Abstract
The basic reproduction number of an infectious agent is the average number of infections one case can generate over the course of the infectious period, in a naïve, uninfected population. It is well-known that the estimation of this number may vary due to several methodological issues, including different assumptions and choice of parameters, utilized models, used datasets and estimation period. With the spreading of the novel coronavirus (2019-nCoV) infection, the reproduction number has been found to vary, reflecting the dynamics of transmission of the coronavirus outbreak as well as the case reporting rate. Due to significant variations in the control strategies, which have been changing over time, and thanks to the introduction of detection technologies that have been rapidly improved, enabling to shorten the time from infection/symptoms onset to diagnosis, leading to faster confirmation of the new coronavirus cases, our previous estimations on the transmission risk of the 2019-nCoV need to be revised. By using time-dependent contact and diagnose rates, we refit our previously proposed dynamics transmission model to the data available until January 29th, 2020 and re-estimated the effective daily reproduction ratio that better quantifies the evolution of the interventions. We estimated when the effective daily reproduction ratio has fallen below 1 and when the epidemics will peak. Our updated findings suggest that the best measure is persistent and strict self-isolation. The epidemics will continue to grow, and can peak soon with the peak time depending highly on the public health interventions practically implemented.
Keywords: Novel coronavirus, Emerging and reemerging pathogens, Mathematical modeling, Basic reproduction number, Effective daily reproduction ratio
Introduction
Coronaviruses are a group of enveloped viruses with a positive-sense, single-stranded RNA and viral particles resembling a crown – from which the name derives. They belong to the order of Nidovirales, family of Coronaviridae, and subfamily of Orthocoronavirinae (Carlos, Dela Cruz, Cao, Pasnick, & Jamil, 2020). They can affect mammals, including humans, causing generally mild infectious disorders, sporadically leading to severe outbreaks clusters, such as those generated by the “Severe Acute Respiratory Syndrome” (SARS) virus in 2003 in mainland China, and by the “Middle East Respiratory Syndrome” (MERS) virus in 2012 in the Kingdom of Saudi Arabia and in 2015 in South Korea (Gralinski & Menachery, 2020).
Currently, there exist no vaccines or anti-viral treatments officially approved for the prevention or management of the disease. Anti-retroviral drugs belonging to the class of protease inhibitors, including Lopinavir and Ritonavir, usually utilized for the treatment of HIV/AIDS patients, seem to exert anti-viral effects against coronaviruses. GS-734 (Remdesivir), a nucleotide analogue pro-drug, originally developed against the Ebola and the Marburg viruses, has been recently suggested to be effective also against coronaviruses. Other potential pharmaceuticals include nucleoside analogues, neuraminidase inhibitors, and RNA synthesis inhibitors. Also, Umifenovir (Abidol), used for treating severe influenza cases, anti-inflammatory drugs and EK1 peptide have been proposed as possible drugs against coronaviruses (Lu, 2020).
A recent coronavirus outbreak has started since December 29th, 2019 in Wuhan, Hubei province, People’s Republic of China, and has progressively expanded to various parts of China and has reached as well other countries, including Japan, South Korea, Thailand, Vietnam, Malaysia, Singapore, Nepal, Cambodia, the Philippines, Russia, the United Arab Emirates, Australia, Canada, the United States of America and Europe (France, Germany, Italy, UK, Finland and Sweden). So far, the new virus has infected more than 31,000 people and killed at least 636 of them (National Health Commission of the People’s Republic of China, 2020).
While considerable progress has been achieved with respect to seventeen years ago, when the world had to face, completely unprepared, the SARS pandemics, several issues still remain to cope with. In order to respond swiftly and properly to the outbreak, public health decision- and policy-makers need timely and accurate epidemiological information, concerning, for example, how long it may take from exposure to the virus to illness/symptoms onset or which individuals, with specific characteristics or co-morbidities, are at higher risk of a poor prognosis. However, many data are still lacking and available data may not be accurate or reliable and may contain substantial uncertainty, concerning, for instance, the precise timing and natural history of cases. Simulating different scenarios with evolving knowledge and gradually improved data quality present significant challenges for modelers. On the other hand, scenario analysis could help ruling out some (unrealistic or over optimistic) assumptions, enabling to test different hypotheses.
As recognized by the World Health Organization (WHO), mathematical models, especially those devised in a timely fashion, can play a key role in providing health decision- and policy-makers with evidence-based information. Modeling can, indeed, better help understanding: i) how transmissible the disease is, ii) when the infectiousness is highest during the course of infection, iii) how severe the infection is, and iv) how effective interventions have been and ought to be.
The international community of modelers has accepted the challenge of designing mathematical models of coronavirus dynamics and transmission and has swiftly reacted to the current coronavirus outbreak. Several models have been produced, resulting, sometimes, in different estimates. Previously, our group has devised a deterministic compartmental (SEIR) model (Tang et al., 2020). In the present article, we update this model, based on the latest available data and information.
Methods
Time-dependent dynamic model
On January 23rd, 2020, Wuhan, the epicenter of the current coronavirus outbreak, announced the implementation of travel restriction as strategy for controlling the infection. Following this announcement, many other cities and provinces of China decided to enforce similar measures. In the meantime, many other control measures have been adopted, like convincing all the residents to stay at home and avoid contacts as much as possible. From the mathematical point of view, this can significantly contribute to decreasing the contact rate among the persons. On the other hand, also from January 23rd, 2020, gradually increasing numbers of 2019-nCoV testing kits were sent to Wuhan from other provinces, gradually shortening the time period of diagnosis (i.e. the value of increases greatly). Considering these control strategies, we adapted our previous model (Tang et al., 2020) as time-dependent dynamic system, by taking January 23rd, 2020 as the newly initial time:
Here, we assume that the contact rate is a decreasing function with respect to time t, which is given by
where is the contact rate at the initial time (i.e. January 23rd, 2020), is the minimum contact rate under the current control strategies, and is the exponential decreasing rate of the contact rate. Definitely, there are and with . This is basically to assume the contacts are decreasing and the change rate per contact is . This constant provides a measure of public health intervention improvement in terms of self-isolation of all including susceptible individuals in the period.
Similarly, we set to be an increasing function with respect to time t, equivalently, the period of diagnosis is a decreasing function of t with the following form:
here, is the diagnose rate at the initial time with , is the fastest diagnose rate with , and is the exponential decreasing rate. This rate is highly relevant to the resources available in the epicenter.
Using the formula we derived in (Tang et al., 2020) but replacing the constant contact rate c and with the aforementioned time-dependent coefficients to reflect the evolving public health interventions and resources available, we defined
as the effective daily reproduction ratio, to measure the ‘daily reproduction number’, the number of new infections induced by a single infected individual during his/her infectious period per day.
The data
We obtained the updated data of the cumulative number of laboratory-confirmed 2019-nCov cases from the National Health Commission of the People’s Republic of China (National Health Commission of the People’s Republic of China, 2020). The data information includes the cumulative confirmed cases, the cumulative number of deaths, newly confirmed cases and the cumulative number of cured cases, which are reported daily by the National Health Commission of the People’s Republic of China.
Parameter estimation process
Under the gradually enhanced control strategies since January 23rd, 2020, the parameter values with substantial changes include the contact rate, the diagnose rate and the quarantined rate q. Therefore, we fixed the parameter values except these three as the estimated values in our previous study (Tang et al., 2020). The initial contact rate is assumed to be the average contact rate between January 10th, 2020 and January 22nd, 2020, hence . With the same assumption, we set . Note that, the initial conditions can be obtained by solving our previous model (Tang et al., 2020) from January 10th, 2020 to January 23rd, 2020. Thus, the main task is to estimate the parameter values .
We use the Markov Chain Monte Carlo (MCMC) method to fit the model to the data, and adopt an adaptive Metropolis-Hastings (M-H) algorithm to carry out the MCMC procedure. The algorithm is run for 70,000 iterations with a burn-in of the first 50,000 iterations, and the Geweke convergence diagnostic method is employed to assess convergence of chains. The estimation results are given in Table 1.
Table 1.
Parameter values.
| Parameter | Definitions | Estimated mean value | Standard deviation | Data source |
|---|---|---|---|---|
| Contact rate at the initial time | 1 | |||
| Minimum contact rate under the current control strategies | MCMC | |||
| Exponential decreasing rate of contact rate | MCMC | |||
| Probability of transmission per contact | 1 | |||
| Quarantined rate of exposed individuals | MCMC | |||
| Transition rate of exposed individuals to the infected class | – | 2 | ||
| Rate at which the quarantined uninfected contacts were released into the wider community | – | 3,4 | ||
| Probability of having symptoms among infected individuals | 1 | |||
| Initial transition rate of symptomatic infected individuals to the quarantined infected class | 1 | |||
| The shortest period of diagnosis | MCMC | |||
| Exponential decreasing rate of diagnose rate | MCMC | |||
| Transition rate of quarantined exposed individuals to the quarantined infected class | 1 | |||
| Recovery rate of symptomatic infected individuals | 1 | |||
| Recovery rate of asymptomatic infected individuals | 1 | |||
| Recovery rate of quarantined infected individuals | 1 | |||
| Disease-induced death rate | 1 |
Results
Since Wuhan was locked down on January 23rd, 2020, almost all regions across the country have imposed travel restriction and, at the same time, the case confirmation speed has been improved due to development of new coronavirus nucleic acid-based detection technologies. Under the scenario of adopting the strongest prevention and control strategy and improving the level of detection and treatment in China, the previously estimated basic reproduction number is no longer suitable for evaluating the epidemic trend in the near future. Therefore, we use the updated data to parameterize the proposed model (Tang et al., 2020) and re-estimate the 2019-nCov transmission risk.
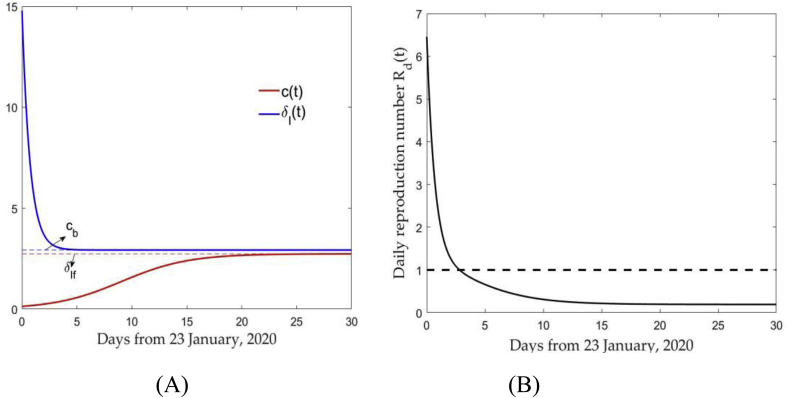
To estimate the effective daily reproduction ratio, we initially get the time-dependent contact rate and as Fig. 1(a). Using the discrete values of contact rate and diagnose rate , we can calculate the effective daily reproduction ratio, shown in Fig. 1(b). It follows that under the strict prevention and control measures, the effective daily reproduction ratio has been less than 1 since January 26th, 2020, that is, the number of new infections has begun to decline. Note that the effective daily reproduction ratio declined from January 23rd, 2020 to January 25th, 2020, as a combination of the restrictive measures, including the lock-down of Wuhan, contact tracing followed by quarantine and isolation, that have been implemented. In practice, this time variation of the contact and diagnose rates leads to sub-exponential rather than exponential growth dynamics, and hence provides better estimates of epidemic size compared to fully exponential growth models. We refer to (Pell, Kuang, Viboud, & Chowell, 2018; Smirnova & Chowell, 2017) for earlier studies on sub-exponential growth of modern epidemics.
Fig. 1.
(A) Time-dependent contact rate and diagnose rate ; (B) Effective daily reproduction ratio , declining due to reduction of c(t) and increase of .
Near-casting in a rapidly evolving situation requires timely information of the implementation of public health interventions. We emphasize that this information is not only about the policy and decision, but also the implementation which is highly dependent on the resources available. We illustrate this with two simulated predictions: one based on the assumption that the interventions implemented during January 23rd, 2020 to January 29th, 2020 will be sustained, and another one based on additional data beyond January 29th, 2020.
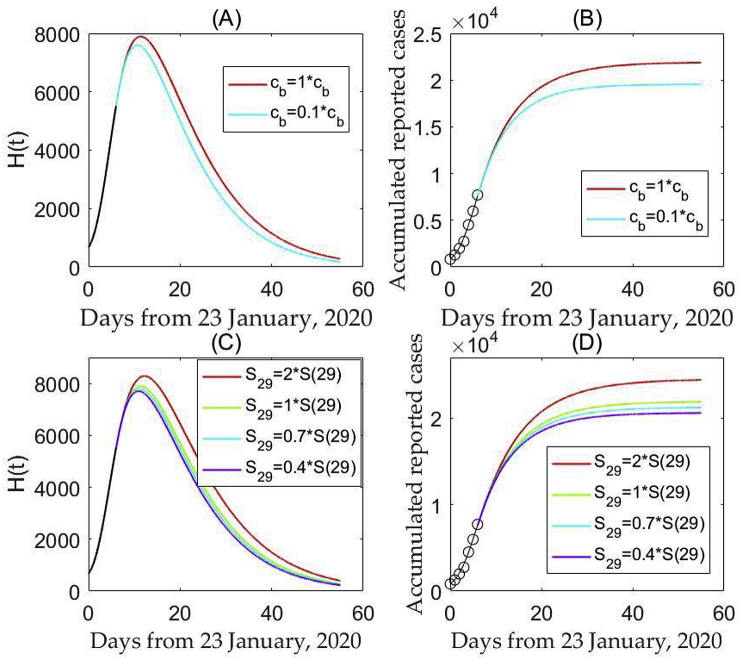
We first plot the time series on the predicted number of reported cases, i.e., the number of hospitalized individuals H(t) and the predicted cumulative cases based on the updated parameters listed in Table 1, and shown in Fig. 2. It shows that the number of hospitalized individuals will peak on around February 4th, 2020 (Fig. 2(A, C)), while the predicted number of cumulative cases will continue to grow for some duration but with a slower growth rate (Fig. 2(D)). Moreover, sensitivity analysis revealed that further enhanced measures can reduce the peak value and hence decrease the predicted cumulative case numbers ((Fig. 2(A and B)). We caution that increasing the number of susceptible individuals may lead to an increase in the peak value and enlarge the predicted cumulative case numbers ((Fig. 2(C and D)), emphasizing the importance of sustaining the implemented control strategies such as self-isolation in order to reduce the susceptibility. We emphasize that the peak time is defined here as the time when the number of confirmed cases reaches the maximum, so sustaining the intervention measures is critical.
Fig. 2.
Predictions and effect of control measures on infection based on assumption that parameters obtained from fitting the data from January 23rd to January 29th, 2020 (and hence the interventions) remain unchanged. (A–B) Decreasing the minimum contact rate after January 29th, 2020; (C–D) Decreasing/increasing the susceptible population size as of January 29th, 2020.
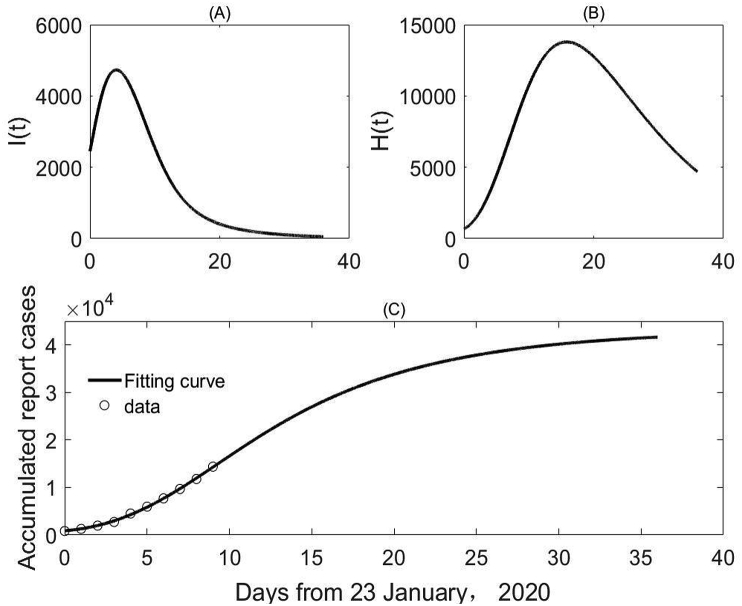
We repeated the procedure as above but fitted our model to the data of confirmed cases between January 23rd and February 1st, 2020 (Fig. 3) and observed the improved . As a result, in comparison with the results in Fig. 2, we obtained higher projected cumulative confirmed cases and delayed peak time.
Fig. 3.
Best fitting of the model to the data of cumulative confirmed cases between January 23rd and February 1st, 2020: the projected number of infected (A), quarantined infected (B), and cumulative confirmed cases (C).
Discussion
In our previous article (Tang et al., 2020), we had estimated a reproduction number of 6.47 (95% CI 5.71–7.23), which represents a higher value than those so far computed. For instance, the WHO has estimated a reproduction number of 1.4–2.5, Li and colleagues (Li et al., 2020) have computed a slightly higher value of 2.2 (95% CI 1.4 to 3.9), while in several other mathematical models which have been so far devised and released as pre-prints or undergone peer-review and published, the reproduction number varies from 1.3 to 4.7.
Within the existing literature, two studies estimated under-estimation of coronavirus cases: the investigations by Zhao and collaborators (Zhao et al., 2020) and by Read et al. (Read, Bridgen, Cummings, Ho, & Jewell, 2020). Specifically, Zhao and coworkers (Zhao et al., 2020) have assessed from a quantitative standpoint the under-reporting rate of coronavirus cases, modeling the epidemic growth curve using the exponential growing Poisson process. Authors computed the number of under-reported coronavirus cases to be 469 (95% CI 403–540). Based on this estimate, the basic reproduction number was found to be 2.56 (95% CI 2.49–2.63). In a previous version of the investigation, released in the bioRxiv pre-print server, the reproduction number was computed to oscillate between 3.30 (95% CI 2.73–3.96) and 5.47 (95% CI 4.16–7.10). The basic reproduction number was also found to be associated with 0-fold–2-fold increase in the reporting rate. More in detail, with report rates increasing over the time, the mean value was statistically likely to be higher than 3 but less than 5. Read and coauthors (Read et al., 2020) used a deterministic SEIR model, assuming coronavirus cases being Poisson distributed and with parameter inference being achieved by maximum likelihood estimation utilizing the Nelder-Mead optimization approach. According to the dynamics transmission model, an ascertainment rate of 5.0% (95% CI 3.6–7.4) was computed and, based on this, authors estimated that as of January 22nd, 2020 in Wuhan there were 14,464 (95% CI 6510–25095) infected individuals, and 21,022 (95% CI 11,090–33,490) infections. The basic reproduction number was computed to be 3.11 (95% CI 2.39–4.13). In terms of public health implications, in order to stop the outbreak, at least 58–76% of transmissions should be blocked.
There are three further models incorporating data from international travels: the models of Imai and coauthors (Imai et al., 2020), of Kucharski et al. (Kucharski et al., 2020) and of Wu and collaborators (Wu, Leung, & Leung, 2020). In particular, Imai and coworkers (Imai et al., 2020) estimated a reproduction number of 2.6 (uncertainty range: 1.5–3.5). Depending on the different scenarios and levels of zoonotic exposure, the reproduction number was found to vary from 1.7 to 2.6 to 1.9–4.2. Depending on the different estimates of generation time, the reproduction number oscillated from 1.3 to 2.7 to 1.7–4.3. Based on the level of infectiousness, the reproduction number varied in the range of 1.6–2.9. Finally, assuming that the novel virus would cause more mild-to-moderate cases than the SARS virus, the reproduction number would be 2.0 (uncertainty 1.4–2.3). Moreover, authors found that only public health interventions blocking over 60% of transmission would be really effective in controlling and containing the coronavirus outbreak. Partially based on the findings of Imai and coworkers (Imai et al., 2020) and building on a SIR model, Yu (Yu, 2020) has computed a basic reproduction number of 3.5 and has estimated that only a quarantine rate of infectious population higher than 90% would enable to effectively control the coronavirus outbreak. Kucharski and colleagues (Kucharski et al., 2020) designed a stochastic SEIR model, based on the Euler-Maruyama algorithm with a 6-h time-step and with the transmission rate following a geometric Brownian motion. Time-varying reproduction number was estimated using the sequential Monte-Carlo approach. Authors utilized several datasets to overcome the issue of unreliability of some data sources and to provide real-time estimates, relying on the Poisson probability calculation. Transmission was modeled as a random process, fluctuating and varying over the time. Similar to the model of Imai and coworkers (Imai et al., 2020), the risk of transmission and the risk of causing a large outbreak were modeled based on a negative binomial offspring distribution, with incubation and infectious period being Erlang distributed. Median reproduction number was found to oscillate between 1.6 and 2.9 before the introduction and implementation of travel restriction. The study by Wu and collaborators (Wu et al., 2020), based on nowcasting and forecasting approach, estimated a basic reproductive number of 2.68 (95% credible interval or CrI 2.47–2.86) with 75,815 individuals (95% CrI 37,304–130,330) being infected in Wuhan as of January 25th, 2020. The epidemics doubling time was found to be 6.4 days (95% CrI 5.8–7.1). The dynamics transmission model by Shen and coworkers (Shen, Peng, Xiao, & Zhang, 2020) predicted 8042 (95% CI 4199–11,884) infections and 898 (95% CI 368–1429) deaths, with a fatality rate of 11.02% (95% CI 9.26–12.78%). Authors computed a basic reproduction number of 4.71 (95% CI 4.50–4.92), which decreased to 2.08 (95% CI 1.99–2.18) on January 22nd, 2020. Based on these estimates, the pandemics outbreak is expected to significantly decrease within 77 [95% CI 75–80] days from its beginning. Furthermore, authors found that every one-day reduction in the duration of the period from illness/symptom onset to isolation would reduce the peak population size by 72–84% and the cumulative infected cases and deaths both by 68–80%. The study by Majumder and Mandl (Majumder & Mandl, 2020) utilized the “Incidence Decay and Exponential Adjustment” (IDEA) model and led to an estimate of the reproduction number in the range of 2.0–3.1. Finally, Riou and Althaus (Riou & Althaus, 2020), using a stochastic model simulating epidemics trajectories, computed a reproduction number of 2.2 (90% high density interval 1.4–3.8). Using statistical approaches, namely exponential growth and maximum likelihood techniques, Liu and colleagues (Liu et al., 2020) estimated the value of the reproduction number ranging from 2.90 (95% CI 2.32–3.63) to 2.92 (95% CI 2.28–3.67). Zhang and Wang (Zhang & Wang, 2020), employing a Bayesian framework to infer time-calibrated phylogeny from 33 available genomic sequences, found that the time of the most recent common ancestor (MRCA) was December 17th, 2019 (95% highest posterior density interval from December 7th, 2019 to December 23rd, 2019) and that the value of the reproduction number oscillated between 1.1 and 1.6.
These different findings may be due to several methodological issues, including different assumptions and choice of parameters, utilized models (stochastic versus deterministic, compartmental versus IDEA, etc.), used datasets and estimation period. Furthermore, by comparing the various updated versions of the above-mentioned investigations, the reproduction number was found to vary, reflecting the dynamics of transmission of the coronavirus outbreak as well as the dynamics of case reporting. More in detail, the reproduction number tended to increase over the time in parallel with the increase in cases being reported and the findings were highly sensitive and dependent on the period in which the estimate was made and on the data available at that given time. It should be mentioned that much of the model frameworks and data fitting and analysis have been developed from earlier studies about the SARS outbreak (Chowell et al., 2004; Gumel et al., 2004). It was believed that the SARS outbreak was characterized by a large basic reproduction number within hospitals (nosocomial infection) and a relatively small basic reproduction number in the general community, leading to a moderate basic production number overall. We argue that the current 2019 n-Cov situation is similar to what was observed about the SARS, except that the entire city of Wuhan is the epicenter with a population of over 11 million inhabitants, and the community is the entire country due to the travels done before the shutdown of the epidemic center.
The outbreak situation is fast evolving both in the epicenter of Wuhan, the Hubei province and throughout China. Our simulations show that the control outcome depends highly on the interventions implemented in the field, which depend on the resources provided to the frontline workers and patients infected. The size of peak value and peak time depends on a number of factors including in particular the speed of diagnoses and hospitalization of confirmed cases.
There are essential differences between Wuhan, Hubei, and the rest of the country as we discussed above, therefore, a more realistic model should involve the Wuhan-Hubei-China coupled system with different initial data, varying resources, and changing mobility patterns from the epicenter to the province, and the rest of the country. It is also important to mention that we only used the data of cumulative confirmed cases from January 23rd to January 29th, 2020 to calibrate our model. Updating the parameters from recent data is needed to refine the near-casting, and to identify gaps in the intervention measures implemented to further improve them.
Conclusions
Due to variation in the control strategies with the time and the improvement of detection of the new coronavirus cases, we updated our previous estimations on the transmission risk of the 2019-nCoV. By using time-dependent contact and diagnose rates we refit our proposed dynamics transmission model to the data available until January 29th, 2020 and re-estimated the daily reproduction number, which implies the evolving transmission risk. We observed that the effective daily reproduction ratio has already fallen below 1, therefore while the epidemics will continue to grow, the epidemic will peak soon.
Accurately near-casting the epidemic trend and projecting the peak time require real-time information of the data and the knowledge about the implementation and the resources available to facilitate the implementation, not only the policy and decision, of major public health interventions.
Funding
This research was funded by the National Natural Science Foundation of China (grant numbers: 11631012 (YX, ST), 61772017(ST)), and by the Canada Research Chair Program (grant number: 230720 (JW) and the Natural Sciences and Engineering Research Council of Canada (Grant number:105588-2011 (JW).
Declaration of competing interest
None.
Acknowledgement
We all appreciate the joint support by the Tianyuan Mathematical Center in Northwest China, the Fields Institute for Research in Mathematical Sciences, and the Mathematics for Public Health (MfPH) Laboratory (Fields-CQAM).
Handling Editor: Dr Y. Shao
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Sanyi Tang, Email: sytang@snnu.edu.cn.
Yanni Xiao, Email: yxiao@mail.xjtu.edu.cn.
Jianhong Wu, Email: wujh@yorku.ca.
References
- Carlos W.G., Dela Cruz C.S., Cao B., Pasnick S., Jamil S. Novel Wuhan (2019-nCoV) coronavirus. American Journal of Respiratory and Critical Care Medicine. 2020 doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- Chowell G., Castillo-Chavez C., Fenimore P.W., Kribs-Zaleta C.M., Arriola L., Hyman G.M. Model parameters and outbreak control for SARS. Emerging Infectious Diseases. 2004;10(7):1258–1263. doi: 10.3201/eid1007.030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2) doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumel A.B., Ruan S.G., Day T. Modelling strategies for controlling SARS outbreaks. Proceedings of the Royal Society of London B. 2004;271:2223–2232. doi: 10.1098/rspb.2004.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai N., Cori A., Dorigatti I., Baguelin M., Donnelly C.A., Riley S. 2020. Report 3: Transmissibility of 2019-nCoV.https://www.imperial.ac.uk/media/imperial-college/medicine/sph/ide/gida-fellowships/Imperial-2019-nCoV-transmissibility.pdf Available at: [Google Scholar]
- Kucharski A., Russell T., Diamond C., CMMID nCoV working group, Funk S., Eggo R. Analysis of early transmission of 2019-nCoV and implications for outbreaks in new locations. 2020. https://cmmid.github.io/ncov/wuhan_early_dynamics/index.html Available at:
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Hu J., Kang M., Lin L., Zhong H., Xiao J. Transmission dynamics of 2019 novel coronavirus (2019-nCoV) bioRxiv. 2020 [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Bioscience Trends. 2020 doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Majumder M., Mandl K.D. Early transmissibility assessment of a novel coronavirus in Wuhan, China. Social Science Research Network. 2020 [Google Scholar]
- National Health Commission of the People’s Republic of China 2020. http://www.nhc.gov.cn/ Available at: [DOI] [PMC free article] [PubMed]
- Pell B., Kuang Y., Viboud C., Chowell G. Using phenomenological models for forecasting the 2015 Ebola challenge. Epidemics. 2018;22:62–70. doi: 10.1016/j.epidem.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Read J.M., Bridgen J.R., Cummings D.A., Ho A., Jewell C.P. Novel coronavirus 2019-nCoV: Early estimation of epidemiological parameters and epidemic predictions. medRxiv. 2020 doi: 10.1098/rstb.2020.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveillance. 2020;25(4) doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Peng Z., Xiao Y., Zhang L. Modelling the epidemic trend of the 2019 novel coronavirus outbreak in China. bioRxiv. 2020 doi: 10.1016/j.xinn.2020.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova A., Chowell G. A primer on stable parameter estimation and forecasting in epidemiology by a problem-oriented regularized least squares algorithm. Infectious Diseases Modelling. 2017;2:268–275. doi: 10.1016/j.idm.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Wang X., Li Q., Bragazzi N.L., Tang S., Xiao Y. Estimation of the transmission risk of 2019-nCov and its implication for public health interventions. Journal of Clinical Medicine. 2020;9:462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. Updated estimating infected population of Wuhan coronavirus in different policy scenarios by SIR model. 2020. http://uni-goettingen.de/en/infectious+diseases/619691.html Available at:
- Zhang C., Wang M. MRCA time and epidemic dynamics of the 2019 novel coronavirus. bioRxiv. 2020 [Google Scholar]
- Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. International Journal of Infectious Diseases. 2020 doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]