Abstract
Background:
Fibroblast growth factor 18 (FGF18) functions in the development of several tissues, including the lung, limb bud, palate, skeleton, central nervous system, and hair follicle. Mice containing a germline knockout of Fgf18 (Fgf18−/−) die shortly after birth. Postnatally, FGF18 is being evaluated for pathogenic roles in fibrosis and several types of cancer. The specific cell types that express FGF18 have been difficult to identify, and the function of FGF18 in postnatal development and tissue homeostasis has been hampered by the perinatal lethality of Fgf18 null mice.
Results:
We engineered a floxed allele of Fgf18 (Fgf18flox) that allows conditional gene inactivation and a CreERT2 knockin allele (Fgf18CreERT2) that allows the precise identification of cells that express Fgf18 and their lineage. We validated the Fgf18flox allele by targeting it in mesenchymal tissue and primary mesoderm during embryonic development, resulting in similar phenotypes to those observed in Fgf18 null mice. We also use the Fgf18CreERT2 allele, in combination with a conditional fluorescent reporter to confirm known and identify new sites of Fgf18 expression.
Conclusion:
These alleles will be useful to investigate FGF18 function during organogenesis and tissue homeostasis, and to target specific cell lineages at embryonic and postnatal time points.
Keywords: bone development, Cre-lox recombination, FGF signaling, knock in mice, reporter gene
1 ∣. BACKGROUND
The Fibroblast Growth Factor (FGF) family consists of 18 signaling proteins that regulate embryonic and postnatal development, and in the adult function as homeostatic and tissue repair factors. FGF18 is in the FGF8 subfamily, comprised of FGF8, FGF17, and FGF18.1-4 This family of FGFs activate FGFR4 and the mesenchymal splice variants of FGFRs 1, 2, and 3.5,6
Mice containing a germline knockout of Fgf18 (Fgf18−/−) die shortly after birth. The likely deleterious phenotypes include cleft palate, saccular stage lung immaturity, and abnormal nerve terminals in the diaphragm.7-10 During embryonic development, Fgf18 primarily signals to mesenchymal tissue in developing bone and cartilage, lung, and brain.8-13 The function of FGF18 in postnatal tissue is poorly understood due to the perinatal lethality of the Fgf18−/− mouse.
In neonatal rats and mice, Fgf18 expression increases in the lung during alveologenesis and then returns to lower levels in the adult.14,15 Cell sorting experiments suggest that Fgf18 is expressed in myofibroblasts of the postnatal lung.16 In skin, Fgf18 expression is found in inner root sheath cells of the hair follicles where it is thought to maintain stem cells in a quiescent state.17-20 In adult joint tissue, Fgf18 is expressed in the articular surface where it is thought to maintain cartilage homeostasis and protect against osteoarthritis.21 Fgf18 is also expressed in the postnatal growth plate.22,23 In the postnatal brain, Fgf18 is transiently expressed in neurons in the cerebral cortex and hippocampus24 and in adult brain tanycytes lining the ventricular wall.25
To facilitate functional studies of Fgf18 in specific tissues and at postnatal and adult time points we have engineered a floxed allele of Fgf18 (Fgf18flox). Targeting this allele in mesenchymal tissue during embryonic development results in similar phenotypes to those observed in Fgf18−/− null mice. To allow the precise identification of cells that express Fgf18 and to follow their lineage, we have engineered a knockin CreERT2 allele, Fgf18CreERT2. Activation of fluorescent reporter alleles in embryonic, neonatal, and adult mice confirm known and identify new sites of Fgf18 expression. These studies validate these novel alleles of Fgf18 for future functional and lineage studies in mice.
2 ∣. EXPERIMENTAL PROCEDURES
2.1 ∣. Animals
All animals were housed in pathogen-free barrier facilities. All studies were performed under a protocol approved by the Institutional Animal Care at Washington University in St. Louis (Approval No. 20160013) or in accordance with the recommendations in the Guide for Care and Use of Laboratory Animals of the National Institutes of Health under a protocol approved by the Animal Care and Usage Committee of NCI-Frederick (NIH) (Animal Study Proposal: 17-069). Mice of both sexes were utilized. Mice were maintained on a mixed C57BL/6J x 129X1/SvJ or outbred genetic background. Fgf18CreERT2 mice and Fgf18flox were generated in house. Fgf18− allele was generated by recombining the Fgf18flox allele in the germline with Sox2-Cre.26
All other mouse strains, including, ROSAtdTomato, Dermo1Cre, TCre, Sox2-Cre, and FLPeR have been previously described.26-30 Postnatal day 0 (P0) was assigned as the day of birth. Embryonic day 0.5 (E0.5) was assigned as the day the vaginal plug was found in timed-mating experiments. Adult mice were 8 weeks of age or older.
2.2 ∣. Genetic crosses
To generate Fgf18−/− fetuses, TCre+/+; Fgf18−/+ mice were intercrossed. To generate TCre+; Fgf18flox/− mice, Fgf18flox/flox females were crossed to TCre+/+; Fgf18−/+ males. Mice were maintained on an outbred background. The TCre line has been described previously.29 To investigate the effectiveness of Cre mediated recombination, we generated Fgf18CreERT2/+; ROSAtdTomato/tdTomato males and mated them to females to generate experimental litters. This cross was used to assess both GFP signal and Cre-mediated recombination. Fgf18CreERT2/+; ROSAtdTomato/+ when exposed to tamoxifen are referred to as Fgf18Lineage mice.
2.3 ∣. Southern blot
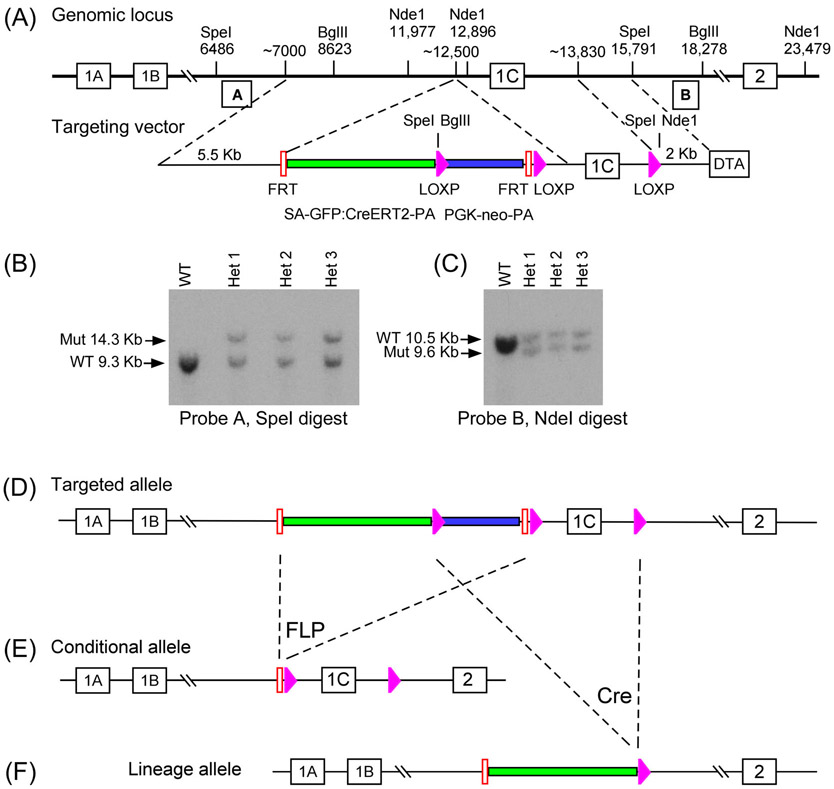
Genomic DNA digested with SpeI was screen by DNA analysis with probes designed against the 5′ end and generated by PCR amplification (Probe A: Forward, gctgaatatgttcagatcttggctctgtgc; Reverse, cacaggtgggtagcagatgcagcatgatac). Genomic DNA digested with NdeI was screen by DNA analysis with probes designed against the 3′ end and generated by PCR amplification (Probe B: Forward, tgtgagtattctgaatgactgtggtcccac; Reverse, gacagccagctcatgtgctcatgatgtctg).
2.4 ∣. Sample preparation: Skeleton
Skeletons were prepared as described previously.31,32 For postnatal day P0 skeletal preparations, carcasses were skinned and eviscerated, and then soaked in acetone for 12-24 hours, cleared in 2% KOH (12-24 hours), stained with alizarin red S (Millipore-Sigma, 58 005) and alcian blue (Millipore-Sigma, 74 240) for 12-24 hours, cleared in 1% KOH/20% glycerol, and stored in glycerol. For embryo skeleton preparations, fetuses were skinned and eviscerated, stained with alizarin red S and alcian blue (12-24 hours), cleared in 1%KOH/20% glycerol, and stored in glycerol.
2.5 ∣. Sample preparation: Visceral organs
At designated collection times, mice were sacrificed with an overdose of a cocktail containing ketamine and xylazine, perfused with PBS through the right ventricle, and indicated organs were fixed with submersion in 10% neutral buffered formalin (VWR International, Radnor, Pennsylvania; 89 370). Lungs were fixed via intratracheal inflation at a pressure of 20 cm H2O of 10% neutral buffered formalin. Organs were fixed overnight at 4°C with gentle agitation. Samples were washed twice in PBS, cryoprotected in 15% sucrose in PBS overnight at 4°C with gentle agitation, and then 30% sucrose overnight at 4°C with gentle agitation. Organs were patted dry, equilibrated in Tissue-Tek O.C.T. compound (VWR, 4583) for 20 minutes, embedded, and frozen on dry ice. Frozen sections were cut at 6 μm with a cryostat, dried at room temperature, and stored at −80°C until use. Slides were warmed for 10 minutes at room temperature and washed in PBS before being permeabilized in PBS + 0.5% Triton X-100 (Sigma, X-100) for 15 minutes. Slides were washed three times in PBS, mounted in Vectashield antifade mounting medium with DAPI (ThermoFisher, NC9524612), sealed with nail polish, and stored at 4°C until imaged.
2.6 ∣. Tamoxifen administration
For embryonic experiments tamoxifen (80 mg/kg) and progesterone (40 mg/kg) were co-injected intraperitoneally (IP) into pregnant dams 24 hours before collection of embryos. To prepare, 100 mg of tamoxifen (Millipore-Sigma, T5648) was dissolved in 5 mL of corn oil (Millipore-Sigma C8267) to which was added 5 mL of 50 mg/mL progesterone dissolved in sesame oil (Watson Pharma Inc., Parsippany, New Jersey; NDC 0591-3128-79). This was then sterilized through a 0.22 μm filter and stored at 4°C before use. For early neonatal experiments, tamoxifen was administered by IP injection at a dose of 150 μg on four sequential days beginning at postnatal day 5 (P5). Tamoxifen was prepared at a concentration of 20 μg/μl dissolved in corn oil or sunflower seed oil for injection. For juvenile and adult experiments tamoxifen was administered by chow (400 mg/kg tamoxifen citrate) (Envigo, Huntingdon, United Kingdom; TD.130860) for 10-14 days (juvenile), 7 days (adult visceral organs), or 14 days (adult skeleton). Tamoxifen was administered by chow in longer-term juvenile and adult experiments to avoid unnecessary injections.33 IP injections were unavoidable in embryonic and neonatal experiments. Differences in recombination efficiency between the two methods of tamoxifen administration were not directly assessed.
2.7 ∣. In situ hybridization
Whole mount in situ hybridization was performed according to published protocols.34 The Fgf18 probe was generated from a previously published plasmid linearized with SacII and transcribed using SP6 polymerase.35 Prolonged development of the stain was required for E8.0 embryos. Samples at E9.5 and E12.5 were cleared in 50% glycerol:PBS for several days prior to imaging.
2.8 ∣. Imaging
Except adult brain analysis, eGFP and tdTomato were detected by native fluorescence. Images are representative of at least two mice. For brain analysis, serial 60 μm thick coronal vibratome sections were generated and immunolabeled with anti-Tomato dsred as well as anti-Glial Fibrillary Acidic Protein (GFAP) antibodies, performed according to published protocols.25,36 Immunolabeled sections were counterstained with Hoechst stain. Atlas images are from the Mouse Brain Library (http://www.mbl.org/mbl_main/atlas.html).37
Fluorescent microscopy of slides was performed using either Zeiss LSM-700 Confocal Microscope Carl Zeiss AG, Oberkochen, Germany) (single plane) and or on a Zeiss Axio Imager M2 with respective ZEN software. Differential interference contrast microscopy of slides was performed using Zeiss Axio Imager M2. Whole-mount brightfield images were performed using an Olympus SZX12 Stereozoom microscope (Olympus Corporation, Tokyo, Japan) or Olympus MVX10 microscope. Whole mount fluorescent images were performed using an Olympus MVX10 microscope excited using an X-Cite Turbo XT600-T fluorescence light source (Excelitas Technologies Corp, Waltham, Massachusetts) at 475 and 575 nm wavelengths. Images were processed using either Canvas X (Canvas GFX), ImageJ, or Photoshop (Adobe Inc., San Jose, California).
2.9 ∣. Measurements, statistical analysis, and plotting
Length of the axis was measured from the atlas to the tip of the tail. Data were found to be normally distributed by a Shapiro-Wilk test. Significant differences in mean values between two sets of data were calculated by using an unpaired t-test. A P value of less than .05 was considered significant. Sample size (n) represents the number of animals. Data are represented as mean ± SD. * indicates P < .05. All figures were made in Canvas X. Plots were generated in Prism 7 (GraphPad Software, San Diego, California).
3 ∣. RESULTS AND DISCUSSION
3.1 ∣. Generation of an Fgf18CreERT2 knockin allele and a Fgf18flox allele
We designed a novel multifunctional targeting vector to generate a conditional (eGFP:CreERT2) lineage tracing allele and knockout (flox) allele from a single gene-targeting event in embryonic stem cells. The allele design was modeled after the design used by the International Mouse Phenotyping Consortium (IMPC) “knockout first” conditional allele,38 in which the splice acceptor (SA) LacZ was replaced with CreERT2.
The targeting vector design for the Fgf18 genomic locus placed a splice acceptor (SA)-eGFP:CreERT2 fusion gene39 into the intron located between Fgf18 exons 1B and 1C, along with a PGK-neo selection cassette (Figure 1A). The SA contains 875 bp of the En2 intron, the En2 SA, and 40 bp of En2 exon 2.38 The SA sequence is followed by a T2A sequence and a 15 bp linker sequence placed in frame with eGFP:CreERT2. Integration of the targeting vector into the genome was validated by Southern blot of the 5′ and 3′ ends (Figure 1B,C).
FIGURE 1.
Generation and validation of the Fgf18CreERT2 and Fgf18flox mouse lines. A: Scheme displaying targeting vector design and insertion into the Fgf18 genomic locus. B, C: Southern blot analysis of the Fgf18 targeted allele, with DNA digested with SpeI (left) or NdeI (right) in WT and het animals. Probes were amplified by PCR from indicated regions. D: Scheme of the successfully inserted intermediate targeted allele. E, F: Scheme displaying generation of a conditional allele (Fgf18flox; E) and a tamoxifen-inducible eGFP:CreERT2 lineage allele (Fgf18CreERT2, F) by crossing the targeted allele to germline expressed FLP recombinase and CRE recombinase, respectively. Pink arrows indicate LoxP sites; Red bars indicate FRT sites; Green bars indicate SA-GFP:CreERT2-pA cassette; Blue bars indicate PGK-neo-pA cassette. WT, wild type; Het, Fgf18Targeted allele/+
We strategically included Frt and LoxP recombination sites in the targeted allele (Figure 1D) to allow the generation of two independent alleles by crossing the parental mouse line to either a germline expressed FLP recombinase or a germline expressed Cre recombinase mouse (Figure 1E,F).26,27 This generated a conditional allele, with the critical exon 1C flanked by loxP sites (Fgf18flox) and a tamoxifen inducible eGFP:CreERT2 allele driven by the endogenous Fgf18 promoter (Fgf18CreERT2) that can be used for lineage tracing. Importantly, the Fgf18CreERT2 allele has exon 1C deleted and is predicted to behave as a functional null allele.
3.2 ∣. Functional testing of the Fgf18flox allele
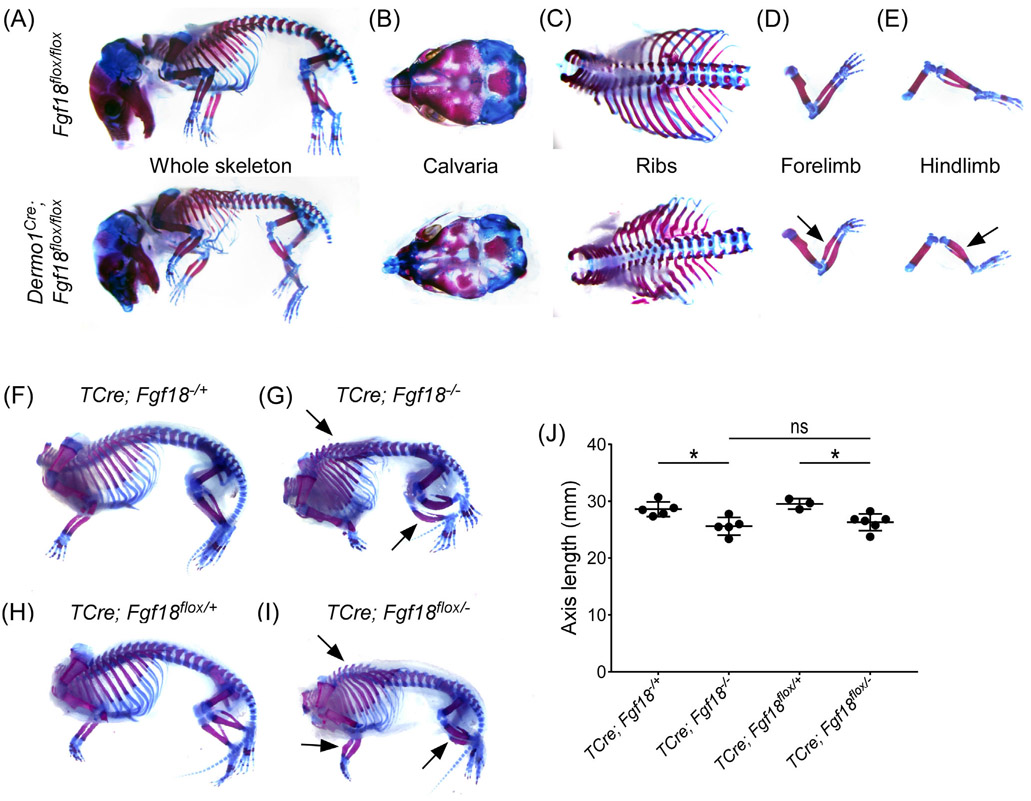
We tested the functionality of the Fgf18flox allele by crossing Fgf18flox/flox mice to Dermo1Cre (Twist2tm1.1[cre]Dor) mice to generate Dermo1Cre; Fgf18flox/flox conditional knockout mice. Dermo1Cre targets the mesenchymal condensations giving rise to both osteoblast and chondrocyte lineages.30 Like Fgf18−/− mice, Dermo1Cre; Fgf18flox/flox mice are not viable. Analysis of E18.5 Dermo1Cre; Fgf18flox/flox embryos revealed similar phenotypic features to those observed in Fgf18−/− embryos (Figure 2A).8,9 Specifically, Dermo1Cre; Fgf18flox/flox mice show incomplete suture closure of the calvaria, deformities in the ribs, a bend in the radius, and bowing of the tibia (Figure 2B-E). In contrast to Fgf18−/− mice, Dermo1Cre; Fgf18flox/flox animals do not have a cleft palate defect (data not shown), suggesting this phenotype is due to FGF18 signaling outside of the Dermo1Cre targeted mesenchyme lineage. This further suggests that the lethality evident in the Fgf18−/− mice is not solely due to a cleft palate defect.
FIGURE 2.
Recapitulation of known skeletal phenotypes upon removal of Fgf18 using the Fgf18flox allele. Alizarin red and Alcian blue stained whole skeleton of E18.5 mice following mesenchyme specific Fgf18 inactivation with Dermo1Cre (A-E) or TCre (F-I). A-E: Top, Fgf18flox/flox controls; Bottom, Dermo1Cre; Fgf18flox/flox conditional knockouts. Skeletal defects are seen in (A) whole skeleton, (B) calvaria, (C) ribs, (D) forelimb, and (E) hindlimb. Arrows indicate a characteristic bend in the radius and bowed hindlimb zeugopods. F, H: controls, (TCre; Fgf18−/+, TCre; Fgf18flox/+, respectively), G: homozygous Fgf18 knockout (TCre; Fgf18−/−), I: mesenchyme specific inactivation of Fgf18 (TCre; Fgf18flox/−). Arrows indicate kyphosis, bend in the radius, and bowed hindlimb zeugopods. J: Quantification of the length of the axial skeleton in TCre; Fgf18−/+, TCre; Fgf18−/−, TCre; Fgf18flox/+, and TCre; Fgf18flox/− animals. Student's t-test, *P < .05. Error bars, mean ± SD. n = 5 (TCre; Fgf18−/+, TCre; Fgf18−/−), n = 3 (TCre; Fgf18flox/+), n = 6 (TCre; Fgf18flox/−)
To further examine the effects of widespread FGF18 inactivation within the embryonic mesoderm, we generated TCre (Tg[T-cre]1Lwd) ;Fgf18flox/− conditional knockout mice. TCre targets progenitors cells in the primitive streak and tailbud.29 Like Dermo1Cre; Fgf18flox/flox mice, TCre; Fgf18flox/− mice were not viable. Analysis of the E18.5 embryos revealed TCre; Fgf18flox/− mice had a distinct hunchbacked appearance (kyphosis) and misshapen limbs (Figure 2H,I; arrows). TCre; Fgf18flox/− phenocopied Fgf18−/− mice (Figure 2F,G).8,9,12 Heterozygous control mice did not exhibit skeletal defects (Figure 2 F,H). Quantification of length of the axis revealed no difference between the germline and conditional knockouts, but both were significantly shorter than their littermate controls (Figure 3J). TCre; Fgf18flox/− conditional knockouts also did not have a cleft palate defect (data not shown). This is presumably due to absence of TCre expression in the first and second pharyngeal arches from which the palate is derived. These data suggest that, upon Cre recombination, the floxed allele described here abolishes Fgf18 activity.
FIGURE 3.
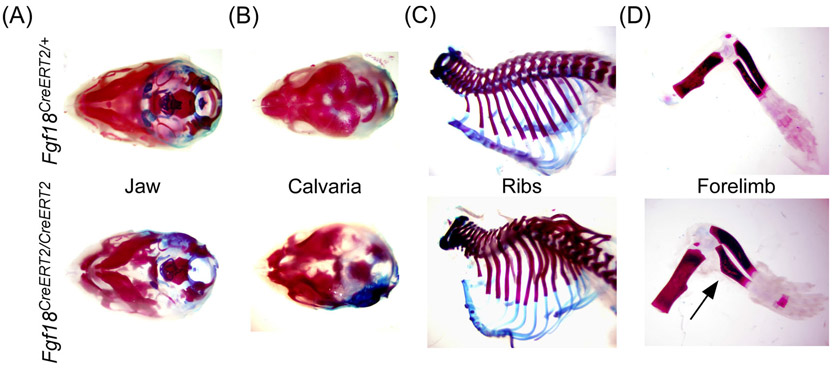
Validation that Fgf18CreERT2 behaves as a functional null allele. A-D: Alizarin red and Alcian blue stained whole skeleton of P0 Fgf18CreERT2/CreERT2 and control (Fgf18CreERT2/+) mice showing defects in (A) jaw, (B) calvaria, (C) ribs, and (D) limbs. Arrow indicates a characteristic bend in the radius
3.3 ∣. Fgf18CreERT2 is a functional null allele
To determine whether the Fgf18CreERT2 allele behaves as a functional null allele, heterozygous mice were intercrossed, and homozygous mice were examined at P0 (Figure 3). Fgf18CreERT2/CreERT2 mice resulted in perinatal lethality. Phenotypic analysis of Fgf18CreERT2/CreERT2 skeletons displayed abnormalities characteristic of Fgf18−/− mice including defects in the jaw, incomplete suture closing of the calvaria, deformities of the ribs, and the characteristic bend in the radius8,9,13 (Figure 3A-D). This demonstrates that Fgf18CreERT2 behaves as a functional null and suggests it can be crossed to the Fgf18flox allele to generate heterozygous CreERT2/flox (Fgf18CreERT2/flox) mice, in which Fgf18CreERT2 can be used to conditionally inactivate the Fgf18flox allele. This self-knockout strategy should effectively inactivate Fgf18 in all Fgf18-expressing cells and their lineage descendants when mice are exposed to tamoxifen.
3.4 ∣. Fgf18CreERT2 activity during embryonic development
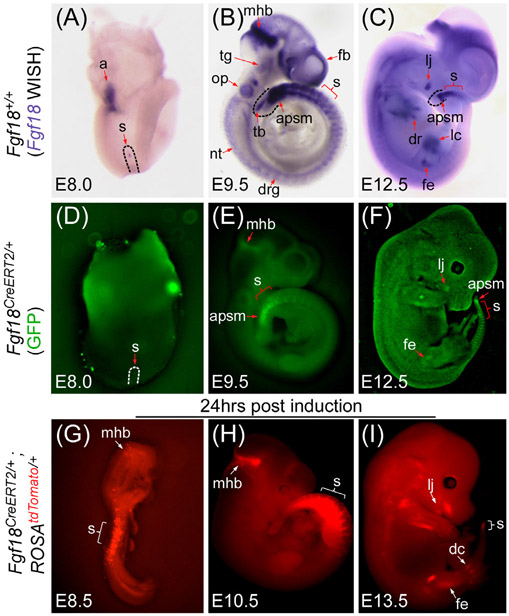
To determine the expression of Fgf18CreERT2 and validate its ability to recombine floxed alleles during embryogenesis, we compared the endogenous Fgf18 mRNA pattern by whole-mount in situ hybridization (WISH) (Figure 4A-C) with the GFP fluorescence pattern of the Fgf18 driven eGFP:CreERT2 fusion protein (Figure 4D-F). We then crossed the Fgf18CreERT2/+ allele with a Cre-dependent ROSAtdTomato/+ reporter allele to generate Fgf18Lineage mice, administered tamoxifen at similar embryonic time points and assayed for recombination mediated tdTomato fluorescence 24 hours later (Figure 4G-I).
FIGURE 4.
Fgf18CreERT2 expression in during embryogenesis. A-C: Whole-mount in situ hybridization (WISH) for Fgf18 mRNA in E8.0 (A), E9.5 (B), and E12.5 (C) wild type embryos. D-F: Whole mount eGFP fluorescence in E8.0 (D), E9.5 (E), and E12.5 (F) Fgf18CreERT2/+ embryos. Note: The Fgf18CreERT2 allele contains an eGFP fused upstream of CreERT2. G-I: Whole mount tdTomato fluorescence of mice induced 24 hours before collection at E8.5 (G), E10.5 (H), and E13.5 (I) in Fgf18CreERT2/+; ROSAtdTomato/+ embryos. All embryos are shown laterally and are of the stage indicated. Arrows and brackets indicate localized expression. a, allantois; apsm, anterior presomitic mesoderm; dc, digit condensation; dr, digit ray; drg, dorsal root ganglion; fb, forebrain; fe, femur; lc, Limb paddle condensation; lj, lower jaw; mhb, mid-hindbrain; nt, neural tube; op, otic pit; s, somite or somites; tb, tailbud; tg, trigeminal ganglion
Expression of Fgf18 at E8.0 has been reported in the somites and the presomitic mesoderm.40 At this stage, weak Fgf18 expression was observed in the allantois and in a few cells within the emerging first somite (Figure 4A). However, the GFP signal in Fgf18CreERT2/+ embryos was undetectable, suggesting very low levels of expression (Figure 4D). After inducing at E7.5 and harvesting at E8.5, Fgf18Lnease expressing cells were found in the somites (Figure 4G) and in a few cells in the mid-hindbrain junction (mhb) and allantois (data not shown).
At E9.5 Fgf18 expression displayed a more widespread pattern. As reported,40 Fgf18 mRNA was strongly detected in the somites, mhb, and anterior presomitic mesoderm (apsm). Lower levels were detected in the otic pit, trigeminal ganglion, dorsal root ganglia, tailbud, forebrain, and neural tube (Figure 4B). Clear GFP signal was observed only in the somites, mhb, and apsm (Figure 4E). Fgf18Lineage expressing cells were observed in the somites and mhb (Figure 4H). Low levels of tdTomato were seen in the forebrain as well as individual recombined cells in a salt-and-pepper pattern in the tailbud and future jaw, consistent with incomplete recombination (data not shown).
At E12.5, Fgf18 mRNA was still present in the somites, brain, and apsm. New sites of expression were detected in the future jaw, and in aspects of the limbs (Figure 4C). Fgf18CreERT2 embryos showed weak GFP signal in the somites, the apsm, the limbs, and the jaw (Figure 4F). Fgf18Lineage expressing cells were extensive in the limbs and digits, in the nascent somites, and the jaw (Figure 4I). tdTomato+ cells were also present in the interdigit mesenchyme and in tissues in the back of the embryo, consistent with previously reported sites of Fgf18 expression.40,41 These data suggest that the Fgf18CreERT2 allele faithfully recapitulates the endogenous expression pattern, however detection of the eGFP:CreERT2 fusion protein by native fluorescence is not sensitive enough for lowly expressed tissues or tissues with high autofluorescence.
3.5 ∣. Fgf18CreERT2 lineage during postnatal skeletal development
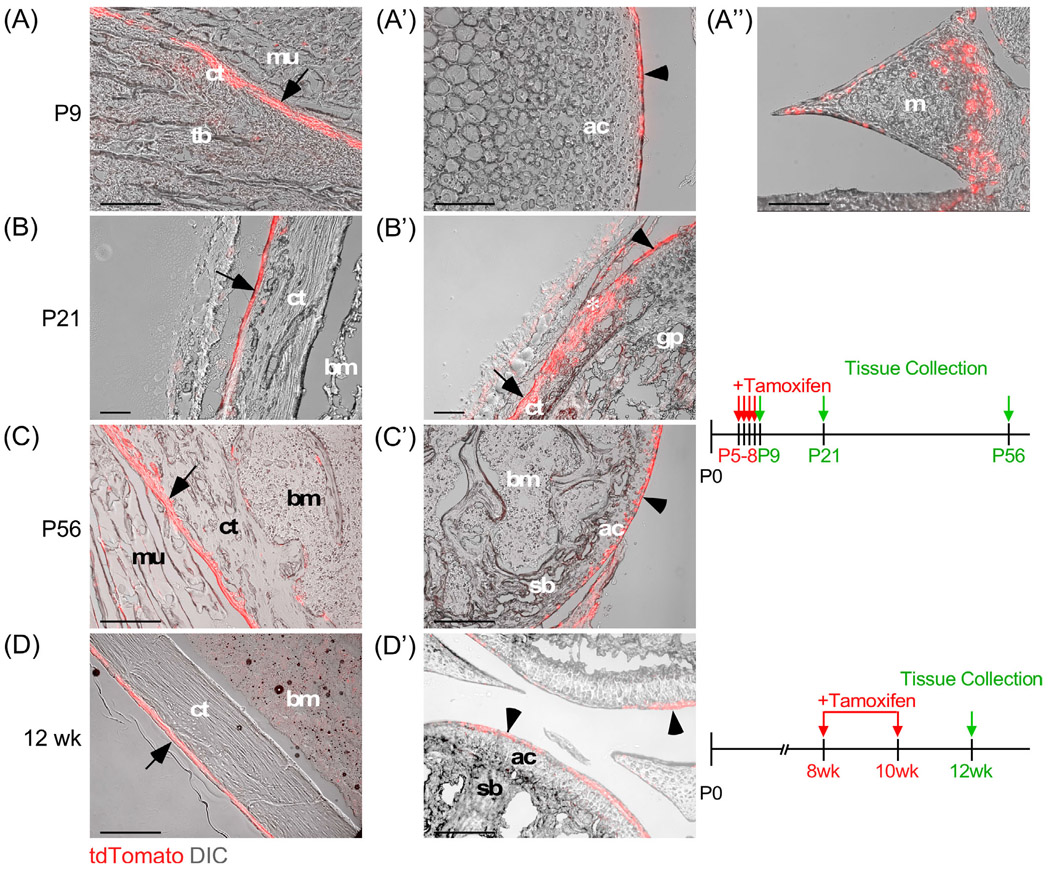
Fgf18 is expressed in proliferative zone chondrocytes in 1-week-old postnatal rodents and in articular cartilage of adult animals, assayed by qPCR, in situ hybridization, and immunofluorescence.21,23,42 To identify in situ the cells expressing Fgf18CreERT2 in the postnatal and adult skeleton, Fgf18Lineage neonatal mice were injected with tamoxifen and then harvested at different time points for analysis (Figure 5A-C). In neonatal mice, Fgf18Lineage expressing cells were found in the periosteum of cortical bone, the articular surface of the synovial joint, synovial tissue, and in cells within the perichondrial groove of Ranvier. Endosteal Fgf18Lineage expression was also observed, but less uniformly compared to periosteal expression (data not shown). The lineage of these Fgf18Lineage expressing cells was traced into adult animals (P56) and displayed a similar pattern to the neonatal animals, demonstrating that these cells are maintained. Fgf18Lineage animals induced as adults showed tdTomato+ cells in the periosteum, articular cartilage, and cells lining the meniscus (Figure 5D).
FIGURE 5.
Fgf18CreERT2 expression in skeletal tissue. A-C: Fgf18CreERT2/+; ROSAtdTomato/+ lineage mice were injected with tamoxifen daily from P5 to P8 and were collected at P9 (A-A″), P21 (B, B′), or P56 (C, C′) and imaged for tdTomato epifluorescence and histology using differential interference contrast (DIC) microscopy. tdTomato epifluorescence was observed in the periosteum (A, B, C), articular surface (A′, C′), meniscus (A″), and the groove of Ranvier (B′, asterisk). D: Fgf18CreERT2/+; ROSAtdTomato/+ lineage mice were given tamoxifen chow at 8 weeks of age for 14 days and collected at 12 weeks of age. tdTomato epifluorescence and DIC (grey) microscopy showing tdTomato+ cells in the periosteum (D) and articular surface (D′). Arrow indicates tdTomato+ cells in the periosteum. Arrowhead indicates tdTomato+ cells at the articular surface. ac, articular chondrocytes; bm, bone marrow; ct, cortical bone; gp, growth plate; m, meniscus; mu, muscle; p, periosteum; sb, subchondral bone; s, synovium; tb, trabecular bone. *indicates groove of Ranvier. Scale bar, 100 μm
These studies show that Fgf18CreER is active in adult articular chondrocytes indicating that the Fgf18 gene is expressed in this tissue in adult mice and may therefore have a role in maintaining joint tissue homeostasis or priming the tissue for injury response. These expression data are consistent with immunostaining for FGF18 in rat articular chondrocytes.21 Since FGF18 is a secreted growth factor it could act directly on chondrocytes at the joint surface and/or it could be secreted into synovial fluid and signal to any of the tissues lining the joint space.
3.6 ∣. Fgf18CreERTT lineage in postnatal visceral organ development
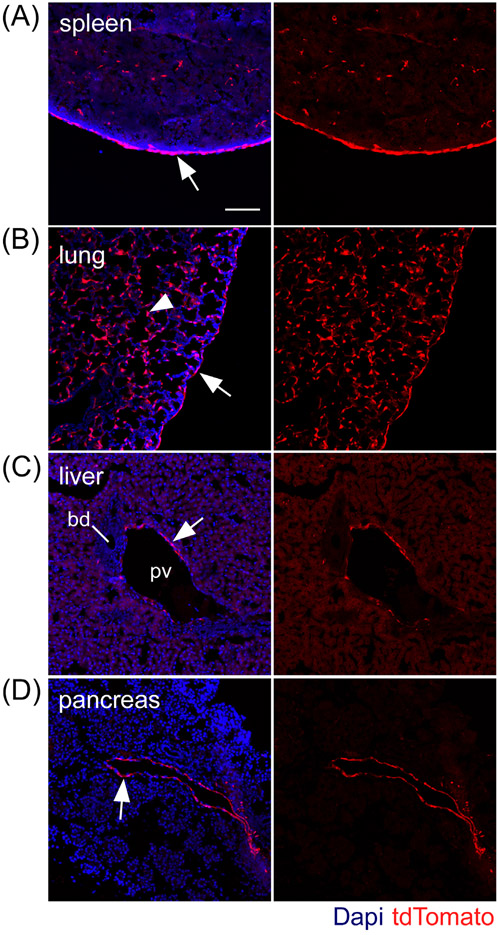
In rodent postnatal visceral organ development Fgf18 expression has been observed in the lung,14,15 but its expression in other organs has not been reported. To identify cells expressing Fgf18 during postnatal visceral organogenesis, Fgf18Lineage mice were given daily tamoxifen doses from postnatal day 5 (P5) to P8 and collected on P9. Fgf18Lineage expressing cells were detected in the spleen, lung, liver, and pancreas (Figure 6). The bladder, kidney, and small intestine were analyzed but did not display tdTomato+ cells. In the spleen, tdTomato+ cells were seen in the sub-mesothelial connective tissue capsule (Figure 6A, arrow) as well as sparse cells in the parenchyma. The lung shows the greatest amount of tdTomato+ cells, located in the single-cell mesothelium (Figure 6B, arrow) and many cells in the parenchyma (Figure 6B, arrowhead). Cells in the parenchyma were determined to be predominantly alveolar myofibroblasts (data not shown, manuscript in submission). Expression in the liver and pancreas was more restricted with tdTomato+ cells located in perivascular regions, likely smooth muscle cells (Figure 6C,D). These results suggest that Fgf18, based on expression, has a functional role in postnatal organogenesis of the spleen, lung, liver, and pancreas.
FIGURE 6.
Fgf18CreERT2 expression in postnatal visceral organs. A-D: Fgf18CreERT2/+; ROSAtdTomato/+ mice were injected with tamoxifen daily from P5 to P8 and were collected at P9. tdTomato+ expressing cells include: (A) splenic capsule (arrow); (B) pulmonary mesothelial cells (arrow) and alveolar mesenchyme (arrowhead); (C) liver and (D) pancreas perivascular cells (arrows). Bd, bile duct; pv, portal vein. Dapi (Blue). Scale bar, 100 μm
3.7 ∣. Fgf18CreERT2 lineage in adult visceral organ homeostasis
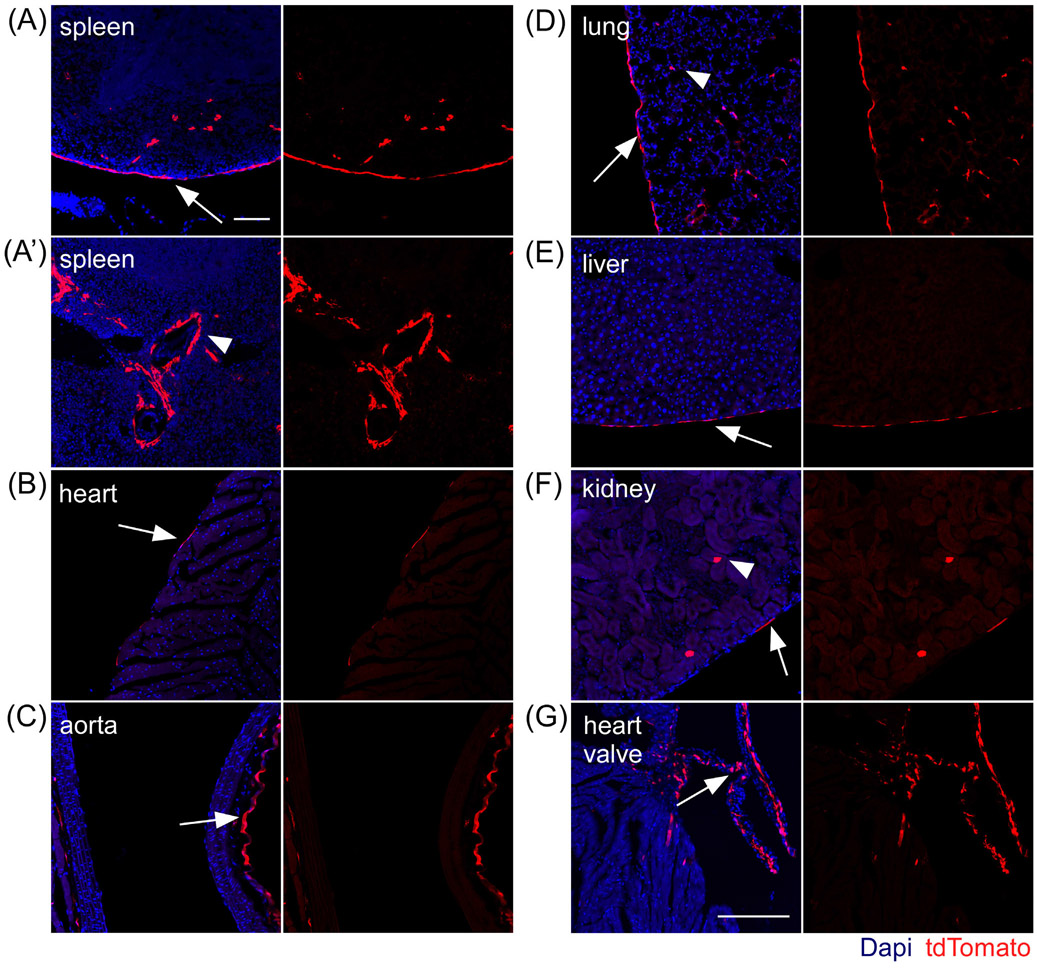
In adult murine tissue Fgf18 expression has been reported in the lungs and kidneys at high levels with lower-level expression in the heart, testis, spleen, skeletal muscle, and brain.43 To identify cells expressing Fgf18 during visceral organ homeostasis, adult Fgf18Lineage mice were induced for 1-week with tamoxifen chow followed by a 1-week washout before tissue collection. Fgf18Lineage expressing cells were detected in the spleen, heart, aorta, lung, liver, kidney, and cardiac valve (Figure 7). The pancreas, testis, and white visceral fat depot were analyzed but did not show any tdTomato+ cells. In the spleen Fgf18Lineage labeled cells showed a similar capsule expression pattern as observed in the postnatal animal (Figure 7A), but exhibited additional tdTomato+ cells located in the perivascular region around large vessels (Figure 7A′). In the aorta, tdTomato+ cells were located in the outermost mesenchymal derived interstitial cells (adventitia) of the vascular wall (Figure 7C). In the heart, lung, liver, and kidney, tdTomato+ cells were found, to differing degrees, in visceral mesothelial cells (Figure 7B,D,L,K). The strongest mesothelial expression was seen in the lung followed by the liver. Fgf18Lineage cells were found in sparse parenchyma cells in lung and kidney. tdTomato+ cells were abundant in cardiac valve interstitial cells (Figure 7G). Taken together these results indicate that Fgf18CreERT2 is able to mark diverse cell types in multiple adult organs under homeostatic conditions. Future work to define the fate and function of these cells during injury response should be informative.
FIGURE 7.
Fgf18CreERT2 expression in adult visceral organs. A-G: Fgf18CreERT2/+; ROSAtdTomato/+ mice were given tamoxifen chow at 8 weeks of age for 7 days and collected at 10 weeks of age. tdTomato+ expressing cells include: (A, A′) splenic capsule (arrow) and perivascular (arrowhead) cells; (B) select epicardial cells (arrow); (C) aorta interstitial cells in the vascular wall (arrow); (D) pulmonary mesothelial (arrow) and alveolar interstitial cells (arrowhead); (E) hepatic mesothelial cells (arrow); (F) kidney mesothelial cells (arrow) and occasional epithelial tubule cells (arrowhead); (G) cardiac valve interstitial cells. Dapi (Blue). Scale bar, 100 μm
3.8 ∣. Fgf18CreERT2 lineage in the adult brain
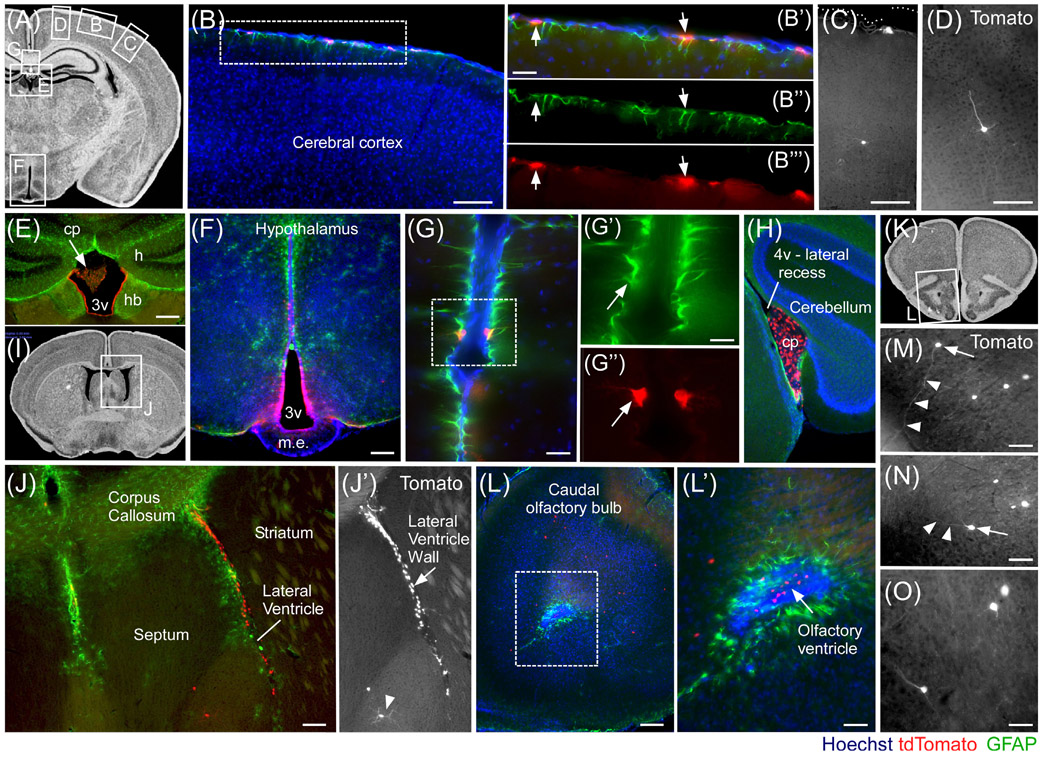
To gain better insight into the expression of Fgf18 and cells derived from them in the brain, adult Fgf18Lineage mice were induced for 2 weeks with tamoxifen chow followed by a 1-week washout before tissue collection. Tissue from multiple bregma coordinates was immunolabeled to detect astrocytes (GFAP+) as well as Fgf18Lineage expressing cells (Tomato+) (Figure 8). Fgf18Lineage expressing cells were detected in hypothalamic tanycytes and the ependymal lining of the dorsal third ventricle (3 V) (Figure 8F). Fgf18Lineage expressing cells were additionally detected in a subset of cortical pial astrocytes (Figure 8B); in rare neurons in the cerebral cortex (Figure 8C,D); the choroid plexus, and ependymal lining of the dorsal third ventricle (3 V) and forth ventricle recess (4 V) (Figure 8E,H); pial astrocytes (Figure 8G); septum (Figure 8J), and amygdala (not shown). Interestingly, Fgf18Lineage expressing cells were also found in the walls of the lateral ventricles. These could be followed anteriorly into the olfactory bulbs, where Fgf18Lineage expressing olfactory bulb (OB) neurons were clearly evident, reminiscent of neuronal supply to the OB via the rostral migratory stream (Figure 8J,L-O).44 In these mice, it is likely the long period between initial tamoxifen treatment and tissue collection resulted in the Fgf18Lineage expressing cells including both cells expressing Fgf18CreERT2 and their descendants at the time of harvest. This conclusion is supported by a more restricted number and distribution of Fgf18Lineage labeled cells in the hypothalamus of animals pulsed more briefly with tamoxifen and harvested acutely.25
FIGURE 8.
Fgf18CreERT2 expressing cells and their putative descendants in the adult brain. Fgf18CreERT2/+; ROSAtdTomato/+ mice were given tamoxifen chow at 9 weeks of age for 2 weeks and collected at 12 weeks of age. A, I, K: Photomicrographs from brain atlases to aid regional visualization. B-H, J, L-O: tdTomato+ expressing cells include: GFAP+ astrocytes of the Glia limitans (B-B″′); rare non-pyramidal (C) and pyramidal cortical neurons (D); ventricular ependymal cells, adjacent to the medial habenula (E), with no expression in the hippocampus; hypothalamic tanycytes and sparse ependymal cells of the third ventricle (3v) wall (F); pair of GFAP+ pial astrocytes (G-G″); strong and specific expression in the choroid plexus of the forth ventricle's (4v) lateral recess (H), near the cerebellum; medial and lateral walls of the lateral ventricles and rare septal neurons (J, J′), shown in hemisection; anterior olfactory bulbs (L, L′), near rostral migratory stream cells; scattered neurons in the anterior olfactory cortex (M-O). Arrows and arrow heads in (M, N) indicate tdTomato+ neurons and their neurite trajectory. Lines indicate structures. Scale bars: 100 μm in B-F, J, and L; 50 μm in B′, G and L′,: 25 μm in G′ and M-O. v, ventricle; h, hippocampus; cp, choroid plexus; m. e., median eminence; hb, habenula. Atlas images are from the Mouse Brain Library (http://www.mbl.org/mbl_main/atlas.html)
3.9 ∣. Conclusion
This study demonstrates the generation and validation of two novel alleles targeting the Fgf18 locus that will be useful for the investigation of the tissue specific function of the gene (Fgf18flox) and the lineage of its expressing cells (Fgf18CreERT2). Using Fgf18flox, it was confirmed that the majority of the skeletal phenotypes of Fgf18−/− animals, including deformities of the calvaria, ribs, hindlimb, forelimb, and axis, are dependent on Fgf18 expression originating from the mesenchymal compartment. Using Fgf18CreERT2, expression of Fgf18 was confirmed in known and identified in new sites in the embryo in multiple organs including the somites and mid-hindbrain junction; the postnatal and juvenile skeleton; in postnatal and adult visceral organs; and in adult brain. These tools will be useful to investigate FGF18 function during organogenesis and tissue homeostasis, and to target specific cell lineages at various embryonic and postnatal time points.
ACKNOWLEDGMENTS
We thank W. Lewis and E. Truffer for animal caretaking and M. Brown for technical assistance.
Funding information
American Heart Association, Grant/Award Number: 18PRE34030091; Biotechnology and Biological Sciences Research Council, Grant/Award Number: BBL/003406/1; Center for Cancer Research at the National Cancer Institute; Intramural Research Program of the National Institutes of Health; National Cancer Institute; National Institutes of Health, Grant/Award Number: R01HL111190
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interests.
REFERENCES
- 1.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn. 2008;237:18–27. [DOI] [PubMed] [Google Scholar]
- 3.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ornitz DM, Marie PJ. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29:1463–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ornitz DM, Xu J, Colvin JS, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006; 281:15694–15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito K, Ohkawara B, Yagi H, et al. Lack of Fgf18 causes abnormal clustering of motor nerve terminals at the neuromuscular junction with reduced acetylcholine receptor clusters. Sci Rep. 2018;8:434 http://www.nature.com/articles/s41598-017-18753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohbayashi N, Shibayama M, Kurotaki Y, et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usui H, Shibayama M, Ohbayashi N, Konishi M, Takada S, Itoh N. Fgf18 is required for embryonic lung alveolar development. Biochem Biophys Res Commun. 2004;322:887–892. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa H, Ashigaki S, Takamatsu M, et al. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J Neurosci. 2004;24:8711–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung IH, Schoenwolf GC, Lewandoski M, Ornitz DM. A combined series of Fgf9 and Fgf18 mutant alleles identifies unique and redundant roles in skeletal development. Dev Biol. 2016;411:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol. 2007;302:80–91. [DOI] [PubMed] [Google Scholar]
- 14.Chailley-Heu B, Boucherat O, Barlier-Mur A-M, Bourbon JR. FGF-18 is upregulated in the postnatal rat lung and enhances elastogenesis in myofibroblasts. Am J Physiol Lung Cell Mol Physiol. 2005;288:L43–L51. [DOI] [PubMed] [Google Scholar]
- 15.Franco-Montoya M-L, Boucherat O, Thibault C, et al. Profiling target genes of FGF18 in the postnatal mouse lung: possible relevance for alveolar development. Physiol Genomics. 2011;43:1226–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGowan SE, McCoy DM. Fibroblast growth factor signaling in myofibroblasts differs from lipofibroblasts during alveolar septation in mice. Am J Physiol Lung Cell Mol Physiol. 2015;309:L463–L474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. [DOI] [PubMed] [Google Scholar]
- 18.Kawano M, Komi-Kuramochi A, Asada M, et al. Comprehensive analysis of FGF and FGFR expression in skin: FGF18 is highly expressed in hair follicles and capable of inducing Anagen from Telogen stage hair follicles. J Inves Dermatol. 2005;124:877–885. [DOI] [PubMed] [Google Scholar]
- 19.Kimura-Ueki M, Oda Y, Oki J, et al. Hair cycle resting phase is regulated by cyclic epithelial FGF18 signaling. J Investig Dermatol. 2012;132:1338–1345. [DOI] [PubMed] [Google Scholar]
- 20.Leishman E, Howard JM, Garcia GE, et al. Foxp1 maintains hair follicle stem cell quiescence through regulation of Fgf18. Development. 2013;140:3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori Y, Saito T, Chang SH, et al. Identification of fibroblast growth Factor-18 as a molecule to protect adult articular cartilage by gene expression profiling. J Biol Chem. 2014;289:10192–10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karuppaiah K, Yu K, Lim J, et al. FGF signaling in the osteoprogenitor lineage non-autonomously regulates postnatal chondrocyte proliferation and skeletal growth. Development. 2016;143:1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarus JE, Hegde A, Andrade AC, Nilsson O, Baron J. Fibroblast growth factor expression in the postnatal growth plate. Bone. 2007;40:577–586. [DOI] [PubMed] [Google Scholar]
- 24.Hoshikawa M, Yonamine A, Konishi M, Itoh N. FGF-18 is a neuron-derived glial cell growth factor expressed in the rat brain during early postnatal development. Mol Brain Res. 2002;105:60–66. [DOI] [PubMed] [Google Scholar]
- 25.Kaminskas B, Goodman T, Hagan A, Bellusci S, Ornitz DM, Hajihosseini MK. Characterization of endogenous players in fibroblast growth factor-regulated functions of hypothalamic tanycytes and energy-balance nuclei. J Neuroendocrinol. 2019; https://onlinelibrary.wiley.com/doi/abs/10.1111/jne.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119:S97–S101. [DOI] [PubMed] [Google Scholar]
- 27.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000; 28:106–110. [PubMed] [Google Scholar]
- 28.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perantoni AO, Timofeeva O, Naillat F, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. [DOI] [PubMed] [Google Scholar]
- 30.Yu K, Xu J, Liu Z, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. [DOI] [PubMed] [Google Scholar]
- 31.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. [DOI] [PubMed] [Google Scholar]
- 32.Nagy A, Gertsenstein M, Vintersten K, Behringer R. In: University of Michigan, ed. Manipulating the Mouse Embryo: A Laboratory Manual 3rd Ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 33.Whitfield J, Littlewood T, Soucek L. Tamoxifen Administration to Mice. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2015. Tamoxifen Administration to Mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson DG, Nieto MA. [22] detection of messenger RNA by in situ hybridization to tissue sections and whole mounts Methods in Enzymology. Vol 225 Cambridge, Massachusetts: Academic Press; Guide to Techniques in Mouse Development; 1993:361–373. http://www.sciencedirect.com/science/article/pii/007668799325025W. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. [DOI] [PubMed] [Google Scholar]
- 36.Haan N, Goodman T, Najdi-Samiei A, et al. Fgf10-expressing Tanycytes add new neurons to the appetite/energy-balance regulating centers of the postnatal and adult hypothalamus. J Neurosci. 2013;33:6170–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams R Mapping genes that modulate mouse brain development: a quantitative genetic approach In: Goffinet AF, Rakic P, eds. Mouse Brain Development. New York: Springer; 2000:21–49. [DOI] [PubMed] [Google Scholar]
- 38.Skarnes WC, Rosen B, West AP, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mugford JW, Sipilä P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruoka Y, Ohbayashi N, Hoshikawa M, Itoh N, Hogan BLM, Furuta Y. Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mech Dev. 1998;74:175–177. [DOI] [PubMed] [Google Scholar]
- 41.Mok GF, Cardenas R, Anderton H, Campbell KHS, Sweetman D. Interactions between FGF18 and retinoic acid regulate differentiation of chick embryo limb myoblasts. Dev Biol. 2014;396:214–223. [DOI] [PubMed] [Google Scholar]
- 42.Ellsworth JL, Berry J, Bukowski T, et al. Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthr Cartil. 2002;10:308–320. [DOI] [PubMed] [Google Scholar]
- 43.Hu MC-T, Qiu WR, Wang Y, et al. FGF-18, a novel member of the fibroblast growth factor family, stimulates hepatic and intestinal proliferation. Mol Cell Biol. 1998;18:6063–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obernier K, Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146:dev156059. [DOI] [PMC free article] [PubMed] [Google Scholar]