Iron oxides are important components of our soil, water supplies, and ecosystems, as they sequester nutrients, carbon, and metals. Microorganisms can form iron oxides, but it is unclear whether this is a significant mechanism in the environment. Unlike other major microbial energy metabolisms, there is no marker gene for iron oxidation, hindering our ability to track these microbes. Here, we investigate a promising possible iron oxidation gene, cyc2, in iron-rich hydrothermal vents, where iron-oxidizing microbes dominate. We pieced together diverse Zetaproteobacteria genomes, compared these genomes, and analyzed expression of cyc2 and other hypothetical iron oxidation genes. We show that cyc2 is widespread among iron oxidizers and is highly expressed and potentially regulated, making it a good marker for the capacity for iron oxidation and potentially a marker for activity. These findings will help us understand and potentially quantify the impacts of neutrophilic iron oxidizers in a wide variety of marine and terrestrial environments.
KEYWORDS: Cyc2 Fe oxidation pathway, biogeochemistry, environmental microbiology, hydrothermal vent, iron cycling, iron oxidizers, metagenomics, metatranscriptomics, microbial ecology, Zetaproteobacteria
ABSTRACT
Zetaproteobacteria create extensive iron (Fe) oxide mats at marine hydrothermal vents, making them an ideal model for microbial Fe oxidation at circumneutral pH. Comparison of neutrophilic Fe oxidizer isolate genomes has revealed a hypothetical Fe oxidation pathway, featuring a homolog of the Fe oxidase Cyc2 from Acidithiobacillus ferrooxidans. However, Cyc2 function is not well verified in neutrophilic Fe oxidizers, particularly in Fe-oxidizing environments. Toward this, we analyzed genomes and metatranscriptomes of Zetaproteobacteria, using 53 new high-quality metagenome-assembled genomes reconstructed from Fe mats at Mid-Atlantic Ridge, Mariana Backarc, and Loihi Seamount (Hawaii) hydrothermal vents. Phylogenetic analysis demonstrated conservation of Cyc2 sequences among most neutrophilic Fe oxidizers, suggesting a common function. We confirmed the widespread distribution of cyc2 and other model Fe oxidation pathway genes across all represented Zetaproteobacteria lineages. High expression of these genes was observed in diverse Zetaproteobacteria under multiple environmental conditions and in incubations. The putative Fe oxidase gene cyc2 was highly expressed in situ, often as the top expressed gene. The cyc2 gene showed increased expression in Fe(II)-amended incubations, with corresponding increases in carbon fixation and central metabolism gene expression. These results substantiate the Cyc2-based Fe oxidation pathway in neutrophiles and demonstrate its significance in marine Fe-mineralizing environments.
IMPORTANCE Iron oxides are important components of our soil, water supplies, and ecosystems, as they sequester nutrients, carbon, and metals. Microorganisms can form iron oxides, but it is unclear whether this is a significant mechanism in the environment. Unlike other major microbial energy metabolisms, there is no marker gene for iron oxidation, hindering our ability to track these microbes. Here, we investigate a promising possible iron oxidation gene, cyc2, in iron-rich hydrothermal vents, where iron-oxidizing microbes dominate. We pieced together diverse Zetaproteobacteria genomes, compared these genomes, and analyzed expression of cyc2 and other hypothetical iron oxidation genes. We show that cyc2 is widespread among iron oxidizers and is highly expressed and potentially regulated, making it a good marker for the capacity for iron oxidation and potentially a marker for activity. These findings will help us understand and potentially quantify the impacts of neutrophilic iron oxidizers in a wide variety of marine and terrestrial environments.
INTRODUCTION
Neutrophilic Fe-oxidizing microbes are common in marine and terrestrial environments (1), precipitating reactive Fe oxyhydroxides that sequester organic carbon, phosphate, arsenic, and many other metals (2–4). However, it has been difficult to study the effects of neutrophilic Fe oxidation in natural systems due to myriad challenges that have slowed the discovery of genetic markers of neutrophilic Fe oxidation. These Fe oxidizers are difficult to culture, so only recently have we obtained enough isolate genomes to deduce hypothetical neutrophilic Fe-oxidizing pathways. Comparative genomics has led to multiple proposed pathways, each involving an outer membrane cytochrome (5–7). However, only one pathway is present in all well-established neutrophilic Fe-oxidizing isolates (Zetaproteobacteria and Gallionellaceae), centering on a fused cytochrome-porin, Cyc2 (7–10). Yet, beyond comparative genomics, we lack evidence of the Cyc2 pathway function in neutrophilic Fe oxidizers, particularly the uncultured Fe oxidizers that dominate natural Fe systems.
The Cyc2 pathway in the neutrophilic Fe oxidizers is modeled after the Fe oxidation pathways found in acidophilic Fe oxidizers Acidithiobacillus ferrooxidans and Leptospirillum sp., where Cyc2 Fe oxidase function has been verified (11, 12). Weak homologs to cyc2 from these organisms were first found in the genomes of the Gallionellaceae Sideroxydans lithotrophicus ES-1 and Gallionella capsiferriformans ES-2 (13). The genome of Zetaproteobacteria type strain Mariprofundus ferrooxydans PV-1, on the other hand, lacked homologs to known Fe oxidation genes until a proteome study discovered that cyc2 was in fact carried by PV-1 but missing from the draft genome (14). Subsequently, cyc2 homologs were found within the few Zetaproteobacteria lineages with genomic representation (9, 15). Despite the identification of cyc2-like genes, low amino acid sequence homology (20% sequence identity between PV-1 and A. ferrooxidans Cyc2) suggests that their function is speculative and needs to be validated. Without a means of testing this function biochemically or genetically, we focus on more comprehensive comparative genomics and expression in Fe-oxidizing environments.
To this end, we turned to Zetaproteobacteria in natural Fe microbial mats. The Zetaproteobacteria discovered to date are all considered to be Fe oxidizers, since every isolate grows by Fe oxidation and uncultured Zetaproteobacteria are typically found in Fe-oxidizing environments (10, 16–21). The Zetaproteobacteria are often the dominant organisms in marine hydrothermal Fe mats (22–25), where they play a key role in mat formation (26). This abundance and ubiquity in Fe-oxidizing mats make Zetaproteobacteria ideal for study through metagenomic and metatranscriptomic approaches. Furthermore, their taxonomic diversity allows for a robust comparative genomics study. We sampled paired metagenomes and metatranscriptomes from three hydrothermal venting regions: Loihi Seamount, the Mid-Atlantic Ridge (MAR) (Rainbow, TAG, and Snake Pit vents), and the Mariana Backarc (Urashima vent field). Recovery of high-quality metagenome-assembled genomes (MAGs) allowed us to improve the limited genomic representation of the Zetaproteobacteria (see reference 10 for a summary of genomic representation prior to this study). Using the MAGs for reference mapping, we explored in situ environmental expression of the Zetaproteobacteria within undisturbed natural Fe mats. In addition, we examined expression responses to Fe(II) using shipboard Fe(II) amendment experiments. With these results, we assess and update the model neutrophilic Fe oxidation pathway expressed in natural environments.
This article was submitted to an online preprint archive (27).
RESULTS
Microbial Fe mat sampling and geochemistry.
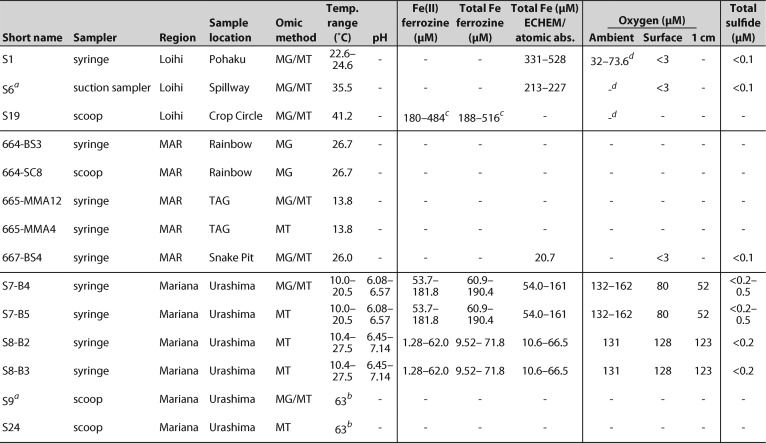
Over three expeditions, we sampled a wide diversity of Fe microbial mats (see Table S1 in the supplemental material). Sampled mats varied in their physical setting, with meter-scale Loihi mats found in direct and indirect flow from vent fissures, mat mounds on the scale of tens of centimeters at the MAR at the diffuse-venting periphery of black smoker fields, and the mats at the Mariana Backarc Urashima vent fields covering a 7-m-tall Fe chimney (Fig. S1 and Text S1). Temperatures ranged from 10 to 63°C, while geochemical conditions also varied widely, notably concentrations of Fe(II) from 1.3 to 190 μM and O2 from <3 to 123 μM within the mats (Table 1). Mariana mats had shallow O2 gradients, while at Loihi, O2 was undetectable (<3 μM) at 1 cm below the mat surface. These Fe(II) and O2 conditions favor biotic Fe oxidation (10). At Mariana, total dissolved Fe was depleted by 49% to 74% in our low-temperature mats relative to the conservative mixing of the local high-temperature zero-Mg endmember (Fig. S2), which suggests that a substantial amount of Fe is being oxidized and precipitated within these mats.
TABLE 1.
Summary of key geochemistry for each samplee
16S and MG samples taken from pre-T0 time point.
Temperature ∼0.5 m within mat.
Data from vent fluids collected by major sampler.
Ambient O2 concentrations at Loihi Seamount approximately 60 μM.
Symbols and abbreviations: −, no data; MAR, Mid-Atlantic Ridge; MG, metagenome; MT, metatranscriptome.
Supplemental methods, text, and references. Download Text S1, PDF file, 0.8 MB (803.9KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Photographs of sampling locations for samples chosen for metagenome and metatranscriptome sequencing from Loihi Seamount (A), Mid-Atlantic Ridge (B), and Mariana Backarc (C). Images on the left show context for the specific sampling location, which is marked by an arrowhead and depicted on the right. Download FIG S1, PDF file, 2.4 MB (2.4MB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Urashima Vent Field (Mariana) geochemistry, plotted to highlight mixing of a single endmember with seawater. Mn and Si are conservative, or unreactive, during mixing (A). Total dissolved Fe is depleted in low-temperature fluids compared to conservative mixing (B; enlarged in panel C). This is particularly pronounced within the Golden Horn Chimney Fe mats. Baltan is a high-temperature vent in the Urashima Vent Field. Saipanda is another low-temperature vent that was not sampled for this study. Download FIG S2, PDF file, 0.7 MB (696.6KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample names, origin, and type for this study. Download Table S1, XLSX file, 0.05 MB (51.7KB, xlsx) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Zetaproteobacteria abundance and diversity.
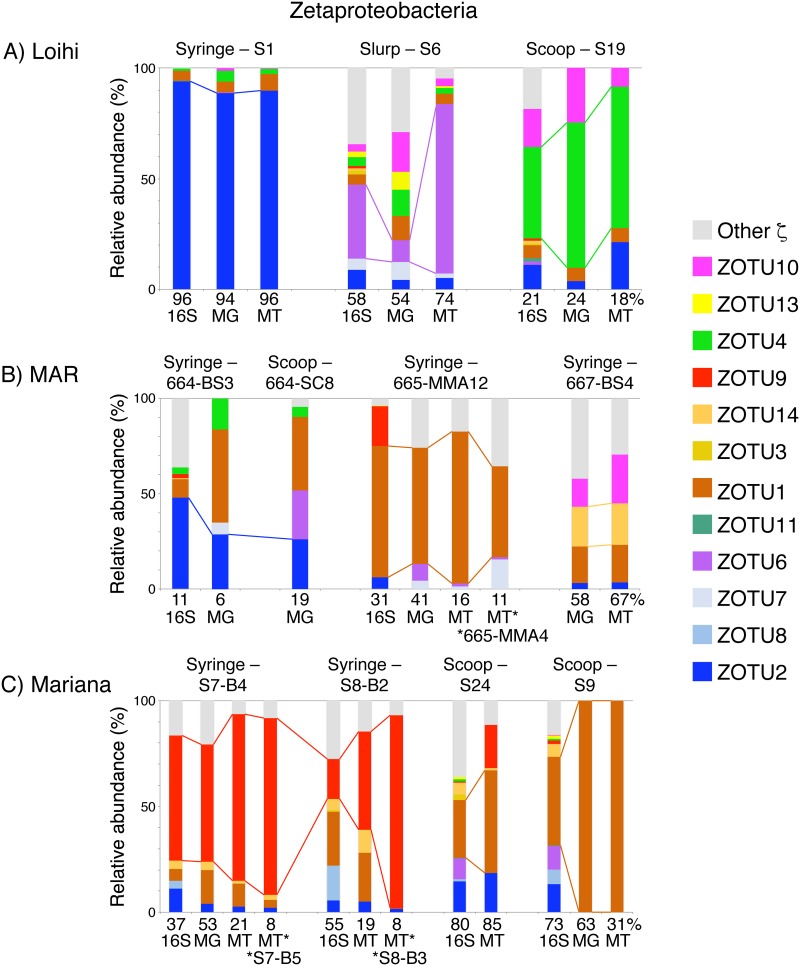
We initially assessed Zetaproteobacteria abundance and diversity using a 16S rRNA gene survey (Fig. S3). Fe mat communities at all sites hosted abundant Zetaproteobacteria populations, from 16.4 to 95.9% of the total bacterial community at Loihi, 10.7 to 31.3% at the Mid-Atlantic Ridge, and 37.1 to 79.9% at Mariana. Many samples were dominated by the Zetaproteobacteria, notably sample S1 (96% Zetaproteobacteria), a centimeter-scale sample of actively growing Fe mat surface. In addition to Zetaproteobacteria, the mats hosted variable flanking microbial communities that differed between the three sites (Fig. S3 and S4) but were similar to previous studies (9, 22, 23, 28, 29). Overall, the relatively simple, Zetaproteobacteria-rich composition of these marine Fe mats makes them good systems for studying neutrophilic Fe oxidation mechanisms.
PacBio 16S rRNA gene survey of Bacteria (A) and Zetaproteobacteria (B) microbial communities from Fe mats at Loihi Seamount, the Mid-Atlantic Ridge, and Mariana Backarc. The abundance of Zetaproteobacteria is highlighted (A). Blue asterisks denote samples chosen for metagenomics. Red asterisks denote samples chosen for metatranscriptomics. Numbers at the bottom of the bar charts denote the number of total 16S rRNA gene sequences sampled. Sample short name and Fe mat type are also given. Download FIG S3, PDF file, 0.7 MB (735.8KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of 16S rRNA gene, metagenome, and metatranscriptome relative abundance for the microbial communities at Loihi Seamount (A), the Mid-Atlantic Ridge (B), and Mariana Backarc (C). 16S rRNA gene plots represent the bacterial population only. The relative abundance of the Zetaproteobacteria is tracked for samples from the same Fe mat location and/or for MT samples mapped to the same metagenomes. Asterisks show MT samples that were mapped to a reference MG from a different sample. Download FIG S4, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
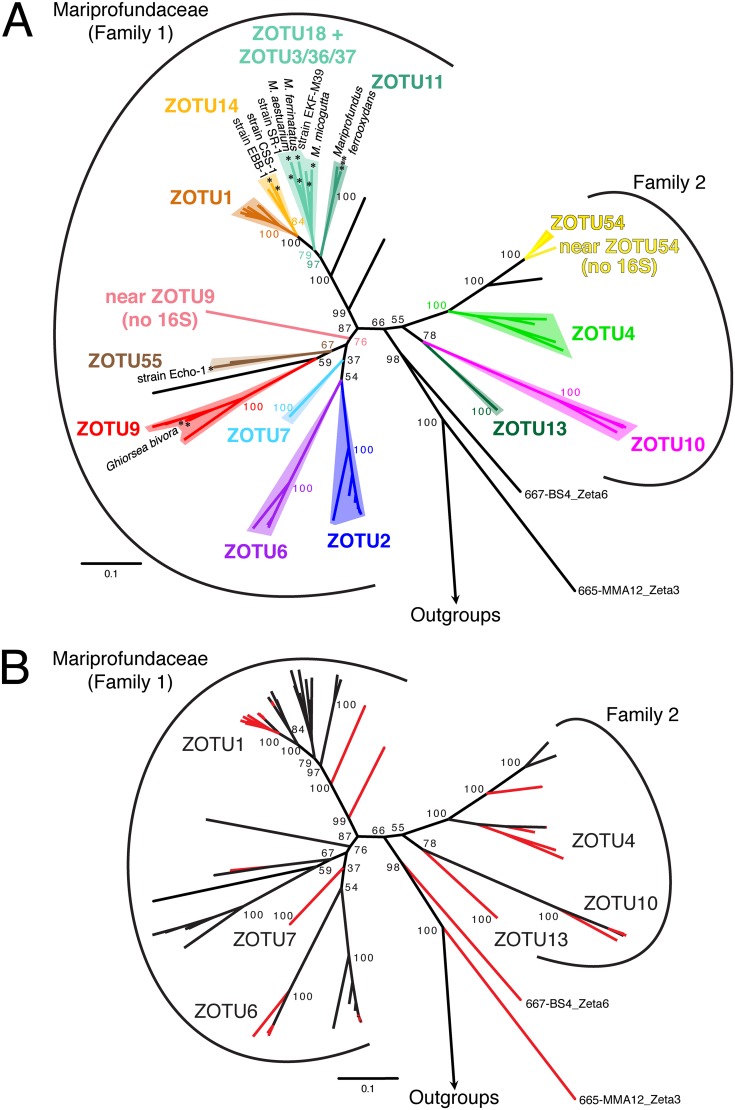
We used the 16S rRNA gene community profiling results to choose metagenomic (MG) and metatranscriptomic (MT) samples, aiming to recover high-abundance and diverse Zetaproteobacteria to produce high-quality genomes with sufficient MT read depth (Fig. S3). We recovered 126 total high-quality MAGs from our samples (>70% complete, <10% redundant) (see Tables S4 and S5 at https://doi.org/10.6084/m9.figshare.c.4646336) along with 79 improved MAGs by reanalyzing a Loihi metagenomic data set from the work of Fullerton et al. (9) (Text S1; see also Table S5 at https://doi.org/10.6084/m9.figshare.c.4646336). Of these, 53 MAGs belonged to the Zetaproteobacteria (selected genomes in Table S2), which were compared to a collection of published high-quality genomes (Text S1). MAGs from this study improve the representation of nine different Zetaproteobacteria operational taxonomic units (ZOTUs) spanning the Zetaproteobacteria phylogenetic tree by providing 2 to 13 additional high-quality MAGs for each of these ZOTUs (Fig. 1; Table S2). Many of these ZOTUs previously had poor genome representation (labeled ZOTUs in Fig. 1B). These diverse ZOTUs were abundant and active within our Fe mats (abundance by 16S rRNA gene and MG; activity by MT) (Fig. 2). MAG relative abundance generally matched relative activity, with the exception of MAG S6_Zeta1 (ZOTU6), which had higher activity than expected, likely in response to the shipboard incubation conditions. By substantially improving Zetaproteobacteria genome representation and pairing this with metatranscriptomes, we are poised to investigate genetic commonalities and diversity across the Zetaproteobacteria, particularly of the Fe oxidation mechanism.
FIG 1.
Zetaproteobacteria concatenated ribosomal protein reference maximum likelihood tree (100 bootstraps) showing the most commonly sampled ZOTUs (A) and highlighting genomes produced by this study (B). (A) All isolates of the Zetaproteobacteria are marked with an asterisk. Deep-branching genomes 665-MMA12_Zeta3 and 667-BS4_Zeta6 were classified as Zetaproteobacteria, though they are more deeply rooted than any prior lineage. (B) Genomes produced by this study are highlighted in red. Six ZOTUs that lacked sufficient depth for comparative genomics prior to this study are labeled.
FIG 2.
Comparison of 16S rRNA gene, metagenome (MG), and metatranscriptome (MT) relative abundance for the Zetaproteobacteria from various mats at Loihi Seamount (A), the Mid-Atlantic Ridge (B), and Mariana Backarc (C). The relative abundance of the most abundant Zetaproteobacteria operational taxonomic unit (ZOTU) by 16S rRNA gene is tracked across similar Fe mat samples from the same region. Asterisks denote MTs from different samples that were mapped to the indicated MG. Percentages are shown at the bottom of each bar graph to indicate the relative proportion of Zetaproteobacteria in each sample (see Fig. S4).
Zetaproteobacteria genomes used in comparative genomics, concatenated ribosomal protein phylogenetic tree, and gene expression estimates. Download Table S2, XLSX file, 0.02 MB (21KB, xlsx) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogeny of the putative Fe oxidase Cyc2.
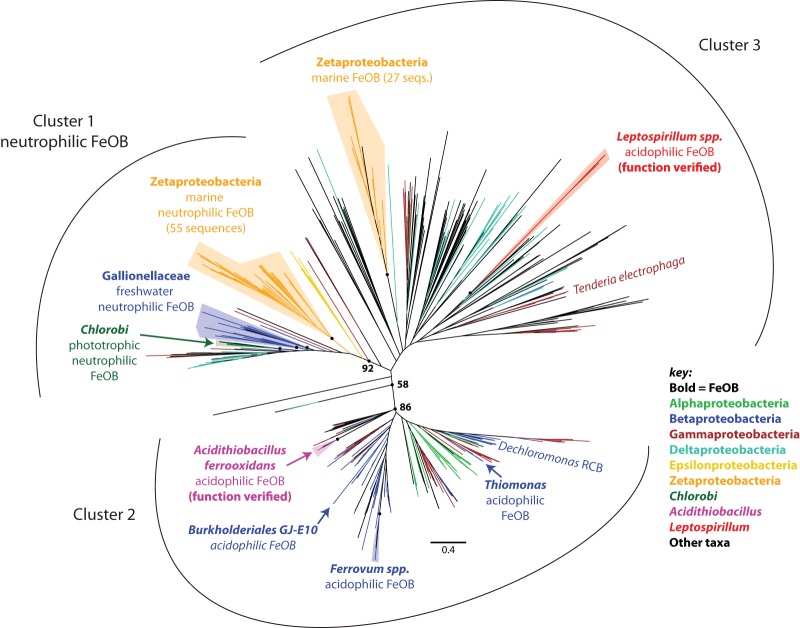
The key component of the proposed neutrophilic Fe oxidation pathway is Cyc2, which has been shown to oxidize Fe(II) in acidophiles. Our preliminary analyses showed that some Zetaproteobacteria genomes have multiple cyc2 copies that were not closely related. To investigate these, we developed a comprehensive Cyc2 phylogeny. This phylogeny includes sequences from terrestrial to marine and circumneutral to acidic environments, as well as both known Fe oxidizers and organisms not known to oxidize Fe (Fig. 3; see Fig. S8 for detailed tree with sequence names at https://doi.org/10.6084/m9.figshare.c.4646336). Cyc2 sequences form three clusters, but the Fe-oxidizing function has been verified only for the Cluster 2 Acidithiobacillus ferrooxidans Cyc2 (11) and the Cluster 3 Cyc2 homolog Cyt572 of Leptospirillum sp. (12). However, most of the neutrophilic Fe oxidizers fall within Cluster 1, a well-supported group (93% bootstrap) that is largely comprised of the Zetaproteobacteria, Gallionellaceae, and Chlorobium ferrooxidans. This strongly suggests that Cluster 1 Cyc2s share a function.
FIG 3.
Cyc2 maximum likelihood phylogenetic tree (100 bootstraps), showing three distinct clusters. All groups of Fe-oxidizing bacteria are labeled, in addition to the electrode-oxidizing Tenderia electrophaga. Zetaproteobacteria and Gallionellaceae cluster with other neutrophilic Fe oxidizers in Cluster 1 (92% cluster support). Fe oxidation has been demonstrated for Cyc2 from Cluster 2 Acidithiobacillus ferrooxidans and Cluster 3 Leptospirillum sp. Unlabeled circles at nodes correspond to the following bootstrap values in parentheses: Zetaproteobacteria Cluster 1 (97%), Gallionellaceae (87%/63%), Chlorobi (99%), Acidithiobacillus ferrooxidans (100%), Ferrovum spp. (100%), Zetaproteobacteria Cluster 3 (99%), and Leptospirillum spp. (100%).
Putative Fe oxidation pathway distribution revealed by comparative genomics of the Zetaproteobacteria.
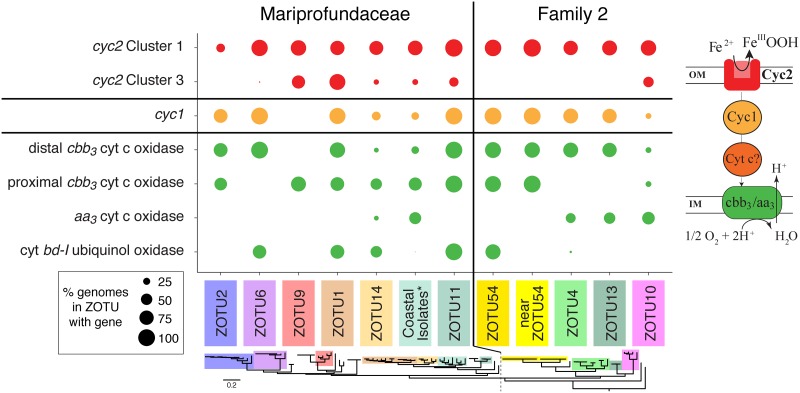
Our Cyc2 phylogeny shows that Zetaproteobacteria possess Cyc2 from both Clusters 1 and 3. All ZOTUs possess a Cluster 1 cyc2 gene; with our new genomes, this includes four additional ZOTUs that are now known to possess cyc2 (ZOTUs 1, 7, 13, and 14; Fig. 4). In contrast, fewer ZOTUs have the Cluster 3 cyc2 gene, and 65% of genomes with Cluster 3 cyc2 (n = 15) also have Cluster 1 cyc2. This suggests that both Cyc2 types have a use in the Zetaproteobacteria, though it is unknown how Cluster 1 and 3 Cyc2s may differ in function. ZOTU2 is unusual in that only 3 of the 10 genomes appear to have cyc2, though this may be due to assembly issues specific to ZOTU2 (Text S1). In any case, the presence of cyc2 in all Zetaproteobacteria OTUs suggests its centrality to these neutrophilic Fe oxidizers.
FIG 4.
Dot plot showing the distribution of genes from the putative Fe oxidation pathway between major ZOTUs. Each dot represents the percentage of genomes in the ZOTU which possess the gene of interest. Genes are colored by their relative position within the core putative Fe oxidation pathway, shown at right. ZOTUs are ordered by the reference ribosomal protein tree (bottom), separated into the two families of the Zetaproteobacteria. The coastal isolate group (see asterisk) includes isolates Mariprofundus aestuarium CP-5, Mariprofundus ferrinatatus CP-8, Mariprofundus sp. strain SR1, Mariprofundus sp. strain EKF-M39, and Mariprofundus micogutta ET2.
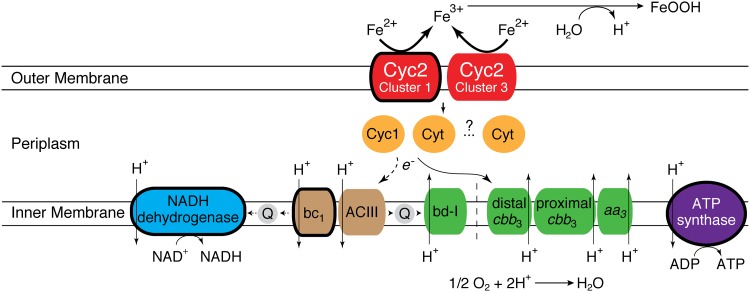
In addition to cyc2, other proposed genes for the Fe oxidation pathway were also widely distributed in Zetaproteobacteria genomes. Homologs of cyc1 were present in the genomes of all ZOTUs except ZOTU9 (Fig. 4); cyc1 encodes a diheme c-type cytochrome thought to be a periplasmic electron carrier in the A. ferrooxidans Fe oxidation pathway (11). ZOTU9, which includes the Fe- and H2-oxidizing Ghiorsea spp. (30), must use another periplasmic electron carrier. Indeed, many other putative periplasmic cytochromes can be found in Zetaproteobacteria genomes (see below). Cyc1 or another electron carrier likely passes electrons to a terminal oxidase or to complex I via complex III (reverse electron transport). Genes for the bc1 complex were found in all ZOTUs, whereas we found alternative complex III (ACIII) genes in only a few Zetaproteobacteria, primarily in ZOTU11 and Family 2 (ZOTUs 4, 10, and 13). We found three types of aerobic terminal oxidases: (i) cbb3-type cytochrome c oxidase, (ii) aa3-type cytochrome c oxidase, and (iii) cytochrome bd-I ubiquinol oxidase. Further, two distinct forms of the cbb3-type cytochrome c oxidase were found, clustering in the proximal and distal cbb3 subtrees defined by Ducluzeau et al. (31) (Fig. S5). All ZOTUs possess genes for one or more of these terminal oxidases, suggesting that all Zetaproteobacteria are aerobic Fe oxidizers (Fig. 4). Taken together, these findings allow us to update the neutrophilic Fe oxidation pathway model (Fig. 5).
FIG 5.
Proposed Fe oxidation pathway model showing variation in the genetic capability of all Zetaproteobacteria. Components that are conserved in all Zetaproteobacteria are outlined with a thick line. Components of the pathway from the same module have the same color.
Maximum likelihood phylogenetic tree of CcoN, showing that the Zetaproteobacteria possess two forms of cbb3-type cytochrome c oxidase: proximal and distal. Only the distal version is found within the conserved cassette discovered by Field et al. (E. K. Field, A. Sczyrba, A. E. Lyman, C. C. Harris, et al., ISME J 9:857–870, 2015, https://doi.org/10.1038/ismej.2014.183). Download FIG S5, PDF file, 0.7 MB (693.4KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In situ expression of the putative Fe oxidation pathway.
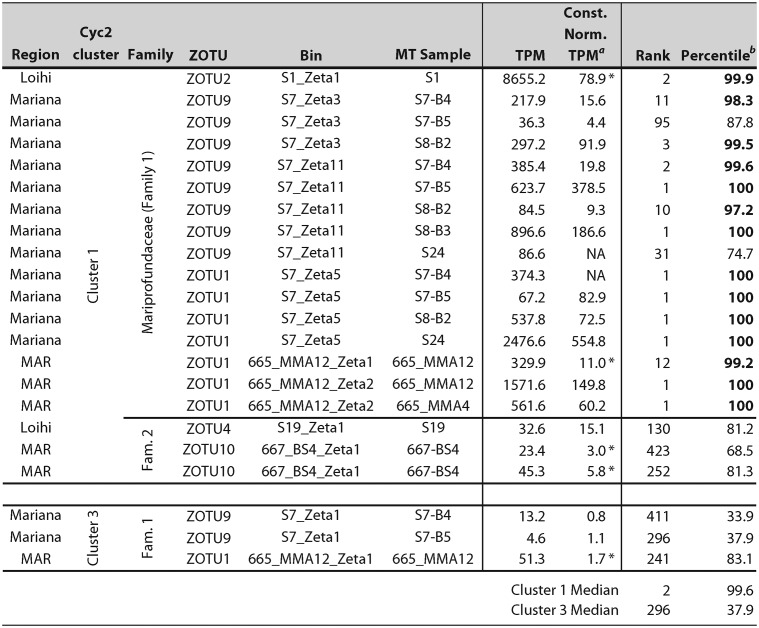
Our next step was to determine whether the putative Fe oxidation pathway genes are expressed in the environment; high expression would lend support for the model. In situ expression from six unique ZOTUs in 10 different samples (total of 21 observations) shows that cyc2 genes from Cluster 1 are highly expressed in all Zetaproteobacteria and samples, ranging from 3.0× to 555× baseline constitutive gene expression (Table 2). Cluster 1 cyc2 was frequently the highest-expressed gene in the genome, particularly in the Mariprofundaceae (Family 1). Interestingly, cyc2 expression levels differed between Zetaproteobacteria Families 1 and 2, though expression was still high in all Zetaproteobacteria. The cyc1 and cbb3-type terminal oxidase genes are expressed up to 17.3× and 56.6× constitutive gene expression, respectively. On average, this places the cyc1 and terminal oxidase genes in the 73.4 and 75.4 percentile range in Zetaproteobacteria metatranscriptomic expression, respectively (Fig. S6). This gene expression is consistent with protein expression levels of M. ferrooxydans PV-1, where the corresponding proteins were expressed at or above the 87th percentile (14). In combination, our data suggest that genes in the core model Fe oxidation pathway are highly expressed in situ by diverse Zetaproteobacteria under various environmental conditions.
TABLE 2.
Expression and relative importance of cyc2 genes from in situ samples
Const. Norm. TPM, constitutive normalized TPM. Asterisks denote constitutive normalized TPM from bins with sufficient read depth (see Table S7 at https://doi.org/10.6084/m9.figshare.c.4646336). NA, constitutive normalized expression cannot be calculated.
High cyc2 expression above the 90th percentile indicated in boldface.
Heat map showing the percentile expression for key genes in the Fe oxidation pathway, including genes thought to be involved in electron transport from Fe(II) to O2 and from Fe(II) to the quinone pool for reverse electron transport (RET). Download FIG S6, PDF file, 0.7 MB (715.2KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variation in gene expression may help us resolve which modules are most commonly used during Fe oxidation in the environment. For example, expression of complex III module genes further supports the importance of bc1 over ACIII for reverse electron transport. Average expression of bc1 was 6.6× constitutive expression, while ACIII expression was much lower at 0.6×. In ZOTUs with both complexes, bc1 genes were expressed 1.5× to 18.6× higher than the ACIII complex. The limited distribution of ACIII in only a few Zetaproteobacteria lineages, despite our deep sampling with near-complete genomes, combined with its low expression suggests that ACIII is not required for Fe oxidation under the sampled conditions.
Comparison of relative in situ expression may also help identify genes that may be involved as intermediate electron carriers, particularly in ZOTU9, which lacks cyc1. We identified at least 14 different putative periplasmic c-type cytochrome genes (PCs) with high expression (>90th percentile) in one or more Zetaproteobacteria genomes (see Table S6A at https://doi.org/10.6084/m9.figshare.c.4646336). Some of these genes (cyc1, PC12, PC61, PC16, and PC38) were more highly expressed in some genomes than cyc2 in the Mariprofundaceae, and all were found in at least one genome where they were more highly expressed than cyc1. Interestingly, these putative periplasmic cytochromes were found and expressed at different levels in different Zetaproteobacteria lineages, with some unique to a single ZOTU (e.g., PC73 in ZOTU2) and most found in several ZOTUs. The cytochromes c previously found on a conserved cassette identified in Zetaproteobacteria isolates and single amplified genomes (SAGs) (including cyc1, PC2, and PC3) (15) are most highly expressed in genomes from ZOTUs 1 and 2. These results help us narrow the list of potential electron carriers in the Zetaproteobacteria.
Expression of Fe oxidation pathway genes in Fe-amended mat incubations.
To link gene expression more specifically with Fe oxidation, we added Fe(II) to mat samples and analyzed metatranscriptomic responses over time. We performed shipboard incubations at Loihi Seamount and Mariana, using fresh Fe mats, live and killed, while monitoring Fe oxidation. Microbes within the mat were actively oxidizing Fe(II) faster than abiotic processes, with the pseudo-first-order Fe oxidation rate constants 3.7× (Loihi) and 5.3× (Mariana) higher in live samples than azide-killed ones (Fig. S7). These results show that we stimulated biotic Fe oxidation, which should lead to increased expression of Fe oxidation genes.
Plot of Fe(II) addition experiment results from Loihi (A) and Mariana (B) Fe mats, showing a higher living (orange; total) than killed (black; abiotic-only) Fe oxidation rate. Fe(II) was added to dormant Fe mat samples at 0 min. Pseudo-first-order rate constants were calculated from the log-linear best fit from each experimental condition. Download FIG S7, PDF file, 0.7 MB (686.3KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
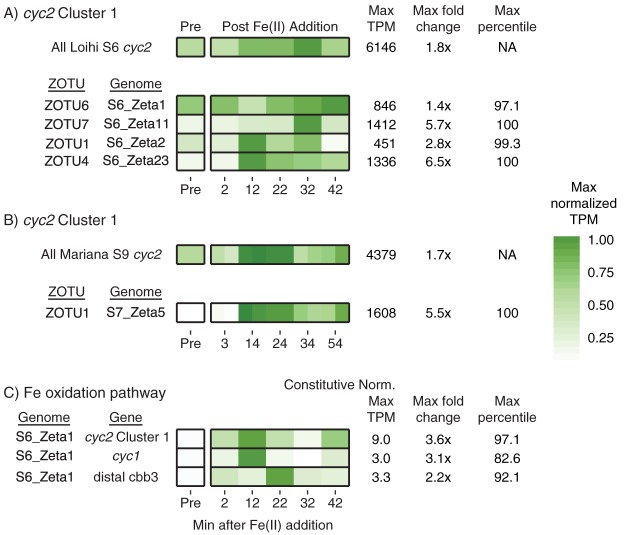
As with the in situ samples, cyc2 was highly expressed in the Zetaproteobacteria during the time series experiments, reaching a maximum of 97.1st to 100th percentile in the four most active Zetaproteobacteria lineages. After Fe(II) was added, there was an increase in total cyc2 expression (sum of all cyc2 genes), as well as cyc2 expression by each individual MAG (Fig. 6A and B). Expression increased at different rates for each ZOTU, with some peaking earlier and others peaking at the end of the experiment. Expression of cyc2 increased less for the most abundant Zetaproteobacteria (e.g., S6_Zeta1), which already had high expression of cyc2 prior to Fe(II) amendment. Less-abundant Zetaproteobacteria (S6_Zeta11/S6_Zeta23) were also expressing cyc2 prior to Fe(II) amendment but had a much larger change in expression (5.7× to 6.5×), reached their maximum quickly, and maintained a higher expression over the course of the experiment. Overall, while the degree and timing of response differed, all Zetaproteobacteria increased their cyc2 expression in response to Fe(II) amendment.
FIG 6.
Normalized TPM expression changes for cyc2 and other Fe oxidation pathway genes in the Fe(II) amendment experiments, showing increases after Fe(II) addition. (A and B) TPM expression changes are shown for all Cluster 1 cyc2 and for cyc2 from specific Zetaproteobacteria MAGs at Loihi (A) and Mariana (B) (duplicates shown). NA, not applicable. (C) Constitutive normalized expression changes shown for the Fe oxidation pathway in MAG S6_Zeta1. S9 MT data mapped to the S7 MG for expression estimates of MAGs in panel B. TPM values were maximally normalized to emphasize peak expression.
Other genes in the Fe oxidation pathway also generally followed this trend, with cyc1 expression in 4 of 5 genomes and cbb3-type terminal oxidase genes in 2 of 4 genomes also increasing after Fe(II) addition. However, low read recruitment depth led to substantial noise. To correct for this noise, we normalized expression using six constitutively expressed genes (Text S1; also see Table S7 at https://doi.org/10.6084/m9.figshare.c.4646336) and focused on expression patterns in S6_Zeta1, which had high read depth (Fig. 6C). Constitutive normalized expression of S6_Zeta1 shows a similar pattern of cyc2 expression change over the time series compared to expression patterns prior to constitutive normalization, with a maximum increase of 3.6× after Fe(II) amendment. The genes encoding Cyc1 and the cbb3-type terminal oxidase also increased after Fe(II) addition, reaching a maximum fold change of 1.5× to 3×. This trend was also observed for 7 of 8 putative periplasmic cytochromes more highly expressed than cyc1, increasing 2.1× to 4.5× over the course of the experiment (see Table S6B at https://doi.org/10.6084/m9.figshare.c.4646336). These results suggest that the Fe(II) amendment increased the expression of many genes thought to be in the Fe oxidation pathway.
If the Zetaproteobacteria represented by the S6_Zeta1 genome is an autotrophic Fe oxidizer, Fe(II) amendment should stimulate genes for carbon fixation, central metabolism, and growth. Like cyc2 expression, genes for central metabolic pathways, including the tricarboxylic acid (TCA) cycle, increased in expression 2.0× to 3.4× over 1 h in S6_Zeta1 from ZOTU6 (see Fig. S9 at https://doi.org/10.6084/m9.figshare.c.4646336). Similarly, expression of genes for glycogen synthesis increased 1.8× in the first 2 min after Fe(II) addition. Carbon fixation genes increased 1.9× in the first 12 min. Some of the highest fold changes after Fe(II) amendment were seen in genes related to proper protein folding (molecular chaperones groEL, groES, and dnaK) and membrane protein quality control (htpX-type protease) (32). For example, groEL increased 119× after Fe(II) addition. Though these gene responses may correspond with shock to the cell after Fe(II) amendment, these genes were also highly expressed under in situ conditions in S1_Zeta1, which suggests that they may be necessary for promoting active Fe oxidation in the environment. Together, these data suggest that Fe(II) amendment stimulated genes for both Fe oxidation and growth.
DISCUSSION
The main objective of this study was to assess the Cyc2-based Fe oxidation pathway in neutrophilic Fe-oxidizing bacteria (FeOB), using environmental metagenomics and metatranscriptomics of marine Fe mats. This work contributes 53 new nearly complete Zetaproteobacteria genomes paired with expression profiles, both in situ and from incubations, along with a comprehensive Cyc2 phylogeny. Using these, we have characterized the distribution and usage of the model Fe oxidation pathway across the full range of known Zetaproteobacteria in Fe mats at three geographically distinct venting regions. The emerging pattern shows that the pathway as a whole is highly expressed, with increased expression in all Fe pathway genes following Fe(II) amendment. The cyc2 gene is among the highest expressed and is the only gene in the pathway shared by all Zetaproteobacteria, suggesting that it plays a key role in Fe oxidation.
Assessing the Fe oxidation pathway model through Zetaproteobacteria comparative genomics.
In this study, we assessed the current model for Fe oxidation by comparing genomes representing the full diversity of the Zetaproteobacteria (Fig. 1B). Since the Zetaproteobacteria are thought to be an entire class of Fe-oxidizing bacteria, all genomes should have an Fe-oxidizing pathway, which may be conserved. Our results show that overall, the basic model of neutrophilic Fe oxidation in the Zetaproteobacteria holds (Fig. 4). All Zetaproteobacteria lineages, including novel ones presented in this study, possess genes encoding the putative Fe oxidase Cyc2, an intermediate electron carrier, and a terminal oxidase. However, our survey shows that each of these components can have multiple versions, suggesting that the pathway contains interchangeable modules, as depicted in our updated model (Fig. 5). Across all Zetaproteobacteria, there are two types of Cyc2, multiple potential periplasmic cytochrome electron carriers, and four terminal oxidases. The variations are likely linked with specific adaptations related to niche, with some genomes possessing multiple versions of certain components, perhaps to span multiple niches (33). Within each ZOTU, individual genomes possessed pathway gene variations consistent with the ZOTU as a whole, even though some genomes were missing genes that a majority of others in the ZOTU possessed. These false negatives could result from incomplete MAGs, which is why we focused on ZOTUs. Accounting for modularity and genome variability within ZOTUs, comparative genomics has confirmed that this Fe oxidation pathway is common to all Zetaproteobacteria lineages.
Support for cyc2 as an Fe oxidation gene in neutrophilic Fe oxidizers.
Cyc2 homologs in neutrophilic FeOB are commonly referred to as “putative Fe oxidases” based on homology to the functionally characterized A. ferrooxidans Cyc2, though sequence similarity is low. Indeed, our phylogenetic analysis shows that Cyc2 sequences are highly diverse, and most of the neutrophilic FeOB Cyc2 homologs fall into Cluster 1 (Fig. 3). This cluster forms a distinct group from the clusters containing biochemically characterized Fe oxidases: Cyc2 from A. ferrooxidans (Cluster 2) and Cyt572 from Leptospirillum sp. (Cluster 3). Although none of the Cluster 1 Cyc2s have been biochemically characterized, the high bootstrap support suggests a common function, and the prevalence of sequences from Fe-oxidizing isolates strongly suggests involvement in Fe oxidation.
Another clue to function lies in the expression of cyc2 in Fe mat environments, where Fe oxidation by the Zetaproteobacteria is required for carbon fixation and growth. Genes central to fitness are often highly expressed (34, 35). We measured cyc2 expression in five different active Fe mats from three different hydrothermal vent fields and confirmed that cyc2 is frequently the highest-expressed gene. This is true in diverse Zetaproteobacteria lineages (Table 2), suggesting that the pathway is important to Zetaproteobacteria fitness in many different environments. The level of cyc2 expression in the Zetaproteobacteria is consistent with the high expression in Fe-oxidizing isolates of Acidithiobacillus sp. (often above microarray detection limits) and Ferrovum sp. (8× above average expression) (36–38). In the environment, the neutrophilic Gallionellaceae have been shown to express cyc2 highly, up to the 100th percentile in an Fe-rich aquifer (39). Together with the expression of cyc1 and terminal oxidase genes, this shows that the putative Fe oxidation pathway is consistently expressed under Fe-oxidizing conditions. The especially high expression of cyc2 across various Fe-oxidizing taxa and Fe-oxidizing environments supports its role in Fe oxidation in both acidophiles and neutrophiles.
To link cyc2 to Fe oxidation, we followed its expression when Fe(II) was added to Fe mat samples. In separate experiments at Loihi and Mariana, Fe(II) amendment resulted in both active biotic Fe oxidation (see Fig. S7 in the supplemental material) and increased cyc2 gene expression. This increase in expression was found not only for the whole sample but also in every Zetaproteobacteria genome detected within these samples (Fig. 6). Although there was variation in the timing and magnitude of the response, which may be lineage specific, the fact that expression increased in all Zetaproteobacteria suggests that cyc2 expression is stimulated by the presence of Fe(II). The Fe(II) amendment also resulted in increases in carbon fixation and central metabolism gene expression, suggesting a link between cyc2 expression, neutrophilic Fe oxidation, and growth (see Fig. S9 at https://doi.org/10.6084/m9.figshare.c.4646336).
Can the cyc2 gene be used as a marker for Fe oxidation?
Unlike many other energy metabolisms, neutrophilic Fe oxidation is challenging to track in the environment due to the lack of an isotopic signature and difficulties distinguishing biotic from abiotic Fe oxides. Until recently, there have not been any candidates for a widely applicable Fe oxidation genetic marker; instead, it seemed that there were many different potential Fe oxidases, with various levels of functional verification (e.g., references 6, 7, and 40). Our work adds to the mounting evidence that Cyc2 is an Fe oxidase. The cyc2 gene is widely distributed across many Fe-oxidizing lineages, with homologs in acidophiles and neutrophiles. Specifically for neutrophiles, cyc2 is common across the well-studied neutrophilic chemolithotrophs Gallionellaceae and Zetaproteobacteria. As we have sequenced more of these neutrophilic FeOB genomes, this association has held true (9, 13, 15, 21, 41). However, our Cyc2 phylogeny has identified a substantial number of cultured and uncultured organisms that have not yet been shown or tested to be capable of Fe oxidation, work that could bolster confidence. In all, the cyc2 gene is a promising genomic marker of the capacity for Fe oxidation across many different Fe-oxidizing lineages, including neutrophiles.
Not only is cyc2 common to all well-established neutrophilic Fe oxidizers, it is also highly expressed in environments where neutrophilic Fe oxidizers predominate (this study and reference 39). This opens the possibility of cyc2 expression levels as an indicator of microbial Fe oxidation activity. Indeed, when we stimulated Zetaproteobacteria Fe oxidation in incubations, expression of cyc2 increased along with an increase in carbon fixation and central metabolism genes. This is consistent with Fe-oxidation-fueled chemolithoautotrophic growth, and so relative cyc2 expression levels can correspond to increases in Fe oxidation activity. However, our results suggest that cyc2 expression levels may not be easily related to Fe oxidation activity in the environment. All Zetaproteobacteria in our samples were expressing cyc2 prior to Fe(II) amendment, when there was no detectable dissolved Fe(II). This could represent baseline expression by obligate Fe oxidizers, which always need to be prepared for Fe oxidation. In this case, relative changes in cyc2 expression would remain more informative for activity. Alternatively, cyc2 expression before Fe(II) amendment could result from cryptic cycling of Fe between Fe oxidizers and reducers (42). Such cryptic cycling would make developing a genetic marker for activity even more important for tracking Fe oxidation activity. Because of these potential complications, further transcriptomics experiments should focus on isolates or microcosms without Fe reducers. In combination with our results, such experiments will help us understand how to use cyc2 expression levels to interpret Fe oxidation activity in the environment.
Conclusions.
Using paired metagenomes and metatranscriptomes from the Zetaproteobacteria, we have been able to demonstrate that the Cyc2-based neutrophilic Fe oxidation pathway is widespread and highly expressed in the environment, validating the environmental importance of the pathway. We have shown that the Cluster 1 cyc2 gene, conserved in the Zetaproteobacteria and other neutrophilic Fe oxidizers, is highly expressed in multiple Fe mat environments and is stimulated by Fe(II) addition, suggesting it may be regulated. This makes cyc2 an excellent marker of Fe oxidation capability and may allow us to detect and monitor the activity of Fe oxidizers in the environment. However, to correlate expression with activity, further efforts should focus on testing the regulation of cyc2 in diverse organisms and simple communities. The phylogeny of Cyc2 shows at least three distinct clusters, with some neutrophilic Fe oxidizers possessing multiple copies (e.g., Clusters 1 and 3 in the Zetaproteobacteria). This may be akin to an example in Pseudomonas aeruginosa, which has multiple cbb3-type cytochrome c oxidases that are optimized for high and low O2 concentrations and for resistance to respiratory inhibitors cyanide and nitrite and thus allow growth under different conditions (43). If Cyc2 variants are similarly optimized, they may enable Fe oxidation under various conditions, a hypothesis that could be tested by independently monitoring cyc2 from different clusters in diverse habitats and growth conditions. Without a marker of activity, the roles of neutrophilic Fe oxidizers have been virtually invisible outside model Fe-oxidizing environments, like Fe microbial mats. By applying our findings to other environments, we can start to reveal how Fe-oxidizing microbes drive key biogeochemical cycles in the varied marine and freshwater habitats where they thrive.
MATERIALS AND METHODS
Biological sample collection.
Samples were collected from various vent fields on three separate cruises to the Mid-Atlantic Ridge (2012), Loihi Seamount (2013), and the Mariana Backarc (2014) (see Table S1 in the supplemental material). To preserve in situ expression, 18 samples were collected using devices half-filled with 2× RNAlater (Invitrogen, Carlsbad, CA, USA). Samples collected using a syringe sampler device (44) provided ∼10 to 30 ml of mat material representing a discrete microbial population, as opposed to 2 liters (scoop) or >5 liters (suction sample) of bulk sample. After settling for a few hours at 4°C, the overlying supernatant was removed and samples were stored at −80°C.
Geochemical measurements and sampling.
At the Mid-Atlantic Ridge and Loihi Seamount, geochemistry was measured in situ using cyclic voltammetry (ECHEM), as described in the work of MacDonald et al. (45) (MAR) and Chan et al. (26) (Loihi). The detection limits were 3 μM O2, 7 to 10 μM Fe2+, and 0.1 μM sulfide (45, 46).
At Mariana, geochemistry samples were collected using the hydrothermal fluid and particle sampler (HFPS) (47) or the microbial mat sampler (44). The HFPS pulls fluid through a titanium inlet nozzle at 1 to 4 liters/min. The fluid flows through a continuously flushed titanium and Teflon manifold and is diverted into sample containers or to a SeaBird (Bellevue, WA) SBE 63 oxygen sensor and an AMT (Rostock, Germany) deep-sea glass pH electrode. In extremely low-outflow vent environments, seawater will be entrained in the HFPS and dilute the in situ fluid. We collected temperature, pH, and O2 concentrations for ambient water, at the surface of the chimney, and with the nozzle inserted into the microbial mat. HFPS chemistry results represent the fluid composition at the measured temperature, including any entrained seawater that occurs during sampling. The microbial mat sampler has a much lower intake rate (<0.2 liter min−1) and is better able to capture chemical microenvironments. The microbial mat sampler was equipped with 0.22-μm inline filtering and a check valve for chemical analysis. Fe(II) and total Fe concentrations were assayed using the ferrozine method with 40 mM sulfamic acid to stabilize Fe(II) (48, 49); the detection limit was estimated at 0.12 μM Fe(II). Samples recovered with the HFPS were processed as described previously (47) and analyzed shipboard for pH by glass electrode and on shore for total dissolved iron by atomic absorption at National Oceanic and Atmospheric Administration/Pacific Marine Environmental Laboratory (NOAA/PMEL) and by inductively coupled plasma mass spectrometry (ICPMS) at the University of Washington Department of Civil Engineering.
Fe(II) amendment experiments.
Shipboard Fe(II) amendment experiments were conducted on bulk mat samples from Loihi (J2-677-SSyellow) and Mariana (J2-801-SC8). Samples were transported to the ship after 2 h (Loihi) and 11 h (Mariana) of remotely operated vehicle (ROV) operations and allowed to settle at 4°C for 1 h prior to removing the majority of the supernatant and starting the experiment. One sample was taken immediately prior to Fe(II) amendment [pre-Fe(II) addition]. Water bath temperature and initial Fe(II) amendment concentration were set to mimic environmental conditions. Fe oxidation pseudo-first-order rate constants (k1) were calculated using a log-linear fitted trend line.
At Loihi Seamount, Fe mat floc was added to two 250-ml vessels; one remained alive while the other was killed using 1 mM azide, which has been shown to interact with Fe(II), though not at this concentration and time interval (50). Both vessels were shaken by hand several times a minute in a 35°C water bath. To initiate the experiment, 100 μM FeCl2 was added. After this, starting after 2 min and subsequently at 10-min intervals, samples from each vessel were removed for Fe(II) and total Fe measurements by the ferrozine method (48). Concurrently, 30 ml from the living vessel was mixed 1:1 with 2× RNAlater (Invitrogen). This mixture was held at 4°C for a few hours prior to freezing at −80°C.
At Mariana, Fe mat material was sparged with a 2% O2 gas mix (pH 5.9/6.2 before/after sparge). Each time point (n = 5) and treatment (duplicate living and 3 mM azide killed) had its own 125-ml reaction vessel with 30 ml mat material. In addition to a pre-Fe(II) addition sample, one sample was taken at the end which did not experience any Fe(II) addition. Both of these nonamended samples had low Fe(II) concentrations (below detection [BD] and 0.3 μM, respectively). Each reaction vessel was amended with 333 μM FeCl2 and suspended in a 28°C water bath with frequent mixing by hand. Starting after 4 to 6 min and subsequently at 10-min intervals, one vessel was sacrificed at each time point, for sampling for Fe(II), total Fe, and pH and mixing of 25 ml of material 1:1 with 2× RNAlater.
DNA and RNA extraction.
DNA samples were extracted using the FastDNA Spin kit for soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions, except that 250 μl of a 0.5 mM sodium citrate solution (pH 5.8) was added prior to lysis. RNA samples were extracted using the NucleoSpin RNA kit (Macherey-Nagel, Bethlehem, PA, USA), with modifications detailed in Text S1. Prior to library preparation, an internal in vitro-transcribed RNA standard, pTXB1, was added (Text S1; see also Table S3 at https://doi.org/10.6084/m9.figshare.c.4646336). RNA extractions were used for metatranscriptome library preparation after nondegraded total RNA (visible 16S and 23S rRNA peaks) was confirmed by a fragment analyzer (2.2 to 9.3 RNA quality number [RQN]; median 5.7 RQN) (Advanced Analytical, Ankeny, IA, USA).
16S rRNA, metagenome, and metatranscriptome sequencing.
Microbial community composition was first estimated using a PacBio-based 16S rRNA gene survey, with SILVAngs used for taxonomic classification (see Text S1) (75). Zetaproteobacteria operational taxonomic units (ZOTUs) were classified from these 16S rRNA gene sequences using ZetaHunter (51). Samples were chosen for metagenomic (MG) and metatranscriptomic (MT) sequencing based on the microbial community composition and Zetaproteobacteria diversity. MG and MT libraries were prepared and sequenced at the University of Delaware Sequencing and Genotyping Center. Sequencing details are provided in Text S1.
Metagenome assembly, binning, and annotation.
Raw sequence reads were trimmed to remove adaptors, poor-quality regions, and short sequences (Trimmomatic) (52), and paired reads were merged if overlapping (Flash) (53). Metagenome libraries were assembled from quality-controlled (QC’ed) reads using metaSPAdes v3.10, with read error correction disabled to improve recovery of real community genomic variation (54). Only contigs mapping ≥1× read coverage over 90% of their length were utilized in downstream analysis (∼92% remained).
Metagenome-assembled genomes (MAGs) were binned using four binning programs: MaxBin (55), MetaBAT (superspecific and very sensitive settings) (56), CONCOCT (57), and BinSanity (58). The resulting bins were combined and dereplicated using DAS Tool (59). Manual taxonomic and outlier (guanine-cytosine (GC) content/coverage) curation of bins was performed in ggkbase (https://ggkbase.berkeley.edu/), with additional curation performed using Anvi’o v3 (60). Finalized curated bins were tested for completeness and redundancy using CheckM (61) and classified using PhyloSift (62), and gene calling and SEED annotation were performed using RAST (63). RAST gene calls were used for Clusters of Orthologous Groups (COG) annotation within Anvi’o (60, 64), and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was performed through BlastKOALA (65). Genes of interest (e.g., cyc2, cyc1, and terminal oxidases) were further manually curated based on evidence using NCBI BLASTp (66) against Zetaproteobacteria protein references. Gene annotation was assessed with maximum likelihood phylogenetic trees built from alignments using RAxML (67). The Cyc2 phylogenetic tree was constructed from an alignment of 634 unique Cyc2 protein sequences identified from NCBI and IMG databases using BLASTp (66, 68, 69). Additional information on the Cyc2 sequences and tree construction is provided in Text S1.
RNA read recruitment and expression estimates.
Raw total RNA reads were quality controlled (see above) using Trimmomatic (average 99% of reads passed) (52). rRNA reads were removed using SortMeRNA (v2.1b) (70). The resulting non-rRNA reads, primarily mRNA, were used for subsequent recruitment for expression estimates. MT reads were recruited to the MG from the same sample, with the following exceptions: 665-MMA4 was recruited to 665-MMA12; S7_B5, S8_B2, S8_B3, S9, and S24 were recruited to S7_B4 MG. Reads were mapped using Bowtie 2, with default parameters (71).
To determine gene read recruitment, we used BEDTools to extract the read count from each gene coordinate region (72). We used three normalization methods for estimating gene expression: (i) transcripts per million (TPM), normalizing for sequencing effort and gene and read lengths (73); (ii) TPM values further normalized to the average expression of six constitutively expressed genes (adk, gyrA, recA, rpoB, rpoC, and secA) (74) to correct for changes in organism relative abundance (constitutive normalized expression); and (iii) TPM values normalized to the maximum expression for the time series for visual representation.
Data accessibility.
High-quality full-length reads (20-pass minimum) from the PacBio 16S rRNA gene survey were submitted to GenBank (MK048478 to MK048944). Raw metagenome and metatranscriptome reads, as well as 5-pass-filtered PacBio 16S rRNA gene reads, were submitted to the NCBI SRA under BioProject accession PRJNA555820. Metagenome assemblies from this study and reassembled metagenome assemblies from the work of Fullerton et al. (9) were submitted to the JGI IMG database (sequence project IDs Gp0295814 to Gp0295821 and Gp0295823 [this study]; analysis project IDs Ga0256915 and Ga0257019 to Ga0257023 [Fullerton et al. {9}]). Zetaproteobacteria MAGs were also submitted to the JGI IMG database (see sequence project IDs listed above). Specific accession numbers per sample are shown in Table S4 at https://doi.org/10.6084/m9.figshare.c.4646336.
ACKNOWLEDGMENTS
S.M.M. and C.S.C. drafted the manuscript. All authors contributed to experimental design and editing. D.A.B., B.T.G., and S.M.M. performed geochemical analysis. S.M.M., J.B.S., B.T.G., and C.S.C. implemented shipboard Fe(II) amendment experiments. S.M.M., S.W.P., and C.S.C. conducted bioinformatics analysis.
We acknowledge George W. Luther, III, for generous ship time and geochemical data from the MAR. We thank the captains and crew of the R/Vs Knorr, Thompson, and Revelle and ROV Jason II. We also acknowledge Anna Leavitt, Arne Sturm, Angelos Hannides, and Karyn Rogers for their assistance with the shipboard experiments; Kevin Roe for chemical analyses of fluid samples; Vadesse Noundou for assistance with Zetaproteobacteria central metabolism; Ryan Moore and Karol Miaskiewicz for bioinformatics assistance; and Jennifer Biddle, David Emerson, Thomas Hanson, and Jessica Keffer for their comments on the manuscript. We also thank the University of Delaware Sequencing and Genotyping Center for their help with sample preparation and sequencing, in particular Bruce Kingham, Summer Thompson, and Olga Shevchenko.
This work was funded by NSF OCE-1155290 and ONR N00014-17-1-2640 (to C.S.C.); C-DEBI (contribution no. 501) (to J.B.S.); NSF OCE-1061827 and OCE-1031947 (to B.T.G.); NOAA/PMEL (contribution no. 5003), NOAA Ocean Exploration and Research, and JISAO (contribution no. 2019-1017) (to D.A.B.); NSF EAR-1833525 (to C.S.C. and S.W.P.); and NIGMS P20 GM103446 (to S.W.P.), in addition to two Delaware Space Grant Fellowships (NASA grant NNX10AN63H) and the University of Delaware Dissertation Fellowship to S.M.M. Computational infrastructure support by the University of Delaware CBCB Core Facility was funded by Delaware INBRE (NIH NIGMS P20 GM103446) and the Delaware Biotechnology Institute.
The authors declare no conflicts of interest.
REFERENCES
- 1.Emerson D, Fleming EJ, McBeth JM. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 2.Chan CS, Fakra SC, Emerson D, Fleming EJ, Edwards KJ. 2011. Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J 5:717–727. doi: 10.1038/ismej.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laufer K, Nordhoff M, Halama M, Martinez RE, Obst M, Nowak M, Stryhanyuk H, Richnow HH, Kappler A. 2017. Microaerophilic Fe(II)-oxidizing Zetaproteobacteria isolated from low-Fe marine coastal sediments: physiology and characterization of their twisted stalks. Appl Environ Microbiol 83:e03118-16. doi: 10.1128/AEM.03118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kendall B, Anbar AD, Kappler A, Konhauser KO. 2012. The global iron cycle, p 65–92. In Knoll AH, Canfield DE, Konhauser KO (ed), Fundamentals of geobiology, 1st ed. Blackwell Publishing Ltd, Oxford, United Kingdom. [Google Scholar]
- 5.Liu J, Wang Z, Belchik SM, Edwards MJ, Liu C, Kennedy DW, Merkley ED, Lipton MS, Butt JN, Richardson DJ, Zachara JM, Fredrickson JK, Rosso KM, Shi L. 2012. Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front Microbiol 3:37. doi: 10.3389/fmicb.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White GF, Edwards MJ, Gomez-Perez L, Richardson DJ, Butt JN, Clarke TA. 2016. Mechanisms of bacterial extracellular electron exchange. Adv Microb Physiol 68:87–138. doi: 10.1016/bs.ampbs.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 7.He S, Barco RA, Emerson D, Roden EE. 2017. Comparative genomic analysis of neutrophilic iron(II) oxidizer genomes for candidate genes in extracellular electron transfer. Front Microbiol 8:1584. doi: 10.3389/fmicb.2017.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato S, Ohkuma M, Powell DH, Krepski ST, Oshima K, Hattori M, Shapiro N, Woyke T, Chan CS. 2015. Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol 6:1265. doi: 10.3389/fmicb.2015.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fullerton H, Hager KW, McAllister SM, Moyer CL. 2017. Hidden diversity revealed by genome-resolved metagenomics of iron-oxidizing microbial mats from Lo’ihi Seamount, Hawai’i. ISME J 11:1900–1914. doi: 10.1038/ismej.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAllister SM, Moore RM, Gartman A, Luther GW, Emerson D, Chan CS. 2019. The Fe(II)-oxidizing Zetaproteobacteria: historical, ecological, and genomic perspectives. FEMS Microbiol Ecol 95:fiz015. doi: 10.1093/femsec/fiz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castelle C, Guiral M, Malarte G, Ledgham F, Leroy G, Brugna M, Giudici-Orticoni M-T. 2008. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J Biol Chem 283:25803–25811. doi: 10.1074/jbc.M802496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeans C, Singer SW, Chan CS, VerBerkmoes NC, Shah M, Hettich RL, Banfield JF, Thelen MP. 2008. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J 2:542–550. doi: 10.1038/ismej.2008.17. [DOI] [PubMed] [Google Scholar]
- 13.Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, Nolan M, Woyke T. 2013. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4:254. doi: 10.3389/fmicb.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barco RA, Emerson D, Sylvan JB, Orcutt BN, Jacobson Meyers ME, Ramírez GA, Zhong JD, Edwards KJ. 2015. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81:5927–5937. doi: 10.1128/AEM.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field EK, Sczyrba A, Lyman AE, Harris CC, Woyke T, Stepanauskas R, Emerson D. 2015. Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J 9:857–870. doi: 10.1038/ismej.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C, Moyer CL. 2007. A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2:e667. doi: 10.1371/journal.pone.0000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makita H, Kikuchi S, Mitsunobu S, Takaki Y, Yamanaka T, Toki T, Noguchi T, Nakamura K, Abe M, Hirai M, Yamamoto M, Uematsu K, Miyazaki J, Nunoura T, Takahashi Y, Takai K. 2016. Comparative analysis of microbial communities in iron-dominated flocculent mats in deep-sea hydrothermal environments. Appl Environ Microbiol 82:5741–5755. doi: 10.1128/AEM.01151-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barco RA, Hoffman CL, Ramírez GA, Toner BM, Edwards KJ, Sylvan JB. 2017. In-situ incubation of iron-sulfur mineral reveals a diverse chemolithoautotrophic community and a new biogeochemical role for Thiomicrospira. Environ Microbiol 19:1322–1337. doi: 10.1111/1462-2920.13666. [DOI] [PubMed] [Google Scholar]
- 19.Probst AJ, Castelle CJ, Singh A, Brown CT, Anantharaman K, Sharon I, Hug LA, Burstein D, Emerson JB, Thomas BC, Banfield JF. 2017. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ Microbiol 19:459–474. doi: 10.1111/1462-2920.13362. [DOI] [PubMed] [Google Scholar]
- 20.Meyer JL, Jaekel U, Tully BJ, Glazer BT, Wheat CG, Lin H-T, Hsieh C-C, Cowen JP, Hulme SM, Girguis PR, Huber JA. 2016. A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci Rep 6:22541. doi: 10.1038/srep22541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrison CE, Price KA, Field EK. 2019. Environmental evidence for and genomic insight into the preference of iron-oxidizing bacteria for more-corrosion-resistant stainless steel at higher salinities. Appl Environ Microbiol 85:e00483-19. doi: 10.1128/AEM.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAllister SM, Davis RE, McBeth JM, Tebo BM, Emerson D, Moyer CL. 2011. Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl Environ Microbiol 77:5445–5457. doi: 10.1128/AEM.00533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hager KW, Fullerton H, Butterfield DA, Moyer CL. 2017. Community structure of lithotrophically-driven hydrothermal microbial mats from the Mariana Arc and Back-Arc. Front Microbiol 8:1578. doi: 10.3389/fmicb.2017.01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander Roost J, Thorseth IH, Dahle H. 2017. Microbial analysis of Zetaproteobacteria and co-colonizers of iron mats in the Troll Wall Vent Field, Arctic Mid-Ocean Ridge. PLoS One 12:e0185008. doi: 10.1371/journal.pone.0185008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Roost J, Daae FL, Steen IH, Thorseth IH, Dahle H. 2018. Distribution patterns of iron-oxidizing Zeta- and Beta-Proteobacteria from different environmental settings at the Jan Mayen Vent Fields. Front Microbiol 9:3008. doi: 10.3389/fmicb.2018.03008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. 2016. The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol 7:796. doi: 10.3389/fmicb.2016.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAllister SM, Polson SW, Butterfield DA, Glazer BT, Sylvan JB, Chan CS. 2019. Validating the Cyc2 neutrophilic Fe oxidation pathway using meta-omics of Zetaproteobacteria iron mats at marine hydrothermal vents. bioRxiv 722066. doi: 10.1101/722066. [DOI] [PMC free article] [PubMed]
- 28.Scott JJ, Glazer BT, Emerson D. 2017. Bringing microbial diversity into focus: high-resolution analysis of iron mats from the Lō’ihi Seamount. Environ Microbiol 19:301–316. doi: 10.1111/1462-2920.13607. [DOI] [PubMed] [Google Scholar]
- 29.Scott JJ, Breier JA, Luther GW III, Emerson D. 2015. Microbial iron mats at the Mid-Atlantic Ridge and evidence that Zetaproteobacteria may be restricted to iron-oxidizing marine systems. PLoS One 10:e0119284. doi: 10.1371/journal.pone.0119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori JF, Scott JJ, Hager KW, Moyer CL, Küsel K, Emerson D. 2017. Physiological and ecological implications of an iron- or hydrogen-oxidizing member of the Zetaproteobacteria, Ghiorsea bivora, gen. nov., sp. nov. ISME J 11:2624–2636. doi: 10.1038/ismej.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducluzeau AL, Ouchane S, Nitschke W. 2008. The cbb3 oxidases are an ancient innovation of the domain Bacteria. Mol Biol Evol 25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 32.Sakoh M, Ito K, Akiyama Y. 2005. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J Biol Chem 280:33305–33310. doi: 10.1074/jbc.M506180200. [DOI] [PubMed] [Google Scholar]
- 33.McAllister SM. 2019. The Zetaproteobacteria: ecology and metabolic functions of a model neutrophilic Fe-oxidizing clade. Dissertation. University of Delaware, Newark, DE. [Google Scholar]
- 34.Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF. 2008. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A 105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urich T, Lanzén A, Stokke R, Pedersen RB, Bayer C, Thorseth IH, Schleper C, Steen IH, Øvreas L. 2014. Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ Microbiol 16:2699–2710. doi: 10.1111/1462-2920.12283. [DOI] [PubMed] [Google Scholar]
- 36.Yarzábal A, Appia-Ayme C, Ratouchniak J, Bonnefoy V. 2004. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology 150:2113–2123. doi: 10.1099/mic.0.26966-0. [DOI] [PubMed] [Google Scholar]
- 37.Quatrini R, Appia-Ayme C, Denis Y, Ratouchniak J, Veloso F, Valdes J, Lefimil C, Silver S, Roberto F, Orellana O, Denizot F, Jedlicki E, Holmes D, Bonnefoy V. 2006. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 83:263–272. doi: 10.1016/j.hydromet.2006.03.030. [DOI] [Google Scholar]
- 38.Ullrich SR, Poehlein A, Levicán G, Mühling M, Schlömann M. 2018. Iron targeted transcriptome study draws attention to novel redox protein candidates involved in ferrous iron oxidation in “Ferrovum” sp. JA12. Res Microbiol 169:618–627. doi: 10.1016/j.resmic.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Jewell TNM, Karaoz U, Brodie EL, Williams KH, Beller HR. 2016. Metatranscriptomic evidence of pervasive and diverse chemolithoautotrophy relevant to C, S, N, and Fe cycling in a shallow alluvial aquifer. ISME J 10:2106–2117. doi: 10.1038/ismej.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilbert M, Bonnefoy V. 2013. Insight into the evolution of the iron oxidation pathways. Biochim Biophys Acta 1827:161–175. doi: 10.1016/j.bbabio.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Crowe SA, Hahn AS, Morgan-Lang C, Thompson KJ, Simister RL, Llirós M, Hirst M, Hallam SJ. 2017. Draft genome sequence of the pelagic photoferrotroph Chlorobium phaeoferrooxidans. Genome Announc 5:e01584-16. doi: 10.1128/genomeA.01584-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emerson D. 2009. Potential for iron-reduction and iron-cycling in iron oxyhydroxide-rich microbial mats at Loihi Seamount. Geomicrobiol J 26:639–647. doi: 10.1080/01490450903269985. [DOI] [Google Scholar]
- 43.Hirai T, Osamura T, Ishii M, Arai H. 2016. Expression of multiple cbb3 cytochrome c oxidase isoforms by combinations of multiple isosubunits in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:12815–12819. doi: 10.1073/pnas.1613308113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breier JA, Gomez-Ibanez D, Reddington E, Huber JA, Emerson D. 2012. A precision multi-sampler for deep-sea hydrothermal microbial mat studies. Deep Sea Res Part I Oceanogr Res Pap 70:83–90. doi: 10.1016/j.dsr.2012.10.006. [DOI] [Google Scholar]
- 45.MacDonald DJ, Findlay AJ, McAllister SM, Barnett JM, Hredzak-Showalter P, Krepski ST, Cone SG, Scott J, Bennett SK, Chan CS, Emerson D, Luther GW III. 2014. Using in situ voltammetry as a tool to identify and characterize habitats of iron-oxidizing bacteria: from fresh water wetlands to hydrothermal vent sites. Environ Sci Process Impacts 16:2117–2126. doi: 10.1039/c4em00073k. [DOI] [PubMed] [Google Scholar]
- 46.Glazer BT, Rouxel OJ. 2009. Redox speciation and distribution within diverse iron-dominated microbial habitats at Loihi Seamount. Geomicrobiol J 26:606–622. doi: 10.1080/01490450903263392. [DOI] [Google Scholar]
- 47.Butterfield DA, Roe KK, Lilley MD, Huber JA, Baross JA, Embley RW, Massoth GJ. 2013. Mixing, reaction and microbial activity in the sub-seafloor revealed by temporal and spatial variation in diffuse flow vents at axial volcano In Wilcock WS, Delong EF, Kelley DS, Baross JA, Cary SC (ed), The subseafloor biosphere at mid-ocean ridges. American Geophysical Union, Washington, DC. [Google Scholar]
- 48.Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal Chem 42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- 49.Klueglein N, Kappler A. 2013. Abiotic oxidation of Fe(II) by reactive nitrogen species in cultures of the nitrate-reducing Fe(II) oxidizer Acidovorax sp. BoFeN1—questioning the existence of enzymatic Fe(II) oxidation. Geobiology 11:180–190. doi: 10.1111/gbi.12019. [DOI] [PubMed] [Google Scholar]
- 50.Hendrix K, Bleyen N, Mennecart T, Bruggeman C, Valcke E. 2019. Sodium azide used as microbial inhibitor caused unwanted by-products in anaerobic geochemical studies. Appl Geochem 107:120–130. doi: 10.1016/j.apgeochem.2019.05.014. [DOI] [Google Scholar]
- 51.McAllister SM, Moore RM, Chan CS. 2018. ZetaHunter, a reproducible taxonomic classification tool for tracking the ecology of the Zetaproteobacteria and other poorly resolved taxa. Microbiol Resour Announc 7:e00932-18. doi: 10.1128/MRA.00932-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang DD, Froula J, Egan R, Wang Z. 2015. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alneberg J, Bjarnason BS, De Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 58.Graham ED, Heidelberg JF, Tully BJ. 2017. BinSanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 5:e3035. doi: 10.7717/peerj.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. 2018. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. 2014. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 66.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 68.Chen IA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, Ratner A, Huang J, Andersen E, Huntemann M, Varghese N, Hadjithomas M, Tennessen K, Nielsen T, Ivanova NN, Kyrpides NC. 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 71.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 74.Rocha DJP, Santos CS, Pacheco LG. 2015. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek 108:685–693. doi: 10.1007/s10482-015-0524-1. [DOI] [PubMed] [Google Scholar]
- 75.Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W. 2017. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol 261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, text, and references. Download Text S1, PDF file, 0.8 MB (803.9KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Photographs of sampling locations for samples chosen for metagenome and metatranscriptome sequencing from Loihi Seamount (A), Mid-Atlantic Ridge (B), and Mariana Backarc (C). Images on the left show context for the specific sampling location, which is marked by an arrowhead and depicted on the right. Download FIG S1, PDF file, 2.4 MB (2.4MB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Urashima Vent Field (Mariana) geochemistry, plotted to highlight mixing of a single endmember with seawater. Mn and Si are conservative, or unreactive, during mixing (A). Total dissolved Fe is depleted in low-temperature fluids compared to conservative mixing (B; enlarged in panel C). This is particularly pronounced within the Golden Horn Chimney Fe mats. Baltan is a high-temperature vent in the Urashima Vent Field. Saipanda is another low-temperature vent that was not sampled for this study. Download FIG S2, PDF file, 0.7 MB (696.6KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample names, origin, and type for this study. Download Table S1, XLSX file, 0.05 MB (51.7KB, xlsx) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PacBio 16S rRNA gene survey of Bacteria (A) and Zetaproteobacteria (B) microbial communities from Fe mats at Loihi Seamount, the Mid-Atlantic Ridge, and Mariana Backarc. The abundance of Zetaproteobacteria is highlighted (A). Blue asterisks denote samples chosen for metagenomics. Red asterisks denote samples chosen for metatranscriptomics. Numbers at the bottom of the bar charts denote the number of total 16S rRNA gene sequences sampled. Sample short name and Fe mat type are also given. Download FIG S3, PDF file, 0.7 MB (735.8KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of 16S rRNA gene, metagenome, and metatranscriptome relative abundance for the microbial communities at Loihi Seamount (A), the Mid-Atlantic Ridge (B), and Mariana Backarc (C). 16S rRNA gene plots represent the bacterial population only. The relative abundance of the Zetaproteobacteria is tracked for samples from the same Fe mat location and/or for MT samples mapped to the same metagenomes. Asterisks show MT samples that were mapped to a reference MG from a different sample. Download FIG S4, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Zetaproteobacteria genomes used in comparative genomics, concatenated ribosomal protein phylogenetic tree, and gene expression estimates. Download Table S2, XLSX file, 0.02 MB (21KB, xlsx) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maximum likelihood phylogenetic tree of CcoN, showing that the Zetaproteobacteria possess two forms of cbb3-type cytochrome c oxidase: proximal and distal. Only the distal version is found within the conserved cassette discovered by Field et al. (E. K. Field, A. Sczyrba, A. E. Lyman, C. C. Harris, et al., ISME J 9:857–870, 2015, https://doi.org/10.1038/ismej.2014.183). Download FIG S5, PDF file, 0.7 MB (693.4KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map showing the percentile expression for key genes in the Fe oxidation pathway, including genes thought to be involved in electron transport from Fe(II) to O2 and from Fe(II) to the quinone pool for reverse electron transport (RET). Download FIG S6, PDF file, 0.7 MB (715.2KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plot of Fe(II) addition experiment results from Loihi (A) and Mariana (B) Fe mats, showing a higher living (orange; total) than killed (black; abiotic-only) Fe oxidation rate. Fe(II) was added to dormant Fe mat samples at 0 min. Pseudo-first-order rate constants were calculated from the log-linear best fit from each experimental condition. Download FIG S7, PDF file, 0.7 MB (686.3KB, pdf) .
Copyright © 2020 McAllister et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
High-quality full-length reads (20-pass minimum) from the PacBio 16S rRNA gene survey were submitted to GenBank (MK048478 to MK048944). Raw metagenome and metatranscriptome reads, as well as 5-pass-filtered PacBio 16S rRNA gene reads, were submitted to the NCBI SRA under BioProject accession PRJNA555820. Metagenome assemblies from this study and reassembled metagenome assemblies from the work of Fullerton et al. (9) were submitted to the JGI IMG database (sequence project IDs Gp0295814 to Gp0295821 and Gp0295823 [this study]; analysis project IDs Ga0256915 and Ga0257019 to Ga0257023 [Fullerton et al. {9}]). Zetaproteobacteria MAGs were also submitted to the JGI IMG database (see sequence project IDs listed above). Specific accession numbers per sample are shown in Table S4 at https://doi.org/10.6084/m9.figshare.c.4646336.