Abstract
Macrolide antibiotics, such as erythromycin and josamycin, are natural polyketide products harboring 14- to 16-membered macrocyclic lactone rings to which various sugars are attached. These antibiotics are used extensively in the clinic because of their ability to inhibit bacterial protein synthesis. More recently, some macrolides have been shown to also possess anti-inflammatory and other therapeutic activities in mammalian cells. To better understand the targets and effects of this drug class in mammalian cells, we used a genome-wide shRNA screen in K562 cancer cells to identify genes that modulate cellular sensitivity to josamycin. Among the most sensitizing hits were proteins involved in mitochondrial translation and the mitochondrial unfolded protein response, glycolysis, and the mitogen-activated protein kinase signaling cascade. Further analysis revealed that cells treated with josamycin or other antibacterial agents exhibited impaired oxidative phosphorylation and metabolic shifts to glycolysis. Interestingly, we observed that knockdown of the mitogen-activated protein kinase kinase kinase 4 (MAP3K4) gene, which contributes to p38 mitogen-activated protein kinase signaling, sensitized cells only to josamycin but not to other antibacterial agents. There is a growing interest in better characterizing the therapeutic effects and toxicities of antibiotics in mammalian cells to guide new applications in both cellular and clinical studies. To our knowledge, this is the first report of an unbiased genome-wide screen to investigate the effects of a clinically used antibiotic on human cells.
Keywords: antibiotics, shRNA, drug action, mitochondria, natural product, erythromycin, josamycin, mitochondrial translation
Introduction
A subgroup of macrolide antibiotics, characterized by 14- to 16-membered macrocycles to which various sugar substituents are attached, are among the most widely used antibacterial agents to date (Fig. 1A). In the early 1950s, erythromycin, a 14-membered macrolide, was isolated from Saccharopolyspora erythraea and became the first macrolide adopted clinically (1). Usually serving as bacteriostatic agents, macrolides bind the bacterial 50S ribosomal subunit within the polypeptide exit tunnel and inhibit protein synthesis in the pathogen (2). In addition to broad-spectrum activity against Gram-positive and atypical bacteria, these antibiotics have excellent tissue penetration, especially newer antibacterial agents, such as azithromycin and clarithromycin (1, 3). They accumulate in lung epithelial fluid and immune cells at concentrations of up to several hundred times greater than in extracellular fluid, granting these molecules rapid access to sites of infection (1, 4). For these reasons, they are widely prescribed for respiratory tract infections as well as skin, gastrointestinal, sexually transmitted, and other infections (3, 5) and play important roles in veterinary medicine (6). In addition to the three main macrolide antibiotics approved for human use in the United States, at least five others, including spiramcyin and josamycin (also known as leucomycin A3), are used clinically in other parts of the world (Fig. 1A) (7).
Figure 1.
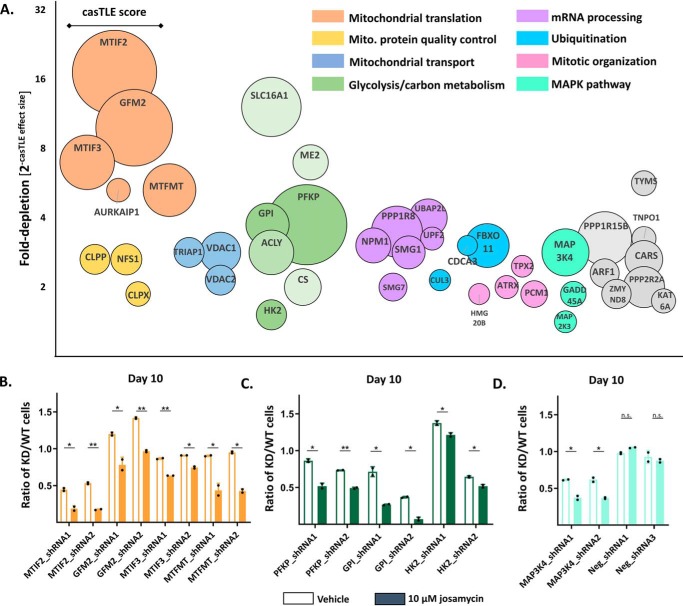
Identifying genetic modifiers of cellular sensitivity to josamycin in K562 cells through an shRNA screen. A, structures of common macrolide antibacterial agents (erythromycin, clarithromycin, and josamycin) and the nonmacrolide antibacterial chloramphenicol. B, josamycin dose–response in K562 cells after 48 h. Points indicate means ± S.D.; n = 3. C, overview of the shRNA screening process in K562 cells.
Beyond antibacterial activity, macrolides have been extensively investigated and are sometimes prescribed for their immunomodulatory and other therapeutic properties, especially for respiratory conditions (1, 8–13). In the 1980s, low-dose erythromycin led to impressive survival benefits in patients with diffuse panbronchiolitis by a mechanism seemingly unrelated to its antibacterial effect (11, 14, 15). Since then, macrolides have been evaluated in many clinical studies of respiratory conditions such as asthma (16), cystic fibrosis (17), and chronic obstructive pulmonary disease (11, 18). In parallel, targeted in vitro analyses of immune and other cells exposed to macrolides have ascribed the observed immunomodulatory effects to changes in cytokine production, adhesion molecule expression, neutrophil chemotaxis, and more; mechanisms proposed to explain these changes include shifts in intracellular calcium levels, mitogen-activated protein kinase (MAPK)5 signaling, and the activity of transcription factors such as NF-κB (1, 9). More recently, researchers have conducted more extensive, unbiased characterizations of the effects of antibiotics in mammalian cells. Examples range from examining global gene expression changes in human cells treated with azithromycin or doxycycline (19, 20) to characterizing tetracycline toxicities in various model organisms (21). These works are timely, given a growing appreciation for and cautious interest in repurposing antibiotics for their alternative therapeutic effects (11–13, 22). More research regarding the targets and effects of macrolides in mammalian cells can guide new applications in cellular and clinical studies.
Over the past few years, many unbiased proteomic and genomic methods have been advanced to interrogate the interactions of bioactive small molecules with cellular protein targets. These approaches can generally be classified into affinity-based biochemical methods, computational inference methods based on gene expression or ligand similarity, and functional genomic methods (23). Within the last category, high-throughput screens utilizing genome-wide shRNA knockdown (KD) and CRISPR-Cas9 KO libraries have become especially popular (24). By characterizing how gene KD or KO affect cellular sensitivity to a drug, one can glean insights into both a drug's direct subcellular binding targets as well as its indirect interactions and effects on cellular signaling and processes. We previously conducted parallel shRNA and CRISPR-Cas9 screens to successfully identify genetic modifiers of cellular sensitivity to the host-targeting antiviral molecule GSK983 (25). In that study, the primary direct binding target of GSK983 was identified among the top sensitizing genes of the shRNA screen rather than the CRISPR-Cas9 screen (25). Other drug targets and mechanisms of action identified through shRNA screens include those of the anti-leukemic agent STF-118804 (26) and the anti-depressive agent ISRIB (27). shRNA screens have also helped to map out genes that modify cellular sensitivity to anti-cancer drugs, including the MDM2 inhibitor Nutlin-3 (28), the BCR-ABL inhibitor imatinib (29), the topoisomerase inhibitor etoposide (30), and the bioactive natural product aurilide B (31).
We chose josamycin, a first-line treatment for respiratory infection in Russia and a second-line treatment for Chlamydia trachomatis in Europe (32), as our primary target for characterization on the basis of its unusual immunomodulatory characteristics (33, 34) and impressive host-targeting antiviral activity (35). Here we conducted a genome-wide shRNA screen with josamycin in the well-studied K562 human erythroleukemia cell line. Although human targets of macrolide antibiotics have been investigated for over 50 years, to our knowledge, our study is the first to address this question using an unbiased functional genomics platform.
Results
Unbiased genome-wide shRNA screen for targets of josamycin in K562 cells
We previously optimized a high-density shRNA library (∼25 shRNAs per protein-coding gene and ∼10,000 negative controls) and validated its performance in pooled genetic screens (36, 37). Using this shRNA library, we conducted a genome-wide screen to identify gene knockdowns that greatly altered the sensitivity of the human K562 erythroleukemia cell line to josamycin. Because sensitivity to macrolide antibiotics varies based on drug and cell type (35, 38), josamycin was titrated into a K562 cell culture and found to reduce cell growth with an IC50 of 39 μm (Fig. 1B). Upon stable infection of K562 cells with the shRNA library, these cells were split and cultured in the absence or presence of 10 μm josamycin for 14 days (Fig. 1C). To identify which cells (and therefore genes) were selectively depleted or enriched in the josamycin-treated population, genomic DNA was isolated from each population after 14 days. The shRNA-encoding cassettes from each population were amplified and subjected to deep sequencing (39). The individual genes conferring sensitivity to or protection from josamycin were ranked according to a maximum likelihood estimator that incorporates results across the entire set of shRNAs targeting each gene (Table S1) (25). To improve statistical confidence in the identified hits, a subsequent batch shRNA screen was performed on a smaller scale with a custom-synthesized library that was comprised of the top ∼500 genes identified from the genome-wide screen to obtain higher coverage (Table S2).
Genes involved in mitochondrial translation, central carbon metabolism, and MAPK signaling sensitize K562 cells to josamycin
Gene knockdowns that modify cellular sensitivity to josamycin provide insight into the drug's direct subcellular binding targets as well as its indirect effects on cellular signaling and processes. Analysis of the top 40 sensitizing genes identified in the secondary screen revealed that major pathways sensitizing K562 cells to josamycin were involved in mitochondrial function and central carbon metabolism (Fig. 2A and Fig. S1A). In fact, the genes that most strongly sensitized cells to josamycin included those responsible for critical steps in mitochondrial protein translation, including initiation (MTIF2 and MTIF3), formylation (MTFMT), and termination (GFM2). Other macrolide antibacterial agents as well as nonmacrolide inhibitors of bacterial translation (e.g. chloramphenicol, Fig. 1A) are known to affect mitochondrial function, presumably by directly binding to mitochondrial ribosomes, given their strong similarity to bacterial ribosomes (40, 41). Therefore, it is almost certain that the mitochondrial ribosome is a direct binding target of josamycin, given prior literature and the top hits in our screen.
Figure 2.
Results of the shRNA screen for genes that sensitize cells to josamycin. A, the top 40 sensitizing hits from the batch shRNA screen. Circle size is proportional to the casTLE score, a measure of statistical confidence in each gene. The y axis is 2(−casTLE effect size), which describes the normalized -fold depletion of a specific gene in the treated population. The x axis does not represent any quantitative feature. B, individual shRNA validations of the mitochondrial translation hits MTIF2, GFM2, MTIF3, and MTFMT using a competitive growth assay. The ratio of KD/WT cells is the proportion of knockdown to WT K562 cells after 10 days of coculture initiated at a 1:1 ratio in an untreated (unshaded columns) versus a 10 μm josamycin–treated population (shaded columns). C, individual shRNA validation of the glycolysis hits Gpi, Pfkp, and Hk2 using a competitive growth assay. D, individual shRNA validation of Map3k4 and nontargeting shRNAs using a competitive growth assay. In B–D, error bars indicate mean ± S.D., n = 2, and dots represent individual values. Differences in the KD/WT ratio between DMSO- and drug-treated cells are indicated: *, p < 0.05; **, p < 0.01; n.s., p > 0.05, calculated as detailed under “Experimental procedures.”
Other notable mitochondrial hits included genes involved in mitochondrial protein quality control and unfolded protein response (CLPP and CLPX) as well as anion (VDAC1 and VDAC2) and phosphatidic acid (TRIAP1) transport. A second major category of sensitizing hits was genes involved in central carbon metabolism, including glycolytic enzymes (GPI, PFKP, and HK2) as well as enzymes responsible for oxidative carbon metabolism (ME2, CS, and ACLY) and monocarboxylate transport (SLC16A1). This suggests that josamycin greatly alters cellular metabolism. Components of the MAPK signaling cascade (MAP3K4, MAP2K3, and GADD45A) also emerged among the most sensitizing genes; this pathway has been implicated previously in the mode of action of macrolide antibiotics in mammalian systems (9). Various genes involved in mRNA surveillance (SMG1 and SMG7), ribosome biogenesis (NPM1 and ARF1), ubiquitination (FBOX11 and UBAP2L), mitotic organization (TPX2, ATRX, PCM1, and NPM1), and phosphatase activity (PPP1R15B and PPP2R2A) also appeared prominently among the top 40 sensitizing hits. Some gene knockdowns showed a protective effect and were enriched in the josamycin-treated population (Fig. S1A and Table S2). These included genes involved in DNA damage and repair (PRKDC), cytosolic protein translation (EIF2AK1 and EIF4G1), RNA processing (DAZAP1, STRAP, and HNRPA1), transcription initiation by RNA polymerase II (C14ORF166, TAF3, and TAF4), and oxidative phosphorylation (ATP5O, ATP5H, ATP5F1, ATP5B, and NDUFB8).
Individual shRNA validation
To validate our findings on an individual gene level, we performed competitive growth assays to compare the growth rate of a given KD cell line relative to its WT counterpart under untreated and josamycin-treated conditions. We established mCherry-positive K562 KD cell lines using the top two sensitizing shRNA constructs for each gene (Table S3). Suppression of mRNA of target genes in KD cells was confirmed by quantitative reverse transcription PCR (RT-qPCR) (Fig. S2, A and B, and Table S4).We then initiated cocultures of a given KD cell line with WT K562 cells at a 1:1 ratio under untreated and josamycin-treated conditions. By comparing the KD/WT ratio in the two cocultured populations over time, we observed whether a KD cell line was specifically depleted under josamycin treatment. This design detects and corrects for any non-drug-related growth defects of a construct relative to WT K562 cells in a controlled fashion over the many days it takes for some phenotypes to emerge. We confirmed that MTIF2, GFM2, MTIF3, MTFMT, PFKP, GPI, HK2, and MAP3K4 knockdown cells were specifically depleted under josamycin treatment relative to untreated cocultured controls after 10 days (Fig. 2, B–D). These results confirm the findings of our pooled genetic screens. In contrast, KD cells harboring nontargeting shRNA constructs were not depleted under josamycin treatment (Fig. 2D). Additionally, the effects of knocking down two highly protective genes, PRKDC and EIF2AK1, were also validated in this assay (Fig. 1B). These KD cell lines were specifically enriched under josamycin treatment relative to untreated cocultured controls (Fig. 1B).
Differential cellular effects of josamycin compared with other antibiotics
To determine whether these hits were unique to josamycin, we tested whether knocking down MTIF2, GPI, and MAP3K4 similarly sensitized cells to another macrolide, clarithromycin, as well as a structurally unrelated inhibitor of the bacterial ribosome, chloramphenicol (Fig. 1A). Josamycin was a slightly more potent inhibitor of K562 cell growth than clarithromycin or chloramphenicol (Fig. 1B). We therefore conducted the KD/WT assay at three drug concentrations. Interestingly, although knockdown of MTIF2 and GPI sensitized cells to josamycin, clarithromycin, and chloramphenicol at the three tested concentrations in a dose-dependent manner (Fig. 3, A and B), knockdown of MAP3K4 sensitized cells to only josamycin and was not dose-dependent (Fig. 3C). This suggests that MAP3K4 is a unique target of josamycin. In the MAP3K4 coculture experiments, the KD/WT ratio increased unexpectedly with josamycin concentration. At high concentrations, we speculate that josamycin-mediated alterations in MAPK signaling, especially in MAP3K4 KD cells, may lead to compensatory cellular signaling and feedback, as observed previously in this pathway (42). Whether josamycin directly binds a MAPK protein or indirectly induces changes in cellular processes that impact MAPK signaling and cell growth remains unknown. None of the antibiotics showed a sensitizing effect in cells infected with a nontargeting shRNA construct (Fig. 3D).
Figure 3.
Testing the relevance of gene hits identified with josamycin across a panel of bacterial ribosome inhibitors. A, comparison of knockdown of MTIF2 on cellular sensitivity to josamycin, clarithromycin, and chloramphenicol after 9 days. The KD/WT ratio is the proportion of knockdown to WT K562 cells in an untreated versus drug-treated population at concentrations of 6, 12.5, and 25 μm after 9 days of coculture initiated at a 1:1 ratio. At these concentrations, all drugs caused decreases in cell growth, as assessed by flow cytometry. B, comparison of knockdown of GPI on cellular sensitivity to various drugs after 9 days. C, comparison of knockdown of MAP3K4 on cellular sensitivity to various drugs after 9 days. D, comparison of the nontargeting shRNA construct (negative control) on cellular sensitivity to various drugs after 9 days. In A–D, error bars indicate mean ± S.D., n = 2, and dots represent individual values.
Josamycin inhibits oxidative phosphorylation and induces a metabolic shift to glycolysis
Although most mitochondrial proteins are encoded by nuclear genes, 13 crucial proteins in respiration, including COX1, COX2, and COX3 are mitochondrially encoded and translated (43, 44). Because our data suggested that josamycin directly binds the mitochondrial ribosome, we tested whether josamycin induced mitochondrial dysfunction at early time points. To do so, we conducted real-time measurements of the oxygen consumption rate using a Seahorse XFp analyzer. Treatment of K562 cells with the genome-wide screening concentration of 10 μm josamycin for 16 h induced significant inhibition of mitochondrial basal and maximal respiration (Fig. S3, A and B).
We next treated K562 cells with josamycin, clarithromycin, and chloramphenicol at a common concentration of 15 μm, observed previously to cause growth defects for all three drugs. Basal and maximal respiration were significantly inhibited by josamycin and chloramphenicol after 16 h and by clarithromycin to a lesser extent (Fig. 4, A and B). Overnight treatment with josamycin or chloramphenicol also caused marked decreases in the total ATP production rate, attributable to oxidative phosphorylation (Fig. 4C). Furthermore, josamycin-treated cells demonstrated a significant increase in the ATP production rate, attributable to glycolysis (Fig. 4C), with its percentage of the overall ATP production rate also rising sharply, from 38% to 59%, indicating a strong cellular metabolic shift to glycolysis (Fig. 4C). Although cancer cells rely heavily on glycolysis, the Seahorse ATP rate assay has previously determined ATP rates and breakdowns commonly in this range (45). Examining mitochondrial translation more specifically, these drugs caused decreases in the levels of mitochondrially encoded COX2 after 48 h but not in the levels of nuclearly encoded COX4 (Fig. 4D). This result underscores the strong mitochondrial translation signature found in our unbiased genetic screen. To extend our results beyond this cancer cell line, we also found that josamycin decreased basal and mitochondrial respiration in primary human umbilical vein endothelial cells (HUVECs) (Fig. 4E). Josamycin did not cause major growth phenotypes in HUVECs at 48 h (Fig. 4F) and caused only small changes in mitochondrial membrane potential (Fig. S3C) or superoxide generation (Fig. S3D) in K562 cells. Additionally, overnight josamycin treatment did not alter cytoplasmic pH levels in K562 cells (Fig. S3E). These results demonstrate that josamycin alters cellular energy production and mitochondrial translation in a relatively nontoxic fashion at early time points in both cancer and primary cells.
Figure 4.
Josamycin impairs oxidative phosphorylation and induces a metabolic shift to glycolysis in K562 cells. A, real-time monitoring of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in K562 cells after 16 h of pretreatment with 0 or 15 μm josamycin. Oligomycin (a complex V inhibitor), FCCP (a mitochondrial uncoupler), and a rotenone/antimycin A mixture (respiratory chain inhibitors) were added at 20-min intervals to assess effects on mitochondrial function. B, basal and maximal respiration in K562 cells treated with 0 (gray columns) or 15 μm josamycin, clarithromycin, and chloramphenicol (green columns) after 16 h. Basal respiratory rates were estimated by the response of cells to oligomycin, whereas maximal respiratory capacity was estimated by the difference between the FCCP-treated state and the rotenone/antimycin-treated state. C, ATP production rates attributable to glycolysis or respiration in untreated K562 cells or cells treated with 15 μm josamycin, clarithromycin, or chloramphenicol after 16 h. The percentage of the total ATP production rate attributable to glycolysis is indicated within the respective columns. D, protein levels of mitochondrially encoded COX2 and nuclearly encoded COX4 after 48 h of treatment with 0 or 15 μm josamycin, clarithromycin, or chloramphenicol. E, basal and maximal respiration in HUVECs treated with 0 or 15 μm josamycin after 16 h. F, josamycin dose–response in HUVECs after 48 h. In B, C, and E, error bars indicate mean ± S.D., n = 3, and dots represent individual values. Differences between DMSO- and drug-treated cells are indicated: *, p < 0.05; **, p < 0.01; n.s., p > 0.05, as detailed under “Experimental procedures.”
Discussion
Identifying host targets of macrolide antibacterial agents is a prerequisite for understanding the mechanisms of their nonantibiotic effects or uses (46). Here we report the first unbiased and comprehensive genome-wide screen in a human cell line to determine cellular modulators of sensitivity to a clinically used antibiotic, josamycin.
Illustrating the power of genome-wide approaches to drug target identification, the most sensitizing genes to emerge from our screen were involved in mitochondrial translation; an expected mammalian target, given the similarity between the mitochondrial and bacterial ribosome (Fig. 2A) (44). The mitochondrial DNA genome encodes 22 tRNAs, two rRNAs, and 13 of the 90 proteins in the electron transport chain; these generally hydrophobic proteins are translated in the mitochondrial matrix (44, 47). Although mitochondrial ribosomes utilize tRNAs and translation factors that are distinct from bacteria, the core structure of the peptidyl transferase center is conserved between their ribosomes (44). It is key amino acid differences in the polypeptide exit tunnel that increase the selectivity of macrolide antibiotics (48). However, bacterial protein synthesis inhibitors, including macrolides, tetracyclines, and chloramphenicol, can distinctly impact mitochondrial translation and function in vitro (49, 50), in cells (38, 40, 51), in vivo (52), and, in the case of aminoglycosides, in the clinic (53). In K562 cells, the oxazolidinone eperezolid slowed growth exclusively though this mechanism (54). Previously, josamycin itself has been shown to inhibit protein synthesis in vitro in a bovine mitochondrial translation system (IC50 = 12.3 μm) (50) and also to down-regulate mitochondrially encoded proteins in transformed human liver epithelial cells after 96 h (43). We also observed changes in mitochondrial function upon antibiotic treatment in both cancer and primary cells (Fig. 4, A–E). Given all of this, it is almost certain that the mitochondrial ribosome is a direct binding target of josamycin and that this interaction is at least partially responsible for its effects on mitochondrial function.
Our genetic approach clearly implicated mitochondrial translation initiation (MTIF2 and MTIF3), formylation (MTFMT), and termination (GFM2) factors as key modulators of K562 growth sensitivity to josamycin (Fig. 2A). The mitochondrial elongation and release factors (TSFM, TUFM, GFM1, MTRF1, MTRF1L, and MRRF) were not genetic modulators. Interestingly, although a previous yeast haploinsufficiency profiling screen conducted with the antibiotics tigecycline and chloramphenicol primarily identified mitochondrial ribosomal proteins within the large subunit, our top hits were all translation factors (47). These results suggest that the exact genetic factors identified in antibiotic target identification screens may be distinct to drug and/or cell type.
Our shRNA screen also identified knockdown of glycolytic genes as highly sensitizing to antibiotic treatment, even in a cancer cell line like K562 (Fig. 2A). Previous research has shown that doxycycline can induce glycolysis in MCF12A cells (20). Duewelhenke et al. (40) also reported that bacterial protein synthesis inhibitors impaired mitochondrial energetics and increased extracellular lactate production. Here we observed a shift toward glycolysis, as measured by real-time respiration, in antibiotic-treated K562 cells (Fig. 4C) and impaired respiration in primary HUVECs (Fig. 4E). Interestingly, we also observed that knockdown of components of the electron transport chain, including ATP5O, ATP5H, ATP5F1, ATP5B, and NDUFB8, slightly protected cells from josamycin (Fig. S1A and Table S2). We hypothesize that knockdown in the electron transport chain may promote a stronger glycolytic state. Beyond josamycin, knockdown of GPI also broadly sensitized cells to clarithromycin and chloramphenicol (Fig. 3B).
Clinically useful antibiotics like josamycin, chloramphenicol and erythromycin are not potent inhibitors of cell growth (>50 μm growth inhibition 50 values (GI50) for cells in the NCI-60 database) (55); however, our 14-day shRNA screen also identified interesting classes of human targets with subtle growth phenotypes, some of which are highlighted below. In addition to altered cellular bioenergetics, the antiproliferative effects of mitochondrial ribosome-targeting antibiotics have been attributed to retrograde mitochondrion-to-nucleus signaling (51). In our study, knockdown of the mitochondrial protein quality control genes CLPP and CLPX sensitized cells to josamycin (Fig. 2A). Previously, inhibition of the mitochondrial ATP-dependent proteasome CLPX/CLPP complex has been shown to impair oxidative phosphorylation (56); conversely, overexpression of CLPX can stimulate a mitochondrial unfolded protein response that up-regulates mitochondrial metabolic genes (57). Especially because our screen only highlights a distinct subset of mitochondrial proteins, our results suggest that josamycin induces the mitochondrial unfolded protein response and that this affects growth.
Our screen also identified a growth phenotype associated with impaired cytoplasmic protein translation. Specifically, knockdown of PPP1R15B was sensitizing, whereas knockdown of EIF2AK1 was protective (Fig. 2A and Fig. S1A). Because EIF2AK1 slows cytosolic protein translation in response to stress in opposition to the constitutive phosphatase PPP1R15B, our screen suggests that impaired cytoplasmic translation further sensitizes cells to josamycin.
The most protective shRNAs were against PRKDC, a DNA damage response gene with emerging roles in mitochondrial genome maintenance and function (Fig. S1B) (58–60). Interestingly, a proteomics study exploring the antibiotic doxycycline (another mitochondrial translation inhibitor) as a potential cancer therapy found that PRKDC was the most down-regulated protein upon 3 days of treatment with doxycycline in MCF7 cells (59). As researchers cautiously attempt to repurpose antibiotics for cancer treatment because of their off-target activity inhibiting mitochondrial biogenesis, understanding the cellular modifiers of sensitivity to this process may help delineate mechanisms of response or resistance (22).
In terms of anti-inflammatory activities, josamycin-mediated inhibition of p38 MAPK signaling may partially explain its effects on neutrophils and other immune cells (34) and its anti-influenza effects in vivo (35). Knockdown of the MAPK signaling components GADD45A, MAP3K4, and MAP2K3 sensitized cells to josamycin (Fig. 2A); these genes can act together to activate p38 MAPK signaling (61, 62). Whether josamycin directly binds a MAPK protein or indirectly induces changes in cellular processes that impact MAPK signaling and cell growth remains a subject of further study.
The p38 MAPK pathway has known roles in pulmonary inflammation (63, 64) and influenza infection (65–67). Additionally, other macrolide antibacterial agents are known to affect MAPK activation, leading to changes in the release of cytokines like IL-1, IL-6, IL-8, tumor necrosis factor α, and granulocyte/macrophage colony–stimulating factor (1, 9). Importantly, the sensitizing effect of MAP3K4 was not observed in K562 cells treated with chloramphenicol, suggesting that this effect is macrolide-specific, although it may be cell type–specific (Fig. 3C) (68). Downstream physiological effects of p38 MAPK signaling will differ between immune cells and K562 cells, an erythroid and megakaryocytic progenitor. Potential compensatory cell signaling and feedback in response to josamycin-mediated alterations of MAPK signaling may also drive cell growth and inflammation phenotypes (Fig. 3C) (42). Additionally, the inhibitory effects of josamycin on neutrophil degranulation (34, 69–71) and inflammation (72–74) are somewhat variable, hinting that a more complicated mechanism may be at work. Of course, it remains possible that josamycin exerts its anti-inflammatory and anti-influenza activities in vivo through cell type interactions that cannot be recapitulated using cell culture.
As increasing numbers of bioactive natural products and preclinical drug candidates are identified, determining their biological mechanism of action remains a challenge. This work demonstrates the power of genome-wide screens as a robust platform for drug target identification. Our study also represents the first systematic effort to identify mammalian targets of macrolide antibiotics, which possess interesting biological activity attractive for various therapeutic applications (11, 13, 35). The unique effect on p38 MAPK signaling with josamycin also suggests a path forward to using genetic screens to distinguish host targets for different drugs with the same primary modes of action.
Experimental procedures
Chemicals and reagents
Josamycin and clarithromycin were from Sigma-Aldrich. Chloramphenicol was from Gold Biotechnologies. Chemicals were dissolved in DMSO, aliquoted, and stored at −20 °C.
Cell culture and dose–response curves
K562 cells were cultured in RPMI medium (Gibco) supplemented with 10% fetal bovine serum (Hyclone), penicillin (10,000 IU/ml), streptomycin (10,000 IU/ml), and l-glutamine (2 mm) (25). HUVECs were cultured in EGM-2 endothelial cell growth medium (Lonza, CC-3162). Cells were maintained in log phase in a humidified incubator kept at 37 °C and 5% CO2. To test the effects of various drugs on cell growth, K562 cells were seeded in 24-well plates at 100,000 cells/well and HUVECs at 50,000 cells/well. Density and viability at various time points were assessed by flow cytometry with a BD Accuri C6 flow cytometer as described previously (25).
Genome-wide and batch shRNA analyses
A genome-wide shRNA screen was conducted as described previously (25). Briefly, K562 cells were infected with a genome-wide shRNA library (25). When cells were appropriately selected and recovered to ensure that knockdown constructs were expressed, this pooled population was split and grown in the presence or absence of 10 μm josamycin for 14 days. This concentration leads to a ∼30% reduction in cell growth after 48 h. Each day, both populations were diluted to 500,000 cells/ml as needed. At the end of the screen, genomic DNA from each population was isolated and sequenced (25). The ratio of the frequency of a particular shRNA-encoding construct in untreated and josamycin-treated cells was determined using the casTLE framework developed in our laboratory (25). The results of this analysis are summarized in Table S1. A similar screen was repeated in K562 cells using the top ∼500 genes identified from the genome-wide screen and is summarized in Table S2.
Individual shRNA validation
To validate results from the genome-wide screen, individual shRNA hits were retested with the two most sensitizing shRNAs from the batch screen; their sequences are listed in Table S3. Oligonucleotides corresponding to these shRNAs (Elim Biopharma) were ligated into pMCB309 using restriction digest cloning as described previously (25). pMCB309 harbors two BstXI sites, a puromycin resistance cassette, and an mCherry fluorescent reporter gene. The resulting constructs were combined with third-generation lentiviral packaging plasmids to produce the lentivirus, as described previously (25). To generate cell lines harboring knockdowns in a particular gene, K562 cells were infected with the appropriate lentivirus and selected, as detailed previously (25).
For competitive growth assays, 50%–50% mixtures of WT K562 cells and K562 cells expressing a nontargeting or a targeting shRNA were seeded into 24-well plates at 500,000 cells/ml. These mixtures were grown in the presence or absence of 10 μm josamycin for 9–10 days and maintained at roughly their starting density of 500,000 cells/ml. Monitoring the percentage of mCherry-positive cells under treatment and control conditions allowed us to compute the ratio of KD/WT cells. For comparisons between different antibiotics, experiments were performed at three drug concentrations with a single shRNA, and the extent of mCherry depletion was similarly monitored in the three populations across drugs. The most sensitizing MTIF2, GPI, and MAP3K4 shRNAs from the batch screen were used along with the control shRNA NegCtrl_3. The statistical significance of the difference in the KD/WT ratio between DMSO- and drug-treated cells were calculated using a t test adjusted for multiple comparisons (Holm–Sidak) in GraphPad Prism 8.0.2.
Mitochondrial membrane potential measurements
The JC-1 dye (Thermo Fisher) is membrane-permeable and undergoes a fluorescence emission shift from red to green upon mitochondrial depolarization, typically an early feature of apoptosis (75). K562 cells were plated in 24-well plates (1 ml) and exposed to josamycin or a control drug (antimycin A or carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP)) at various concentrations and times. JC-1 was added to cells at 2.5 μg/ml with slight vortexing, and the cells were incubated for 25 min at 37 °C. Subsequently, cells were centrifuged, washed with drug-containing PBS, and resuspended in 200 μl of drug-containing PBS. Fluorescence shifts were measured on a BD Accuri C6. Statistical comparisons of log-transformed red/green (585/533 nm) ratios under different conditions were compared with an untreated control using one-way ANOVA, Dunnett's test in GraphPad Prism 8.0.2.
Mitochondrial superoxide generation and acidity assays
The MitoSox (Molecular Probes; Thermo Fisher) dye selectively accumulates and detects superoxide generated in the mitochondria of live cells (76). As this localization depends on mitochondrial potential, we confirmed that drug-treated cells exhibited no changes in mitochondrial potential at early time points. Briefly, K562 cells were resuspended in RPMI at a density of 106 cells/ml. The MitoSox dye was then added to cells at 5 μm for 20 min at 37 °C in the dark. Cells were washed twice in Hank's Balanced Salt Solution containing Ca+2, Mg+2 and the drug of interest, and resuspended in 300 μl of this buffer prior to flow cytometric analysis. Antimycin A was used to confirm that mitochondrial superoxide production generated a strong fluorescence signal shift. A cell-permeable SNARF-1 AM ester dye (Molecular Probes; C-1272) was used to study the effects of josamycin on cytoplasmic and endosomal acidification. K562 cells were plated overnight (16 h) with drug. The next day, cells were resuspended in PBS containing 7 μm of SNARF-1 AM ester dye for 30 min. For calibration, the ionophore nigericin was used in a high K+ buffer, as previously described (77). Using a BD Accuri C6, we detected 670 nm/585 nm fluorescence ratios; lower ratios correspond to lower intracellular pH. Statistical comparisons of log-transformed base/acid (670/585 nm) ratios under different conditions were compared with an untreated control using one-way ANOVA, Dunnett's test in GraphPad Prism 8.0.2.
Western blot reagents and procedures
For mitochondrial translation blot experiments, Mtco2 (Abcam, 110258) and Cox4 (sc-517553) were used, along with the secondary antibody HRP goat anti-mouse (minimal cross-reactivity) Poly 4053 clone (BioLegend, 405306). Cells were washed twice in cold PBS and lysed in radioimmune precipitation assay buffer (Fisher Scientific) containing HALT protease and phosphatase inhibitor mixture (Fisher Scientific) on ice for 20 min. After centrifugation, lysates were boiled for 5–10 min in Laemmli buffer containing reducing agent, and ∼15 μg/sample per well was loaded onto two parallel SDS-PAGE gels. Semidry transfer to a PVDF membrane was performed with the Trans-Blot Turbo System (Bio-Rad). Membranes were blocked with 5% BSA in phosphate buffer saline, 0.1% Tween 20 (PBST) for 1 h at room temperature and stained with primary antibody (generally 1:1000 concentration) overnight at 4 °C. Membranes were washed (three times, 5 min) in PBST before and after secondary antibody incubation (1:2000 concentration for 1 h, room temperature). Membranes were incubated with Pierce ECL Plus Western blot substrate (Thermo Fisher).
RT-qPCR reagents and procedures
RT-qPCR was used to confirm the efficacy of shRNA-mediated knockdown of target mRNA levels. Total RNA was isolated from multiple nontargeting shRNA control cell lines and knockdown cell lines in parallel using the RNEasy Mini Kit (Qiagen) with the RNase-free DNase set (Qiagen). Briefly, 150 ng of total RNA was reverse-transcribed and amplified using the QuantiTect SYBR Green RT-PCR Kit in 20-μl reactions containing total RNA (3 μl), 2× SYBR Green Mix (10 μl), 0.5 μm gene-specific primers (0.2 μl), QuantiTect RT Mix (0.2 μl), and RNase-free water (5.6 μl), following the manufacturer's recommended protocols. One-step RT-qPCR was carried out on a QuantStudio3 (Applied Biosystems) under the following conditions: 30 min at 50 °C followed by 95 °C for 15 min and then 40 cycles of 15 s denaturation at 94 °C, 30 s of annealing at 53.5 °C, and 30 s extension at 72 °C. Reaction products were confirmed by DNA gel electrophoresis.
Target gene primers were designed in PrimerQuest (IDT), and sequences are given in Table S4. Gene expression across nontargeting shRNA control cell lines and knockdown cell lines was first normalized to levels of the ribosomal protein RPL19 using a primer and method described previously (25). Normalized, log-transformed gene expression values (cycle thresholds) were used for statistical analyses. Expression of a target gene in the knockdown cell lines was compared with expression in the two negative control cell lines using one-way ANOVA, Dunnett's test in GraphPad Prism 8.0.2.
Cellular bioenergetic assays
The Seahorse XFp analyzer (Agilent) allows real-time measurements of the cellular oxygen consumption rate and extracellular acidification rate. The Cell Mito Stress Test Kit and Real-Time ATP Rate Assay Kit were used to probe cellular mitochondrial function and metabolic changes, respectively, upon macrolide administration. All drug and medium reagents were from Agilent. Calibration, and procedures were performed as specified by the manufacturer. Briefly, K562 or HUVECs were treated with drug (or vehicle control) for 16 h overnight in RPMI medium or EGM-2. Cells were then washed twice in PBS containing the appropriate drug or control and resuspended in Seahorse XF RPMI medium or DMEM (pH 7.4) supplemented with 10 mm glucose, 1 mm sodium pyruvate, 4 mm glutamate, and the appropriate drug or control. K562 cells were seeded in triplicate at 50,000 cells/well in poly-l-lysine–coated Seahorse XFp cell culture miniplates. HUVECs were seeded and spun down at 30,000 cells/well in noncoated plates. Adhesion was confirmed visually, and the plate was incubated in a non-CO2 incubator for at least 30 min. For the Mito Stress Test Kit, all drugs (oligomycin, FCCP, rotenone, and antimycin A) were used at 1 μm. The real-time ATP rate assay was conducted in the presence of 1.5 μm oligomycin and 0.5 μm rotenone or antimycin A. The online Seahorse Report Generator aided graphical analysis of mitochondrial parameters. The statistical significance of the difference in basal and maximal respiration between DMSO- and drug-treated cells was calculated using a t test adjusted for multiple comparisons (Holm–Sidak) in GraphPad Prism 8.0.2. The statistical significance of differences in the individual ATP production rates attributable to glycolysis or oxidative phosphorylation between DMSO- and drug-treated cells was calculated similarly.
Author contributions
A. G., A. Ö.-A., and D. M. formal analysis; A. G. and A. Ö.-A. investigation; A. G. and A. Ö.-A. methodology; A. G. writing-original draft; A. Ö.-A. and D. M. data curation; D. M. software; D. M. visualization; M. C. B. and C. K. conceptualization; M. C. B. supervision; M. C. B. and C. K. writing-review and editing; C. K. funding acquisition.
Supplementary Material
Acknowledgments
We thank Khanh Nguyen (Glenn laboratory) and members of the C. K., M. C. B., and Vollrath laboratories for technical advice.
This work was funded by NIAID, National Institute of Health grant U19-AI109662; NIGMS, National Institutes of Health Grant R01-GM087934; NIH, Director's New Innovator Award Grant 1DP2HD084069-01; and a seed grant from Stanford Chemistry, Engineering and Medicine for Human Health (ChEM-H). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1–S4.
The raw sequencing files were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA561630.
- MAPK
- mitogen-activated protein kinase
- KD
- knockdown
- HUVEC
- human umbilical vein endothelial cell
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- ANOVA
- analysis of variance
- casTLE
- Cas9 high-throughput maximum likelihood estimator
- RT-qPCR
- quantitative reverse transcription PCR.
References
- 1. Culić O., Eraković V., and Parnham M. J. (2001) Anti-inflammatory effects of macrolide antibiotics. Eur. J. Pharmacol. 429, 209–229 10.1016/S0014-2999(01)01321-8 [DOI] [PubMed] [Google Scholar]
- 2. Hansen J. L., Ippolito J. A., Ban N., Nissen P., Moore P. B., and Steitz T. A. (2002) The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10, 117–128 10.1016/S1097-2765(02)00570-1 [DOI] [PubMed] [Google Scholar]
- 3. Zuckerman J. M., Qamar F., and Bono B. R. (2011) Review of macrolides (azithromycin, clarithromycin), ketolids (telithromycin) and glycylcyclines (tigecycline). Med. Clin. North Am. 95, 761–791, viii 10.1016/j.mcna.2011.03.012 [DOI] [PubMed] [Google Scholar]
- 4. Bearden D. T., and Rodvold K. A. (1999) Penetration of macrolides into pulmonary sites of infection. Infect. Med. 16, 480–484A [Google Scholar]
- 5. Hicks L. A., Bartoces M. G., Roberts R. M., Suda K. J., Hunkler R. J., Taylor T. H. Jr., and Schrag S. J. (2015) US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect. Dis. 60, 1308–1316 [DOI] [PubMed] [Google Scholar]
- 6. Villarino N., Brown S. A., and Martín-Jiménez T. (2013) The role of the macrolide tulathromycin in veterinary medicine. Vet. J. 198, 352–357 10.1016/j.tvjl.2013.07.032 [DOI] [PubMed] [Google Scholar]
- 7. Zarogoulidis P., Papanas N., Kioumis I., Chatzaki E., Maltezos E., and Zarogoulidis K. (2012) Macrolides: from in vitro anti-inflammatory and immunomodulatory properties to clinical practice in respiratory diseases. Eur. J. Clin. Pharmacol. 68, 479–503 10.1007/s00228-011-1161-x [DOI] [PubMed] [Google Scholar]
- 8. Asano K., Tryka E., Cho J. S., and Keicho N. (2012) Macrolide therapy in chronic inflammatory diseases. Mediators Inflamm. 2012, 692352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanoh S., and Rubin B. K. (2010) Mechanisms of action and clinical application of macrolides as immunomodulatory medications. Clin. Microbiol. Rev. 23, 590–615 10.1128/CMR.00078-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steel H. C., Theron A. J., Cockeran R., Anderson R., and Feldman C. (2012) Pathogen- and host-directed anti-inflammatory activities of macrolide antibiotics. Mediators Inflamm. 2012, 584262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crosbie P. A., and Woodhead M. A. (2009) Long-term macrolide therapy in chronic inflammatory airway diseases. Eur. Respir. J. 33, 171–181 10.1183/09031936.00042208 [DOI] [PubMed] [Google Scholar]
- 12. Pillozzi S., Masselli M., Gasparoli L., D'Amico M., Polletta L., Veltroni M., Favre C., Basso G., Becchetti A., and Arcangeli A. (2016) Macrolide antibiotics exert antileukemic effects by modulating the autophagic flux through inhibition of hERG1 potassium channels. Blood Cancer J. 6, e423 10.1038/bcj.2016.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Nuffel A. M., Sukhatme V., Pantziarka P., Meheus L., Sukhatme V. P., and Bouche G. (2015) Repurposing drugs in oncology (ReDO): clarithromycin as an anti-cancer agent. Ecancermedicalscience. 9, 513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poletti V., Casoni G., Chilosi M., and Zompatori M. (2006) Diffuse panbronchiolitis. Eur. Respir. J. 28, 862–871 10.1183/09031936.06.00131805 [DOI] [PubMed] [Google Scholar]
- 15. Kudoh S., Azuma A., Yamamoto M., Izumi T., and Ando M. (1998) Improvement of survival in patients with diffuse panbronchiolitis treated with low-dose erythromycin. Am. J. Respir. Crit. Care Med. 157, 1829–1832 10.1164/ajrccm.157.6.9710075 [DOI] [PubMed] [Google Scholar]
- 16. Wong E. H., Porter J. D., Edwards M. R., and Johnston S. L. (2014) The role of macrolides in asthma: current evidence and future directions. Lancet Respir. Med. 2, 657–670 10.1016/S2213-2600(14)70107-9 [DOI] [PubMed] [Google Scholar]
- 17. Equi A., Balfour-Lynn I. M., Bush A., and Rosenthal M. (2002) Long term azithromycin in children with cystic fibrosis: a randomised, placebo-controlled crossover trial. Lancet 360, 978–984 10.1016/S0140-6736(02)11081-6 [DOI] [PubMed] [Google Scholar]
- 18. Donath E., Chaudhry A., Hernandez-Aya L. F., and Lit L. (2013) A meta-analysis on the prophylactic use of macrolide antibiotics for the prevention of disease exacerbations in patients with chronic obstructive pulmonary disease. Respir. Med. 107, 1385–1392 10.1016/j.rmed.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 19. Ribeiro C. M. P., Hurd H., Wu Y., Martino M. E. B., Jones L., Brighton B., Boucher R. C., and O'Neal W. K. (2009) Azithromycin treatment alters gene expression in inflammatory, lipid metabolism, and cell cycle pathways in well-differentiated human airway epithelia. PLoS ONE 10.1371/journal.pone.0005806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahler E., Sullivan W. J., Cass A., Braas D., York A. G., Bensinger S. J., Graeber T. G., and Christofk H. R. (2013) Doxycycline alters metabolism and proliferation of human cell lines. PLoS ONE 8, e64561 10.1371/journal.pone.0064561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E. G., Mottis A., Jovaisaite V., Frochaux M. V., Quiros P. M., Deplancke B., Houtkooper R. H., and Auwerx J. (2015) Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691 10.1016/j.celrep.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamb R., Ozsvari B., Lisanti C. L., Tanowitz H. B., Howell A., Martinez-Outschoorn U. E., Sotgia F., and Lisanti M. P. (2015) Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget 6, 4569–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schenone M., Dančík V., Wagner B. K., and Clemons P. A. (2013) Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 9, 232–240 10.1038/nchembio.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jost M., and Weissman J. S. (2018) CRISPR approaches to small molecule target identification. ACS Chem. Biol. 13, 366–375 10.1021/acschembio.7b00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deans R. M., Morgens D. W., Ökesli A., Pillay S., Horlbeck M. A., Kampmann M., Gilbert L. A., Li A., Mateo R., Smith M., Glenn J. S., Carette J. E., Khosla C., and Bassik M. C. (2016) Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 12, 361–366 10.1038/nchembio.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matheny C. J., Wei M. C., Bassik M. C., Donnelly A. J., Kampmann M., Iwasaki M., Piloto O., Solow-Cordero D. E., Bouley D. M., Rau R., Brown P., McManus M. T., Weissman J. S., and Cleary M. L. (2013) Next-generation NAMPT inhibitors identified by sequential high-throughput phenotypic chemical and functional genomic screens. Chem. Biol. 20, 1352–1363 10.1016/j.chembiol.2013.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sidrauski C., Tsai J. C., Kampmann M., Hearn B. R., Vedantham P., Jaishankar P., Sokabe M., Mendez A. S., Newton B. W., Tang E. L., Verschueren E., Johnson J. R., Krogan N. J., Fraser C. S., Weissman J. S., et al. (2015) Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife 4, e07314 10.7554/eLife.07314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brummelkamp T. R., Fabius A. W., Mullenders J., Madiredjo M., Velds A., Kerkhoven R. M., Bernards R., and Beijersbergen R. L. (2006) An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol. 2, 202–206 10.1038/nchembio774 [DOI] [PubMed] [Google Scholar]
- 29. Luo B., Cheung H. W., Subramanian A., Sharifnia T., Okamoto M., Yang X., Hinkle G., Boehm J. S., Beroukhim R., Weir B. A., Mermel C., Barbie D. A., Awad T., Zhou X., Nguyen T., et al. (2008) Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 105, 20380–20385 10.1073/pnas.0810485105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burgess D. J., Doles J., Zender L., Xue W., Ma B., McCombie W. R., Hannon G. J., Lowe S. W., and Hemann M. T. (2008) Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 9053–9058 10.1073/pnas.0803513105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takase S., Kurokawa R., Arai D., Kanto K. K., Okino T., Nakao Y., Kushiro T., Yoshida M., and Matsumoto K. (2017) A quantitative shRNA screen identifies ATP1A1 as a gene that regulates cytotoxicity by aurilide B. Sci. Rep. 7, 1–9 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arsic B., Barber J., ikoš A., Mladenovic M., Stankovic N., and Novak P. (2018) 16-Membered macrolide antibiotics: a review. Int. J. Antimicrob. Agents 51, 283–298 10.1016/j.ijantimicag.2017.05.020 [DOI] [PubMed] [Google Scholar]
- 33. Morikawa K., Oseko F., Morikawa S., and Iwamoto K. (1994) Immunomodulatory effects of three macrolides, midecamycin acetate, josamycin, and clarithromycin, on human T-lymphocyte function in vitro. Antimicrob. Agents Chemother. 38, 2643–2647 10.1128/AAC.38.11.2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Labro M. T., and el Benna J. (1990) Synergistic bactericidal interaction of josamycin with human neutrophils in vitro. J. Antimicrob. Chemother. 26, 515–524 10.1093/jac/26.4.515 [DOI] [PubMed] [Google Scholar]
- 35. Sugamata R., Sugawara A., Nagao T., Suzuki K., Hirose T., Yamamoto K., Oshima M., Kobayashi K., Sunazuka T., Akagawa K. S., mura S., Nakayama T., and Suzuki K. (2014) Leucomycin A3, a 16-membered macrolide antibiotic, inhibits influenza A virus infection and disease progression. J. Antibiot. 67, 213–222 10.1038/ja.2013.132 [DOI] [PubMed] [Google Scholar]
- 36. Kampmann M., Horlbeck M. A., Chen Y., Tsai J. C., Bassik M. C., Gilbert L. A., Villalta J. E., Kwon S. C., Chang H., Kim V. N., and Weissman J. S. (2015) Next-generation libraries for robust RNA interference-based genome-wide screens. Proc. Natl. Acad. Sci. U.S.A. 112, E3384–E3391 10.1073/pnas.1508821112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morgens D. W., Deans R. M., Li A., and Bassik M. C. (2016) Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 34, 634–636 10.1038/nbt.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viluksela M., Vainio P. J., and Tuominen R. K. (1996) Cytotoxicity of macrolide antibiotics in a cultured human liver cell line. J. Antimicrob. Chemother. 38, 465–473 10.1093/jac/38.3.465 [DOI] [PubMed] [Google Scholar]
- 39. Bassik M. C., Kampmann M., Lebbink R. J., Wang S., Hein M. Y., Poser I., Weibezahn J., Horlbeck M. A., Chen S., Mann M., Hyman A. A., Leproust E. M., McManus M. T., and Weissman J. S. (2013) A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922 10.1016/j.cell.2013.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duewelhenke N., Krut O., and Eysel P. (2007) Influence on mitochondria and cytotoxicity of different antibiotics administered in high concentrations on primary human osteoblasts and cell lines. Antimicrob. Agents Chemother. 51, 54–63 10.1128/AAC.00729-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Riesbeck K., Bredberg A., and Forsgren A. (1990) Ciprofloxacin does not inhibit mitochondrial functions but other antibiotics do. Antimicrob. Agents Chemother. 34, 167–169 10.1128/AAC.34.1.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Braicu C., Buse M., Busuioc C., Drula R., Gulei D., Raduly L., Rusu A., Irimie A., Atanasov A. G., Slaby O., Ionescu C., and Berindan-Neagoe I. (2019) A comprehensive review on MAPK: A promising therapeutic target in cancer. Cancers 11, pii: E1618 10.3390/cancers11101618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nadanaciva S., Dillman K., Gebhard D. F., Shrikhande A., and Will Y. (2010) High-content screening for compounds that affect mtDNA-encoded protein levels in eukaryotic cells. J. Biomol. Screen. 15, 937–948 10.1177/1087057110373547 [DOI] [PubMed] [Google Scholar]
- 44. Ott M., Amunts A., and Brown A. (2016) Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 85, 77–101 10.1146/annurev-biochem-060815-014334 [DOI] [PubMed] [Google Scholar]
- 45. Mookerjee S. A., Gerencser A. A., Nicholls D. G., and Brand M. D. (2017) Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 292, 7189–7207 10.1074/jbc.M116.774471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cameron E. J., McSharry C., Chaudhuri R., Farrow S., and Thomson N. C. (2012) Long-term macrolide treatment of chronic inflammatory airway diseases: risks, benefits and future developments. Clin. Exp. Allergy. 42, 1302–1312 10.1111/j.1365-2222.2012.03979.x [DOI] [PubMed] [Google Scholar]
- 47. Skrtić M, Sriskanthadevan S., Jhas B., Gebbia M., Wang X., Wang Z., Hurren R., Jitkova Y., Gronda M., Maclean N., Lai C. K., Eberhard Y., Bartoszko J., Spagnuolo P., Rutledge A. C., et al. (2011) Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20, 674–688 10.1016/j.ccr.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Böttger E. C., Springer B., Prammananan T., Kidan Y., and Sander P. (2001) Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2, 318–323 10.1093/embo-reports/kve062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McKee E. E., Ferguson M., Bentley A. T., and Marks T. A. (2006) Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob. Agents Chemother. 50, 2042–2049 10.1128/AAC.01411-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L., Ging N. C., Komoda T., Hanada T., Suzuki T., and Watanabe K. (2005) Antibiotic susceptibility of mammalian mitochondrial translation. FEBS Lett. 579, 6423–6427 10.1016/j.febslet.2005.09.103 [DOI] [PubMed] [Google Scholar]
- 51. Richter U., Lahtinen T., Marttinen P., Myöhänen M., Greco D., Cannino G., Jacobs H. T., Lietzén N., Nyman T. A., and Battersby B. J. (2013) A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr. Biol. 23, 535–541 10.1016/j.cub.2013.02.019 [DOI] [PubMed] [Google Scholar]
- 52. Kalghatgi S., Spina C. S., Costello J. C., Liesa M., Morones-Ramirez J. R., Slomovic S., Molina A., Shirihai O. S., and Collins J. J. (2013) Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci. Transl. Med. 5, 192ra85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bindu L. H., and Reddy P. P. (2008) Genetics of aminoglycocide-induced and prelingual non-syndromic mitochondrial hearing impairment: a review. Int. J. Audiol. 47, 702–707 10.1080/14992020802215862 [DOI] [PubMed] [Google Scholar]
- 54. Nagiec E. E., Wu L., Swaney S. M., Chosay J. G., Ross D. E., Brieland J. K., and Leach K. L. (2005) Oxazolidinones inhibit cellular proliferation via inhibition of mitochondrial protein synthesis. Antimicrob. Agents Chemother. 49, 3896–3902 10.1128/AAC.49.9.3896-3902.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shankavaram U. T., Varma S., Kane D., Sunshine M., Chary K. K., Reinhold W. C., Pommier Y., and Weinstein J. N. (2009) CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics 10, 1–10 10.1186/1471-2164-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cole A., Wang Z., Coyaud E., Voisin V., Gronda M., Jitkova Y., Mattson R., Hurren R., Babovic S., Maclean N., Restall I., Wang X., Jeyaraju D. V., Sukhai M. A., Prabha S., et al. (2015) Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 27, 864–876 10.1016/j.ccell.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Al-Furoukh N, Ianni A., Nolte H., Hölper S., Krüger M., Wanrooij S., and Braun T. (2015) ClpX stimulates the mitochondrial unfolded protein response (UPR mt) in mammalian cells. Biochim. Biophys. Acta 1853, 2580–2591 10.1016/j.bbamcr.2015.06.016 [DOI] [PubMed] [Google Scholar]
- 58. Papeta N., Zheng Z., Schon E. A., Brosel S., Altintas M. M., Nasr S. H., Reiser J., D'Agati V. D., and Gharavi A. G. (2010) Prkdc participates in mitochondrial genome maintenance and prevents Adriamycin-induced nephropathy in mice. J. Clin. Invest. 120, 4055–4064 10.1172/JCI43721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lamb R., Fiorillo M., Chadwick A., Ozsvari B., Reeves K. J., Smith D. L., Clarke R. B., Howell S. J., Cappello A. R., Martinez-Outschoorn U. E., Peiris-Pagès M., Sotgia F., and Lisanti M. P. (2015) Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: implications for more effective radiation therapy. Oncotarget 6, 14005–14025 10.18632/oncotarget.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park S. J., Gavrilova O., Brown A. L., Soto J. E., Bremner S., Kim J., Xu X., Yang S., Um J. H., Koch L. G., Britton S. L., Lieber R. L., Philp A., Baar K., Kohama S. G., et al. (2017) DNA-PK promotes the mitochondrial, metabolic, and physical decline that occurs during aging. Cell Metab. 25, 1135–1146.e7 10.1016/j.cmet.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wingert S., Thalheimer F. B., Haetscher N., Rehage M., Schroeder T., and Rieger M. A. (2016) DNA-damage response gene GADD45A induces differentiation in hematopoietic stem cells without inhibiting cell cycle or survival. Stem Cells 34, 699–710 10.1002/stem.2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Abell A. N., Granger D. A., and Johnson G. L. (2007) MEKK4 stimulation of p38 and JNK activity is negatively regulated by GSK3β. J. Biol. Chem. 282, 30476–30484 10.1074/jbc.M705783200 [DOI] [PubMed] [Google Scholar]
- 63. Otterbein L. E., Otterbein S. L., Ifedigbo E., Liu F., Morse D. E., Fearns C., Ulevitch R. J., Knickelbein R., Flavell R. A., and Choi A. M. (2003) MKK3 mitogen-activated protein kinase pathway mediates carbon monoxide-induced protection against oxidant-induced lung injury. Am. J. Pathol. 163, 2555–2563 10.1016/S0002-9440(10)63610-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mannam P., Zhang X., Shan P., Zhang Y., Shinn A. S., Zhang Y., and Lee P. J. (2013) Endothelial MKK3 is a critical mediator of lethal murine endotoxemia and acute lung injury. J. Immunol. 190, 1264–1275 10.4049/jimmunol.1202012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Börgeling Y., Schmolke M., Viemann D., Nordhoff C., Roth J., and Ludwig S. (2014) Inhibition of p38 mitogen-activated protein kinase impairs influenza virus-induced primary and secondary host gene responses and protects mice from lethal H5N1 infection. J. Biol. Chem. 289, 13–27 10.1074/jbc.M113.469239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shin H.-B., Choi M.-S., Yi C.-M., Lee J., Kim N.-J., and Inn K.-S. (2015) Inhibition of respiratory syncytial virus replication and virus-induced p38 kinase activity by berberine. Int. Immunopharmacol. 27, 65–68 10.1016/j.intimp.2015.04.045 [DOI] [PubMed] [Google Scholar]
- 67. Marchant D., Singhera G. K., Utokaparch S., Hackett T. L., Boyd J. H., Luo Z., Si X., Dorscheid D. R., McManus B. M., and Hegele R. G. (2010) Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. J. Virol. 84, 11359–11373 10.1128/JVI.00804-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li C. H., Cheng Y. W., Liao P. L., Yang Y. T., and Kang J. J. (2010) Chloramphenicol causes mitochondrial stress, decreases ATP biosynthesis, induces matrix metalloproteinase-13 expression, and solid-tumor cell invasion. Toxicol. Sci. 116, 140–150 10.1093/toxsci/kfq085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Abdelghaffar H., Vazifeh D., and Labro M. T. (1997) Erythromycin A-derived macrolides modify the functional activities of human neutrophils by altering the phospholipase D-phosphatidate phospohydrolase transduction pathway: L-cladinose is involved both in alterations of neutrophil functions and modulation of this transductional pathway. J. Immunol. 159, 3995–4005 [PubMed] [Google Scholar]
- 70. Labro M. T., el Benna J., and Babin-Chevaye C. (1989) Comparison of the in vitro effect of several macrolides on the oxidative burst of human neutrophils. J. Antimicrob. Chemother. 24, 561–572 10.1093/jac/24.4.561 [DOI] [PubMed] [Google Scholar]
- 71. Labro M. T., and el Benna J. (1991) Effects of anti-infectious agents on polymorphonuclear neutrophils. Eur. J. Clin. Microbiol. Infect. Dis. 10, 124–131 10.1007/BF01964424 [DOI] [PubMed] [Google Scholar]
- 72. Yamanaka Y., Tamari M., Nakahata T., and Nakamura Y. (2001) Gene expression profiles of human small airway epithelial cells treated with low doses of 14- and 16-membered macrolides. Biochem. Biophys. Res. Commun. 287, 198–203 10.1006/bbrc.2001.5550 [DOI] [PubMed] [Google Scholar]
- 73. Shimizu T., Shimizu S., Hattori R., Gabazza E. C., and Majima Y. (2003) In vivo and in vitro effects of macrolide antibiotics on mucus secretion in airway epithelial cells. Am. J. Respir. Crit. Care Med. 168, 581–587 10.1164/rccm.200212-1437OC [DOI] [PubMed] [Google Scholar]
- 74. Takizawa H., Desaki M., Ohtoshi T., Kawasaki S., Kohyama T., Sato M., Tanaka M., Kasama T., Kobayashi K., Nakajima J., and Ito K. (1997) Erythromycin modulates IL-8 expression in normal and inflamed human bronchial epithelial cells. Am. J. Respir. Crit. Care Med. 156, 266–271 10.1164/ajrccm.156.1.9612065 [DOI] [PubMed] [Google Scholar]
- 75. Cossarizza A., and Salvioli S. (2001) Flow cytometric analysis of mitochondrial membrane potential using JC-1. Curr. Protoc. Cytom. Chapter 9, Unit 9.14 10.1002/0471142956.cy0914s13 [DOI] [PubMed] [Google Scholar]
- 76. Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., and Pacher P. (2007) Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 358, 203–208 10.1016/j.bbrc.2007.04.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chow S., Hedley D., and Tannock I. (1996) Flow cytometric calibration of intracellular pH measurements in viable cells using mixtures of weak acids and bases. Cytometry 24, 360–367 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.