Abstract
The evidence base in health psychology is vast and growing rapidly. These factors make it difficult (and sometimes practically impossible) to consider all available evidence when making decisions about the state of knowledge on a given phenomenon (e.g., associations of variables, effects of interventions on particular outcomes). Systematic reviews, meta-analyses, and other rigorous syntheses of the research mitigate this problem by providing concise, actionable summaries of knowledge in a given area of study. Yet, conducting these syntheses has grown increasingly laborious owing to the fast accumulation of new evidence; existing, manual methods for synthesis do not scale well. In this article, we discuss how semi-automation via machine learning and natural language processing methods may help researchers and practitioners to review evidence more efficiently. We outline concrete examples in health psychology, highlighting practical, open-source technologies available now. We indicate the potential of more advanced methods and discuss how to avoid the pitfalls of automated reviews.
Keywords: Machine learning, natural language processing, semi-automation, evidence synthesis, health psychology, systematic review
Systematic reviews of evidence, including statistical meta-analyses, have become key tools in modern psychological research. When performed well, such reviews provide rigorous, comprehensive, and up-to-date synopses of all evidence pertaining to a given health psychology question. Such questions generally take the form of addressing whether certain factors are predictive of health outcomes (e.g., Does perceived stress reduce quality of life? Does it cause premature aging? Do providers with racial bias harm their minority patients?); or, whether certain interventions succeed in improving a health-related outcome (e.g., Does mindfulness training reduce perceived stress? Does aerobic or resistance exercise for cancer survivors improve their mental health?). Such questions are addressed in a multitude of primary research designs, ranging from correlational to experimental studies. One of the reasons that synopses of the literature are so valuable is the vast and fast-growing volume of published evidence (see Figure 1).
Figure 1.
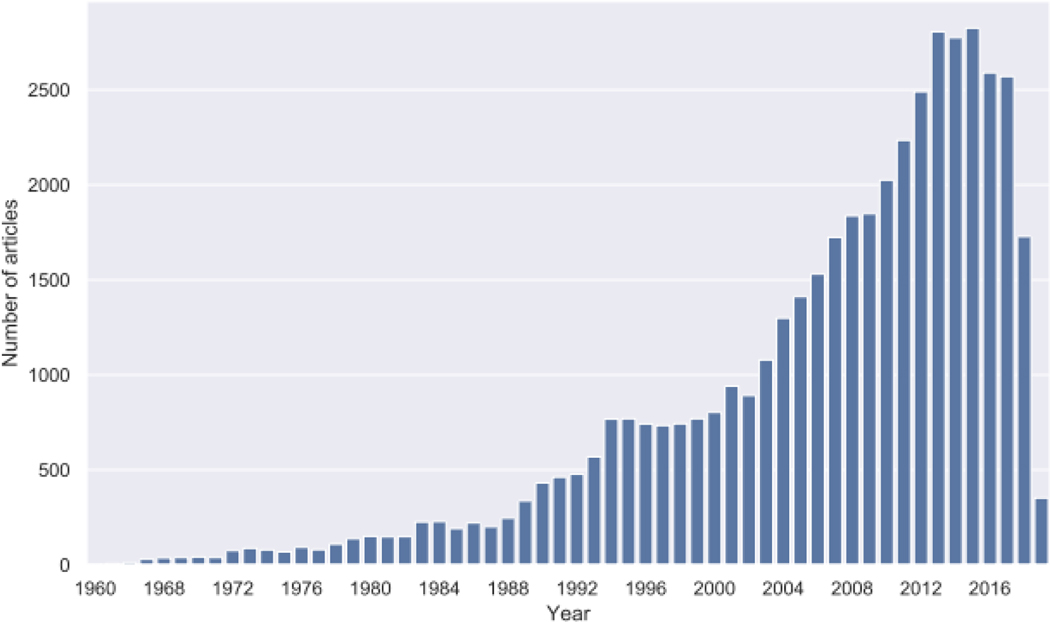
The number of clinical trials relevant to health psychology is increasing rapidly over time. Numbers of articles retrieved from search of PubMed for randomized controlled trials (RCTs) including the MeSH term ‘Behavioral Disciplines and Activities.’ (Note. The drop in numbers from 2016 is artefactual, and due to time lag until manual indexing at PubMed.)
Yet, if systematic reviews are not conducted with sufficient methodological rigor (which in turn incurs additional manual effort), then they are susceptible to bias and represent poor allocations of resources (cf. Glasziou et al., 2014; Johnson & Hennessy, 2019). Frameworks for evaluating these potential biases have been established, including the ROBIS tool (Whiting et al., 2016) and the AMSTAR inventory (Shea et al., 2018). These inventories examine facets of methodological quality, such as whether methods were preregistered, whether study intake was done in duplicate, or whether the methodological quality of the included research was scrutinized and brought to bear in judging the outcomes of the research. Standards such as PRISMA (e.g., Liberati et al., 2009) seek to improve the reporting quality of meta-analyses and systematic reviews, but do not, in and of themselves, make recommendations as to methodological conduct (Johnson & Hennessy, 2019).
While the increasing volume of published evidence heightens the need for rigorous summaries of the same, it simultaneously imposes increased workloads on the domain experts performing these syntheses. This workload in turn motivates the need for technologies that can expedite synthesis. In this article, we provide an overview of methods and tools for semi-automation of evidence syntheses in health psychology. We discuss where in the review process such systems may fit, and we briefly review the machine learning (ML) and natural language processing (NLP) methods underlying semi-automation technologies. We also highlight current limitations of semi-automation and suggest future directions.
The Systematic Review of the Near Future?
We commence this overview with an illustrative example of how a systematic review might use automation technologies in practice given the current state of technology (see Figure 2). First, the researcher crafts a search in the conventional way, choosing keywords relating to the topic of interest. A randomized controlled trial (RCT) classification machine learning system then automatically retains articles describing RCTs, and discards everything else (we note that most automation research has focused on reviews of RCTs; other research designs have not been much considered).
Figure 2.
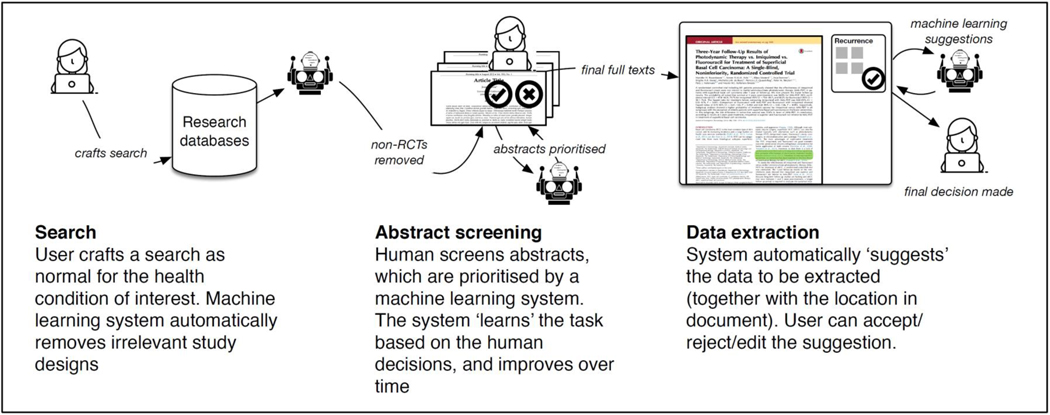
How contemporary automation technologies might aid systematic reviews.
Second, the researcher uses a semi-automatic machine learning system to screen abstracts (one such system is abstrackr: http://abstrackr.cebm.brown.edu/). Abstracts are presented to the researcher by the system initially in a random order, and the researcher indicates whether they should be included or not. After the system has recorded a certain number of user decisions, it is able to ‘learn’ which abstracts are likely to be included. Thereafter, the abstracts are ordered automatically by predicted relevance. The user may choose to stop screening early, after it appears that no more relevant abstracts are in the set.
Finally, the researcher uses a semi-automatic data extraction system (one such system is RobotReviewer, by IJM and BCW, www.robotreviewer.net). The system presents the full text article (as a PDF) in the user interface, together with some suggestions about relevant data to extract. For example, when the system might automatically show data relating to the trial population or risks of bias, the researcher is able to see relevant text highlighted in the original document, and then validate whether the machine learning suggestions were correct (or not). Where the machine learning is wrong, the researcher can correct the extracted data using the user interface.
Two of the current authors (IJM and BCW) have published a recent survey on automation tools that is currently available for use on each step of the systematic review process (Marshall & Wallace, 2019), but given the rapid changes in the field, we also refer the interested reader to SRToolbox (http://systematicreviewtools.com/), which is frequently updated with new examples.
Automation and Semi-Automation
Because of the complexities involved in high-quality systematic reviews, technology capable of fully and reliably automating systematic reviews is unlikely to appear in the near future. A more fruitful avenue for the immediate future, in our view, is semi-automation, which refers to scenarios in which domain experts are aided by ML and NLP systems. The idea is to reduce the human workload by, for example, reducing the number of irrelevant articles that must be assessed, or by automatically highlighting snippets in articles that seem likely to contain relevant information to be extracted.
Some steps of evidence synthesis are more amenable to automation than others. For example, screening citations for relevance may be framed as a standard text classification problem for which highly effective methods exist. Yet, something like extracting odds ratios from full-text articles while recognizing which interventions and outcomes these describe (and identifying an associated estimate of dispersion) is considerably harder, though potentially feasible. Analytic tasks, such as reasoning about the importance of results for clinical practice, are beyond the capabilities of the current generation of artificial intelligence methods.
It is useful to consider, for each step in the review process, the level of performance a model would need to achieve before being practically useful. For this goal, it is important to note that the output of an automated system may be used in different ways. One extreme would be to rely entirely on automation. For example, identifying relevant evidence (classifying articles) would entail simply trusting that whatever a machine learning model returned is relevant, and further, that whatever it failed to return is not. In other words, relying entirely on a machine learning model means that full automation effectively assumes perfect (or human-level) model accuracy.
A less extreme approach would be to use the model as a kind of “second opinion.” Again, considering the case of evidence retrieval, this method would be akin to performing “double screening,” but where one human is replaced by a model. Disagreements between the model and the human screener could be resolved manually. A further alternative would be to use the model to assist humans in extractive tasks, for example, by highlighting sections of an article that might contain sought after information. These latter two modes of operation do not require a model to be perfect in order to be useful. Figure 3 depicts this approach to setting tolerable accuracy standards.
Figure 3.
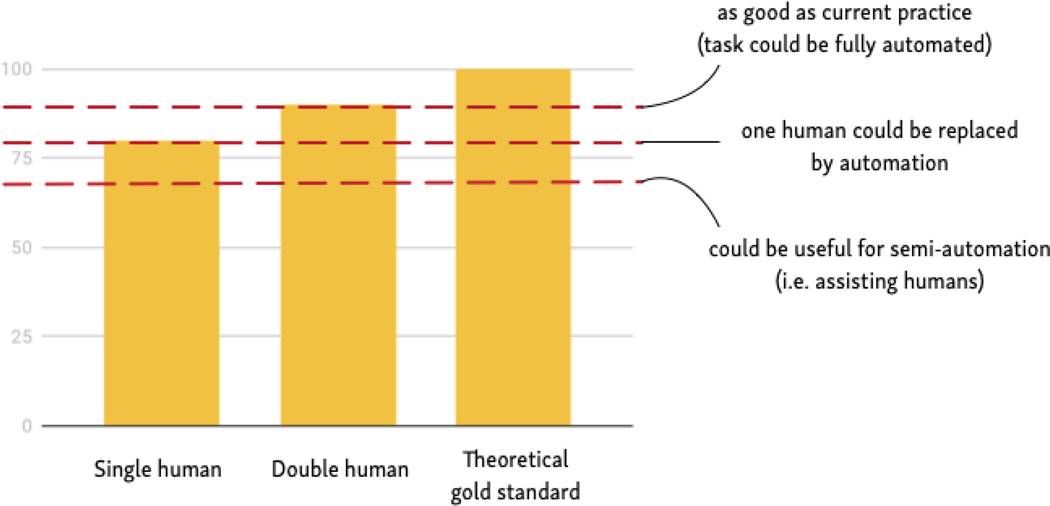
Hypothetical evaluation thresholds for automatic synthesis methods: When are they good enough to use?
Technologies and Tasks (for Conventional Reviews)
We now discuss more concretely some of the steps (or “tasks”) in the systematic review pipeline for which automation technologies have been proposed and the specific ML/NLP methods upon which these rely.
Search and Abstract Screening
Evidence syntheses seek rigorous strategies to summarize the entirety of the available evidence relevant to a particular question of interest. After crafting a well-formed question, the first step in conducting a formal evidence synthesis is typically to search for reports describing relevant research. Relevance is usually determined as a function of pre-specified inclusion criteria that define the attributes that studies must have in order to be included in the review at hand.
To identify a set of candidate articles (i.e., those which may meet the inclusion criteria), one crafts a query with which to search relevant databases. Such queries are often difficult to build, test, and implement; given the importance accorded to not missing any relevant studies, involving an information specialist or librarian is commonly recommended (cf. Johnson & Hennessy, 2019; Shea et al., 2018). The query will retrieve a set of candidate articles that will usually be designed to have near perfect sensitivity (the set must contain all or practically all of the relevant articles) but low precision (being synonymous with positive predictive value; most of the retrieved articles will not be relevant). Consequently, researchers must screen candidate articles, manually determining whether they meet the inclusion criteria. Usually screening is first done based on citation data (i.e., mainly titles and abstracts). This process can be extremely time-consuming, especially for large reviews of complex interventions and/or outcomes. Such complexity is particularly common in health psychology studies (see, e.g., Hennessy et al., in press); thus, literature searching imposes a particularly hefty burden on reviewers in this field.
Machine learning (ML) provides a potential means of reducing the number of citations that must be manually screened for a given review, without necessarily sacrificing comprehensiveness (Cohen, Hersh, Peterson, & Yen, 2006; O’Mara-Eves, Thomas, McNaught, Miwa, & Ananiadou, 2015; Wallace, Small, Brodley, Lau, & Trikalinos, 2012).
In general, ML involves specifying a model f with parameters θ that maps from inputs x and outputs y, and then identifying model parameters (θ) that maximize agreement with training examples. Examples comprise input/output pairs: (xi, yi), where xi is a vector representation of an input i (e.g., a representation of study citation i), and yi is a “target” (e.g., whether study i “meets inclusion criteria” or not). This process raises the question of how to map citations (text) to vectors. A classic representation involves creating a long, sparse vector for a particular document with |V| dimensions, where V denotes the entire vocabulary under consideration (e.g., it might contain 50,000 unique English words). To represent a given abstract i, we can construct a |V| dimensional vector xi such that the entry in position j, xij, is 1 if and only if the word corresponding to j in V is present in the abstract text. This representation scheme is called binary bag of words (BoW).
A simple (but often surprisingly effective) model for text classification is logistic regression performed over these representations. In this case, we might set y to 1 to encode “meets inclusion criterion” as 0 otherwise. Then p(y=1) = σ(θ • xi), where • denotes a dot product and σ is the Sigmoid function.
Variants of this approach and similar ones can achieve strong predictive performance for some text classification tasks. Still, BoW representations are clearly suboptimal; in addition to discarding word order, BoW represents different words as independent 0–1 scalers in the vector, so that the similarity between words is lost. That is, a naïve indicator representation of “dog” is just as similar to that for “cat” as it is to “banana” (specifically, these will all be orthogonal to one another). In part for these reasons, more recent work in NLP has focussed on variants of neural networks, which allow models to capitalize on dense vector representations of words that preserve semantic similarities.
Neural network architectures such as recurrent neural networks (RNNs) can in turn compose these representations in order to induce a final input (document) representation used to make a prediction. Concretely, the RNN may be viewed as learning to yield representations that are in turn fed to, for example, a logistic regression. This model is trained end-to-end; that is, RNN parameters are estimated in tandem with the “top-level” logistic regression parameters. An in-depth discussion of conventional neural network based NLP models is beyond the scope of the current article, but Goldberg (2017) provides further technical details. We note that this area is experiencing very rapid growth, with new state-of-the-art models emerging every year.
Recently, Devlin (2018) and Vaswani (2017) introduced Bidirectional Encoder Representations from Transformers (BERT) for language understanding. The details of BERT are beyond the scope of this article, but we point the interested reader to Devlin (2018). BERT has been extensively used in NLP, for example, for text sentiment analysis, automatic sentence completion, question answering, or even in Google search and translations. Such pre-trained “encoders” are likely to be used increasingly in the context of automated evidence synthesis systems.
Beyond aiding citation screening for particular clinical questions (or reviews), there are many potential places in evidence synthesis where text classification may be gainfully applied. To name one example, one can use such techniques to classify articles as reports of RCTs (or not) on the basis of their text (Marshall, Noel-Storr, Kuiper, Thomas, & Wallace, 2018).
Categorizing reports of RCTs with respect to their risks of bias or for other characteristics is another example of text classification, as it entails mapping articles to one of two or three categories (high, unknown and low; the former two are sometimes grouped). But, in this case, one typically also wants to extract snippets of text that support these categorizations (Figure 4). This support is therefore also an example of data extraction, which we discuss next.
Figure 4.
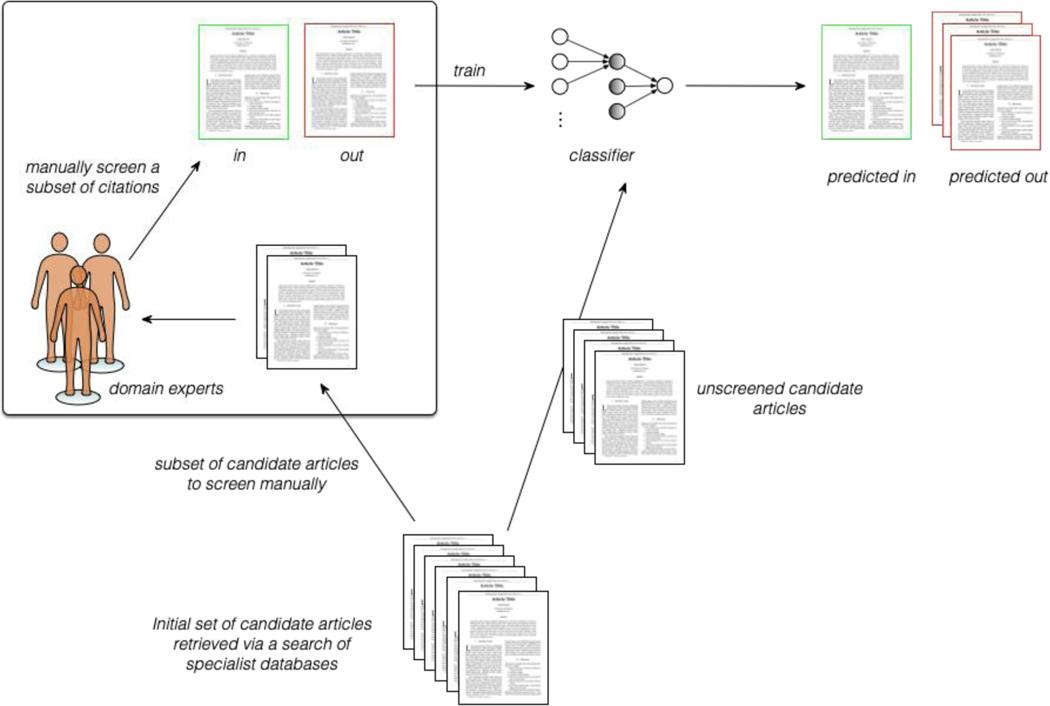
Schematic depicting how one might semi-automate the citation screening process via machine learning. The first step is to retrieve a set of candidate articles that might be relevant to one’s clinical question from appropriate sources (e.g., PubMed). A subset of these must then be manually assessed for relevance, or “screened.” This screened set can then be used to train a classifier. Subsequently, the classifier can be applied to the remaining (unscreened) citations. The predictions for these can be used in different ways to speed up the screening process.
Data Extraction
A critical but laborious step in evidence synthesis is extracting structured data from trial reports that is to be included in the review at hand. This step sometimes entails identifying snippets, say in the case of risk of bias assessment, where one extracts the rationale text supporting a risk of bias judgment (Figure 5). Other times, one aims to extract specific quantities such as study sample sizes or reported effect estimates. From an ML perspective, these tasks may be viewed as classifying individual words in a document. For example, we may classify each word as belonging to a rationale snippet supportive of a risk of bias determination (or not) or classify every word as belonging to a span describing the study sample size.
Figure 5.
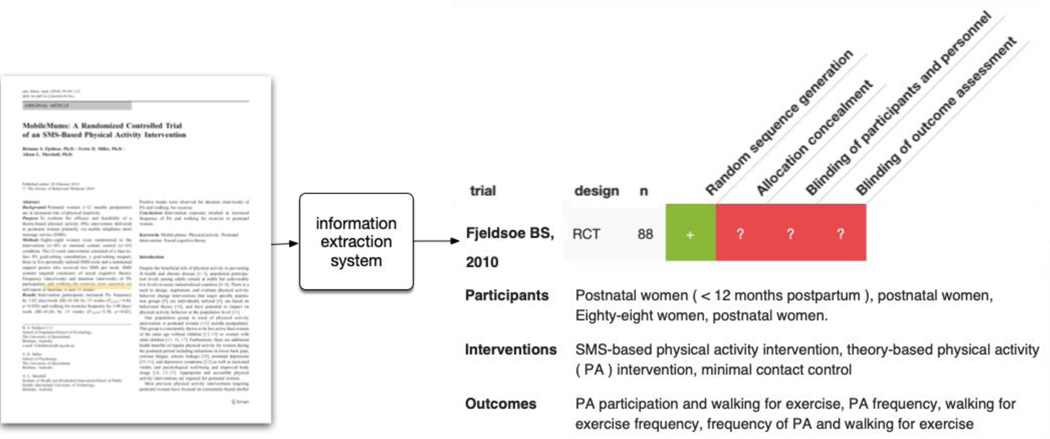
Example of an automatic data extraction system. Here, the system takes a trial report (in PDF format) as input, and automatically produces more structured output, comprising text snippets (here, information on the trial PICO), structured data (e.g., the number of participants), or analytical tasks (e.g., assessing risks of bias).
Word-level classification tasks differ from typical text classification settings (described above) in that adjacent predictions will tend to be strongly correlated: If a particular word in a document is relevant to a risk of bias assessment, it increases the likelihood that subsequent words are also relevant. This intuition motivates ML model variants that explicitly take structure into account to improve predictions. Such approaches are often labelled sequence tagging models because they make predictions over sequences of inputs.
An in-depth technical discussion of sequence tagging methods is beyond the scope of the present article. Briefly, the sequence tagging analogue of logistic regression is the conditional random field (CRF; Sutton & McCallum, 2012), which explicitly models correlations between outputs. In the case of NLP, these are typically correlations between the predictions for adjacent words. This formulation is often referred to as a “linear-chain” CRF. Older realizations of CRF sequence tagging models consumed sparse representations of input words (as described above) in order. More recent models add a CRF on top of word-level feature vectors induced by neural network architectures (Lample, Ballesteros, Subramanian, Kawakami, & Dyer, 2016; Ma & Hovy, 2016), such as BERT (discussed above).
All that these approaches have in common is that they are supervised (or in other words are dependent on manually labelled data in order to ‘learn’ to do a task). Yet, in health research (and particularly acutely in specialized domains such as health psychology) there is a dearth of training data. It is expensive to pay individuals with the requisite level of expertise to annotate full-text health psychology articles. At the same time, these articles tend to be somewhat technical, which means the work does not naturally lend itself to use of crowdsourcing platforms via which one can field annotation tasks to workers (typically laypersons) across the world, usually at relatively low cost. We note that annotating documents is done routinely in the conduct of conventional systematic reviews, which highlights a key problem: that the work in conducting systematic reviews is not done or recorded in a standardised way.
Recent work has aimed to address this data-availability problem, at least in the case of RCTs, by combining redundant crowd worker labels and domain expert annotations. Using this strategy, Nye et al. (2018) have released EBM-NLP, a corpus with sequence annotations that demarcate descriptions of trial following PICO elements: Populations, Interventions/Comparators, and Outcomes.
In some cases, a text classification model trained once might be generically useful. For example, pre-trained systems that reliably identify RCTs have been developed; these could be used in any systematic review that includes this study design without further training.
By contrast, some tasks might be idiosyncratic to a particular review question (for example, determining whether a particular psychological outcome measure was assessed), which might require targeted training data to build an automated system.
Another approach, called distant supervision, may be used to train models when manually labelled data are sparse (Mintz, Bills, Snow, & Jurafsky, 2009). Distant supervision refers to deriving labels (such as on words) that use existing structured resources and rules. For example, the free-text descriptions of trial PICO elements stored in the Cochrane Database of Systematic Reviews can be heuristically matched to snippets in the corresponding full-text articles to induce “labels” codifying whether individual tokens belong to PICO element descriptions. These labels will be imperfect (or “noisy”) but may nonetheless contain sufficient signal to train accurate models. Additionally, one can use strategies to mitigate noise using a small amount of direct, manual annotation (Wallace, Kuiper, Sharma, Zhu, & Marshall, 2016).
More recently, Grames et al. (2019) proposed a 5-step search-term-identifying algorithm for systematic reviews using keyword co-occurrence networks. (a) First, the research team writes a naïve search to locate a group of relevant reports. Then (b), they remove duplicates from the search results and (c) represent each report in a BoW manner. After that (d), the algorithm uses a document-feature matrix (also known as a keyword co-occurrence network), using the dictionary terms as features in which less important keywords are omitted. In this fashion, the possible search terms central to the topic will be suggested, and then are manually sorted into concept groups. The final step (e) would be redundant term removal and keyword translation, in which Boolean search strings can be used for the review team when searches are conducted.
There have been several explorations of automated data extraction from reports of clinical trials (Kiritchenko, de Bruijn, Carini, Martin, & Sim, 2010; Marshall, Kuiper, Banner, & Wallace, 2017; Summerscales, Argamon, Bai, Hupert, & Schwartz, 2011; Thomas, McNaught, & Ananiadou, 2011; for a survey, see Jonnalagadda, Goyal, & Huffman, 2015). These works have aimed to extract population size and characteristic descriptions, text snippets indicating risks of bias, and in some cases, reported outcome measures. Yet, despite progress, these technologies are not yet mature, either due to difficulties in modelling the long-term dependencies in texts or due to the specificity of clinical trial texts that conventional data extraction cannot generalize well.
Similarly, most of the work on data extraction from published evidence has been for RCT reports; extracting data from other types of research (e.g., non-randomized treatment studies or cross-sectional observational studies) may be particularly important in health psychology. An automated strategy for coding the methodological quality of meta-analyses and systematic reviews would be quite valuable (e.g., one that applies the AMSTAR to individual evidence synthesis reports). Similarly, for research in experimental medicine, intervention content has been an important consideration for decades, but only in the last decade has a refined behaviour change technique (BCT) taxonomy emerged (Michie et al., 2013). Currently, considerable human training is necessary to code BCTs, and this task is quite labour intensive, making the potential payoff of developing automated (or semi-automated) data extraction a rich prospect. An additional hurdle for this application is that often, published reports lack all the details about the BCTs that were included in the experimental arm (and even more so for the control arm(s)). Thus, there is a need for research on approaches for extracting data from such reports and also highlights the need to collect additional training data in this space.
Models trained to perform data extraction from articles will be imperfect, but they can nonetheless be useful. In our view, the most promising mode of use for data extraction models is for semi-automation in which they are used to expedite manual extraction. There has not yet been much in the way of evaluation of such technologies in this assistive setting; we discuss the evaluations that have been performed below.
From Research to Practice: Evaluating (Semi-)Automation Technologies
To date, much of the research on automating extraction of data from articles has evaluated performance of systems retrospectively. Specifically, reviewers hold out a set of data during model development and training (the “test set”), and then make predictions on it using a model trained using the rest of the data. These predictions can then be evaluated against the human labels, to calculate sensitivity and specificity. We note that many superficially objective review tasks are complex and involve human judgement. For example, human reviewers often disagree on whether a clinical trial was adequately blinded (Marshall et al., 2016). Evaluations of analytic complexity tasks will often, therefore, require consideration of how closely human experts are in agreement with the results.
Note that labels will be associated with either articles/abstracts (for classification tasks) or individual words (for tagging/extraction tasks). Performance is usually measured via sensitivity (also known as recall), precision, and their combination. For the latter, a standard choice is F1, which is the harmonic mean of precision and sensitivity. For evaluating classifier performance in the context of citation screening (as described above), custom metrics have been defined that attempt to better capture the specific trade-offs involved (Cohen et al., 2006; Wallace, Small, Brodley, & Trikalinos, 2010). These metrics tend to emphasize sensitivity, given that the notion of comprehensiveness is central to the evidence synthesis process.
Quantifying model performance in this way provides a sense of model predictive performance but is just a proxy outcome for the most important outcome: Will the model actually be helpful in practice? Simulation studies can be used to assess this concern to some degree, especially for text classification tasks such as citation screening or RCT identification because the number of articles automatically screened out is a natural measure of effective labour reduction. But, the degree to which automation saves work is more difficult to assess for data extraction tasks. It is not obvious how, for example, specific F1 scores for sequence tagging models might actually translate to practical utility. For technologies that seek to aid data extraction, we therefore argue that user studies are key to understanding whether extraction models are, in fact, useful to end-users synthesizing evidence.
Researchers have recently performed one such study that assesses the use of semi-automation in aiding risk of bias extraction from RCTs (Soboczenski et al., 2019). The authors enrolled individuals (mostly researchers experienced with evidence synthesis) to perform risk of bias assessments on four articles, which were randomly drawn from a superset of RCT reports constructed to be topically diverse. The authors provided predictions from our automated risk of bias system for two of the four articles, including extracted text snippets supporting bias assessments across four domains. Participants could opt to delete or edit these snippets. For the other two articles, risk of bias assessment proceeded entirely manually. The order in which individuals were provided model predictions as opposed to performing fully manual assessment was random. The authors found that using ML reduced the time to perform risk of bias extraction by about 25% and that users found the semi-automation system helpful in general. The research community might aspire to perform additional such user studies in future work to realistically estimate the utility of ML for aiding data extraction in practice.
To summarize this section: Text classification and data extraction models may automate or semi-automate some aspects of evidence synthesis in health psychology. Yet, a lack of labelled, validated “training data” for health psychology research, specifically, is a current obstacle to developing such models. Furthermore, once developed, models should be evaluated in practice to realistically assess their usefulness (e.g., via prospective user studies).
A Look into the Future
We now turn to more speculative, innovative uses of ML and “machine reading” technologies, highlighting how such methods might be used going forward. A theme in these potential uses is a departure from traditional evidence syntheses and toward faster “rapid” reviews of the evidence.
Real-time Literature Surveillance
One potential use of automated technologies that can process the whole of the literature is to enable real-time evidence surveillance, which can be accomplished by alerting interested parties automatically through text classification methods when new evidence is published. Furthermore, using extraction models, key aspects of newly published evidence could be inferred and stored in a structured format. As an example, one can imagine a scenario in which newly published evidence that seems relevant to a previously conducted systematic review is automatically recognized and data are extracted from it and used to auto-complete an extraction template or draft. Then, a relevant individual is notified to edit this draft accordingly (because automated extraction will be imperfect), which would potentially facilitate living systematic reviews (Bagg & McAuley, 2018; Maas, 2018; Thomas et al., 2017).
Exploring the Evidence
Automated categorization and extraction of data from published literature might also power novel browsing tools, which one could use to search for evidence relevant to their clinical questions. For example, in the case of RCTs, identifying snippets corresponding to Population, Interventions/Comparators and Outcomes (PICO; as discussed above) elements can in turn support faceted search—that is, search over these specific aspects. Faceted search may allow, for example, one to retrieve all evidence relevant to a particular condition (P), regardless of the interventions (I) and outcomes (O) studied (Soto, Przybyla, & Ananiadou, 2018). More generally, automatic tagging of studies (e.g., by study type) and their categorization into high-level sets reflecting clinical topics may afford interactive visualizations of the evidence that are not currently feasible (Parikh et al., 2019).
Automated Synthesis
Often, the aim of evidence synthesis is to use reliable methods to assess whether a particular treatment is the best option for a given condition with respect to particular outcome(s). Moving beyond just extraction, then, an audacious aim is to build a model that tries to infer the findings that an article reports directly—or, better, one that accurately gathers the information and then standardizes it according to a pre-established coding scheme, including both (a) information about the studies or interventions (e.g., population, sample size, recruitment strategies, intervention content and dose, follow-up time, types of outcome measures); and (b) quantitative information about the association(s) examined in the studies on the measures in question (e.g., effect size, sampling error). These elements define the database for a systematic review with meta-analysis. Surely, this vision is one that can save considerable human work if the automated system is accurate, reliable, and valid. Yet, the vision is also a daunting one, given the myriad details in both qualitative and quantitative coding.
One notion of this concept has been recently operationalized in preliminary work that aims to infer whether a given intervention is reported to significantly increase, have no significant effect, or significantly decrease a particular outcome with respect to a specified comparator (Lehman, DeYoung, Barzilay, & Wallace, 2019). This work assumes a full-text report of an RCT is given in order to permit inferences. Another assumption is that one should extract a snippet of text from the article that supports the determination made. An annotated dataset comprising about 10,000 “prompts” (interventions, comparators, and outcomes) and matched full-text RCT reports has been released, and initial models have been proposed (Lehman et al., 2019). Current performance is modest (about 0.5 F1 score, averaged across the three classes) but far better than chance. It is possible that modelling innovations will yield fast progress on this task; this process will open the door to fully automated “rapid” syntheses that will, of course, lack the rigor of manually performed analyses, but may nonetheless prove useful in providing fast overviews of the evidence for particular interventions.
Another ambitious contemporary effort to harness the rapidly accumulating body of intervention evidence is The Human Behaviour-Change Project (HBCP), which is amid a multi-year plan to develop and evaluate a behaviour change intervention knowledge system (Michie et al., 2017). The automated system aims to deliver comprehensive, high quality, accessible syntheses and interpretations of evidence, and to do so in a timely fashion. The system relies on machine learning and natural language technologies (similar to those we have reviewed above) in order to automate or semi-automate the process. The hope is to deliver an outward facing software interface that can answer versions of the question “What works, compared with what, how well, with what exposure, with what behaviours (for how long), for whom, in what settings and why?” (Michie et al., 2017). Clearly, to the extent that such a system succeeds, it can improve human welfare and public health considerably. As one indication of the complexity of the problem, HBCP’s strategy rests on first developing ontologies of behaviour change interventions that then help to guide machine learning. In this context, ontologies are data structures of organized, unique identifiers to represent particular types of entities (usually objects, attributes, processes, and mixtures of these); they define and label these elements and specify relationships between the entities (Larson et al., 2017). Norris, Finnerty, Hastings, Stokes, and Michie’s (2019) recent scoping review found 15 extant ontologies, and these authors judged nearly all to be logical, but they found none captured the breadth and detail of behaviour change sufficiently (e.g., most focused on only a single theoretical perspective). Thus, more work is needed to establish a stronger ontology before HBCP can succeed in routine problems of setting out the most optimal intervention for a particular behaviour change problem in a particular population. As well, even semi-automated reviewing faces many daunting challenges and must overcome certain risks, as we discuss in the next section.
Risks in Systematic Reviews and How Automation May Ameliorate or Exaggerate Them
Some of the challenges that traditional systematic reviewing faces can be minimized or entirely overcome using automatized or semi-automatized strategies, such as those that we have listed here. Chief among them is the ease with which such systems can sort through millions of citations to find reports that meet the selection criteria. If automation can also code reports for relevant qualitative and quantitative information of studies—and do so with great accuracy—then the amount of human effort needed is greatly reduced. Importantly, it also means that it will soon be feasible to tackle systematic review topics that previously were not practically possible to undertake. There are thousands of trials evaluating health promotion or medical interventions; synthesizing these manually at scale is simply not feasible, but automation may one day harness this mass of unstructured knowledge.
Nevertheless, challenges remain. Assumptions that reviewers make regarding automation technology may profoundly affect any conclusions reached. For example, if an automated screening model relies on only title and abstract screening, it may miss reports that would match selection criteria but that only include the relevant information in the full-text body of the report. Of course, the same risk occurs in purely human-based systematic reviews, and it is usually addressed with common sense, with the recognition from experience that titles and abstracts typically lack the necessary information. Now, with automation, it is possible to check how much literature exists that would be excluded if the reviewers only completed title and abstract screening. Another “solution” is to train models explicitly to have high recall (sensitivity), which will increase workload but ensure (as a goal) comprehensiveness.
Similarly, most methods to date have focused on extracting data from RCTs specifically, but this focus may be overly restrictive, omitting (by construction) non-randomised and other uncontrolled trials. In fact, it is possible that the results from excluded trials are even more valuable than those that enter a review. By their very nature, RCTs have aspects that may often reduce the effects of an intervention, such as incentives for people to participate and teams to track participants’ locations over time (so that they may be contacted later for follow-up measures). Such aspects led Flay (1986) to label RCTs as efficacy trials, which he contrasted with effectiveness trials—those done without the trappings of RCTs but also often in cooperation with communities. Some interventions demand community involvement and therefore become very difficult to evaluate in controlled trials (even in a community-randomized fashion). In turn, these interventions might have the greatest benefits because they join an intervention effort with the support of the community. A well-designed and well-run automated systematic review could readily evaluate such potential to the extent that relevant literature exists. The same point can be made about grey literature, which is literature that is unpublished or any work that databases have not located. Automated search could in theory make use of very large numbers of repositories and registries scattered over the internet; such an approach would not be currently feasible when a handcrafted search strategy is needed for each database.
Automation is likely to lead to increasingly sophisticated analyses of evidence pertaining to human behaviour and efforts to reduce the risk of illness and improve quality of life. If the information in studies can be abstracted efficiently and accurately, then it becomes possible to examine not only the treatment and control groups at one point in time but also how these groups change over time. In this fashion, it becomes possible to see the degree to which intervention group members achieve clinically significant outcomes over time (e.g., markedly reduced depression) and whether that is the natural course for those in the control group. Arithmetic means observed in studies are only rarely meta-analysed yet may yield extremely valuable information.
The process of systematic reviewing requires a long series of interrelated steps, and all must be performed with high rigor in order to ensure valid conclusions (cf. Borenstein et al., 2009; Higgins, 2008; Johnson & Hennessy, 2019). Systematic review teams routinely return to earlier steps when they discover that their pre-designed methods are not capturing the realities encountered during the review. Although pre-registering a detailed methodological protocol is regarded as good practice, review teams often (legitimately) change their approach as they consider the literature they find and learn from this process. Mundane tasks (such as abstract screening) might also allow opportunity for careful thought and serendipitous insights that then change the direction of a review. Care should be taken that automation enables more time (rather than less) for thoughtful analysis.
A final risk is that the automated process will not capture qualitative information about the phenomenon being studied. Yet, at the outset of training the computers to do the work, human interactions with the software and the literature may reduce this risk. Another way is for humans to examine a random sample of studies that are both included in the sample reviewed and excluded from it.
Conclusions
Machine learning and natural language processing techniques have the potential to help domain experts in health psychology more efficiently make sense of and synthesize the evidence that is rapidly accumulating in published articles. Models for text classification (e.g., to aid citation screening) and data extraction (e.g., identifying descriptions of trial participants) are relatively mature, with some prototypes already available for use. We emphasize that currently, such technologies should be thought of as fundamentally assistive, rather than providing full-fledged automation of synthesis. So far as we are aware, no review automation technologies have yet been specifically tested in the health psychology literature, despite HBCP’s efforts to date (though these are at least somewhat promising). Furthermore, existing approaches have predominantly focused on processing reports of RCTs, but models designed to consume and process literature describing other study types may be particularly important for health psychology.
The preceding observations indicate a need for research in applying, existing, and perhaps, developing, new models for semi-automation in the context of health psychology. Doing so will require creation of labelled datasets (i.e., annotated articles in this domain) to facilitate the training and evaluation of models. We hope the promise of semi-automation technologies spurs development of such resources to facilitate further research. There is a current lack of off-the-shelf systems for training and using machine learning for research synthesis. Health psychology researchers who are interested in trying such technologies will now have to contend with using technically difficult research code sometimes; training new systems (for the moment) will likely depend on collaborations with computer scientists.
Acknowledgments
The authors are supported by the National Institutes of Health (NIH) under the National Library of Medicine (NLM) award R01LM012086 (Wallace and Marshall), the Science of Behavior Change Common Fund Program through an award administered by the National Institute on Aging, U.S. PHS grant 5U24AG052175 (Johnson), the U.S. National Science Foundation grants 1743418 and 1843025 (Rajasekaran), and the UK Medical Research Council (MRC), through its Skills Development Fellowship program, grant MR/N015185/1 (Marshall). The views presented here are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or other governing bodies.
Contributor Information
Iain J. Marshall, King’s College London
Blair T. Johnson, University of Connecticut
Zigeng Wang, University of Connecticut.
Sanguthevar Rajasekaran, University of Connecticut.
Byron C. Wallace, Northeastern University
References
- Bagg MK, & McAuley JH (2018). Correspondence: Living systematic reviews. Journal of Physiotherapy, 64(2), 133 10.1016/j.jphys.2018.02.015 [DOI] [PubMed] [Google Scholar]
- Cer D, Yang Y, Kong SY, Hua N, Limtiaco N, John RS, ... & Sung YH (2018). Universal sentence encoder. arXiv preprint arXiv:1803.11175. [Google Scholar]
- Cohen AM, Hersh WR, Peterson K, & Yen PY−. (2006). Reducing workload in systematic review preparation using automated citation classification. Journal of the American Medical Informatics Association,13(2), 206–219. 10.1197/jamia.M1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J, Chang MW, Lee K, & Toutanova K. (2018). Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805. [Google Scholar]
- Flay BR (1986). Efficacy and effectiveness trials (and other phases of research) in the development of health promotion programs. Preventive Medicine, 15(5), 451–474. 10.1016/0091-7435(86)90024-1 [DOI] [PubMed] [Google Scholar]
- Goldberg Y. (2017). Neural network methods for natural language processing. Synthesis Lectures on Human Language Technologies, 10, 1–309. 10.2200/s00762ed1v01y201703hlt037 [DOI] [Google Scholar]
- Grames EM, Stillman AN, Tingley MW, & Elphick CS (2019). An automated approach to identifying search terms for systematic reviews using keyword co‐occurrence networks. Methods in Ecology and Evolution. 10.1111/2041-210X.13268 [DOI] [Google Scholar]
- Hennessy EA, Johnson BT, Acabchuk RL, McCloskey K, & Stewart-James J. (2019). Self-regulation mechanisms in health behaviour change: A systematic meta-review of meta-analyses, 2006–2017. Health Psychology Review. 10.1080/17437199.2019.1679654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyyer M, Manjunatha V, Boyd-Graber J, & Daumé III H. (2015). Deep unordered composition rivals syntactic methods for text classification. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers) (Vol. 1, pp. 1681–1691). [Google Scholar]
- Johnson BT, & Hennessy EA (2019). Systematic reviews and meta-analyses in the health sciences: Best practice methods for research syntheses. Social Science & Medicine, 233, 237–251. 10.1016/j.socscimed.2019.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonnalagadda SR, Goyal P, & Huffman MD (2015). Automating data extraction in systematic reviews: A systematic review. Systematic Reviews, 78, 4 10.1186/s13643-015-0066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiritchenko S, de Bruijn B, Carini S, Martin J, & Sim I. (2010). ExaCT: Automatic extraction of clinical trial characteristics from journal publications. BMC Medical Informatics and Decision Making, 10, 56 10.1186/1472-6947-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lample G, Ballesteros M, Subramanian S, Kawakami K, & Dyer C. (2016, June). Neural Architectures for Named Entity Recognition, San Diego, California. 10.18653/v1/n16-1030 [DOI] [Google Scholar]
- Larsen KR, Michie S, Hekler EB, Gibson B, Spruijt-Metz D, Ahern D, ... & Yi J. (2017). Behavior change interventions: the potential of ontologies for advancing science and practice. Journal of B, 40(1), 6–22. [DOI] [PubMed] [Google Scholar]
- Lehman E, DeYoung J, Barzilay R, & Wallace BC (2019). Inferring which medical treatments work from reports of clinical trials. 2019 Annual Conference of the North American Chapter of the Association for Computational Linguistics Minneapolis, MN. [Google Scholar]
- Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, ... & Moher D. (2009). The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PLoS Medicine, 6(7), e1000100. 10.1371/journal.pmed.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas A. (2018). Living systematic reviews: A novel approach to create a living evidence base. Journal of Neurotrauma. 10.1089/neu.2018.6059 [DOI] [PubMed] [Google Scholar]
- Marshall IJ, Kuiper J, Banner E, & Wallace BC (2017). Automating biomedical evidence synthesis: RobotReviewer. Proceedings of the Conference. Association for Computational Linguistics. Meeting, 2017, 7–12. 10.18653/v1/P17-4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall IJ, Noel-Storr A, Kuiper J, Thomas J, & Wallace BC (2018). Machine learning for identifying randomized controlled trials: An evaluation and practitioner’s guide. Research Synthesis Methods, 9(4), 602–614. 10.1002/jrsm.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall IJ, Kuiper J, Wallace BC (2016). RobotReviewer: Evaluation of a system for automatically assessing bias in clinical trials. Journal of the American Medical Informatics Association, 23(1), 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall IJ, & Wallace BC (2019). Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Systematic Reviews, 8(1), 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, & Hovy E. (2016). End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF. Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 1064–1074. 10.18653/v1/p16-1101 [DOI] [Google Scholar]
- Michie S, Richardson M, Johnston M, Abraham C, Francis J, Hardeman W, . . . Wood CE (2013). The behavior change technique taxonomy (v1) of 93 hierarchically clustered techniques: Building an international consensus for the reporting of behaviour change interventions. Annals of Behavioral Medicine, 46, 81–95. doi: 10.1007/s12160-013-9486-6 [DOI] [PubMed] [Google Scholar]
- Michie S, Thomas J, Johnston M, Mac Aonghusa P, Shawe-Taylor J, Kelly MP, ... & West R. (2017). The human behaviour-change project: Harnessing the power of artificial intelligence and machine learning for evidence synthesis and interpretation. Implementation Science, 12(1), 121 10.1186/s13012-017-0641-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz M, Bills S, Snow R, & Jurafsky D. (2009). Distant supervision for relation extraction without labeled data. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP (Volume 2 - ACL-IJCNLP ‘09) 10.3115/1690219.1690287 [DOI] [Google Scholar]
- Norris E, Finnerty AN, Hastings J, Stokes G, & Michie S. (2019). A scoping review of ontologies related to human behaviour change. Nature Human Behaviour, 3(2), 164. [DOI] [PubMed] [Google Scholar]
- Nye B, Jessy Li J, Patel R, Yang Y, Marshall IJ, Nenkova A, & Wallace BC (2018). A corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. Proceedings of the Conference. Association for Computational Linguistics. Meeting, 2018, 197–207. [PMC free article] [PubMed] [Google Scholar]
- O’Mara-Eves A, Thomas J, McNaught J, Miwa M, & Ananiadou S. (2015). Using text mining for study identification in systematic reviews: A systematic review of current approaches. Systematic Reviews, 4, 5 10.1186/2046-4053-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S, Conrad E, Agarwal O, Marshall I, Wallace BC, & Nenkova A. (2019). Browsing health: Information extraction to support new interfaces for accessing medical evidence. Proceedings of the workshop on extracting structured knowledge from scientific publications Minneapolis, MN. [Google Scholar]
- Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, ... & Stoyanov V. (2019). Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692. [Google Scholar]
- Shea BJ, Reeves BC, Wells G, Thuku M, Hamel C, Moran J, ... & Henry DA (2017). AMSTAR 2: A critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ, 358, j4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboczenski F, Trikalinos TA, Kuiper J, Bias RG, Wallace BC, & Marshall IJ (2019). Machine learning to help researchers evaluate biases in clinical trials: a prospective, randomized user study. BMC Medical Informatics and Decision Making, 19(1), 96 10.1186/s12911-019-0814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto AJ, Przybyla P, & Ananiadou S. (2018). Thalia: Semantic search engine for biomedical abstracts. Bioinformatics, 35(10), 1799–1801. 10.1093/bioinformatics/bty871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerscales RL, Argamon S, Bai S, Hupert J, & Schwartz A. (2011). Automatic summarization of results from clinical trials. 2011 IEEE International Conference on Bioinformatics and Biomedicine. 10.1109/bibm.2011.72 [DOI] [Google Scholar]
- Sutton C, & McCallum A. (2012). An introduction to conditional random fields. Foundations and Trends in Machine Learning, 4(4), 267–373. 10.1561/2200000013 [DOI] [Google Scholar]
- Thomas J, McNaught J, & Ananiadou S. (2011). Applications of text mining within systematic reviews. Research Synthesis Methods, 2(1), 1–14. 10.1002/jrsm.27 [DOI] [PubMed] [Google Scholar]
- Thomas J, Noel-Storr A, Marshall I, Wallace B, McDonald S, Mavergames C, … Living Systematic Review Network. (2017). Living systematic reviews: 2. Combining human and machine effort. Journal of Clinical Epidemiology, 91, 31–37. 10.1016/j.jclinepi.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, ... & Polosukhin I. (2017). Attention is all you need. In Advances in Neural Information Processing Systems (pp. 5998–6008). [Google Scholar]
- Vyas J, Nowling RJ, Meusburger T, Sargeant D, Kadaveru K, Gryk MR, Kundeti V, Rajasekaran S, and Schiller MR (2010). MimoSA: a system for minimotif annotation. BMC Bioinformatics, 11:328, http://www.biomedcentral.com/1471-2105/11/328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BC, Kuiper J, Sharma A, Zhu MB, & Marshall IJ (2016). Extracting PICO sentences from clinical trial reports using supervised distant supervision. Journal of Machine Learning Research, 17, 132 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/27746703 [PMC free article] [PubMed] [Google Scholar]
- Wallace BC, Small K, Brodley CE, Lau J, & Trikalinos TA (2012). Deploying an interactive machine learning system in an evidence-based practice center. Proceedings of the 2nd ACM SIGHIT Symposium on International Health Informatics - IHI ‘12 10.1145/2110363.2110464 [DOI] [Google Scholar]
- Wallace BC, Small K, Brodley CE, & Trikalinos TA (2010). Active learning for biomedical citation screening. Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ‘10 10.1145/1835804.1835829 [DOI] [Google Scholar]
- Whiting P, Savović J, Higgins JP, Caldwell DM, Reeves BC, Shea B, ... & Churchill R. (2016). ROBIS: A new tool to assess risk of bias in systematic reviews was developed. Journal of Clinical Epidemiology, 69, 225–234. 10.1016/j.jclinepi.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov R, & Le QV (2019). XLNet: Generalized Autoregressive Pretraining for Language Understanding. arXiv preprint [Google Scholar]


