FIG 8.
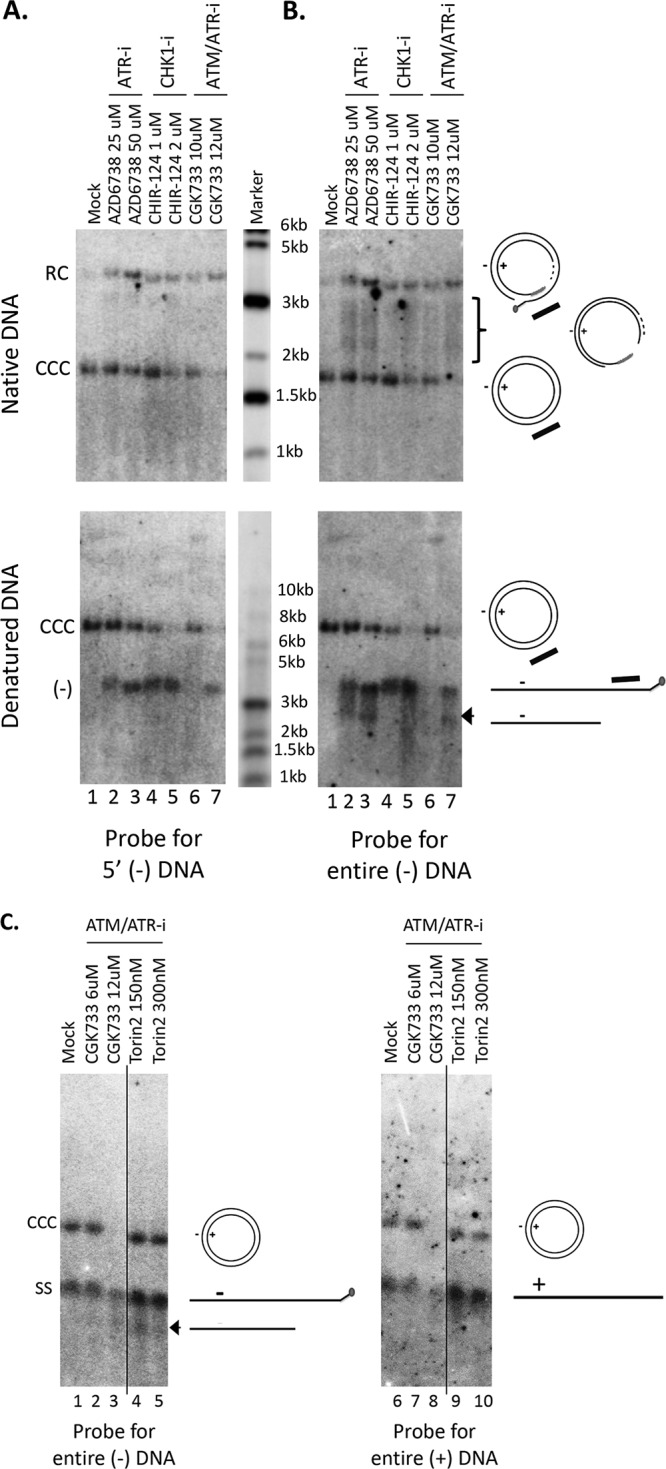
Accumulation of 5′ truncated minus strands under conditions of ATR-CHK1 inhibition. HBV PF DNA extracted from HBV-infected HepG2-NTCP cells, either mock treated or treated with the indicated inhibitors, was analyzed by Southern blotting using a riboprobe hybridizing to the 5′ end of the viral minus strand DNA (A), to the full-length minus strand DNA (B; C, lanes 1 to 5), or to the full-length plus strand DNA (C, lanes 6 to 10). Native DNA (undenatured) was used for the results shown in the top of panels A and B and heat denatured (95°C, 10 min) DNA at the bottom of panels A and B and in panel C. The diagrams to the right of the images depict the various HBV DNA forms detected on the Southern blots, including the RC DNA, CCC DNA, full-length minus strand DNA (–), full-length plus strand DNA (+), RC DNA processing products missing the 5′ end of the minus strand (bracket), and the 5′ truncated minus strand DNA (arrowhead, B, bottom; and C). The bracket indicates the RC DNA processing products accumulating under conditions of ATR-CHK1 inhibition. The short black bar represents the riboprobe for the specific detection of the 5′ end of the minus strand DNA; it specifically hybridizes to the minus strand DNA sequence from nt 1377 to 1805. The vertical thin lines in the images denote where the different parts of the same gel, and with the same exposure, were spliced together in order to remove other parts of the gel that are not presented here.
