Abstract
Chitin degradation is important for biomass conversion and has potential applications for agriculture, biotechnology, and the pharmaceutical industry. Chitinase A from the Gram-negative bacterium Serratia marcescens (SmChiA) is a processive enzyme that hydrolyzes crystalline chitin as it moves linearly along the substrate surface. In a previous study, the catalytic activity of SmChiA against crystalline chitin was found to increase after the tryptophan substitution of two phenylalanine residues (F232W and F396W), located at the entrance and exit of the substrate binding cleft of the catalytic domain, respectively. However, the mechanism underlying this high catalytic activity remains elusive. In this study, single-molecule fluorescence imaging and high-speed atomic force microscopy were applied to understand the mechanism of this high-catalytic-activity mutant. A reaction scheme including processive catalysis was used to reproduce the properties of SmChiA WT and F232W/F396W, in which all of the kinetic parameters were experimentally determined. High activity of F232W/F396W mutant was caused by a high processivity and a low dissociation rate constant after productive binding. The turnover numbers for both WT and F232W/F396W, determined by the biochemical analysis, were well-replicated using the kinetic parameters obtained from single-molecule imaging analysis, indicating the validity of the reaction scheme. Furthermore, alignment of amino acid sequences of 258 SmChiA-like proteins revealed that tryptophan, not phenylalanine, is the predominant amino acid at the corresponding positions (Phe-232 and Phe-396 for SmChiA). Our study will be helpful for understanding the kinetic mechanisms and further improvement of crystalline chitin hydrolytic activity of SmChiA mutants.
Keywords: chitinase, processivity, single-molecule biophysics, Serratia marcescens, chitin, biomass conversion, biotechnology, chitin degradation, high-speed atomic force microscopy, single-molecule fluorescence imaging
Introduction
Chitin is one of the most abundant polysaccharides found in nature, only secondary to cellulose. Chitin is a water-insoluble polysaccharide consisting of β-1,4 glycosidic linkages of the monomer GlcNAc, and it is mostly found in the shells of crustaceans, the exoskeleton of insects, and the cell walls of some fungi (1–3). Chitin degradation is important not only in nature, but also in industrial applications such as biomass conversion, and has potential applications in agriculture, biotechnology, and the pharmaceutical industry (4, 5). Due to its stable crystalline structure, chitin is very durable, only decomposing at very high temperatures and under high pressure (6).
There are many naturally occurring organisms that possess the ability to degrade chitin under mild conditions by using enzymes called chitinases (7). According to the Carbohydrate-Active enZYmes database (CAZY: http://www.cazy.org/)2 (52), chitinase is classified in the glycoside hydrolase family 18 (EC 3.2.1.14) and functions to catalyze the hydrolysis of the β-1,4 glycosidic bonds of chitin. This hydrolysis occurs on the crystalline surface of chitin. Moreover, the chitinases of the bacterium Serratia marcescens are a well-known model for the study of chitin degradation. S. marcescens chitinase A (SmChiA),3 an exo-processive enzyme, is the most powerful enzyme among the chitinases of Serratia in the hydrolysis of crystalline chitin (8, 9). SmChiA is a linear molecular motor enzyme that hydrolyzes chitin from the reducing end and works in the extracellular environment without the need for ATP. SmChiA is composed of two domains: a catalytic domain (CD) and a chitin-binding domain (CBD) (Fig. 1A) (10). Both the chitin-binding cleft of CD and the chitin-binding surface of CBD have aromatic residues lined along them. These aromatic residues play important roles in both substrate binding and the hydrolytic activity and processivity of the SmChiA (11, 12).
Figure 1.
Schematic model of SmChiA. A, crystal structure of SmChiA complexed with (GlcNAc)7 (sphere model) and crystalline chitin (stick model). The schematic shows the CD and CBD. Phe-232 and Phe-396 (sphere model, colored with cyan) are located at the entrance and exit of the chitin-binding cleft, respectively. B and C, structural superimposition to compare the aromatic residues inside the chitin-binding cleft of SmChiA (blue and side chain highlighted with cyan; PDB entry 1CTN) and OfChi-h bound with (GlcNAc)7 (pink; PDB entry 5GQB) in side view (B) and bottom view (C). The red boxes indicate the two different aromatic residues in SmChiA and OfChi-h. The numbers (−6 to +2) represent the chitin-binding subsites of SmChiA according to the standard nomenclature (38, 39). The red dashed lines in B and C show the position of the bond cleavage.
Processive enzymes play the important roles in various biological activities such as DNA/RNA synthesis (13, 14), cargo transport (15, 16), and protein (17, 18) and polysaccharide (19–21) degradations. Once processive enzymes bind to their substrates, they can repeat multiple cycles of catalysis without dissociation (22, 23). Processive cellulases and chitinases are well-known examples of the processive enzymes that perform multiple rounds of hydrolytic cleavage of cellulose and chitin, respectively (19–21). Processivity prevents the dissociation/reassociation process once the enzyme binds to the end of a single polymer chain, thereby reducing the number of times the enzyme rebinds to the end of the same substrate chain (20, 24). Processive cellulases and chitinases share the similar feature of a long and deep substrate-binding cleft and substrate-binding surface, which contain aromatic amino acid residues (20, 21, 25). These aromatic residues play an important role in the carbohydrate-protein interaction by which hydrophobic stacking (CH-π interaction) is formed between the aromatic side chain and sugar ring. This interaction is thought to be beneficial for processivity by reducing the sliding energy of the polymer carbohydrate chain (20, 26–29).
Studies on processivity of the cellulases and chitinases using biochemical methods, such as the fluorescence labeling of the substrate (30, 31) and 14C-labeled chitin (32), or via the use of biosensors (33) have been performed extensively. However, they often involve complicated procedures and have some limitations. Furthermore, processivity cannot be directly measured using a biochemical assay as it requires interpretations and is usually estimated from the dissociation rate. Recently, single-molecule imaging methods with fluorescence microscopy or high-speed atomic force microscopy (HS-AFM) have been used to directly visualize the processive movement of the enzymes because they are more straightforward than biochemical methods (34–37). In our previous study (37), we reported not only the processivity but also the kinetic parameters of SmChiA WT (His6-tagged), including the binding rate constant (kon), dissociation rate constant (koff), translational velocity (ktr), and productive binding ratio, obtained using single-molecule fluorescence imaging.
Recently, Liu et al. (38) reported the structure alignment of the substrate-binding cleft of Ostrinia furnacalis chitinase-h (OfChi-h) and SmChiA. As a result, two different aromatic residues, Phe-232 and Phe-396 for SmChiA and Trp-225 and Trp-389 for OfChi-h, were found in the beginning and end of the cleft (at the chitin-binding subsites −6 and +2) (38, 39), respectively, as shown in Fig. 1 (B and C). In addition, the mutation of these two positions of SmChiA into those of OfChi-h (F232W/F396W) showed a higher hydrolytic activity against crystalline chitin compared with the SmChiA WT. However, the details of the mechanism for improved activity have not yet been reported. In the present study, we performed detailed biochemical analysis and single-molecule imaging analysis using fluorescence microscopy and HS-AFM to understand the mechanism by which F232W/F396W mutant showed a higher catalytic activity than WT. A reaction scheme including processive catalysis was used to explain the properties of SmChiA WT and F232W/F396W, in which all the kinetic parameters were experimentally determined. We also performed amino acid sequence alignment of 258 SmChiA-like proteins and revealed the predominant aromatic amino acid residues responsible for the chitin binding.
Results
SmChiA F232W/F396W mutant showed higher kcat than WT
The previous study by Liu et al. (38) compared the hydrolytic activity of OfChi-h, SmChiA WT, and SmChiA F232W/F396W. They found that the SmChiA F232W/F396W mutant showed higher hydrolytic activities against several insoluble chitin substrates than SmChiA WT; however, these results were obtained at only one substrate concentration (38). In our previous study (37), we used a range of crystalline chitin concentrations (0–2 mg/ml) to estimate the kcat and Km values of SmChiA WT. In the present study, to confirm the high catalytic activity of the F232W/F396W mutant and estimate kcat and Km, we measured the hydrolytic activity of SmChiA WT and F232W/F396W at various crystalline chitin concentrations (0–6 mg/ml) using a liquid-handling robot, developed in our previous report (40). The measurement was performed in two independent experiments carried out in triplicate. It is worth noting that contrary to the previous study (37), in the present study, His6 tags were removed from the C terminus of the constructs during purification. This was done as positive charges of His6 may change the binding/dissociation dynamics of SmChiA against the crystalline chitin immobilized on the surface of a negatively charged glass surface. Furthermore, fluorescently labeled enzymes (Cy3-D415C WT and F232W/F396W) were used for all biochemical measurements as these enzymes were also used for the single-molecule imaging analysis described later. Note that in our previous study (37), we confirmed that there is no significant difference between hydrolytic activities of Cy3-labeled and unlabeled enzymes.
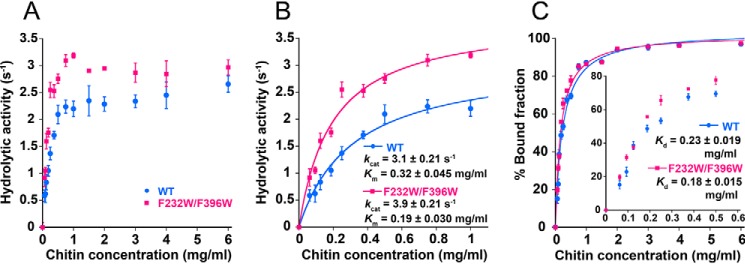
As a result, F232W/F396W was found to show higher hydrolytic activity than WT in all of the crystalline chitin concentrations tested (Fig. 2A). At a high crystalline chitin concentration (defined as more than 1 mg/ml), the hydrolytic activities of WT and F232W/F396W tended to show a degree of inhibition and were not appropriate within the Michaelis–Menten equation. Thus, we used the hydrolytic activity at a low chitin concentration range (0–1 mg/ml) to ensure fit within the Michaelis–Menten equation (Fig. 2B). The turnover number (kcat) and Michaelis constant (Km) for WT and F232W/F396W on crystalline chitin were subsequently estimated to be 3.1 ± 0.21 and 3.9 ± 0.21 s−1 and 0.32 ± 0.045 and 0.19 ± 0.030 mg/ml, respectively (Table 1). Furthermore, the values of kcat/Km for WT and F232W/F396W were 10 and 21 ml mg−1 s−1, respectively. This result suggests a 2 times larger rate constant of productive binding for F232W/F396W than that for WT.
Figure 2.
Biochemical analysis. A, hydrolytic activity of SmChiA WT (blue) and F232W/F396W (pink) at various concentrations of crystalline chitin (0–6 mg/ml). B, same as A at a low concentration range (0–1 mg/ml). The data points were fitted with the Michaelis–Menten equation to estimate kcat and Km of WT and F232W/F396W. Hydrolytic activity was measured in 50 mm sodium phosphate (pH 6.0) at 25 °C. C, the bound fraction of WT and F232W/F396W at various concentrations of crystalline chitin (0–6 mg/ml). Inset, the low chitin concentration range (<0.6 mg/ml). The amount of free enzymes was used to calculate the bound fraction percentage. The distribution was fitted with the Langmuir equation to estimate the dissociation constant (Kd). Error bars, S.D. of the sextupled (A and B) or triplicate (C) measurements.
Table 1.
Summary of kinetic parameters determined by biochemical analysis
| Kinetic parameters | kcata | Kma | kcat/Km | Kdb |
|---|---|---|---|---|
| s−1 | mg/ml | ml mg−1 s−1 | mg/ml | |
| WT | 3.1 ± 0.21 | 0.32 ± 0.045 | 10 | 0.23 ± 0.019 |
| F232W/F396W | 3.9 ± 0.21 | 0.19 ± 0.030 | 21 | 0.18 ± 0.015 |
a kcat and Km were estimated from the biochemical activity measurement at a low chitin concentration range (0–1 mg/ml) with the fitting by the Michaelis–Menten equation.
b Kd was estimated from the bound fraction analysis with the fitting by Langmuir's equation.
In addition, we performed a biochemical binding assay to compare the ratio of bound fractions between WT and F232W/F396W at various crystalline chitin concentrations. The free enzymes in the solution were used to calculate the percentage of the bound fraction. The plot was fitted using Langmuir's equation to estimate the dissociation constant (Kd) (Fig. 2C). At a crystalline chitin concentration below 0.6 mg/ml, the binding affinity of F232W/F396W was found to be slightly higher than that of the WT (Fig. 2C, inset). At a concentration of 2 mg/ml crystalline chitin, almost all of the enzymes of both the WT and F232W/F396W were found to be bound to the crystalline chitin surface (over 90%). The values of Kd for WT and F232W/F396W were 0.23 ± 0.019 and 0.18 ± 0.015 mg/ml, respectively (Table 1). These results indicate that the binding affinity increased slightly as a result of the mutation of two phenylalanine residues into tryptophan residues.
No significant differences in binding and dissociation rate constants and productive binding ratio for WT and F232W/F396W
To further clarify the mechanism responsible for the higher hydrolytic activity in the F232W/F396W mutant compared with the WT, we first performed single-molecule fluorescence imaging according to the methods described in our previous study (37). Note that in the single-molecule fluorescence imaging and HS-AFM observation, it is difficult to define the chitin concentrations because the chitin microfibrils are attached on the glass or mica surface. Both the kon and koff of the WT and F232W/F396W were found to be similar (Fig. 3). The distributions of kon were fitted using the double Gaussian function (Fig. 3, top). The values of the peak positions for the WT were 8.2 ± 0.30 × 108 and 1.4 ± 0.43 × 109 m−1 μm−1 s−1, whereas those of the F232W/F396W were 9.1 ± 0.43 × 108 and 1.8 ± 0.29 × 109 m−1 μm−1 s−1, respectively. The multiple peaks of kon were related to the bundles of chitin microfibrils, wherein the first peak was represented by kon for a single crystalline chitin microfibril, as explained in our previous study (37). Essentially, no significant differences in overall kon between WT and F232W/F396W were obtained (Table 2).
Figure 3.
Comparison of the distribution of kon and koff of SmChiA WT (blue) and F232W/F396W (pink) obtained by single-molecule fluorescence imaging analysis. Top, the distributions of kon were fitted with the double Gaussian function. The first peak corresponds to the kon against the single crystalline chitin microfibril. The sample number (N) in the distribution of kon represents the number of crystalline chitins observed. Bottom, the distributions of koff were fitted with the double-exponential decay function. A slow dissociation is associated with the binding of the enzyme to the hydrophobic crystalline chitin surfaces, whereas the fast dissociation is associated with the binding of the enzyme to the hydrophilic crystalline chitin surfaces. The sample number (N) in the distribution of koff represents the number of SmChiA molecules observed.
Table 2.
Summary of kinetic parameters determined by single-molecule analysis and reaction scheme
| Kinetic parameters | kona | koffNPa,b | konP/konNPa,c | ktrd | kpce | Pd,f | koffPd | kcatg |
|---|---|---|---|---|---|---|---|---|
| m−1 μm−1 s−1 | s−1 | nP/nNP | nm/s | s−1 | s−1 | s−1 | ||
| WT | 8.2 × 108 ± 3.0 × 107 | 1.2 | 0.074 ± 0.0041 | 52 ± 0.53 | 50 | 30 | 1.4 ± 0.26 | 2.9 |
| F232W/F396W | 9.1 × 108 ± 4.3 × 107 | 0.99 | 0.076 ± 0.0089 | 51 ± 0.72 | 49 | 52 | 0.82 ± 0.089 | 4.1 |
a kon, konP/konNP, and koffNP values were obtained from single-molecule fluorescence imaging analysis.
b koffNP, which was the average koff, was assumed to only contain nonproductive binding.
c konP/konNP or nP/nNP was denoted as the productive binding ratio, where nP and nNP represent the number of moving and non-moving molecules for a total of 40 s observation, respectively. For the analysis, we defined moving molecules as the molecules which showed movements larger than 20 nm for 3 or more frames.
d ktr, processivity (P), and koffP were obtained by HS-AFM observation. The molecules that showed movements for 3 or more frames were analyzed.
e ktr was used to denote the translational velocity. kpc was calculated by dividing the ktr value by the step size (a product size, 1.04 nm).
f P was estimated from the run length divided by the step size (a product size, 1.04 nm).
g kcat was calculated from Equation 3.
The distribution of koff was fitted using a double-exponential decay function (Fig. 3, bottom). The slow and fast dissociations were associated with the bindings of the SmChiA to the hydrophobic and hydrophilic surfaces of crystalline chitin, respectively (37). The ratios of the slow and fast dissociations were calculated from the area under the fitting curves. The slow dissociation rate constants for WT and F232W/F396W were 0.089 ± 0.15 s−1 (14%) and 0.058 ± 0.12 s−1 (15%), respectively. The fast dissociation rate constants for WT and F232W/F396W were 1.4 ± 0.089 s−1 (86%) and 1.2 ± 0.058 s−1 (85%), respectively. Subsequently, the average value of koff was calculated from summation of the koff and the ratio of the slow dissociation fraction and that of the fast dissociation fraction. As a result, the average values of koff for WT and F232W/F396W were 1.2 and 0.99 s−1, respectively (Table 2). As described below, we used these values of koff as those of koff for nonproductive binding (koffNP).
Next, we measured the productive binding ratio. Productive binding only occurs when SmChiA binds to the reducing end of the chitin chain on the hydrophobic surface of a crystalline chitin. Therefore, the productive binding ratio of SmChiA is low, as was determined by single-molecule imaging analysis in our previous study (37). In the present study, the productive binding ratio was determined from the ratio of the number of moving molecules (nP) and nonmoving molecules (nNP) after the binding to the chitin surface. For measurement, we further improved the localization precision of single-molecule fluorescence imaging to identify the slowly moving molecules more precisely (4.3 and 3.8 nm in the x and y direction at 3 frames per second (fps), with a laser at 1 μW/μm2 power). The productive binding ratios for WT and F232W/F396W were 0.074 ± 0.0041 and 0.076 ± 0.0089, respectively, and approximately the same (Table 2).
F232W/F396W showed high processivity and low dissociation rate after productive binding
As no significant difference was found between the WT and F232W/F396W using single-molecule fluorescence imaging analysis, we then applied single-molecule imaging with HS-AFM to improve the localization precision. Several chitin microfibrils were observed to avoid heterogeneity on the crystalline chitin surface. At least 10 molecules per chitin were observed in order to estimate the translational velocity (ktr), run length, and moving time (Fig. 4). The distributions of ktr were fitted with the single Gaussian function (Fig. 4, left). As a result, the WT and F232W/F396W were found to have a similar ktr, 52 ± 0.53 and 51 ± 0.72 nm/s, respectively. Then the processive catalysis rate constant (kpc) was calculated from the ktr divided by the step size 1.04 nm, which is same as the size of the reaction product, chitobiose (36). The values of kpc for WT and F232W/F396W were 50 and 49, respectively. The distributions of run length (Fig. 4, center) and moving time (Fig. 4, right) were fitted with the single-exponential decay function. Note that the first bins (gray bars) of the run length were not included for fitting, because precise measurement of short run length was difficult. The values of run length for WT and F232W/F396W were 31 ± 2.6 and 54 ± 4.1 nm, respectively, and F232W/F396W showed a longer run length. Then the values of processivity (P, run length divided by step size) were calculated as 30 and 52 for WT and F232W/F396W, respectively. Because F232W/F396W showed longer run length than WT, its moving time was also longer. The values of moving time for WT and F232W/F396W were 0.69 ± 0.12 and 1.2 ± 0.13 s, respectively. Because we only analyzed moving molecules in the HS-AFM observation, the inverse of the moving time corresponds to the productive dissociation rate constant, koffP. The values of koffP for WT and F232W/F396W were 1.4 ± 0.26 and 0.82 ± 0.089 s−1, respectively (Table 2).
Figure 4.
Comparison of the distribution of ktr, run length, and moving time of WT (blue) and F232W/F396W (pink) obtained by HS-AFM. Left, the distributions of ktr were fitted with the single Gaussian function. The processive catalysis rate constant (kpc) was calculated by dividing ktr with the step size (product size: 1.04 nm). Center and right, the distributions of run length and moving time were fitted with the single-exponential decay function. The processivity (P) was estimated by dividing the run length by the step size. The inverse of the moving time was determined as the productive dissociation rate constant (koffP), because all of the molecules analyzed by HS-AFM were moving molecules. The N and N chitin represent the number of SmChiA molecules and chitin microfibrils, respectively. The first bins (gray bars) of the run length were not included for fitting, because precise measurement of short run length was difficult.
Reaction scheme and calculation of kcat from kinetic parameters obtained by single-molecule analysis
To calculate the kcat from the kinetic parameters determined by single-molecule imaging analysis, a reaction scheme of SmChiA, including productive binding, nonproductive binding, and processive catalysis, is considered (Fig. 5). As also described above, SmChiA has two binding modes: productive and nonproductive bindings. In the reaction scheme shown in Fig. 5, these two modes are represented by four rate constants, the productive binding rate constant (konP), the productive dissociation rate constant (koffP), the nonproductive binding rate constant (konNP), and the nonproductive dissociation rate constant (koffNP). Furthermore, because SmChiA is a processive enzyme, another kinetic parameter is included, kpc. From this reaction scheme, we can derive the following equations to estimate kcat and Km.
| (Eq. 1) |
| (Eq. 2) |
Figure 5.
Reaction scheme of the processive catalysis of SmChiA described with the kinetic parameters obtained experimentally. The green, blue, and red arrows denote the productive binding/dissociation, nonproductive binding/dissociation, and processive catalysis (hydrolysis cycle), respectively. E, enzyme (SmChiA); S, substrate (crystalline chitin); ES, enzyme-substrate complex after productive binding; E′S, enzyme-substrate complex after nonproductive binding; P, product (chitobiose).
As described, the values of koffP, koffNP, and kpc for WT and F232W/F396W in Equation 1 were already determined by the single-molecule imaging analysis. Because the ratio of productive binding was very low in the single-molecule fluorescence imaging, we approximated the values of koffNP with those of koff (Fig. 3 and Table 2). The values of koffP were obtained as the inverse of the moving time in the HS-AFM observation (Fig. 4 and Table 2). The values of kpc were also estimated from the ktr divided by the step size (1.04 nm) in the HS-AFM observation (Fig. 4 and Table 2).
On the other hand, quantitative determinations of absolute values of konP and konNP were difficult even with single-molecule imaging analysis. The values of kon that were obtained from the single-molecule fluorescence imaging included both productive and nonproductive bindings. We could not distinguish productive and nonproductive bindings because of low excitation laser power (0.14 μW/μm2) and low localization precision (8.6 and 8.4 nm in x and y directions, respectively) under the experimental conditions used in the kon and koff measurements. Therefore, to calculate the values of kcat from the kinetic parameters obtained, we modified Equation 1 and used the ratio of konP and konNP as follows.
| (Eq. 3) |
The ratio of konP and konNP in Equation 3 corresponds to the productive binding ratio (nP/nNP) (Table 2), determined from the ratio of nP and nNP in the single-molecule fluorescence imaging with high excitation laser power (1 μW/μm2) and high localization precision (∼4 nm for both x and y directions). The obtained values of kcat for WT and F232W/F396W were 2.9 and 4.1 s−1, respectively (Table 2). These values showed good agreement with those determined by the biochemical assay, which were 3.1 and 3.9 s−1 for WT and F232W/F396W, respectively (Table 1).
Discussion
Our biochemical analysis confirmed that the F232W/F396W mutant of SmChiA has a higher hydrolytic activity than the WT, as shown previously (38) (Fig. 2). We found that F232W/F396W showed higher hydrolytic activity than WT at all chitin concentrations ranging from 0.063 to 6 mg/ml. Hydrolytic activities were slightly inhibited for both WT and F232W/F396W at chitin concentrations higher than 1 mg/ml and could not be fitted using the Michaelis–Menten equation (Fig. 2A). This inhibition can be either the product or substrate inhibition, and additional experiments are required to understand the mechanism.
At low chitin concentration range (0–1 mg/ml), the Michaelis–Menten equation could be applied to obtain an estimate of the kcat and Km (Fig. 2B). The kcat value for F232W/F396W (3.9 s−1) was higher than that for WT (3.1 s−1) (Table 1). The Km value for F232W/F396W (0.19 mg/ml) was lower than for WT (0.32 mg/ml), indicating that the affinity of the productive binding to the crystalline chitin of this mutant was higher than that of the WT. Actually, the F232W/F396W slightly increased the binding affinity to crystalline chitin, including productive and nonproductive bindings, as demonstrated by the results of biochemical bound fraction analysis, especially at low chitin concentrations (Fig. 2C). The Kd values for WT and F232W/F396W were consistent with the Km values (Table 2). Because Phe was mutated to Trp at two positions, the surface area that engages the CH-π interactions and the electronegativity of the π-system can be increased (41).
Among the various kinetic parameters analyzed, processivity was the only one parameter for which F232W/F396W showed a higher value than the WT. The processivity of WT obtained by HS-AFM using β-chitin as a substrate was almost identical with that estimated in a previous report using HS-AFM (34) (values of 30 and 29 for the present and previous studies, respectively) (Fig. 4). Moreover, the processivity of WT was also similar to the apparent processivity (36 ± 5) estimated by biochemical analysis using α-chitin (32).
For Phe-232 and Phe-396 of SmChiA, single point mutations have been reported previously. The mutation of Phe-232 to Ala dramatically decreased the hydrolytic activity. However, the binding affinity of F232A and WT against crystalline chitin were similar (12). In addition, the Trp-231 of Vibrio harveyi chitinase A (VhChiA) corresponding to the Phe-232 in SmChiA (Fig. 6), was mutated to Phe (W231F), which resulted in nondetectable levels of activity against the crystalline chitin substrate (42). Phe-232 may be important in guiding the chitin chain into the substrate binding cleft not only in SmChiA but also in chitinase A in other organisms. Moreover, the mutation of F396A decreased the hydrolytic activity against the crystalline chitin. This F396A mutation increased the Kd value 2-fold and slightly reduced the processivity compared with WT (11, 43). The slightly decreased Km was also observed when Phe-396 was mutated to Trp with D313N (44, 45). Taken together, F232W/F396W mutation in SmChiA would be better in guiding the chitin chain, whereby the binding affinity was increased and the degree of processivity was improved as a result of the larger aromatic surface area of Trp compared with Phe.
Figure 6.
Comparison of the aromatic amino acid residues responsible for the binding to the crystalline chitin surface and chain sliding into the chitin binding cleft. The amino acid sequences of 258 SmChiA-like proteins were aligned and visualized by Clustal-Omega and WebLogo3. The aromatic amino acids of SmChiA are shown in cyan. The alignment shows the conservation of several aromatic amino acid residues involved in chitin binding. The structural alignment image was constructed by superimposing the crystal structure of SmChiA (PDB entry 1CTN; blue and side chain highlighted with cyan), OfChi-h (PDB entry 5GQB; pink), and VhChiA (PDB entry 3B8S; orange) using PyMOL software. The side chains of aromatic amino acid residues are shown as stick models.
In previous studies, the hydrolytic activity of SmChiA against crystalline chitin was found to decrease after the introduction of an alanine mutation into the aromatic residues inside the substrate binding cleft (W167A, W275A, or F396A). These mutants showed reduced levels of processivity on the crystalline chitin (11, 32). On the other hand, interestingly, these mutants were found to outperform SmChiA WT in the hydrolysis of soluble chitin and soluble chitosan (11, 12). Furthermore, recently, the F232W/F396W SmChiA mutant was found to have an increased hydrolytic activity for insoluble chitin (insect cuticle, α-chitin, and chitin nanowhisker), but a decreased activity in ethylene glycol chitin, which is a water-soluble chitin substrate (38). In the present study, we found that high hydrolytic activity of the F232W/F396W against crystalline chitin resulted from high processivity. Our results indicate that processivity is an important factor for the hydrolysis of crystalline substrates. On the other hand, it remains elusive why the SmChiA alanine mutants (W167A, W275A, or F396A) and F232W/F396W, respectively, showed higher and lower hydrolytic activities than the SmChiA WT against the soluble substrates. To understand the mechanism and correlation with the processivity, measurements of the processivity of SmChiA WT and these mutants against the soluble substrates will be required.
The processivity (P) of SmChiA is related to koffP (Fig. 4). If the value of P is high, that of koffP is low (Table 2). Several studies have reported largely different values of the dissociation rate (30–33). The koff from the chitin nanowhiskers was 0.012 ± 0.002 s−1, whereas the koff from the α-chitin after 10 min and 2 h of incubation was 0.0028 ± 0.0003 and 0.0015 ± 0.0005 s−1, respectively (32). These values were much lower than those found in the present study and our previous study (37). Because the previous studies of processive cellulases reported different koff values measured with different methods (31, 33, 46–51), this difference could also occur in the case of processive chitinase. For chitinase, biochemical analysis revealed that the koff value is also dependent on the type of substrate and the reaction time (32). It is worth noting that in biochemical analysis, it is very difficult to distinguish productive and nonproductive bindings. On the other hand, in our single-molecule analysis, we have analyzed only productively bound SmChiA molecules moving on the surface of crystalline chitin.
To gain further insight into the aromatic amino acid residues in the substrate-binding cleft, we then performed bioinformatics analysis. The amino acid sequence of SmChiA and those of 258 SmChiA-like proteins obtained by the Blast analysis were aligned (Fig. 6). As a result, we found that Phe-232 and Phe-396 of SmChiA were not conserved, and the Trp was predominant and higher than 80% at both positions. Interestingly, Hudson et al. (41) previously reported that, for most proteins that interact with carbohydrates, aromatic side chains are involved, and the most preferable amino acid residue is tryptophan. Furthermore, we also found that not only are the aromatic residues within the substrate binding cleft, but also many other amino acid residues in the substrate binding cleft are not conserved in SmChiA (data not shown). Our results strongly suggest that although SmChiA is the most studied processive chitinase, the amino acid sequence is not optimized for high hydrolytic activity. Bioinformatics analysis used in this study will be helpful to find the amino acid residues that are not conserved in SmChiA, and these amino acid residues can be targets of the mutation for further improvement of the crystalline chitin hydrolytic activity. Furthermore, our single-molecule imaging analysis and reaction scheme will make it possible to understand the mechanisms of the highly active mutants.
To conclude, the F232W/F396W mutant of SmChiA showed high processivity and low productive dissociation rate constant, which resulted in a kcat value higher than that for WT. The values of kcat for F232W/F396W and WT obtained by biochemical analysis were well-reproduced by the kinetic parameters obtained by the single-molecule analysis, indicating the validity of the proposed reaction scheme. Our results highlight the importance of integration of biochemical analysis and single-molecule analysis to understand the mechanisms of SmChiA. To further clarify the mechanism in more detail, single-molecule analysis of the SmChiA mutants with higher time resolution and higher localization precision probed with a gold nanoparticle will be very important (36).
Experimental procedures
Preparation of enzymes
The SmChiA WT gene (including the D415C mutation for fluorescent labeling) (37) in the expression plasmid pET27b with the C-terminal Factor Xa (FaXa) recognition sequence and His6 tag was used as the template to introduce the F232W/F396W mutation by PCR. The PCR product was treated with 1 μl of DpnI (New England Biolabs) to reduce the template background at 37 °C for 15 min. After DpnI treatment, 1% agarose gel electrophoresis was performed. The target fragments were extracted and purified using a gel extraction kit (Promega). The purified DNA fragments were ligated using the NEBuilder Assembly Tool (New England Biolabs). The DNA fragments and reagent were mixed at a ratio of 1:1 (v/v) and then incubated at 50 °C for 30 min. After the ligation reaction, the samples were immediately used for transformation using E. coli (TunerTM DE3) as a host. Transformed cells were incubated for 1 h at 37 °C and spread on an LB plate with 25 μg/ml kanamycin. Single colonies were inoculated into 10 ml of LB medium with 25 μg/ml kanamycin and incubated overnight at 37 °C and 250 rpm. The plasmid containing mutant gene was purified from the harvested cell and the sequence was verified. The Escherichia coli colonies carrying SmChiA F232W/F396W or WT expression plasmids were cultured in 10 ml of LB medium with 25 μg/ml kanamycin at 37 °C and 250 rpm until A600 = 1. Then 5 ml of the culture was added to 1 liter of LB medium with 25 μg/ml kanamycin in a 3-liter flask and cultured at 37 °C and 130 rpm until A600 = 1.8. Then the media were cooled on ice water for 10 min, isopropyl β-d-1-thiogalactopyranoside was added at a final concentration of 500 μm, and cells were further cultured at 20 °C and 130 rpm overnight. The culture was then centrifuged at 6000 × g for 10 min. Ten times the volume of the cell weight of 50 mm sodium phosphate (pH 7.0), containing 100 mm NaCl, was added and supplemented with protease inhibitor mixture (cOmplete Mini, EDTA-free, Roche Applied Science). The cell suspension was sonicated on ice for 20 min at 3-s intervals. The disrupted cells were then centrifuged at 4 °C and 30,000 × g for 10 min. The supernatant was incubated with Ni-NTA Superflow (Qiagen) and equilibrated with 50 mm sodium phosphate (pH 7.0) containing 100 mm NaCl for 15 min at room temperature under gentle rotation. Then the Ni-NTA resin was packed into an open column and washed with 0 and 50 mm imidazole in 50 mm sodium phosphate (pH 7.0) containing 100 mm NaCl and eluted with 100 mm imidazole in 50 mm sodium phosphate (pH 7.0) containing 100 mm NaCl. The eluted fractions were pooled and concentrated to 500 μl using a 30,000 molecular weight cut-off VIVASPIN Turbo 50 (Sartorius). The sample was then injected into a Superdex 200 10/300 GL column (GE Healthcare) and eluted with 50 mm Tris-HCl (pH 8.0) containing 100 mm NaCl. The fractions were collected at a flow rate of 0.5 ml/min. The eluted fractions were mixed and concentrated to 200 μl using a 30-kDa molecular mass cut-off VIVASPIN Turbo 15 (Sartorius). Protein concentrations were estimated from the absorbance at 280 nm and the molar extinction coefficients (ϵ280 = 107,050 and 118,050 m−1 cm−1 for WT and F232W/F396W, respectively). The molar extinction coefficients were calculated by using the ProtParam in the Expasy bioinformatics resource portal web service (https://web.expasy.org/protparam/)2. One hundred microliters of 100 μm sample were incubated with 5 μl of 1 mg/ml FaXa protease (New England Biolabs) and 2 μl of 100 mm calcium chloride at 23 °C overnight to digest C-terminal His6. Then 10 μl of sodium phosphate (1 m, pH 7.0) was added to the FaXa-treated sample, and the sample was centrifuged at 4 °C and 16,000 × g for 10 min to precipitate calcium phosphate. The supernatant was applied to the Ni-NTA column to remove the cleaved His6 tag and undigested samples. The column was washed, and the flow-through fractions were collected with 50 mm sodium phosphate (pH 7.0) containing 100 mm NaCl. The collected fractions were mixed and concentrated to 500 μl using a 30-kDa molecular mass cut-off membrane (VIVASPIN Turbo 15, Sartorius). DTT at a final concentration of 10 mm was added to the sample to prevent the formation of disulfide bonds between D415C. The sample was then loaded onto a Superdex 200 10/300 GL column (GE Healthcare) with 50 mm sodium phosphate (pH 7.0) containing 100 mm NaCl to remove FaXa protease and DTT. Cy3-maleimide (GE Healthcare) was dissolved in DMSO and mixed with the sample at the same molar concentration as the enzyme before incubating for 1 h at room temperature. The unreacted Cy3-maleimide was removed using a NAP-5 column (GE Healthcare). The labeling ratio of Cy3 to enzyme (97% for WT and 83% for F232W/F396W mutant) was calculated from the absorbance at 280 and 550 nm, the molar extinction coefficient of the enzyme as described above, and the Cy3-maleimide (ϵ280 = 12,000 m−1 cm−1 and ϵ550 = 150,000 m−1 cm−1). The samples were then stored at −80 °C until further use. In this study, WT (D415C-Cy3) and F232W/F396W/D415C-Cy3 are described as WT and F232W/F396W, respectively.
Crystalline β-chitin was purified from Satsuma tube worms (Lamellibrachia satsuma), as described in a previous study (34).
Biochemical measurement of chitinase hydrolytic activity
We used a liquid-handling robot, Beckman Coulter Biomek 4000, to measure the hydrolytic activity of all of the samples (blank, WT, and F232W/F396W). The samples were measured in triplicate simultaneously in 96-well plates. The purified enzymes were diluted to 100 nm using 100 mm sodium phosphate (pH 6.0) in a low-protein-binding microtube. In 96-well reaction plates, the diluted enzymes were incubated with crystalline chitin (0–6 mg/ml) at 25 °C for 30 min in a reaction mixture volume of 150 μl (1:1 (v/v) enzyme/substrate ratio) without shaking. The reactions were stopped with 200 μl of the Schales' reagent (500 mm sodium carbonate, 1.5 mm potassium ferricyanide). Insoluble chitin was separated on 96-well 1.2-μm hydrophilic low-protein-binding Durapore® membrane filter plates (Merck Millipore). The filtered solution was heated at 95 °C for 15 min, and 100 μl of the samples were transferred to 384-well clear plates. Absorbance at 420 nm was measured using a multimode microplate reader (SpectraMax iD3, Molecular Devices). The amounts of soluble products were calculated from the standard curve with chitobiose. The error bars shown in Fig. 2 (A and B) represent the S.D. values of the sextupled experiments.
Biochemical bound fraction analysis
Bound fraction analysis was performed manually using a multichannel pipette. Each measurement condition was measured in triplicate. SmChiA WT and F232W/F396W were diluted to 100 nm using 100 mm sodium phosphate (pH 6.0) in a low-protein-binding microtube and transferred to a 96-well low-binding plate (Eppendorf). Then 100 nm enzyme was incubated with various concentrations of crystalline chitin (final concentration 0–6 mg/ml) at a 1:1 (v/v) enzyme/substrate ratio at 25 °C for 15 min. The 96-well plate was then centrifuged at 4400 rpm for 15 min at 25 °C. Then 100 μl of the supernatant (unbound fraction) was transferred to a 96-black well plate to measure the fluorescence. The fluorescence intensity was measured using a microplate reader (SpectraMax® iD3, Molecular Devices). The excitation and emission wavelengths were 550 and 610 nm, respectively, optimized by fluorescence spectral scanning. The intensities of the blank (no enzyme) were subtracted, and signal intensities were compared with that of the positive control (enzymes without chitin, 100% intensity). The percentages of the bound fractions were calculated, plotted, and fitted using Langmuir's equation. The error bars shown in Fig. 2C represent the S.D. values of the triplicate experiments.
Single-molecule fluorescence imaging analysis
The coverslips used for the single-molecule fluorescence imaging were cleaned with ethanol and sonicated for 10 min before rinsing with MilliQ water and cleaned with 10 m potassium hydroxide overnight to remove any contaminants on the glass surface. Before using, the coverslips were rinsed with MilliQ water. Then 60 μl of 0.01 mg/ml crystalline chitin suspension was spin-coated on the coverslip before placement on the microscope stage. The observation area was determined using bright-field microscopy. Then 20 μl of 50 pm SmChiA WT or F232W/F396W in 50 mm sodium phosphate (pH 6.0) was dropped onto the coverslip. For kon and koff analysis, fluorescence images of single molecules were recorded at 4 fps at a laser power of 0.14 μW/μm2 with localization precisions of 8.6 and 8.4 nm in x and y directions, respectively. After observation, 10 μl of 10 nm WT was dropped onto the coverslip to strain the crystalline chitin.
The values of kon were calculated from the number of SmChiA molecules bound to the single crystalline chitin divided by the SmChiA concentration, chitin length, and observation time (m−1 μm−1 s−1). Binding events were counted for 40 s after focusing. The length of the microfibrils was measured from the fluorescence images of crystalline chitin stained with 10 nm WT using ImageJ. The distributions of kon were fitted using double Gaussian functions.
The binding time distribution was fitted with the double exponential decay functions, according to the following equation,
| (Eq. 4) |
where a, b, c, and d are the fitting parameters.
The productive binding ratio was estimated from the ratio of the number of moving molecules and the number of nonmoving molecules in the initial 40-s movies after focusing. The observation condition was 1 μW/μm2 with 3 fps to improve the moving molecule–finding efficiency. The localization precisions in the x and y directions were 4.3 and 3.8 nm, respectively. For the analysis, we defined moving molecules as the molecules that showed movements larger than 20 nm (∼5 times larger than the localization precision) for 3 or more frames. Eight independent images were used to analyze and calculate the average value of the productive binding ratio. Before performing the detailed analysis of the image sequences, the trajectory of nonmoving molecules was verified to confirm whether there was any drift in the image sequences. If the image sequences showed any drift, they were not used for analysis.
HS-AFM
HS-AFM observations were carried out using the system described previously (34), except the preparation of stage. The mica surface on the stage was freshly cleaved and coated with 2 μl of fluoro surf FS-1010S135-0.5 (Fluoro Technology) to make the surface hydrophobic and to have high affinity to the crystalline chitin. Then 10 μl of crystalline chitin suspension was dropped onto the surface before incubating at room temperature (∼25 °C) for 10 min and covered with a moisture cap. The surface was rinsed twice with 80 μl of 50 mm sodium phosphate (pH 6.0). Immobilized crystalline chitins were initially observed without the SmChiA in 78 μl of 50 mm sodium phosphate (pH 6.0). After that, 2 μl of chitinase was added to obtain an enzyme at a final concentration of 2 μm. The solution was then mixed gently, and the images were recorded at 5 fps.
Home-built software based on Igor Pro (WaveMetrics) was used for the visualization and analysis of the HS-AFM images. We analyzed moving molecules that showed movements for 3 or more frames. A linear tracking function was used to track the moving molecules. First, the region of interest was selected manually at the first and last frame of each individual moving molecule. The center of the region of interest was calculated using the software to obtain the position at each frame of tracking molecules. The translational velocity (ktr) of the moving molecules was calculated using the following equation,
| (Eq. 5) |
where Δx and Δy denote the difference between the start and the end positions of a moving molecule in the x and y direction, respectively.
The distributions of ktr were fitted with the Gaussian function. The processive catalysis rate constant (kpc) was calculated by dividing ktr by the step size of SmChiA (1.04 nm, the length of chitobiose). The distribution of the run length was fitted with the single-exponential decay function. The processivity was estimated from the obtained run length constant on the assumption that the SmChiA step size was 1.04 nm. The distribution of the moving time was fitted with the single-exponential decay function, and the inverse of the obtained moving time constant was used as the productive dissociation rate constant (koffP).
Structure and sequence alignment by bioinformatics
The sequence of SmChiA and SmChiA-like proteins from the different organisms were obtained using the Protein Blast tool in the NCBI database (http://blast.ncbi.nlm.nih.gov). After the download of sequences, the signal sequences were removed according to the prediction of SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP)2 (53), with the appropriate organism group for prediction. The sequences for which SignalP could not predict the signal sequence were excluded from the alignment. The amino acid sequences of 258 chitinases were aligned and visualized using Clustal Omega (Clustalω: http://www.ebi.ac.uk/Tools/msa/clustalo)2 (54) and WebLogo (http://weblogo.threeplusone.com)2 (55), respectively. The structure of SmChiA (PDB entry 1CTN) was superimposed with OfChi-h (PDB entry 5GQB) and VhChiA (PDB entry 3B8S) using the alignment function of PyMOL.
Author contributions
A. V. data curation; A. V. and P. V. formal analysis; A. V., A. N., H. W., T. U., and R. I. methodology; A. V. writing-original draft; A. N. and R. I. conceptualization; A. N. resources; R. I. supervision; R. I. funding acquisition; R. I. project administration; R. I. writing-review and editing.
Acknowledgments
We thank the Japan Agency for Marine Earth Science and Technology (JAMSTEC) for providing the tubes of L. satsuma for preparing the crystalline chitin. We also thank Yasuko Okuni and Mayuko Yamamoto for technical help and advice in sample preparation. We also thank all of the laboratory members for kind advice and discussion.
This study was supported by Grant-in-Aid for Scientific Research on Innovative Areas “Molecular Engine” JP18H05424 (to R. I.) and Grants-in-Aid for Scientific Research JP18H02418 and JP18H04755 (to R. I.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SmChiA
- Serratia marcescens chitinase A
- CBD
- chitin-binding domain
- CD
- catalytic domain
- FaXa
- Factor Xa
- HS-AFM
- high-speed atomic force microscopy
- koffNP
- non-productive dissociation rate constant
- koffP
- productive dissociation rate constant
- konNP
- non-productive binding rate constant
- konP
- productive binding rate constant
- kpc
- processive catalysis rate constant
- ktr
- translational rate (velocity)
- Ofchi-h
- Ostrinia furnacalis chitinase-h
- VhChiA
- Vibrio harveyi chitinase A
- μW
- microwatt
- fps
- frames per second
- LB
- Luria broth
- Ni-NTA
- nitrilotriacetic acid
- PDB
- Protein Data Bank.
References
- 1. Langner T., and Göhre V. (2016) Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Curr. Genet. 62, 243–254 10.1007/s00294-015-0530-x [DOI] [PubMed] [Google Scholar]
- 2. Merzendorfer H., and Zimoch L. (2003) Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 206, 4393–4412 10.1242/jeb.00709 [DOI] [PubMed] [Google Scholar]
- 3. Hamed I., Özogul F., and Regenstein J. M. (2016) Industrial applications of crustacean by-products (chitin, chitosan, and chitooligosaccharides): a review. Trends Food Sci. Technol. 48, 40–50 10.1016/j.tifs.2015.11.007 [DOI] [Google Scholar]
- 4. Songsiriritthigul C., Pantoom S., Aguda A. H., Robinson R. C., and Suginta W. (2008) Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism. J. Struct. Biol. 162, 491–499 10.1016/j.jsb.2008.03.008 [DOI] [PubMed] [Google Scholar]
- 5. Yan N., and Chen X. (2015) Sustainability: don't waste seafood waste. Nature 524, 155–157 10.1038/524155a [DOI] [PubMed] [Google Scholar]
- 6. Deguchi S., Tsujii K., and Horikoshi K. (2015) In situ microscopic observation of chitin and fungal cells with chitinous cell walls in hydrothermal conditions. Sci. Rep. 5, 11907 10.1038/srep11907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Einbu A., and Vårum K. M. (2008) Characterization of chitin and its hydrolysis to GlcNAc and GlcN. Biomacromolecules 9, 1870–1875 10.1021/bm8001123 [DOI] [PubMed] [Google Scholar]
- 8. Mekasha S., Byman I. R., Lynch C., Toupalová H., Anděra L., Næs T., Vaaje-Kolstad G., and Eijsink V. G. H. (2017) Development of enzyme cocktails for complete saccharification of chitin using mono-component enzymes from Serratia marcescens. Process Biochem. 56, 132–138 10.1016/j.procbio.2017.02.021 [DOI] [Google Scholar]
- 9. Nakagawa Y. S., Eijsink V. G., Totani K., and Vaaje-Kolstad G. (2013) Conversion of α-chitin substrates with varying particle size and crystallinity reveals substrate preferences of the chitinases and lytic polysaccharide monooxygenase of Serratia marcescens. J. Agric. Food Chem. 61, 11061–11066 10.1021/jf402743e [DOI] [PubMed] [Google Scholar]
- 10. Perrakis A., Tews I., Dauter Z., Oppenheim A. B., Chet I., Wilson K. S., and Vorgias C. E. (1994) Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2, 1169–1180 10.1016/S0969-2126(94)00119-7 [DOI] [PubMed] [Google Scholar]
- 11. Zakariassen H., Aam B. B., Horn S. J., Vårum K. M., Sørlie M., and Eijsink V. G. (2009) Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 284, 10610–10617 10.1074/jbc.M900092200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uchiyama T., Katouno F., Nikaidou N., Nonaka T., Sugiyama J., and Watanabe T. (2001) Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J. Biol. Chem. 276, 41343–41349 10.1074/jbc.M103610200 [DOI] [PubMed] [Google Scholar]
- 13. Bambara R. A., Uyemura D., and Choi T. (1978) On the processive mechanism of Escherichia coli DNA polymerase I. Quantitative assessment of processivity. J. Biol. Chem. 253, 413–423 [PubMed] [Google Scholar]
- 14. Bonderoff J. M., and Lloyd R. E. (2010) Time-dependent increase in ribosome processivity. Nucleic Acids Res. 38, 7054–7067 10.1093/nar/gkq566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rock R. S., Ramamurthy B., Dunn A. R., Beccafico S., Rami B. R., Morris C., Spink B. J., Franzini-Armstrong C., Spudich J. A., and Sweeney H. L. (2005) A flexible domain is essential for the large step size and processivity of myosin VI. Mol. Cell 17, 603–609 10.1016/j.molcel.2005.01.015 [DOI] [PubMed] [Google Scholar]
- 16. Shastry S., and Hancock W. O. (2010) Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 20, 939–943 10.1016/j.cub.2010.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schrader E. K., Harstad K. G., and Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 10.1038/nchembio.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snoberger A., Anderson R. T., and Smith D. M. (2017) The proteasomal ATPases use a slow but highly processive strategy to unfold proteins. Front. Mol. Biosci. 4, 18 10.3389/fmolb.2017.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henrissat B., and Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 10.1016/S0959-440X(97)80072-3 [DOI] [PubMed] [Google Scholar]
- 20. Sørlie M., Zakariassen H., Norberg A. L., and Eijsink V. G. H. (2012) Processivity and substrate-binding in family 18 chitinases. Biocatal. Biotransform. 30, 353–365 10.3109/10242422.2012.676282 [DOI] [Google Scholar]
- 21. Rouvinen J., Bergfors T., Teeri T., Knowles J. K., and Jones T. A. (1990) Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249, 380–386 10.1126/science.2377893 [DOI] [PubMed] [Google Scholar]
- 22. Breyer W. A., and Matthews B. W. (2001) A structural basis for processivity. Protein Sci. 10, 1699–1711 10.1110/ps.10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Von Hippel P. H., Fairfield F. R., and Dolejsi M. K. (1994) On the processivity of polymerases. Ann. N.Y. Acad. Sci. 726, 118–131 10.1111/j.1749-6632.1994.tb52803.x [DOI] [PubMed] [Google Scholar]
- 24. Beckham G. T., and Crowley M. F. (2011) Examination of the α-chitin structure and decrystallization thermodynamics at the nanoscale. J. Phys. Chem. B 115, 4516–4522 10.1021/jp200912q [DOI] [PubMed] [Google Scholar]
- 25. Divne C., Ståhlberg J., Teeri T. T., and Jones T. A. (1998) High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325 10.1006/jmbi.1997.1437 [DOI] [PubMed] [Google Scholar]
- 26. Hu G., Oguro A., Li C., Gershon P. D., and Quiocho F. A. (2002) The “cap-binding slot” of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap. Biochemistry 41, 7677–7687 10.1021/bi0201926 [DOI] [PubMed] [Google Scholar]
- 27. Quiocho F. A. (1989) Protein-carbohydrate interactions: basic molecular features. Pure Appl. Chem. 61, 1293–1306 10.1351/pac198961071293 [DOI] [Google Scholar]
- 28. Williams S. J., and Davies G. J. (2001) Protein-carbohydrate interactions: learning lessons from nature. Trends Biotechnol. 19, 356–362 10.1016/S0167-7799(01)01699-7 [DOI] [PubMed] [Google Scholar]
- 29. Meyer J. E., and Schulz G. E. (1997) Energy profile of maltooligosaccharide permeation through maltoporin as derived from the structure and from a statistical analysis of saccharide-protein interactions. Protein Sci. 6, 1084–1091 10.1002/pro.5560060515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jalak J., Kurašin M., Teugjas H., and Väljamäe P. (2012) Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 287, 28802–28815 10.1074/jbc.M112.381624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurasin M., and Väljamäe P. (2011) Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177 10.1074/jbc.M110.161059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurašin M., Kuusk S., Kuusk P., Sørlie M., and Väljamäe P. (2015) Slow off-rates and strong product binding are required for processivity and efficient degradation of recalcitrant chitin by family 18 chitinases. J. Biol. Chem. 290, 29074–29085 10.1074/jbc.M115.684977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cruys-Bagger N., Tatsumi H., Ren G. R., Borch K., and Westh P. (2013) Transient kinetics and rate-limiting steps for the processive cellobiohydrolase Cel7A: effects of substrate structure and carbohydrate binding domain. Biochemistry 52, 8938–8948 10.1021/bi401210n [DOI] [PubMed] [Google Scholar]
- 34. Igarashi K., Uchihashi T., Uchiyama T., Sugimoto H., Wada M., Suzuki K., Sakuda S., Ando T., Watanabe T., and Samejima M. (2014) Two-way traffic of glycoside hydrolase family 18 processive chitinases on crystalline chitin. Nat. Commun. 5, 3975 10.1038/ncomms4975 [DOI] [PubMed] [Google Scholar]
- 35. Nakamura A., Tasaki T., Ishiwata D., Yamamoto M., Okuni Y., Visootsat A., Maximilien M., Noji H., Uchiyama T., Samejima M., Igarashi K., and Iino R. (2016) Single-molecule imaging analysis of binding, processive movement, and dissociation of cellobiohydrolase Trichoderma reesei Cel6A and its domains on crystalline cellulose. J. Biol. Chem. 291, 22404–22413 10.1074/jbc.M116.752048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakamura A., Okazaki K. I., Furuta T., Sakurai M., and Iino R. (2018) Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin. Nat. Commun. 9, 3814 10.1038/s41467-018-06362-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakamura A., Tasaki T., Okuni Y., Song C., Murata K., Kozai T., Hara M., Sugimoto H., Suzuki K., Watanabe T., Uchihashi T., Noji H., and Iino R. (2018) Rate constants, processivity, and productive binding ratio of chitinase A revealed by single-molecule analysis. Phys. Chem. Chem. Phys. 20, 3010–3018 10.1039/C7CP04606E [DOI] [PubMed] [Google Scholar]
- 38. Liu T., Chen L., Zhou Y., Jiang X., Duan Y., and Yang Q. (2017) Structure, catalysis, and inhibition of OfChi-h, the lepidoptera-exclusive insect chitinase. J. Biol. Chem. 292, 2080–2088 10.1074/jbc.M116.755330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papanikolau Y., Prag G., Tavlas G., Vorgias C. E., Oppenheim A. B., and Petratos K. (2001) High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis. Biochemistry 40, 11338–11343 10.1021/bi010505h [DOI] [PubMed] [Google Scholar]
- 40. Kawai F., Nakamura A., Visootsat A., and Iino R. (2018) Plasmid-based one-pot saturation mutagenesis and robot-based automated screening for protein engineering. ACS Omega 3, 7715–7726 10.1021/acsomega.8b00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hudson K. L., Bartlett G. J., Diehl R. C., Agirre J., Gallagher T., Kiessling L. L., and Woolfson D. N. (2015) Carbohydrate-aromatic interactions in proteins. J. Am. Chem. Soc. 137, 15152–15160 10.1021/jacs.5b08424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pantoom S., Songsiriritthigul C., and Suginta W. (2008) The effects of the surface-exposed residues on the binding and hydrolytic activities of Vibrio carchariae chitinase A. BMC Biochem. 9, 2 10.1186/1471-2091-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jana S., Hamre A. G., Wildberger P., Holen M. M., Eijsink V. G., Beckham G. T., Sørlie M., and Payne C. M. (2016) Aromatic-mediated carbohydrate recognition in processive Serratia marcescens chitinases. J. Phys. Chem. B 120, 1236–1249 10.1021/acs.jpcb.5b12610 [DOI] [PubMed] [Google Scholar]
- 44. Zakariassen H., Hansen M. C., Jøranli M., Eijsink V. G., and Sørlie M. (2011) Mutational effects on transglycosylating activity of family 18 chitinases and construction of a hypertransglycosylating mutant. Biochemistry 50, 5693–5703 10.1021/bi2002532 [DOI] [PubMed] [Google Scholar]
- 45. Madhuprakash J., Dalhus B., Rani T. S., Podile A. R., Eijsink V. G. H., and Sørlie M. (2018) Key residues affecting transglycosylation activity in family 18 chitinases: insights into donor and acceptor subsites. Biochemistry 57, 4325–4337 10.1021/acs.biochem.8b00381 [DOI] [PubMed] [Google Scholar]
- 46. Kari J., Olsen J., Borch K., Cruys-Bagger N., Jensen K., and Westh P. (2014) Kinetics of cellobiohydrolase (Cel7A) variants with lowered substrate affinity. J. Biol. Chem. 289, 32459–32468 10.1074/jbc.M114.604264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakamura A., Watanabe H., Ishida T., Uchihashi T., Wada M., Ando T., Igarashi K., and Samejima M. (2014) Trade-off between processivity and hydrolytic velocity of cellobiohydrolases at the surface of crystalline cellulose. J. Am. Chem. Soc. 136, 4584–4592 10.1021/ja4119994 [DOI] [PubMed] [Google Scholar]
- 48. Shibafuji Y., Nakamura A., Uchihashi T., Sugimoto N., Fukuda S., Watanabe H., Samejima M., Ando T., Noji H., Koivula A., Igarashi K., and Iino R. (2014) Single-molecule imaging analysis of elementary reaction steps of Trichoderma reesei cellobiohydrolase I (Cel7A) hydrolyzing crystalline cellulose Iα and IIII. J. Biol. Chem. 289, 14056–14065 10.1074/jbc.M113.546085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moran-Mirabal J. M., Bolewski J. C., and Walker L. P. (2011) Reversibility and binding kinetics of Thermobifida fusca cellulases studied through fluorescence recovery after photobleaching microscopy. Biophys. Chem. 155, 20–28 10.1016/j.bpc.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 50. Jung J., Sethi A., Gaiotto T., Han J. J., Jeoh T., Gnanakaran S., and Goodwin P. M. (2013) Binding and movement of individual Cel7A cellobiohydrolases on crystalline cellulose surfaces revealed by single-molecule fluorescence imaging. J. Biol. Chem. 288, 24164–24172 10.1074/jbc.M113.455758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luterbacher J. S., Walker L. P., and Moran-Mirabal J. M. (2013) Observing and modeling BMCC degradation by commercial cellulase cocktails with fluorescently labeled Trichoderma reesei Cel7A through confocal microscopy. Biotechnol. Bioeng. 110, 108–117 10.1002/bit.24597 [DOI] [PubMed] [Google Scholar]
- 52. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nielsen H. (2017) Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 10.1007/978-1-4939-7015-5_6 [DOI] [PubMed] [Google Scholar]
- 54. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]