Abstract
The genomes of most cellulolytic clostridia do not contain genes annotated as transaldolase. Therefore, for assimilating pentose sugars or for generating C5 precursors (such as ribose) during growth on other (non-C5) substrates, they must possess a pathway that connects pentose metabolism with the rest of metabolism. Here we provide evidence that for this connection cellulolytic clostridia rely on the sedoheptulose 1,7-bisphosphate (SBP) pathway, using pyrophosphate-dependent phosphofructokinase (PPi-PFK) instead of transaldolase. In this reversible pathway, PFK converts sedoheptulose 7-phosphate (S7P) to SBP, after which fructose-bisphosphate aldolase cleaves SBP into dihydroxyacetone phosphate and erythrose 4-phosphate. We show that PPi-PFKs of Clostridium thermosuccinogenes and Clostridium thermocellum indeed can convert S7P to SBP, and have similar affinities for S7P and the canonical substrate fructose 6-phosphate (F6P). By contrast, (ATP-dependent) PfkA of Escherichia coli, which does rely on transaldolase, had a very poor affinity for S7P. This indicates that the PPi-PFK of cellulolytic clostridia has evolved the use of S7P. We further show that C. thermosuccinogenes contains a significant SBP pool, an unusual metabolite that is elevated during growth on xylose, demonstrating its relevance for pentose assimilation. Last, we demonstrate that a second PFK of C. thermosuccinogenes that operates with ATP and GTP exhibits unusual kinetics toward F6P, as it appears to have an extremely high degree of cooperative binding, resulting in a virtual on/off switch for substrate concentrations near its K½ value. In summary, our results confirm the existence of an SBP pathway for pentose assimilation in cellulolytic clostridia.
Keywords: pentose phosphate pathway (PPP); phosphofructokinase; mass spectrometry (MS); enzyme kinetics; bacterial metabolism; Clostridium thermocellum; Clostridium thermosuccinogenes; pyrophosphate; sedoheptulose 1,7-bisphosphate; transaldolase
Introduction
Transaldolase plays a key role in the non-oxidative pentose phosphate pathway (PPP).3 Together with transketolase it is responsible for the interconversion of C5 and C3/C6 metabolites, as depicted in Fig. 1. Specifically, transaldolase transfers a three-carbon ketol unit from sedoheptulose 7-phosphate (S7P) to glyceraldehyde 3-phosphate (G3P), forming erythrose 4-phosphate (E4P) and fructose 6-phosphate (F6P). Transketolase is responsible for the transfer of a two-carbon ketol unit from xylulose 5-phosphate, either to ribose 5-phosphate, yielding the S7P and G3P used by transaldolase, or to E4P, one of the products of transaldolase, yielding G3P and F6P. In contrast to the oxidative part, the non-oxidative PPP is reversible and essential both for catabolism of pentoses (e.g. xylose) and for the production of the C5 metabolites required for anabolism during growth on other substrates. The latter can also be facilitated by the oxidative PPP, but is then accompanied with the formation of NADPH, another important role of the PPP in many organisms.
Figure 1.
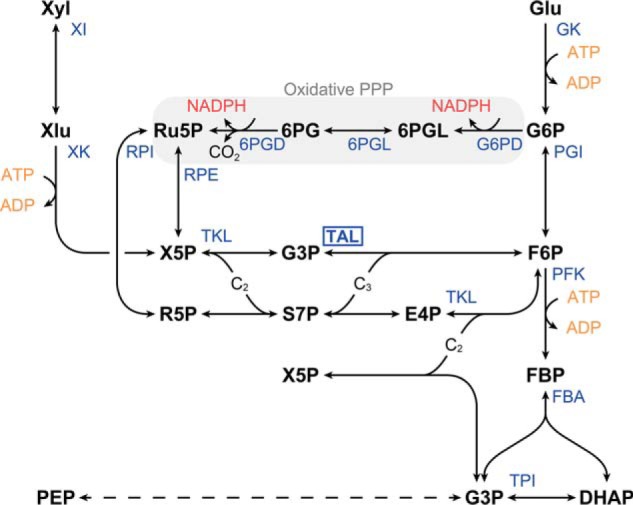
The reactions of the pentose phosphate pathway and its connection to the C3/C6 metabolites of the Embden-Meyerhof-Parnas (EMP) pathway. 6PG, 6-phosphogluconate; 6PGD, 6-phosphogluconate dehydrogenase; 6PGL, 6-phosphogluconolactone; FBA, fructose bisphosphate aldolase; GK, glucokinase; Glu, glucose; G6P, glucose 6-phosphate; G6PD, glucose-6-phosphate dehydrogenase; Pi, orthophosphate; PEP, phosphoenolpyruvate; PGI, phosphoglucoisomerase; PPi, pyrophosphate; R5P, ribose 5-phosphate; RPI, ribose 5-phosphate isomerase; Ru5P, ribulose 5-phosphate; RPE, ribulose 5-phosphate 3-epimerase; TAL, transaldolase; TKL, transketolase; TPI, triose-phosphate isomerase; Xyl, xylose; XI, xylose isomerase; XK, xylulokinase; Xlu, xylulose; X5P, xylulose 5-phosphate. The dashed arrow represents the trunk or lower part of the glycolysis, which consist of several reactions. X5P is shown in two different locations in the diagram for clarity. The gray box indicates reactions in the oxidative PPP.
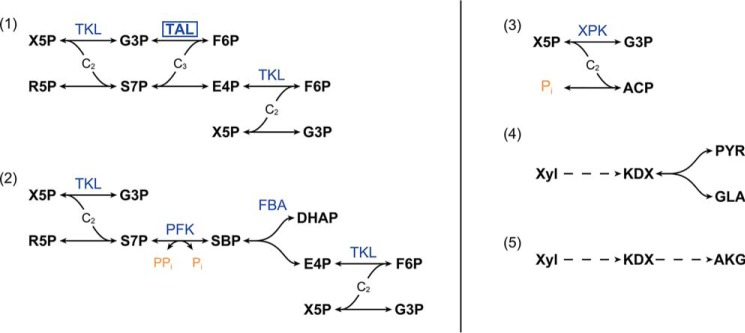
Several cellulolytic clostridia4 have been reported to lack an annotated transaldolase gene, whereas at least a few of those are able to grow very efficiently on pentose sugars, including Clostridium thermosuccinogenes and Clostridium cellobioparum subsp. termitidis, and Clostridium stercorarium (1–3). This implies that an alternative route to link C5 to the rest glycolysis must be present in those organisms. In C. thermosuccinogenes only the genes in the PPP responsible for the conversions of C5 sugars to xylulose 5-phosphate (i.e. xylose transporters, xylose isomerase, and xylulokinase) were up-regulated during growth on xylose versus glucose (1). In Clostridium termitidis, a transketolase was found to be up-regulated as well, during growth on xylose or xylan versus cellobiose (2). Yet, neither of those transcriptome studies resulted in an obvious candidate for an alternative pathway. Clostridium thermocellum also lacks an annotated transaldolase, but in contrast to the other cellulolytic clostridia, it cannot grow on xylose (4). However, it lacks the oxidative PPP as well (present in C. thermosuccinogenes) (5), so C. thermocellum would still be expected to have an alternative route, to produce the C5 metabolites required for anabolism. Although most organisms use transketolase/transaldolase, several alternative pathways to convert C5 to C3/C6 intermediates are known to exist (Fig. 2).
The phosphoketolase pathway (PKP). In the PKP, xylulose 5-phosphate is directly cleaved with orthophosphate into acetyl-P and G3P by phosphoketolase (6). The PKP is responsible for the degradation of pentose sugars in lactic acid bacteria, but more recently, it was shown that PKP is also, at least in part, responsible for pentose utilization in a variety of other bacteria, including Clostridium acetobutylicum and cyanobacteria (7–9).
The Weimberg pathway and Dahms pathway. The Weimberg pathway is a 5-step, oxidative, nonphosphorylating pathway that converts pentoses into 2-oxoglutarate, an intermediate in the TCA cycle (10, 11). In the Dahms pathway 2-keto-3-deoxy-d-xylonate or 2-keto-3-deoxy-l-arabinonate, intermediates in the Weimberg pathway, are cleaved by an aldolase into pyruvate and glycolaldehyde (11, 12). Both variants of the pathway occur in prokaryotes.
The sedoheptulose 1,7-bisphosphate pathway. In the amoebozoan Entamoeba histolytica, which lacks both glucose 6-phosphate dehydrogenase and transaldolase, S7P was shown to be the substrate of a pyrophosphate (PPi)-dependent 6-phosphofructokinase (PFK). The resulting sedoheptulose 1,7-bisphosphate (SBP) is subsequently cleaved into dihydroxyacetone phosphate (DHAP) and E4P by fructose-bisphosphate aldolase, effectively replacing the function of transaldolase, shown in Fig. 2 (13). PPi-PFK is physiologically reversible, in contrast to ATP-dependent PFK, allowing it to function in the PPP. Although, a number of reports exists where S7P kinase activity with high affinity is documented for PPi-PFKs from methanotrophs (14–17), the study discussed above is the only one where the SBP pathway was shown to exist in a WT metabolism. A double transaldolase knockout in Escherichia coli (ΔtalAB) resulted in xylose degradation via the SBP pathway, in conjunction with its native ATP-PFK and fructose-bisphosphate aldolase enzymes. In addition, S7P and SBP were observed to accumulate (18).
There is no real indication (genomic or physiological) for any of the routes other than the SBP pathway, based on the reliance of cellulolytic clostridia on PPi-PFKs, analogous to E. histolytica. Hence, the SBP pathway is generally assumed to be the responsible pathway (19, 20), but this has never been experimentally verified. This is not a trivial exercise, as metabolites required for enzyme assays are difficult to acquire and stable isotopic labeling studies are complicated by the recursive nature of the PPP and the typically low thermodynamic driving force in anaerobic metabolism, leading to relative high reverse fluxes (21, 22).
Figure 2.
Overview of the different pathways known for the interconversion of C5 and C3/C6 metabolites. (1) Typical pentose phosphate pathway involving transketolase and transaldolase. (2) Sedoheptulose 1,7-bisphosphate pathway. (3) Phosphoketolase pathway. (4), Dahms pathway. (5) Weimberg pathway. ACP, acetyl phosphate; AKG, α-ketoglutarate; FBA, fructose-bisphosphate aldolase; GLA, glycolaldehyde; KDX, 2-keto-3-deoxy-d-xylonate; PPi, pyrophosphate; PYR, pyruvate; R5P, ribose 5-phosphate; TAL, transaldolase; TKL, transketolase; X5P, xylulose 5-phosphate; XPK, xylulose 5-phosphate phosphoketolase; Xyl, xylose. Dashed arrows represent more than one reaction.
SBP should be a rather uncommon metabolite in bacteria without the Calvin cycle, the only known pathway with SBP as intermediate. In this cycle, SBP is formed from E4P and DHAP by aldolase and cleaved by sedoheptulose bisphosphatase. Besides algae and plants, some nonphotosynthetic eukaryotes have also been reported to possess this enzyme (23), but it has not been annotated in any of the cellulolytic clostridia. Therefore, presence of an SBP pool alone in C. thermosuccinogenes would already be a strong, albeit indirect, indication for the presence of the SBP pathway. The first aim of this study was therefore to investigate the possible presence of SBP in C. thermosuccinogenes, using high mass resolution Orbitrap MS. Unfortunately, an SBP reference standard was not commercially available, due to poor chemical stability. Instead, we constructed an E. coli ΔtalAB strain with a double transaldolase knockout, which accumulated SBP (18). We were able to use metabolite extracts from this strain as an SBP reference.
The accumulation of S7P in addition to SBP in E. coli ΔtalAB suggests that E. coli PFK has a low affinity for S7P. However, this is expected, because activity toward SBP is not necessary in E. coli because of the presence of transaldolase. If the PPi-PFKs of cellulolytic clostridia are indeed natively responsible for the conversion of S7P in the PPP, one would expect a much higher affinity for S7P. Therefore, the second aim of the study was to confirm in vitro the ability of PFKs of phosphorylate S7P and to compare their affinity for S7P versus F6P.
Results
Metabolomics
Presence of SBP in metabolite extracts of cellulolytic clostridia would give a preliminary indication of the presence of the SBP pathway. To unambiguously confirm the presence of SBP it is crucial to have a reference standard. Nakahigashi et al. (18) showed the accumulation of S7P and a molecular ion conform with SBP (m/z 369.0) in extracts of E. coli harboring a double transaldolase knockout (ΔtalAB). We decided to use the above mentioned strain, which presumably accumulated SBP as well as S7P, to subsequently function as a molecular reference for SBP. The strain used by Nakahigashi et al. (18) was recreated as described under “Materials and methods.” The growth rate of the ΔtalAB derivative on a minimal medium with xylose was only marginally lower compared with that of the WT: 0.33 ± 0.01 h−1 versus 0.39 ± 0.01 h−1, when grown in shake flasks with 50 ml of M9 medium (in triplicate, with the standard deviation reported), in line with the findings of Nakahigashi et al. (18). Furthermore, the growth rate on minimal medium with glucose was in fact slightly higher for the mutant: 0.52 ± 0.01 h−1 versus 0.49 ± 0.01 h−1.
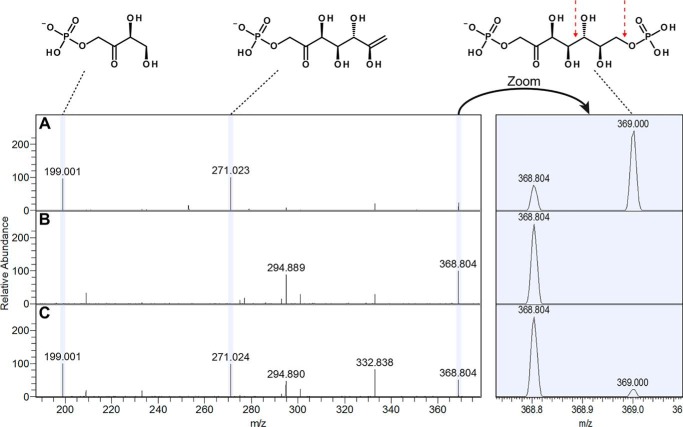
Mass spectrometry of metabolome extracts from E. coli ΔtalAB and WT grown on xylose showed the accumulation of S7P and a compound with a m/z 369.0 for the ΔtalAB strain. This peak showed (i) the expected accurate mass of SBP, (ii) the retention behavior relative to S7P corresponding to the chemical composition of SBP (i.e. slightly later and with a higher tendency for tailing, due to higher acidity), and (iii) the higher-energy collisional dissociation fragments exactly matching those expected for SBP (i.e. phosphate ester loss and β-bond cleavage), as shown in Fig. 3. Based on these observations it was concluded that SBP was indeed produced and that it could successfully be used as a reference.
Figure 3.
Identification of SBP (m/z 368.99) via targeted monitoring of fragments for the precursor ion at m/z 368–370. A, E. coli ΔtalAB extract; B, blank run; C, xylose grown C. thermosuccinogenes extract. A clear peak is present in A and C at m/z 369 corresponding to SBP, confirmed by indicative fragment peaks at m/z 199 and 271, which are further absent in the blank (B). The red arrow indicates the bonds that, when broken, result in the fragment at m/z 199 and 271. The identity of the peak at m/z 368.804 is unknown. Presence of this peak in the blank run (B) indicates that it is a background signal, whereas the nearby peak at m/z 369 is not.
Nevertheless, we could only successfully detect SBP at a relatively strong signal (comparable to S7P), when the metabolic extract was analysed without any preceding purification/enrichment steps, further illustrating its very low stability.
Traces of SBP were also detected in the cell extract of WT E. coli, suggesting that even in the WT metabolism a small fraction of the S7P is converted to SBP by PFK. This flux is amplified after the double transaldolase deletion, causing S7P to accumulate. This provides further support for our assumption that the E. coli PFKs have a low affinity for S7P.
Next, C. thermosuccinogenes was grown on xylose versus glucose, to try to detect SBP, and determine if the SBP pool increases during growth on xylose, as would be expected, because virtually the entire flux of carbon will have to be channeled through SBP into glycolysis. The results of the metabolome extract analysis are shown in Fig. 4. SBP was found to be present and normalized to the optical density of the cultures at 600 nm (OD600), the SBP concentration increased 4-fold. Similarly, the S7P concentration increased 2.5-fold during growth on xylose.
Figure 4.
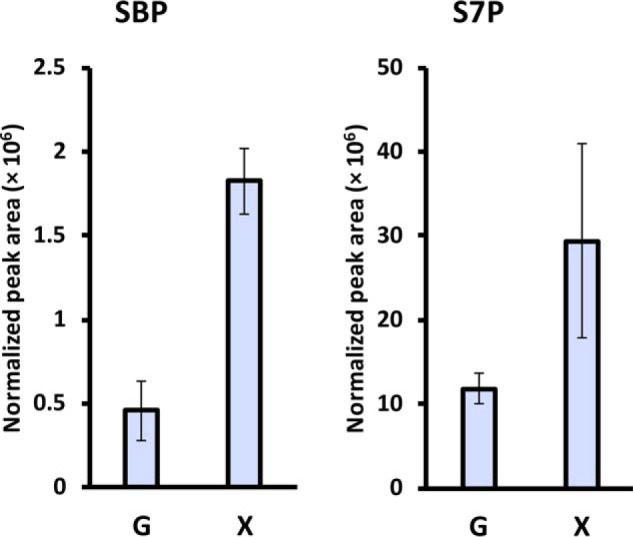
Relative pools of SBP and S7P in C. thermosuccinogenes grown on xylose (X), compared with glucose (G). Mass spectrometry peak area is normalized to OD600 of the culture. The error bars represent the mean ± S.D. of biological triplicates.
In E. coli ΔtalAB grown on xylose, the S7P concentration was roughly 6-fold higher compared with that of C. thermosuccinogenes grown on xylose. For SBP this difference was roughly 20-fold. Although many factors could explain the difference in concentrations between the two organisms, the higher accumulation in E. coli suggests that the PFK and the fructose-bisphosphate aldolase enzymes of C. thermosuccinogenes have higher affinities for S7P and SBP, respectively, compared with those of E. coli. A higher affinity, in turn, suggests evolutionary pressure toward the use of those substrates. For this reason, we studied the in vitro affinities of the cellulolytic clostridia PFKs toward F6P and S7P.
In vitro phosphofructokinase assays
C. thermosuccinogenes and C. thermocellum contain two PFKs: the PPi-dependent PFK and another one that was shown to function with both ATP and GTP in C. thermosuccinogenes (24). A third PFK has been annotated in the genome of C. thermosuccinogenes for which no activity had been detected, and which is absent in C. thermocellum. From assays with cell-free extract, it was already clear that PPi-dependent PFK is the dominant isoform (1, 25). We repeated the aforementioned assays with C. thermosuccinogenes cell-free extract in the presence of NH4, which we had serendipitously found to activate ATP/GTP-PFK, but were still unable to detect ATP-dependent activity (data not shown), confirming that PPi-dependent PFK is the dominant isoform in this organism as well.
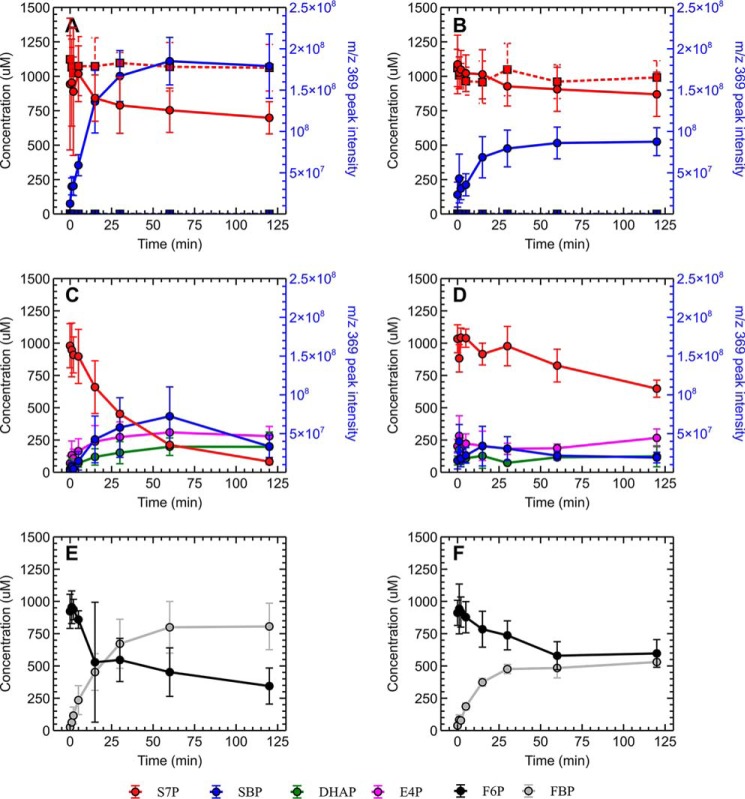
To determine whether the PPi-PFK proteins participated in the non-oxidative pentose phosphate pathways of their respective organisms, we investigated whether the PPi-PFKs were in fact capable of interconversion of S7P and SBP. In vitro time course experiments showed that S7P concentrations decreased over time in an assay mixture where both PPi-PFK and PPi were also present (Fig. 5); this decrease in S7P concentrations was concomitant with an increase in signal intensity at the m/z 369, which corresponds to the presence of SBP (Fig. 3). In the absence of PPi, S7P concentrations remained relatively stable, and no increase in signal intensity at m/z 369 was observed (Fig. 5), providing further evidence that the PPi-PFK proteins were using PPi as a cofactor to phosphorylate of S7P. As expected, assay reactions containing S7P and PPi, but no PPi-PFK protein, did not show any decrease in S7P, nor increase in peak intensity at m/z 369 (Fig. S1).
Figure 5.
In vitro time course assay of C. thermocellum (A, C, and E) and C. thermosuccinogenes (B, D, and F) PPi-PFK proteins' abilities to convert S7P to SBP. A and B, conversion of S7P (red) in the presence (circled data points on solid lines) or absence (square data points on dotted lines) of 5 mm pyrophosphate, with corresponding increase in a compound (SBP) with a m/z of 369 (blue). SBP peak intensities in assays lacking PPi were, in general, between the range of 10,000 and 20,000 arbitrary units throughout the assay. C and D, conversion of S7P (red) to SBP (blue), and SBP's subsequent conversion to DHAP (green) and E4P (pink). E and F, control reactions for the purified PPi-PFK proteins, demonstrating their ability to function as 6-PFKs and convert F6P (black) to FBP (gray). Error bars represent 1 S.D. (n ≥ 2).
Further confirmation of the identity of the m/z 369 compound as SBP was obtained by repeating the assay with the inclusion of fructose-bisphosphate aldolase. The SBP pathway would result in the formation of E4P and DHAP (both commercially available compounds) from SBP via the action of the aldolase (Fig. 2). In this additional set of assays, a similar pattern of decreasing the S7P concentration coupled to an increase in peak intensity at m/z 369 was observed; in addition, it was also observed that E4P and DHAP concentrations increased over time as well, where accumulation of these compounds was not detected in the reactions without the added aldolase. Notably, the peak intensities at m/z 369 in the aldolase-containing reactions were lower than that observed in the corresponding assay reactions that did not contain aldolase, in line with the conversion of SBP to E4P and DHAP. Furthermore, S7P concentrations in the aldolase-containing reactions trended to be lower than in their corresponding aldolase-free reactions; this is likely due to the consumption of SBP by aldolase, producing DHAP and E4P, which promotes further conversion of S7P to SBP by delaying the chemical equilibrium. It was also observed that the DHAP concentrations tended to be lower than those of E4P, despite the fact that they should be produced in equimolar amounts, as the stoichiometry in Fig. 2 would suggest; one explanation is that the added rabbit aldolase contains triose-phosphate isomerase as a trace contaminant, which would catalyze the interconversion of DHAP to G3P. Nonetheless, the results support the proposed SBP pathway.
For the determination of the enzyme kinetics for F6P and S7P, analysis via MS is impractical, as the response is not obtained in real-time. Instead, the formation of FBP and SBP can be coupled to oxidation of NADH via auxiliary fructose-bisphosphate aldolase and glycerol-3-phosphate dehydrogenase (both from rabbit), as illustrated in Fig. 5. We confirmed that rabbit aldolase was able to convert the formed SBP to E4P and DHAP, validating the coupled assay method, as shown in Fig. 6. Note that commonly, triose-phosphate isomerase is used additionally for such assays (to convert glyceraldehyde 3-phosphate to DHAP, increasing the signal and the driving force), which we excluded, as this would not function with E4P, making it easier to directly compare the two different substrates.
Figure 6.
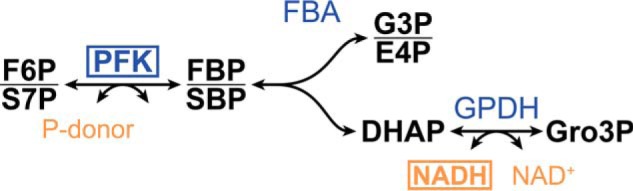
Enzyme assay to couple FBP/SBP formation by PFK to NADH oxidation, allowing the study of the PFK enzyme kinetics. FBA, fructose-bisphosphate aldolase; GPDH, glycerol-3-phosphate dehydrogenase; FBP, fructose 1,6-bisphosphate; G3P, glyceraldehyde 3-phosphate; Gro3P, glycerol 3-phosphate. Boxes highlight the investigated enzyme (i.e. PFK) and detected metabolite (i.e. NADH).
The results of the kinetics assays are presented in Table 1. For clarity, we use the term affinity to discuss Km values, although strictly speaking this is incorrect, as Km is not equal to the dissociation constant. Although the two tested PPi-dependent PFKs showed 2–3-fold lower maximal activity (kcat) with S7P versus F6P, the affinity (Km) for both substrates was comparably high (Km of ∼0.1 mm). This is in stark contrast with the PfkA from E. coli, for which the affinity constant for S7P is almost 2 orders of magnitude larger (i.e. lower affinity; Km of 2.5 mm) than that for F6P; the latter being on par with the affinities of the other tested PFKs. In exponentially growing E. coli cells on glucose, the concentrations of F6P and S7P are 2.2 and 0.9 mm, respectively (26), which means that in vivo PfkA is highly saturated with F6P, but not with S7P. The intracellular concentrations of those metabolites are not known in any of the cellulolytic clostridia, but with the equally high affinities for both F6P and S7P it is reasonable to assume that both metabolites are saturating, and thus physiologically relevant substrates for the PPi-dependent PFKs. These results strongly suggest that the PPi-dependent PFKs of cellulolytic clostridia, lacking a transaldolase, evolved for the use of S7P as a substrate, whereas the (ATP-dependent) PfkA of E. coli, which does possess a transaldolase, did not; although PfkA is still able to use S7P as a substrate at higher, nonphysiological concentrations. The latter can explain why traces of SBP were found in WT E. coli grown on xylose, and why S7P accumulates in the ΔtalAB strain. The data of the assays and the fitted kinetic models can be found in Figs. S2–S5.
Table 1.
Kinetics of the PFKs tested
Parameters of Michaelis-Menten (or Hill) kinetics were approximated by minimizing the sum of the squared vertical difference. The plots with the data points can be found in Figs. S2–S5. ATP/GTP-PFK did not show any activity with S7P, and due to the high cooperativity (n = 24) with F6P, it was not possible to fit the Michaelis-Menten equation.
| Km | kcat | n | kcat/Km | |||
|---|---|---|---|---|---|---|
| mm | s−1 | s−1 m−1 | ||||
| CDQ83_11320 C.ts PPi-PFK | PPi | F6P | 0.070 | 182 | NAa | 2.6 × 106 |
| S7P | 0.11 | 49 | 0.46 × 106 | |||
| Clo1313_1876 C.tc PPi-PFK | F6P | 0.046 | 127 | NA | 2.8 × 106 | |
| S7P | 0.10 | 80 | 0.80 × 106 | |||
| CDQ83_07225 C.ts A/GTP-PFK | ATP | F6P | 0.688 | 41 | 24 | 59 × 103 |
| S7P | – | – | – | – | ||
| PfkA E.co PFK | F6P | 0.065 | 1.2 | NA | 19 × 103 | |
| S7P | 2.5 | 0.25 | 0.15 × 103 |
a NA, not applicable. The Hill equation is only used in the case of CDQ83_07225, where n is the Hill coefficient, and Km is replaced by K½.
The ATP/GTP-dependent PFK of C. thermosuccinogenes was able to use F6P, but did not show any activity with S7P, at least, not in the range of tested S7P concentrations, up to 4 mm. Interestingly, it showed an extreme degree of cooperativity with F6P, reflected by a Hill coefficient (n) of 24. The high degree of cooperativity results in a virtual on/off switch of the enzyme, activating it at F6P concentrations above the K½ of 0.7 mm, as can be seen in Fig. 7.
Figure 7.
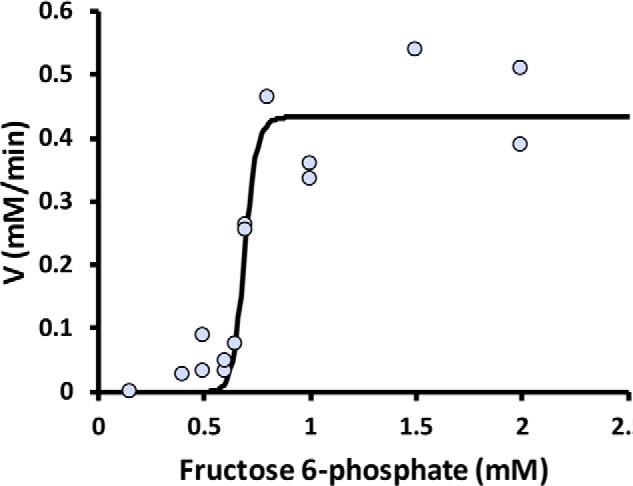
Fitted Hill kinetics to the results of the coupled assays with ATP/GTP-PFK (CDQ83_07225) of C. thermosuccinogenes. V represents the rate of NADH oxidation per minute, which is plotted against the corresponding fructose 6-phosphate concentration.
Discussion
The SBP pathway in cellulolytic clostridia
A considerable pool of SBP is shown to be present in C. thermosuccinogenes, which, together with the S7P pool, increases severalfold when C. thermosuccinogenes is grown on xylose versus glucose. This increase demonstrates the role for SBP in the pentose metabolism, and agrees with the hypothesis that the SBP pathway is present instead of transaldolase. In the SBP pathway, PFK and fructose-bisphosphate aldolase together convert S7P to E4P and DHAP (via SBP, as shown in Fig. 2). These two enzymes are known to convert F6P to glyceraldehyde 3-phosphate and DHAP (via fructose 1,6-bisphosphate as intermediate). In E. coli, it was already shown that these enzymes could take over the role of transaldolase after a double transaldolase knockout, and in E. histolytica it was shown that the SBP pathway likely exists in the WT metabolism (13, 18).
If in C. thermosuccinogenes and other cellulolytic clostridia, in the absence of a transaldolase, the SBP pathway is really the native pathway to connect pentose with hexose metabolism, their affinities for these “alternative” substrates should reflect that. Indeed, here we show that the PPi-PFKs of both C. thermosuccinogenes and C. thermocellum can use S7P, and have an affinity similar to that for F6P. The same was previously found for E. histolytica PPi-PFK, where the Km for S7P is 0.064 mm and 0.038 mm for F6P (13). On the contrary, here we show that the affinity of E. coli PfkA for S7P is almost 2 orders of magnitude lower compared with its affinity for F6P, such that the affinity constant for S7P is much higher than its typical intracellular concentration. Considering the fact that E. coli has a transaldolase to facilitate the interconversion of pentoses and hexoses, it makes sense that the affinity of PfkA for S7P is such that in vivo this reaction does not proceed; there is no need for S7P kinase activity and the resultant SBP pathway.
The SBP pathway versus transaldolase
A question that remains is whether there is an advantage to having either the transaldolase or the SBP pathway? A crucial aspect of the SBP pathway is the physiological reversibility of the PPi-PFK in contrast to the ATP-dependent variant (27), because the non-oxidative PPP should be able to operate in both directions. It therefore seems a prerequisite to rely on PPi-PFK, and the associated PPi-generating metabolism to use the SBP pathway. If this is not the case and PFK is ATP-dependent, transaldolase would still be required to facilitate the reverse (anabolic) direction in the PPP. As such, it seems that transaldolase might simply be or become obsolete in the case PPi-PFK is used in glycolysis. The underlying question therefore is why a PPi-dependent PFK is used at all, instead of an ATP-dependent one?
The irreversibility (i.e. large decrease in Gibbs free energy) of ATP-dependent PFK grants it a lot of control over the metabolism, but comes at the cost of about half-available free energy (26). The trade-off between control and energy conservation could perhaps be the main factor behind the use of a PPi-dependent PFK versus an ATP-dependent one. Organisms that almost exclusively rely on substrate level phosphorylation for ATP generation, such as the strictly anaerobic cellulolytic clostridia, might prioritize energy conservation over control, whereas respiring organisms might have done the opposite.
PPi is a by-product of many anabolic reactions, often operating close to equilibrium. Many organisms hydrolyze PPi to orthophosphate using inorganic pyrophosphatase, to drive these anabolic reactions forward, releasing heat (28). Using the otherwise “wasted” PPi instead of ATP for the phosphorylation of F6P should therefore allow the conservation of metabolic energy. It was already calculated for C. thermocellum, however, that the formation of PPi as by-product of anabolism alone is not enough to sustain the PFK reaction in glycolysis as it accounted for less than 10% of the flux (25), meaning that there must be another, dedicated source of PPi.
Unusual kinetics for ATP/GTP-PFK
ATP/GTP-PFK was previously found to have similar affinities for ATP versus GTP (24), and here we show that it has an extreme degree of cooperativity for F6P. The extreme cooperativity effectively means that below ∼0.7 mm F6P there is no activity, whereas above this concentration the enzyme operates at maximum activity. At this point we can only speculate on the function behind this peculiar characteristic, and doing so it seems wholly plausible that it could function as a kind of relief valve that prevents the intracellular concentration of F6P from rising above 0.7 mm.
It might in fact be detrimental for organisms relying on the SBP pathway to accumulate a large intracellular concentration of F6P (relative to S7P/SBP), due to the competition between S7P/SBP and F6P/fructose 1,6-bisphosphate for PPi-PFK. For example, a 10-fold higher F6P concentration compared with S7P means that only ∼9% of the PPi-PFK enzyme pool is available for S7P-depdendent activity. The other way around, when S7P accumulates relative to F6P, which prevents F6P phosphorylation by PPi-PFK, presence of ATP/GTP-PFK will still enable this reaction to occur, but with ATP or GTP instead of PPi.
In the case one enzyme is responsible for two separate (metabolic) reactions (via the same active site), it becomes crucial for the cell to regulate those relative pools, for both reactions to be able to occur. Our hypothesis is that the ATP/GTP-dependent PFK is responsible for a fail-safe mechanism relieving the negative effects caused by the perturbation of the S7P and F6P pools. How and if the enzyme's activation by NH4+ relates to this hypothesized function is not clear.
Widespread occurrence of the SBP pathway?
It is common for both PPi-dependent and ATP-dependent PFKs to coexist in one organism. In such cases, it was previously thought that PPi-PFK might have an alternative, unknown function (29). Here we show that the PPi-PFK has a dual function in glycolysis and the PPP, whereas the ATP-dependent PFK might have an alternative function. Similarly, in Amycolatopsis methanolica PPi-PFK is used in glycolysis during growth on glucose, whereas its ATP-PFK is used in the ribulose monophosphate cycle, during growth on one-carbon compounds (30). The widespread occurrence of PPi-PFK could therefore suggest that the SBP pathway is also more widespread than is currently recognized, particularly when the presence of PPi-PFK coincides with the absence of a transaldolase. The latter might also be underestimated due to F6P aldolases being wrongly annotated as transaldolase, resulting from their high similarity (31, 32).
Of all the 45 cellulolytic clostridia (Hungateiclostridiaceae) genomes in the JGI database, only the two Ruminiclostridium papyrosolvens genomes contain annotated transaldolases; two per genome, of which one contains the characteristic Glu and Tyr residues associated with transaldolase activity, rather than F6P aldolase activity (32). Except for the C. thermosuccinogenes genomes, none of the genomes harbors a complete oxidative PPP, and besides the C. thermocellum genomes almost all harbor xylose isomerase and xylulokinase (required for growth on pentoses), meaning that cellulolytic clostridia in general rely on the SBP pathway for the PPP, with the possible exception of R. papyrosolvens.
How widespread the SBP pathway is outside the cellulolytic clostridia would require further research, which is outside the scope of this study. The spread of PPi-PFKs in a wide variety of organisms, the proved existence of the SBP pathway in cellulolytic clostridia as well as the eukaryotic E. histolytica, and the high affinity of methylotrophic PFKs for S7P does hint at a much more widespread occurrence of the pathway.
SBP identification
In some metabolomics studies, the identification of SBP is simply omitted, because the standard reagent was not available (33). In others, SBP was synthesized in vitro using purified enzymes (34), which is laborious and expensive; and due to the low stability the product cannot be stored for longer periods. The method described here for the identification of SBP, relying on the E. coli ΔtalAB strain is simple and cheap, and might therefore benefit other researchers studying pentose metabolism. Furthermore, we noticed that commonly used practices for metabolomics studies, such as prolonged storage, and enrichment of extracts (e.g. via vacuum evaporation) will decrease the chance of detecting SBP in the extracts enormously.
Conclusion
An E. coli double transaldolase mutant was shown here to accumulate SBP, verified by Orbitrap MS. A metabolite extract from this E. coli mutant was used as an SBP reference for analysis (because SBP is not commercially available), and enabled us to show that a significant pool of SBP is present in C. thermosuccinogenes, an uncommon metabolite in organisms without the Calvin cycle. Moreover, the SBP pool was elevated during growth on xylose, confirming its relevance in pentose assimilation.
In vitro assays showed that PPi-PFK of C. thermosuccinogenes and C. thermocellum is able to convert S7P to SBP, and that they have similar affinity for S7P and F6P, the canonical substrate. In contrast, PfkA of E. coli showed a very poor affinity for S7P, which explains the high accumulation of S7P and SBP in the E. coli mutant. Furthermore, the enzyme kinetics suggest that the PPi-PFK enzymes of cellulolytic clostridia may have evolved for the use of S7P.
Additionally, we found that the ATP/GTP-dependent PFK of C. thermosuccinogenes shows an extremely high degree of cooperative binding toward F6P, resulting in a virtual on/off switch for substrate concentrations near its K½ value. We hypothesize that this PFK might represent a fail-safe mechanism that regulates the relative pools of F6P and S7P to prevent competition for the active site of PPi-PFK between the two parallel substrate pools causing one substrate to dominate the other. Overall, these results verify the existence of the SBP pathway in cellulolytic clostridia instead of the canonical transaldolase, connecting pentose metabolism with the rest of the metabolism.
Materials and methods
Growth medium and cultivation
For strain construction, plasmid construction, and protein purification, E. coli strains were grown on LB medium containing per liter 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl.
For metabolome extraction, E. coli BW25113 was grown on M9 minimal medium, made with M9 Minimal Salts (×5) from Sigma, containing additionally 0.4% xylose, 1 mm MgSO4, 0.3 mm CaCl2, 1 mg/liter of biotin, and 1 mg/liter of thiamine hydrochloride, which were all separately sterilized. Cells were grown at 37 °C in shaker flasks containing 50 ml of medium, inoculated with 0.5 ml of overnight culture grown in LB.
C. thermosuccinogenes was grown anaerobically in adapted CP medium (35), as described previously (1), which contained per liter 0.408 g of KH2PO4, 0.534 g of Na2HPO4·2H2O, 0.3 g of NH4Cl, 0.3 g of NaCl, 0.1 g of MgCl2·6H2O, 0.11 g of CaCl2·2H2O, 4.0 g of NaHCO3, 0.1 g of Na2SO4, 1.0 g of l-cysteine, 1.0 g of yeast extract (BD Pharmingen, BD Bacto), 1 ml of vitamin solution, 1 ml of trace elements solution I, and 1 ml of trace elements solution II. No resazurin was added to eliminate the possibility of it interfering with the metabolomics, as it appeared to adsorb to the nylon filter used for making the metabolome extracts.
The vitamin solution, which was ×1,000 concentrated, contained per liter 20 mg of biotin, 20 mg of folic acid, 100 mg of pyridoxine-HCl, 50 mg of thiamine-HCl, 50 mg of riboflavin, 50 mg of nicotinic acid, 50 mg of Ca-D-pantothenate, 1 mg of vitamin B12, 50 mg of 4-aminobenzoid acid, and 50 mg of lipoic acid.
Trace elements solution I, which was ×1,000 concentrated, contained per liter 50 mm HCl, 61.8 mg of H3BO4, 99.0 mg of MnCl2·4H2O, 1.49 g of FeCl2·4H2O, 119 mg of CoCl2·6H2O, 23.8 mg of NiCl2·6H2O, 68.2 mg of ZnCl2, and 17.0 mg of CuCl2·2H2O. Trace elements solution II, which was ×1,000 concentrated, contained per liter 10 mm NaOH, 17.3 mg of Na2SeO3, 33.0 mg of Na2WO4·2H2O, and 24.2 mg of Na2MoO4·2H2O.
Construction of E. coli ΔtalAB double knockout strain
E. coli BW25113, a K-12 derivative, which has been used for the Keio collection, was used to make the double transaldolase knockout, as was done by Nakahigashi et al. (18). First, the strain was transformed with pKD46, a temperature-sensitive plasmid containing the Lambda Red recombination system and an ampicillin-resistance marker. Cells were then grown at 30 °C and transformed with a linear knockout cassette derived from pKD3, containing a kanamycin-resistance marker flanked by FRT sites and 50-base pair arms homologous to the chromosome, such that talB would be removed, save for the start codon, and the last seven codons. The knockout cassette was generated from pKD4 with primers AGACCGGTTACATCCCCCTAACAAGCTGTTTAAAGAGAAATACTATCATGGTGTAGGCTGGAGCTGCTTC and GACCGACTTCCCGGTCACGCTAAGAATGATTACAGCAGATCGCCGATCATCATATGAATATCCTCCTTAGTTCCTATTCC. Transformed cells were grown on LB + kanamycin, at 37 °C, to select mutants and simultaneously cure pKD46. Following the selection of a correct mutant, pCP20, a temperature-sensitive plasmid containing the yeast flippase (flp) recombinase gene was transformed to remove the kanamycin marker from the genome by recombining the FRT sites. pCP20 was cured by growing the cells at 37 °C. The whole process was repeated to remove the talA gene as well. Primers GAATTAACGCACTCATCTAACACTTTACTTTTCAAGGAGTATTTCCTATGGTGTAGGCTGGAGCTGCTTC and TTCGGGACATATAACACTCCGTGGCTGGTTTATAGTTTGGCGGCAAGAAGCATATGAATATCCTCCTTAGTTCCTATTCC were used to generate the linear knockout cassette for talA, using pKD4 as a template.
Plasmid construction and heterologous expression of 6-phosphofructokinases
The two 6-phosphofructokinases of C. thermosuccinogenes (CDQ83_11320 and CDQ83_07225) were cloned previously into pET-28b(+) (24). The 6-phosphofructokinase of E. coli (BW25113_3916) was cloned in identical fashion using primers TACTTCCAATCCAATGCAATTAAGAAAATCGGTGTGTTGACAAGC and TTATCCACTTCCAATGTTAATACAGTTTTTTCGCGCAGTCC. The pET-28b(+)-based vectors were constructed in E. coli DH5α and then transformed to E. coli Rosetta for heterologous expression.
The pyrophosphate-dependent 6-phosphofructokinase of C. thermocellum strain DSM 1313 (locus_tag Clo1313_1876) was amplified using primers XSH0718 (sequence 5′-CATCACCACCACCACCATATGAGCCGTTTAGAAGGTG-3′) and XSH0719 (sequence 5′-GCGGCCGCGAGACCCTAACCTTATTTTCTTGCAAGAACC-3′), and then cloned into the pD861-NT expression vector (DNA 2.0 Inc., Menlo Park, CA) using isothermal assembly (36), to create plasmid pSH157. The assembled plasmid pSH157 was then cloned into T7 Express lysY/Iq Competent E. coli (New England Biolabs catalog number C3013I), using 50 μg/ml of kanamycin for selection of transformants.
Expression of the 6-phosphofructokinases was done by growing the E. coli strains in 0.5–2 liters of LB medium containing the appropriate antibiotics up to an OD600 of around 0.6, after which the cultures were place on ice for 20 min and expression was induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside for the pET-28b(+)-based vectors and with 4 mm rhamnose for the pD861-NT-based vector. Cells were grown for another 3–4 h at 37 °C, after which the cells were harvested for protein purification.
Metabolite extraction for MS
Biomass was isolated by rapid vacuum filtration of 5–20 ml of culture broth, adapted from the method by Sander et al. (37). For this, 0.2-μm nylon filters (Whatman®) were used. After filtration, the filter was immediately placed upside down in 3 to 10 ml of solvent, which was kept in a polystyrene Petri dish (50 mm diameter, Falcon®) placed on an aluminum block precooled at −80 °C. The extraction solvent consisted of a mixture of acetonitrile, methanol, and water, mixed at a ratio of 2:2:1 (v/v). The filter was kept in the extraction solvent for 5 min, after which the extract was kept on dry ice until transferred to a freezer for storage at −80 °C. All aliquots were stored until being further processed in low protein binding collection tubes (Thermo ScientificTM). The entire process was carried out aerobically.
20 ml of the E. coli cultures grown in M9 medium at OD600 of 0.5–0.8 was used, in combination of 5 ml of extraction solvent. For C. thermosuccinogenes, up to 10 ml of culture was used, depending on the OD600, in combination with 3 ml of extraction solvent. The higher the OD600, the smaller the culture that could be filtered without clogging the filter. It is not clear why cellulolytic clostridia cultures tend to clog the filters so quickly.
HILIC mass spectrometry for detection of sedoheptulose 7-phosphate and sedoheptulose 1,7-bisphosphate in metabolite cell extracts
Identification of sedoheptulose 7-phosphate and sedoheptulose 1,7-bisphosphate was performed using a LC-MS setup as described by Schatschneider et al. (38). Briefly, LC-Orbitrap-MS analysis was performed using an ACQUITY UPLC M-Class chromatography system coupled to a high-resolution Orbitrap mass spectrometer (Q-Exactive plus, Thermo Fisher Scientific). For chromatographic separation, a ZIC-HILIC column (1.0 × 150 mm, 5-μm particle size, Merck KGaA, Germany) was operated at room temperature using 20 mm ammonium carbonate in water (pH 9.1) as mobile phase A and 100% acetonitrile as mobile phase B. A gradient was maintained at 40 μl/min from 25% A to 55% A over 15 min and further to 30% A over 2.5 min. Samples were taken from −80 °C immediately before injection, brought to 4–8 °C on ice, and mixed with injection buffer (85% solvent B in solvent A, including 20 mm sodium citrate) at a ratio of 1:1 (v/v). The reaction mixture was centrifuged at 14,000 rpm for 3 min at 4–8 °C and 2.5 μl were subsequently injected onto the separation column. The mass spectrometer was operated alternating in full scan and PRM mode. Full scan was acquired from 260–700 m/z in ESI negative mode (−2.5 kV), at a resolution of 70,000. Parallel reaction monitoring was performed for m/z 289.03 ([M-H]−, S7P) and 368.99 ([M-H]−, S1,7BP) precursor ions at an isolation window of 2.0 m/z. High-energy collisional dissociation fragmentation was performed using a NCE of 27 and which fragment ions were measured at a resolution of 17,000. Raw data were analyzed using XCalibur 4.1 (Thermo) and areas were integrated using Skyline 4.1.0. The fragmentation pattern and elution time for sedoheptulose 7-phosphate was compared with a commercial standard (Sigma Aldrich) and the peak obtained from the E. coli double transaldolase mutant and WT metabolite extracts were used as control. The fragmentation pattern and elution position for sedoheptulose 1,7-bisphosphate was compared with the metabolite extract E. coli ΔtalAB (see above).
In vitro phosphofructokinase assays with analysis by MS
Protein purification
50 ml of LB cultures of E. coli strains overexpressing the C. thermocellum or C. thermosuccinogenes PPi-PFK were grown, induced, and harvested as described above.
Protein purification for the purposes of demonstrating in vitro conversion of sedoheptulose 7-phosphate to sedoheptuoloase 1,7-bisphosphate by the purified PPi-PFK in the presence of PPi was performed as previously described (39). To obtain purified PPi-PFK protein, the induced E. coli cells were resuspended in 100 mm Tris-HCl buffer (pH 7 at 55 °C). Approximately 70,000 units of Readylyse enzyme (Lucigen catalog number R1802) was added to the cell suspension, and the mixture was incubated for 10 min at room temperature. 5 units of DNase I (Thermo Fisher Scientific catalog number 90083) were then added to reduce the viscosity of the cell lysate; the sample was incubated for another 10 min at room temperature. The resulting solution was at >20,000 × g for 5 min, and the cell extract supernatant was used in future steps. E. coli proteins in the cell extract were denatured by incubating the cell extract at 55 °C for 30 min; the denatured proteins were removed by centrifugation at >20,000 × g for 5 min. His tag purification of the PPi-PFKs from the heat-treated cell extracts was done using the HisSpinTrap kit (GE Healthcare catalog number 28-4013-53). Eluted PPi-PFK proteins were desalted using a 10-kDa molecular mass cutoff filter (Millipore catalog number UFC501024) and 100 mm Tris-HCl buffer, to reduce the imidazole concentration to <1 mm.
Assay conditions
Enzyme assays were performed in an anaerobic chamber, with an atmospheric composition of 85% nitrogen, 10% carbon dioxide, and 5% hydrogen. Assay chemicals were purchased from Sigma Aldrich. All samples were incubated at 55 °C in a heat block for the entirety of the experiment. Assay reaction composition was based off previously described assay conditions, and comprised 100 mm Tris-HCl (pH 7 at 55 °C), 5 mm MgCl2, 5 mm sodium PPi, 1 mm of either sedoheptulose 7-phosphate or fructose 6-phosphate, 4 units of rabbit aldolase where used (Sigma catalog number A8811), and purified PPi-PFK protein (see next paragraph for more information on enzyme loading). In all cases, the reactions were started upon addition of sodium PPi, or an equivalent volume of buffer for assay reactions that did not contain PPi. The initial assay volume was 400 μl.
The specific PPi-PFK activity was first determined for each of the purified PPi-PFKs on the day of the experiment. The amount of purified PPi-PFK used for the assay was then adjusted for each sample and replicate, such that each assay reaction would contain an enzyme loading corresponding to ∼0.01 μmol/min of PPi-PFK activity.
Samples from the enzyme reactions were collected in the following manner: the tube containing a given assay mixture was removed from the heat block and briefly vortexed to mix the contents. 20 μl of assay mixture was then collected and then quickly added to 80 μl of very cold (≤ −30 °C) 1:1, acetonitrile:methanol mixture to quench enzyme activity, and then vortexed briefly to mix; quenching solution was kept at ≤ −30 °C by putting them in contact with a metal heat block sitting atop a 4-inch thick aluminum block, both of which had been pre-chilled at −80 °C for at least 48 h prior to use (40). The quenched sample was then stored at −80 °C until analysis. Standards of S7P, F6P, FBP, E4P, and DHAP at three different concentrations each were also prepared to allow for quantification of these compounds in the assay samples.
Mass spectrometry analyses of assay samples
Assay samples were analyzed as previously described (39), using an LC-MS/MS system a Thermo Scientific Vanquish UHPLC coupled by heated electrospray ionization (HESI) to a hybrid quadrupole-high resolution mass spectrometer (Q Exactive Orbitrap, Thermo Scientific). Liquid chromatography separation was performed using an ACQUITY UPLC BEH C18 (2.1 × 100 mm column, 1.7-μm particle size), with a flow rate of 0.2 ml/min. For the instrument run, Solvent A was 97:3, water:methanol with 10 mm tributylamine and ∼9.8 mm acetic acid (pH ∼ 8.2); solvent B was 100% methanol. Total run time was 24.5 min with the following gradient: 0 min, 5% B; 2.5 min, ramp from 5% B to 95% B over 14.5 min; hold at 95% B for 2.5 min; return to 5% B over 0.5 min; hold at 5% B for 5 min. MS scans consisted of full negative mode MS scanning for m/z between 70 and 1000 from time 0 to 18.5 min. Sample preparation involved first evaporating the solvents with a nitrogen blowdown evaporator, and then resuspending the dried samples in Solvent A.
Metabolite peaks were identified using the open source software, El-MAVEN (46) (https://elucidatainc.github.io/ElMaven/)5. Response factors for S7P, F6P, FBP, E4P, and DHAP standards were used to determine the concentrations of these five compounds in the assay samples.
Phosphofructokinase kinetics assays
The harvested cells were washed with cold 50 mm MOPS buffer (pH 7.4 at room temperature) containing 20 mm imidazole and resuspended in the same buffer with cOmpleteTM, mini, EDTA-free protease inhibitor mixture (Roche) added; 1 tablet per ∼10 ml. Cells were lysed using a French press at ∼120 kilopascal. Lysate was centrifuged at 20,000 × g for 10 min at 4 °C. A HisTrapTM HP column (GE Healthcare, optimal at pH 7.4) with an ÄKTA pure FPLC system were used for the purification. Elution was done over a gradient with 50 mm MOPS buffer (pH 7.4 at room temperature) containing 500 mm imidazole. The buffer of the eluted protein was then exchanged with 50 mm MOPS (pH 7.0 at room temperature) using an Amicon® ultracentrifugal filter (Merck) with a nominal molecular mass limit of 10,000 Da. SDS-PAGE was used to verify purity.
The 6-phosphofructokinase assay was adapted from Zhou et al. (25) and contained 50 mm MOPS (pH 7.0 at room temperature), 5 mm MgCl2, 2 mm ATP or 1 mm pyrophosphate, 0.15 mm NADH, 4 units/ml of aldolase (lyophilized, rabbit), and 2 units/ml of glycerol-3-phosphate dehydrogenase (lyophilized, rabbit). The reaction was carried out at 55 °C. Fructose 6-phosphate or sedoheptulose 7-phosphate (Ba-salt, Carbosynth) was added to start the reaction, at varied concentrations. The final volume was 1 ml for reactions with fructose 6-phosphate and 0.5 ml for reactions with sedoheptulose 7-phosphate. For the PfkA of E. coli, 0.25 mm ADP was added to the assay, as it is known to be an allosteric activator (41). For the ATP/GTP-dependent 6-phosphofructokinases of C. thermosuccinogenes, 20 mm NH4Cl2 was added to the reaction, as it was found to be an absolute requirement for its activity. Previously, auxiliary enzymes from an ammonium sulfate suspension were used (24), but with sedoheptulose 7-phosphate as a Ba-salt, this was not possible due to precipitation of BaSO4, which is how we found out that the enzyme requires ammonium. The reaction was followed in a Hitachi U-2010 spectrophotometer with a thermoelectric cell holder, by measuring the decreasing absorbance of reduced NADH at 340 nm. The reaction was run up to 5 min and a window of 10 to 40 s was used for determining the initial rates.
The Michaelis-Menten equation and the Hill equation were fitted to the data by minimizing the sum of the squares of the vertical differences, to find Km/K½, kcat, and n. The data and the fitted models can be found in the Figs. S2–S5.
Author contributions
J. G. K., S. H., L. R. L., D. G. O., and R. v. K. conceptualization; J. G. K., S. H., M. P., R. H., D. M. S., J. C., and D. A.-N. investigation; J. G. K., S. H., and M. P. visualization; J. G. K., S. H., M. P., R. H., D. M. S., J. C., D. A.-N., L. R. L., D. G. O., and R. v. K. methodology; J. G. K. and S. H. writing-original draft; J. G. K., S. H., M. P., D. A.-N., L. R. L., D. G. O., and R. v. K. writing-review and editing; L. R. L. and R. v. K. resources; L. R. L., D. G. O., and R. v. K. supervision.
Supplementary Material
Acknowledgment
We thank the Center for Bioenergy Innovation, a United States Department of Energy Bioenergy Research Center, supported by the Office of Biological and Environmental Research in the DOE Office of Science.
This work was supported by European Union Marie Skłodowska-Curie Innovative Training Networks (ITN) Contract 642068 (to J. G. K.) Lee R. Lynd is a founder of Enchi Corporation, which has a financial interest in C. thermocellum.
This article contains Figs. S1–S5.
“Cellulolytic clostridia” refers to a large group of mesophilic and thermophilic bacteria of which most are able to grow on (hemi)cellulosic substrates, and of which C. thermocellum and C. cellulolyticum are probably the most well-studied. Recently, they have been placed within a newly named family called Hungateiclostridiaceae, containing Genus names such as Hungateiclostridium and Pseudoclostridium (42), and for a brief time when some were also referred to cellulolytic clostridia as the Ruminiclostridium genus (43, 44). However, even more recently, the family name Hungateiclostridiaceae and the corresponding Genus names were suggested to be illegitimate and placement within the Acetivibrio genus was proposed (45). In light of this ongoing discussion, we have decided to refer to them as cellulolytic clostridia, and use the commonly used names C. thermocellum and C. thermosuccinogenes for the organisms of interest in this study (instead of Hungateiclostridium thermocellum and Pseudoclostridium thermosuccinogenes). Note that C. thermosuccinogenes ferments a wide range of C5 and C6 sugars, and sugar polymers, but not crystalline cellulose.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PPP
- pentose phosphate pathway
- SBP
- sedoheptulose 1,7-bisphosphate
- DHAP
- dihydroxyacetone phosphate
- PKP
- phosphoketolase pathway
- PFK
- phosphofructokinase
- F6P
- fructose 6-phosphate
- S7P
- sedoheptulose 7-phosphate
- E4P
- erythrose 4-phosphate
- G3P
- glyceraldehyde 3-phosphate.
References
- 1. Koendjbiharie J. G., Wiersma K., and van Kranenburg R. (2018) Investigating the central metabolism of Clostridium thermosuccinogenes. Appl. Environ. Microbiol. 84, e00363–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Munir R. I., Spicer V., Krokhin O. V., Shamshurin D., Zhang X., Taillefer M., Blunt W., Cicek N., Sparling R., and Levin D. B. (2016) Transcriptomic and proteomic analyses of core metabolism in Clostridium termitidis CT1112 during growth on α-cellulose, xylan, cellobiose and xylose. BMC Microbiol. 16, 91 10.1186/s12866-016-0711-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schellenberg J. J., Verbeke T. J., McQueen P., Krokhin O. V., Zhang X., Alvare G., Fristensky B., Thallinger G. G., Henrissat B., Wilkins J. A., Levin D. B., and Sparling R. (2014) Enhanced whole genome sequence and annotation of Clostridium stercorarium DSM8532T using RNA-seq transcriptomics and high-throughput proteomics. BMC Genomics 15, 567 10.1186/1471-2164-15-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lynd L. R., Weimer P. J., van Zyl W. H., and Pretorius I. S. (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 10.1128/MMBR.66.3.506-577.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamed R., and Zeikus J. G. (1980) Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J. Bacteriol. 144, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki R., Katayama T., Kim B.-J., Wakagi T., Shoun H., Ashida H., Yamamoto K., and Fushinobu S. (2010) Crystal structures of phosphoketolase. J. Biol. Chem. 285, 34279–34287 10.1074/jbc.M110.156281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu L., Zhang L., Tang W., Gu Y., Hua Q., Yang S., Jiang W., and Yang C. (2012) Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J. Bacteriol. 194, 5413–5422 10.1128/JB.00713-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerlach E. S., Sund C. J., Hurley M. M., Liu S., Servinsky M. D., and Germane K. L. (2015) Phosphoketolase flux in Clostridium acetobutylicum during growth on l-arabinose. Microbiology 161, 430–440 10.1099/mic.0.000008 [DOI] [PubMed] [Google Scholar]
- 9. Xiong W., Lee T.-C., Rommelfanger S., Gjersing E., Cano M., Maness P.-C., Ghirardi M., and Yu J. (2015) Phosphoketolase pathway contributes to carbon metabolism in cyanobacteria. Nat. Plants 2, 15187 [DOI] [PubMed] [Google Scholar]
- 10. Weimberg R. (1961) Pentose oxidation by Pseudomonas fragi. 236, 629–635 [PubMed] [Google Scholar]
- 11. Brouns S. J., Walther J., Snijders A. P., van de Werken H. J., Willemen H. L., Worm P., de Vos M. G., Andersson A., Lundgren M., Mazon H. F., van den Heuvel R. H., Nilsson P., Salmon L., de Vos W. M., Wright P. C., Bernander R., and van der Oost J. (2006) Identification of the missing links in prokaryotic pentose oxidation pathways. J. Biol. Chem. 281, 27378–27388 10.1074/jbc.M605549200 [DOI] [PubMed] [Google Scholar]
- 12. Dahms A. S., and Anderson R. L. (1969) 2-Keto-3-deoxy-l-arabonate aldolase and its role in A new pathway of l-arabinose degradation. Biochem. Biophys. Res. Commun. 36, 809–814 10.1016/0006-291X(69)90681-0 [DOI] [PubMed] [Google Scholar]
- 13. Susskind B. M., Warren L. G., and Reeves R. E. (1982) A pathway for the interconversion of hexose and pentose in the parasitic amoeba Entamoeba histolytica. Biochem. J. 204, 191–196 10.1042/bj2040191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karadsheh N. S., Tejwani G. A., and Ramaiah A. (1973) Sedoheptulose-7-phosphate kinase activity of phosphofructokinase from the different tissues of rabbit. Biochim. Biophys. Acta Enzymol. 327, 66–81 10.1016/0005-2744(73)90104-6 [DOI] [PubMed] [Google Scholar]
- 15. Reshetnikov A. S., Rozova O. N., Khmelenina V. N., Mustakhimov I. I., Beschastny A. P., Murrell J. C., and Trotsenko Y. A. (2008) Characterization of the pyrophosphate-dependent 6-phosphofructokinase from Methylococcus capsulatus Bath. FEMS Microbiol. Lett. 288, 202–210 10.1111/j.1574-6968.2008.01366.x [DOI] [PubMed] [Google Scholar]
- 16. Khmelenina V. N., Rozova O. N., and Trotsenko Y. A. (2011) Characterization of the recombinant pyrophosphate-dependent 6-phosphofructokinases from Methylomicrobium alcaliphilum 20Z and Methylococcus capsulatus Bath. Methods Enzymol. 495, 1–14 10.1016/B978-0-12-386905-0.00001-2 [DOI] [PubMed] [Google Scholar]
- 17. Rozova O. N., Khmelenina V. N., and Trotsenko Y. A. (2012) Characterization of recombinant PPi-dependent 6-phosphofructokinases from Methylosinus trichosporium OB3b and Methylobacterium nodulans ORS 2060. Biochemistry 77, 288–295 [DOI] [PubMed] [Google Scholar]
- 18. Nakahigashi K., Toya Y., Ishii N., Soga T., Hasegawa M., Watanabe H., Takai Y., Honma M., Mori H., and Tomita M. (2009) Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol. Syst. Biol. 5, 306 10.1038/msb.2009.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rydzak T., McQueen P. D., Krokhin O. V., Spicer V., Ezzati P., Dwivedi R. C., Shamshurin D., Levin D. B., Wilkins J. A., and Sparling R. (2012) Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol. 12, 214 10.1186/1471-2180-12-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dash S., Khodayari A., Zhou J., Holwerda E. K., Olson D. G., Lynd L. R., and Maranas C. D. (2017) Development of a core Clostridium thermocellum kinetic metabolic model consistent with multiple genetic perturbations. Biotechnol. Biofuels 10, 108 10.1186/s13068-017-0792-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson B. E., and McInerney M. J. (2002) Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415, 454–456 10.1038/415454a [DOI] [PubMed] [Google Scholar]
- 22. Dash S., Olson D. G., Joshua Chan S. H., Amador-Noguez D., Lynd L. R., and Maranas C. D. (2019) Thermodynamic analysis of the pathway for ethanol production from cellobiose in Clostridium thermocellum. Metab. Eng. 55, 161–169 10.1016/j.ymben.2019.06.006 [DOI] [PubMed] [Google Scholar]
- 23. Olson W. J., Stevenson D., Amador-Noguez D., and Knoll L. J. (2018) Dual metabolomic profiling uncovers Toxoplasma manipulation of the host metabolome and the discovery of a novel parasite metabolic capability. bioRxiv 10.1101/463075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koendjbiharie J. G., Wevers K., and van Kranenburg R. (2019) Assessing cofactor usage in Pseudoclostridium thermosuccinogenes via heterologous expression of central metabolic enzymes. Front. Microbiol. 10, 1162 10.3389/fmicb.2019.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou J., Olson D. G., Argyros D. A., Deng Y., van Gulik W. M., van Dijken J. P., and Lynd L. R. (2013) Atypical glycolysis in Clostridium thermocellum. Appl. Environ. Microbiol. 79, 3000–3008 10.1128/AEM.04037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park J. O., Rubin S. A., Xu Y.-F., Amador-Noguez D., Fan J., Shlomi T., and Rabinowitz J. D. (2016) Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 12, 482–489 10.1038/nchembio.2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mertens E. (1991) Pyrophosphate-dependent phosphofructokinase, an anaerobic glycolytic enzyme? FEBS Lett. 285, 1–5 10.1016/0014-5793(91)80711-B [DOI] [PubMed] [Google Scholar]
- 28. Tommi K., Juho K., Adrian G., Gabibov A., Skulachev V., Wieland F., and Just W. (2013) Inorganic pyrophosphatases: one substrate, three mechanisms. FEBS Lett. 587, 1863–1869 [DOI] [PubMed] [Google Scholar]
- 29. Bapteste E., Moreira D., and Philippe H. (2003) Rampant horizontal gene transfer and phospho-donor change in the evolution of the phosphofructokinase. Gene 318, 185–191 10.1016/S0378-1119(03)00797-2 [DOI] [PubMed] [Google Scholar]
- 30. Alves A. M., Euverink G. J., Santos H., and Dijkhuizen L. (2001) Different physiological roles of ATP- and PPi-dependent phosphofructokinase isoenzymes in the methylotrophic actinomycete Amycolatopsis methanolica. J. Bacteriol. 183, 7231–7240 10.1128/JB.183.24.7231-7240.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schurmann M., and Sprenger G. A. (2001) Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 276, 11055–11061 10.1074/jbc.M008061200 [DOI] [PubMed] [Google Scholar]
- 32. Sautner V., Friedrich M. M., Lehwess-Litzmann A., and Tittmann K. (2015) Converting transaldolase into aldolase through swapping of the multifunctional acid-base catalyst: common and divergent catalytic principles in F6P aldolase and transaldolase. Biochemistry 54, 4475–4486 10.1021/acs.biochem.5b00283 [DOI] [PubMed] [Google Scholar]
- 33. Sato S., Arita M., Soga T., Nishioka T., and Tomita M. (2008) Time-resolved metabolomics reveals metabolic modulation in rice foliage. BMC Syst. Biol. 2, 51 10.1186/1752-0509-2-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clasquin M. F., Melamud E., Singer A., Gooding J. R., Xu X., Dong A., Cui H., Campagna S. R., Savchenko A., Yakunin A. F., Rabinowitz J. D., and Caudy A. A. (2011) Riboneogenesis in yeast. Cell 145, 969–980 10.1016/j.cell.2011.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plugge C. M. (2005) Anoxic media design, preparation, and considerations. Methods Enzymol. 397, 3–16 10.1016/S0076-6879(05)97001-8 [DOI] [PubMed] [Google Scholar]
- 36. Gibson D. G. (2011) Enzymatic assembly of overlapping DNA fragments. in Methods Enzymol. 498, 349–361 10.1016/B978-0-12-385120-8.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sander K., Asano K. G., Bhandari D., Van Berkel G. J., Brown S. D., Davison B., and Tschaplinski T. J. (2017) Targeted redox and energy cofactor metabolomics in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. Biotechnol. Biofuels 10, 270 10.1186/s13068-017-0960-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schatschneider S., Abdelrazig S., Safo L., Henstra A. M., Millat T., Kim D.-H., Winzer K., Minton N. P., and Barrett D. A. (2018) Quantitative isotope-dilution high-resolution-mass-spectrometry analysis of multiple intracellular metabolites in Clostridium autoethanogenum with uniformly 13C-labeled standards derived from spirulina. Anal. Chem. 90, 4470–4477 10.1021/acs.analchem.7b04758 [DOI] [PubMed] [Google Scholar]
- 39. Tian L., Lo J., Shao X., Zheng T., Olson D. G., and Lynd L. R. (2016) Ferredoxin:NAD+ oxidoreductase of Thermoanaerobacterium saccharolyticum and its role in ethanol formation. Appl. Environ. Microbiol. 82, 7134–7141 10.1128/AEM.02130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olson D. G., Hörl M., Fuhrer T., Cui J., Zhou J., Maloney M. I., Amador-Noguez D., Tian L., Sauer U., and Lynd L. R. (2017) Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab. Eng. 39, 169–180 [DOI] [PubMed] [Google Scholar]
- 41. Blangy D., Buc H., and Monod J. (1968) Kinetics of the allosteric interactions of phosphofructokinase from Escherichia coli. J. Mol. Biol. 31, 13–35 10.1016/0022-2836(68)90051-X [DOI] [PubMed] [Google Scholar]
- 42. Zhang X., Tu B., Dai L. R., Lawson P. A., Zheng Z. Z., Liu L.-Y., Deng Y., Zhang H., and Cheng L. (2018) Petroclostridium xylanilyticum gen. nov., sp. nov., a xylan-degrading bacterium isolated from an oilfield, and reclassification of clostridial cluster III members into four novel genera in a new Hungateiclostridiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 68, 3197–3211 10.1099/ijsem.0.002966 [DOI] [PubMed] [Google Scholar]
- 43. Koeck D. E., Wibberg D., Maus I., Winkler A., Albersmeier A., Zverlov V. V., Liebl W., Pühler A., Schwarz W. H., and Schlüter A. (2016) Corrigendum to “Complete genome sequence of the cellulolytic thermophile Ruminoclostridium cellulosi wild-type strain DG5 isolated from a thermophilic biogas plant.” [J. Biotechnol. 188 (2014) 136–137] J. Biotechnol. 237, 35 10.1016/j.jbiotec.2016.08.020 [DOI] [PubMed] [Google Scholar]
- 44. Yutin N., and Galperin M. Y. (2013) A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol. 15, 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tindall B. J. (2019) The names Hungateiclostridium Zhang et al. 2018, Hungateiclostridium thermocellum (Viljoen et al. 1926) Zhang et al. 2018, Hungateiclostridium cellulolyticum (Patel et al. 1980) Zhang et al. 2018, Hungateiclostridium aldrichii (Yang et al. 1990) Zhang et al. 2018, Hungateiclostridium alkalicellulosi (Zhilina et al. 2006) Zhang et al. 2018, Hungateiclostridium clariflavum (Shiratori et al. 2009) Zhang et al. 2018, Hungateiclostridium straminisolvens (Kato et al. 2004) Zhang et al. 2018 and Hungateiclostridium saccincola (Koeck et al. 2016) Zhang et al. 2018 contravene Rule 51b of the International Code of Nomenclature of Prokaryotes and require replacement names in the genus Acetivibrio Patel et al. 1980. Int. J. Syst. Evol. Microbiol. 69, 3927–3932 [DOI] [PubMed] [Google Scholar]
- 46. Sharma A. (2019) El-MAVEN 10.5281/zendo.3583792 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.