Abstract
Formalin-fixed paraffin-embedded (FFPE) tissues provide an important resource for genomic research. However, variability in the integrity or quality of RNA obtained from archival FFPE specimens can lead to unreliable data and wasted resources, and standard protocols for measuring RNA integrity do not adequately assess the suitability of FFPE RNA. The main goal of this study was to identify improved methods for evaluating FFPE RNA quality for whole genome sequencing. We examined RNA quality metrics conducted prior to RNA-sequencing in paired frozen and FFPE samples with varying levels of quality based on age in block and time in formalin. RNA quality was measured by the RNA integrity number (RIN), a modified RIN called the paraffin-embedded RNA metric (PERM), the percentage of RNA fragments >100–300 nucleotides in size (DV100-300), and two quantitative PCR-based methods. This information was correlated to sequencing read quality, mapping, and gene detection. Among fragmentation-based methods, DV and PCR-based metrics were more informative than RIN or PERM in determining sequencing success. Across low- and high-quality FFPE samples, a minimum of 80% of RNA fragments >100 nucleotides (DV100>80) provided the best indication of gene diversity and read counts upon sequencing. The PCR-based methods further showed quantitative reductions in amplifiable RNA of target genes based on sample age and time in formalin that inform input quantity of RNA for sequencing. These results should aid in screening and prioritizing archival FFPE samples for retrospective analyses of gene expression.
Keywords: FFPE, transcriptomics, RNA integrity, RNA-sequencing, DV, PCR, biorepositories
INTRODUCTION
Biorepositories offer a vast untapped source of samples for the acquisition of genomic data in translational and clinical research. However, use of these resources for genomic analyses requires better methods for mitigating the effects of tissue fixation. The vast majority of archival tissue specimens are stored as formalin-fixed paraffin-embedded (FFPE) blocks. The combination of formalin fixation and age in block induces a wide range of nucleic acid modifications beyond simple fragmentation (Hoffman et al., 2015; Hester et al., 2016), and RNA obtained from archival FFPE tissue samples can vary widely in quality and suitability for genomic analysis, both within and across studies (Bass et al., 2014; Greytak et al., 2015). This issue may confound transcriptomic analyses and result in wasted time and resources. The standard method for assessing quality of RNA from fresh or frozen specimens is the RNA integrity number (RIN), which quantifies RNA intactness based on specific features from the RNA electropherogram trace (Imbeaud et al., 2005; Schroeder et al., 2006). However, RIN values alone do not adequately discern between low and high quality FFPE RNA or provide a reliable metric to guide sequencing decisions (Hester et al., 2016). Better methods are thus needed for evaluating FFPE RNA quality and determining the appropriate amount of RNA to be used in sequencing.
Several alternative analyses have been proposed in recent years for assessing FFPE RNA quality based on different features of the electropherogram trace. One approach involves quantifying the percent of RNA fragments greater than 200 nucleotides (DV200). This metric was first described in a technical report by Illumina which showed that higher DV200 values correspond with greater consistency in RNA-sequencing (RNA-seq) read counts among replicate FFPE samples (Illumina 2015). The report also proposed that samples with DV200 < 70% (70% of RNA fragments within the sample were less than 200 nucleotides in length)should have at least twice as much input RNA for sequencing library preparation and that those with DV200 < 30% should not be sequenced at all because of the short average RNA fragment sizes. However, few the peer-reviewed reports in literature have used this method (e.g., Landolt et al., 2016; Millier et al., 2017), and to our knowledge there are no studies that have systematically evaluated its performance in predicting RNA- seq success. A related method for assessing FFPE RNA quality is called the paraffin-embedded RNA metric (PERM) (Chung et al., 2016). This modified RIN is calculated from the area under the electropherogram trace weighted for more intact fragments. To date, the PERM method has been compared to RIN and quantitative reverse transcription polymerase chain reaction (RT-PCR) results, but not RNA-seq outcomes.
RNA from FFPE samples is not only highly fragmented but also biochemically modified by adducts and other cross-links that impede cDNA synthesis (Masuda et al., 1999; von Ahlfen et al., 2007; Evers et al., 2011). These changes can result in lower effective RNA inputs and potentially reduce sequencing fidelity for transcriptomic applications. Quantitative real-time-PCR (qPCR) and, more recently, digital droplet PCR (ddPCR) platforms have been used to study this issue in degraded samples (e.g., Nolan et al., 2006; von Ahlfen et al., 2007; Millier et al., 2017). However, these methods have not been directly compared to RIN, DV, and PERM methods in the assessment of sequencing results from FFPE samples.
In the present experiment, we evaluated RNA from archival FFPE samples ranging from high to low quality based on age in block and fixation protocol. We begin by presenting retrospective analyses of two studies, one with FFPE samples collected relatively recently (Study 1), and the other with older FFPE samples (Study 2) to investigate the influence of age in paraffin block on RNA quality. We conclude with a controlled prospective investigation into the effects of time in formalin on FFPE sample RNA quality (Study 3). Methods of measuring RNA quality included RIN, fragment analysis, PERM, and real-time qPCR and ddPCR amplification and quantitation. These metrics were compared to RNA-seq results from the same samples across each study. Our goals were to evaluate and compare pre-sequencing RNA metrics for predicting RNA-seq quality derived from a heterogeneous batch of archival FFPE samples.
MATERIALS AND METHODS
Experimental overview
Quality metrics for FFPE RNA were evaluated in corresponding frozen (FR) and FFPE sample pairs from three different RNA-seq experiments, described previously in Hester et al. (2016) and Wehmas et al. (2018). All studies used liver samples from male B6C3F1 mice, which were housed in an AAALAC-accredited animal facility located in Research Triangle Park, NC. All procedures involving animals were approved by the U.S. Environmental Protection Agency (U.S. EPA) Institutional Animal Care and Use Committee.
In Study 1 (Hester et al., 2016), we used “high quality” FFPE samples that had been archived for <2 years. At collection, liver tissue was either snap frozen or fixed in fresh 4% paraformaldehyde (methanol-free 10% formalin) for 18–24 hours before being transferred to 70% ethanol. Mice had been treated for 7 days with di-2-ethylhexyl phthalate (DEHP) in the feed at 0, 1500, 3000, or 6000 ppm (n=16 FR, n=16 FFPE, n=4 per dose group). RNA-seq data from this experiment showed 35% lower total counts overall in FFPE compared to FR samples but high concordance in fold-change values for differentially expressed genes (DEGs), enriched pathways, and benchmark dose estimates for target genes resulting from DEHP treatment
In Study 2 (Hester et al., 2016), we used “low quality” FFPE samples that had been stored at room temperature in a biorepository for >21 years. At collection, liver tissue was either frozen or fixed in 10% buffered formalin for 18–24 hours before being transferred to 70% ethanol. Mice had been treated for 6 days with dichloroacetic acid (DCA) in the drinking water at 0, 1.0, 2.0, or 3.5 g/L (n=24 FR, n=24 FFPE, n=6 per dose group). RNA-seq data from this experiment showed 97% lower total counts in FFPE compared to FR samples and poor concordance in global DEGs and target pathways. Only the most highly-induced DEGs correlated between FR and FFPE sample pairs.
In Study 3 (Wehmas et al., 2018), we used FFPE samples of varying quality based on fixation time in formalin. Frozen liver specimens were divided into four preservation groups as follows: (1) FR; (2) 70% ethanol fixation for 3 months (OH); (3) 10% neutral buffered formalin for 18 hours followed by 70% ethanol to 3 months (18F); and (4) 10% neutral buffered formalin for 3 months (3F). All samples from OH, 18F, and 3F groups were maintained at 4°C until 3 months and then processed by standard histological methods into paraffin blocks. The 18-hour fixation time was selected to represent a standard protocol for clinical and experimental samples (Fox et al., 1985). The 3-month formalin fixation time point was selected as an arbitrary long-term fixation scenario to increase formalin-induced RNA degradation. FFPE blocks were stored at room temperature for <4 months until sectioning for RNA isolation. Mice had been treated for 7 days with phenobarbital (PB) in the drinking water at 0 or 600 ppm (n=12 FR, OH, 18F, and 3F FFPE groups; within each condition, n=6 for control and PB treatment groups). RNA-seq metrics varied by fixation protocol; among FFPE groups, data quality was generally highest for OH and lowest for 3F compared to FR (Wehmas et al., 2018). Total counts were around 23% and 48% lower for 18F and 3F groups respectively compared to FR. While concordance with FR DEGs and significantly enriched signaling pathways were lower with time in formalin, R2 values remained high between FR and 18F or 3F groups (>0.979 for both).
RNA isolation and demodification
For all studies, total RNA was isolated from ~30–50 mg of frozen liver tissue following homogenization in RNAzolRT (Molecular Research Center, Cincinnati, OH) and then purified by RNeasy MinElute column according to manufacturer recommendations (Qiagen GmbH, Hilden, Germany). For FFPE samples, total RNA was isolated from two to four paraffin sections at 10 μm in thickness. The number of sections used for each isolation was adjusted based on the area of liver tissue preserved in paraffin to obtain approximately equal amounts of RNA per specimen. Sections were collected with a Historange or Leica microtome under RNase-free conditions, deparaffinized (Qiagen Deparaffinization Solution, cat. #19093), digested with proteinase K, and purified according to the Qiagen AllPrep® DNA/RNA kit protocol (Qiagen, cat. #80234). The concentration of RNA obtained from all samples was measured using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Qubit fluorometer (ThermoFisher Scientific, Waltham, MA). All RNA samples were stored at −80°C.
Pre-sequencing RNA quality evaluation
RNA integrity (RIN), PERM, and DV values were based on electropherogram traces from an Agilent 2100 Bioanalyzer (Agilent Technologies GmbH, Berlin, Germany). The RIN is derived from a mathematical formula that assimilates features of the RNA electropherogram plot, including 18S and 28S peaks (Schroeder et al., 2006). The PERM is a weighted sum of the fluorescent units at 5-second intervals from 25–70 seconds on the Agilent 2100 Bioanalyzer trace (Chung et al., 2016). The DV analysis is based on the percentage of RNA fragments greater than 100, 150, 200, 250, or 300 (DV100 – DV300) using the Smear Analysis function on the Agilent 2100 Bioanalyzer, as reported by Illumina (Illumina 2015). Both DV and PERM can easily be calculated from the Bioanalyzer output when the RIN is measured.
RNA was also evaluated by real-time qPCR and ddPCR to estimate FFPE mRNA integrity, based on methods adapted from Nolan et al. (2006). Briefly, three sets of primers and probes targeting different regions across the housekeeping gene beta-actin (Actb) were designed using Integrated DNA Technologies (IDT) PrimerQuest software, confirmed in Primer-BLAST (NCBI), and synthesized by IDT, Inc. (Coralville, IA). The housekeeping gene was used as a surrogate indicator of overall RNA quality. Primer and probe sequences, amplicon lengths, and relative locations can be found in Fig. 1A. Oligo(dT)-primed cDNA was prepared using iScript™ Select cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA) according to manufacturer specifications. For ddPCR, 5’-Actb/Mid-Actb/3’-Actb primers and 5’-Actb-FAM/Mid-Actb-HEX/3’-Actb-FAM probes were multiplexed in a single reaction. All samples were run in duplicate with the QX200 Droplet Digital PCR System using ddPCR Supermix for Probes (no dUTP) according to the manufacturer’s instructions (Bio-Rad Laboratories, Inc.). The ddPCR method provides absolute quantification of copy number with no standard curve required. Data was analyzed using QuantaSoft software (Bio-Rad Laboratories, Inc.). The limit of quantification was set by study, identifying the lowest concentration samples where the coefficient of variation between technical replicates was generally less than 25–30%. Samples with coefficients of variation consistently higher than 25–30% or concentration within 95% CI of the no template controls were assigned 0.0001 copies. Missing samples not tested were identified as NA. For real-time qPCR, primers for 5’-Actb/Mid-Actb/3’-Actb and 5’-Actb-FAM/Mid-Actb-HEX/3’-Actb-TexasRed hydrolysis probes were multiplexed into a single reaction prepared using PrimeTime® Gene Master Mix (IDT, Inc.) and run according to manufacturer instructions in triplicate with recommended adjustments from Nolan et al. (2006). Copy numbers of each amplicon were based on 10-fold dilution standard curves run in triplicate with each reaction. Real-time qPCR analysis was performed using CFX Manager Software (v3.1) (Bio-Rad Laboratories). The limit of quantification was set by study, assessing the standard curve and identifying the lowest concentration samples where the coefficient of variation between technical replicates was generally less than 40%. Samples with concentrations below this threshold or samples with Cq values greater than 36 were assigned 0.0001 copies. Missing samples not tested were identified as NA.
Figure 1.
(A) Location and sizes of amplicons targeting Actb with primer and probe sequences and assay concentrations. (B) Locations and sizes of B2m amplicons with primer sequences and assay concentrations.
Following absolute quantification of 5’-Actb and 3’-Actb by real-time qPCR and ddPCR, the 5’:3’ ratio of each sample, was calculated by dividing copies of 5’-Actb by −3’-Actb. Mid-Actb was only to be used in the 5’:3’calculation if no 3’-Actb amplification occurred, however mid-Actb was not used to calculate the 5’:3’ ratio for any of the studies. According to Nolan et al. (2006), ratios of 1–5 are considered high quality as there is near equal amounts of the 5’-most and 3’-most amplicons, indicating long intact RNA fragments. Samples with ratios of 6–10 are considered of moderate quality with some RNA fragmentation, and samples with ratios >10 are considered poor quality as there is much more 5’-most amplicon than 3’, indicating shorter RNA. Of note, when Nolan et al. (2006) calculated the 5’:3’ ratio on recent paraffin embedded tissue (<1 yr. old) fixed previously in formalin for 18 hrs. (i.e. high quality FFPE tissue samples), they obtained values around ~100.
A second ddPCR-based method, measured three regions of varying size (80, 127, and 429 base pairs) and location across the housekeeping gene beta-2 microglobin (B2m). Primers were designed using IDT PrimerQuest software, confirmed in Primer-BLAST (NCBI), and synthesized by IDT, Inc. Primer sequences, amplicon lengths, and relative locations can be found in Fig. 1B Oligo(dT) and random hexamer primed cDNA was prepared using iScript™ Advanced cDNA Synthesis Kit (Bio-Rad Laboratories, Inc.) according to manufacturer specifications. Each B2m amplicon was run separately across all samples in duplicate with the QX200 Droplet Digital PCR System using EvaGreen ddPCR Supermix according to the manufacturer’s instructions (Bio-Rad Laboratories, Inc.). Data was analyzed using QuantaSoft software (Bio-Rad Laboratories, Inc.). The limit of quantification was set by study, identifying the lowest concentration samples where the coefficient of variation between technical replicates was generally less than 25–30%. Samples with coefficients of variation consistently higher than 25–30% or concentration within 95% CI of the no template controls were assigned 0.0001 copies.
RNA-sequencing and analysis
RNA library preparation and sequencing were completed at Expression Analysis (EA Genomic Services, Q2 Solutions -- a Quintiles Quest Joint Venture, Durham, NC), as described previously (Hester et al., 2016; Wehmas et al., 2018). Degraded FFPE RNA underwent reduced or no fragmentation during library preparation depending on Bioanalyzer profiles. RNA was ribodepleted with Ribo-Zero Gold Library Prep Kit and cDNA libraries were synthesized using the TruSeq Stranded Total RNA (Illumina, San Diego, CA; cat. #RS-122–2303). Each sample was sequenced to at least 25 million paired-end reads using 50 base pair sequencing on Illumina HiSeq instruments. Average sequencing depth for FR and FFPE samples were 32.0 and 30.4 million reads per sample, respectively, for Study 1, and 25.9 and 18.1 million reads per sample for Study 2. In Study 3, average sequencing depth for FR, 18F and 3F samples were 34.2, 33.3, and 33.0 million reads per sample, with read quality (Q) scores >33.0 regardless of preservation method (Table S1).
RNA-seq data were analyzed as described previously (Hester et al., 2016; Wehmas et al., 2018). Briefly, basecall files were transformed into FASTQ files, and reads were trimmed and quality-filtered. For Studies 1 and 2, FASTQ files were aligned to the Mus musculus reference genome (GRCm38/mm10) and transcriptome (RefSeq Transcripts – 2014-10-17) using Star (v 2.4) aligner in Partek Flow (v4.0.15 or v5.0.16, St. Louis, MO). For Study 3, FASTQ files were aligned to the Mus musculus reference genome (GRCm38/mm10) and transcriptome (RefSeq Transcripts - 2015-02-02) using STAR (v 2.4.1d) aligner in Partek Flow (v 5.0.16.0719). Gene and transcript counts were generated using Partek Expectation Maximization algorithm. Gene features in Studies 1 and 2 were normalized to reads per kilobase of exon per million mapped reads (RPKM) and filtered for low expression (RPKM ≤ 10). Gene features in Study 3 were filtered by geometric average < 1 across all samples. An offset of 0.0001 was applied to compensate for zero counts in the data set prior to total count.
Other statistical methods
Due to the variability in normality and homogeneity of variance across much of the data, statistical comparisons were made using nonparametric tests. As the major focus of this manuscript is the influence of preservation methods on RNA quality, the Wilcoxon signed rank test was used to determine how RNA quality metrics were influenced by preservation. However, to ensure that chemical treatment was not confounding preservation effects, we also used the Wilcoxon signed rank test to investigate the effect of chemical treatment within each experiment. Supplementary information on potential chemical treatment effects may be found in Table S2. There are many quality metrics associated with RNA-seq data that check library preparation, sequencer functionality, sample contamination, and read integrity, but it can be difficult to identify one metric that comprehensively captures sequencing success as well as data integrity. To help address this issue, we applied a summary metric, generated a priori, that measures the quantity and variety of genes identified by multiplying total gene counts by the number of unique genes detected, which we refer to as the quantity diversity index, or QDI. We also appropriated a well-established metric from ecology to quantify gene abundance and uniqueness across samples through calculating a Hill Number, which we refer to as effective gene number (Lou 2007). Total RNA concentration, DV, PERM, RIN, and qPCR results were correlated with RNA-seq global outputs (alignment and mapping parameters) and gene-level outputs, including the QDI and effective gene number, using the Spearman method with rho indicating the correlation value. Analyses were conducted using the stats package in R (v 3.3.1) (R Core Team 2016). All group values presented in results indicate mean ± standard error (SE) unless otherwise indicated. Statistical significance was defined as a p-value < 0.05.
RESULTS
Study 1: High-quality (<2-year-old) FFPE samples
Total and amplifiable RNA concentrations.
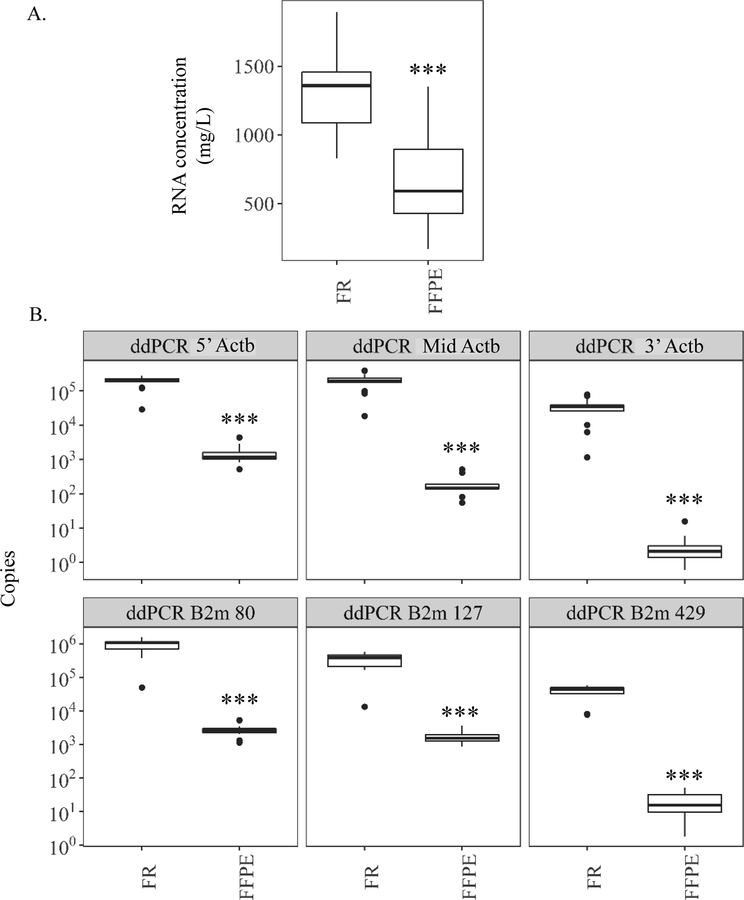
Prior to sequencing, we detected that higher quality FFPE samples (<2 yrs. in block) had ~2-fold less concentrated RNA than paired FR samples (1306 ± 65.4 mg/L) (Fig. 2A). Measuring polymerase-amplifiable RNA revealed even greater differences in concentration between FFPE and FR controls. While the absolute copies of Actb detected at each amplicon by ddPCR was generally higher than real-time qPCR in both FFPE and FR samples, the fold change differences between FFPE and FR samples across 5’-Actb and Actb2 showed similar trends (Fig. 2B). For instance, ddPCR and real-time qPCR detected the smallest difference between the two preservation methods at 5’-Actb (124 and 2,499-fold, respectively), the amplicon nearest the 5’ end of oligo dT primed cDNA. The fold change differences between FFPE and FR increased in magnitude for Mid-Actb, which is located farther 3’. Digital droplet PCR and real-time qPCR methods detected 1,089 and 34,130-fold fewer copies of Mid-Actb, respectively, in FFPE samples vs. FR. For 3’-Actb, ddPCR measured an order of magnitude increase in the disparity between FFPE and FR samples compared to ddPCR amplified Mid-Actb. This translated into 10,876-fold fewer copies of 3’-Actb measured by ddPCR in FFPE samples compared to FR. real-time-based qPCR quantified 10780-fold fewer copies 3’-Actb in FFPE samples compared to FR (Table S3).
Figure 2.
Study 1 quantification of total and amplifiable RNA within mouse liver samples aged in formalin-fixed paraffin-embedded (FFPE) blocks 2 years or paired frozen (FR) controls. (A) RNA concentration as measured by fluorometric methods using Qubit. (B – Top) Digital drop PCR (ddPCR) amplification of two common housekeeping genes (Actb and B2m) was used as surrogates for overall RNA quality. Amounts of Actb measured by ddPCR across three similarly sized amplicons. Amplicons located across the gene body 5’ to 3’ (5’-Actb = 5’ most amplicon based on cDNA; Mid-Actb = middle amplicon; and 3’-Actb = 3’ most amplicon based on cDNA). (B – Bottom) Amounts of three differently sized (80, 127, and 429 base pairs) B2m amplicons spanning the gene body. Amplicon 80 spans exons 2–3. Amplicon 127 spans exons 1–2, and amplicon 429 spans exons 2–4. *** p value <0.001, paired Wilcoxon signed rank test.
Substantial size-dependent measurement variations were observed between FFPE and FR samples for amplicons size 80, 127, and 429 across the second housekeeping gene B2m. The location of the amplicon was not as important for B2m detection because the cDNA was prepared with random hexamer and oligo dT primers as opposed to the oligo dT only primed cDNA synthesized for Actb detection. The most copies of B2m across FFPE and FR were detected within the shortest amplicon, B2m 80, with a little less detected within the slightly larger B2m 127 (Table S3). Fold differences detected between FFPE and FR across B2m 80 and B2m 127 (335 and 204-fold, respectively) were relatively similar to 5’-Actb and these changes were modest compared to those seen between FFPE and FR (1836-fold) for the largest amplicon, B2m 429 (Fig. 2B, Table S3).
Pre-sequencing RNA quality.
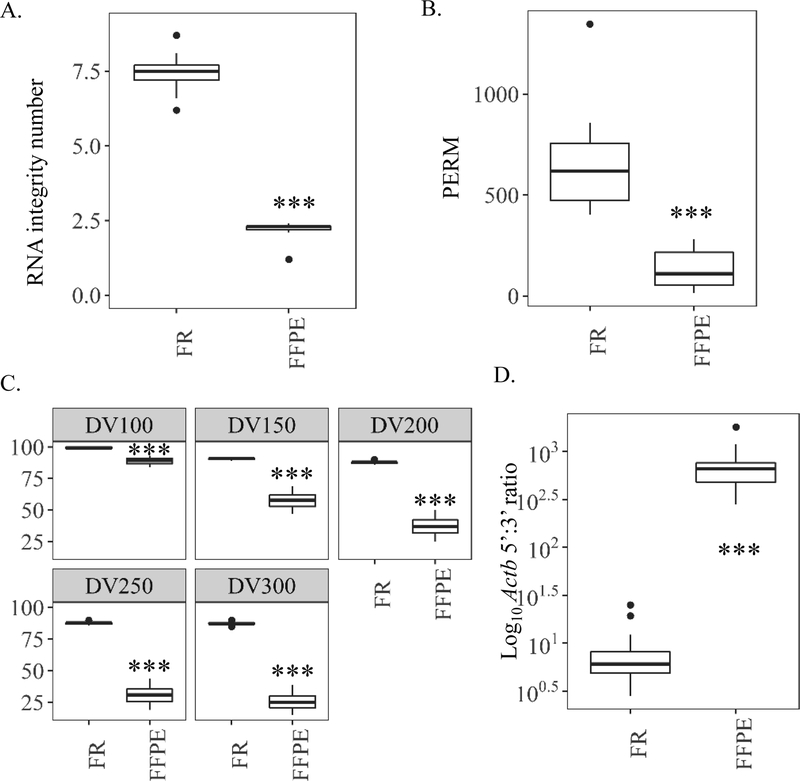
Measurements of RNA quality by RIN, PERM, and DV fragment analysis (all Bioanalyzer-based metrics) showed clear differences between FFPE and FR samples. Processing to FFPE block significantly reduced the RIN by 3.4-fold relative to FR (7.5 ± 0.1) (Fig. 3A), while a more dramatic reduction was seen in PERM values (5.0-fold lower vs. FR) (Fig. 3B). In FFPE samples, DV analyses detected a larger proportion of RNA at smaller size cutoffs. For instance, only 37% of FFPE samples was > 200 nucleotides (DV200 =37%) whereas DV200 = 88% for paired FR samples. For those same samples DV100 = 89% for FFPE while DV100 = 99% for FR (Fig. 3C). Generally, the DV100–300 remained relatively consistent (99–87%) for FR samples. The ddPCR 5’:3’ ratio also showed marked differences between paired FR and FFPE samples. For example, the average 5’:3’ Actb ratio (5’-Actb/3’-Actb) was much higher in FFPE samples (709 ± 113) compared to FR (9 ± 2), indicating enhanced degradation of Actb mRNA from FFPE samples (Fig. 3D, Table S3).
Figure 3.
Study 1 RNA quality measurements from mouse liver samples aged in formalin-fixed paraffin-embedded (FFPE) blocks 2 years relative to paired frozen (FR) controls. (A) RNA quality methods based on Bioanalyzer electropherogram traces: RNA integrity number (RIN). (B) Paraffin embedded RNA metric (PERM) to quantify RNA quality using a weighted area under the curve via Bioanalyzer spectropherogram output. (C) Fragment analysis (DV) to quantify the percent of RNA fragments greater than a specified value (100, 150, 200, 250, or 300 nucleotides) by Bioanalyzer smear test. (D) Ratio of similarly sized 5’ most Actb amplicon to 3’ most amplicon copies (5’-Actb/3’-Actb) as measured by digital drop PCR. Values of less than 10 indicate more intact, higher quality RNA. *** p value <0.001, paired Wilcoxon signed rank test.
Sequencing results.
We observed several preservation-related differences in RNA-seq results, as described previously in Hester et al. (2016). Dramatic differences were seen for read duplications (1.2-fold), deletion rates (3.7-fold), and percent reads mapping to ribosomal RNA (5.4-fold), which were significantly higher in 2-year-old FFPE samples compared to FR. Metrics that were significantly reduced in FFPE vs. FR samples included the percent of aligned reads (1.1-fold), and total gene counts (1.7-fold) (Hester et al., 2016; Table S3). The QDI was about 1.7-fold lower in FFPE samples compared to FR (260 ± 12.8 billion). In contrast, the effective gene number was slightly higher (1.1 fold) in FFPE samples relative to FR (1245 ± 24.2) (Table S3). Total sequencing clusters, total reads, average read lengths, average read quality, and average coverage depth did not show large differences in FFPE compared to FR samples (Table S1, Table S3).
Correlating quantity and quality with RNA-sequencing success.
There was poor correlation between total RNA and amplifiable RNA measured by qPCR among FFPE samples except for Mid-Actb measured using the real-time qPCR method. However, it correlated negatively with total RNA concentration (rho = −0.654) (Table S4). Similarly, there was poor concordance between total RNA and QDI or effective gene number (Table S4). There were some improvements for comparisons between ddPCR amplified RNA and QDI or effective gene number (Fig. S1, Table S4). The Actb amplicon nearest the 5’ end of cDNA (5’-Actb) trended toward a significant correlation with QDI (rho = 0.489), and B2m 127, near in size to 5’-Actb, significantly correlated with QDI (rho = 0.654) (Fig. S1, Table S4). Similar but slightly better correlation values were seen between amplifiable RNA from Actb and effective gene number but not with B2m and effective gene number (Fig. S2). Both real-time qPCR and ddPCR methods identified significant correlations between 5’-Actb and Mid-Actb with effective gene number (rho ≥ 0.577) (Table S4). All metrics to assess RNA quality prior to sequencing (RIN, PERM, DV, and 5’:3’ ratio) correlated poorly with QDI with RIN, PERM, and 5’:3’ ratio possessing negative correlation coefficients (Fig. S3A–B). These association did not improve much relative to effective gene number with PERM correlating negatively (Fig. S4A–B). However, all DV100–300 positively associated with QDI (Fig. S3c) and significantly so with effective gene number (max rho = 0.604) (Fig. S4c, Table S4).
Study 2: Low-quality (>21 year-old) FFPE samples
Total and amplifiable RNA concentrations.
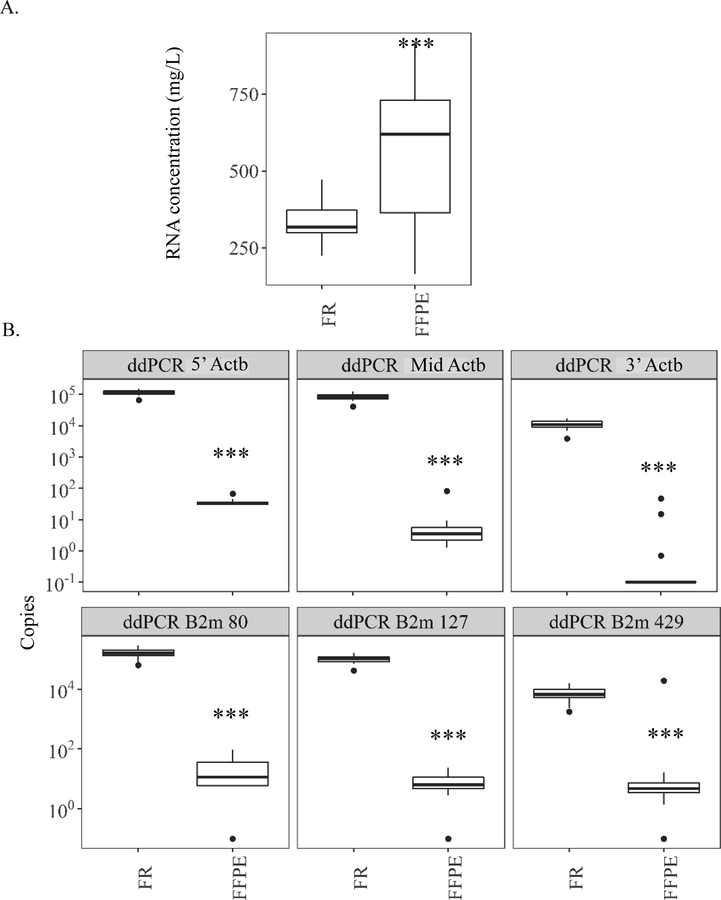
Older low-quality FFPE samples in block for >21 years showed mixed results on RNA amounts. Total RNA concentration was higher in FFPE sections by 1.7-fold compared to FR samples (Fig. 4A), while amplifiable RNA was dramatically lower Significantly fewer copies of the housekeeping genes Actb and B2m were measured in FFPE compared to FR samples (Fig. 4B). However, the magnitude of the difference depended upon location of the amplicon for Actb and size of amplicon for B2m. As with Study 1, the absolute amounts of Actb measured in paired FFPE and FR samples by real-time qPCR and ddPCR were not the same (Table S5). Many of the FFPE samples > 21yr. old fell outside real-time qPCR based detection limits, unlike ddPCR, making comparisons between FFPE and FR samples less reliable. Relative to FR, FFPE samples had 3318-fold fewer copies of 5’-Actb and 11460-fold fewer copies of Mid-Actb compared to FR when measured by ddPCR (Fig. 4B). Unlike Study 1, this trend did not continue with 3’-Actb, which had 3,959-fold fewer copies of FFPE compared to paired FR sample (Fig. 4B, Table S5).
Figure 4.
Study 2 on effects of long age in block on quantification of amplifiable RNA within mouse liver samples stored as formalin-fixed paraffin-embedded (FFPE) blocks for over 21 years relative to paired frozen (FR) controls. Digital drop PCR (ddPCR) amplification of two common housekeeping genes (Actb and B2m) was used as surrogates for overall RNA quality. (A) RNA concentration as measured by fluorometric methods using Qubit. (B – Top) Amounts of Actb measured by ddPCR across three similarly sized amplicons. Amplicons located across the gene body 5’ to 3’ (5’-Actb = 5’ most amplicon based on cDNA; Mid-Actb = middle amplicon; and 3’-Actb = 3’ most amplicon based on cDNA). (B – Bottom) Amounts of three differently sized (80, 127, and 429 base pairs) B2m amplicons spanning the gene body. Amplicon 80 spans exons 2–3. Amplicon 127 spans exons 1–2, and amplicon 429 spans exons 2–4. An offset of 0.01 was applied to zero copies detected for Actb and B2m amplicons so that samples with zero could still appear in the log10 scale of the figure. *** p value <0.001, paired Wilcoxon signed rank test.
B2m also showed large contrasts in the amount of amplicon detected in aged FFPE vs. FR samples. The degree of these changes depended mainly upon length of the amplicon. The moderately long B2m 127 amplicon had 13,256-fold fewer copies detected in FFPE samples compared to matching FR. The shortest amplicon, B2m 80, had a 7,385.6-fold difference between FFPE and FR samples, which was more similar to that of 5’-Actb ddPCR results. The longest amplicon, B2m 429, had only 9-fold more RNA in FR compared to FFPE (Fig. 4B, Table S5).
Pre-sequencing RNA quality.
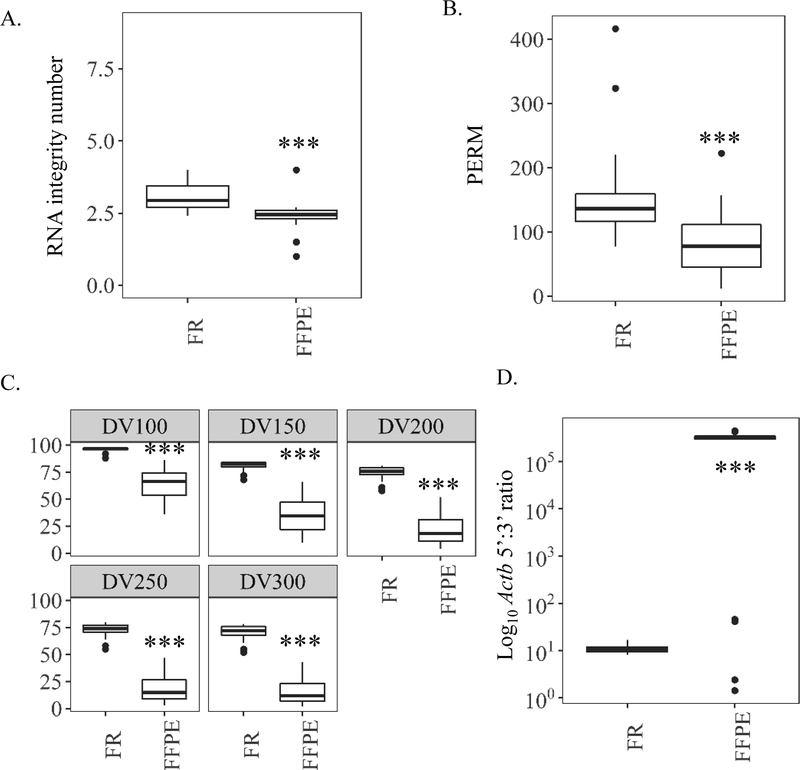
Differences between FR and older FFPE samples were discernable across all RNA quality metrics. For example, FR samples had 1.3-fold greater RIN values and ~1.7-fold greater PERM values compared to paired FFPE samples (Fig 5A–B). Larger differences between paired FFPE and FR samples were observed for DV values across all nucleotide lengths. For example, DV200 = 22% for FFPE samples and DV200 = 75% for FR. For the same FFPE samples, DV100 = 64% and DV100 = 93% for matching FR indicating a much larger proportion of the RNA from the FFPE samples is within 100–200 nucleotides in length (Fig. 5C, Table S5). The 5’:3’ ratios calculated from ddPCR showed high degradation for older FFPE (>281k) compared to matching FR (11) samples (Fig. 5D). Real-time qPCR-based methods could not calculate the 5’:3’ ratio for older FFPE samples due to limited signal detection in 19/22 samples; ratios in corresponding FR samples were modestly lower than those obtained by ddPCR (6) (Table S5).
Figure 5.
Study 2 on the influence of >21 years in formalin fixed paraffin embedded (FFPE) block relative to paired frozen (FR) controls on mouse liver RNA concentration and quality. (A) RNA quality methods based on Bioanalyzer electropherogram traces: RNA integrity number (RIN). (B) Paraffin embedded RNA metric (PERM) to quantify RNA quality using a weighted area under the curve via Bioanalyzer spectropherogram output. (C) Fragment analysis (DV) to quantify the percent of RNA fragments greater than a specified value (100, 150, 200, 250, or 300 nucleotides) by Bioanalyzer smear test. (D) Ratio of similarly sized 5’ most Actb amplicon to 3’ most amplicon copies (5’-Actb/3’-Actb) as measured by digital drop PCR. Values of less than 10 indicate more intact, higher quality RNA. *** p value <0.001, paired Wilcoxon signed rank test.
Sequencing results.
RNA-seq metrics for aged FFPE samples showed enhanced indicators of degradation compared to <2-year-old samples, as summarized in Hester et al. (2016, Table S1 and Table S5). Similar to Study 1, there was a significant but greater level of read deletion rates (defined as the total number of deleted bases, divided by the total number of mapped bases), (2.1-fold) as well as % read duplications (7.2-fold) in aged FFPE samples compared to FR. The % reads mapped to ribosomal RNAs was 1.5-fold higher in FR compared to FFPE samples. However, these FFPE samples also had significantly lower number of mapped reads (2.2 fold), and total gene counts (7.4 fold) compared to paired FR samples (Hester et al., 2016; Table S5). The QDI and effective gene number across FFPE samples was 13.2- and 11.1-fold lower, respectively, vs. FR (Table S5).
Correlating quantity and quality with RNA-sequencing success.
Total RNA from FFPE samples correlated poorly with most amplicons across both housekeeping genes except the 5’ most Actb amplicon and the smallest B2m amplicon (5’-Actb and B2m 80). These two amplicons had modest but significant correlations with total RNA concentration (rho = 0.497 and 0.481, respectively) (Table S6). Correlations between total RNA and QDI or effective gene number were slightly better in older low-quality FFPE samples (rho = 0.545 and 0.468, respectively) than younger Study 1 FFPE samples (rho = −0.165 and −0.394) (Table S6). The association with QDI was best for 5’-Actb (rho = 0.422) among Actb amplicons but not significant (Fig. S5A). This was also the case for effective gene number vs. 5’-Actb (rho = 0.395) (Fig. S5B, Table S6). However, QDI and effective gene numbers significantly correlated with B2m 80, the shortest B2m amplicons (rho = 0.837 and 0.578, respectively) (Fig. S6A–B, Table S6). Other B2m amplicons (127 and 429) showed poor correlations with QDI and effective gene number (Fig. S6A–B). PERM correlated slightly better with QDI and effective gene number (rho = 0.675 and 0.563) than RIN (Fig. S7). Both amplicons had significant correlations with QDI and effective gene number; however, the correlation with RIN was negative. Of all the Bioanalyzer based methods of assessing RNA quality, DV100 correlated best with QDI (rho = 0.78) and effective gene number (rho = 0.689) (Fig. S7). The 5’:3’ Actb ratio showed essentially no association with QDI and effective gene number (Table S6). All correlation values may be found in Table S6.
Study 3: Variable-quality FFPE samples based on time in formalin
Total and amplifiable RNA concentrations.
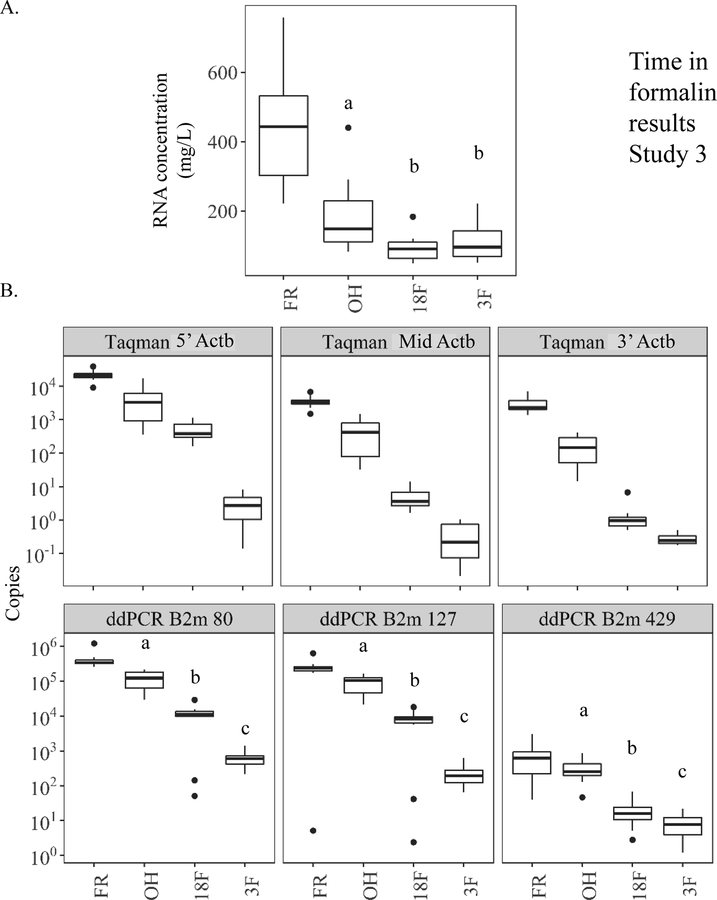
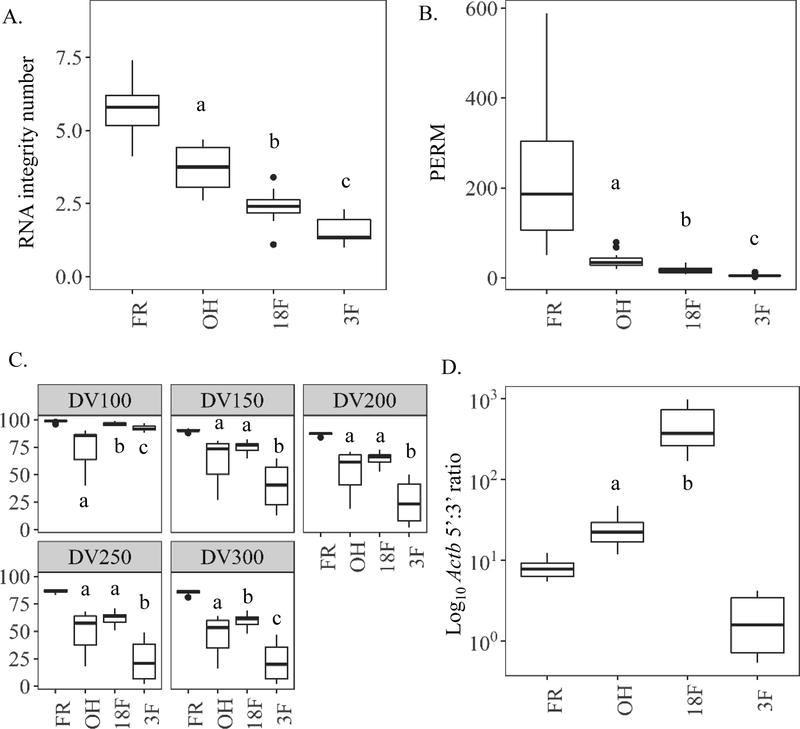
Use of ethanol as a fixative resulted in a 2.4-fold reduction in total RNA yield but short (18F) and long-term formalin fixation (3F) had a more dramatic impact, reducing yields by 4.7- and 4.1-fold respectively compared to paired FR control samples (441 ± 48.8 mg/L) (Fig. 6A). Due to sample limitations for groups 18F and 3F, only real-time qPCR Actb results are explored in Study 3. At the individual Actb amplicon level there was a systematic decrease in amount detected across each amplicon with fixation (OH) and time in formalin (18F and 3F) compared to FR (Fig. 6B). Like Studies 1 and 2, fewer copies of each amplicon were detected with distance from the 5’ end of cDNA (3’ end of mRNA). Therefore, the smallest, yet significant, fold difference between each preservation group (OH, 18F, and 3F) relative to FR was measured for 5’-Actb (4.6, 43.7, and 6659-fold). These fold changes across preservation groups increased in magnitude for Mid-Actb (7.0, 712, and 9261-fold) and 3’-Actb (16.5 and 1991- fold) compared to FR; however, several samples within 3F fell outside the real-time qPCR assay limits of detection for 3’-Actb (Table S7).
Figure 6.
Study 3 on the effects of time in formalin on quantification of amplifiable RNA within paired mouse liver samples preserved in 70% ethanol (OH), 18 hr. formalin then 10% formalin to 3 months (18F), or 3 months in 10% formalin (3F) prior to paraffin-embedding relative to frozen (FR) controls. Real-time qPCR and digital drop PCR (ddPCR) amplification of two common housekeeping genes (Actb and B2m respectively) was used as surrogates for overall RNA quality. (A) RNA concentration as measured by fluorometric methods using Qubit. (B – Top) Amounts of Actb measured by Real-time qPCR qPCR across three similarly sized amplicons. Amplicons located across the gene body 5’ to 3’ (5’-Actb = 5’ most amplicon based on cDNA; Mid-Actb = middle amplicon; and 3’-Actb = 3’ most amplicon base on cDNA). (B – Bottom) Amounts of three differently sized (80, 127, and 429 base pairs) B2m amplicons measured by ddPCR spanning the gene body. Amplicon 80 spans exons 2–3. Amplicon 127 spans exons 1–2, and amplicon 429 spans exons 2–4. a, b, and c indicate p value <0.05, paired, pairwise Wilcoxon signed rank test.
The amounts of amplifiable RNA measured for each B2m amplicon trended similarly to Actb across preservation groups with FR>OH>18>3F (Fig. 6B). Unlike the other studies, the fold differences in amplified RNA detected between preservation groups and FR were not always dependent upon amplicon length for B2m. For instance, OH and 18F had very consistent fold changes compared to FR across each B2m amplicon ranging from 2.8 to 3.4-fold for OH and 29.9 to 38.3-fold for 18F. On the other hand, the comparison of 3F with FR across B2m amplicons was more similar to Studies 1 and 2. For instance, the fold difference between the B2m amplicons nearest in size (B2m 80 and B2m 127) and FR were quite similar (639 and 1077-fold.) The smallest fold difference between 3F and FR was measured for the longest amplicon B2m 429 (89-fold) (Table S7).
Pre-sequencing RNA quality.
For RIN and PERM, formalin fixation resulted in time-dependent negative effects on sample quality. Eighteen hours in formalin (18F) reduced RIN and PERM by 2.4- and 13.1-fold, respectively, compared to FR. Extended time in formalin (3F) lowered RIN and PERM values more (3.6- and 37.8-fold) compared to FR. Ethanol fixation (OH) reduced RIN and PERM the least vs. FR (Fig. 7A–B). Fragment analysis values for FR remained consistent across the five size ranges (98–85% for DV100-300) (Fig. 7C). This was not the case for the fixation groups. Fixation and time in formalin tended to result in more RNA fragments less than 100–300 nucleotides and extended time in formalin (3F) caused the most fragmentation with DV100–300 values significantly less than all other groups (Fig. 7C). Real-time qPCR 5’:3’ ratios for Actb remained low for FR (7.8 ± 0.6) indicating minimal degradation whereas ratios for OH and 18F samples were significantly higher (26 and 496, respectively). Extended time in formalin (3F) resulted in no significant differences as many of these samples were at the limits of detection for the real-time qPCR assay (Fig. 7D, Table S7).
Figure 7.
Study 3 on the effects of time in formalin on RNA quality within paired mouse liver samples preserved in 70% ethanol (OH), 18 hr. formalin then 10% formalin to 3 months (18F), or 3 months in 10% formalin (3F) prior to paraffin-embedding relative to frozen (FR) controls. (A) RNA integrity number (RIN) to quantify RNA quality by Bioanalyzer. (B) Paraffin embedded RNA metric (PERM) to quantify RNA quality using a weighted area under the curve via Bioanalyzer spectropherogram output. (C) Fragment analysis (DV) to quantify the percent of RNA fragments greater than a specified value (100, 150, 200, 250, or 300 nucleotides) by Bioanalyzer smear test. (D) Ratio of similarly sized 5’ most Actb amplicon to 3’ most amplicon copies (5’-Actb/3’-Actb) as measured by digital drop PCR. Values of less than 10 indicate more intact, higher quality RNA. a, b, and c indicate p value <0.05, paired, pairwise Wilcoxon signed rank test.
Sequencing results.
The use of ethanol for tissue preservation had the least impact on post-sequencing results relative to FR as previously reported in Wehmas et al. (2018). As with Study 1 and 2, formalin fixation for 18 hr. increased deletion rates across reads (3.6-fold) and increased % read duplicates (1.2 fold) compared to FR. Samples from 18F also had increased % reads mapping to rRNA (2.3 fold) relative to FR like Study 1. Extended time in formalin (3F) enhanced read deletion rates and % read duplication but not % reads mapping to rRNA. 18F and 3F groups had 1.3- and 1.7-fold reductions in total gene counts while each had minor changes in total reads mapped, % aligned reads, and unique genes detected (Wehmas et al., 2018; Table S7). Ethanol fixation reduced QDI modestly but significantly (1.1-fold) compared to FR. Time in formalin significantly reduced QDI by 1.3- and 1.9-fold for 18F and 3F, respectively vs. FR (231 ± 5.2 bil); whereas, the opposite effect was seen on effective gene number. However, only OH and 18F samples had significantly higher effective gene numbers (by 1.1 and 1.3-fold, respectively) compared to FR (1212 ± 26.0) (Table S7).
Correlating quantity and quality with RNA-sequencing success.
The correlation of total RNA with amplifiable RNA (Actb and B2m) varied across the different preservations. For example, total RNA from OH samples significantly correlated with all Actb and the shorter B2m amplicons, but the correlations were all negative (Table S8). Across 18F samples, total RNA significantly associated with only 3’-Actb (rho = 0.782) whereas 3F sample total RNA significantly correlated with B2m 429 (rho = 0.709) (Table S8). Limited sample quantities for 18F and 3F groups made any correlations with ddPCR Actb and B2m results difficult to interpret (Table S8). Total RNA correlated poorly with QDI across all preservation methods and only slightly better for effective gene number (Table S8).
Unlike Studies 1 and 2, no discernable pattern emerged between amplifiable RNA amplicon locations or sizes and QDI or effective gene number. The associations of real-time qPCR amplified Actb and B2m with QDI were poor across all preservation groups (Fig. S8, Table S8). Comparisons of amplifiable RNA with effective gene number were not much improved. Within OH samples, B2m 127 significantly associated with effective gene number but negatively (rho = −0.664). Otherwise, only 3F samples had significant correlations between B2m 80 and B2m 429 with effective gene number (rho = 0.800 and 0.770, respectively) (Fig. S8B).
RIN values correlated poorly with QDI across all preservation methods (OH, 18F, and 3F) (rho <0 .500) (Fig 13A). However, correlations between RIN and QDI were better than that of PERM, which had poor or negative correlations with QDI across all preservation groups (Fig. 13B, Table S8). We observed modest improvements in correlations of DV100-300 with QDI for some preservation groups, but none that were significant. Associations between RIN, PERM and DV with effective gene number across the different preservation groups were also poor. Only OH samples had significant correlations between RIN and effective gene number and DV100–300 values and effective gene number, but these were negative (rho = −0.574 to −0.655) (Fig. 13C). When relating real-time qPCR 5’:3’ ratio to QDI or effective gene number, we observed insignificant correlations. All correlation values may be found in Table S8.
Studies 1–3: Composite analysis
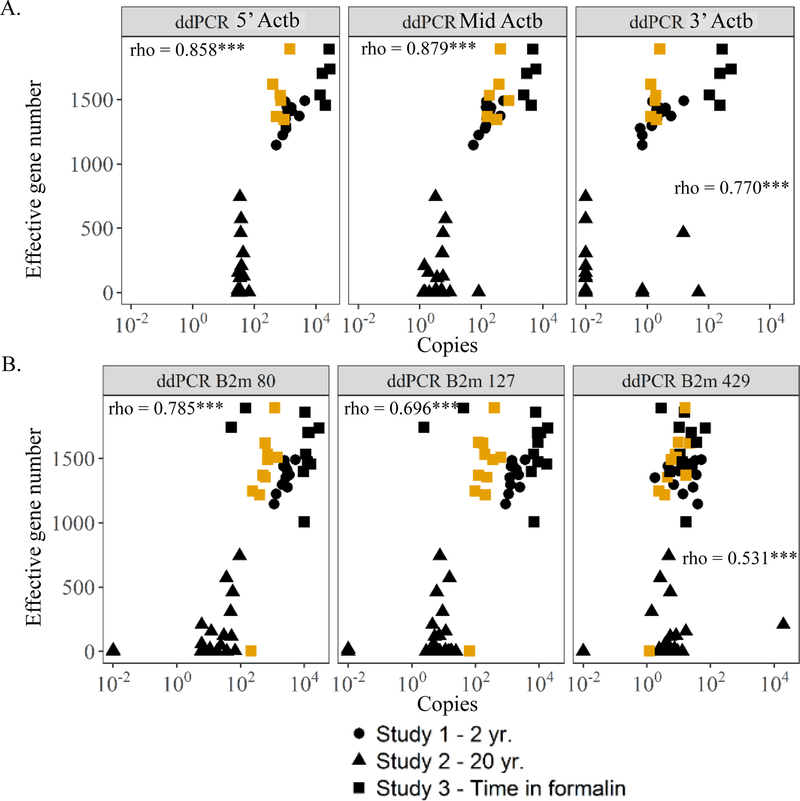
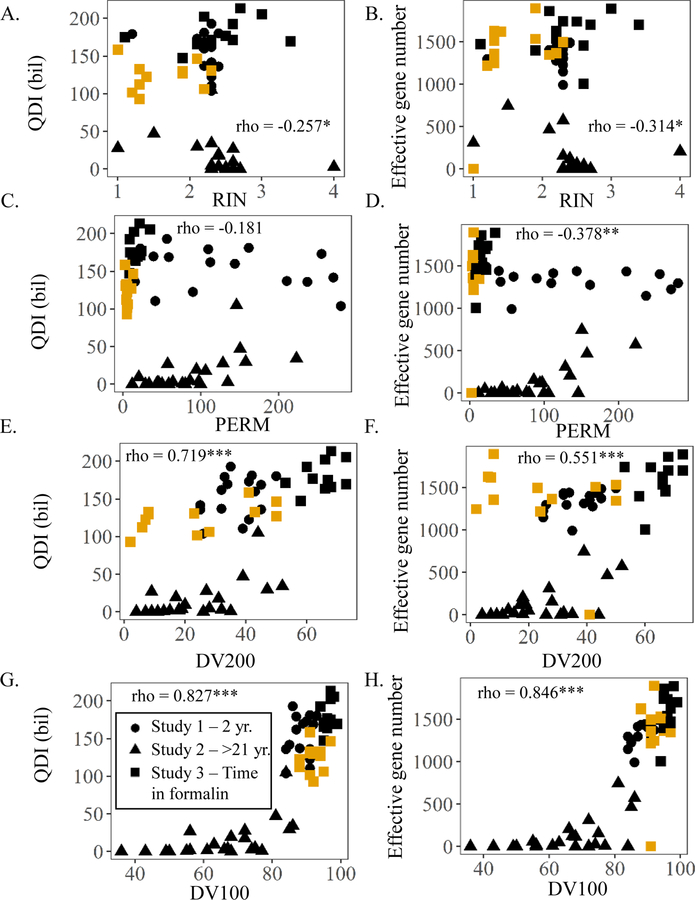
Comparison of RIN, PERM, DV100-300, Actb, and B2m results with eight key sequencing metrics (% reads aligned, % unique paired end reads mapped, deletion rates, % ribosomal RNA, total gene counts, unique genes detected, QDI, and effective gene number) across FFPE samples from Studies 1–3 showed the best performance for ddPCR of 5’-Actb (rho > 0.8 for 7/8 sequencing metrics), B2m amplicon 80 (rho > 0.8 for 6/8), and ddPCR of Mid-Actb (rho > 0.8 for 5/8), and DV100 (rho > 0.8 for 5/8) (Table S9). For instance, the 5’ most Actb amplicon, 5’-Actb, and the shortest B2m amplicon, B2m 80, measured with ddPCR had correlations with QDI at 0.898 and 0.889 respectively (Fig 8A–B). Correlations between 5’-Actb and B2m 80 by ddPCR with effective gene number were high as well at 0.858 and 0.785 respectively (Fig. 9A–B). RIN and PERM, on the other hand, had correlations with QDI and effective gene number that were significant but negative (Fig. 10A–D). RIN, PERM, and total RNA did not correlate much better with the other RNA sequencing metrics (Table S9). DV200 correlated better than RIN, PERM, 5’:3’ ratio, and total RNA (rho > 0.7 for 4/8 metrics) but not as well as DV100 or the qPCR methods (Table S9). This is best illustrated when looking at the association of DV100 with QDI or effective gene number and DV200 with QDI or effective gene number. DV100 vs. QDI or effective gene number was 0.827 and 0.846, respectively as opposed to DV200 vs. QDI or effective gene number, which was lower at 0.719 and 0.551, respectively (Fig. 10E–F).
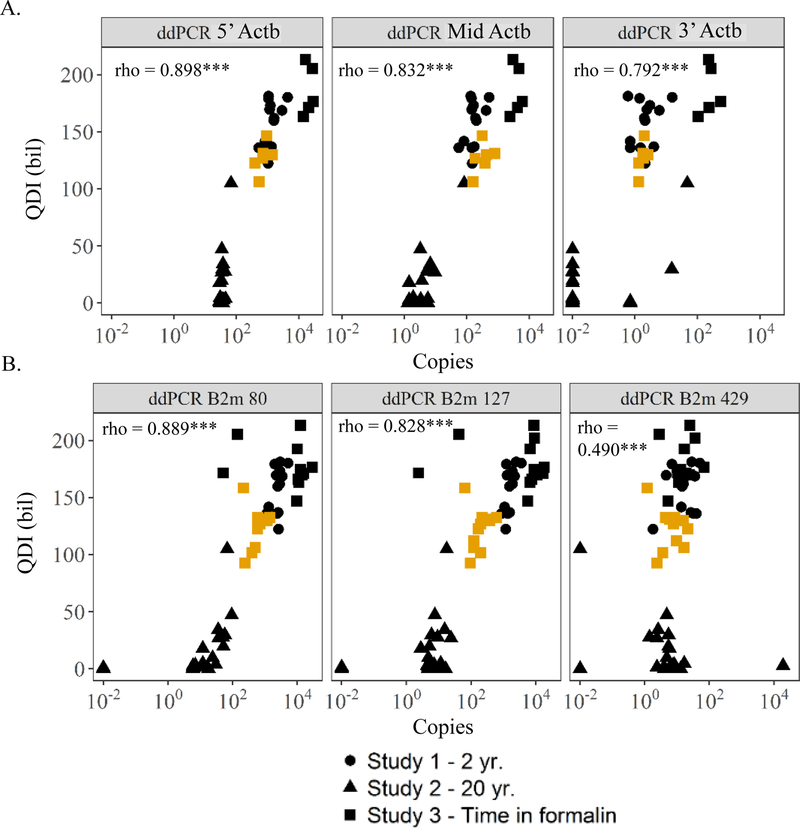
Figure 8.
Correlations between Study 1, 2, and 3 metrics and quantity diversity index (QDI) as an indicator of RNA sequencing success. (A) Correlations between similarly sized Actb amplicons located at increasing distances from the 3’ end of Actb mRNA (5’-Actb<Mid-Actb<3’-Actb) with QDI. (B) Correlations between copies of B2m amplicons ranging in size (80, 127, and 429 base pairs) and location of amplification across B2m mRNA with QDI. Offset of 0.01 added to zero copy number values so samples are not excluded from the figures following log10 scaling. rho indicates correlation coefficient for spearman test on only the fixed and embedded samples across all three studies (*** indicates p-value <0.001). Symbols filled with BLACK indicates samples fixed for 18 hr. in formalin. Symbols filled with YELLOW indicates samples fixed for 3 mo. in formalin.
Figure 9.
Correlations between Study 1, 2, and 3 metrics and effective gene number as an indicator of RNA sequencing success. (A) Correlations between similarly sized Actb amplicons located at increasing distances from the 3’ end of Actb mRNA (5’-Actb<Mid-Actb<3’-Actb) with effective gene number. (B) Correlations between copies of B2m amplicons ranging in size (80, 127, and 429 base pairs) and location of amplification across B2m mRNA with effective gene number. Offset of 0.01 added to zero copy number values so samples are not excluded from the figures following log10 scaling. rho indicates correlation coefficient for spearman test on only the fixed and embedded samples across all three studies (*** indicates p-value <0.001). Symbols filled with BLACK indicates samples fixed for 18 hr. in formalin. Symbols filled with YELLOW indicates samples fixed for 3 mo. in formalin.
Figure 10.
Correlations between Study 1, 2, and 3 metrics and quantity diversity index (QDI) or effective gene number as an indicator of sequencing success. (A) Correlation between RNA integrity number (RIN) and QDI. (B) Correlation between RNA integrity number (RIN) and effective gene number. (C) Correlation between paraffin embedded RNA metric (PERM) and QDI. (D) Correlation between paraffin embedded RNA metric (PERM) and effective gene number. (E) Correlation between DV200 and QDI. (F) Correlation between DV200 and effective gene number. (G) Correlation between % RNA fragments greater than 100 (DV100) and QDI. (H) Correlation between % RNA fragments greater than 100 (DV100) and effective gene number. rho indicates correlation coefficient for spearman test on only the fixed and embedded samples across all three studies (*, **, and *** indicates p-value <0.05; 0.01; and 0.001 respectively). Symbols filled with BLACK indicates samples fixed for 18 hr. in formalin. Symbols filled with YELLOW indicates samples fixed for 3 mo. in formalin.
Effectiveness of DV200>30% threshold on Studies 1–3 FFPE samples
While the DV200 metric, (was used as a way of assessing quality even though it was not fully vetted by Illumina for this library prep), proposed by Illumina for identifying FFPE samples worth sequencing correlated well with RNA-seq metrics in higher-quality samples that had been fixed ~18 hours in formalin and aged <2 years in block (Study 1 FFPE and Study 3 18F), it performed less well with more degraded samples (i.e. extensive time in formalin (Study 3 3F) or many years in paraffin-embedded blocks Study 2 FFPE). The 18hr formalin fixed samples and <2 year old blocks had relatively good levels of read mapping, gene counts, diversity, and lower error rates; therefore, the recommended DV200>30% threshold distinguished 13/15 FFPE samples which demonstrated minimal change in QDI (~41% loss) and effective gene number (~7% gain) compared to FR from Study 1. There were 2 potential false negatives, which had ~48% loss in QDI and 7% gain in effective gene number vs. FR. However, for Study 2, the DV200 cutoff of 30% did not perform as well as in Study 1 in delineating older lower-quality (>21-year-old) FFPE samples suitable for RNA-seq. It identified 17/24 FFPE samples not worth sequencing (97% and 95% loss in QDI and effective gene number relative to FR), but there were 7 potential false positives, which had large losses in QDI and effective gene number (84% and 76%, respectively) compared to FR. For Study 3, the threshold identified 12/12, 18-hour formalin-fixed samples from Study 3 for sequencing. These 18F samples had minimal loss in QDI (22%) compared to FR with an increase in effective gene number (32%). DV200 > 30 performs less well for extended time in formalin. Losses in QDI (46%) and gains in effective gene number (11%) vs. FR were similar to Study 1 FFPE samples, yet the 30% threshold would have resulted in 8 potential false negatives (Table S10).
By setting a revised threshold for DV100, which correlated better than DV200 with several RNA seq outputs, better categorization of FFPE samples for sequencing was achieved. For example, DV100 >80% distinguished FFPE with moderate changes in QDI (<46% loss) and effective gene number (12–32% gain) vs. FR compared to high (92 and 91% losses, respectively). This revised DV threshold correctly identified 16/16 FFPE samples with modest changes in QDI and effective gene number relative to FR samples from Study 1. Furthermore, it distinguished 20/24 FFPE samples not worth sequencing (97 and 96% losses in QDI and effective gene numbers vs. FR) from Study 2. Of the 4 samples recommended for sequencing (potential false positives), 3 had ~83% and 45% losses in QDI and effective gene numbers while one sample had 55% and 99% loses each, respectively. In Study 3, DV100>80 would have identified 12/12 and 12/12 of 18F and 3F samples for sequencing. As mentioned previously, these samples had modest losses in QDI and some gains in effective gene number relative to FR. There is one potential false positive that would not have been identified by this threshold which was later identified as an outlier by principal component analysis on differentially expressed genes from group 3F. This sample exhibited a modest loss in QDI (27%) but was the only sample in Study 3 to show a substantial loss in effective gene number (99%) (Table S10).
DISCUSSION
Current methods for assessing RNA integrity in FFPE specimens do not adequately identify samples that will yield poor or unusable RNA-seq data. Here, we examined several FFPE RNA quality metrics in relation to key sequencing outputs. Our findings showed variability in the usefulness of different RNA quality measures across different types of FFPE samples. Overall, fragment analysis (DV) and qPCR-based methods outperformed RIN and PERM in determining sequencing read quality for mapping and gene detection. qPCR amplification of housekeeping genes correlated best with major RNA-seq outputs such as mapping and gene detection, which are necessary for differential gene expression analysis. ddPCR provided better absolute quantification of housekeeping genes Actb and B2m across samples compared to real-time qPCR. Among Bioanalyzer-based methods, the DV100 metric showed the highest correlation with primary indicators of RNA-seq quality, including overall gene diversity across all FFPE samples analyzed. Furthermore, a DV100 threshold of >80% correctly distinguished most FFPE samples (24/28) with higher total gene counts and diversity (as indicated by QDI values and effective gene number) and performed better than the DV200 threshold of >30% recommended previously (Illumina, 2015).
A major challenge in identifying better metrics to evaluate FFPE RNA is the variation in pre-analytical variables across studies. Here we looked across different study types to investigate two key variables, age in paraffin block and time in formalin. Retrospective analysis of Studies 1 and 2 suggested that quantifying amplifiable RNA at smaller size ranges may help indicate whether a sample should be sequenced or not, especially for longer-term fixed tissues or older FFPE blocks. However, these trends were not very strong or disappeared in prospective Study 3, which looked specifically at fixation type and time. This evidence suggests that age in block has an important amplifying effect on RNA modifications induced by initial formalin fixation. Collectively, the summary analysis of all three studies highlights the idea that different quality metrics may be needed to capture different types of nucleic acid degradation (e.g. fragmentation vs. biochemical modification).
These results raise several issues and considerations for evaluating FFPE sample quality prior to sequencing. Total RNA amounts extracted from preserved tissue did not correlate well with amplifiable RNA results and therefore should not be used as a sole indicator of how much FFPE RNA is required for library preparations. Fragment analysis provided a more informative and efficient screen to help predict sequencing viability based on sample intactness. The DV100 > 80 threshold alone may be sufficient to screen higher quality FFPE samples recently archived with brief formalin fixation times (<24 hr.). For lower quality FFPE samples (stored long-term or fixed for extended times in formalin), or those on the borderline of DV100 > 80 threshold, fragment analysis combined with qPCR amplification of housekeeping genes may be needed to provide information on RNA intactness as well as modifications, which could impede cDNA synthesis. Generally, extended times in formalin (>18–24 hrs) resulted in more interference with cDNA synthesis and lower quality qPCR data, presumably due to formaldehyde-induced modification of nucleic acids. Longer times in FFPE block also interfered with cDNA synthesis and qPCR results, likely due to a combination RNA fragmentation and modifications.
Our data indicates that measuring RNA amplification of shorter housekeeping gene amplicons (~80–100 base pairs) was most informative in delineating FFPE samples with the highest QDI and effective gene number. Sample fragmentation and extensive adducts likely make it difficult to detect longer gene amplicons. The location of the amplicon across the housekeeping gene body may not be as important if a combination of random hexamer and oligo dT primers are used in cDNA synthesis as seen with the B2m results. The 5’-Actb amplicon nearest the 3’ end of Actb mRNA had the best amplification results because only oligo dT primers were used for those specific cDNA synthesis reactions to allow for 5’:3’ ratio calculations. Of the qPCR methods, ddPCR provided superior results compared to real-time qPCR for absolute quantification. This is based on ease of implementation (i.e. no need for large standard curves with each reaction), better limits of detection, improved precision, and greater accuracy.
When determining RNA quality from FFPE tissue samples, initial considerations should be time in formalin and age in block, which are the two major known factors leading to poor sequencing data. Second, isolation of >100 ng total RNA per sample (the minimal amount required for library preparation) is recommended. Third, a Bioanalyzer electropherogram and screen of FFPE RNA for DV100>80% should be conducted to determine fragmentation distribution of samples. If samples were fixed in formalin for ~18 hr. and stored under controlled conditions for ~2 yrs. or less block, then fragment analysis may be sufficient to adequately determine RNA quality. In contrast, if FFPE samples are old, underwent extended formalin fixation times (>24 hrs.) or have DV100 values that border the 80% threshold, qPCR on well-established housekeeping genes will enable an estimate of the degree of RNA fragmentation. A working threshold of >500 copies of Actb and B2m from FFPE samples corresponded with better gene detection and diversity data from RNA-seq. Finally, extended heated incubation with chemical modification is recommended during the RNA isolation process to improve overall yield and certain RNA-seq parameters (Wehmas et al. 2017). Collectively, these findings indicate that RNA fragmentation analysis and PCR-based methods are more informative than RIN for characterizing FFPE RNA quality for sequencing but also highlight the need for additional criteria or metrics.
Our findings indicate that RNA fragmentation analysis and PCR-based methods are more informative than RIN for characterizing FFPE RNA quality for sequencing but also highlight the need for additional criteria or metrics. This work should help advance the use of archival FFPE samples for genomic analyses, enhancing retrospective analysis, reducing the need for de novo mechanistic studies, and facilitating cross-species comparisons.
The expansion of genomic and genetic data analytics have fostered new strategies for health care and clinical research, allowing for more individualized approaches to disease prevention and treatment. This type of “precision medicine” will rely heavily on our understanding of human genetic variation and gene-by-environment interactions. The methods and approaches defined within this manuscript provide ways to improve genomic analysis of FFPE samples, increasing access an important resource for clinical laboratories where frozen tissue specimens are often limited or not available. By advancing the use of archival FFPE samples for retrospective analysis, this effort should also reduce the need for de novo mechanistic studies and streamline translational cross-species comparisons.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank staff at EA for generation of RNA-seq data and bioinformatics support; the HESI Genomics Committee; HESI and U.S. EPA reviewers for constructive comments on this manuscript; and Jeanene Olin for technical histology assistance. The research described in this article has been reviewed by the U.S. EPA and approved for publication. Approval does not signify that the contents necessarily reflect the views or policies of the Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
FUNDING INFORMATION
Funding was provided by the U.S. EPA Office of Research and Development and the Health and Environmental Science Institute (HESI) scientific initiative, which is primarily supported by in-kind contributions (from public and private sector participants) of time, expertise, and experimental effort. HESI contributions are supplemented by direct funding (that largely supports program infrastructure and management) provided by HESI’s corporate sponsors. A list of supporting organizations (public and private) is available at http://hesiglobal.org/application-of-genomics-to-mechanism-based-risk-assessment-technical-committee/.
REFERENCES
- Bass BP, Engel KB, Greytak SR, and Moore HM (2014). A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen? Archives of pathology & laboratory medicine 138(11), 1520–30, 10.5858/arpa.2013-0691-RA. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol 57, 289–300. [Google Scholar]
- Chung JY, Cho H, and Hewitt SM (2016). The paraffin-embedded RNA metric (PERM) for RNA isolated from formalin-fixed, paraffin-embedded tissue. Biotechniques 60, 239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers DL, Fowler CB, Cunningham BR, Mason JT, and O’Leary TJ (2011). The Effect of Formaldehyde Fixation on RNA: Optimization of Formaldehyde Adduct Removal. J Molec Diagn 13, 282–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CH, Johnson FB, Whiting J, and Roller PP (1985). Formaldehyde fixation. J Histochem Cytochem 33, 845–53. [DOI] [PubMed] [Google Scholar]
- Greytak SR, Engel KB, Bass BP, and Moore HM (2015). Accuracy of Molecular Data Generated with FFPE Biospecimens: Lessons from the Literature. Cancer Res 75, 1541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester SD, Bhat V, Chorley BN, Carswell G, Jones W, Wehmas LC, and Wood CE (2016). Dose-Response Analysis of RNA-Seq Profiles in Archival Formalin-Fixed Paraffin-Embedded Samples. Toxicol Sci 154, 202–13. [DOI] [PubMed] [Google Scholar]
- Hoffman EA, Frey BL, Smith LM, and Auble DT (2015). Formaldehyde crosslinking: a tool for the study of chromatin complexes. J Biol Chem 290, 26404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, and Auffray C (2005). Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 33, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina (2015). Technical Note: Expression Analysis of FFPE Samples. https://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/evaluating-rna-quality-from-ffpe-samples-technical-note-470-2014-001.pdf. Last accessed, April 8, 2018.
- Lou J (2007). Partitioning diversity into independent alpha and beta components. Ecology 88(10), 2427–2439, doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- Landolt L, Marti HP, Beisland C, Flatberg A, and Eikrem OS (2016). RNA extraction for RNA sequencing of archival renal tissues. Scand J Clin Lab Invest 76, 426–34. [DOI] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, and Okubo K (1999). Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 27, 4436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millier MJ, Stamp LK, and Hessian PA (2017). Digital-PCR for gene expression: impact from inherent tissue RNA degradation. Sci Rep 7, 17235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Hands RE, and Bustin SA (2006). Quantification of mRNA using real-time RT-PCR. Nat Protoc 1, 1559–82. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2016). R: A language and environment for statistical computing doi: R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, and Ragg T (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahlfen S, Missel A, Bendrat K, and Schlumpberger M (2007). Determinants of RNA quality from FFPE samples. PLoS One 2, e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmas LC, Wood CE, Gagne R, Williams A, Yauk C, Gosink MM, and Hester S (2018). Demodifying RNA for Transcriptomic Analyses of Archival Formalin-Fixed Paraffin-Embedded Samples. Toxicol Sci 162, 535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.