Abstract
Background
Metabolic reprogramming, immune evasion and tumor-promoting inflammation are three hallmarks of cancer that provide new perspectives for understanding the biology of cancer. We aimed to figure out the relationship of tumor glycolysis and immune/inflammation function in the context of breast cancer, which is significant for deeper understanding of the biology, treatment and prognosis of breast cancer.
Methods
Using mRNA transcriptome data, tumor-infiltrating lymphocytes (TILs) maps based on digitized H&E-stained images and clinical information of breast cancer from The Cancer Genome Atlas projects (TCGA), we explored the expression and prognostic implications of glycolysis-related genes, as well as the enrichment scores and dual role of different immune/inflammation cells in the tumor microenvironment. The relationship between glycolysis activity and immune/inflammation function was studied by using the differential genes expression analysis, gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, gene set enrichment analyses (GSEA) and correlation analysis.
Results
Most glycolysis-related genes had higher expression in breast cancer compared to normal tissue. Higher phosphoglycerate kinase 1 (PGK1) expression was associated with poor prognosis. High glycolysis group had upregulated immune/inflammation-related genes expression, upregulated immune/inflammation pathways especially IL-17 signaling pathway, higher enrichment of multiple immune/inflammation cells such as Th2 cells and macrophages. However, high glycolysis group was associated with lower infiltration of tumor-killing immune cells such as NKT cells and higher immune checkpoints expression such as PD-L1, CTLA4, FOXP3 and IDO1.
Conclusions
In conclusion, the enhanced glycolysis activity of breast cancer was associated with pro-tumor immunity. The interaction between tumor glycolysis and immune/inflammation function may be mediated through IL-17 signaling pathway.
Keywords: Breast cancer, Glycolysis, Immune/inflammation function, PGK1, IL-17 signaling pathway, TCGA, Prognosis
Background
Breast cancer is the leading cause of cancer death among women [1]. Breast cancer can be classified as Normal-like, Luminal A, Luminal B, HER2-enriched and Basal-like subtypes. In addition to surgery, systemic therapies including chemotherapy, hormonal therapy, and molecular targeted therapy can be chosen based on the molecular characteristics to combat cancer [2]. Although these systemic therapies have improved patients’ outcomes, many patients do not respond to these existing treatments, which leads to poor prognosis [3, 4]. On this account, some researches have been carried out to explore new effective therapies in breast cancer such as Immunotherapy and metabolic therapy [5, 6]. However, only a minority of patients benefit from these emerging therapies [7]. Exploring the interplay between tumor cells and tumor microenvironment could lead to deeper understanding of breast cancer initiation, progression, and therapeutic resistance, possibly provide prospective strategies for cancer prevention and treatment [8, 9].
Metabolic reprogramming is a key hallmark of cancer [10]. The most frequently mentioned way of metabolic reprogramming is aerobic glycolysis. Aerobic glycolysis, also known as the “Warburg effect”, is a general way of glucose metabolism in cancer cells. In this way, glucose is mainly processed into lactate even when oxidative capacity is intact. This will lead to a highly acidic microenvironment. According to current research, tumor aerobic glycolysis can contribute to malignant transformation and tumor progression [11]. Therefore, tumor aerobic glycolysis has possible implications for prognosis judgment and cancer treatment [12]. Exploiting tumor glycolysis for clinical application requires figuring out how intrinsic and extrinsic factors to be integrated to modify the metabolic phenotype [13].
Tumor microenvironment (TME), cancer cells’ supporting hotbed, has complicated and changeable composition including tumor infiltrating lymphocytes (TILs), other immune and inflammatory cells, fibroblasts, the blood and lymphatic vascular networks, the extracellular matrix (ECM) and so on [14]. Numerous evidences suggest that the immune cells infiltration in the TME could interact with tumor cells, which may affect tumor progression and the efficacy of existing anticancer therapies [15, 16]. The immune cells recruited to the tumor site have dual characters, some can restrain carcinogenesis and tumor progression while others may play a tumor-promoting role [17]. Thus, it is important to figure out the cellular heterogeneity composition of the immune cells infiltration and the cause behind it. The results may be significant for optimizing existing treatments and identifying novel therapeutic targets [18].
Several studies have investigated the relationship between tumor glycolysis and immune/inflammation function [19–21]. A highly acidic microenvironment due to tumor glycolysis may differentially influence immune cells infiltration, ultimately leading to immune escape and cancer progression [22]. Exploration of the associations is providing a deeper look into cancer biological processes and can lead to more effective therapy selection. So far, however, there has been little comprehensive analysis focusing on the relationships between the tumor glycolysis, immune/inflammation function and the clinical features based on clinical data in the field of breast cancer. Given this, we implemented studies with transcriptome and clinical data of breast cancer from The Cancer Genome Atlas (TCGA) projects to explore the landscape of tumor glycolysis and immunity in breast cancer, to identify the relationship between the tumor glycolysis and immune and inflammatory cells infiltration, and to figure out the impact of the two on breast cancer prognosis.
Methods
The preprocessing step for transcriptome and clinical data
We obtained mRNA transcriptome data of breast cancer from TCGA. The RNA sequence data of breast cancer was downloaded directly from TCGA data portal (https://portal.gdc.cancer.gov/). The obtained expression data of 19657 genes from 1208 samples including 112 normal tissues and 1096 tumor tissues were used for follow-up research. The corresponding clinicopathological features including ER, PR, Her2 status, tumor stage, survival time and survival status were downloaded from TCGA data portal.
Identification of differentially expressed genes and functional enrichment analysis
The tumor samples were divided into low glycolysis group and high glycolysis group based on the median expression of PGK1. Gene expression profiles were derived based on differential genes expression analysis using limma R package. Differentially expressed genes (DEGs) were identified through volcano plot filtering. The thresholds for DEGs were |log2FC| ≥ 1 and adjusted P value < 0.05. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed using DAVID (https://david.ncifcrf.gov/) and WebGestalt (http://www.webgestalt.org/) [23, 24], using P < 0.05 as the cut-off criterion. Gene set enrichment analysis (GSEA) was conducted to find out the significantly upregulated and downregulated pathways between high and low glycolysis group using WebGestalt, using FDR < 0.05 as the cut-off criterion.
Cell type enrichment analysis
To evaluate the cellular heterogeneity of the tumor microenvironment, the xCell tool, a gene signature-based method, was used to figure out the abundance of cell types in tumor microenvironment, based on the tissue transcriptome profiles [25]. Total 64 cell types including adaptive and innate immune cells, stem cells, epithelial cells and extracellular matrix cells and their enrichment scores were obtained. The present study focused on immune landscape of breast cancer. Therefore, 34 adaptive and innate immune cell types were retained for the further analysis.
TIL density analysis
Mappings of tumor-infiltrating lymphocytes (TILs), based on digital scanned H&E images from TCGA breast cancer samples were downloaded from The Cancer Imaging Archive (TCIA) [26]. TIL maps were identified by a convolutional neural network trained computational stain developed by Saltz et al. [27]. Spearman correlation coefficient of glycolysis and TIL percentage from spatial estimates of TIL maps was calculated.
Statistical analysis
SPSS 23.0 and R 3.4.4 were used for statistical analysis. Differences of continuous data between groups were analyzed by the ANOVA test. Categorical data were compared using the χ2 or Fisher exact test. Spearman correlation analysis was used to assess the potential relevance. Overall survival rate of each group was estimated using the Kaplan–Meier method and differences between groups were assessed by using the log-rank test. Cox proportional hazard regression model was used to analyze the prognostic significance of glycolysis-related genes. P value < 0.05 was considered statistically significant.
Results
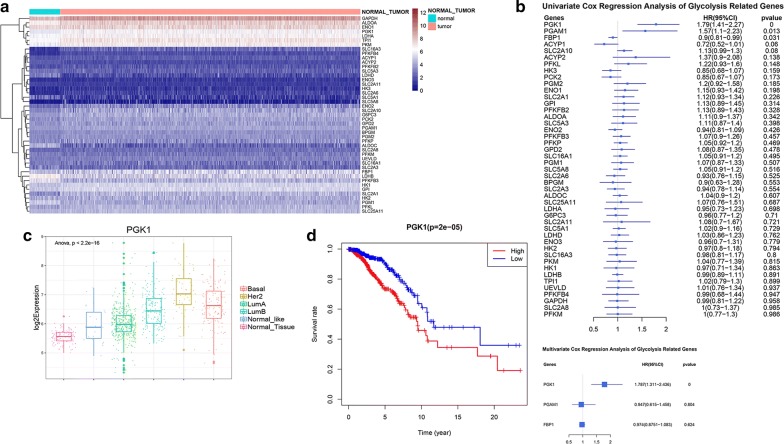
Expression of glycolysis-related genes and the prognostic significance
The catabolic glycolysis and anabolic gluconeogenesis are two reciprocal pathways of glucose metabolism, which are catalyzed by a series of enzymes. Referring to published literature, 45 glycolysis and gluconeogenesis related genes, which can encode enzymes and metabolite transporters in the glucose metabolism pathways, were selected [28]. We compared the expression profile of these glycolysis and gluconeogenesis related genes between normal mammary tissues and breast cancer tissues. A heatmap was illustrated to display the results (Fig. 1a). The results revealed that the expression of 29 glycolysis and gluconeogenesis related genes including SLC2A1, HK2, GPI, PFKP, ALDOA, TPI1, GAPDH, PGK1, PGAM1, ENO1, PKM, LDHA, etc. were significantly higher in tumor tissues compared with normal mammary tissues. This result was consistent with the finding that tumors are characterized by enhanced glycolysis activity and glucose uptake for energy production and macromolecular biosynthesis [29].
Fig. 1.
Expression of glycolysis-related genes and its prognostic significance. a Expression of glycolysis and gluconeogenesis related genes in normal and tumor tissues. b Univariate Cox regression and multivariate Cox regression were applied to calculate hazard ratios (HRs) of glycolysis-related genes. c The boxplot showed expression of PGK1 in normal tissues and different subtypes of breast cancer. d Kaplan–Meier survival analysis of low and high PGK1 expression was shown
Trying to screen out the candidate enzymes and metabolite transporters in glucose metabolism pathway that may play key roles in breast cancer progression, we explored the association between the expression of glycolysis and gluconeogenesis related genes and clinical manifestation of breast cancer based on TCGA database. Univariate and multivariate Cox proportional-hazards regression analysis were conducted to evaluate gene-associated hazard ratios (HRs) for overall survival. The results indicated that PGK1 overexpression was an independent risk factor for overall death in breast cancer (Fig. 1b).
The 1096 breast cancer patients derived from TCGA were then divided into two groups based on the median expression of PGK1. Higher PGK1 expression was positively related to larger tumor size, higher risk of lymphatic and distant metastasis, more advanced tumor stage and higher overall death. Moreover, the patients with higher PGK1 expression had a lower positive rate of ER and PR, but a higher positive rate of Her2 (Table 1). Compared with Luminal A and Luminal B subtypes, HER2-enriched and Basal-like subtypes had remarkable higher PGK1 expression (Fig. 1c). Survival analysis showed high PGK1 expression group was significantly associated with shorter overall survival, which confirmed the prognostic significance of PGK1 (Fig. 1d).
Table 1.
Clinical characteristics of 1096 samples according to PGK1 low or high expression
| Characteristics | PGK1 | ||
|---|---|---|---|
| Low expression | High expression | P-value | |
| Age | 57.84 ± 12.81 | 58.77 ± 13.57 | 0.243 |
| T stage | 0.002 | ||
| T1 | 156 | 124 | |
| T2 | 304 | 331 | |
| T3 | 77 | 62 | |
| T4 | 10 | 29 | |
| Tx | 1 | 2 | |
| N stage | 0.047 | ||
| N0 | 270 | 247 | |
| N1 | 187 | 176 | |
| N2 | 44 | 75 | |
| N3 | 38 | 39 | |
| Nx | 9 | 11 | |
| M stage | 0.007 | ||
| M0 | 453 | 457 | |
| M1 | 4 | 17 | |
| Mx | 91 | 74 | |
| Tumor stage (AJCC 8th version) | 0.001 | ||
| Stage I | 110 | 72 | |
| Stage II | 309 | 315 | |
| Stage III | 117 | 131 | |
| Stage IV | 3 | 16 | |
| Stage X | 6 | 6 | |
| NA | 3 | 8 | |
| ER status | < 0.001 | ||
| Positive | 461 | 345 | |
| Negative | 64 | 176 | |
| NA | 23 | 27 | |
| PR status | < 0.001 | ||
| Positive | 410 | 288 | |
| Negative | 113 | 232 | |
| NA | 25 | 28 | |
| Her2 status | < 0.001 | ||
| Positive | 50 | 111 | |
| Negative | 288 | 276 | |
| NA | 210 | 161 | |
| Overall survival | < 0.001 | ||
| Alive | 495 | 453 | |
| Dead | 53 | 95 | |
PGK1 expression can reflect the activity of glycolysis to some extent
Phosphoglycerate kinase 1 (PGK1), the first ATP-generating enzyme in glycolytic pathway, catalyzes the conversion process from 1,3-diphosphoglycerate to 3-phosphoglycerate and from ADP to ATP. Given the important role of PGK1 in the glycolytic pathway and breast cancer progression, we selected PGK1 for further study. We then explored whether the expression of PGK1 could represent the activity of glycolysis. For this purpose, we compared the expression of other glycolysis related genes between normal tissues, low PGK1 expression group and high PGK1 expression group. The results showed that key genes in the glycolytic pathway including SLC2A1, HK2, GPI, PFKP, ALDOA, TPI1, GAPDH, PGAM1, ENO1, PKM, LDHA had remarkable higher expression in high PGK1 expression group (Fig. 2). What’s more, the expression level of carbonic anhydrase 9 (CA9) in high PGK1 expression group was seven times higher compared with low PGK1 expression group (Fig. 2). CA9 catalyzes the reversible hydration of carbon dioxide and participates in pH regulation. CA9 overexpression could prevent the accumulation of acidic metabolites in tumor cells at the expense of acidifying the extracellular tumor microenvironment. Thus, the overexpression of CA9 suggested higher glycolysis activity, lactic acid production and lactic acid output [30]. All the results above indicated that PGK1 expression can reflect the activity of glycolysis to some extent, therefore, low PGK1 expression group and high PGK1 expression group could be considered as low glycolysis group and high glycolysis group, respectively.
Fig. 2.
PGK1 expression can reflect the activity of glycolysis to some extent. The boxplots represented the expression of glycolytic pathway-specific genes SLC2A1, LDHA, ENO1, PFKP, PGAM1, PKM, HK2, GPI, TPI1, ALDOA, GAPDH and pH-regulated gene CA9 in normal tissue, low PGK1 expression and high PGK1 expression group
Glycolysis activity was closely related with immune function
To further investigate the biological difference between low glycolysis group and high glycolysis group, we analyzed the differential gene profiles between them 470 DEGs including 255 upregulated and 215 downregulated genes were identified to be significantly associated with the glycolysis activity (Fig. 3a). We found that numerous cytokines, chemokines, inflammatory mediators and related receptors were dysregulated in the high glycolysis group and involved in immune cell recruitment and function (Fig. 3b).
Fig. 3.
Glycolysis activity was closely related with immune function. a The volcano plot showed the DEGs between low glycolysis group and high glycolysis group. The blue color represented the downregulated genes, while the red showed the upregulated genes. b The heatmap displayed dysregulated inflammation-related genes, the glycolysis group and molecular subtypes group at the top of the heatmap. c, d GO and KEGG enrichment analysis of 470 DEGs. The top 30 most significant GO and KEGG terms were illustrated
GO and KEGG enrichment analysis were conducted to uncover the gene function and biological pathways of DEGs. The results showed that immune-related functions were noticeable as shown in Fig. 3c, d. More specifically, the immune-related functions included GO:0061844 antimicrobial humoral immune response mediated by antimicrobial peptide, GO:0002548 monocyte chemotaxis, GO:0048247 lymphocyte chemotaxis, GO:0070098 chemokine-mediated signaling pathway, GO:0030593 neutrophil chemotaxis, GO:0071621 granulocyte chemotaxis, GO:0071674 mononuclear cell migration, GO:1990266 neutrophil migration, GO:0097530 granulocyte migration, GO:0097529 myeloid leukocyte migration, GO:0030595 leukocyte chemotaxis, GO:0050900 leukocyte migration, GO:0006954 inflammatory response, etc. (Fig. 3c). The enriched signaling pathways were hsa04657 IL-17 signaling pathway, hsa04060: Cytokine-cytokine receptor interaction, hsa04062: Chemokine signaling pathway and some metabolism-related signaling pathways (Fig. 3d).
Immune-related pathways were upregulated in the high glycolysis group
Given the finding above, we then conducted GSEA to explore whether immune-related pathways were upregulated or downregulated in high glycolysis group. FDR < 0.05 was considered statistically significant. Top 30 upregulated pathways and seven downregulated pathways were shown in Fig. 4a. The results revealed that numerous immune-related KEGG pathways were significantly upregulated, which included hsa04657 IL-17 signaling pathway (Fig. 4b), hsa04612 Antigen processing and presentation, hsa05332 Graft-versus-host disease, hsa05323 Rheumatoid arthritis, hsa05330 Allograft rejection, hsa05169 Epstein-Barr virus infection, hsa04650 Natural killer cell mediated cytotoxicity, hsa04621 NOD-like receptor signaling pathway, etc. Glucose metabolism pathways were also upregulated as expected. Not only the immune-related pathways and glucose metabolism pathways but also some other pathways such as cell cycle, DNA replication and ovarian steroidogenesis were influenced by glycolysis activity, which were shown in Fig. 4a. Altogether, these findings indicated that high glycolysis activity was associated with enhanced immune/inflammation activity in breast cancer.
Fig. 4.
Immune-related pathways were upregulated in the high glycolysis group. a Top 30 upregulated pathways and seven downregulated pathways in high glycolysis group were obtained by GSEA. The red bars represented the upregulated pathways, while the blue bars showed the downregulated pathways. b GSEA showed that IL-17 signaling pathway was significantly upregulated in high glycolysis group. The value of normalizedEnrichmentScore and FDR were displayed in the figure
Correlation between immune/inflammation cells infiltration and glycolysis activity
In order to verify the association between immune/inflammation function and glycolysis activity, immune cell types enrichment scores were compared between low glycolysis group and high glycolysis group. High glycolysis group showed significantly higher immunescore. Most of the immune cell type enrichment scores were remarkable higher in high glycolysis group including CD4+ memory T-cells, CD8+ naive T-cells, CD8+ Tem, Th1 cells, Th2 cells, NK cells, pro B-cells, Plasma cells, Monocytes, Macrophages, Macrophages M1, DC, pDC, iDC and aDC. Instead, CD4+ Tcm, NKT cell and Mast cells enrichment scores were lower in high glycolysis group (Fig. 5a). What’s more, PGK1 expression was positively correlated with Th1 cells (r = 0.285, P < 0.001), Th2 cells (r = 0.480, P < 0.001), Monocytes (r = 0.246, P < 0.001), Macrophages (r = 0.304, P < 0.001), Macrophages M1 (r = 0.302, P < 0.001), Pro B-cells (r = 0.314, P < 0.001) and pDC (r = 0.263, P < 0.001), but negatively correlated with NKT cells (r = − 0.226, P < 0.001) (Fig. 5b). TILs density also showed positive correlation with tumor glycolysis (r = 0.194, P < 0.001) (Fig. 5c). HER2-enriched and Basal subtypes had the highest fraction of TILs, which was in line with PGK1 expression in breast cancer subtypes (Fig. 5d).
Fig. 5.
Correlation between immune/inflammation cells infiltration and glycolysis activity. a A heatmap showed the immune cell types enrichment scores of all samples. Groups based on glycolysis activity and molecular subtypes were shown for each sample (above the heatmap). Note that ** represented the cell types which showed significantly different enrichment between high and low glycolysis groups. b Correlation between PGK1 expression and individual immune cell enrichment scores were analyzed. The Spearman correlation coefficient between PGK1 expression and Th1 cells, Th2 cells, Monocytes, Macrophages, Macrophages M1, Pro B-cells, NKT cells were remarkable. c The scatter plot demonstrated that the Spearman correlation coefficient between PGK1 expression and TIL percentage. d TIL density in different subtypes of breast cancer was shown in the boxplot
High glycolysis may be associated with suppressed antitumor immunity
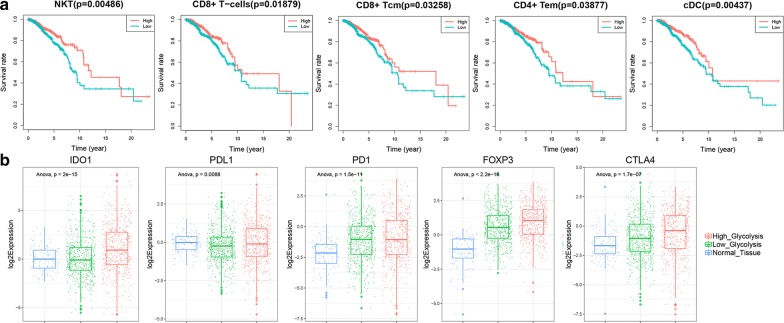
These immune and inflammatory cells may play a conflicting role in both tumor antagonism and tumor promotion. To identify the tumor-antagonizing and tumor-promoting immune and inflammatory cells, we conduct survival analysis to explore association between clinical outcome and enrichment scores of immune cell types in the tumor microenvironment. The results revealed that highly enriched cDC, NKT, CD8+ T-cells, CD8+ Tcm, CD4+ Tem cells led to better overall survival in breast cancer, while the immunescore and other immune cell types enrichment scores didn’t show prognosis significance (Fig. 6a). However, these immune cells which may play an antitumor role weren’t enriched in high glycolysis group. Instead, NKT cell enrichment scores were significantly lower in high glycolysis group.
Fig. 6.
High glycolysis may be associated with suppressed antitumor immunity. a Survival analysis of each type of immune cell showed that groups with highly enriched cDC, NKT, CD8+ T-cells, CD8+ Tcm, CD4+ Tem cells had higher survival rates. b The expression of immunosuppressive factors including IDO1, PDL1, PD1, FOXP3 and CTLA4 in normal tissue, low glycolysis group and high glycolysis group
Among the 255 upregulated genes, we found that the expression of Indoleamine 2,3-dioxygenase 1 (IDO1) in high glycolysis group is three times higher than that in low glycolysis group (Fig. 6b). IDO1 encodes an enzyme that catalyzes the rate-limiting step of tryptophan catabolism to the kynurenine and acts as a suppressor of anti-tumor immunity. Tryptophan shortage and kynurenine accumulation caused by IDO1 overexpression inhibit T lymphocytes proliferation and induce apoptosis of cytotoxic T lymphocytes (CTLs) [31, 32]. Based on this finding, we further compared the expression of other well-known immunosuppressive factors including PD1, PDL1, CTLA4 and FOXP3 [33]. Similar with IDO1, we found that PDL1, CTLA4 and FOXP3 also had higher expression in high glycolysis group (Fig. 6b). Overexpression of immunosuppressive factors and lack of NKT cells in high glycolysis group both indicated that high glycolysis may be associated with suppressed antitumor immunity.
Discussion
Sufficient energy and metabolic intermediates for biosynthesis are the foundation of tumor cells initiation, proliferation and metastasis. Thus, many types of cancer are characterized by enhanced level of glycolysis and suppressed mitochondrial metabolism. In this way, tumor cells gain an advantage over stromal and immune cells in the competition to share limited nutrients and acquire ATP and intermediate metabolites faster [11]. The present study demonstrated that the key enzymes in glycolytic pathways had higher expression in breast cancer tissue, which indicated increased glycolysis activity in tumors. Among these glycolysis related genes, PGK1 has attracted our attention for its significant correlation with advanced tumor stage, and poor prognosis. By regulating ATP and 3-phosphoglycerate levels, PGK1 plays a critical role in coordinating energy production with biosynthesis and redox balance [34]. A recent comprehensive analysis of pan-cancer metabolic transformation also indicated that PGK1 was the most frequently upregulated gene in glycolytic pathway and had 100% frequency of upregulation in different types of cancer [28]. It has been demonstrated that high expression and activity of PGK1 was associated with poor prognosis in several types of cancer [35, 36]. Metabolism remodeling is an exquisite process regulated by oncogenic signaling pathways and transcription factors [11]. PGK1 functions as downstream targets of multiple oncogenic signal pathways and directly regulated by MYC,HIF-1α and other major regulator in metabolic reprogramming [37]. What’s more, PGK1 can translocate to Mitochondria and function as a protein kinase to inhibit pyruvate dehydrogenase complex, and promote tumor progress by increasing glycolysis and suppressing mitochondrial tricarboxylic acid cycle [38]. All of the above evidence indicated that PGK1 played a key role in metabolism reprogramming. Our study confirmed this hypothesis as glycolytic pathway-specific genes and pH-regulating genes had remarkable higher expression and glucose metabolism pathways and cell proliferation-related pathways such as cell cycle and DNA replication significantly upregulated in high PGK1 expression group. Therefore, PGK1 expression was regarded to represent the activity of glycolysis in the present study.
Much work so far has focused on the tumor glycolysis. However, the biological events affected by the tumor glycolysis, especially its immune/inflammation regulatory role, haven’t been well clarified. we compared gene expression profile, immune cell type enrichment scores and TIL percentage between low and high glycolysis groups and conducted functional enrichment analysis. The results suggested that numerous cytokines, chemokines, related receptors and immune/inflammation-related pathways were upregulated in the high glycolysis group. Consistent with this, high glycolysis group had a higher immunescore and higher TIL percentage. However, NKT, CD8+ T-cells, CD8+ Tcm, CD4+ Tem, cDC cells which may have an anti-tumor effect in breast cancer weren’t enriched in high glycolysis group. What’s more, immunosuppressive factors including IDO1, PD1, PDL1, CTLA4 and FOXP3 showed remarkable higher expression in high glycolysis group.
The upregulated immune factors and increased infiltration of immune cells did not bring a good outcome to the patients in the high glycolysis group. It seemed to be an inflammatory condition. Inflammation is another key hallmark for cancer initiation and progression, which refers to the local immune response involving cytokines, chemokines, small inflammatory protein mediators, and infiltrating immune cells in the TME [15]. The presence of tumor-antagonizing CTLs and NK cells is not surprising, however the list of tumor-promoting inflammatory cells now includes monocytes, macrophages, DCs, mast cells and neutrophils, as well as T and B lymphocytes [39]. Our study also indicated that high glycolysis contributed to an immunosuppressive tumor microenvironment. The phenomenon that tumor glycolysis and immune/inflammation function influence each other was also observed in other studies. One study found that IL-6 secreted by macrophages can phosphorylate PGK1 to promote glycolysis and tumorigenesis [40]. Several excellent studies have revealed that the metabolic microenvironment of tumor cells including glucose deprivation, lactic acid accumulation and acidification of tumor microenvironment may selectively disable tumor-killing T and NK cell activation and promote immune evasion, while attracting tumor-promoting inflammatory cells [19, 22, 41–45]. Lactic acid accumulation was demonstrated to induce macrophages toward an inflammatory pro-tumor phenotype [46]. In addition to the one-way effect, some studies have shown that cytokines from inflammatory cells and lactic acid from tumor cells can form a positive feedback loop to promote tumor progression [20, 47].
In our study, the most enriched immune/inflammation-related pathway in high glycolysis group was IL-17 signaling pathway. Emerging evidence indicated the tumor-promoting role of IL-17 in colorectal cancer, pancreatic cancer and lung cancer [48–50]. The upregulation of IL-17 signaling pathway recruited myeloid suppressive cells, which could induce angiogenesis and restrain anti-tumor immunity [51]. Interestingly enough one study showed that increased glucose uptake and metabolism reprogramming mediated by IL-17 signaling pathway was necessary for the activation of lymph node stromal cells [52]. Our study revealed that IL-17 mediated metabolism reprogramming existed not only in lymph node stromal cells but also in breast cancer tissues. In addition to macrophages which have been the research focus of pro-tumor inflammation, T helper cells especially Th2 cells showed remarkable correlation with tumor glycolysis in our study (r = 0.480, P < 0.001). Previous studies indicated that tumor-associated antigen and HLA-G expressed in tumors can promote immune evasion by strongly favoring Th2 development [53, 54]. However, the relationships between Th2 cells and tumor glycolysis have not been elucidated. Since our study was entirely based on public database and bioinformatic methods, the results should be interpreted with caution. Further in vitro and in vivo studies are needed to confirm our point.
Taken together, these evidences indicated that tumor glycolysis and pro-tumor immunity interdependently promoted each other. Thus, targeting glycolysis may refresh anti-tumor immunity and improve responsiveness to existing treatment such as chemotherapy and Her2-targeted therapy [55]. Since high glycolysis group had higher expression of immune checkpoint and higher TILs density which were both promising predictor for immunotherapy [56], they may exhibit a better immunotherapy response. In line with our research, tumor metabolic parameters on fluorodeoxyglucose positron emission tomography (FDG-PET) showed positive correlation with PD1/PDL1 expression and presence of TILs in non-small lung cancer [57, 58]. Some literature have also suggested the value of FDG-PET in predicting response to immune checkpoint inhibitors in non-small cell lung cancer [59], which partially supported our hypotheses. In our study, Her2-enriched and basal subtypes had higher glycolysis activity and higher immune inflammation cells infiltration in our study, which was consistent with its higher proliferative ability and higher malignant potential [2]. Thus, FDG-PET as the noninvasive detection method of tumor glycolysis may have the potential value to identify molecular subtypes of breast cancer and predict response rate to neoadjuvant chemotherapy since TIL is already known as a good predictor of response to neoadjuvant chemotherapy [60–62].
Given the critical role of PGK1 in metabolism remodeling and tumorigenesis, it may be a potential therapeutic target. However, targeting key metabolic enzyme can lead to severe systemic toxicity, because they are also essential for sustaining the normal cell function [6]. Attenuating the enzymatic activity of PGK1 by targeting its post-translational modifications (including phosphorylation, acetylation, ubiquitination, etc.) which have been shown significantly altered in cancer might reduce systemic toxicity and is an alternative approach [35, 40, 63, 64]. Despite all this, there is an urgent requirement for clinical application to establish a selective targeted delivery system to inhibit PGK1 activity in cancer cells precisely [6].
Numerous studies have revealed that the initiation and progression of cancer are dependent on a complex interplay between tumor cells and tumor microenvironment. Our study demonstrated that tumor glycolysis and immune inflammatory response may be the entry points for exploring the communication between tumor cells and tumor microenvironment. Deeper insight of the correlation between tumor glycolysis and immune inflammatory infiltration may lead to a revolution in cancer prevention and treatment.
Conclusion
In conclusion, the enhanced glycolysis activity of breast cancer was associated with enriched inflammatory cell infiltration and enhanced inflammation response but higher expression of immunosuppressive factors and inhibition of antitumor immunity. The interaction between tumor glycolysis and immune/inflammation function may be mediated through IL-17 signaling pathway. Thus, drugs targeting metabolism may have an anti-inflammatory effect and vice versa. The level of glycolysis activity may guide the selection of checkpoint inhibitor immunotherapy. PGK1, as an important enzyme in glycolytic pathways, had obvious influence on breast cancer patients’ clinical outcome, which revealed that PGK1 played a critical role in the progression of breast cancer and may be a potential therapeutic target.
Acknowledgements
Not applicable.
Abbreviations
- TME
Tumor microenvironment
- TILs
Tumor infiltrating lymphocytes
- ECM
Extracellular matrix
- TCGA
The Cancer Genome Atlas
- DEGs
Differentially expressed genes
- GSEA
Gene set enrichment analysis
- TCIA
The Cancer Imaging Archive
- HRs
Hazard ratios
- PGK1
Phosphoglycerate kinase 1
- CA9
Carbonic anhydrase 9
- IDO1
Indoleamine 2,3-dioxygenase 1
- CTLs
Cytotoxic T lymphocytes
- FDG-PET
Fluorodeoxyglucose positron emission tomography
Authors’ contributions
WHL, MX and PFY designed the study. YL and KHL collected the mRNA transcriptome data and clinical information from TCGA. WHL and YL performed analyses on TCGA data. WHL and MX performed statistical analyses. WHL wrote the manuscript, JZ, QYZ, ZWH, FD and CW reviewed and revised the manuscript. All authors read and approved the final manuscript.
Funding
The research was supported from Grants 2017CKB899 and 2015CKB733 funded by Hubei Provincial Natural Science Foundation of China.
Availability of data and materials
All data was obtained from The Cancer Genome Atlas (TCGA, https://tcgadata.nci.nih.gov/tcga/). Other data generated or analyzed during this study were included in this published article and its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenhui Li and Ming Xu contributed equally to this work
References
- 1.Global Burden of Disease Cancer C. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso F, Harbeck N, Barrios CH, Bergh J, Cortes J, El Saghir N, et al. Research needs in breast cancer. Ann Oncol. 2017;28(2):208–217. doi: 10.1093/annonc/mdw571. [DOI] [PubMed] [Google Scholar]
- 4.DeMichele A, Yee D, Esserman L. Mechanisms of resistance to neoadjuvant chemotherapy in breast cancer. N Engl J Med. 2017;377(23):2287–2289. doi: 10.1056/NEJMcibr1711545. [DOI] [PubMed] [Google Scholar]
- 5.McCoach CE, Bivona TG. Engineering multidimensional evolutionary forces to combat cancer. Cancer Discov. 2019;9:587–604. doi: 10.1158/2159-8290.CD-18-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emens LA. Breast cancer immunotherapy: facts and hopes. Clin Cancer Res. 2018;24(3):511–520. doi: 10.1158/1078-0432.CCR-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med. 2015;13:45. doi: 10.1186/s12916-015-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 16.Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, et al. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin Cancer Biol. 2015;35(Suppl):S199–s223. doi: 10.1016/j.semcancer.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terlizzi M, Casolaro V, Pinto A, Sorrentino R. Inflammasome: cancer’s friend or foe? Pharmacol Ther. 2014;143(1):24–33. doi: 10.1016/j.pharmthera.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cascone T, McKenzie JA, Mbofung RM, Punt S, Wang Z, Xu C, et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 2018;27(5):977–987.e4. doi: 10.1016/j.cmet.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 21.Justus CR, Sanderlin EJ, Yang LV. Molecular connections between cancer cell metabolism and the tumor microenvironment. Int J Mol Sci. 2015;16(5):11055–11086. doi: 10.3390/ijms160511055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill KS, Fernandes P, O’Donovan TR, McKenna SL, Doddakula KK, Power DG, et al. Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim Biophys Acta. 2016;1866(1):87–105. doi: 10.1016/j.bbcan.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saltz JGR, Hou L, Kurc T, Singh P, Nguyen V, 2018. Tumor-infiltrating lymphocytes maps from TCGA H&E whole slide pathology images. Cancer Imaging Arch. [DOI]
- 27.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(1):181–193. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 30.Swietach P, Vaughan-Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev. 2007;26(2):299–310. doi: 10.1007/s10555-007-9064-0. [DOI] [PubMed] [Google Scholar]
- 31.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9(10):1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 32.Carvajal-Hausdorf DE, Mani N, Velcheti V, Schalper KA, Rimm DL. Objective measurement and clinical significance of IDO1 protein in hormone receptor-positive breast cancer. J Immunother Cancer. 2017;5(1):81. doi: 10.1186/s40425-017-0285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol. 2015;33(9):983–991. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 34.Wang S, Jiang B, Zhang T, Liu L, Wang Y, Wang Y, et al. Insulin and mTOR pathway regulate HDAC3-mediated deacetylation and activation of PGK1. PLoS Biol. 2015;13(9):e1002243. doi: 10.1371/journal.pbio.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, et al. MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 2017;77(21):5782–5794. doi: 10.1158/0008-5472.CAN-17-0671. [DOI] [PubMed] [Google Scholar]
- 36.Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology. 2017;65(2):515–528. doi: 10.1002/hep.28887. [DOI] [PubMed] [Google Scholar]
- 37.Qing G, Skuli N, Mayes PA, Pawel B, Martinez D, Maris JM, et al. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1alpha. Cancer Res. 2010;70(24):10351–10361. doi: 10.1158/0008-5472.CAN-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell. 2016;61(5):705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15(11):e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Yu G, Chu H, Wang X, Xiong L, Cai G, et al. Macrophage-Associated PGK1 Phosphorylation Promotes Aerobic Glycolysis and Tumorigenesis. Mol Cell. 2018;71(2):201–215.e7. doi: 10.1016/j.molcel.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Fu D, Geschwind JF, Karthikeyan S, Miller E, Kunjithapatham R, Wang Z, et al. Metabolic perturbation sensitizes human breast cancer to NK cell-mediated cytotoxicity by increasing the expression of MHC class I chain-related A/B. Oncoimmunology. 2015;4(3):e991228. doi: 10.4161/2162402X.2014.991228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber V, Camisaschi C, Berzi A, Ferro S, Lugini L, Triulzi T, et al. Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol. 2017;43:74–89. doi: 10.1016/j.semcancer.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Scott KE, Cleveland JL. Lactate wreaks havoc on tumor-infiltrating T and NK cells. Cell Metab. 2016;24(5):649–650. doi: 10.1016/j.cmet.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Sun L, Suo C, Li ST, Zhang H, Gao P. Metabolic reprogramming for cancer cells and their microenvironment: beyond the Warburg effect. Biochim Biophys Acta Rev Cancer. 2018;1870(1):51–66. doi: 10.1016/j.bbcan.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Villalba M, Rathore MG, Lopez-Royuela N, Krzywinska E, Garaude J, Allende-Vega N. From tumor cell metabolism to tumor immune escape. Int J Biochem Cell Biol. 2013;45(1):106–113. doi: 10.1016/j.biocel.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Paolini L, Adam C, Beauvillain C, Preisser L, Blanchard S, Pignon P, et al. Lactic acidosis together with GM-CSF and M-CSF induces human macrophages toward an inflammatory protumor phenotype. Cancer Immunol Res. 2020 doi: 10.1158/2326-6066.CIR-18-0749. [DOI] [PubMed] [Google Scholar]
- 47.Netea-Maier RT, Smit JWA, Netea MG. Metabolic changes in tumor cells and tumor-associated macrophages: a mutual relationship. Cancer Lett. 2018;413:102–109. doi: 10.1016/j.canlet.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 48.Wang K, Kim MK, Di Caro G, Wong J, Shalapour S, Wan J, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity. 2014;41(6):1052–1063. doi: 10.1016/j.immuni.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Zoltan M, Riquelme E, Xu H, Sahin I, Castro-Pando S, et al. Immune cell production of interleukin 17 induces stem cell features of pancreatic intraepithelial neoplasia cells. Gastroenterology. 2018;155(1):210–23.e3. doi: 10.1053/j.gastro.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998–1013.e16. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Bechara R, Zhao J, McGeachy MJ, Gaffen SL. IL-17 receptor-based signaling and implications for disease. Nat Immunol. 2019;20(12):1594–1602. doi: 10.1038/s41590-019-0514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majumder S, Amatya N, Revu S, Jawale CV, Wu D, Rittenhouse N, et al. IL-17 metabolically reprograms activated fibroblastic reticular cells for proliferation and survival. Nat Immunol. 2019;20(5):534–545. doi: 10.1038/s41590-019-0367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziegler A, Heidenreich R, Braumüller H, Wolburg H, Weidemann S, Mocikat R, et al. EpCAM, a human tumor-associated antigen promotes Th2 development and tumor immune evasion. Blood. 2009;113(15):3494–3502. doi: 10.1182/blood-2008-08-175109. [DOI] [PubMed] [Google Scholar]
- 54.Agaugue S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood. 2011;117(26):7021–7031. doi: 10.1182/blood-2010-07-294389. [DOI] [PubMed] [Google Scholar]
- 55.Bénéteau M, Zunino B, Jacquin MA, Meynet O, Chiche J, Pradelli LA, et al. Combination of glycolysis inhibition with chemotherapy results in an antitumor immune response. Proc Natl Acad Sci USA. 2012;109(49):20071–20076. doi: 10.1073/pnas.1206360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takada K, Toyokawa G, Okamoto T, Baba S, Kozuma Y, Matsubara T, et al. Metabolic characteristics of programmed cell death-ligand 1-expressing lung cancer on F-fluorodeoxyglucose positron emission tomography/computed tomography. Cancer Med. 2017;6(11):2552–2561. doi: 10.1002/cam4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopci E, Toschi L, Grizzi F, Rahal D, Olivari L, Castino GF, et al. Correlation of metabolic information on FDG-PET with tissue expression of immune markers in patients with non-small cell lung cancer (NSCLC) who are candidates for upfront surgery. Eur J Nucl Med Mol Imaging. 2016;43(11):1954–1961. doi: 10.1007/s00259-016-3425-2. [DOI] [PubMed] [Google Scholar]
- 59.Grizzi F, Castello A, Lopci E. Is it time to change our vision of tumor metabolism prior to immunotherapy? Eur J Nucl Med Mol Imaging. 2018;45(6):1072–1075. doi: 10.1007/s00259-018-3988-1. [DOI] [PubMed] [Google Scholar]
- 60.Antunovic L, De Sanctis R, Cozzi L, Kirienko M, Sagona A, Torrisi R, et al. PET/CT radiomics in breast cancer: promising tool for prediction of pathological response to neoadjuvant chemotherapy. Eur J Nucl Med Mol Imaging. 2019;46(7):1468–1477. doi: 10.1007/s00259-019-04313-8. [DOI] [PubMed] [Google Scholar]
- 61.Kitajima K, Fukushima K, Miyoshi Y, Nishimukai A, Hirota S, Igarashi Y, et al. Association between (1)(8)F-FDG uptake and molecular subtype of breast cancer. Eur J Nucl Med Mol Imaging. 2015;42(9):1371–1377. doi: 10.1007/s00259-015-3070-1. [DOI] [PubMed] [Google Scholar]
- 62.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19(1):40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 63.Nie H, Ju H, Fan J, Shi X, Cheng Y, Cang X, et al. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat Commun. 2020;11(1):36. doi: 10.1038/s41467-019-13601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qian X, Li X, Shi Z, Xia Y, Cai Q, Xu D, et al. PTEN Suppresses glycolysis by dephosphorylating and inhibiting autophosphorylated PGK1. Mol Cell. 2019;76:516–527. doi: 10.1016/j.molcel.2019.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Saltz JGR, Hou L, Kurc T, Singh P, Nguyen V, 2018. Tumor-infiltrating lymphocytes maps from TCGA H&E whole slide pathology images. Cancer Imaging Arch. [DOI]
Data Availability Statement
All data was obtained from The Cancer Genome Atlas (TCGA, https://tcgadata.nci.nih.gov/tcga/). Other data generated or analyzed during this study were included in this published article and its additional files.