Abstract
Gibberellins (GA) are phytohormones controlling major aspects of plant lifecycle including seed germination, growth and flower development. GA signaling is also involved in resistance to adverse conditions, thus providing a mechanism for environmentally responsive growth regulation. We recently characterized the function of a core component of the GA signal transduction pathway: RGL3. RGL3 belongs to the DELLA family of negative GA response regulators. Jasmonate (JA) rapidly induces RGL3 expression, which in turn enhances the expression of JA-responsive genes by inhibiting the activity of key repressors of JA signaling, the JAZ proteins. JA and ethylene (ET) are well known to play synergistic roles in plant disease resistance. Accordingly, we showed that RGL3 regulates plant defense responses by modulating JA/ET-mediated defense signaling pathway.
Keywords: gibberellin, jasmonate, ethylene, RGL3, plant defense
GA plays critical roles in modulating plant growth by integrating both developmental and environmental stimuli. Key regulators of the GA signaling pathway are the nuclear-localized DELLA proteins (DELLAs), a subset of the GRAS family of transcriptional regulators that repress all aspect of GA response. GA promotes growth by stimulating the proteasome-dependent destruction of DELLAs.1 Recently, DELLAs have been reported to be implicated in plant disease resistance. Upon pathogen attack, an interplay between the phytohormones salicylic acid (SA), JA and ET activates distinct defense response signaling cascades, depending on the lifestyle of the invading pathogen.2 Whereas SA is generally associated with resistance against biotrophs, JA/ET are associated with resistance to necrotrophs. Moreover, the interaction between these two types of resistance is antagonistic and the activation of one attenuates the other.2 Recent studies showed that DELLAs enhance resistance against necrotrophic pathogens while attenuating resistance against biotrophs by modulating the SA/JA balance.3,4 This is consistent with findings of DELLAs being able to interact with and inhibit the activity of JAZs, repressors of JA response, thus providing a mechanism for appropriate defense responses.5 Indeed, JAZs bind and inhibit the activity of a wide range of transcription factors including the basic helix-loop-helix JIN1/MYC2, MYC3 and MYC4 involved in JA-mediated expression of wound response genes,6-10 and EIN3 / EIL proteins involved in JA/ET-mediated expression of pathogenesis-related genes.11-14 In this addendum we discuss the role of the Arabidopsis DELLA protein RGL3 in regulating JA/ET-dependent defense responses through direct competitive binding to JAZs.
JA Induces RGL3 Expression via Direct Binding of MYC2 to its Promoter Region
The Arabidopsis thaliana genome encodes five DELLAs, GAI, RGA, RGL1, RGL2 and RGL3, that play distinct but also overlapping functions in repressing GA response.1,15,16 Studies of loss-of-function alleles of DELLAs have revealed a specific role for each DELLA, except RGL3. This functional specificity is conferred by their expression pattern rather than by their molecular activity.17 We recently showed that JA positively regulates the expression of RGL3 in contrast to the other DELLA genes.18 The induction is dependent on a functional JA-signaling cascade as it is abolished in the JA-receptor coi1-1 mutant and in myc2 myc3 myc4 triple mutant plants. Furthermore, chromatin immunoprecipitation (ChIP) analysis and electrophoretic mobility shift assays (EMSA) demonstrated that MYC2 regulates the transcription of RGL3 through direct association with its promoter region (to a CACATG G-box-like motif located near the transcription start site).18 Thus RGL3 expression is directly induced by JA through its signaling pathway.
RGL3 Enhances MYC2 and EIN3 Activity via Competitive Binding to JAZs
Given the ability of DELLAs to interact and inhibit the activity of JAZs, repressors of MYC2, JA-mediated accumulation of RGL3 protein should enhance the expression of MYC2-regulated genes. Indeed, our results showed that induction of MYC2-regulated genes including VSP2, TAT1 and LOX2, is reduced in rgl3-5 mutant and enhanced in transgenic lines overexpressing RGL3. Thus RGL3 competes with MYC2 for binding to JAZs.5,18
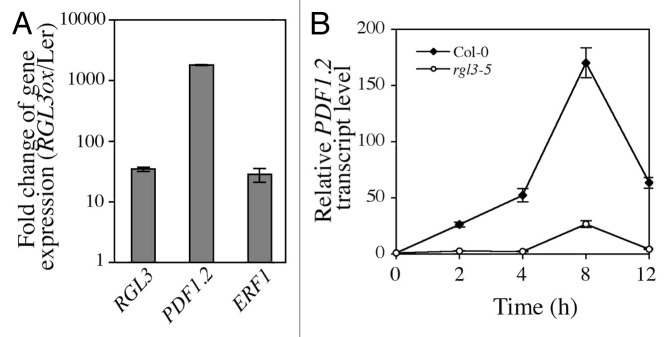
The JAZ proteins also interact with and inhibit EIN3 transcriptional activity to mediate JA/ET crosstalk.13 Whereas ET enhances EIN3 protein stability, JA releases EIN3 from the JAZs, thereby activating ET/JA-regulated processes.13 We next investigated whether DELLA function also modulates EIN3 activity via competitive binding to JAZs, in a similar manner than with MYC2. To this end, we monitored the interaction between JAZ8 and EIN3 by fluorescence lifetime imaging microscopy (FLIM) analysis of nuclei in transiently transformed Nicotiana benthamiana cells, in the presence and in the absence of a DELLA protein (RGA). Whereas without RGA, the fluorescence resonance energy transfer (FRET) between GFP-EIN3 and JAZ8-RFP was of 12.2%, expression of RGA significantly decreased the FRET to 9.0% (26.2% loss) (Fig. 1). This result demonstrates that the interaction between EIN3 and JAZ8 is diminished in the presence of DELLA. Consistent with this, overexpression of RGL3 enhances EIN3-regulated genes, including ERF1 and PDF1.2, in six week old Arabidopsis plants (Fig. 2A).12,14 In addition, JA-mediated induction of PDF1.2 is reduced in rgl3-5 mutant in comparison to wild-type plants (Fig. 2B). Taken together, our results indicate that RGL3 plays a critical role in modulating JA action by enhancing both MYC2 and EIN3 transcriptional activity.
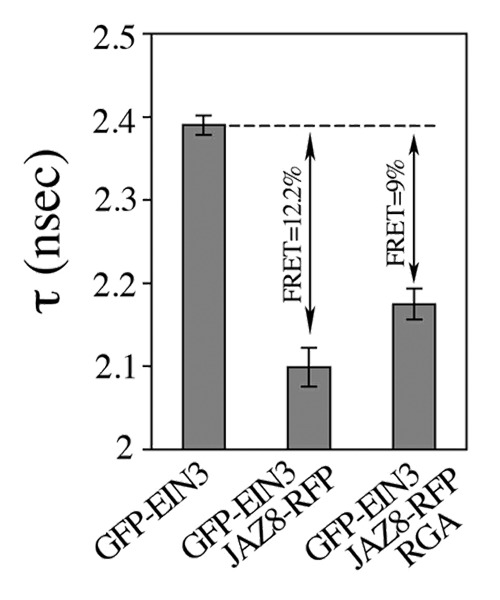
Figure 1. DELLA competes with EIN3 for binding to JAZ. Fluorescence lifetime analyses (in nsec) of GFP-EIN3, GFP-EIN3/JAZ8-RFP alone or together with RGA, and mean (± se) FRET value (%) in Nicotiana benthamiana agro-infiltrated leaves.
Figure 2. RGL3 enhances EIN3-dependent JA-responsive gene expression. (A) Relative transcript levels of RGL3, PDF1.2 and ERF1 in 6-week-old wild-type (Ler) and a line overexpressing RGL3. Data (mean ± SD) are represented as fold change in gene expression (RGL3ox/Ler). (B) Time-course induction (mean ± SD) of PDF1.2 transcripts in 3-week-old wild-type (Col-0) and rgl3-5 mutant plants treated with 50 µM of MeJA for the time indicated.
RGL3 Contributes Positively to JA/ET-Mediated Plant Defense Responses
JA and ET play a crucial role in plant defense and are generally involved in the activation of defense responses against necrotrophic pathogens.2 Our results showed that rgl3-5 mutants are significantly more susceptible to the necrotrophic fungus Botrytis cinerea as represented by larger lesions correlated with increased pathogen growth as compared with wild-type.18 Conversely, rgl3-5 mutants are more resistant to the hemibiotroph Pseudomonas syringae pv tomato DC3000 (Pst) compared with wild-type infected plants.18 This increased resistance is based on the capacity of Pst to activate the JA pathway via the production of a JA-mimicking phytotoxin, coronatine, therefore attenuating the defense response by exploiting the SA/JA antagonism.2,19 Consistent with this, induction of JA/ET-mediated expression of pathogenesis-related genes is compromised in rgl3-5 infected leaves in comparison with wild-type infected leaves.18 Collectively, these results showed that RGL3 contributes positively to the JA/ET-defense pathway.
Previous studies have indicated that DELLAs can also interact with EIN3 and MYC2 to regulate apical hook development and the synthesis of sequiterpenes in inflorescences, respectively.20,21 Further analyses will be required to assess the significance of these interactions in regulating plant defense responses.
Material and Methods
Fluorescence-lifetime imaging microscopy (FLIM)
FLIM analysis was performed using a Nikon TE2000 microscope connected to a LiFA FLIM system. Fluorescence-lifetime was measured using the LiFLIM software version 1.2.8. on N. benthamiana agro-infiltrated leaves expressing GFP-EIN3 (control) and co-expressing various combinations of GFP-EIN3, JAZ8-RFP and RGA proteins. At least 60 nuclei per condition were analyzed.
Gene expression analyses
qRT-PCR were performed on at least two biological repeats as previously described.18
Glossary
Abbreviations:
- GAI
GA INSENSITIVE
- RGA
REPRESSOR OF GA1-3
- RGL1
RGL2 and RGL3, RGA-LIKEs
- JIN1/MYC2
JASMONATE INSENSITIVE1
- EIN3
ETHYLENE INSENSITIVE3
- EIL
EIN3-Like
- JAZ
JA ZIM-domain
- COI
CORONATINE INSENSITIVE1
- ERF1
ETHYLENE RESPONSE FACTOR1
- PDF1.2
PLANT DEFENSIN1.2
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Thomas Potuschak for EIN3 pENTRY vector. The authors’ research group is currently supported by the Centre National de la Recherche Scientifique and Bayer.
References
- 1.Achard P, Genschik P. . Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 2009; 60:1085 - 92; http://dx.doi.org/ 10.1093/jxb/ern301; PMID: 19043067 [DOI] [PubMed] [Google Scholar]
- 2.Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. . Pathological hormone imbalances. Curr Opin Plant Biol 2007; 10:372 - 9; http://dx.doi.org/ 10.1016/j.pbi.2007.06.003; PMID: 17646123 [DOI] [PubMed] [Google Scholar]
- 3.Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. . Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 2008; 18:656 - 60; http://dx.doi.org/ 10.1016/j.cub.2008.04.034; PMID: 18450450 [DOI] [PubMed] [Google Scholar]
- 4.Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, et al. . DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 2008; 18:650 - 5; http://dx.doi.org/ 10.1016/j.cub.2008.03.060; PMID: 18450451 [DOI] [PubMed] [Google Scholar]
- 5.Hou X, Lee LYC, Xia K, Yan Y, Yu H. . DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 2010; 19:884 - 94; http://dx.doi.org/ 10.1016/j.devcel.2010.10.024; PMID: 21145503 [DOI] [PubMed] [Google Scholar]
- 6.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. . JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004; 16:1938 - 50; http://dx.doi.org/ 10.1105/tpc.022319; PMID: 15208388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, et al. . The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666 - 71; http://dx.doi.org/ 10.1038/nature06006; PMID: 17637675 [DOI] [PubMed] [Google Scholar]
- 8.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. . JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007; 448:661 - 5; http://dx.doi.org/ 10.1038/nature05960; PMID: 17637677 [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, et al. . The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011; 23:701 - 15; http://dx.doi.org/ 10.1105/tpc.110.080788; PMID: 21335373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauwels L, Goossens A. . The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 2011; 23:3089 - 100; http://dx.doi.org/ 10.1105/tpc.111.089300; PMID: 21963667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. . Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997; 89:1133 - 44; http://dx.doi.org/ 10.1016/S0092-8674(00)80300-1; PMID: 9215635 [DOI] [PubMed] [Google Scholar]
- 12.Solano R, Stepanova A, Chao Q, Ecker JR. . Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 1998; 12:3703 - 14; http://dx.doi.org/ 10.1101/gad.12.23.3703; PMID: 9851977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, An F, Feng Y, Li P, Xue L, A M, et al. . Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 2011; 108:12539 - 44; http://dx.doi.org/ 10.1073/pnas.1103959108; PMID: 21737749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. . Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998; 10:2103 - 13; PMID: 9836748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, et al. . The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 1997; 11:3194 - 205; http://dx.doi.org/ 10.1101/gad.11.23.3194; PMID: 9389651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverstone AL, Ciampaglio CN, Sun T. . The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998; 10:155 - 69; PMID: 9490740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. . Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 2010; 27:1247 - 56; http://dx.doi.org/ 10.1093/molbev/msq012; PMID: 20093430 [DOI] [PubMed] [Google Scholar]
- 18.Wild M, Davière J-M, Cheminant S, Regnault T, Baumberger N, Heintz D, et al. . The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012; 24:3307 - 19; http://dx.doi.org/ 10.1105/tpc.112.101428; PMID: 22892320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurie-Berry N, Joardar V, Street IH, Kunkel BN. . The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 2006; 19:789 - 800; http://dx.doi.org/ 10.1094/MPMI-19-0789; PMID: 16838791 [DOI] [PubMed] [Google Scholar]
- 20.An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, et al. . Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 2012; 22:915 - 27; http://dx.doi.org/ 10.1038/cr.2012.29; PMID: 22349459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong G-J, Xue X-Y, Mao Y-B, Wang L-J, Chen X-Y. . Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012; 24:2635 - 48; http://dx.doi.org/ 10.1105/tpc.112.098749; PMID: 22669881 [DOI] [PMC free article] [PubMed] [Google Scholar]