Abstract
The AGTR1 gene encodes angiotensin II receptor type 1, which is involved in cardiovascular diseases such as coronary heart disease (CHD). In the current study, we analyzed AGTR1 promoter methylation level in a Han Chinese population by SYBR green-based quantitative methylation-specific PCR (qMSP). We collected blood samples from 761 CHD patients and 398 non-CHD controls at the Ningbo First Hospital. A data mining analysis was also performed to explore the association between AGTR1 methylation and AGTR1 gene expression, using datasets from the cBioPortal for Cancer Genomics and the Gene Expression Omnibus (GEO) database. Our results showed a significantly higher percentage of methylated reference (PMR) of AGTR1 in male CHD patients compared with male non-CHD controls (median PMR: 2.12% vs. 0.59%, p = 0.037). The data mining analysis showed that AGTR1 expression was significantly increased in human hepatoma HepG2 cells treated with the demethylation agent 5-aza-2’-deoxycytidine (fold = 3.12, p = 0.009). Further data mining analysis using the cholangiocarcinoma (TCGA, PanCancer Atlas) data indicated an inverse association between AGTR1 methylation and AGTR1 expression (r = -0.595, p = 1.29E-04). Overall, our results suggest that AGTR1 methylation is involved in the regulation of AGTR1 gene expression and that AGTR1 hypermethylation is associated with CHD in males. These findings may provide new clues about the pathogenesis of CHD.
Keywords: Coronary heart disease, CHD, AGTR1, DNA methylation, males, angiotensin II receptor type 1, HepG2 cells, 5-aza-2’-deoxycytidine
INTRODUCTION
Coronary heart disease (CHD) is a type of cardiovascular disease, which remains a significant public health problem globally [1,2]. CHD is a complex disease influenced by both environmental and genetic factors. Previous studies suggested that epigenetic modifications are involved in the pathophysiology of CHD [3,4].
An important epigenetic mechanism of gene expression regulation is DNA methylation, which represents the methylation of cytosine to 5-methylcytosine, primarily at the CpG sites [5]. Recent studies showed that the methylation level of FOXP3, BAX, and PON1 is significantly associated with CHD [6-8]. Interestingly, CDKN2B promoter methylation was significantly associated with the risk of CHD in women but not in men [9], suggesting its sex-dependent effects in the pathogenesis of CHD.
Angiotensin II is a potent vasopressor hormone that plays a vital role in blood pressure regulation and is associated with the pathogenesis of CHD [10]. Angiotensin II mainly acts through angiotensin II receptor type 1 (AGTR1) and angiotensin II receptor type 2 (AGTR2), which mediate its cardiovascular effects [11,12]. AGTR1 is a member of the G-protein-coupled receptor superfamily and is encoded by a gene on chromosome 3q [12]. Previous genetic studies found that AGTR1 variants are associated with essential hypertension in Polish and Finnish populations [13,14]. Moreover, AGTR1 rs3772622 polymorphism increases the risk of CHD in Northern Han Chinese patients with non-alcoholic fatty liver disease (NAFLD) [15]. On the other hand, another study on Chinese patients showed no correlation between CHD and 8 AGTR1 haplotypes with a frequency greater than 3% [16]. Similarly, in our previous study, we did not find an association between four CpG-SNPs on the AGTR1 promoter and CHD in a Han Chinese population [17].
Aberrant AGTR1 methylation is associated with multiple diseases. AGTR1 hypermethylation increases the risk of oral cancer [18] and non-small-cell lung carcinoma [19]. A previous study found a significantly lower AGTR1 methylation in hypertensive patients than in healthy controls [20].
The relationship between AGTR1 methylation and CHD is still not clear. Therefore, in this study, we aimed to explore the association of AGTR1 promoter methylation with CHD in a Han Chinese population.
MATERIALS AND METHODS
Patients
We collected blood samples from 761 CHD patients (501 men and 260 women, median age: 62 years) and 398 non-CHD controls (220 men and 178 women, median age: 60 years) at the Ningbo First Hospital. CHD patients were diagnosed with coronary artery stenosis greater than 50% in one or more of the major coronary arteries. The control group consisted of inpatients with coronary artery stenosis less than 50% in the major coronary arteries and without any atherosclerotic vascular disease. Standardized coronary angiography with Seldinger’s technique was performed in all patients [21,22]. The results of the angiography were independently evaluated by at least two cardiologists. None of the control inpatients had histories of congenital heart disease, cardiomyopathy, and severe liver or kidney disease. The Institutional Review Board of Ningbo First Hospital and Ningbo University approved the study (NBU-CHD-20180305). All participants provided written informed consent form.
SYBR green-based quantitative methylation-specific PCR (qMSP)
We extracted genomic DNA from peripheral blood samples as previously described [23]. The details of bisulfite conversion and qMSP procedures are available from our previous studies [24,25]. To measure the methylation of AGTR1 promoter we calculated the percentage of methylated reference (PMR) [24,25]. The primer sequences used in qMSP were forward 5’-GGAGGAGGAGGGAATGTAA-3’ and reverse 5’-CCTATCACTCGCTACTACCT-3’.
Data mining analysis
A data mining analysis was performed to explore the association between AGTR1 methylation and AGTR1 gene expression. We obtained the cholangiocarcinoma dataset (TCGA, PanCancer Atlas) from the cBioPortal for Cancer Genomics (http://www.cbioportal.org) [26]; AGTR1 methylation and AGTR1 gene expression were determined with Infinium HumanMethylation450 BeadChip and RNAseq, respectively. In addition, we used the data for human hepatoma HepG2 cells from the Gene Expression Omnibus (GEO) database (GSE5230).
Statistical analysis
PASW statistics 18.0 software (SPSS Inc., Somers, NY, USA) was used for statistical analysis. We compared continuous variables between the two groups by a t-test or a non-parametric test. A univariate linear regression analysis was used to determine the individual risk factors, then a multivariate regression analysis was performed to explore the association between these factors and CHD. A Spearman’s correlation test was used to assess the association between AGTR1 methylation and each of the 8 metabolic parameters, as follows low-density lipoprotein (LDL), total cholesterol (TC), triglyceride (TG), apolipoprotein A1 (ApoA1), apolipoprotein B (ApoB), apolipoprotein E (ApoE), lipoprotein A [Lp(a)], and high-sensitivity C-reactive protein (hs-CRP). A two-tailed p < 0.05 was considered to be significant.
RESULTS
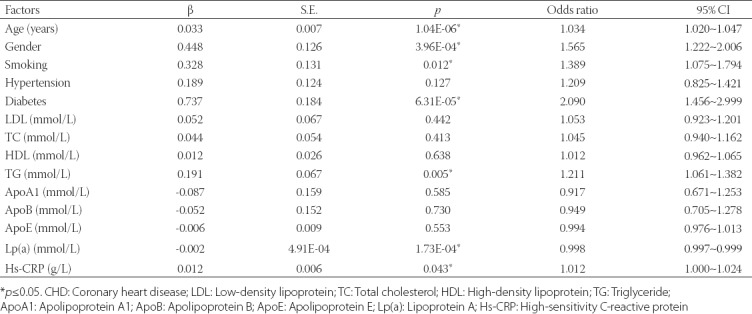
In this study, we included 761 CHD patients (mean age: 58.52 ± 9.52) and 398 controls [mean age: 61.41 ± 9.29] (Table 1). As shown in Figure 1, the selected region for the AGTR1 methylation assay is located in a transcription factor-binding site. The methylation level of an AGTR1 CpG was measured, representing a 142-bp amplified fragment in the CpG island of AGTR1 (Figure 1).
TABLE 1.
Univariate logistic regression analysis of environmental factors that may influence CHD in the Han Chinese population
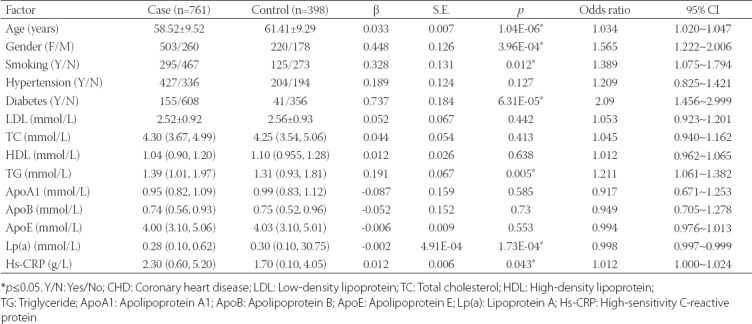
FIGURE 1.
DNA fragment tested in the AGTR1 methylation assay. (A) The genomic position of the amplified fragment in AGTR1x gene CpG island; F and R stand for forward and reverse primer, respectively. (B) DNA sequencing indicated a good bisulfite conversion. (C) Gel electrophoresis showed the correct size of the amplified DNA fragment (142 bp).
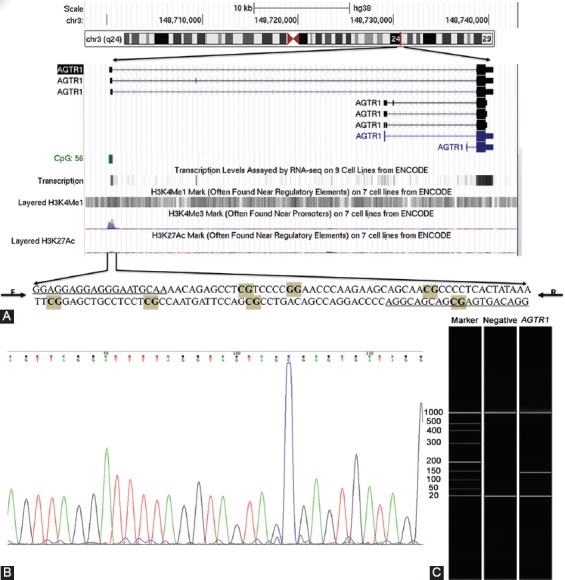
Demographic characteristics and laboratory parameters, including LDL, TC, TG, ApoA1, ApoB, ApoE, Lp(a), and hs-CRP are shown in Table 1. Our univariate regression analyses showed that age, male sex, cigarette smoking, diabetes, TG, and hs-CRP were the risk factors of CHD (all p < 0.05, Table 1). A multivariate regression analysis showed that 4 factors (age, male sex, diabetes, and TG) were the risk factors and Lp(a) was the protective factor of CHD (all p < 0.05, Table 2). However, AGTR1 methylation was not associated with CHD in the subgroup analyses by diabetic status (p > 0.05, data not shown). We found no significant relationships between AGTR1 methylation and clinical indexes, including age, TG, and Lp(a) (p > 0.05, data not shown).
TABLE 2.
Multivariate logistic regression analysis of environmental factors that may influence CHD in the Han Chinese population
AGTR1 methylation was shown to be significantly higher in male CHD patients than in male controls (mean PMR: 2.12 vs. 0.59, p = 0.037, Table 3). Furthermore, GEO data analysis showed a significantly decreased AGTR1 expression in HepG2 cells treated with a demethylation agent (5-aza-2’-deoxycytidine [5-AZA], fold = 3.12, p = 0.009, Figure 2). Further bioinformatics analysis using the cholangiocarcinoma (TCGA, PanCancer Atlas) dataset indicated a significant inverse association between AGTR1 methylation and AGTR1 gene expression (r = -0.595, p = 1.29E-04, Figure 3). All the above results suggested a pivotal role of AGTR1 methylation in the regulation of AGTR1 gene expression.
TABLE 3.
A comparison of AGTR1 methylation between CHD and non-CHD groups in the Han Chinese population
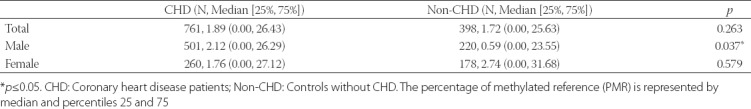
FIGURE 2.
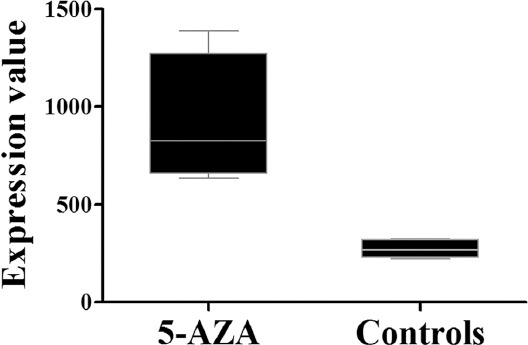
The data mining analysis showed that the demethylation agent 5-aza-2’-deoxycytidine significantly increased AGTR1 gene expression in treated human hepatoma HepG2 cells vs. controls (fold change = 3.12, p = 0.009).
FIGURE 3.
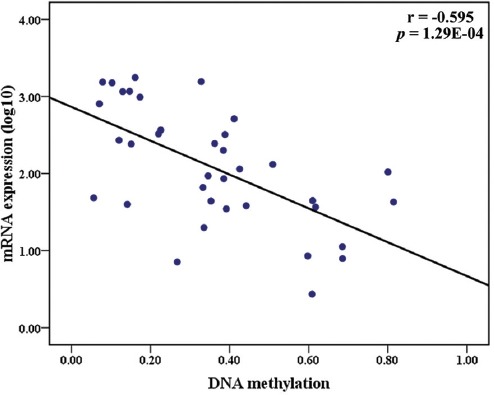
The Spearman’s rank correlation test using the cholangiocarcinoma (TCGA, PanCancer Atlas) data indicated an inverse association between AGTR1 methylation and AGTR1 expression (r = -0.595, p = 1.29E-04). The x-axis shows AGTR1 methylation levels. The y-axis shows log-transformed AGTR1 gene expression values.
In our previous study, we found no association of CHD with CpG-SNP rs275653 on the AGTR1 promoter [17]. Here, we performed an interaction test between rs275653 and AGTR1 methylation. We did not observe a significant interaction between the rs275653 SNP and methylation level (data not shown).
DISCUSSION
In the present study, we explored the association between AGTR1 methylation and CHD. We found four risk factors (age, male sex, diabetes, and TG) and one protective factor [Lp(a)] of CHD. Although there was no association of AGTR1 methylation with CHD in the total sample, we found elevated AGTR1 methylation in male CHD patients compared with male non-CHD controls. Moreover, the data mining analyses showed that a demethylation agent could induce AGTR1 expression and that AGTR1 expression was inversely associated with AGTR1 methylation. These results suggest that AGTR1 methylation contributes to the risk of CHD in males through the regulation of AGTR1 gene expression.
Sex differences exist in the incidence and development of CHD as well as in the frequency of CHD-related polymorphisms [27]. The incidence of CHD is higher in men than in women, in all age groups [28]. Men have a higher prevalence of ST-elevation myocardial infarction but are less likely to have heart failure compared with women [29]. The difference in the distribution of CHD between males and females is possibly due to different lifestyles, including smoking, alcohol consumption, high sodium diet, and physical activity [30-33].
A previous study showed that males with the deletion/deletion (D/D) angiotensin-I converting enzyme (ACE) gene polymorphism have a higher risk of premature myocardial infarction compared with women [34], suggesting a sex difference in the effect of ACE gene polymorphisms on CHD risk. In addition, sex hormones were suggested to regulate DNA methylation in cardiac calsequestrin 2 gene (CASQ2) promoter in mice [35]. A study investigating the association of AGTR1 promoter methylation with the risk of essential hypertension showed that CpG1 hypomethylation in the AGTR1 promoter is likely associated with the risk of essential hypertension, with significantly lower CpG1 methylation levels in males than in females [20]. In the current study, we showed that male sex was a risk factor of CHD, and we detected AGTR1 hypermethylation only in male patients with CHD. Therefore, the sex differences in AGTR1 methylation levels in CHD patients may reflect the differences in lifestyle and sex hormone levels. AGTR1 mediates the cardiovascular effects of angiotensin II [36], and AGTR1 promoter hypermethylation suppresses the gene expression. Our results indicated that sex-dependent difference exist in CHD and that AGTR1 hypermethylation increases the risk of CHD by regulating AGTR1 gene expression.
Some studies showed that an elevated Lp(a) level is an independent risk factor for CHD [37,38]. High levels of Lp(a) are independently associated with CHD-induced recurrent heart failure in patients with chronic heart failure [39]. Interestingly, contrary to the previous findings, our results suggest that Lp(a) may be a protective factor for CHD. Therefore, the role of Lp(a) in CHD should be further explored in larger-sample studies.
Although we demonstrated an association between AGTR1 methylation and CHD in males, the cause-effect relationship remains unclear. In addition, we measured AGTR1 methylation only in peripheral blood samples and DNA methylation profiles may vary among different tissues. Further studies with longitudinal cohort design and that include different tissues should confirm our findings.
CONCLUSION
Overall, our results suggest that AGTR1 methylation is involved in the regulation of AGTR1 gene expression and that AGTR1 hypermethylation is associated with CHD in males. These findings may provide new clues about the pathogenesis of CHD.
ACKNOWLEDGMENTS
This study was supported by K. C. Wong Magna Fund in Ningbo University, Ningbo Health Branding Subject Found (grant no. PPXK2018-01), and the Natural Science Foundation of Zhejiang Province (grant no. LY19H020003).
Footnotes
Conflict of interest statement: The authors declare no conflict of interests
REFERENCES
- 1.Sharma P, Garg G, Kumar A, Mohammad F, Kumar SR, Tanwar VS, et al. Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene. 2014;541(1):31–40. doi: 10.1016/j.gene.2014.02.034. https://doi.org/10.1016/j.gene.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 2.Kanu JS, Gu Y, Zhi S, Yu M, Lu Y, Cong Y, et al. Single nucleotide polymorphism rs3774261 in the AdipoQ gene is associated with the risk of coronary heart disease (CHD) in Northeast Han Chinese population:a case-control study. Lipids Health Dis. 2016;15:6. doi: 10.1186/s12944-015-0173-4. https://doi.org/10.1186/s12944-015-0173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guay SP, Legare C, Houde AA, Mathieu P, Bosse Y, Bouchard L. Acetylsalicylic acid, aging and coronary artery disease are associated with ABCA1 DNA methylation in men. Clin Epigenetics. 2014;6(1):14. doi: 10.1186/1868-7083-6-14. https://doi.org/10.1186/1868-7083-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guay SP, Brisson D, Munger J, Lamarche B, Gaudet D, Bouchard L. ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics. 2012;7(5):464–72. doi: 10.4161/epi.19633. https://doi.org/10.4161/epi.19633. [DOI] [PubMed] [Google Scholar]
- 5.Haochang Hu, Xiaoying Chen, Cheng Wang, Yuting Jiang, Jingjing Li, Xiuru Ying, et al. The role of TFPI2 hypermethylation in the detection of gastric and colorectal cancer. Oncotarget. 2017;8(48):84054–65. doi: 10.18632/oncotarget.21097. https://doi.org/10.18632/oncotarget.21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu L, Jia L, Liu Z, Zhang Y, Wang J, Yuan Z, et al. Elevated methylation of FOXP3 (Forkhead Box P3)-TSDR (Regulatory T-Cell-Specific Demethylated Region) is associated with increased risk for adverse outcomes in patients with acute coronary syndrome. Hypertension. 2019;74(3):581–9. doi: 10.1161/HYPERTENSIONAHA.119.12852. https://doi.org/10.1161/HYPERTENSIONAHA.119.12852. [DOI] [PubMed] [Google Scholar]
- 7.Su J, Li J, Yu Q, Xu X, Wang J, Yang J, et al. Association of PON1 gene promoter DNA methylation with the risk of Clopidogrel resistance in patients with coronary artery disease. J Clin Lab Anal. 2019;33(2):e22867. doi: 10.1002/jcla.22867. https://doi.org/10.1002/jcla.22867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Ji H, Huang Y, Hu H, Li B, Yang Y, et al. Association of BAX hypermethylation with coronary heart disease is specific to individuals aged over 70. Medicine (Baltimore) 2019;98(4):e14130. doi: 10.1097/MD.0000000000014130. https://doi.org/10.1097/MD.0000000000014130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Jiang D, Xu L, Han L, Hu H, Huang Y, et al. Elevated methylation of cyclin dependent kinase inhibitor 2B contributes to the risk of coronary heart disease in women. Exp Ther Med. 2019;17(1):205–13. doi: 10.3892/etm.2018.6920. https://doi.org/10.3892/etm.2018.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pousada G, Baloira A, Valverde D. Molecular and clinical analysis of TRPC6 and AGTR1 genes in patients with pulmonary arterial hypertension. Orphanet J Rare Dis. 2015;10:1. doi: 10.1186/s13023-014-0216-3. https://doi.org/10.1186/s13023-014-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagami T, Guo DF, Kitami Y. Molecular biology of angiotensin II receptors:an overview. J Hypertens Suppl. 1994;12(10):S83–94. [PubMed] [Google Scholar]
- 12.Takayanagi R, Ohnaka K, Sakai Y, Nakao R, Yanase T, Haji M, et al. Molecular cloning, sequence analysis and expression of a cDNA encoding human type-1 angiotensin II receptor. Biochem Biophys Res Commun. 1992;183(2):910–6. doi: 10.1016/0006-291x(92)90570-b. https://doi.org/10.1016/0006-291X(92)90570-B. [DOI] [PubMed] [Google Scholar]
- 13.Dzida G, Sobstyl J, Puzniak A, Golon P, Mosiewicz J, Hanzlik J. Polymorphisms of angiotensin-converting enzyme and angiotensin II receptor type 1 genes in essential hypertension in a Polish population. Med Sci Monit. 2001;7(6):1236–41. [PubMed] [Google Scholar]
- 14.Kainulainen K, Perola M, Terwilliger J, Kaprio J, Koskenvuo M, Syvänen AC, et al. Evidence for involvement of the type 1 angiotensin II receptor locus in essential hypertension. Hypertension. 1999;33(3):844–9. doi: 10.1161/01.hyp.33.3.844. https://doi.org/10.1161/01.HYP.33.3.844. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Lu LL, Yuan DX, Geng N, Xuan SY, Xin YN. AGTR1 rs3772622 gene polymorphism increase the risk of nonalcoholic fatty liver disease patients suffer coronary artery disease in Northern Chinese Han population. Lipids Health Dis. 2016;15:113. doi: 10.1186/s12944-016-0279-3. https://doi.org/10.1186/s12944-016-0279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan LJ, Wang XD. Analysis of correlations between coronary heart disease and haplotypes of the angiotensin II receptor type 1 (AGTR1) gene. Genet Mol Res. 2016;15(1) doi: 10.4238/gmr.15017457. https://doi.org/10.4238/gmr.15017457. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Chen X, Ye H, Hong Q, Xu M, Duan S. Association of four CpG-SNPs in the vascular-related genes with coronary heart disease. Biomed Pharmacother. 2015;70:80–3. doi: 10.1016/j.biopha.2015.01.014. https://doi.org/10.1016/j.biopha.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Foy JP, Pickering CR, Papadimitrakopoulou VA, Jelinek J, Lin SH, William WN Jr, et al. New DNA methylation markers and global DNA hypomethylation are associated with oral cancer development. Cancer Prev Res (Phila) 2015;8(11):1027–35. doi: 10.1158/1940-6207.CAPR-14-0179. https://doi.org/10.1158/1940-6207.aCAPR-14-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo S, Yan F, Xu J, Bao Y, Zhu J, Wang X, et al. Identification and validation of the methylation biomarkers of non-small cell lung cancer (NSCLC) Clin Epigenetics. 2015;7:3. doi: 10.1186/s13148-014-0035-3. https://doi.org/10.1186/s13148-014-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan R, Mao S, Zhong F, Gong M, Yin F, Hao L, et al. Association of AGTR1 promoter methylation levels with essential hypertension risk:a matched case-control study. Cytogenet Genome Res. 2015;147(2-3):95–102. doi: 10.1159/000442366. https://doi.org/10.1159/000442366. [DOI] [PubMed] [Google Scholar]
- 21.Ye H, Zhou A, Hong Q, Chen X, Xin Y, Tang L, et al. Association of seven thrombotic pathway gene CpG-SNPs with coronary heart disease. Biomed Pharmacother. 2015;72:98–102. doi: 10.1016/j.biopha.2015.04.009. https://doi.org/10.1016/j.biopha.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhong J, Chen X, Wu N, Shen C, Cui H, Du W, et al. Catechol-O-methyltransferase promoter hypomethylation is associated with the risk of coronary heart disease. Exp Ther Med. 2016;12(5):3445–9. doi: 10.3892/etm.2016.3757. https://doi.org/10.3892/etm.2016.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen W, Liu Y, Li L, Zhang Y, Zhou W. Negative moods correlate with craving in female methamphetamine users enrolled in compulsory detoxification. Subst Abuse Treat Prev Policy. 2012;7:44. doi: 10.1186/1747-597X-7-44. https://doi.org/10.1186/1747-597X-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong Q, Chen X, Ye H, Wu X, Wang X, Kong L, et al. Chemotherapy-induced hypomethylation of N-myc downstream-regulated gene 4 in the bone marrow of patients with acute myeloid leukemia. Oncol Lett. 2017;13(5):3309–13. doi: 10.3892/ol.2017.5839. https://doi.org/10.3892/ol.2017.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Chen X, Jiang Y, Yang Y, Zhong J, Zhou C, et al. CCL2 promoter hypomethylation is associated with gout risk in Chinese Han male population. Immunol Lett. 2017;190:15–9. doi: 10.1016/j.imlet.2017.06.011. https://doi.org/10.1016/j.imlet.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal:an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. https://doi.org/10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolovou G, Damaskos D, Anagnostopoulou K, Cokkinos DV. Apolipoprotein E gene polymorphism and gender. Ann Clin Lab Sci. 2009;39(2):120–33. https://doi.org/10.1016/j.ijcard.2007.11.024. [PubMed] [Google Scholar]
- 28.Hyvarinen M, Qiao Q, Tuomilehto J, Soderberg S, Eliasson M, Stehouwer CD. The difference between acute coronary heart disease and ischaemic stroke risk with regard to gender and age in Finnish and Swedish populations. Int J Stroke. 2010;5(3):152–6. doi: 10.1111/j.1747-4949.2010.00423.x. https://doi.org/10.1111/j.1747-4949.2010.00423.ax. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Hu X, Xue Q, Zhao Y, Wang Y, Gao L. Influence of sex on outcomes after percutaneous coronary intervention in patients over 75 years of age with coronary heart disease. Clin Interv Aging. 2014;9:1831–7. doi: 10.2147/CIA.S62643. https://doi.org/10.2147/CIA.S62643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. https://doi.org/10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation:27K discovery and replication. Am J Hum Genet. 2011;88(4):450–7. doi: 10.1016/j.ajhg.2011.03.003. https://doi.org/10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philibert RA, Plume JM, Gibbons FX, Brody GH, Beach SR. The impact of recent alcohol use on genome wide DNA methylation signatures. Front Genet. 2012;3:54. doi: 10.3389/fgene.2012.00054. https://doi.org/10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rönn T, Volkov P, Davegårdh C, Dayeh T, Hall E, Olsson AH, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9(6):e1003572. doi: 10.1371/journal.pgen.1003572. https://doi.org/10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovic D, Bregar D, Guzic-Salobir B, Skof E, Span M, Terzić R, et al. Sex difference in the effect of ACE-DD genotype on the risk of premature myocardial infarction. Angiology. 2004;55(2):155–8. doi: 10.1177/000331970405500207. https://doi.org/10.1177/000331970405500207. [DOI] [PubMed] [Google Scholar]
- 35.Sebag IA, Gillis MA, Calderone A, Kasneci A, Meilleur M, Haddad R, et al. Sex hormone control of left ventricular structure/function:mechanistic insights using echocardiography, expression, and DNA methylation analyses in adult mice. Am J Physiol Heart Circ Physiol. 2011;301(4):H1706–15. doi: 10.1152/ajpheart.00088.2011. https://doi.org/10.1152/ajpheart.00088.2011. [DOI] [PubMed] [Google Scholar]
- 36.Murphy TJ, Alexander RW, Griendling KK, Runge MS, Bernstein KE. Isolation of a cDNA encoding the vascular type-1 angiotensin II receptor. Nature. 1991;351(6323):233–6. doi: 10.1038/351233a0. https://doi.org/10.1038/351233a0. [DOI] [PubMed] [Google Scholar]
- 37.Tmoyan NA, Ezhov MV, Afanasieva OI, Klesareva EA, Razova OA, Kukharchuk VV, et al. The association of lipoprotein(a) and apolipoprotein(a) phenotypes with peripheral artery disease. Ter Arkh. 2018;90(9):31–6. doi: 10.26442/terarkh201890931-36. https://doi.org/10.26442/terarkh201890931-36. [DOI] [PubMed] [Google Scholar]
- 38.Lamina C, Kronenberg F Lp(a)-GWAS-Consortium. Estimation of the required lipoprotein(a)-lowering therapeutic effect size for reduction in coronary heart disease outcomes:a Mendelian randomization analysis. JAMA Cardiol. 2019;4(6):575–9. doi: 10.1001/jamacardio.2019.1041. https://doi.org/10.1001/jamacardio.2019.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J, Pan Y, Xiao J, Ma W, Li L, Zhong M, et al. High level of lipoprotein(a) as predictor for recurrent heart failure in patients with chronic heart failure:a cohort study. Arq Bras Cardiol. 2019;113(2):197–204. doi: 10.5935/abc.20190120. https://doi.org/10.5935/abc.20190120. [DOI] [PMC free article] [PubMed] [Google Scholar]


