Abstract
Many researchers believe that personalized medicine is a potential solution to the well-known challenges facing the pharmaceutical industry worldwide today. Achieving the full promise of personalized medicine requires a new, systems approach to biomedicine that will only be enabled by novel, interoperable IT infrastructures that facilitate simpler data access, data sharing and enhanced collaboration. This article will describe how the technology developed in the cancer Biomedical Informatics Grid® (caBIG®) program (http://cabig.cancer.gov/) from the National Cancer Institute (NCI) is already enabling many research organizations to implement personalized medicine approaches for their basic and clinical research programs.
Keywords: caBIG®, personalized medicine, interoperability, data standards
An Infrastructure for Interconnecting Research Institutions
Many researchers believe that personalized medicine is a potential solution to the challenges facing the pharmaceutical industry worldwide today - traditional “one size fits all” approaches for discovering new therapeutics are failing, research costs are rising, fewer drug candidates are making it to market and fewer blockbuster drugs are available to offset the cost of research, estimated at $897M in a 2006 study carried out by Tufts University [1]. A straightforward application of the personalized medicine approach to research can already be seen at many pharmaceutical companies, such as the development of comparative diagnostic tests and the release of drugs that are highly effective in specific, limited patient populations. A few examples include Gleevec® (Novartis) for chronic myelogenous leukemia (CML), Vectibix® (Amgen) for patients that have EGFR-expressing metastatic colon cancer and Selzentry (Pfizer) for patients with CCR5-tropic HIV1 [2].
The potential benefits of personalized medicine approaches for drug discovery, development and commercialization are numerous [3], including:
Improved biological targets, based on validated biomarkers
“Enriched” clinical trials in which the patient population is pre-disposed to respond, thereby leading to higher efficacy rates
Reduction in the time, cost and failure rate of clinical trials
Avoidance of adverse side effects (ADRs) during clinical trial or later when widespread use leads to ADRs and product recalls
Revival of drugs that failed clinical trials or had been withdrawn from the market
Fewer withdrawals of marketed drugs
Higher compliance with drug regimens
To achieve the full promise of personalized medicine will require a new, systems approach to biomedicine. This systems approach will be predicated on the widespread use of 21st century information technology (IT). In this article I will present an IT infrastructure solution that directly addresses some of the challenges in implementing a personalized medicine approach to drug discovery. The technology developed in the cancer Biomedical Informatics Grid® (caBIG®) program (http://cabig.cancer.gov/) from the National Cancer Institute (NCI) is already enabling many research organizations to implement personalized medicine approaches for their basic and clinical research programs.
caBIG®: An Infrastructure for Connecting Research Organizations
caBIG® was launched in 2004 by the NCI as part of its mission to advance research on cancer and to improve clinical outcomes for patients. The ultimate goal of caBIG® is the creation of a virtual web of interconnected data, individuals and organizations that fundamentally redefines how treatment-focused research is conducted, resulting in improved patient/participant interaction with biomedical organizations and, ultimately, improved patient outcomes. Like the cancer centers of caBIG’s ® starting focus, the same unmet needs for data management and interoperability exist within most pharmaceutical companies. In both cases, the organizations have traditionally worked as self-contained entities, but that culture of isolationism no longer meets imperatives in a ”flat”, fast-moving and unforgiving environment. The caBIG® program was born in the cancer research community, but its technology and capabilities are applicable to almost any therapeutic area and to any organization, public or private, academic or commercial.
It’s about the Data and the “Disconnects”
For personalized medicine to come to fruition, research organizations need to generate and manipulate huge quantities of data, build biologically accurate models of disease states and work with clinicians to map those models to observed patient outcomes. Diverse, high-throughput technologies (e.g., next-generation DNA sequencing, DNA microarrays, digital imaging) can generate terabytes or more of data per experiment, so there is rarely a problem getting enough data. Because it is difficult for a single researcher to have the skills necessary to generate all the data he/she needs, or to exploit fully this sizable quantity of very diverse information, there is a growing trend toward collaboration between and among teams that utilise varied expertise (e.g., molecular biologists, bioinformaticians, protein chemists, clinicians and so on). Such collaborative research can potentially include multiple research groups in geographically dispersed sites: collaborators at academic research centers or contract research organizations. Clearly, the ability to exchange data and use it in meaningful ways and the availability of interoperable tools that provide end-to-end support for research workflows are essential for productive research and fruitful collaborations.
Unlike those technologies that generate data, information technologies that manage and exchange that data have been slow to develop. Traditionally, and for a variety of reasons, laboratories within a single institution (much less between different institutions) have rarely collaborated, thereby forming isolated “data silos” that represent real obstacles for cohesive and comprehensive research. Often, the same types of biomedical data are collected by different research groups using “homegrown” information systems that are based on standards or data structures particular to their own laboratory or institution and which may or may not be shared by other groups. While the current situation encourages competition between groups, potentially resulting in innovations and recognition for those who publish first, it also results in redundant information generated by armies of investigators using a lot of precious and increasingly scarce resources. Consequently, the costs of business as usual continue to rise without significant increase in benefits in terms of medically relevant output. Interconnected, interoperable information technology-based approaches, like caBIG®, can help mitigate these disconnects, enforce standards-based data collection and accelerate large-scale, collaborative research, potentially reducing per capita costs.
Regulators are also starting to add their voices to the calls for greater data standardization. For instance, the Food and Drug Administration Amendments Act (FDAAA), signed into law in September 2007, mandates that the sponsors of clinical trials (often pharmaceutical companies) submit the results of their trials to a nationwide clinical trials registry and results data bank at ClinicalTrials.gov within one year of trial completion [4]. While many questions about implementation and monitoring remain, it is clear that standardized clinical data elements will play a critical role in ensuring that the data provided are accessible and useful [5]. Besides improving collaborative research, an interoperable set of data management tools based on industry-wide standards, like caBIG®, may also be useful to address regulatory compliance issues by providing all the data collected in a clinical trial in industry-standard formats.
Data Interoperability: Overcoming the Tower of Babel
A need for data interoperability underlies all of these diverse data management issues. Simply defined, interoperability is the ability of a system to access and use the parts of another system. This basic definition can be broken down into two equally essential parts:
“Syntactic interoperability” (the ability to access another system)
“Semantic interoperability” (the ability to use the parts (or data) from another system)
Unfortunately, many homegrown and commercial software products use proprietary data formats, which limit or prevent efficient data exchange. In addition, a wide assortment of sometimes competing data standards is already in use. Data formats and software products have also evolved over the years, leaving multiple incompatible formats behind. As such, simply adopting standards-based tools is no guarantee of interoperability.
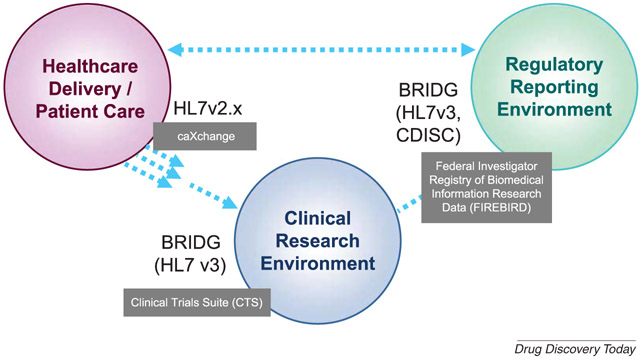
To ensure truly interoperable software, a systematic approach is required adopting widely recognized data standards where they exist and a commitment to working closely with professional associations and organizations to develop data models and define common data elements (CDEs) where such standards do not exist. caBIG® has performed this task since its inception. In partnership with key industry standards organizations such as the Clinical Data Interchange Standards Consortium (CDISC), Health Level 7 (HL7), and FDA, caBIG® has worked closely with the Biomedical Research Integrated Domain Group (BRIDG) (http://bridgmodel.org/). Relationships with other standards organizations ensure that all caBIG® technologies are compliant with widely accepted industry standards across a variety of disciplines (Figure 1).
Figure 1 -. Standards:
The use of accepted industry standards such as BRIDG, and interoperable software applications such as the CTS suite or Firebird, enables the traditionally isolated data silos of Healthcare, Regulatory, and Clinical Care to exchange data in support of collaborative research.
Central Principles of caBIG®
While caBIG® was launched and implemented as a government-sponsored initiative, it has been ”friendly” to the commercial sector throughout its life and that attitude and set of policies are now leading directly to benefits for the pharma and IT industries. Specifically, four fundamental principles underlie the activities of caBIG® and have guided all of its operations since its inception.
Open Access: caBIG® and its associated products are open to all, whether from academic or commercial organizations, enabling and encouraging access to tools, data, and infrastructure by the cancer and greater biomedical research communities.
Open Development: Software development projects are assigned to particular participants, but they are performed iteratively with multiple opportunities for review, comment, further modification, and development by the caBIG® community. By leveraging a large, diverse community of developers and scientists, the tools have been focused on addressing real-world research problems.
Open Source: The software code underlying caBIG® tools, developed with the support of the NCI, is available to all software developers for use and modification. This software is licensed as open source to promote the reuse of existing code, thereby maximizing the benefit of the research dollars spent. An open source license eliminates the expensive and/or complex licensing requirements employed by many commercial products that may limit their application between multiple sites. Furthermore, the open licensing scheme of caBIG® encourages commercial vendors to participate by creating caBIG®-compliant programs without giving up their rights to the intellectual property. This flexibility opens the door for developing interoperability with existing commercial products in use at the pharmaceutical companies, and it potentially reduces the need to “rip and replace” entire existing IT systems. Furthermore, caBIG® recognizes the need for and the importance of commercial software to the biomedical enterprise and accommodates it through non-viral caBIG® licenses.
Federation: caBIG® software and standards enable local organizations, such as Cancer Centers or pharmaceutical companies, to share data resources with the larger research and care community and to use resources contributed by others. caBIG® tools allow these resources, aggregated from multiple sites, to appear as an integrated research dataset while the individual resources remain under the control of the local organizations. This capability is important for pharmaceutical companies who want to have strict control of their data, which may be located in many different physical locations. Support for federated data provides the granularity to manage data access as needed, both internally at the company and externally with collaborators. Federated data management also removes the requirement to build large data warehouses, with the accompanying IT headaches and it allows the organization to access its data no matter where it is stored, thereby eliminating data migration and synchronization issues.
Connecting Organizations: caGrid Makes It Easy
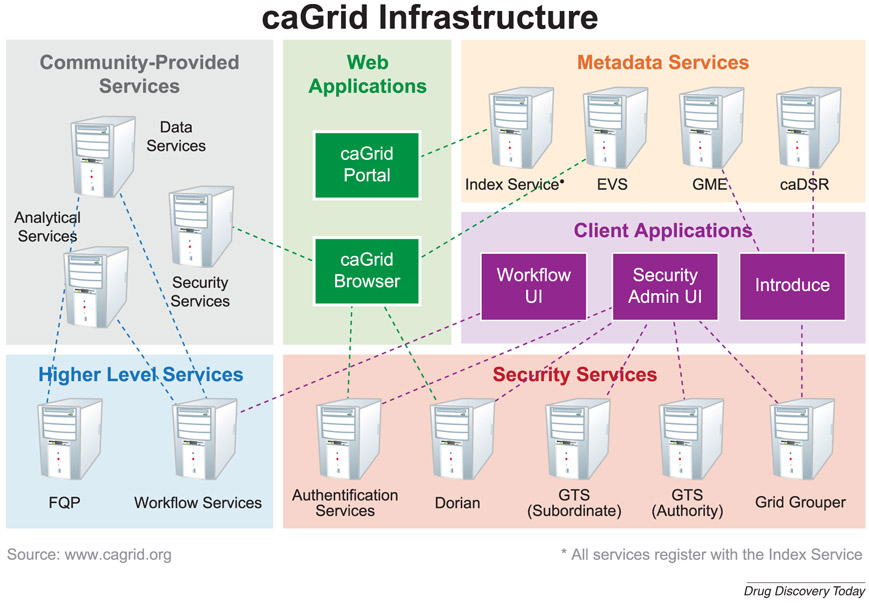
At the heart of the caBIG® program is a grid infrastructure that provides connectivity among people, organizations, data, and analysis tools. A simplified network diagram of the caGrid architecture is given in Figure 2. There are now more than 100 grid nodes currently online at a variety of U.S. government, academic and commercial organizations [6].
Figure 2 -. caGrid:
caGrid provides an interoperable grid infrastructure that supports the specific needs of research organizations by supplying metadata support, security services, web-based applications, and connectivity with client-side and community-provided applications and services facilitating efficient research workflows and collaborative research.
caGrid is a “service-oriented architecture” that works by creating applications from reusable pieces providing specific functionality, called “services.” Because these services are standardized, new applications created from them automatically share identical methods for handling data and can pass data back and forth easily. caBIG® grid technology is based on the Globus toolkit[7], which is the widely recognized standard for open source grid technology, and is used by several other research support networks. Use of this common toolkit makes it simpler to connect caGrid to other research grid architectures or to develop custom grid architectures that are still interoperable [8]. caGrid expands the capabilities of Globus by adding semantic and security services required to meet the needs of biomedicine.
Tools and Applications: Interoperability in Real Time
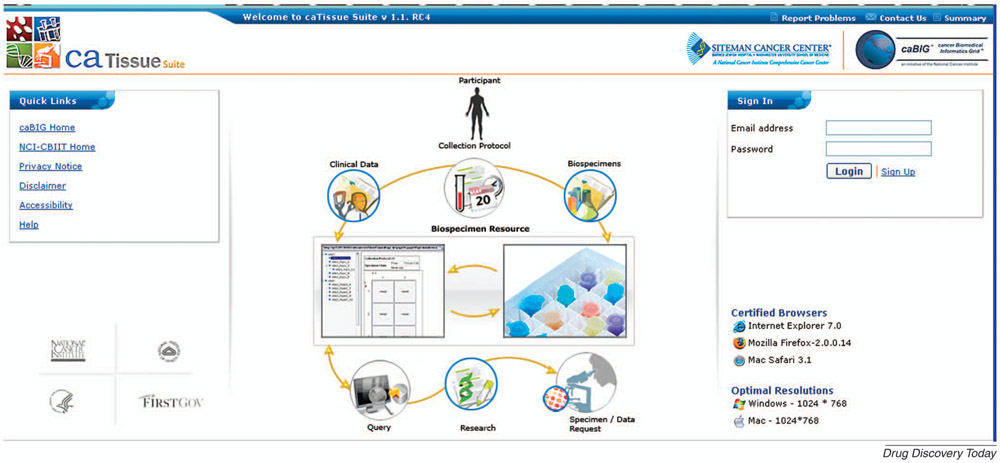
More than 40 analytical tools have been developed as part of caBIG®, supporting integrated workflows for both basic and clinical researchers across the full spectrum of data analysis needs. Currently available tools include software for managing microarray data (caArray), analyzing gene expression patterns and SNPs (geWorkbench), collecting and managing biospecimens (caTissue), managing digital images and their annotations (National Cancer Imaging Archive -NCIA), registering patients in clinical trials (Clinical Participant Registry - C3PR), and monitoring adverse events in clinical protocols (cancer Adverse Events Reporting System - caAERS), among others [9]. Figure 3 is an example screenshot from caTissue which shows the easy to use interface that reflects common workflows and tasks done by researchers.
Figure 3 -. caTissue:
caTissue is one of more than 40 end-user applications developed as part of the caBIG® program that supports scientific workflows for biomedical researchers. caTissue provides support for biospecimen acquisition, annotation, query and sample requests by researchers looking for samples to support their research needs.
caBIG® tools have been designed to address the specific need for interoperable research and clinical care workflows not to compete with, or replace, existing commercial or in-house products solely based on increased functionality. In addition, caBIG® has developed and published guidelines, toolkits and application programming interfaces (APIs) for software developers, whether they are in-house at the research organization, or offsite commercial software vendors, giving organizations interested in caBIG® two paths towards interoperability: 1) adoption of caBIG® tools if those tools meet their research needs or 2) adaptation of existing tools to be caBIG® compatible. This approach allows an organization to utilize the best aspects of caBIG® technology where they fit into the overall enterprise architecture.
Data Security and Privacy for Enterprise and Academia
Researchers, whether working at commercial organizations with the goal of developing a proprietary therapeutic or diagnostic or working at academic institutions to publish a novel finding, are concerned with data security. Individuals and organizations need to protect their intellectual property, comply with state and federal regulations and statutes, determine the terms and conditions under which they can safely share various types of data and have tight data access controls over data they choose to share, both generally and on a case-by-case basis.
Although caBIG® technology facilitates data sharing, the need to provide control over data access, as well as the need for tools to help researchers decide what segments of their data are suitable for sharing, has been a primary consideration from the onset of the program. The Globus toolkit provides a measure of data security functionality, but caGrid needed significantly improved security. As a result, the development team created the Grid Authentication and Authorization with Reliably Distributed Services (GAARDS) security infrastructure, which provides services and tools to administer and enforce security policy in an enterprise Grid. While GAARDS was developed to address the requirements of the cancer research community, similar requirements are common in other fields of biomedical research; therefore the design principles and infrastructure of GAARDS can be employed in security support for a wide range of organizations [10].
Addressing Policy Issues Affecting Data Sharing
Pharmaceutical companies have traditionally functioned as self-contained entities, closely guarding their experimental data as critical intellectual property. With external collaborations, outsourced research arrangements, and requirements for data access by all investigators as a prerequisite for publication in top tier journals becoming more commonplace, data access requirements shift from access prevention to access control. As a result, even though the technology is in place to facilitate data sharing, the need to protect intellectual property, patient privacy and the need to comply with state, federal and regulatory requirements, all introduce complexities in actually sharing the data. Here too, caBIG® has resources to address these issues.
caBIG® provides guidelines and tools to help researchers evaluate the sensitivity of data they may wish to share on the grid. caBIG® has developed the Data Sharing and Security Framework (DSSF), which includes analytical tools to facilitate evaluating the areas of federal privacy regulation, human participant protections, sponsor contract compliance, and proprietary interests. By standardizing access and use contracts as well as policy guidelines around data sharing, the overhead of custom contract development and one-off agreements may be significantly reduced. The Data Sharing and Intellectual Capital (DSIC) Knowledge Center (https://cabig-kc.nci.nih.gov/DSIC/KC), a recently developed support resource for the caBIG® community, provides information and guidance on data sharing and works to further develop the DSSF.
Helping Research Organizations Achieve Their Goals
An infrastructure designed to connect research organizations is only effective if it supports real-world research efforts. In the academic realm, caBIG® is already generating results. A key goal of caBIG® is to connect the 63 NCI-designated Cancer Centers across the U.S., enabling improved data sharing, improved collaboration and improved patient outcomes. As of November 2008, 43 of these centers (including leading institutions such as Baylor, Georgetown, Duke, Johns Hopkins, The Mayo Clinic, and UCSF) were connected to caGrid. More than 15 community cancer centers participating in the NCI National Community Cancer Centers Program (NCCCP) are also getting connected to caGrid. Each of these centers has developed specific implementation plans that leverage caBIG® tools and infrastructure to support a wide variety of basic and clinical research programs within their own institutions, providing proof of the versatility of caBIG® technology and a confidence that caBIG® can provide unique resources to enhance and facilitate research programs. Pharmaceutical companies face many of the same research challenges as these cancer centers. The success of caBIG® in addressing the complex needs of these cancer centers suggests very broad applicability for other research organizations.
Beyond connecting the NCI Cancer Centers, a variety of ongoing cancer research programs are underway that are enabled by caBIG® technology, including:
The Cancer Genome Atlas (TCGA) and the Cancer Molecular Analysis (CMA) Portal: The Cancer Genome Atlas project (TCGA), an integrative, multidisciplinary effort to identify and characterize systematically the genomic changes associated with several cancer types. The CMA portal (https://cma.nci.nih.gov/cma/) provides powerful tools that enable cancer researchers to explore, visualize and integrate genomic characterization, sequencing, and clinical data from a variety of data sets. caBIG® technology integrates TCGA data from a total of 11 different organizations and puts analytical tools developed at three different organizations under one simple interface, clearly demonstrating the effectiveness of a federated, interoperable framework.
Repository of Molecular Brain Neoplasia Data (REMBRANDT): The REMBRANDT project leverages caIntegrator, an application framework for data warehousing, to host and integrate standardized clinical and functional genomics data from clinical trials of brain cancer. This Web portal (https://caintegrator.nci.nih.gov/rembrandt/) provides the ability to query comparable data across multiple domains and allows physician-scientists to understand subtle differences between subclasses of brain tumors, enabling better patient treatment decisions and clearly demonstrating the benefits of cross-discipline research.
Duke University – Department of Defense Breast Cancer Trial: Duke is conducting a genomics-guided adaptive clinical trial on metastatic breast cancer. By using a combination of the caBIG® CDMS system for trial management, caTissue for breast tissue sample management, and caArray for gene expression microarray data management—all connected by caGrid, the Duke researchers can assess a patient’s responsiveness to treatment rapidly and adjust that treatment quickly if needed. In the long run, this approach may not only speed the trial itself, but it should also improve patient outcomes, due to the ability to adjust treatments expeditiously.
Beyond Cancer to All Diseases, Globally
While caBIG® was launched to address the needs of cancer research and care, the tools, standards and infrastructure it has developed are not limited to cancer and can be readily applied to the data management need of any disease area. As a result, caBIG® can provide a common framework to address the data management issues present in many pharmaceutical organizations working on multiple therapeutic areas. For instance, any interested research organization can adopt caGrid and other caBIG® standards and tools. Examples of early adopting organizations include:
The National Heart Lung and Blood Institute (NHLBI): The NHLBI is using technology from both caBIG® and BIRN to develop the CardioVascular Research Grid (CVRG), a distributed community resource in support of cardiovascular data integration and discovery (http://www.cvrgrid.org/).
Northwestern University: Northwestern University is using caGrid to host and connect population and behavioral science data to study tobacco use patterns across different demographic groups.
Nationwide Health Information Network (NHIN): Core caBIG® technology is a vital component of the Nationwide Health Information Network (NHIN) (http://www.nhin.com/), whose goal is to provide secure, nationwide access to health information by connecting researchers, caregivers, providers, and patients in a seamless network.
Furthermore, international organizations are also adopting caBIG® standards and technology. There are ongoing collaborations with the National Cancer Research Initiative (NCRI) in the United Kingdom, the Shanghai Center for Bioinformatics Technology (SCBIT) in China, and the Indian National Knowledge Commission, among others. Despite the expected connectivity challenges that can arise when crossing geographic and language borders, the core interoperable aspects of the technology remain applicable and can be adapted to a wide variety of research applications.
Although there are several other efforts ongoing to connect research organizations, few combine thoroughly developed data interoperability models, extensive collections of analytical tools that span virtually the entire workflow needed by a pharmaceutical or academic researcher and have comprehensive data security frameworks in place to protect the valuable data being generated by research organizations. Rather than cover these programs in detail, please see Lincoln Stein’s excellent review article [11].
Conclusion
Today, all research organizations, whether they are pharmaceutical, biotechnology firms, or academic institutions, need to improve the efficiency and efficacy of their research efforts. Traditional disconnected approaches of the 20th century are neither adequate nor feasible any longer in this increasingly competitive environment with ever-decreasing resources. The next generation of therapeutics and diagnostics can only come from implementing the type of data-driven, collaborative, translational research programs that will lead to effective personalized medicine. Comprehensive, interoperable, and scalable IT infrastructures that can address the extensive data management issues plaguing research organizations today are needed now to enable this type of translational research. The tools, standards and infrastructure developed for the caBIG® program can provide a comprehensive solution to many of these data management issues. . By removing these physical barriers to data sharing, addressing the needs of all constituents of the biomedical research community, and demonstrating the benefits of collaboration, caBIG® also helps break down some of the cultural barriers to collaborative research, leading to improved research efficiency. More than 50 top research institutions across the U.S. are already implementing caBIG® technology to enable their basic and clinical research programs, leading them to the forefront of personalized medicine in the 21st century.
Footnotes
Teaser: A potential solution is proposed to the outdated data management systems common at many research organizations that inhibit the data sharing and collaborative research required to enable to personalized medicine.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tufts Center for the Study of Drug Development (2006) Impact Report, Vol 8, no 5 [Google Scholar]
- 2.Allison M (2008) “Is Personalized Medicine Finally Arriving”, Nature Biotechnology 26, 5. [DOI] [PubMed] [Google Scholar]
- 3.The Case for Personalized Medicine; Personalized Medicine Coalition, November 2006 [Google Scholar]
- 4.Public Law 110-85, September 27, 2007, Food and Drug Administration Amendments Act of 2007 [Google Scholar]
- 5.Messplay Gary C, and Burrell Sarah, E. (2007) Implications of FDAAA 2007. Contract Pharma- November/December 2007, 18–20 [Google Scholar]
- 6.A real-time list of available grid nodes and services is available at http://cagrid-portal.nci.nih.gov
- 7.Additional information about the Globus toolkit can be found on the Globus Alliance Toolkit page: (http://www.globus.org/toolkit/)
- 8.Hung et al. (2008) Integrating caGrid and TeraGrid. Teragrid ’08 conference, June 9-13, 2008 (http://www.teragrid.org/events/teragrid08/Papers/papers/100.pdf) [Google Scholar]
- 9.A complete list of analytical tools can be found on the caBIG® community Web site: (https://cabig.nci.nih.gov/)
- 10.More detailed information about caBIG® grid security can be found at https://cabig.nci.nih.gov/workspaces/Architecture/work-groups/Security%20Work%20Group
- 11.Stein L (2008) Towards a cyberinfrastructure for the biological sciences: progress, visions and challenges. Nat Rev Genet. September;9(9):678–88 [DOI] [PubMed] [Google Scholar]