATA1-ER activity. Overexpressed BRD3, but not BRD4S or BRD4L, could significantly overcome defects associated with BRD2 deficiency, pointing to functional similarity among BRD2 and BRD3. When comparing BRD2 and BRD4S, most of their specific activities were determined not by the bromodomains but by the C-terminal halves of the molecules. We identified a short sequence downstream of the ET domain that contains a putative coiled-coil (CC region) in BRD2 and BRD3 but not in BRD4. Together with the ET domain, the CC region conferred BRD2-like activity to BRD4S, and deletion of the ET or CC domains diminished BRD2 function.
G1E-ER4 cells are a powerful system for the study of BET proteins because they are sensitive to pharmacologic BET inhibition and genetic perturbations of BET proteins (36, 37). Acetylation of GATA1 promotes association with BET proteins (37), which in turn are thought to contribute to GATA1 activity. In this context, it is worth noting that the clinical side effects of BET inhibition include anemia and thrombocytopenia (56, 57). Because GATA1 is essential for erythrocyte and megakaryocyte lineage differentiation, it is likely that the detrimental effects of BET inhibition are linked to GATA1 function.
Over the course of our study, we uncovered a discrepancy in the amount of BRD2 protein compared with that of BRD3 and BRD4S when expressed from the same retroviral vector. BRD2 protein was substantially lower, although transcript levels of BRD2 were in fact higher than that of BRD4S, indicating that the mechanism for the low BRD2 abundance is post-transcriptional. The N-terminal half of BRD2 was responsible for the low protein amounts, as the protein levels of the chimeric construct BRD2-4S were similar to BRD2, whereas the protein levels of the chimeric construct BRD4-2 were similar to BRD4S. By fusing YFP to the N terminus of BRD2, the discrepancy between BRD2 protein and mRNA was reduced, although YFP-BRD2 protein was still lower than that of YFP-BRD3 and YFP-BRD4S. The YFP portion might thus affect BRD2 protein production or turnover. It is interesting to note that a similar discordance between overexpressed BRD2 and BRD3 protein was observed in U2OS cells but not HEK293T cells (9). Cell type–specific mechanisms may exist to tightly regulate BRD2 protein levels. Additional studies are required to elucidate what accounts for this specificity.
Our unexpected finding that the BRD2 bromodomains can be exchanged for those of BRD4 with no substantial loss of BRD2 function suggests that potential differences in their affinities or specificities for acetylated substrates (25–30) do not seem to contribute much to their selective functions. Our results suggest that the distinct functions of BRD2 and BRD4 are instead exerted by their C-terminal halves, in which the extended ET region containing the CC in BRD2 but not BRD4 appears to be the critical element. Gene rescue experiments showed that the ET domain itself was essential for BRD2 function, which is consistent with previous studies demonstrating that the ET domain recruits multiple chromatin regulators, including NSD3, CHD8, ATAD5, and JMJD6 (4–8). However, in our study, although the ET domain was essential, exchanging the BRD2 and BRD4S ET domains did not change the activities of either protein. Instead, the BRD2 ET domain appears to function in conjunction with the adjacent CC. Thus, when the combined ETCC region was grafted into BRD4S, it imparted BRD2-like activity to BRD4S.
Coiled-coils are structural motifs present in a wide variety of proteins and perform a multitude of functions, including directly mediating protein-protein interactions (45). Our results suggest that rather than functioning as independent protein-docking sites, the ET and CC domains act as a single unit to associate with the PAF and CK2 complexes. The CC domain may be modulating the structure of the ET domain, or it may be creating novel interaction surfaces at the junction. Further definition of this element is needed to delineate the amino acids that are required and sufficient for these contacts. Future studies will be necessary to examine the extent to which the ETCC-associated protein complexes co-localize with BRD2 or BRD3, respectively. Nevertheless, these experiments, albeit based on in vitro association, support the idea that the ET and CC domains function as a unit to convey protein contacts specific to BET protein subtypes.
The ETCC region may allow individual BET proteins to incorporate into different transcriptional complexes involved in discrete aspects of the transcription cycle. This may at least in part reconcile how BET proteins can function distinctly on chromatin while having relatively similar genome-wide distribution patterns, as observed in many studies (11, 13, 58–64). Protein interactions mediated by the ET or ETCC regions may also account for bromodomain-independent chromatin binding (32). Thus, the complexes that each BET protein associates with could also account for some of the differences in chromatin occupancy exhibited by each BET protein, as is the case at enhancers, where there is a relative enrichment of BRD4 and at promoters where there is a relative enrichment of BRD2 and BRD3 (13, 36, 63). There is also evidence that the conserved mB domain, which facilitates homo- and heterodimerization of BET proteins, is required for chromatin binding (31). Heterodimerization of BET proteins might further explain their overlapping chromatin occupancy and may suggest that BET proteins function coordinately to assemble multiple factors onto chromatin. Together, these considerations lead to a model in which BET proteins function both independently and interdependently during transcription.
At low doses of BET inhibitors, the expression of only a subset of genes is affected in a manner that is not easily predictable based solely on local chromatin features or levels of BET protein occupancy (36, 65). Predicting whether a gene responds to pharmacologic BET inhibition remains a challenge in part because cells express multiple BET proteins exhibiting mostly overlapping but partially distinct chromatin occupancy patterns. Moreover, partial redundancy among BET proteins, as exemplified by BRD2 and BRD3, might be rooted in structural and functional similarities. Thus, overexpression of BRD3 at least partially compensates for loss of BRD2 (36). This example illustrates that different BET protein dependences can also be a result of the expression levels of each protein. These factors need to be taken into consideration when assessing the effects of targeting BET proteins pharmacologically.
Hence, identifying BET protein-specific mechanisms and predicting what disease processes might be targeted by BET inhibitors remains an important goal. Achieving this level of mechanistic understanding requires consideration of BET protein expression levels, assessment of functional overlap among BET proteins, and the identification of functional domains that are required for BET protein activity in diverse contexts. Our work begins to address these criteria by identifying the ETCC module, which may mechanistically differentiate BRD2 and, given the structural similarities also BRD3, from BRD4. Additional work is required to understand the mechanism by which this domain contributes to transcription, although it may mediate BRD2/3-specific interactions with CK2 and PAF. We speculate that targeting this region with a small-molecule inhibitor might provide a more selective tool for therapeutic BET inhibition.
Experimental procedures
Cell lines, cell culture, and GATA1-ER activation
G1E cells are erythroblasts derived from Gata1-null male mouse embryonic cells (66). G1E-ER4 cells are subcloned from G1E cells after expressing a retrovirally inserted construct that encodes GATA1 fused to the ligand binding domain of the estrogen receptor (GATA1-ER), which allows for conditional activation of GATA1 upon administration of estradiol (39). Brd2-null G1E-ER4 (BRD2 KO1–KO4) cells were derived by single-cell cloning after nucleofection with vectors expressing spCas9 and a guide RNA targeting the first bromodomain of BRD2 as described previously (36). Clones KO1 and KO2 were described previously (67). G1E-ER4–based cells were grown in suspension flasks in a humidified incubator at 37 °C at 5% carbon dioxide in G1E medium. G1E medium consists of the following reagents: Iscove's modified Dulbecco's medium (Corning, 10-016-CV), 15% FBS (Gemini, 100-106), 2% penicillin/streptomycin (Gibco, 15140-122), 140 μm 1-thioglycerol (Sigma, M6145), 2 units/ml epoetin (Amgen), and conditioned medium (titered for optimal cell growth) from a kit ligand-producing CHO cell line. Cells were maintained in culture at a cell density of 0.1–1.0e6 cells/ml. Erythroid maturation experiments were performed by plating G1E-ER4–based cells at a cell density of 0.4–0.5e6 cells/ml in fresh G1E medium containing 100 nm estradiol (Sigma, E2758) for 24 h. For the visualization of hemoglobin production, 40e6 cells were plated as above and matured for 48 h with estradiol. 293T cells used for retrovirus production were grown on 100 mm plasma-treated polystyrene plates in 293T medium: Dulbecco's modified Eagle's medium (Gibco, 11965084), 10% FBS (Gemini, 100-106), 2% penicillin/streptomycin (Gibco, 15140-122), 1% 200 mm l-glutamine (Gibco, 25030-081), and 1% 100 mm sterile-filtered sodium pyruvate. 293T cells were split every 2–3 days using 0.05% trypsin-EDTA (Gibco, 25300-054).
RNA isolation and RT-qPCR
For gene expression analysis, up to 1 million cells were resuspended in 1 ml of TRIzol (Ambion, 15596018). 200 μl of chloroform (Fisher, BP1145-1) was added to each sample. Samples were vigorously inverted for 15 s, rested for 3 min, and spun at 10,000 rpm for 15 min at 4 °C in a microcentrifuge. The aqueous phase (top layer) was applied to purification spin columns from the RNeasy® Mini Kit (Qiagen, 74106). Subsequent purification steps, including an on-column DNase treatment step (Qiagen, 79256), were followed according to the manufacturer's instructions. RNA was eluted from the column in 40 μl of RNase-free water from the kit, and the RNA concentration and purity were quantified using a spectrophotometer (Thermo Scientific, Nanodrop 2000). Reverse transcription was performed on 1 μg of RNA using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, 1708841). The cDNA samples were diluted 5× in sterile 0.2 μm filtered MiliQ water. RT-qPCR was performed using 2 μl of diluted cDNA, 1 μl of 9 μm forward/reverse primers, and 5 μl of Power SYBRTM Green PCR Master Mix (Thermo Fisher Scientific, 4367660) on an ABI Vii7 real-time PCR machine. qPCR primers were designed to target mature transcripts. RT-qPCR primer sequences are found in Table S4. Relative mRNA levels were quantified using the Δ cycle of threshold (Ct) method (relative mRNA level = 2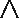 (Ctcontrol gene − Cttarget gene). All gene expression analyses included at least two of the following housekeeping genes as controls for normalization: Pabpc1, Actb, and Gapdh. All RT-qPCR experiments were performed at least in triplicate (three independent GATA1-ER induction replicates). Plots illustrating gene expression were generated in RStudio using ggplot2.
(Ctcontrol gene − Cttarget gene). All gene expression analyses included at least two of the following housekeeping genes as controls for normalization: Pabpc1, Actb, and Gapdh. All RT-qPCR experiments were performed at least in triplicate (three independent GATA1-ER induction replicates). Plots illustrating gene expression were generated in RStudio using ggplot2.
RNA-Seq
For the RNA-Seq experiment, parental G1E-ER4, BRD2 KO1, and BRD2 KO2 cells were matured for 24 h with estradiol prior to harvesting RNA. The experiment was performed in three replicates, yielding a total of nine samples. RNA concentration was determined using the Agilent RNA 6000 Nano kit (Agilent, 5067-1511), and 4 μg of total RNA was enriched for mRNA by poly(A) selection with the Dynabeads mRNA DIRECT Micro Kit (Life Technologies, Inc., 61021) using the mRNA Isolation from Purified Total RNA protocol. The concentration of mRNA-enriched samples was measured using the Agilent RNA 6000 Pico kit (Agilent, 5067-1513) before proceeding to library preparation with the remaining sample volume using the Ion Total RNA-Seq Kit c2 and IonXpress RNA-Seq Barcode 1–16 kit (Life Technologies, 4475936 and 4475485). Final libraries were assessed using the Agilent High Sensitivity DNA kit (Agilent, 5067-4626) before pooling on an equimolar basis as determined by a Bioanalyzer (Agilent) molarity estimate of each library. The pool was subsequently sequenced three times on an Ion Torrent Proton sequencer using the Ion PI Hi-Q OT2 200 Kit, Ion PI Hi-Q Sequencing 200 Kit, and Ion PI Chip Kit version 3 (Life Technologies, A26434, A26433, and A26771).
Computational analysis of RNA-Seq data
Primary analysis of sequence data was performed by Ion Torrent Suite software (version 4.4.3) before being exported into FASTQ format. Mapping to the mm10 genome (GCF_000001635.24_GRCm38.p4_genomic.gff) was performed using the following STAR options: runThreadN 8, readFilesCommand zcat, outSAMtype SAM, chimSegmentMin 32, outFilterType BySJout, outFilterMultimapNmax 20, alignSJoverhangMin 8, alignSJDBoverhangMin 1, outFilterMismatchNmax 999, outFilterMismatchNoverLmax 0.04, alignIntronMin 20, alignIntronMax 1000000, alignMatesGapMax 1000000. The following analyses were performed using the indicated R packages/functions: principal components analysis (stats/prcomp), differential gene expression (DEGandMore/DeWrapper), heat map visualization (stats/pheatmap). The top 10 gene sets enriched in the list of differentially expressed genes were identified from the Molecular Signatures Database hallmark gene sets using the GSEA online tool (http://software.broadinstitute.org/gsea)4 (70, 71) with FDR cutoff of 0.05. Plots illustrating gene expression and gene set enrichment were generated with custom code in RStudio using ggplot2. For the comparison with the previous microarray data set assessing the impact of JQ1 treatment on GATA1-ER activity, we downloaded publicly available data from Gene Expression Omnibus (GSE62709: samples G1E GATA-ER +E2 and G1E GATA-ER +E2 +JQ1 replicates 1–3). Genes were included in the merged data if they had an average microarray measurement of 6 or higher in at least one of the groups and had an average normalized RNA-Seq read count of 6 or higher in at least one of the groups. A correlation between the two data sets was tested by calculating a Pearson correlation coefficient, and a linear regression line was computed. Differentially expressed genes were identified from the microarray based on an average -fold change >2 and FDR <0.01, which were then intersected with differentially expressed genes from the RNA-Seq experiment (using the same criteria) as depicted by the Venn diagrams in Fig. S2G. Statistical significance for each overlap was calculated using Fisher's exact test. For differential isoform analysis, all reads aligning to mouse Brd4 isoforms (Ensembl) by STAR (run with a 2-pass gap alignment mode) were included. Those that map from a common exon to BRD4S- and BRD4L-specific exons were tabulated (union of all three replicates), and the proportion was used to estimate the ratio of isoforms.
Plasmids and cloning
All cloning was performed using standard molecular cloning techniques. PCR was performed using Platinum® Pfx DNA Polymerase (Invitrogen, 11708). All constructs were cloned into the MigR1 murine retroviral vector (with an IRES-GFP selection marker) or a PK1-based N-terminal YFP fusion vector. For constructs requiring assembly of multiple PCR fragments, Gibson Assembly® Master Mix was used (New England Biolabs, E2611L). The details for each construct are provided in Table S5.
Cell growth assay
Differences in cell growth upon transduction with retroviruses expressing BET protein constructs were performed using viral titers that yield 30–40% GFP+ cells. %GFP was measured by flow cytometry and tracked over time while maintaining the cells at a cell density of 0.1–1.0e6 cells/ml. %GFP was normalized by subtracting the starting %GFP. All cell growth assays were performed in triplicate (three independent transduction experiments).
Retrovirus production and transduction
Retroviruses were produced using calcium phosphate co-transfection of 15 μg of construct and 15 μg of pCL-Eco packaging plasmid into 293T cells. 293T cell medium was replaced with G1E medium after 24 h. Virus-containing G1E medium was then harvested after an additional 24 h and passed through a 0.2-μm filter. Viral supernatants were diluted in G1E medium to find titers that yield 30–40% GFP/YFP+ cells. Retroviral transduction was performed by adding 250 μl of titered virus to one million G1E-ER4 (or BRD2 KO) cells in 1 ml of G1E medium supplemented with 4 μl/ml Polybrene and 10 μl/ml 1 m HEPES (Gibco, 15630080) in 12-well plates and spinning at 2000 rpm in a centrifuge for 90 min. The supernatant was removed, and cells were resuspended in 5 ml of fresh G1E medium. Fluorescence was measured 24 h later on a flow cytometer.
TER119 staining, flow cytometry, and cell sorting
Flow cytometry was performed using standard antibody staining protocols and cytometry settings. Briefly, 105 to 106 cells were harvested and washed with PBS. For TER119 expression experiments, cells were infected with low-titer YFP-BET encoding (or empty vector control) virus to guarantee single copy gene integration. After 24 h of rest, estradiol was added to the mixed population of infected/uninfected cells for an additional 24 h. Cells were then resuspended in 100 μl of staining buffer (PBS, 2% FBS) with a 1:200 dilution of anti-TER119-APC (Biolegend, 116211) and incubated on ice for 30 min. Samples were washed three times with PBS and resuspended in 200 μl of staining buffer and analyzed by flow cytometry by gating on YFP− or YFP+ cells. Derivation of stable YFP-BET expressing cell lines was achieved by iterative FACS with narrow sorting gates to achieve similar YFP mean fluorescence intensity between cell lines.
Whole-cell extracts
Whole-cell extracts were prepared using standard laboratory techniques. Briefly, 1–3e6 cells were harvested for each sample. After washing in ice-cold PBS, cells were resuspended in 50–100 μl of radioimmune precipitation assay buffer (150 mm NaCl, 0.5% sodium deoxycholate, 50 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 0.1% SDS, and freshly added 2ul/ml protease inhibitor mixture (Sigma, P8340) and 1 mm phenylmethylsulfonyl fluoride) and incubated on ice for 30 min. Lysed cells were sonicated in a Qsonica sonicator (model Q800R) for 3 × 30-s on/off cycles at maximum amplitude in a water bath chilled to 4 °C. Insoluble material was pelleted by centrifugation at 15,000 rpm for 5 min at 4 °C in a microcentrifuge. The supernatant was transferred to fresh microcentrifuge tubes and used immediately or snap-frozen in liquid nitrogen and stored at −80 °C. Protein concentration was determined using the BCA protein assay (Pierce, 23225) with absorbance measured at 562 nm compared with a BSA protein standard curve.
Western blotting
Western blots were performed using standard laboratory techniques. Briefly, protein samples were resuspended in 4× sample buffer (500 mm Tris-HCl, pH 6.8, 8% SDS, 20% 2-mercaptoethanol, 0.3% bromphenol blue, 30% glycerol) and denatured at 95 °C for 10 min. Equal protein amounts were loaded and separated on 4–15% or 4–20% gradient precast gels (Bio-Rad, 4561084 or 4561096) and transferred to nitrocellulose membranes (Bio-Rad, 1620115). Primary antibodies include Brd2 N-term (Cell Signaling, 5848S), Brd2 C-term (Bethyl, A302-583A), Brd3 (Active Motif, 61489), Brd4L (Bethyl, A301-985A), Brd4 N-term (Abcam, ab128874), HA (monoclonal 12CA5, purified in-house; or Cell Signaling, C29F4, for fluorescent Western blots), GFP/YFP (Abcam, ab290), proliferating cell nuclear antigen (Santa Cruz Biotechnology, Inc., sc-9857), and β-actin-peroxidase (Sigma, A3854). Secondary staining reagents include goat anti-rabbit IgG-HRP (Abcam, ab6721), donkey anti-goat IgG-HRP (Santa Cruz Biotechnology, sc-2020) and Protein A-HRP (Thermo Fisher Scientific, 101023). Stained membranes were imaged using enhanced chemiluminescent substrate (Thermo Scientific, 34080 or 34095) and autoradiography film (Denville Scientific, E3012). For fluorescent Western blots, fluorescent secondary antibodies conjugated to IRDye-680 or IRDye-800 and the Odyssey imaging system and buffers were used (LI-COR).
Coiled-coil prediction
Coiled-coil prediction was performed by inputting the UniProtKB amino acid sequences for the mouse BET proteins BRD2 (Q7JJ13-1), BRD3 (Q8K2F0-1), BRD4S (Q9ESU6-2), and BRD4L (Q9ESU6-1) into a Hidden Markov Model coiled-coil prediction algorithm (https://toolkit.tuebingen.mpg.de/#/tools/marcoil) (44). To generate the plots in Fig. 4A, the output was graphed and scaled to the BET protein diagrams using ggplot2 in R. The sequence alignments of the regions surrounding and containing the ET and CC domains in Fig. 4B were generated using Jalview software (68) for the indicated UniRef entry names and fragments. Sequences were sorted by length, and individual residues were labeled using Clustal X default coloring and thresholding.
Structural assessment of human BRD2/3-CC
The coiled-coil domain of human BRD2(715–757) and that of human BRD3(641–688) were cloned into pGEX6p and expressed with a GST tag. BL21 (DE3) colonies transformed with respective plasmids were inoculated into lysogeny broth and, after overnight incubation, were used to inoculate 1–2 liters of culture. The culture was induced at A600 = 0.75 with 0.5 mm isopropyl β-d-1-thiogalactopyranoside, followed by overnight expression at 25 °C.
The cell pellet was resuspended in 50 mm Tris, pH 7.2, 500 mm NaCl, 0.1% Triton X-100, 1 mm DTT, 10 μg/ml DNase I, 10 μg/ml RNase A, and 1× cOmplete EDTA-free protease inhibitor (Roche Applied Science, 11873580001) at ∼10 ml/g of cells and then lysed via sonication. After clarification at 15,000 × g for 30 min at 4 °C, the clear lysate was incubated with pre-equilibrated GSH-Sepharose 4B beads (GE Healthcare, 17075601) for 1.5 h at 4 °C with mixing. The beads were washed with 3 × 5 column volumes of GST wash buffer containing 10 mm Tris, pH 7.2, 100 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine. The proteins were eluted with wash buffer containing 50 mm reduced GSH. The GST tag was cleaved using HRV-3C protease by incubating with the eluted protein overnight. On the next day, the cleavage protein sample was concentrated and then injected onto a pre-equilibrated (in the GST wash buffer) SuperdexTM 75 16/600 column, and the gel-filtration chromatography was run at 1 ml/min for 1.25× column volumes with 1-ml fractions collected. Fractions containing the desired bands were combined and concentrated to 700 μm for SEC-MALLS analysis.
SEC-MALLS analysis runs were performed on either a SuperdexTM 75 10/300 GL column or a SuperdexTM peptide 10/30 GL column (GE Healthcare) with an in-line MiniDawn MALLS detector with a laser source at 690 nm (Wyatt Technology, Santa Barbara, CA) and Wyatt refractometer. Proteins were eluted in 10 mm Tris, pH 7.2, 100 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine using a flow rate of 0.5 ml/min. The weight-average molecular weight was calculated using the intensity of scattered light at 90° in combination with the change in refractive index. Protein concentration at the detector was determined by the change in refractive index. All experiments were performed at room temperature (25 ºC). Data collection and SEC-MALLS analysis were performed with ASTRA version 6.1 software (Wyatt Technology).
CD spectropolarimetry spectra were recorded on a Jasco J-720 spectropolarimeter using a 1-mm quartz cuvette. In each case, spectra comprised the sum of three successive spectra with a step size of 0.5 nm, a 1-s response time, and a 1-nm bandwidth. Data were acquired at 4 °C. Secondary structure content was estimated using Raussens' method (http://perry.freeshell.org/raussens.html)4 (69).
GST pulldown and proteomic analysis
The sequences from mouse BRD2-mB, BRD2-ET, BRD2-CC, BRD2-CCp, BRD2-CCd, BRD2-ETCC, BRD3-ET, BRD3-ETCC, BRDT-ET, and BRDT-ETCC (Table S5) were cloned into the pGEX-2T GST fusion vector. The GST fusion proteins were expressed and purified as described (37). GST pulldown assays were performed as described using 2.5 μg of GST protein and 500 μg of nuclear extract from MEL cells (37).
LC-MS/MS analysis was performed by the Proteomics and Metabolomics Facility at the Wistar Institute using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled with a Nano-ACQUITY UPLC system (Waters). Samples were digested in-gel with trypsin and injected onto a UPLC Symmetry trap column (180-μm inner diameter × 2 cm packed with 5-μm C18 resin; Waters). Tryptic peptides were separated by reverse-phase HPLC on a BEH C18 nanocapillary analytical column (75-μm inner diameter × 25 cm, 1.7-μm particle size; Waters) using a 95-min gradient formed by solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). A 30-min blank gradient was run between sample injections to minimize carryover. Eluted peptides were analyzed by the mass spectrometer set to repetitively scan m/z from 400 to 2000 in positive-ion mode. The full MS scan was collected at 70,000 resolution followed by data-dependent MS/MS scans at 17,500 resolution on the 20 most abundant ions exceeding a minimum threshold of 20,000. Peptide match was set as preferred, and the “exclude isotopes” and “charge-state screening” options were enabled to reject singly and unassigned charged ions.
Peptide sequences were identified using MaxQuant version 1.5.2.8. MS/MS spectra were searched against a UniProt mouse protein database using full tryptic specificity with up to two missed cleavages, static carboxamidomethylation of Cys, and variable oxidation of Met and protein N-terminal acetylation. Protein identification required a minimum of two unique peptides. False discovery rates for protein and peptide identifications were set at 1%. STRING protein interaction network analysis was performed using the online tool (https://string-db.org/)4 (72) with the highest confidence setting (0.900).
Author contributions
M. T. W. and G. A. B. conceptualization; M. T. W., H. W., N. H., S. C. H., J. A. Y., A. J. S., V. B. and Y. Z. investigation; M. T. W. writing-original draft; M. T. W. and G. A. B. writing-review and editing; J. P. M. resources; J. P. M. methodology; G. A. B. funding acquisition; G. A. B. project administration.
Supplementary Material
Acknowledgments
We thank Zhe (Jim) Zhang and Perry Evans (Children's Hospital of Philadelphia Department of Biomedical and Health Informatics) for bioinformatic support, Tapan Ganguly and Erik Toorens (Perelman School of Medicine Penn Genomics Analysis Core) for assistance with RNA-Seq, Hsin-Yao Tang and Thomas Beer (Wistar Institute Proteomics and Metabolomics Facility) for assistance with mass spectrometry, Kristen Jahn for technical assistance, and members of the Blobel laboratory for critical reading of the manuscript.
Note added in proof
Yichen Zhong was inadvertently omitted as an author on the version of this article that was published as a Paper in Press on December 2,2019. This error has now been corrected.
This work was supported by National Institutes of Health Grants R01DK054937 and R37DK058044 (to G. A. B.) and T32HL007439 and F30DK112573 (to M. T. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The FASTQ files have been deposited with links to BioProject accession number PRJNA560407 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
This article contains Tables S1–S5 and Figs. S1–S8.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- BET
- bromodomain and extraterminal motif
- CC
- coiled-coil
- SEC-MALLS
- size-exclusion chromatography with inline multiangle light scattering
- PAF
- RNA polymerase II–associated factor
- CK2
- casein kinase II
- KO
- knockout
- FDR
- false discovery rate
- YFP
- yellow fluorescent protein
- qPCR
- quantitative PCR
- ET
- extraterminal domain/motif
- IRES
- internal ribosome entry site
- HRP
- horseradish peroxidase.
References
- 1. Marmorstein R., Zhou M. M. (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belkina A. C., Denis G. V. (2012) BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer. 12, 465–477 10.1038/nrc3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taniguchi Y. (2016) The bromodomain and extra-terminal domain (BET) family: functional anatomy of BET paralogous proteins. Int. J. Mol. Sci. 17, E1849 10.3390/ijms17111849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin Y.-J., Umehara T., Inoue M., Saito K., Kigawa T., Jang M. K., Ozato K., Yokoyama S., Padmanabhan B., Güntert P. (2008) Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 17, 2174–2179 10.1110/ps.037580.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rahman S., Sowa M. E., Ottinger M., Smith J. A., Shi Y., Harper J. W., and Howley P. M. (2011) The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 31, 2641–2652 10.1128/MCB.01341-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crowe B. L., Larue R. C., Yuan C., Hess S., Kvaratskhelia M., and Foster M. P. (2016) Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction. Proc. Natl. Acad Sci. 113, 2086–2091 10.1073/pnas.1516813113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Q., Zeng L., Shen C., Ju Y., Konuma T., Zhao C., Vakoc C. R., and Zhou M. M. (2016) Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure 24, 1201–1208 10.1016/j.str.2016.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wai D. C. C., Szyszka T. N., Campbell A. E., Kwong C., Wilkinson-White L. E., Silva A. P. G., Low J. K. K., Kwan A. H., Gamsjaeger R., Chalmers J. D., Patrick W. M., Lu B., Vakoc C. R., Blobel G. A., and Mackay J. P. (2018) The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators. J. Biol. Chem. 293, 7160–7175 10.1074/jbc.RA117.000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambert J. P., Picaud S., Fujisawa T., Hou H., Savitsky P., Uusküla-Reimand L., Gupta G. D., Abdouni H., Lin Z. Y., Tucholska M., Knight J. D. R., Gonzalez-Badillo B., St-Denis N., Newman J. A., Stucki M., et al. (2019) Interactome rewiring following pharmacological targeting of BET bromodomains. Mol. Cell. 73, 621–638.e17 10.1016/j.molcel.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W. B., Fedorov O., Morse E. M., Keates T., Hickman T. T., Felletar I., Philpott M., Munro S., McKeown M. R., Wang Y., Christie A. L., et al. (2010) Selective inhibition of BET bromodomains. Nature 468, 1067–1073 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nicodeme E., Jeffrey K. L., Schaefer U., Beinke S., Dewell S., Chung C. W., Chandwani R., Marazzi I., Wilson P., Coste H., White J., Kirilovsky J., Rice C. M., Lora J. M., Prinjha R. K., Lee K., and Tarakhovsky A. (2010) Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 10.1038/nature09589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith S. G., and Zhou M. M. (2016) The bromodomain: a new target in emerging epigenetic medicine. ACS Chem. Biol. 11, 598–608 10.1021/acschembio.5b00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anders L., Guenther M. G., Qi J., Fan Z. P., Marineau J. J., Rahl P. B., Lovén J., Sigova A. A., Smith W. B., Lee T. I., Bradner J. E., and Young R. A. (2014) Genome-wide localization of small molecules. Nat. Biotechnol. 32, 92–96 10.1038/nbt.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jang M. K., Mochizuki K., Zhou M., Jeong H. S., Brady J. N., and Ozato K. (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 19, 523–534 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- 15. Yang Z., Yik J. H. N., Chen R., He N., Jang M. K., Ozato K., and Zhou Q. (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 19, 535–545 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- 16. Bisgrove D. A., Mahmoudi T., Henklein P., and Verdin E. (2007) Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 13690–13695 10.1073/pnas.0705053104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conrad R. J., Fozouni P., Thomas S., Sy H., Zhang Q., Zhou M. M., and Ott M. (2017) The short isoform of BRD4 promotes HIV-1 latency by engaging repressive SWI/SNF chromatin-remodeling complexes. Mol Cell. 67, 1001–1012.e6 10.1016/j.molcel.2017.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alsarraj J., Faraji F., Geiger T. R., Mattaini K. R., Williams M., Wu J., Ha N. H., Merlino T., Walker R. C., Bosley A. D., Xiao Z., Andresson T., Esposito D., Smithers N., Lugo D., et al. (2013) BRD4 short isoform interacts with RRP1B, SIPA1 and components of the LINC complex at the inner face of the nuclear membrane. PLoS One 8, e80746 10.1371/journal.pone.0080746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deeney J. T., Belkina A. C., Shirihai O. S., Corkey B. E., and Denis G. V. (2016) BET bromodomain proteins Brd2, Brd3 and Brd4 selectively regulate metabolic pathways in the pancreatic β-cell. PLoS One 11, e0151329 10.1371/journal.pone.0151329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andrieu G. P., and Denis G V. (2018) BET proteins exhibit transcriptional and functional opposition in the epithelial-to-mesenchymal transition. Mol. Cancer Res. 16, 580–586 10.1158/1541-7786.MCR-17-0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shang E., Wang X., Wen D., Greenberg D. A., and Wolgemuth D. J. (2009) Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 238, 908–917 10.1002/dvdy.21911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gyuris A., Donovan D. J., Seymour K. A., Lovasco L. A., Smilowitz N. R., Halperin A. L., Klysik J. E., and Freiman R. N. (2009) The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim. Biophys. Acta 1789, 413–421 10.1016/j.bbagrm.2009.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houzelstein D., Bullock S. L., Lynch D. E., Grigorieva E. F., Wilson V. A., and Beddington R. S. P. (2002) Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22, 3794–3802 10.1128/MCB.22.11.3794-3802.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang F., Liu H., Blanton W. P., Belkina A., Lebrasseur N. K., and Denis G. V. (2009) Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem. J. 425, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y., Wang X., Zhang J., Huang H., Ding B., Wu J., and Shi Y. (2008) Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry 47, 6403–6417 10.1021/bi8001659 [DOI] [PubMed] [Google Scholar]
- 26. Morinière J., Rousseaux S., Steuerwald U., Soler-López M., Curtet S., Vitte A. L., Govin J., Gaucher J., Sadoul K., Hart D. J., Krijgsveld J., Khochbin S., Müller C. W., and Petosa C. (2009) Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461, 664–668 10.1038/nature08397 [DOI] [PubMed] [Google Scholar]
- 27. Vollmuth F., Blankenfeldt W., and Geyer M. (2009) Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J. Biol. Chem. 284, 36547–36556 10.1074/jbc.M109.033712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Umehara T., Nakamura Y., Wakamori M., Ozato K., Yokoyama S., and Padmanabhan B. (2010) Structural implications for K5/K12-di-acetylated histone H4 recognition by the second bromodomain of BRD2. FEBS Lett. 584, 3901–3908 10.1016/j.febslet.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Umehara T., Nakamura Y., Jang M. K., Nakano K., Tanaka A., Ozato K., Padmanabhan B., and Yokoyama S. (2010) Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J. Biol. Chem. 285, 7610–7618 10.1074/jbc.M109.062422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J. P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T., Gingras A. C., Arrowsmith C. H., and Knapp S. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 10.1016/j.cell.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Gutierrez P., Mundi M., and Garcia-Dominguez M. (2012) Association of bromodomain BET proteins with chromatin requires dimerization through the conserved motif B. J. Cell Sci. 125, 3671–3680 10.1242/jcs.105841 [DOI] [PubMed] [Google Scholar]
- 32. Hnilicová J., Hozeifi S., Stejskalová E., Dušková E., Poser I., Humpolíčková J., Hof M., and Staněk D. (2013) The C-terminal domain of Brd2 is important for chromatin interaction and regulation of transcription and alternative splicing. Mol. Biol. Cell. 24, 3557–3568 10.1091/mbc.e13-06-0303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dawson M. A., Prinjha R. K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W. I., Robson S. C., Chung C. W., Hopf C., Savitski M. M., Huthmacher C., Gudgin E., Lugo D., Beinke S., Chapman T. D., et al. (2011) Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stonestrom A. J., Hsu S. C., Werner M. T., and Blobel G. A. (2016) Erythropoiesis provides a BRD's eye view of BET protein function. Drug Discov. Today Technol. 19, 23–28 10.1016/j.ddtec.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferreira R., Ohneda K., Yamamoto M., and Philipsen S. (2005) GATA1 Function, a paradigm for transcription factors in hematopoiesis. Mol. Cell. Biol. 25, 1215–1227 10.1128/MCB.25.4.1215-1227.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stonestrom A. J., Hsu S. C., Jahn K. S., Huang P., Keller C. A., Giardine B. M., Kadauke S., Campbell A. E., Evans P., Hardison R. C., and Blobel G. A. (2015) Functions of BET proteins in erythroid gene expression. Blood 125, 2825–2834 10.1182/blood-2014-10-607309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamonica J. M., Deng W., Kadauke S., Campbell A. E., Gamsjaeger R., Wang H., Cheng Y., Billin A. N., Hardison R. C., Mackay J. P., and Blobel G. A. (2011) Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc. Natl. Acad Sci. U.S.A. 108, E159–E168 10.1073/pnas.1102140108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lamonica J. M., Vakoc C. R., and Blobel G. A. (2006) Acetylation of GATA-1 is required for chromatin occupancy. Blood 108, 3736–3738 10.1182/blood-2006-07-032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., and Weiss M. J. (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147 10.1182/blood-2004-04-1603 [DOI] [PubMed] [Google Scholar]
- 40. Paralkar V. R., Mishra T., Luan J., Yao Y., Kossenkov A. V., Anderson S. M., Dunagin M., Pimkin M., Gore M., Sun D., Konuthula N., Raj A., An X., Mohandas N., Bodine D. M., et al. (2014) Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 123, 1927–1937 10.1182/blood-2013-12-544494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lovén J., Hoke H. A., Lin C. Y., Lau A., Orlando D. A., Vakoc C. R., Bradner J. E., Lee T. I., and Young R. A. (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo N., Faller D. V., Denis G. V. (2000) Activation-induced nuclear translocation of RING3. J. Cell Sci. 113, 3085–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukazawa H., and Masumi A. (2012) The conserved 12-amino acid stretch in the inter-bromodomain region of BET family proteins functions as a nuclear localization signal. Biol. Pharm. Bull. 35, 2064–2068 10.1248/bpb.b12-00527 [DOI] [PubMed] [Google Scholar]
- 44. Delorenzi M., and Speed T. (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 10.1093/bioinformatics/18.4.617 [DOI] [PubMed] [Google Scholar]
- 45. Truebestein L., and Leonard T. A. (2016) Coiled-coils: the long and short of it. BioEssays 38, 903–916 10.1002/bies.201600062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen C., Ipsaro J. J., Shi J., Milazzo J. P., Wang E., Roe J. S., Suzuki Y., Pappin D. J., Joshua-Tor L., and Vakoc C. R. (2015) NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell 60, 847–859 10.1016/j.molcel.2015.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olley G., Ansari M., Bengani H., Grimes G. R., Rhodes J., von Kriegsheim A., Blatnik A., Stewart F. J., Wakeling E., Carroll N., Ross A., Park S. M., Deciphering Developmental Disorders Study, Bickmore W. A., Pradeepa M. M., and FitzPatrick D. R. (2018) BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange-like syndrome. Nat. Genet. 50, 329–332 10.1038/s41588-018-0042-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luna-Peláez N., March-Díaz R., Ceballos-Chávez M., Guerrero-Martínez J. A., Grazioli P., García-Gutiérrez P., Vaccari T., Massa V., Reyes J. C., and García-Domínguez M. (2019) The Cornelia de Lange Syndrome-associated factor NIPBL interacts with BRD4 ET domain for transcription control of a common set of genes. Cell Death Dis. 10, 548 10.1038/s41419-019-1792-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luna-Peláez N., and García-Domínguez M. (2018) Lyar-mediated recruitment of Brd2 to the chromatin attenuates Nanog downregulation following induction of differentiation. J. Mol. Biol. 430, 1084–1097 10.1016/j.jmb.2018.02.023 [DOI] [PubMed] [Google Scholar]
- 50. Izumikawa K., Ishikawa H., Yoshikawa H., Fujiyama S., Watanabe A., Aburatani H., Tachikawa H., Hayano T., Miura Y., Isobe T., Simpson R. J., Li L., Min J., and Takahashi N. (2019) LYAR potentiates rRNA synthesis by recruiting BRD2/4 and the MYST-type acetyltransferase KAT7 to rDNA. Nucleic Acids Res. 47, 10357–10372 10.1093/nar/gkz747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vos S. M., Farnung L., Boehning M., Wigge C., Linden A., Urlaub H., and Cramer P. (2018) Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature 560, 607–612 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- 52. Kim J., Guermah M., and Roeder R. G. (2010) The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491–503 10.1016/j.cell.2009.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu M., Yang W., Ni T., Tang Z., Nakadai T., Zhu J., and Roeder R. G. (2015) RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350, 1383–1386 10.1126/science.aad2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ji X., Dadon D. B., Abraham B. J., et al. (2015) Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl. Acad. Sci. U.S.A. 112, 3841–3846 10.1073/pnas.1502971112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., and Greenblatt J. F. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992 10.1128/MCB.22.20.6979-6992.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berthon C., Raffoux E., Thomas X., Vey N., Gomez-Roca C., Yee K., Taussig D. C., Rezai K., Roumier C., Herait P., Kahatt C., Quesnel B., Michallet M., Recher C., Lokiec F., et al. (2016) Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 3, e186–e195 10.1016/S2352-3026(15)00247-1 [DOI] [PubMed] [Google Scholar]
- 57. Amorim S., Stathis A., Gleeson M., Iyengar S., Magarotto V., Leleu X., Morschhauser F., Karlin L., Broussais F., Rezai K., Herait P., Kahatt C., Lokiec F., Salles G., Facon T., et al. (2016) Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 3, e196–e204 10.1016/S2352-3026(16)00021-1 [DOI] [PubMed] [Google Scholar]
- 58. LeRoy G., Chepelev I., DiMaggio P. A., Blanco M. A., Zee B. M., Zhao K., and Garcia B. A. (2012) Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 13, R68 10.1186/gb-2012-13-8-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Asangani I. A., Dommeti V. L., Wang X., Malik R., Cieslik M., Yang R., Escara-Wilke J., Wilder-Romans K., Dhanireddy S., Engelke C., Iyer M. K., Jing X., Wu Y. M., Cao X., Qin Z. S., et al. (2014) Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 10.1038/nature13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen Y., Xu L., Mayakonda A., Huang M. L., Kanojia D., Tan T. Z., Dakle P., Lin R. Y., Ke X. Y., Said J. W., Chen J., Gery S., Ding L. W., Jiang Y. Y., Pang A., et al. (2019) Bromodomain and extraterminal proteins foster the core transcriptional regulatory programs and confer vulnerability in liposarcoma. Nat. Commun. 10, 1353 10.1038/s41467-019-09257-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fong C. Y., Gilan O., Lam E. Y. N., Rubin A. F., Ftouni S., Tyler D., Stanley K., Sinha D., Yeh P., Morison J., Giotopoulos G., Lugo D., Jeffrey P., Lee S. C., Carpenter C., et al. (2015) BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 10.1038/nature14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khoueiry P., Ward Gahlawat A., Petretich M., Michon A. M., Simola D., Lam E., Furlong E. E., Benes V., Dawson M. A., Prinjha R. K., Drewes G., and Grandi P. (2019) BRD4 bimodal binding at promoters and drug-induced displacement at Pol II pause sites associates with I-BET sensitivity. Epigenetics Chromatin 12, 39 10.1186/s13072-019-0286-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fontanals-Cirera B., Hasson D., Vardabasso C., Di Micco R., Agrawal P., Chowdhury A., Gantz M., de Pablos-Aragoneses A., Morgenstern A., Wu P., Filipescu D., Valle-Garcia D., Darvishian F., Roe J. S., Davies M. A., et al. (2017) Harnessing BET inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene. Mol Cell. 68, 731–744.e9 10.1016/j.molcel.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Di Micco R., Fontanals-Cirera B., Low V., Ntziachristos P., Yuen S. K., Lovell C. D., Dolgalev I., Yonekubo Y., Zhang G., Rusinova E., Gerona-Navarro G., Cañamero M., Ohlmeyer M., Aifantis I., Zhou M. M., et al. (2014) Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 9, 234–247 10.1016/j.celrep.2014.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Muhar M., Ebert A., Neumann T., Umkehrer C., Jude J., Wieshofer C., Rescheneder P., Lipp J. J., Herzog V. A., Reichholf B., Cisneros D. A., Hoffmann T., Schlapansky M. F., Bhat P., von Haeseler A., et al. (2018) SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 360, 800–805 10.1126/science.aao2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weiss M. J., Yu C., and Orkin S. H. (1997) Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17, 1642–1651 10.1128/MCB.17.3.1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hsu S. C., Gilgenast T. G., Bartman C. R., Edwards C. R., Stonestrom A. J., Huang P., Emerson D. J., Evans P., Werner M. T., Keller C. A., Giardine B., Hardison R. C., Raj A., Phillips-Cremins J. E., and Blobel G. A. (2017) The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol. Cell. 66, 102–116.e7 10.1016/j.molcel.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. (2009) Jalview version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Raussens V., Ruysschaert J. M., and Goormaghtigh E. (2003) Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: a new scaling method. Anal. Biochem. 319, 114–121 10.1016/S0003-2697(03)00285-9 [DOI] [PubMed] [Google Scholar]
- 70. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mootha V. K., Lindgren C. M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., et al. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 72. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.