Abstract
Background
Fragment-based ligand design is used for the development of novel ligands that target macromolecules, most notably proteins. Central to its success is the identification of fragment binding sites that are spatially adjacent such that fragments occupying those sites may be linked to create drug-like ligands. Current experimental and computational approaches that address this problem typically identify only a limited number of sites as well as use a limited number of fragment types.
Methods
The site-identification by ligand competitive saturation (SILCS) approach is extended to the identification of fragment bindings sites, with the method termed SILCS-Hotspots. The approach involves precomputation of the SILCS FragMaps following which the identification of Hotspots, performed by identifying of all possible fragment binding sites on the full 3D structure of the protein followed by spatial clustering.
Results
The SILCS-Hotspots approach identifies a large number of sites on the target protein, including many sites not accessible in experimental structures due to low binding affinities and binding sites on the protein interior. The identified sites are shown to recapitulate the location of known drug-like molecules in both allosteric and orthosteric binding sites on seven proteins including the androgen receptor, the CDK2 and Erk5 kinases, PTP1B phosphatase and three GPCRs; the β2-adrenergic, GPR40 fatty-acid binding and M2-muscarinic receptors. Analysis indicates the importance of considering all possible fragment binding sites, and not just those accessible to experimental methods, when identifying novel binding sites and performing ligand design versus just considering the most favorable sites. The approach is shown to identify a larger number of known binding sites of drug-like molecules versus the commonly used FTMap and Fpocket methods.
General Significance
The present results indicate the potential utility of the SILCS-Hotspots approach for fragment-based rational design of ligands, including allosteric modulators.
Keywords: allosteric pocket, cryptic pocket, orthosteric, competitive inhibitor, computer-aided drug design, fragment-based drug design
Introduction
While a large number of drugs target the active or orthosteric sites of proteins, current drug discovery efforts are also targeting allosteric sites on proteins. Targeting allosteric sites offers the potential of minimizing unwanted side effects, such as those that may occur when blocking a metabolically essential pathway, while affecting a useful therapeutic outcome.[1, 2] Allosteric modulators may also overcome specificity issues associate with classes of proteins having similar active sites, which occurs with kinases.[3] Examples of drugs on the market that are allosteric inhibitors include the HIV non-nucleoside reverse transcriptase inhibitors, which are non-competitive inhibitors, such as rilpivirine.[4] Recently, allosteric modulators for a number of G-protein coupled receptors (GPCR) have been identified and efforts are ongoing to develop GPCR allosteric modulators into therapeutic agents.[5] As with kinases and reverse transcriptase, this is motivated by the similarity of the orthosteric sites of structurally and biologically similar proteins as well as the need to not completely block the physiological response associated with a specific GPCR but rather to modulate that response.
The rational design of allosteric modulators of proteins when an allosteric site has not been previously identified presents a number of challenges.[6] Proteins often undergo significant conformational changes as they interact with multiple partners including ions, small molecules, lipids, and other proteins. The heterogeneous environment is particularly challenging in the case of membrane bound proteins, such as GPCRs, that typically include hydrophobic transmembrane (TM) regions exposed to the lipid bilayer combined with the more hydrophilic extra- and intracellular regions that are comprised of a range of structural motifs. Accordingly, the ability to initially identify potential novel allosteric sites and also the design of ligands targeting those sites represents a significant challenge to computational methods.
A number of computational approaches have been presented to facilitate binding site identification and ligand design,[7–11] including the widely used FTMap by Vajda and coworkers.[12] Recent examples include the use of Fpockets[13] and Fragment Hotspots[14] to identify allosteric sites on acetylcholinesterase.[15] Efforts from the labs of Barril and Carlson have shown the utility of the CoSolvent or MixMD methodology in identifying allosteric sites.[16, 17] Notable are studies by Astex using both computational techniques, including the PLImap method, and experimental approaches to identify and characterize fragment binding sites.[18, 19] A similar approach has been applied by Caflisch and coworkers.[20, 21] These efforts have led to the concept of “hot” versus “warm” spots associated with the occupancy of the sites by fragments and their binding affinities, where both the hot and weaker affinity warm spots are important for ligand design as the occupancy of warm spots can lead to “substantial increases in affinity.”[18, 19] In the context of ligand design fragment-based approaches have been applied, for example, The plant homeodomain (PHD) Zinc Finger domain and the BRPF1 bromodomain.[22, 23]
The site-identification by ligand competitive saturation (SILCS) methodology[24–26] is an approach in which multiple cosolvent molecules along with water are used simultaneously to map the functional group affinity pattern of proteins, termed FragMaps. The method was developed in the spirit of “site-identification” and has successfully characterized an allosteric site in Erk kinase[27] and identified a novel allosteric site in heme oxygenase[28] as well as been successfully applied in a number of ligand discovery and optimization studies.[29–34] While the SILCS approach can identify important binding sites, including pharmacophore features demarcating those sites,[35] it additionally has the advantage of allowing for large numbers of fragment-like molecules to be rapidly screened against the full 3D structure of a protein, thereby allowing for the characterization of all potential binding sites based on the fragments that occupy those sites and their estimated affinities. As this approach identifies a collection of fragments for each site, it may be used to jump-start a ligand development project.
In the present study, we extend the SILCS approach to map all possible fragment binding sites on a protein, with the method termed SILCS-Hotspots. SILCS-Hotspots combines comprehensive fragment-screening based on the FragMaps and the SILCS-MC docking approach in conjunction with ligand clustering to identify and rank order fragment binding sites, termed Hotspots, on a protein. We note that this approach successfully identifies sites that encompass both the hot and warm spots discussed above. Analysis of the relative spatial locations of those Hotspots may be used to characterize putative allosteric and other ligand binding sites. The method is validated against 7 protein targets for which allosteric as well as orthosteric binding site ligands are known. These include the androgen receptor (AR), Map Kinase 7 (Erk5), Cyclin-dependent Kinase 5 (Cdk5), Protein-tyrosine-phosphatase 1B (Ptp1B), and the GPCRs, the β2 adrenergic receptor, the GPR40 fatty acid binding protein and the M2 muscarinic receptor. As the goal of the present study is to validate SILCS-Hotspots for facilitating the design of allosteric as well as orthosteric ligands with drug-like characteristics, the target systems were selected as they contain such types of ligands in crystallographic-determined orientations. This in contrast to computational methods designed to recapitulate the locations of small molecules used in crystallographic studies to experimentally identify binding sites on proteins.[36] Accordingly, validation of the SILCS-Hotspots method is based on its ability to identify fragment binding sites in the vicinity of those ligand binding sites as well as identify regions of the protein that can relax to allow for covalent linkage of fragments occupying the different Hotspots thereby supplying information that may a priori be used to facilitate the design of drug-like ligands. The power of the SILCS Hotspots approach is identifying all possible fragment binding sites that may be relevant to the design of drug-like molecules, not just those sites to which fragments bind that can be identified experimentally (e.g. those sites for which the affinity of the fragment is favorable enough to be observed). In all cases, the structures used for the SILCS simulations did not contain an allosteric modulator in the site being analyzed indicating the ability of the method to potentially identify novel binding sites for allosteric modulators.
Methods
SILCS-Hotpots Workflow
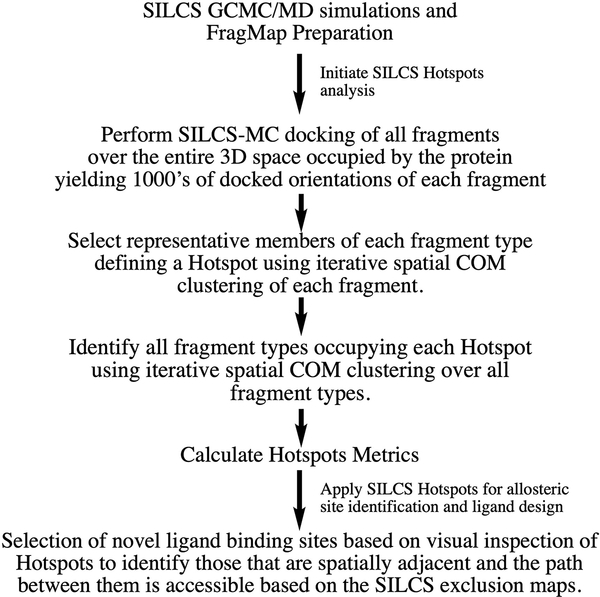
An overview of the SILCS-Hotspots workflow is shown in Scheme 1. The process is initiated by performing the SILCS GCMC/MD simulations from which the SILCS FragMaps are obtained. The SILCS FragMaps, which may be used in a number of ways, are the basis for the fragment docking from which the Hotspots are identified. Fragment docking uses the SILCS-MC approach to sample fragment locations and orientations in the full 3D region occupied by the protein and its surrounding environment. This leads to thousands of docked orientations of each fragment type. Two rounds of spatial clustering are then performed. In the first round a representative member of each fragment type in a region is identified, thereby defining fragment binding sites. In the second round, clustering is performed over all types of fragments, thereby identifying all the fragments that occupy a site, thereby defining a Hotspot. Metrics that may be used to define each Hotspot, as described below, are then calculated. This completes the Hotspots analysis. The final step in Scheme 1 represents a qualitative approach to identify novel allosteric binding sites using the identified Hotpots.
Scheme 1).
Workflow defining the SILCS-Hotspots process
Protein System Preparation
SILCS calculations were initiated with the crystallographic structures listed and described in Table 1. The PDBs associated with soluble proteins AR, Cdk2, Erk5 and Ptp1B were initially processed using the CHARMM-GUI,[37] including all ligands. Missing residues were constructed per the CHARMM-GUI default protocol. Prior to the SILCS simulations all ligands were removed. For all soluble protein subjected to SILCS simulations, the χ1 dihedral of side chains with solvent exposures of 0.5 Å2 or more were randomized by rotating the dihedral in 36° increments yielding 10 initial starting structures that only differed by the selected side chain orientations. The process was not performed for the membrane bound GPCRs. All protein systems, including the GPCRs in the equilibrated bilayers (see below), were then solvated with water, represented by the CHARMM TIP3P model, along with the following solutes at ~0.25 M concentration: benzene, propane, methanol, formamide, imidazole, acetaldehyde, acetate and methylammonium. The water and 8 solutes are simultaneously included in the simulation systems and each protein underwent this procedure 10 times to create 10 individual simulation systems. The simulation systems were created to be 15 Å larger than the largest dimensions of the proteins in the X, Y, and Z directions. The solutes were represented with the CHARMM General Force Field (CGenFF)[38] with the protein modeled using the CHARMM36m force field.[39]
Table 1).
Summary of protein structures and associated ligands.a
| System | PDB | RMSD | Ligand/Comment |
|---|---|---|---|
| Androgen receptor | |||
| 2AM9b | TES (O, testosterone)[51] | ||
| 2PIX | 0.54 | DHT (O, dihydrotestosterone, IC50 ∼10 nM) [52] | |
| FLA (A, flufenamic acid, inhibitor, IC50 ∼ 50 μM) | |||
| CDK2 | |||
| 3MY5b | CDK2/cyclinA complex with DRB (O): active form[53] | ||
| 1PW2b,c | 4.65 | Apo CDK2: inactive form[54] | |
| 3PXF | 4.59 | 2AN (A, 2 2AN molecules bound, a and b, Kd ∼37 μM)[55] | |
| 5FP5 | 4.47 | 1Y6 (A, 2 1Y6 molecules bound, a and b) [56] | |
| 5FP6 | 3.95 | MFZ (A, 2 MFZ molecules bound, a and b)[56] | |
| Erk5 | |||
| 4IC8b | Apo Erk5 (inactive) [57] | ||
| 5BYY | 3.75 | 4WG (O, IC50 =∼0.2 μM, undefined protein conformation)[58] | |
| 4ZSG | 3.45 | 4QX, (A, IC50 =∼5 μM, undefined protein conformation)[58] | |
| PTP1B | |||
| 2F6Fb | S295F mutant with no ligands (Mg+2 and Cl−)[59] | ||
| 1T48 | 2.75 | BB3 (A)[60] | |
| 2NT7 | 1.50 | 9O2 (O)[61] | |
| 3CWE | 1.07 | 825 (O, phosphonic acid analog and Mg2+)[62] | |
| β2 Adrenergic Receptor (GPCR) | |||
| 3SN6b | P0G (O, Gs protein complex, active form) [63] | ||
| 5X7D | 2.75 | CAU (O, carazolol, inactive form) [64] | |
| 8VS (A, inactive form) [64] | |||
| GPR40: Free fatty acid receptor (GPCR) | |||
| 4PHUb | 2YB (A, TAK-875, Partial allosteric agonist, site 1)[65] | ||
| 5KW2b | 6XQ (A, Lilly: Full allosteric agonist, site 2)[66] | ||
| 5TZY | 2.04 vs 4PHU | MK6 (A, Partial positive allosteric agonist, site 1, ∼1 nM)[67] | |
| 1.59 vs 5KW2 | 7OS (A, AgoPAM: Full allosteric agonist, site 2, ∼2 nM) | ||
| M2 muscarinic receptor (GPCR) | |||
| 3UON b | QNB (O, antagonist: inactive form)[68] | ||
| 4MQT | 2.55 | 2CU (A, PAM: ∼1 μM, active form)[69] | |
| IXO (O, active form)[69] | |||
The first structure listed under each protein was used to initiate the SILCS simulations, unless noted. RMSD in Å between protein structures used for the SILCS simulations and the structures used for identification of ligand binding sites are reported (Table S1 supporting information). Comments includes ligands in the structures with allosteric modulators indicated by (A) and active-site or orthosteric ligands indicated by (O).
Used for the SILCS simulations and visualization. These structures do not contain the allosteric binding sites. Two different structures were used to initiate the CDK2 and GPR40 SILCS simulations as described in the text.
Aligned with 3MY5 structure for visualization and analysis.
Preparation of the GPCR structures prior to the SILCS simulations was performed as follows. With the β2-adrenergic receptor, the nucleotide-free Gs heterotrimer was removed from the crystal structure (PDB ID: 3SN6) and the protein processed through the CHARMM-GUI.[37] Missing residues between TM5 and TM6 were constructed using MODELLER.[40] The GPR40 GPCR crystal structures (PDB ID: 4PHU and 5KW2) had the T4 lysozyme fusion protein removed from the structures with the resulting termini simply treated at standard C- and N-termini as the omitted loops were not in the region of the allosteric binding sites. In the case of the M2 muscarinic receptor, the T4 lysozyme fusion protein was removed from the crystal structure (PDB ID: 3UON) and the resulting terminal residues linked via a standard peptide bond. All GPCR proteins were then inserted into a lipid bilayer of 120 × 120 Å containing 90% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 10% cholesterol using the MolCal suite of programs (SilcsBio, LLC).[41] This was followed by a protein-bilayer equilibration protocol adopted from CHARMM-GUI[42] and previously used in our laboratory for studies of the μ-opioid receptor and the β2-adrenergic receptor.[43, 44] Following the equilibration simulation, the aqueous solution was removed and the system, including the bilayer, overlaid with water and the solutes listed above and then subjected to the standard SILCS simulation protocol, as follows.
SILCS simulation protocol
The SILCS simulation protocol used our in-house oscillating μex Grand Canonical Monte Carlo (GCMC) program[45] and GROMACS[46] for energy minimization and molecular dynamics (MD) simulations in conjunction with the MolCal software suite (SilcsBio, LLC). Details of the SILCS simulation protocol have been recently published.[26] Briefly, the 10 systems for each protein were subjected to SILCS oscillating μex GCMC/MD simulations. Following an initial equilibration, the GCMC/MD simulations involved repeated cycles of 200,000 GCMC steps of the water and solutes followed by a 5,000 step steepest descent minimization and a 100 ps MD equilibration followed by a 1 ns production MD simulation of the entire system. Each system was subjected to 100 such GCMC/MD cycles yielding a total of 1 microsecond of simulation trajectories (10 × 100 ns of MD simulation) for each protein.
Calculation of the SILCS FragMaps used snapshots from the MD trajectories saved every 10 ps from which the probability distributions of selected solute atoms that define the FragMaps were determined based on a 1 Å3 grid. The probability distributions were then normalized based on the expected probability distributions of the solute and water in aqueous solution alone. To correct for the presence of the volume occupied by the protein and bilayer, the solute probabilities were calculated relative to the number of water molecules in the system (e.g., 1 solute molecule relative to 55 water molecules corresponds to define 1 M solute), with concentrations calculated based on the individual functional group atom types rather than the solutes themselves as discussed in Ustach et al.[26] The normalized probability-based FragMaps were then converted to Grid Free Energies (GFE) based on a Boltzmann transformation for visualization and subsequent calculations. Selected FragMaps were combined into “generic” maps including Apolar (benzene and propane carbons), Hydrogen Bond Donor (formamide and imidazole protonated N atoms) and Hydrogen Bond Acceptor (formamide O, acetaldehyde O, and imidazole unprotonated N atoms) while the FragMaps for the methanol O, acetate C and methylammonium C or N were used directly. Ligand Grid Free Energies (LGFE), an approximation of the binding affinity of ligands, is based on the summation of the GFE scores for the classified atoms in each ligand.[26]
SILCS-Hotspots Protocol
SILCS-Hotspots is based on the identification and characterization of fragment binding sites, termed Hotspots. Fragments included in the present study are shown in Figure S1 of the supporting information. Identification of the Hotspots is initiated using the SILCS-MC method[47] in which fragment posing and scoring applies Monte Carlo (MC) sampling in the field of the GFE FragMaps with the Metropolis criteria based on the ligand LGFE scores plus intramolecular contributions associated with the CGenFF dihedral, electrostatic and van der Waals (vdW) energies.[26] In SILCS-Hotspots, the system is partitioned into a set of 14.14×14.14×14.14 Å3 sampling boxes that encompass the entire protein and surrounding region. At each sampling box SILCS-MC is performed, in which the fragment is randomly positioned within a sphere of radius 10 Å centered in the sampling box, with one of the rotatable bonds in the fragment randomly varied. The ligand is then subjected to 10,000 MC steps at 300 K of molecular translations and rotations up to 1.0 Å and 180.0°, respectively, and rotation of dihedrals about rotatable bonds of up to 180.0°. This is followed by 40,000 MC simulated annealing steps from 300 to 0 K of molecular translations and rotations up to 0.2 Å and 9.0°, respectively, and rotation of dihedrals about rotatable bonds of up to 9.0°. This process is repeated 1,000 times for each fragment in each sphere yielding thousands of docked orientations of each fragment in and around the entire protein. Pruning of this large number is performed using spatial clustering as described in the following paragraph (Scheme 1).[48]
Center-of-mass (COM) based clustering with 3 Å cluster radius is performed, where identification of the conformer with the largest number of neighbors is performed, with those members removed from the pool of conformations with the process repeated until no conformers remain.[48] The cluster radius of 3 Å is set empirically for this study, but it is adjustable. The sampling boxes in the SILCS MC are designed to overlap and the clustering is performed after pooling the fragment conformations from all sampling boxes together. This yields a collection of poses of each fragment.
A second round of clustering is then performed on all the poses to identify the Hotspots, which may be populated by multiple fragment types. This is performed using the same clustering algorithm and a radius of 4 Å from which the Hotspots are identified that contain one or more members from the collection of fragments under study. The LGFE scores of each of the fragments in each Hotspot are then averaged with the Hotspots ranked based on the mean LGFE scores. For comparison, ranking was also performed on the most favorable LGFE score or ligand efficiency (LE = LGFE/# of non-hydrogen atoms) of the fragments in each Hotspot and the number of fragments in each Hotspot. Hotspots characterization also included the distance to the nearest crystallographic ligand non-hydrogen atom, with relevant Hotspots being defined as within 5 Å of the ligand non-hydrogen atoms, where the Hotspot position is based on the COM of all the fragments in the Hotspot. In the present study, only fragments with an LGFE scores of −2 kcal/mol or more favorable and within 6 Å of any protein Cα atoms were subjected to clustering for Hotspots determination; both metrics are adjustable. It should be noted the Hotspots identification is sensitive to the two clustering radii, where the use of larger clustering radii will typically lead to a decrease in the total number of Hotspots identified with those sites being more spatially separated. The cluster radii as well as the LGFE cutoff of −2 kcal/mol and the 6 Å distance to protein Cα atoms were selected based on empirical observations in the present study.
Results and Discussion
Validation of the capability of the SILCS-Hotspots method to identify binding sites focused on known allosteric sites in seven well-studied proteins (Table 1). In addition, given the availability of orthosteric or active site ligands in some structures, analysis was extended to these species. In all cases, the structures used for the SILCS simulations did not contain an allosteric modulator in the site being analyzed. In the case of GPR40 for which two modulators binding to separate sites are known, to characterize the methodology against both sites, two separate SILCS simulations were performed using a crystal structure in which one of the sites was unoccupied in each case, with the ability of SILCS-Hotspots to characterize that site evaluated. An important part of the present SILCS-Hotspots method is the use of fragments known to occur in drug-like molecules to identify putative binding sites. This approach leads to the identification of binding sites that have a higher potential of being suitable as targets for the design of drug-like molecules. In the present study, the family of fragments was comprised of 90 mono- and bicyclic compounds. These include ring systems that commonly occur in drug molecules.[49] The fragments are shown in Figure S1 of the supporting information. We note that the SILCS-Hotspots procedure may be applied to any collection of fragments, including publicly available[18, 19, 50] and commercial fragment libraries, as well as to full-drug like molecules.
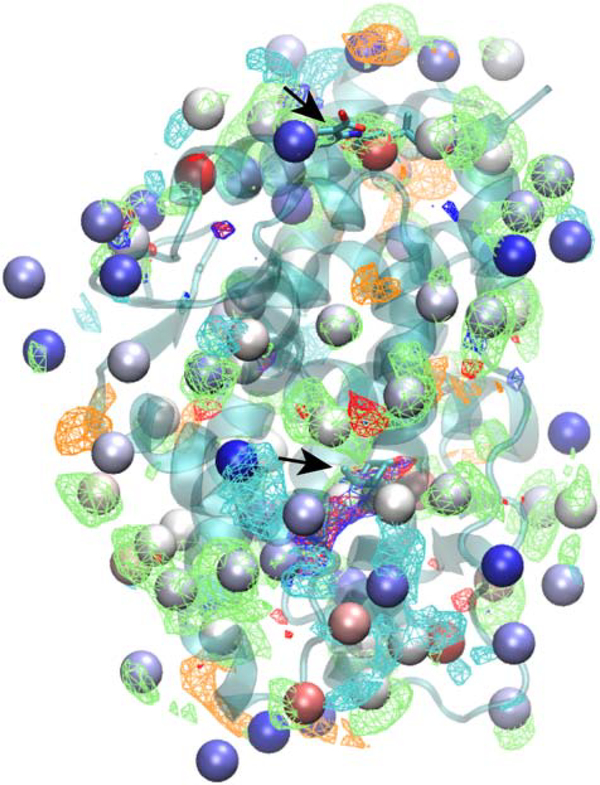
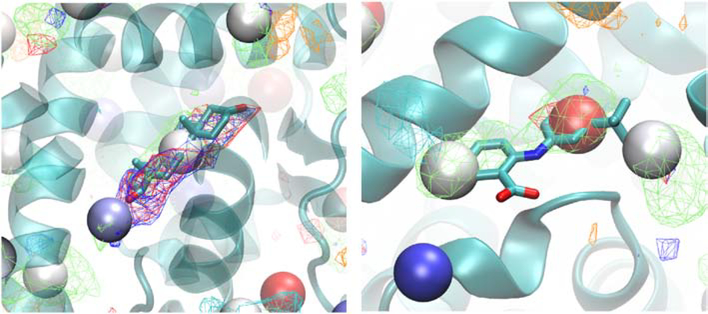
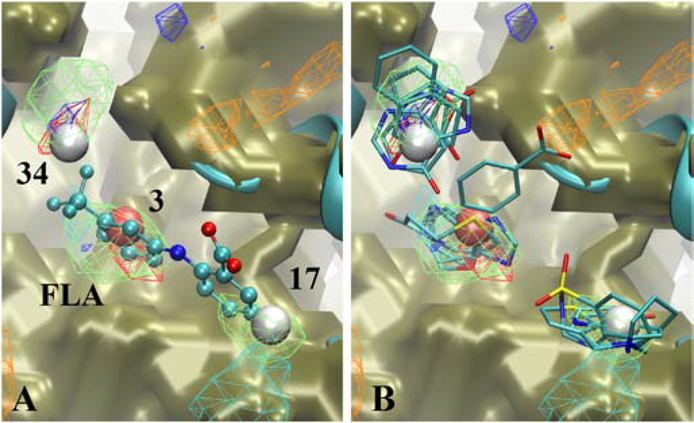
The output from SILCS-Hotspots comprises the location of the centers of the predicted fragment binding sites, termed Hotspots, along with the mean LGFE score for all the ligands in each site. In addition, the pose of each fragment in each Hotspot is output. Initial analysis focused on the Androgen Receptor followed by a more global analysis of all the target systems. Mapping of the Hotspots on that AR is shown on Figure 1. As may be seen the Hotspots encompass the entire protein, including occupying the totally occluded orthosteric site where dihydrotestosterone (DHT) binds, as well as the allosteric site to which flufenamic acid (FLA) binds. As expected, the sites are occupied by the different SILCS FragMaps that are used as the scoring function for the SILCS MC posing of the fragments. Close up views of the local ligand binding sites with the crystallographic orientation of the ligands of the Hotspots are shown in Figure 2. In the case of DHT, there are two Hotspots in the vicinity of the ligand that correspond with apolar, H-bond donor and H-bond acceptor FragMaps. The mean LGFE scores of the two Hotspots sites are −2.82 and −2.48 kcal/mol and these correspond to the 26th and 57th ranked sites, respectively. It should be noted that the starting structure (PDB ID: 2AM9) has testosterone in the orthosteric site, so identification of this site was expected as the use of the GCMC methodology in SILCS allows for sampling of the solutes and water during the SILCS simulations in such occluded sites, as required for proper energetic evaluation in such binding sites, as has been shown with the T4 lysozyme pocket mutant.[45] In the case of the allosteric modulator, FLA, there are three Hotspots that overlap with the ligand. These correspond to apolar, H-bond donor and negative FragMaps. The mean LGFE scores of these sites are −3.47, −2.91 and −2.71 kcal/mol, corresponding to rankings of 3, 17 and 34, respectively. These results show the capability of the SILCS-Hotspots method to identify fragment-binding sites that correspond to known ligand binding sites. However, the identified Hotspots are not typically the highest-ranking sites, as is also the case for the remaining systems, as discussed below.
Figure 1).
Androgen receptor (transparent cartoon representation) showing the SILCS-Hotspots (VDW spheres, colored by LGFE ranking, red: most favorable, blue: least favorable) along with the SILCS FragMaps (mesh representations) and, as indicated by arrows, the orthosteric ligand, dihydrotestosterone (DHT, central region of figure, see arrow) and the allosteric modulator, flufenamic acid (FLA, top of figure, see arrow). SILCS FragMaps are shown for generic apolar (green, −0.9 kcal/mol), generic H-bond donor (blue, −0.9 kcal/mol), generic H-bond donor (red, −0.9 kcal/mol), negative (orange, −1.5 kcal/mol) and positive (cyan, −1.5 kcal/mol) groups.
Figure 2).
Crystallographic orientations of dihydrotestosterone (DHT, left) and flufenamic acid (FLA, Licorice representation, atom colored) overlaid on the Androgen receptor (cartoon) along with the SILCS-Hotspots (VDW spheres, colored by LGFE ranking, red: most favorable, blue: least favorable) and the SILCS FragMaps (see Figure 1 legend).
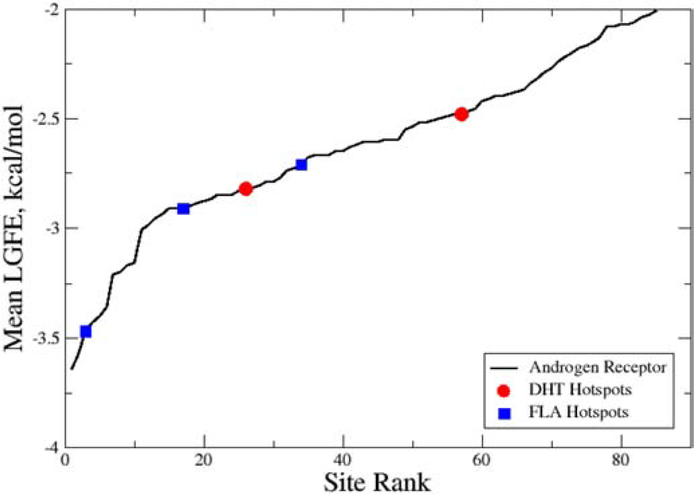
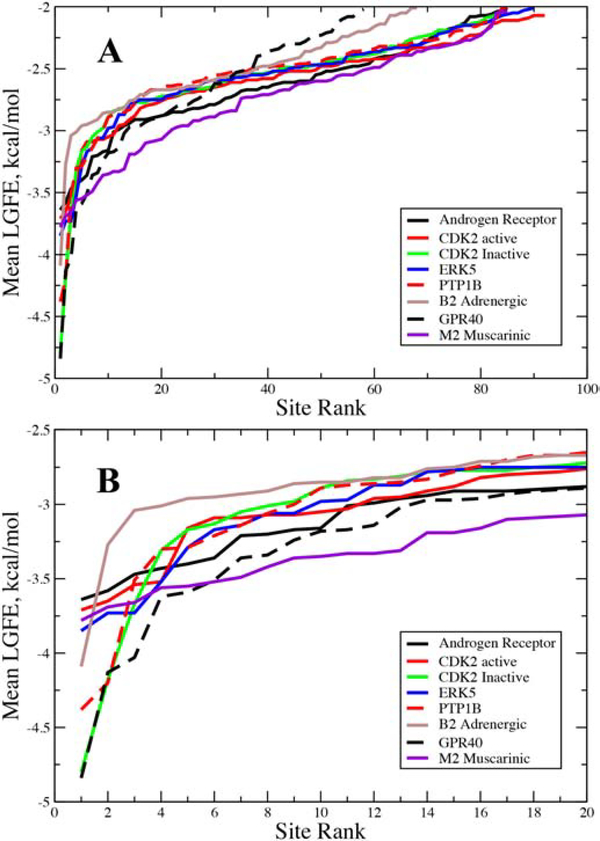
To understand the relevance of the ranking of the identified Hotspots, analysis of the mean LGFE scores of all sites as a function of their rank was undertaken (Figure 3). For the androgen receptor a total of 85 sites with mean LGFEs less then −2.0 kcal/mol were identified, with the most favorable being −3.64 kcal/mol. The top 11 sites have energies of −3.0 or less and these sites appear to comprise a specific class of sites as judged by the slope of that region in Figure 3. Sites less favorable than −3.0 kcal/mol appear to comprise a second regime with an approximately linear slope. While the criteria for defining the class of sites differ the present high- and low-slope sites appear to approximately correspond to the previously discussed hot and warm sites.[18, 19] With respect to the sites that coincide with the ligand binding sites (Table 1), only one that is in the FLA site is in the more favorable set with the rest in the second “low slope” regime (Figure 3). The range of LGFE scores for those sites is relatively small, from −2.48 to −2.82 kcal/mol.
Figure 3).
Hotspots-site mean LGFE score as a function of the site rank order for the Androgen Receptor. Hotspots associated with the dihydrotestosterone (DHT, red circles) and flufenamic acid (FLA, blue squares) ligand binding sites are shown. Hotspots-site mean LGFE scores are the averages over the LGFE scores of all the fragments bound to each hotspot.
The relevance of the ranking of the Hotspots for ligand design was further investigated by considering additional metrics including the number of fragments in each site, the most favorable LGFE of each site and the most favorable LE for the sites (Table 2). This analysis included the top 2 ranked sites as well as the sites in the vicinity of the DHT and FLA ligands. The number of fragments in each site adjacent to the ligands varied from 2 to 55 out of the 90 fragments considered in the study, compared to 11 and 3 for the 2 top ranked sites. The highest-ranking site close to a ligand, site 3, contained the most favorable fragment, with LGFE = −6.2 kcal/mol, more favorable than the values of −4.8 and −3.9 kcal/mol for sites 1 and 2, respectively. A similar trend was observed with the LE metric. Indeed, with respect to the most favorable fragment LGFE scores, sites 17, 26 and 34 have fragments more favorable than site 2 with that of site 34 being more favorable than that in the top ranked site. In combination, this analysis indicates that use of site ranking based on energetics or number of fragments is not sufficient to identify sites appropriate for directing ligand design. Instead, the characteristics and spatial relationship of the individual Hotspots need to be considering when identifying novel ligand binding sites and undertaking fragment-based drug design. Analysis of the additional proteins below yields similar results.
Table 2).
Summary of SILCS-Hotspots sites identified in the Androgen Receptor including the top 2 ranked sites and those adjacent to the orthosteric (DHT) and allosteric (FLA) binding sites.
| Site | Hotspot Rank | Minimum Distance to Ligand | Mean LGFE | # of Fragments | Most favorable LGFE | Most favorable LE |
|---|---|---|---|---|---|---|
| 1 | NA | −3.65 | 11 | −4.8 | −0.77 | |
| 2 | NA | −3.60 | 3 | −3.9 | −0.49 | |
| DHT | 26 | 0.8 | −2.82 | 25 | −4.3 | −0.53 |
| 57 | 3.6 | −2.48 | 2 | −2.9 | −0.34 | |
| FLA | 3 | 0.9 | −3.47 | 47 | −6.2 | −0.86 |
| 17 | 1.2 | −2.91 | 16 | −4.1 | −0.60 | |
| 34 | 1.6 | −2.71 | 55 | −5.1 | −0.59 |
Distances are in Å and energies in kcal/mol. Hotspot rank is based on the Mean LGFE. LE is the ligand efficiency based on the LGFE/# of non-hydrogen atoms. NA indicates that the top 2 ranked sites are not adjacent to the ligands.
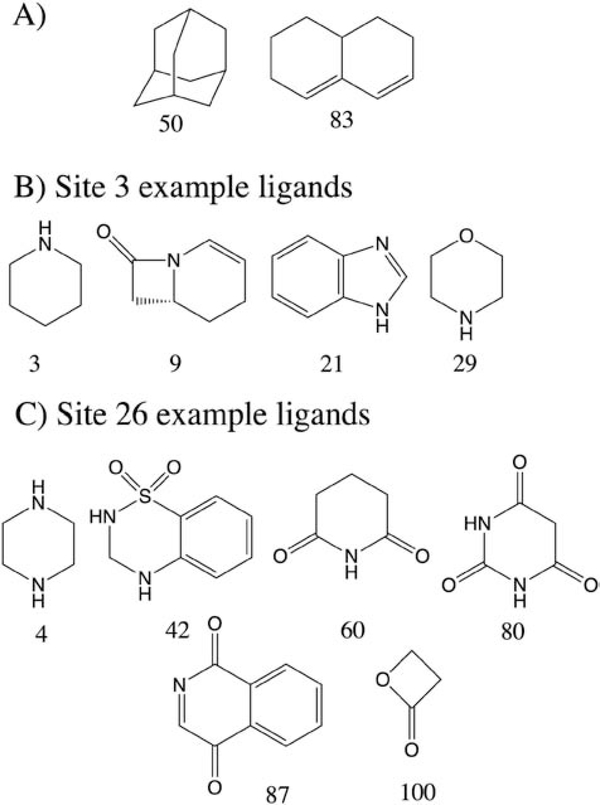
Examination of the specific fragments occupying selected Hotspots was next undertaken. The top scoring DHT and FLA sites, Hotspots 26 and 3, contain 25 and 47 fragments, respectively (Table 2). Of the top fragments in each site, fragments 50 and 83 occurred in both sites (Figure 4 and Figure S1). These fragments, adamantane and hexahydronaphthlene, are hydrophobic molecules and bind favorably to a significant number of sites (45 and 40 Hotspots, respectively) on the AR. However, in addition fragments with significant amounts of polar character are also at those Hotspots. For example, at site 3 fragments 3, 9, 21, 29, and 31 bind with LGFEs of −5.1, −3.9, −2.6, −4.6 and −2.9 kcal/mol, respectively. Similarly, at site 26 fragments included 4, 42, 60, 80, 87 and 100 with LGFE scores −2.9, −4.2, −3.2, −3.5, −3.2 and −2.4 kcal/mol, respectively (Figure 4). Thus, while there is a tendency for nonpolar fragments to be selected for a large number of Hotspots, diverse fragments are being selected for the individual sites dictated by their specific characteristics. This is important as specificity is largely dictated by electrostatic interactions.[70] Importantly, for ionizable groups with pKa values close to 7, multiple protonation states are considered in the Hotspots analysis, thereby allowing for identification of the preferred protonation as well as tautomeric state of a fragment for a given site. Importantly, the SILCS-Hotspots procedure typically identifies multiple fragments at each site, allowing for selection of the most appropriate fragments for ligand design based on synthetic, physiochemical or other considerations.
Figure 4).
Example fragments that bind to the Androgen receptor as identified by SILCS-Hotspots analysis. A) Two nonpolar ligands that bind to a large number of Hotspots. Selected polar ligands that bind to Hotspots 3 B) and 26 C) that comprise part of the FLA and DHT binding sites on the Androgen receptor.
Global analysis based on additional proteins studied
To assure that the observations from the SILCS-Hotspots approach are generalizable, the present study included six proteins of various classes in addition to the AR. These include two kinases, a phosphatase and three GPCRs (Table 1). These represent widely targeted proteins that are comprised of different structural motifs for which crystallographically-characterized allosteric modulators are known. The kinases included both the active and inactive forms of CDK2 along with the inactive form of Erk5 and three GPCRs were studied given the significant role of this class of proteins as drug targets. In the case of the kinases and the β2 adrenergic and M2 muscarinic receptors the SILCS simulations were initially performed on the form (active or inactive) that the allosteric modulator does not bind and, to reiterate, the starting structures did not contain the allosteric modulator being used for validation.
Motivated by the trend seen in the Hotspots site mean LGFE scores as a function of rank for the AR (Figure 3), the same analysis was performed for all the proteins (Figure 5). The same general pattern is observed. The highest-ranking sites fall into the high-slope or hot classification where the differences in the mean LGFE scores changes rapidly with ranking, followed by a larger number of sites in which the differential in the mean LGFE score is lower and the LGFE scores are less favorable, the low-slope or warm classification. Figure 5B shows the same plot for the top 20 ranked sites for each protein. The number of hot sites, sites that have a mean LGFE < −3.0 kcal/mol, for the proteins varies from as low as 3, with the β2 adrenergic receptor, up to 15 with the AR. The majority of proteins have a smaller number of hot sites, typically from 5 to 8. While these sites have more favorable overall affinities for the studied fragments, most sites found in the vicinity of the known ligands are warm sites consistent with the AR results. Visual inspection of the fragments and analysis of the contributions of the different types of FragMaps to the two classes of sites did not reveal any patterns that differentiate them.
Figure 5).
Hotspots-site mean LGFE score as a function of the site rank order for the studied proteins. A) Data for all the sites identified in each protein and B) for the 20 top ranked sites for each protein. Hotspots-site mean LGFE scores are the averages over the LGFE scores of all the fragments bound to each hotspot.
Details of the number of Hotspots within 5.0 Å of each of crystallographic ligand sites are presented in Table 3 for the six additional proteins. The results are similar to those described above for the AR. In all but two cases, there are two or more Hotspots in the vicinity of the binding sites. The exceptions occur with the AN1 Site in the active form of CDK2, with no Hotspots identified and the second MFZ site on the inactive form of CDK2 for which only a single site is identified (see below). Based on the mean LGFE ranking there are a number of Hotspots in the top 10 while the majority of Hotspots are warm sites with mean LGFEs less favorable than −3 kcal/mol. For the majority of Hotspots multiple fragments are present, though in a number of cases less than 5 fragments occupy the respective sites. These results further indicate the ability of the SILCS-Hotspots approach to identify potential allosteric and othosteric binding sites, though it is necessary to take into account all the Hotspots rather than just the most favorable sites, as previously discussed.17
Table 3).
Summary of SILCS-Hotspots sites identified in the kinases CDK2 (active and inactive forms) and ERK5, the PTP1B phosphatase, and GPCRs including the β2-adenergic receptor, GPR40 and the M2 muscarinic receptor.a
| Site | Hotspot Rank | Distance | Mean LGFE | # of Fragments | Min. LGFE | Min. LE |
|---|---|---|---|---|---|---|
| CDK2, active | ||||||
| 2ANa, A, surface | 12 | 2.7 | −3.0 | 29 | −6.3 | −0.74 |
| 49 | 3.5 | −2.5 | 1 | −2.5 | −0.23 | |
| 2ANb, A, buried | No adjacent Hotspots | |||||
| 1Y6a, A, upper surface | 19 | 4.9 | −2.8 | 1 | −2.8 | −0.35 |
| 65 | 2.7 | −2.4 | 11 | −2.7 | −0.51 | |
| 72 | 4.2 | −2.3 | 6 | −2.7 | −0.27 | |
| 78 | 1.1 | −2.2 | 3 | −2.6 | −0.40 | |
| 1Y6b, A, buried | 5 | 1.4 | −3.2 | 54 | −5.6 | −0.76 |
| 92 | 4.2 | −2.1 | 1 | −2.1 | −0.30 | |
| MFZa, A, surface | 8 | 4.9 | −3.1 | 15 | −5.8 | −0.73 |
| 12 | 2.7 | −3.0 | 29 | −6.3 | −0.74 | |
| 49 | 3.0 | −2.5 | 1 | −2.5 | −0.23 | |
| MFZb, A, buried | 5 | 1.7 | −3.2 | 54 | −5.6 | −0.76 |
| 92 | 3.0 | −2.1 | 1 | −2.1 | −0.30 | |
| CDK2, inactive | ||||||
| 2ANa, A, surface | 22 | 3.5 | −2.7 | 3 | −3.0 | −0.43 |
| 33 | 1.5 | −2.6 | 7 | −3.2 | −0.61 | |
| 70 | 0.9 | −2.2 | 1 | −2.2 | −0.28 | |
| 2ANb, A, buried | 6 | 4.2 | −3.1 | 4 | −3.5 | −0.37 |
| 22 | 2.0 | −2.7 | 3 | −3.0 | −0.43 | |
| 1Y6, A, upper near V226 | 50 | 3.2 | −2.5 | 2 | −2.6 | −0.47 |
| 51 | 2.4 | −2.5 | 6 | −3.2 | −0.41 | |
| 1Y6, A, lower near G13 | 34 | 2.5 | −2.6 | 13 | −3.3 | −0.56 |
| 70 | 4.4 | −2.2 | 1 | −2.2 | −0.28 | |
| MFZ, A, surface | 6 | 1.7 | −3.1 | 4 | −3.5 | −0.37 |
| 22 | 4.2 | −2.7 | 3 | −3.0 | −0.43 | |
| MFZ, B, buried | 34 | 1.9 | −2.6 | 13 | −3.3 | −0.56 |
| ERK5 | ||||||
| 4QX, A | 29 | 0.9 | −2.6 | 18 | −3.5 | −0.57 |
| 66 | 3.6 | −2.3 | 2 | −2.5 | −0.25 | |
| 71 | 3.7 | −2.3 | 2 | −2.4 | −0.42 | |
| 4WG, O | 8 | 0.9 | −3.1 | 7 | −4.3 | −0.72 |
| 25 | 4.1 | −2.7 | 14 | −3.6 | −0.45 | |
| 71 | 2.5 | −2.3 | 2 | −2.434 | −0.42 | |
| 78 | 2.6 | −2.2 | 1 | −2.2 | −0.36 | |
| 86 | 0.8 | −2.1 | 2 | −2.1 | −0.35 | |
| PTP1B | ||||||
| BB3, A | 5 | 1.0 | −3.3 | 2 | −3.8 | −0.64 |
| 16 | 3.0 | −2.7 | 6 | −3.1 | −0.45 | |
| 50 | 1.6 | −2.4 | 41 | −3.6 | −0.48 | |
| 65 | 4.5 | −2.3 | 4 | −2.5 | −0.35 | |
| 902, O | 24 | 0.6 | −2.6 | 3 | −2.7 | −0.53 |
| 30 | 2.3 | −2.6 | 7 | −3.2 | −0.54 | |
| 32 | 1.9 | −2.5 | 21 | −4.7 | −0.52 | |
| 825, O | 24 | 1.3 | −2.6 | 3 | −2.7 | −0.53 |
| 32 | 4.8 | −2.5 | 21 | −4.7 | −0.52 | |
| β2 adrenergic receptor (GPCR) | ||||||
| CAU, O | 20 | 1.7 | −2.7 | 1 | −2.7 | −0.53 |
| 31 | 1.5 | −2.6 | 22 | −3.7 | −0.42 | |
| 41 | 1.3 | −2.5 | 2 | −2.5 | −0.25 | |
| 47 | 3.5 | −2.4 | 6 | −2.9 | −0.59 | |
| 56 | 2.0 | −2.2 | 1 | −2.2 | −0.22 | |
| 67 | 3.3 | −2.0 | 1 | −2.0 | −0.29 | |
| 8VS, A | 23 | 3.0 | −2.7 | 17 | −4.3 | −0.47 |
| 58 | 1.2 | −2.2 | 2 | −2.2 | −0.41 | |
| 60 | 2.1 | −2.2 | 2 | −2.2 | −0.23 | |
| GPR40 (GPCR) | ||||||
| MK6, A, Site 1b | 27 | 1.2 | −2.9 | 22 | −4.9 | −0.58 |
| 57 | 1.1 | −2.3 | 8 | −2.7 | −0.45 | |
| 61 | 0.9 | −2.1 | 1 | −2.1 | −0.36 | |
| 7OS, A, Site 2c | 12 | 1.5 | −3.1 | 12 | −5.3 | −0.65 |
| 29 | 2.2 | −2.7 | 9 | −3.7 | −0.57 | |
| M2 muscarinic receptor (GPCR) | ||||||
| 2CU, A | 23 | 2.3 | −3.0 | 12 | −3.9 | −0.65 |
| 28 | 3.2 | −2.9 | 43 | −4.7 | −0.71 | |
| 41 | 2.0 | −2.8 | 2 | −3.3 | −0.33 | |
| 63 | 3.4 | −2.5 | 4 | −2.8 | −0.25 | |
| 75 | 3.2 | −2.4 | 32 | −3.2 | −0.50 | |
| IXO, O | 3 | 2.1 | −3.8 | 2 | −3.9 | −0.65 |
| 10 | 0.8 | −3.3 | 32 | −6.3 | −1.06 | |
| 27 | 3.8 | −3.0 | 57 | −5.1 | −0.68 | |
Sites are defined by the allosteric (A) or orthosteric (O) ligands that occupy those sites in the crystallographic structures (Table 1). For certain ligands there are two sites on the protein, with information on both sites including additional identification information such as adjacent protein residue numbers. Distances are in Å and energies in kcal/mol. LE is the ligand efficiency based on the LGFE/# of non-hydrogen atoms.
SILCS simulation initiated from 5KW2 with ligand in site 2 in the crystal structure.
SILCS simulation initiated from 4PHU with ligand in site 1 in the crystal structure. In both cases the ligands were removed prior to the SILCS simulations.
While the ranking of Hotspots is based on the mean LGFE, by applying the approach to multiple proteins the predictability of other metrics, such as the minimum LGFE or LE and the maximum number of fragments in each site was tested. Presented in Table 4 are the top overall minimum LGFE, minimum LE and mean LGFE scores and the maximum number of fragments for all the Hotspots in each protein along with the top values for those Hotspots in the vicinity of crystallographic identified ligands. While the mean LGFE does not identify the most favorable Hotspot adjacent to any ligand, both the minimum LGFE and minimum LE metrics do in two cases each while the maximum number of fragments in a Hotspot does in one case. Thus, identifying Hotspots with highly favorable LGFE or LE scores for individual fragments or Hotspots with the maximum number of fragments may be of utility for identifying Hotspots to be exploited in ligand design. However, is as evident from the data in Table 3, the majority of the Hotspots located adjacent to known ligands do not have highly favorable values of these metrics, further indicating the need to consider all Hotspots when undertaking ligand design.
Table 4).
Comparison of the minimum LGFE, minimum LE, and mean LGFE scores and the maximum number of fragments over all the fragments that were docked against each protein versus those in the Hotspots in the vicinity of experimentally determined ligands.
| Minimum LGFE | Minimum LE | Mean LGFE | Maximum # of fragments | ||
|---|---|---|---|---|---|
| Androgen Receptor | All fragments | −6.2 | −0.86 | −3.6 | 65 |
| Near ligand | −6.2 | −0.86 | −3.5 | 55 | |
| CDK2, active | All fragments | −6.3 | −0.93 | −3.7 | 56 |
| Near ligand | −6.3 | −0.76 | −3.2 | 54 | |
| CDK2, inactive | All fragments | −6.0 | −0.89 | −4.8 | 40 |
| Near ligand | −3.5 | −0.61 | −3.1 | 13 | |
| Erk5 | All fragments | −4.9 | −0.72 | −3.9 | 37 |
| Near ligand | −4.3 | −0.72 | −3.1 | 18 | |
| PTP1B | All fragments | −6.5 | −1.09 | −4.4 | 41 |
| Near ligand | −4.7 | −0.64 | −3.3 | 41 | |
| β2 adrenergic receptor | All fragments | −5.6 | −0.93 | −4.1 | 45 |
| Near ligand | −4.3 | −0.59 | −2.7 | 22 | |
| GPR40 (4PHU) | All fragments | −5.7 | −0.71 | −4.8 | 52 |
| Near ligand | −5.3 | −0.65 | −3.1 | 12 | |
| GPR40 (5KW2) | All fragments | −6.5 | −0.98 | −3.6 | 62 |
| Near ligand | −4.9 | −0.58 | −2.9 | 22 | |
| M2 muscarinic receptor | All fragments | −6.4 | −0.95 | −3.6 | 62 |
| Near ligand | −4.7 | −0.66 | −2.9 | 22 |
Energies in kcal/mol. Instances when the Hotspots top ranked values correspond to the overall top value are highlighted.
Active versus inactive form of CDK2 kinase and GPCRs
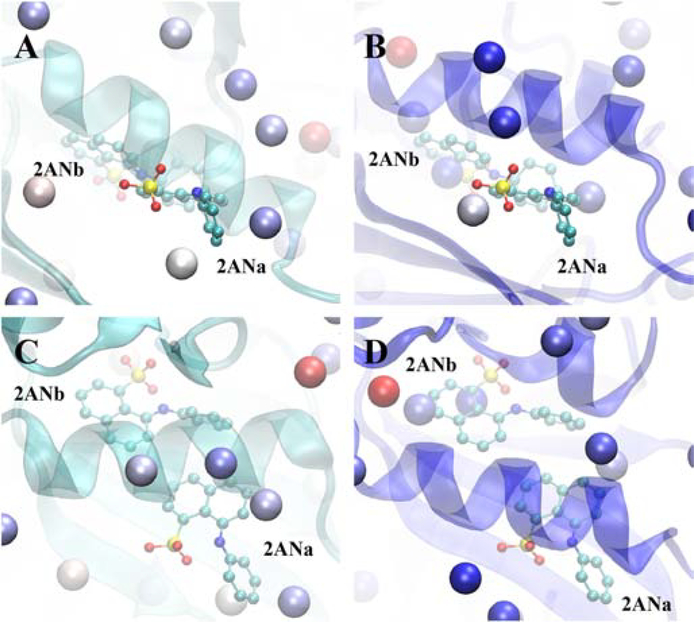
Kinases undergo significant conformational changes upon going from the inactive to active forms associated with in, out, and intermediate conformations of the DFG motif.[71, 72] While for many kinases crystal structures of multiple forms are known, for a large number of kinases crystal structures of only a single form is available. Accordingly, tests were undertaken to determine if application of the SILCS-Hotspots method to one form would yield information suitable for the other form. Towards this, we initially performed the SILCS simulations on the active conformation of CDK2 (PDB ID: 3MY5) with analysis being performed with respect to the allosteric modulators that target the inactive form of the kinase. The results under the CDK2 active section of Table 3 show that for the majority of sites there are two or more Hotspots adjacent to the ligands. However, there were no Hotspots in the vicinity of one of the orientations of the modulator 2ANb. To determine if this was associated with use of the active conformation of the kinase a second set of SILCS FragMaps was calculated starting from an inactive conformation (PDB ID: 1PW2) and subjected to Hotspots analysis. Results in Table 3 for the CDK2 inactive conformation show that Hotspots are identified adjacent to all the allosteric modulators, including 2ANb, though only a single Hotspot adjacent to the MFZb ligand (second conformation of MFZ ligand) is found.
To understand the structural contributions leading to the inability of the active form Hotspots analysis to identify the location of 2ANb, the two crystal conformations of the active and inactive structures and the two orientations of the 2AN ligand are compared. In the active form (Fig. 6A and C), the αC helix of CDK2 (residue 45 to 57) [73] is positioned directly on the 2AN ligands such that no Hotspots can occupy the region adjacent to 2ANb. However, while the second orientation 2ANa is also partially occluded by the αC helix, the phenyl group is on the surface of the protein adjacent to three Hotspots. In the inactive conformation (Fig. 6B and D), the αC helix is shifted, opening the region to which the two conformations 2AN bind such that Hotspots are located adjacent to both the buried (2ANb) and surface exposed (2ANa) conformations. These results indicate that the SILCS-Hotspots approach cannot account for the large conformational change that the DFG motif undergoes such that it is suggested that allosteric ligand design efforts targeting the DFG motif region in the active versus inactive form of kinases need to be based on the respective conformations of the protein. However, as Hotspots are identified in the inactive form of CDK2 that are adjacent to modulators identified in other inactive crystal structures of the protein, the inclusion of protein flexibility in the SILCS methods can account for the structural variability among the individual forms of the protein.
Figure 6).
CDK2 backbone cartoon structures for the A) and C) active conformation (PDB ID: 3MY5, cyan cartoon) and B) and D) inactive conformation (PDB ID: 1PW2, blue cartoon) for two approximately orthogonal orientations of the protein. Included are the two orientations, a and b, of the allosteric modulator 2AN (from PDB ID 3PXF) along with the SILCS-Hotspots for the respective conformations (vdW spheres, colored based on mean LGFE score, red most favorable, blue least favorable). Protein structures were aligned prior to visualization as described in Table S1 of the supporting information.
Similar analysis was performed with the β2 Adrenergic and M2 muscarinic GPCRs. The SILCS FragMaps were calculated for the active form of β2 and the inactive form of M2 with the Hotspots analysis performed on those FragMaps (Table 1). Notably, analysis of the Hotspots adjacent to allosteric modulators on the opposite forms of the respective GPCRs showed that multiple Hotspots are in the vicinity of those ligands (Table 3). In addition, Hotspots are also identified adjacent to the orthosteric ligands when the alternate form of the receptors were used for FragMap generation. These results indicate that the ability to identify allosteric sites is, to some extent, independent of the state of the GPCR on which the FragMaps are calculated. However, we do note in previous studies on the β2 Adrenergic receptor that independent SILCS simulations of both the active and inactive forms yielded two sets of FragMaps that were able to distinguish agonists from antagonists, [44] indicating the potential utility of the method in the development of agonists versus antagonists for GPCRs given the availability of both states of the protein. In addition, the ability to distinguish between agonists and antagonists by active versus inactive form FragMaps, respectively,[44] indicates the need to target the correct form of the GPCR when performing ligand optimization using the SILCS methodology.
Comparison of SILCS-Hotspots with FTMap and Fpockets
Calculations were undertaken on the studied proteins to compare SILCS-Hotspots with the widely used FTMap[12] and Fpocket[13] methods. Presented in Table S2 are results on the number of sites identified by each method within 5 Å of the crystal ligand positions along with the ranking of the sites and the individual site COM to ligand minimum distance for the studied proteins. Overall, SILCS-Hotspots typically identifies more sites in the vicinity of the ligands than both FTMap and Fpocket. FTMap identifies one additional site in 3 cases including identification of a site close to the problematic 2ANb ligand in the active form of CDK2 discussed above. However, the method does not identify sites in 3 cases in which multiple sites are identified by SILCS-Hotspots. Fpockets in no case identifies more sites than SILCS-Hotspots and there are 5 cases where the method does not identify any sites adjacent to the ligands. It is interesting that both FTMap and Fpockets do not identify sites in the vicinity of the allosteric inhibitor FLA on the androgen receptor given that this site is exposed on the surface of the protein (Figure 2). Thus, SILCS-Hotspots identifies a larger number of fragment binding sites in the vicinity of the known ligands and typically identifies more Hotspots in the region of those ligands than FTMap or Fpockets, information that is helpful for ligand design as discussed below. It is noted that FTMap and Fpocket require significantly less CPU time as compared to SILCS due to the need to perform the SILCS simulations prior to the Hotspots analysis. In addition, those methods overall identify a smaller number of sites than SILCS-Hotspots using the default protocols. Modification of those protocol may lead to identification of additional sites with the FTMap and Fpocket approaches.
Towards Ligand Design with SILCS-Hotspots
Ultimately, the utility of Hotspots analysis is to facilitate the design of a ligand with characteristics suitable for potential development into a therapeutic agent with the challenge of no previous knowledge of the location of the binding site of the ligands to be developed. In simple terms, this requires linking the individual fragments located in adjacent Hotspots to create larger ligands with two or more ring systems. Advantages of the SILCS-Hotspots approach are three fold. First, identifying a large number of Hotspots allows for spatially adjacent sites to be selected that may be linked to create drug-like molecules that are comprised of two or more ring systems that occupy multiple Hotspots. Second, the identification of multiple fragments that occupy each Hotspot allows those and related fragments to be considered for ligand development. Third, the inclusion of flexibility of the protein in the SILCS simulations allows for the protein to relax from which unhindered paths between Hotspots may be identified based on the exclusion maps that are not evident in the crystal structures. Such path may be used for covalently linking fragments into larger drug-like molecules. Towards automated ligand development, a number of fragment-linking computational approaches have been presented, many of which would be able to directly utilize the fragments from the Hotspots analysis.[74–77] In the remainder of this section a qualitative overview is presented based on the known crystallographic allosteric ligands and the Hotspots and fragments identified in the present study.
Analysis of the information content in the Hotspots approach was performed on ligands from the Androgen receptor, Erk5, and the GPR40 and M2 muscarinic GPCRs. The allosteric modulator from these systems were some of the largest in the studied systems and the proteins are from three diverse classes. The analysis includes visual presentation of the respective allosteric binding sites with the Hotspots and the ligand or selected fragments from the Hotspots analysis overlaid on the SILCS FragMaps and exclusion maps along with the protein backbone. For each Hotspot, all the fragments are shown if the total number is less than or equal to 5, with 5 selected fragments being shown when the number in that Hotspot was greater than 5. Use of the SILCS exclusion maps instead of the protein surface supplies information on the extent the protein may relax to allow for fragments in the different Hotspots to be linked. The utility of this is especially evident with the GPR40 receptor.
Analysis was first undertaken on the Androgen Receptor allosteric inhibitor FLA (Figure 7). Three Hotspots are present in the vicinity of the ligand with two sites directly in contact with FLA (Table 2). Spatially, the sites are well separated containing a variety of fragments with different functionalities in each site, consistent with the types of FragMaps in each Hotspot. Importantly, the regions between the FragMaps are unobstructed based on the SILCS exclusion map indicating the potential to chemically link those regions, although as this ligand is located on the protein surface the solvent accessible surface also shows accessibility between the Hotspots. Notable is the lack of FragMaps between the Hotspots indicating that these regions, which may be occupied by scaffolding elements in the designed ligands, do not contribute significantly to binding. However, analysis of the SILCS FragMaps at different contour levels may yield information on the types of linkers to insert between ring systems during ligand development.
Figure 7).
Androgen receptor (cartoon, cyan) with A) the allosteric modulator flufenamic acid (FLA, CPK, atom color) or B) selected Fragments for selected Hotspots (labeled and colored by rank). Shown are the SILCS exclusion maps (tan, solid surface), protein backbone (cyan, cartoon representation), selected Hotspots (vdW spheres, coloring based on mean LGFE scores, Table 3), and SILCS FragMaps with cutoff energies for visualization: Positive (cyan, −1.2 kcal/mol), Negative (orange, −1.2 kcal/mol), Apolar (green, −1.2 kcal/mol), H-bond donor (blue, −0.9 kcal/mol) and H-bond acceptor (−0.9 kcal/mol). Fragments shown include 1, 3, 9, 21, and 29, for site 3, 3, 11, 31, 42, 84 for site 17, and 48, 49, 61, 63c and 74 for site 34 (Figure S1 supporting information).
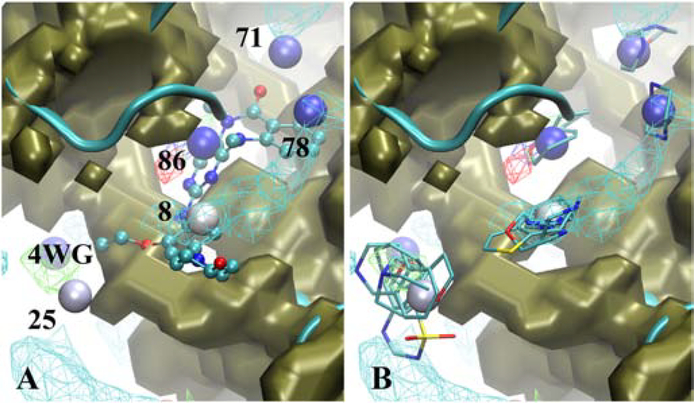
Two Erk5 inhibitors were analyzed in the present study, with the larger, 4WG, being a competitive inhibitor of ATP. Shown in Figure 8A is the crystallographic binding orientation of the ligand along with the Hotspots and SILCS FragMaps and exclusion map. A total of 5 Hotspots are within 5 Å of the ligand encompassing the full binding site. At each of these Hotspots, one or more fragments were present (Figure 8B). Heterocycle or cyclic groups containing polar atoms are present at sites 8 and 86, consistent with the functional groups in ligand 4WG. Hotspot 78 includes a piperazine which is not consistent with the aromatic ring adjacent to the site, though the presence of the positive FragMap consistent with that fragment is evident. This suggests the potential for including a charged moiety in that region. Hotspots 25 and 71 are beyond the extent of the ligand, again suggesting additional modifications that may be used to further improve ligand affinity and/or specificity. Analysis of the binding site in the crystal structure in the presence and absence of the solvent accessible surface (Figure S9) shows the central region of the ligand along with Hotspot 86 are under the surface of the protein, but this region is accessible as defined by the exclusion map. Similar results are seen with the allosteric Erk5 ligand 4QX (Figure S10). Three Hotspots are present in the vicinity of the ligand, with those containing apolar rings systems as well as those that include polar functionality. Again, a portion of the ligand is under the surface of the protein (Figure S11) which would disallow connection between functional groups at the different Hotspots. However, the ability to connect these regions is evident when the SILCS exclusion map along with the Hotspots are analyzed. These results reinforce the utility of the poses of the fragments in the different Hotspots and the ability of the SILCS methodology to identify opening of the protein between those sites allowing them to potentially be combined into larger, drug-like molecules targeting novel allosteric binding sites.
Figure 8).
Erk5 (cartoon, cyan) with A) the competitive inhibitor 4WG (CPK, atom color) or B) selected Fragments for selected Hotspots (labeled and colored by rank). Shown are the SILCS exclusion maps (tan, solid surface), protein backbone (cyan, cartoon representation), selected Hotspots (vdW spheres, coloring based on mean LGFE scores, Table 3), and SILCS FragMaps with cutoff energies for visualization: Positive (cyan, −1.2 kcal/mol), Negative (orange, −1.2 kcal/mol), Apolar (green, −1.2 kcal/mol), H-bond donor (blue, −0.9 kcal/mol) and H-bond acceptor (−0.9 kcal/mol). Fragments shown include 4b, 7b, 29, 71 and 84 for site 8, 38, 41, 45, 76 and 88 for site 25, 7b and 29 for site 71, 4b for site 78, 5 and 6 for site 86, (Figure S1 supporting information).
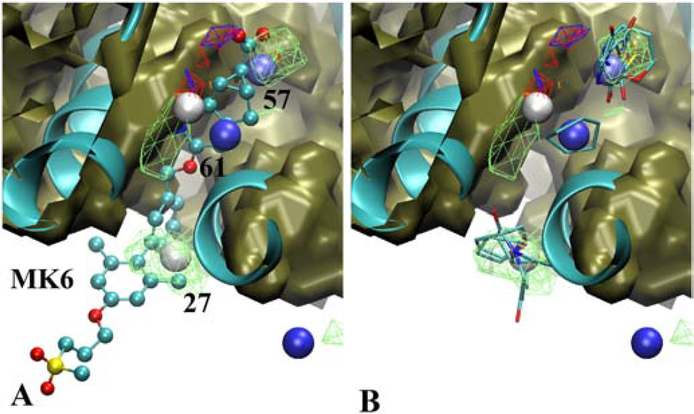
A particular interesting example is the positive allosteric modulator, MK6, at site 1 in the GPCR GPR40. As may be seen in Figure 9 this ligand penetrates into the interior of the GPCR between two transmembrane helices. Three Hotspots are identified that overlay with the crystallographic orientation of the ligand with those sites containing rings with various degrees of polarity consistent with the type of functionality in the ligand. Notably when the ligand is overlaid on the crystal structure used to initiate the SILCS simulations the portion of the ligand that penetrates the interior of the protein is totally under the protein surface (Figure S12). As with Erk5 above, analysis of the exclusion maps reveals that a path between the Hotspots is accessible that would allow for covalent connectivity between functional groups occupying the different Hotspots. The allosteric modulator at site 2 of GPR40, 7OS, occupies a binding pocket on the surface of the protein that is largely accessible in the crystal structure used for the SILCS simulations (Figure S13). Two Hotspots in that site are present, with the fragments occupying those sites corresponding to ring systems in the ligand.
Figure 9).
GPR40 (cartoon, cyan, PDB ID 5KW2) with A) the positive allosteric modulator MK6 (CPK, atom color) or B) selected Fragments for selected Hotspots (labeled and colored by rank). Shown are the SILCS exclusion maps (tan, solid surface), protein backbone (cyan, cartoon representation), selected Hotspots (vdW spheres, coloring based on mean LGFE scores, Table 3), and SILCS FragMaps with cutoff energies for visualization: Positive (cyan, −1.2 kcal/mol), Negative (orange, −1.2 kcal/mol), Apolar (green, −1.2 kcal/mol), H-bond donor (blue, −0.9 kcal/mol) and H-bond acceptor (−0.9 kcal/mol). Fragments shown include 6, 11, 15, 37, and 45 for site 27, 6, 35, 48, 74 and 77 for site 57 and 5 for site 61 (Figure S1 supporting information).
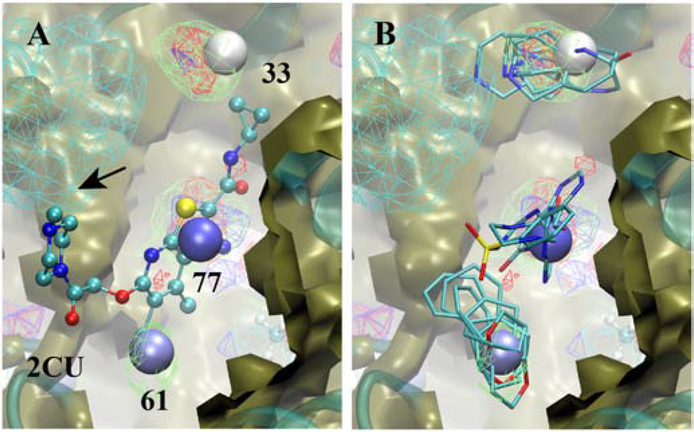
The final system subject to analysis is the allosteric binding site of the M2 muscarinic receptor (Figure 10). The ligand, 2CU, includes a central bicyclic, chlorinated heterocycle linked in an extended fashion through polar chains to cyclopropyl and piperazine moieties at each end. Hotspots are adjacent to the cyclopropyl, the heterocycle and the chlorine atom, identifying the locations of three of the important functional groups that could drive binding. These Hotspots coincide with rings with polar functionalities consistent with the FragMaps as well and the functional groups in the ligand (Figure 10A). Notable is the lack of Hotspots in the vicinity of the piperazine ring. This is due to the omission of Hotspots beyond a cutoff of 6 Å from protein Cα atoms as defined in the present analysis. However, the FragMaps indicated by the arrow in Figure 10A clearly show that a positively charged group would be desirable in this region. This information would indicate the need to adjust the cutoff distance and/or consider positively charged groups during ligand design.
Figure 10).
Allosteric binding site of the M2 muscarinic receptor showing the A) crystallographic orientation of allosteric modulator 2CU (CPK) and B) selected fragments from the Hotspots analysis. Included are the SILCS exclusion maps (tan, solid surface), protein backbone (cyan, cartoon representation), the three Hotspots (vdW spheres, coloring based on mean LGFE scores, Table 3), and selected SILCS FragMaps with cutoff energies for visualization: Positive (cyan, −1.2 kcal/mol), Negative (orange, −1.2 kcal/mol), Apolar (green, −1.2 kcal/mol), H-bond donor (blue, −0.9 kcal/mol) and H-bond acceptor (−0.9 kcal/mol). Fragments shown include 3, 37, 41, 45 and 84 for site 33, 6, 49, 61, 76 and 83 for site 66, and 4b, 52b, 63c and 88 for site 77 (Figure S1 supporting information).
Summary
SILCS-Hotspots analysis identifies a significant number of putative fragment binding sites in and around the entire protein. This represents a significant extension of the SILCS methodology that was previously limited to a single site on a target protein. While a subset of these Hotspots have more favorable mean LGFEs ranging from −3 to −4.8 kcal/mol, the majority of the Hotspots have mean LGFE scores of −3 kcal/mol or higher, which may be considered analogous to the previously described hot and warm spots, respectively.[18, 19] This predicts that proteins have a number of potential sites that may be targeted in fragment-based ligand design that can exploit multiple low affinity fragment binders to create high affinity drug-like ligands. Indeed, such sites may not be accessible to experimental methods used to identify fragment binding sites due to their low affinity,[36] though these sites are indicated to be of utility for ligand design in the present study. Along this line, recent crystallographic studies have identified larger numbers of fragment binding sites than in earlier studies. In these more recent studies, smaller, more polar compounds, termed MiniFrags, were used leading to the identification of an average of 10 fragment binding sites on a collection of 5 proteins.[18, 19] Moreover, those efforts have shown that multiple fragments bind to the same site, consistent with the results of the present study. Indeed, of the proteins selected in the present study, CDK2, has each of its ligands bound to two different sites on the protein. Clearly, to effectively perform fragment-based ligand design it is necessary to identify sites that are in the vicinity of each other allowing for the building of larger, more drug-like molecules. SILCS-Hotspots analysis allows for a range of putative sites to be identified for which a range of fragment affinities are predicted. However, it is noted that the Hotspots analysis does not definitively identify allosteric sites given the large number of sites encompassing the entire protein. At this stage of development of the methodology, user intervention is required to select potential allosteric sites based on the spatial relationship between the Hotspots and the potential to covalently link those sites to create larger, drug-like ligands.
A second aspect of going from individual fragments to larger ligands with drug-like characteristics is the ability to covalently link those fragments. While several of the allosteric ligands analyzed bind to pockets that are already present on the protein surface in the absence of the ligand, a number of the ligands, notably those binding to the kinases and the GPR40 GPCR, are in binding pockets that are not present in the crystal structures used to initiate the SILCS simulations that do not contain those ligands. The inclusion of protein flexibility in SILCS identifies regions of the protein that can undergo local conformational changes, defined in the context of SILCS exclusion maps, information that may be used to identify paths allowing for covalent connections between fragments. With Erk5 paths between fragment binding sites that are accessible in the crystal structures that include the ligands are not accessible in the incorrect form of the protein used in the SILCS simulations. In the case of GPR40, binding site 1 occupied by a positive allosteric modulator is not even present in the crystal structure used for SILCS-Hotspots analysis. These results speak to the ability of SILCS-Hotspots to not only identify a range of putative fragment sites but also to identify paths between those sites that are often not evident in crystal structures allowing the covalent connectivity between fragments to be made as required to design larger, drug-like ligands.
In addition to identification of fragment binding sites, the SILCS-Hotspots approach as applied to an individual fragment may be applied on full ligands that are more drug-like. For example, if no binding site for a known ligand for a protein has been identified SILCS-Hotspots may be used to sample the entire 3D structure to identify possible binding sites for the ligand. Alternatively, it may be of interest to see if alternate sites for a ligand may exist beyond, for example, the orthosteric site of a protein. Emphasizing the importance of such capabilities is the protein CDK2 for which 2 binding sites exist for each of the three allosteric modulators studied (Table 1). However, identification of binding sites for larger ligands is best performed targeting the form of the protein to which they bind (e.g., active or inactive form of a GCPR), as shown in previous studies from our laboratory showing specific SILCS FragMaps for the active and inactive forms of the β2-adrenergic receptor to differentiate between agonists and antagonists, respectively.[44] Such limitations are also evident with the kinases as seen in this study and are likely present when applying SILCS FragMaps to ligand optimization.[26]
Supplementary Material
Highlights.
The Site Identification by Ligand Competitive Saturation (SILCS) is extended to screen 100s of fragment-like molecules to identify fragment binding sites the encompass the entire protein and the specific fragments that bind to those sites.
The SILCS Hotspots method is shown to recapitulate the binding sites of known drug-like ligands to both allosteric and orthosteric sites on seven proteins.
Inclusion of protein flexibility in the SILCS simulations allows for identification of fragment binding sites beneath the protein surface along with accessible regions between fragment-binding sites as required to create covalent links between the individual sites in order to create drug-like ligands.
Both weak and strong fragment binding sites are shown to contribute to the binding sites of known allosteric and orthosteric ligands and, as such, both classes of sites should be considered during ligand design.
Acknowledgment
This work was supported by NIH grants R44GM109635 and R01GM131710 and the Samuel Waxman Cancer Research Foundation. The authors acknowledge computer time and resources from the Computer-Aided Drug Design (CADD) Center at the University of Maryland, Baltimore.
We thank Jeremy for the inspiration he has brought us in so many ways!
Abbreviations
- GCMC
Grand-Canonical Monte Carlo
- GPCR
G-protein coupled receptor
- MD
Molecular Dynamics
- SILCS
Site-identification by Competitive Saturation
Footnotes
Conflict of Interest. A.D.M. Jr. is co-founder and Chief Scientific Officer of SilcsBio, LLC. S.J. is an employee of SilcsBio LLC and S.K.L. was an employee of SilcsBio LLC when the studies were performed.
Supporting Information. Additional Supporting Information can be found online in the supporting information tab for this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wenthur CJ, Gentry PR, Mathews TP, Lindsley CW, Drugs for allosteric sites on receptors, Annu Rev Pharmacol Toxicol 54 (2014) 165–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abdel-Magid AF, Allosteric modulators: an emerging concept in drug discovery, ACS Med Chem Lett 6(2) (2015) 104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].De Smet F, Christopoulos A, Carmeliet P, Allosteric targeting of receptor tyrosine kinases, Nature Biotechnology 32 (2014) 1113. [DOI] [PubMed] [Google Scholar]
- [4].Usach I, Melis V, Peris J-E, Non-nucleoside reverse transcriptase inhibitors: a review on pharmacokinetics, pharmacodynamics, safety and tolerability, Journal of the International AIDS Society 16(1) (2013) 18567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Foster DJ, Conn PJ, Allosteric Modulation of GPCRs: New Insights and Potential Utility for Treatment of Schizophrenia and Other CNS Disorders, Neuron 94(3) (2017) 431–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huang M, Song K, Liu X, Lu S, Shen Q, Wang R, Gao J, Hong Y, Li Q, Ni D, Xu J, Chen G, Zhang J, AlloFinder: a strategy for allosteric modulator discovery and allosterome analyses, Nucleic Acids Res 46(W1) (2018) W451–W458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schmidtke P, Souaille C, Estienne F, Baurin N, Kroemer RT, Large-Scale Comparison of Four Binding Site Detection Algorithms, Journal of Chemical Information and Modeling 50(12) (2010) 2191–2200. [DOI] [PubMed] [Google Scholar]
- [8].Barril X, Druggability predictions: methods, limitations, and applications., WIREs Comput Mol Sci 3 (2013) 327–338. [Google Scholar]
- [9].Joseph-McCarthy D, Campbell AJ, Kern G, Moustakas D, Fragment-based lead discovery and design, J Chem Inf Model 54(3) (2014) 693–704. [DOI] [PubMed] [Google Scholar]
- [10].Broomhead NK, Soliman MEJCB, Biophysics, Can We Rely on Computational Predictions To Correctly Identify Ligand Binding Sites on Novel Protein Drug Targets? Assessment of Binding Site Prediction Methods and a Protocol for Validation of Predicted Binding Sites, 75(1) (2017) 15–23. [DOI] [PubMed] [Google Scholar]
- [11].Kulp JL 3rd, Cloudsdale IS, Kulp JL Jr., Guarnieri F, Hot-spot identification on a broad class of proteins and RNA suggest unifying principles of molecular recognition, PLoS One 12(8) (2017) e0183327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ngan CH, Bohnuud T, Mottarella SE, Beglov D, Villar EA, Hall DR, Kozakov D, Vajda S, FTMAP: extended protein mapping with user-selected probe molecules, Nucleic acids research 40(Web Server issue) (2012) W271–W275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schmidtke P, Le Guilloux V, Maupetit J, Tuff�ry P, fpocket: online tools for protein ensemble pocket detection and tracking, Nucleic Acids Research 38(suppl_2) (2010) W582–W589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Radoux CJ, Olsson TSG, Pitt WR, Groom CR, Blundell TL, Identifying Interactions that Determine Fragment Binding at Protein Hotspots, Journal of Medicinal Chemistry 59(9) (2016) 4314–4325. [DOI] [PubMed] [Google Scholar]
- [15].Roca C, Requena C, Sebastián-Pérez V, Malhotra S, Radoux C, Pérez C, Martinez A, Antonio Páez J, Blundell TL, Campillo NE, Identification of new allosteric sites and modulators of AChE through computational and experimental tools, Journal of enzyme inhibition and medicinal chemistry 33(1) (2018) 1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Seco J, Luque FJ, Barril X, Binding Site Detection and Druggability Index from First Principles, Journal of Medicinal Chemistry 52(8) (2009) 2363–2371. [DOI] [PubMed] [Google Scholar]
- [17].Ghanakota P, Carlson HA, Moving Beyond Active-Site Detection: MixMD Applied to Allosteric Systems, The Journal of Physical Chemistry B 120(33) (2016) 8685–8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].O’Reilly M, Cleasby A, Davies TG, Hall RJ, Ludlow RF, Murray CW, Tisi D, Jhoti H, Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design, Drug Discov Today 24(5) (2019) 1081–1086. [DOI] [PubMed] [Google Scholar]
- [19].Rathi PC, Ludlow RF, Hall RJ, Murray CW, Mortenson PN, Verdonk ML, Predicting “Hot” and “Warm” Spots for Fragment Binding, J Med Chem 60(9) (2017) 4036–4046. [DOI] [PubMed] [Google Scholar]
- [20].Lolli G, Caflisch A, High-Throughput Fragment Docking into the BAZ2B Bromodomain: Efficient in Silico Screening for X-Ray Crystallography, ACS Chem Biol 11(3) (2016) 800–7. [DOI] [PubMed] [Google Scholar]
- [21].Zhu J, Caflisch A, Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions, J Med Chem 59(11) (2016) 5555–61. [DOI] [PubMed] [Google Scholar]
- [22].Amato A, Lucas X, Bortoluzzi A, Wright D, Ciulli A, Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach, ACS Chem Biol 13(4) (2018) 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu J, Zhou C, Caflisch A, Structure-based discovery of selective BRPF1 bromodomain inhibitors, Eur J Med Chem 155 (2018) 337–352. [DOI] [PubMed] [Google Scholar]
- [24].Guvench O, MacKerell AD Jr, Computational Fragment-Based Binding Site Identification by Ligand Competitive Saturation, PLoS Comp. Biol 5 (2009) e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Raman EP, Lakkaraju SK, Denny RA, MacKerell AD Jr., Estimation of relative free energies of binding using pre-computed ensembles based on the single-step free energy perturbation and the site-identification by Ligand competitive saturation approaches, J Comput Chem (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ustach VD, Lakkaraju SK, Jo S, Yu W, Jiang W, MacKerell AD Jr., Optimization and Evaluation of Site-Identification by Ligand Competitive Saturation (SILCS) as a Tool for Target-Based Ligand Optimization, J Chem Inf Model (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Samadani R, Zhang J, Brophy A, Oashi T, Priyakumar UD, Raman EP, St John FJ, Jung KY, Fletcher S, Pozharski E, MacKerell AD, Shapiro PS, Small Molecule Inhibitors of ERK-mediated Immediate Early Gene Expression and Proliferation of Melanoma Cells Expressing Mutated BRaf, Biochem J 467 (2015) 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Heinzl GA, Huang W, Yu W, Giardina BJ, Zhou Y, MacKerell AD Jr., Wilks A, Xue F, Iminoguanidines as Allosteric Inhibitors of the Iron-Regulated Heme Oxygenase (HemO) of Pseudomonas aeruginosa, J Med Chem 59(14) (2016) 6929–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lakkaraju SK, Mbatia H, Hanscom M, Zhao Z, Wu J, Stoica B, MacKerell AD Jr., Faden AI, Xue F, Cyclopropyl-containing positive allosteric modulators of metabotropic glutamate receptor subtype 5, Bioorg Med Chem Lett 25(11) (2015) 2275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cardenas MG, Yu W, Beguelin W, Teater MR, Geng H, Goldstein RL, Oswald E, Hatzi K, Yang SN, Cohen J, Shaknovich R, Vanommeslaeghe K, Cheng H, Liang D, Cho HJ, Abbott J, Tam W, Du W, Leonard JP, Elemento O, Cerchietti L, Cierpicki T, Xue F, MacKerell AD Jr., Melnick AM, Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma, J Clin Invest 126(9) (2016) 3351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lanning ME, Yu W, Yap JL, Chauhan J, Chen L, Whiting E, Pidugu LS, Atkinson T, Bailey H, Li W, Roth BM, Hynicka L, Chesko K, Toth EA, Shapiro P, MacKerell AD Jr., Wilder PT, Fletcher S, Structure-based design of N-substituted 1-hydroxy-4-sulfamoyl-2-naphthoates as selective inhibitors of the Mcl-1 oncoprotein, Eur J Med Chem 113 (2016) 273–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cheng H, Linhares BM, Yu W, Cardenas MG, Ai Y, Jiang W, Winkler A, Cohen S, Melnick A, MacKerell A Jr., Cierpicki T, Xue F, Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design, J Med Chem 61 (2018) 7573–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang H, Jiang W, Chatterjee P, Luo Y, Ranking Reversible Covalent Drugs: From Free Energy Perturbation to Fragment Docking, J Chem Inf Model 59(5) (2019) 2093–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Donohue E, Khorsand S, Mercado G, Varney KM, Wilder PT, Yu W, MacKerell AD, Alexander P, Van QN, Moree B, Stephen AG, Weber DJ, Salafsky J, McCormick F, Second harmonic generation detection of Ras conformational changes and discovery of a small molecule binder, Proceedings of the National Academy of Sciences 116(35) (2019) 17290–17297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yu W, Lakkaraju SK, Raman EP, Fang L, MacKerell AD Jr., Pharmacophore Modeling Using Site-Identification by Ligand Competitive Saturation (SILCS) with Multiple Probe Molecules, J Chem Inf Model 55(2) (2015) 407–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mattos C, Ringe D, Locating and characterizing binding sites on proteins, Nat Biotechnol 14(5) (1996) 595–9. [DOI] [PubMed] [Google Scholar]
- [37].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM, J. Comput. Chem 29(11) (2008) 1859–1865. [DOI] [PubMed] [Google Scholar]
- [38].Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD Jr., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J. Comp. Chem 31(4) (2010) 671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD Jr., CHARMM36m: an improved force field for folded and intrinsically disordered proteins, Nature methods 14 (2017) 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fiser A, Do RKG, Sali A, Modeling of loops in protein structures, Prot. Sci 9 (2000) 1753–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Raman EP, Yu W, Guvench O, Mackerell AD, Reproducing crystal binding modes of ligand functional groups using Site-Identification by Ligand Competitive Saturation (SILCS) simulations, Journal of Chemical Information and Modeling 51(4) (2011) 877–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL 3rd, MacKerell AD Jr., Klauda JB, Im W, CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field, J Chem Theory Comput 12(1) (2016) 405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shim J, Coop A, MacKerell AD Jr., Molecular details of the activation of the mu opioid receptor, J Phys Chem B 117(26) (2013) 7907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lakkaraju SK, Yu W, Raman EP, Hershfeld AV, Fang L, Deshpande DA, MacKerell AD Jr., Mapping Functional Group Free Energy Patterns at Protein Occluded Sites: Nuclear Receptors and G-Protein Coupled Receptors, J Chem Inf Model 55 (2015) 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lakkaraju SK, Raman EP, Yu W, MacKerell AD Jr., Sampling of Organic Solutes in Aqueous and Heterogeneous Environments using Oscillating μex Grand Canonical-like Monte Carlo-Molecular Dynamics Simulations, J Chem Theory Comput 10(6) (2014) 2281–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hess B, Kutzner C, Van Der Spoel D, Lindahl E, Gromacs 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation, J. Chem. Theory Comput 4 (2008) 435–447. [DOI] [PubMed] [Google Scholar]
- [47].Raman EP, Yu W, Lakkaraju SK, Mackerell AD Jr., Inclusion of Multiple Fragment Types in the Site Identification by Ligand Competitive Saturation (SILCS) Approach, J Chem Inf Model 53(12) (2013) 3384–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang Y, Skolnick J, SPICKER: A clustering approach to identify near-native protein folds, 25(6) (2004) 865–871. [DOI] [PubMed] [Google Scholar]
- [49].Taylor RD, MacCoss M, Lawson AD, Rings in drugs, J Med Chem 57(14) (2014) 5845–59. [DOI] [PubMed] [Google Scholar]
- [50].Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG, ZINC: a free tool to discover chemistry for biology, J Chem Inf Model 52(7) (2012) 1757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pereira de Jesus-Tran K, Cote PL, Cantin L, Blanchet J, Labrie F, Breton R, Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity, Protein Sci 15(5) (2006) 987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ, A surface on the androgen receptor that allosterically regulates coactivator binding, Proc Natl Acad Sci U S A 104(41) (2007) 16074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Baumli S, Endicott JA, Johnson LN, Halogen bonds form the basis for selective P-TEFb inhibition by DRB, Chem Biol 17(9) (2010) 931–6. [DOI] [PubMed] [Google Scholar]
- [54].Wu SY, McNae I, Kontopidis G, McClue SJ, McInnes C, Stewart KJ, Wang S, Zheleva DI, Marriage H, Lane DP, Taylor P, Fischer PM, Walkinshaw MD, Discovery of a Novel Family of CDK Inhibitors with the Program LIDAEUS: Structural Basis for Ligand-Induced Disordering of the Activation Loop, Structure 11(4) (2003) 399–410. [DOI] [PubMed] [Google Scholar]
- [55].Betzi S, Alam R, Martin M, Lubbers DJ, Han H, Jakkaraj SR, Georg GI, Schonbrunn E, Discovery of a potential allosteric ligand binding site in CDK2, ACS chemical biology 6(5) (2011) 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ludlow RF, Verdonk ML, Saini HK, Tickle IJ, Jhoti H, Detection of secondary binding sites in proteins using fragment screening, Proceedings of the National Academy of Sciences 112(52) (2015) 15910–15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Glatz G, Gogl G, Alexa A, Remenyi A, Structural mechanism for the specific assembly and activation of the extracellular signal regulated kinase 5 (ERK5) module, J Biol Chem 288(12) (2013) 8596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen H, Tucker J, Wang X, Gavine PR, Phillips C, Augustin MA, Schreiner P, Steinbacher S, Preston M, Ogg D, Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site, Acta Crystallogr D Struct Biol 72(Pt 5) (2016) 682–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Montalibet J, Skorey K, McKay D, Scapin G, Asante-Appiah E, Kennedy BP, Residues distant from the active site influence protein-tyrosine phosphatase 1B inhibitor binding, J Biol Chem 281(8) (2006) 5258–66. [DOI] [PubMed] [Google Scholar]
- [60].Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, McDowell RS, Hansen SK, Allosteric inhibition of protein tyrosine phosphatase 1B, Nat Struct Mol Biol 11(8) (2004) 730–7. [DOI] [PubMed] [Google Scholar]
- [61].Wan ZK, Follows B, Kirincich S, Wilson D, Binnun E, Xu W, Joseph-McCarthy D, Wu J, Smith M, Zhang YL, Tam M, Erbe D, Tam S, Saiah E, Lee J, Probing acid replacements of thiophene PTP1B inhibitors, Bioorg Med Chem Lett 17(10) (2007) 2913–20. [DOI] [PubMed] [Google Scholar]
- [62].Han Y, Belley M, Bayly CI, Colucci J, Dufresne C, Giroux A, Lau CK, Leblanc Y, McKay D, Therien M, Wilson MC, Skorey K, Chan CC, Scapin G, Kennedy BP, Discovery of [(3-bromo-7-cyano-2-naphthyl)(difluoro)methyl]phosphonic acid, a potent and orally active small molecule PTP1B inhibitor, Bioorg Med Chem Lett 18(11) (2008) 3200–5. [DOI] [PubMed] [Google Scholar]
- [63].Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK, Crystal structure of the beta2 adrenergic receptor-Gs protein complex, Nature 477(7366) (2011) 549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu X, Ahn S, Kahsai AW, Meng KC, Latorraca NR, Pani B, Venkatakrishnan AJ, Masoudi A, Weis WI, Dror RO, Chen X, Lefkowitz RJ, Kobilka BK, Mechanism of intracellular allosteric beta2AR antagonist revealed by X-ray crystal structure, Nature 548(7668) (2017) 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Srivastava A, Yano J, Hirozane Y, Kefala G, Gruswitz F, Snell G, Lane W, Ivetac A, Aertgeerts K, Nguyen J, Jennings A, Okada K, High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875, Nature 513(7516) (2014) 124–7. [DOI] [PubMed] [Google Scholar]
- [66].Ho JD, Chau B, Rodgers L, Lu F, Wilbur KL, Otto KA, Chen Y, Song M, Riley JP, Yang HC, Reynolds NA, Kahl SD, Lewis AP, Groshong C, Madsen RE, Conners K, Lineswala JP, Gheyi T, Saflor MD, Lee MR, Benach J, Baker KA, Montrose-Rafizadeh C, Genin MJ, Miller AR, Hamdouchi C, Structural basis for GPR40 allosteric agonism and incretin stimulation, Nat Commun 9(1) (2018) 1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lu J, Byrne N, Wang J, Bricogne G, Brown FK, Chobanian HR, Colletti SL, Di Salvo J, Thomas-Fowlkes B, Guo Y, Hall DL, Hadix J, Hastings NB, Hermes JD, Ho T, Howard AD, Josien H, Kornienko M, Lumb KJ, Miller MW, Patel SB, Pio B, Plummer CW, Sherborne BS, Sheth P, Souza S, Tummala S, Vonrhein C, Webb M, Allen SJ, Johnston JM, Weinglass AB, Sharma S, Soisson SM, Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40, Nature Structural &Amp; Molecular Biology 24 (2017) 570. [DOI] [PubMed] [Google Scholar]
- [68].Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T, Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist, Nature 482(7386) (2012) 547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Korczynska M, Clark MJ, Valant C, Xu J, Moo EV, Albold S, Weiss DR, Torosyan H, Huang W, Kruse AC, Lyda BR, May LT, Baltos JA, Sexton PM, Kobilka BK, Christopoulos A, Shoichet BK, Sunahara RK, Structure-based discovery of selective positive allosteric modulators of antagonists for the M2 muscarinic acetylcholine receptor, Proc Natl Acad Sci U S A 115(10) (2018) E2419–E2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Boggavarapu R, Jeckelmann J-M, Harder D, Ucurum Z, Fotiadis D, Role of electrostatic interactions for ligand recognition and specificity of peptide transporters, BMC Biology 13(1) (2015) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hari SB, Merritt EA, Maly DJ, Sequence determinants of a specific inactive protein kinase conformation, Chem Biol 20(6) (2013) 806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Modi V, Dunbrack RL, Defining a new nomenclature for the structures of active and inactive kinases, Proceedings of the National Academy of Sciences 116(14) (2019) 6818–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Taylor SS, Radzio-Andzelm E, Three protein kinase structures define a common motif, Structure 2(5) (1994) 345–355. [DOI] [PubMed] [Google Scholar]
- [74].Dey F, Caflisch A, Fragment-based de novo ligand design by multiobjective evolutionary optimization, J Chem Inf Model 48(3) (2008) 679–90. [DOI] [PubMed] [Google Scholar]
- [75].Kathryn L, Ian A, Woody S, Computational Approaches for Fragment-Based and De Novo Design, Current Topics in Medicinal Chemistry 10(1) (2010) 14–32. [DOI] [PubMed] [Google Scholar]
- [76].Hao GF, Jiang W, Ye YN, Wu FX, Zhu XL, Guo FB, Yang GF, ACFIS: a web server for fragment-based drug discovery, Nucleic Acids Res 44(W1) (2016) W550–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Batiste L, Unzue A, Dolbois A, Hassler F, Wang X, Deerain N, Zhu J, Spiliotopoulos D, Nevado C, Caflisch A, Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple), ACS Cent Sci 4(2) (2018) 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.