Abstract
Duchenne muscular dystrophy (DMD) is a devastating genetic muscle disease resulting in progressive muscle degeneration and wasting. Glucocorticoids, specifically prednisone/prednisolone and deflazacort, are commonly used by DMD patients. Emerging DMD therapeutics include those targeting the muscle-wasting factor, myostatin (Mstn). The aim of this study was to investigate how chronic glucocorticoid treatment impacts the efficacy of Mstn inhibition in the D2.mdx mouse model of DMD. We report that chronic treatment of dystrophic mice with prednisolone (Pred) causes significant muscle wasting, entailing both activation of the ubiquitin-proteasome degradation pathway and inhibition of muscle protein synthesis. Combining Pred with Mstn inhibition, using a modified Mstn propeptide (dnMstn), completely abrogates the muscle hypertrophic effects of Mstn inhibition independently of Mstn expression or SMAD3 activation. Transcriptomic analysis identified that combining Pred with dnMstn treatment affects gene expression profiles associated with inflammation, metabolism, and fibrosis. Additionally, we demonstrate that Pred-induced muscle atrophy is not prevented by Mstn ablation. Therefore, glucocorticoids interfere with potential muscle mass benefits associated with targeting Mstn, and the ramifications of glucocorticoid use should be a consideration during clinical trial design for DMD therapeutics. These results have significant implications for past and future Mstn inhibition trials in DMD.
Keywords: Muscle Biology, Therapeutics
Keywords: Monogenic diseases, Muscle, Translation
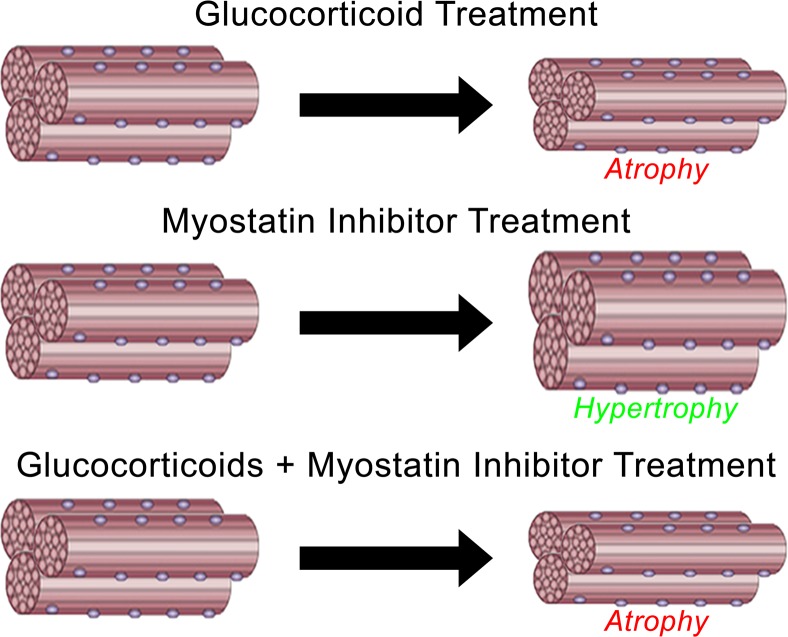
Muscle hypertrophy via myostatin inhibition is blocked by glucocorticoids, suggesting that glucocorticoids interfere with potential muscle mass benefits associated with targeting myostatin.
Introduction
Duchenne muscular dystrophy (DMD) is a lethal X-linked degenerative muscle disease caused by loss of dystrophin, a protein required for stabilization of muscle membranes during contraction (1, 2). Recent advancements in dystrophin-based therapeutic approaches have led to nonsense mutation (3) and exon skipping (4) therapies in clinic, and approval for dystrophin gene replacement and/or editing based therapies are likely on the horizon (5–7). If successful, these therapeutic strategies essentially will transform DMD into a milder disease, known as Becker’s muscular dystrophy (BMD), caused by truncated dystrophin protein products. Effective pharmacological interventions, whether monotherapies or combinational therapies, will therefore remain a major clinical need for these patients to combat the inflammation, fibrosis, and muscle loss associated with even milder forms of dystrophinopathy (8). Currently, the standard DMD therapeutic is the use of glucocorticoids, primarily prednisone/prednisolone (Pred) or deflazacort, which slows disease progression and delays loss of ambulation (9, 10). Prolonged glucocorticoid use, however, does have negative consequences, such as Cushingoid, bone demineralization, hastened cardiomyopathy (11), and — potentially — muscle atrophy (12). In evaluating therapeutics for DMD, the study of potential interactions with glucocorticoid use is an important aspect to consider, as most, if not all, patients in clinical trials are on glucocorticoids.
Myostatin (Mstn; also known as growth and differentiation factor 8) is a member of the TGF-β superfamily of ligands that is most known for its role as a negative regulator of muscle growth (13, 14). Like several other TGF-β ligands, Mstn is secreted as a latent complex consisting of a mature peptide dimer held inactive by an inhibitory propeptide. Upon activation by proteolytic cleavage of the propeptide (15), the mature Mstn dimer can bind to the activin receptor type IIB (ActRIIB, a TGF-β type II receptor), recruit anaplastic lymphoma kinase 4/5 (ALK4/5) (TGF-β type I receptors), and activate the SMAD2/3 signaling pathway to induce target gene transcriptional and repression activity (16). As a result of the potent muscle growth antagonism by Mstn, strategies to inhibit its actions have been heavily investigated, particularly for neuromuscular disease therapeutics. Preclinical work by our group has revealed therapeutic efficacy of Mstn inhibition via a protease-resistant mutant of the Mstn propeptide (dnMstn) and/or a soluble version of ActRIIB (sActRIIB; targets all ActRIIB ligands) in murine spinal muscular atrophy (17) and both murine and canine models of DMD (18–20). Additional studies using genetic ablation (21, 22) and Mstn neutralizing antibodies (23) have also shown beneficial effects of Mstn inhibition for DMD models, and there are currently several Mstn-targeting pharmaceutical products in clinical development (24–29).
The purpose of the current study is to investigate potential interactive effects of Mstn inhibition combined with a chronic glucocorticoid treatment regimen in the mdx mouse model of DMD. Herein, we demonstrate that daily use of Pred induces substantial skeletal muscle atrophy in mdx mice of both the C57BL/10 (B10.mdx) and DBA/2J (D2.mdx) genetic backgrounds, and this cannot be rescued by systemic overexpression of dnMstn. We additionally identify that Pred-induced muscle atrophy strongly impairs muscle protein synthesis, and this occurs independently of both Mstn content and SMAD3 activation.
Results
Daily glucocorticoid treatment induces muscle atrophy in dystrophic mice.
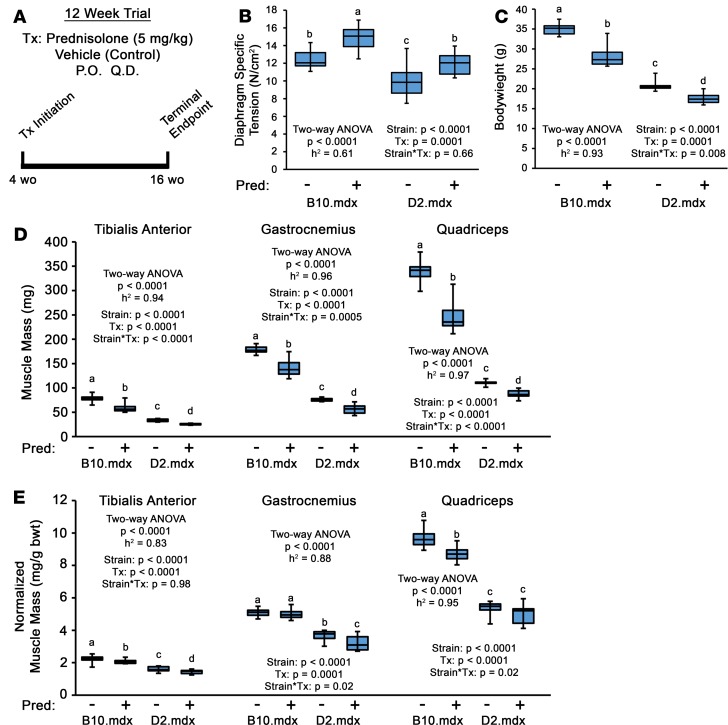
Recently, the D2.mdx mouse has emerged as a potentially better preclinical model for testing DMD therapeutics than traditional B10.mdx mice (30, 31), due to a more severe and fibrotic phenotype attributed to a polymorphism in latent TGF-β binding protein (LTBP) 4 (32). In an experiment to compare drug effects between these mdx lines, we performed a 12-week evaluation of the glucocorticoid, Pred (5 mg/kg, once a day; equates to a dose of 0.41 mg/kg in humans when normalized to body-surface area; ref. 33), in male B10.mdx and D2.mdx mice, with treatment initiation at 4 weeks of age and termination at 16 weeks of age (Figure 1A). While significant (~19%) improvements in diaphragm (Dp) specific tension (SPo; maximum tetanic isometric force production [Po] normalized to cross-sectional area [CSA]) were found with Pred treatment over vehicle controls in both lines (Figure 1B), the most striking phenotype of Pred-treated mice of this experiment — regardless of background strain — was substantial loss of body weight and skeletal muscle mass compared with respective vehicle groups (Figure 1, C–E).
Figure 1. Chronic prednisolone treatment induces muscle wasting in mdx mouse model of DMD regardless of genetic background.
(A) Preclinical trial design consisting of male mdx mice of C57BL/10 (B10.mdx) and DBA/2J (D2.mdx) genetic backgrounds receiving daily oral treatments of vehicle or 5 mg/kg prednisolone (Pred; n = 8–10). Treatments were initiated at 4 weeks of age and terminated at 16 weeks of age (12 weeks of treatment). (B) Ex vivo muscle function of the diaphragm was evaluated at terminal endpoint. (C–E) Body weights (C), absolute muscle masses (D), and body weight-normalized muscle masses (E) measured at terminal endpoint. Data were analyzed using 2-way ANOVA (strain and treatment effects; effect size reported as η2), followed by Tukey’s post hoc tests (α = 0.05). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM. Groups that are significantly different from each other are indicated by nonoverlapping letter designations (P ≤ 0.05).
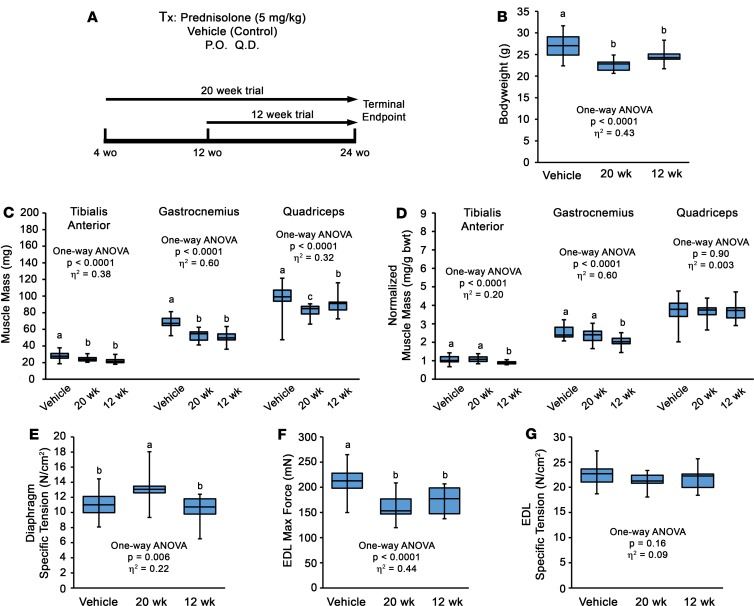
In a follow-up experiment to determine if these effects of Pred treatment are transient and/or dependent on age of initiation, D2.mdx mice were subjected to Pred (or vehicle) treatment initiation at either 4 or 12 weeks of age and terminated at 24 weeks of age (i.e., 20-week and 12-week treatment groups; Figure 2A). Both Pred treatment strategies resulted in comparable losses of body weight and skeletal muscle mass compared with vehicle treatments (Figure 2, B–D); thus, muscle atrophy is a consistent feature of Pred treatment in mdx mice, regardless of treatment duration or age of initiation. Additionally, Pred treatment initiation at 4 weeks of age resulted in significant improvements in Dp SPo, whereas delaying initiation to 12 weeks of age does not procure any functional improvements of the Dp (Figure 2E). The extensor digitorum longus (EDL) muscle demonstrated losses of maximum tetanic isometric force Po and no change in SPo (Figure 2, F and G), consistent with loss of parallel sarcomeres from muscle atrophy. These data are consistent with recent reports of daily glucocorticoid treatment inducing the muscle atrophy program in dystrophic muscle (12, 34).
Figure 2. Delayed prednisolone treatment does not prevent muscle atrophy in D2.mdx mice.
(A) Male D2.mdx mice received daily oral treatments of vehicle or 5 mg/kg prednisolone (Pred) that was initiated at either 4 or 12 weeks of age. (B–G) Terminal endpoint body weights (B), absolute muscle masses (C), body weight-normalized muscle masses (D), diaphragm (E), and extensor digitorum longus (EDL) (F and G) functional evaluations are reported. Data were analyzed using 1-way ANOVA (effect size reported as η2), followed by Tukey’s post hoc tests (α = 0.05). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM. Groups that are significantly different from each other are indicated by nonoverlapping letter designations (P ≤ 0.05).
Glucocorticoids affect muscle protein synthesis.
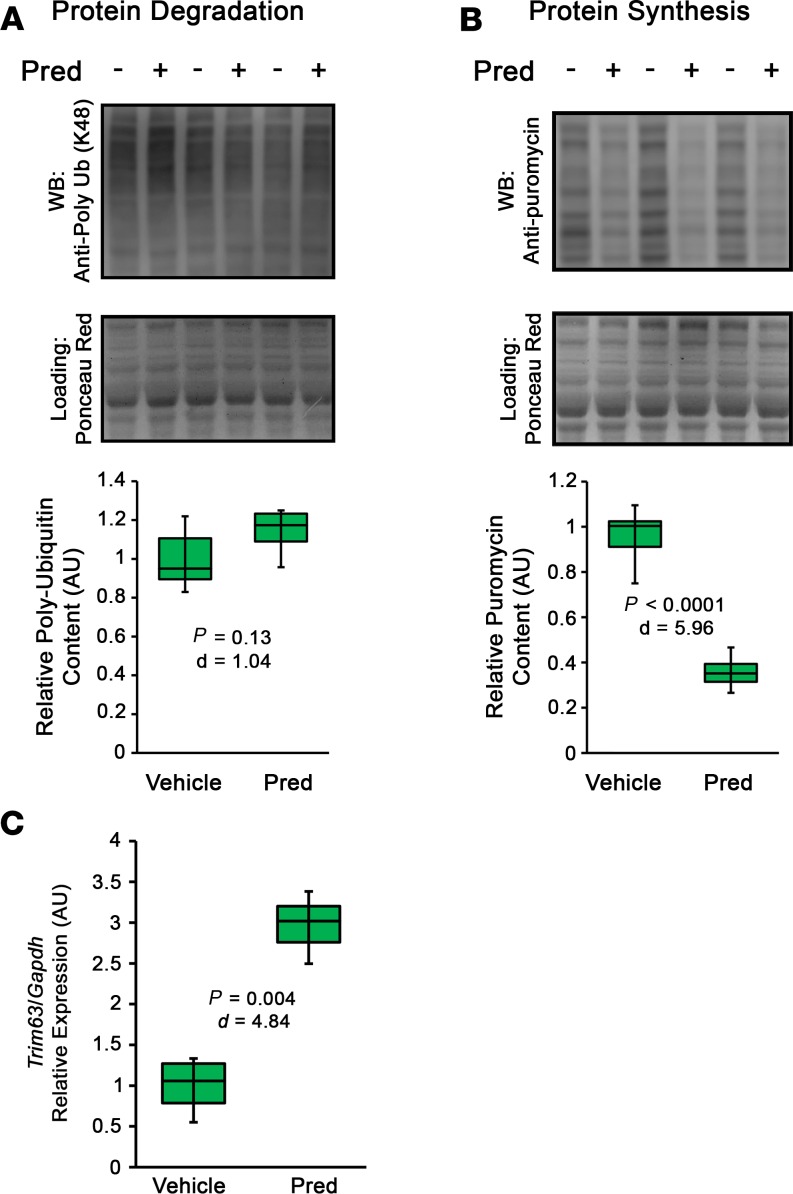
The induction of skeletal muscle atrophy by glucocorticoids has been linked to both the stimulation of catabolism through activation of the ubiquitin-proteasome system (12, 35, 36) and inhibition of muscle protein synthesis (37). To investigate the extent that daily glucocorticoid use affects each of these aspects of protein balance in the absence of a disease setting, we treated male WT DBA/2J mice with vehicle or 5 mg/kg Pred for 10 days and separately assessed accumulation of poly-ubiquitinated proteins (indicative of ubiquitin-proteasome system activity; ref. 38) and relative rates of puromycin incorporation into elongating peptides (indicative of active protein translation; ref. 39). WT mice were used for these assays in order to prevent potentially confounding results of muscle degeneration or regeneration, which spontaneously occur in the muscles of dystrophic animals. While accumulated poly-ubiquitin (K-48 linkage) levels trended toward a ~20% increase in Pred-treated quadriceps muscle (Figure 3A), puromycin incorporation was robustly reduced (~60%) with Pred treatment (Figure 3B). These data suggest that prolonged Pred treatment inhibits rates of muscle protein synthesis more potently than activation of ubiquitin-proteasomal protein degradation. We next sought to verify that Pred can activate gene expression of MuRF1 (product of Trim63 gene) — an E3 ubiquitin ligase involved in degradation of muscle protein (40, 41) — in WT muscle, as reported following administration of dexamethasone (35). Following a single dose of 5 mg/kg Pred, Trim63 gene expression increased by ~3-fold in WT DBA/2J quadriceps (Figure 3C). This suggests that E3 ubiquitin ligase activation is conserved in glucocorticoid-treated skeletal muscle, whether by dexamethasone or Pred.
Figure 3. Chronic prednisolone treatment affects skeletal muscle protein balance in vivo.
Adult male DBA/2J mice received daily oral treatments with vehicle or 5 mg/kg prednisolone (Pred) for 10 days. (A) To assess relative protein degradation, vehicle- and Pred-treated mice (n = 5–6) received a s.c. injection of 20 mg/kg of the proteasome inhibitor, MG-132, 24 hours prior to terminal endpoint. The accumulation of poly-ubiquitinated (K-48 linkage) proteins was assessed by immunoblotting of quadriceps muscle lysates. (B) To assess relative protein synthesis, vehicle- and Pred-treated mice (n = 6) received an i.p. injection of 20 mg/kg puromycin 30 minutes prior to terminal endpoint. The prevalence of puromycin-labeled peptides was assayed by immunoblotting using an anti-puromycin antibody. Ponceau red staining was used to visualize protein loading for immunoblot signal normalization. (C) Relative gene expression of the ubiquitin E3-ligase, Trim63, in quadriceps muscle of DBA/2J mice following a single dose of vehicle or Pred (n = 3), normalized to Gapdh. Data were analyzed using Welch’s t test (α = 0.05), with effect size reported as Cohen’s d (d). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM.
Chronic glucocorticoid treatment blocks muscle hypertrophic effects of Mstn inhibition.
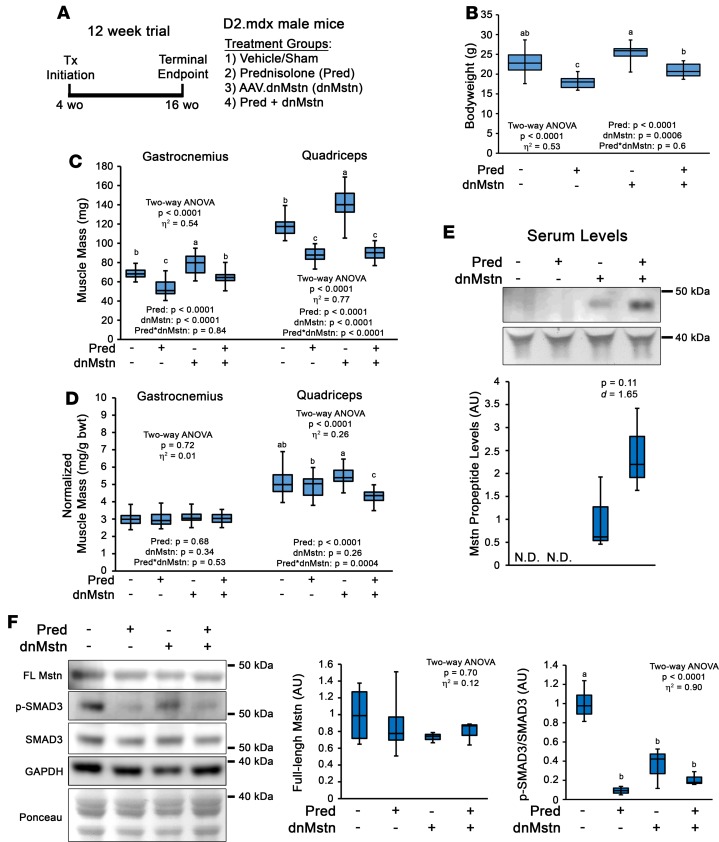
Therapeutics targeting Mstn are currently in advanced development or clinical trial (ref. 25; https://clinicaltrials.gov.; NCT02515669, NCT01524224, NCT01958970, NCT02310763, NCT02145234) and offer a promising approach to increase DMD patient musculature. However, to our knowledge, it has never been reported how glucocorticoid use interacts with the hypertrophic effects of Mstn inhibition. To investigate if Mstn inhibition is capable of rescuing dystrophic muscle from the atrophic effects of chronic Pred treatment, we performed a trial in D2.mdx mice using adeno-associated virus–mediated (AAV-mediated) systemic delivery of a protease-resistant form of dnMstn in combination with vehicle or Pred (15, 19). This 12-week study consisted of Pred (or vehicle) treatment initiated at 4 weeks of age and dnMstn (or PBS control; single i.p. injection) administration performed at 6 weeks of age, with the terminal endpoint at 16 weeks of age (Figure 4A). Consistent with previous experiments in B10.mdx mice (19), Mstn inhibition via dnMstn significantly increases body weight and muscle mass over control treatments (Figure 4, B–D); however, dnMstn treatment does not rescue either from Pred-induced decrements in the combination treatment group. This failure of dnMstn treatment to induce muscle hypertrophy in combination with Pred is not a result of Pred inhibiting dnMstn secretion from the liver, as serum levels of the Mstn propeptide trended toward increased levels (Figure 4E). Previous reports suggest glucocorticoids induce muscle atrophy via upregulation of Mstn (42, 43); therefore, we immunoblotted tibialis anterior muscle lysates for full-length Mstn (using an N-terminal Mstn antibody) and SMAD3 phosphorylation, a pathway stimulated by Mstn signaling (44). Interestingly, Mstn levels were unchanged across treatment groups, while SMAD3 phosphorylation levels are actually decreased with Pred treatment (Figure 4F). This suggests that the atrophic effects of Pred treatment are independent of Mstn and, likely, other ActRIIB ligands, which include Activin A and GDF11 (45, 46).
Figure 4. Chronic prednisolone treatment negates muscle hypertrophic effects of myostatin inhibition in D2.mdx mice.
(A) Preclinical trial design consisting of male D2.mdx mice receiving daily oral treatments of vehicle or 5 mg/kg prednisolone (Pred) initiated at 4 weeks of age. At 6 weeks of age, mice received a single sham control or adeno-associated virus–mediated (AAV-mediated) myostatin (Mstn) inhibitor (AAV8.dnMstn) i.p. injection, and the terminal endpoint was 16 weeks of age (n = 7–23). (B–D) body weights (B), absolute muscle masses (C), and body weight-normalized muscle masses (D) were measured at terminal endpoint. (E) Mstn propeptide levels were measured in serum samples from treatment groups by immunoblotting using Mstn N-terminal antibody (N.D., not detected). (F) Immunoblotting of tibialis anterior muscle lysates for full-length Mstn, phosphorylated SMAD3 (p-SMAD3; S425/427), or total SMAD3 protein levels. Ponceau red staining was used to visualize protein loading for immunoblot signal normalization. GAPDH content is shown for immunoblotting verification of equal loading. Data were analyzed using 2-way ANOVA (strain and treatment effects; effect size reported as η2), followed by Tukey’s post hoc tests (α = 0.05). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM. Groups that are significantly different from each other are indicated by nonoverlapping letter designations (P ≤ 0.05).
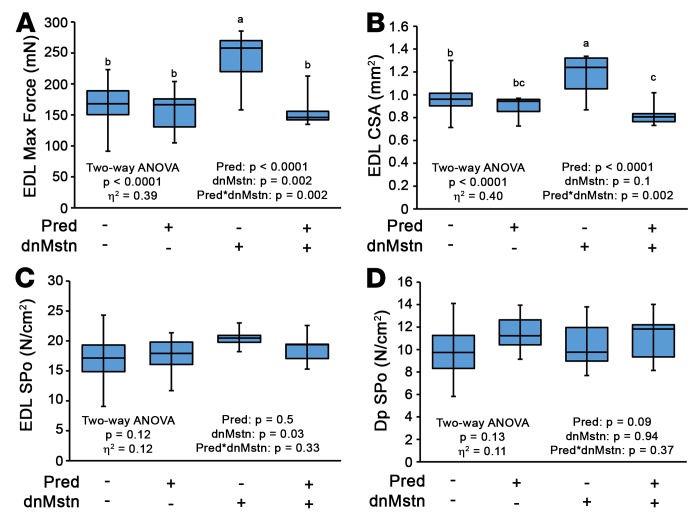
In agreement with our findings in muscle mass among the treatment groups, Po and CSA of the EDL, a muscle composed of primarily type IIB muscle fibers that are most responsive to Mstn activity (17, 47), were significantly increased by dnMstn (Figure 5, A and B). SPo of the EDL and Dp among these treatment groups was unchanged (Figure 5, C and D). These data demonstrate that the muscle hypertrophy elicited by Mstn inhibition is blocked by daily glucocorticoid use and indicate that patient glucocorticoid use will likely be a confounding variable in any Mstn inhibition–based clinical trial for DMD, as many participants will be on Pred or deflazacort (11, 48, 49).
Figure 5. Muscle functional benefits of myostatin inhibition are blocked by chronic prednisolone treatment.
Ex vivo muscle function evaluation of the extensor digitorum longus (EDL) and diaphragm (Dp) from mice of the study depicted in Figure 4A. (A–D) Max force (A), cross-sectional area (CSA) (B), specific tension (SPo) of the EDL (C), and SPo of the Dp (D) are reported. Data were analyzed using 2-way ANOVA (strain and treatment effects; effect size reported as η2), followed by Tukey’s post-hoc tests (α = 0.05). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM. Groups that are significantly different from each other are indicated by nonoverlapping letter designations (P ≤ 0.05).
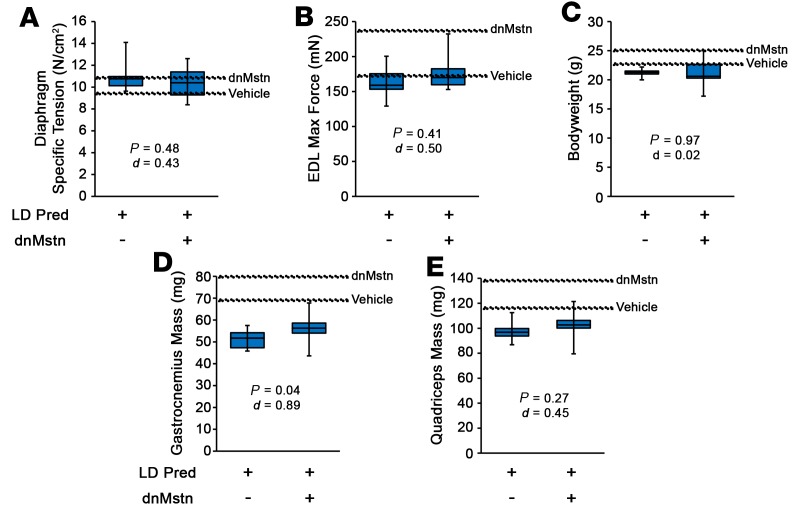
In light of the above data, we sought to determine if a lower dose of Pred may unmask a hypertrophic response to Mstn inhibition. At 1 mg/kg Pred (low-dose Pred [LD-Pred]; equates to 0.08 mg/kg in humans), a dose below that previously reported as being inefficacious in B10.mdx mice (50), male D2.mdx began oral treatments at 4 weeks of age and received a single i.p. injection of either PBS (control) or dnMstn AAV at 6 weeks of age. At 16 weeks of age, the treatment groups did not reveal differences in ex vivo muscle function (Figure 6, A and B). While a modest increase in gastrocnemius mass was found with dnMstn treatment, this value was lower than those of the vehicle-only group of the previous trial, and no other body weight or muscle mass improvements were identified (Figure 6, C–E). These data demonstrate that, even at low, subefficacious doses, daily glucocorticoid treatment blocks skeletal muscle hypertrophy associated with Mstn inhibition.
Figure 6. Low-dose prednisolone treatment does not unmask muscle hypertrophy when combined with myostatin inhibition.
Male D2.mdx mice received daily oral treatments of 1 mg/kg prednisolone (LD Pred) beginning at 4 weeks of age and received a single i.p. injection of either sham of AAV.dnMstn at 6 weeks of age (n = 5–7). (A–E) At terminal endpoint (16 weeks of age), ex vivo muscle function of diaphragm (A) and extensor digitorum longus (EDL) (B) was evaluated, and mouse body weights (C) and muscle masses (D and E) were recorded. Vehicle and dnMstn mean values from Figures 4 And 5 are shown for comparison. Data were analyzed using Welch’s t test (α = 0.05), with effect size reported as Cohen’s d (d). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM.
Distinct transcriptomic signatures are associated with long-term glucocorticoid treatment.
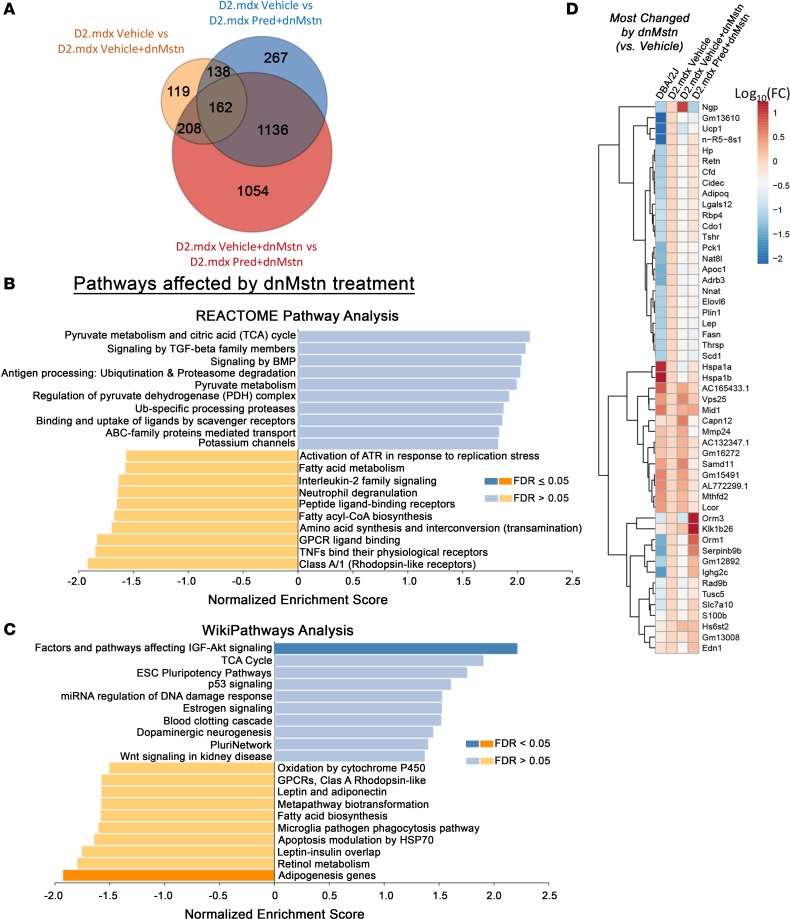
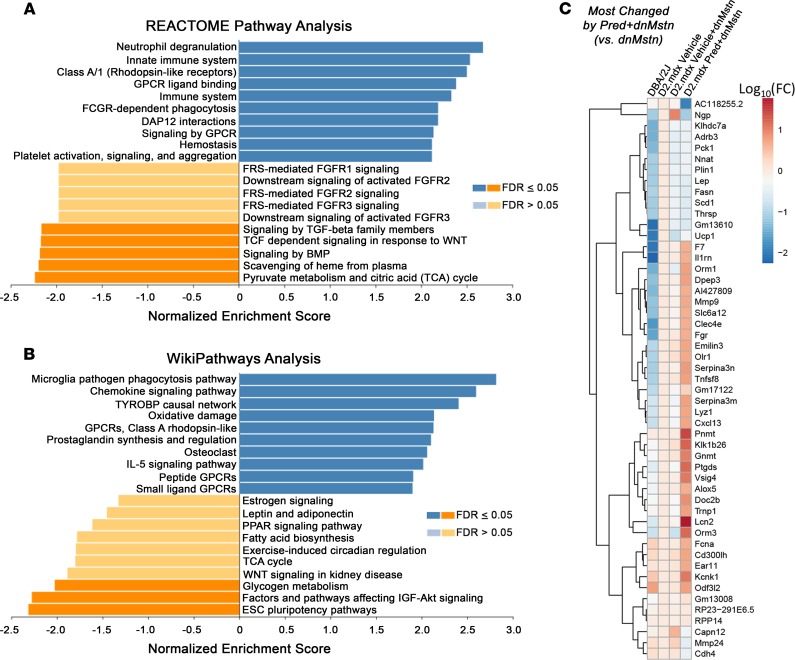
In order to identify the pathways that differ among dnMstn-treated skeletal muscle with and without long-term daily Pred treatment, RNA sequencing (RNA-seq) analysis was performed on quadriceps from these 2 groups, as well as vehicle-only treated D2.mdx and age-matched DBA/2J male mice (Figure 7). As shown in Figure 7A, a large number of transcripts are substantially and distinctly changed by vehicle + dnMstn treatment or by the addition of Pred to dnMstn treatment, compared with vehicle-treated D2.mdx muscle. Unbiased investigation of the most affected cellular pathways associated with dnMstn inhibition in dystrophic muscle was performed using both REACTOME Pathways Knowledgebase (51) and WikiPathways (52) databases (Figure 7, B and C). Interestingly, dnMstn treatment significantly affected genes associated with the activation of the IGF-Akt signaling axis, a potent anabolic pathway (53), whereas adipogenesis was significantly inhibited. Other notable (although not significant) pathways affected by dnMstn treatment include genes changes associated with activation of tricarboxylic acid cycle (TCA) and TGF-β superfamily signaling (Figure 7B). The top genes changed by dnMstn treatment (compared with vehicle-only expression patterns) are depicted in the clustered heatmap in Figure 7D, whereas expression patterns of highly expressed genes most changed by dnMstn can be found in Supplemental Figure 1A (Supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133276DS1).
Figure 7. Transcriptomic analysis of most-changed genes by myostatin inhibition in dystrophic skeletal muscle.
RNA-seq transcriptomic analysis was performed on quadriceps muscles from the study depicted in Figure 4A, as well as those from age-matched DBA/2J WT mice (n = 4). Analysis was performed on WT DBA-2J, vehicle-treated D2.mdx, vehicle-treated D2.mdx with dnMstn, and Pred-treated D2.mdx with dnMstn. (A) Venn diagram depicting number of genes differentially expressed by each D2.mdx treatment group analyzed. Most activated (blue) and inhibited (orange) pathways associated with myostatin inhibition (D2.mdx vehicle + dnMstn vs D2.mdx vehicle-only) identified using (B) REACTOME Pathway Knowledgebase and (C) WikiPathways databases. Pathways having false discover rate (FDR) P values ≤ 0.05 for either direction are indicated by darker coloration. (D) Clustered heatmap analysis of the 50 most-changed genes in D2.mdx vehicle+dnMstn vs. D2.mdx vehicle-only comparison. Gene expression levels were calculated using the DESeq method are displayed as log10 (normalized expression relative to D2.mdx vehicle-only values).
The combination of Pred to dnMstn in dystrophic muscle resulted in the significant activation of pathways associated with immune cell activity and inflammatory cytokine signaling, as demonstrated by both REACTOME (Figure 8A) and WikiPathways (Figure 8B) analyses. Additionally, the combination of Pred also decreases gene expression patterns associated with activation of IGF-Akt pathways, TGF-β superfamiliy signaling, and TCA cycle activity, all of which were pathways identified to be enhanced by dnMstn treatment (Figure 7, B and C). The top genes changed by combining Pred to dnMstn treatment (compared with vehicle + dnMstn expression patterns) are depicted in the clustered heatmap located in Figure 8C, whereas expression patterns of highly expressed genes most changed by combining Pred to dnMstn can be found in Supplemental Figure 1B. Additional clustered heatmaps showing significantly affected gene expression patterns associated with inflammation and TGF-β superfamily activation can be found in Supplemental Figure 1, C and D. These data demonstrate that daily Pred treatment substantially affects muscle transcriptomic profiles that likely account for the drastic phenotype differences in dystrophic mice undergoing chronic Pred treatment.
Figure 8. Transcriptomic analysis of most-changed genes by combining prednisolone with myostatin inhibition.
(A and B) Most activated (blue) and inhibited (orange) pathways associated with combining prednisolone (Pred) with myostatin inhibition (D2.mdx Pred + dnMstn vs. D2.mdx vehicle + dnMstn) identified using REACTOME Pathway Knowledgebase (A) and WikiPathways (B) databases. Pathways having FDR-adjusted P ≤ 0.05 for either direction are indicated by darker coloration. (C) Clustered heatmap analysis of the 50 most-changed genes in D2.mdx vehicle + dnMstn vs. D2.mdx vehicle-only comparison. Gene expression levels were calculated using the DESeq method are displayed as log10 (normalized expression relative to D2.mdx vehicle-only values).
Pred-induced muscle atrophy is not dependent on Mstn expression.
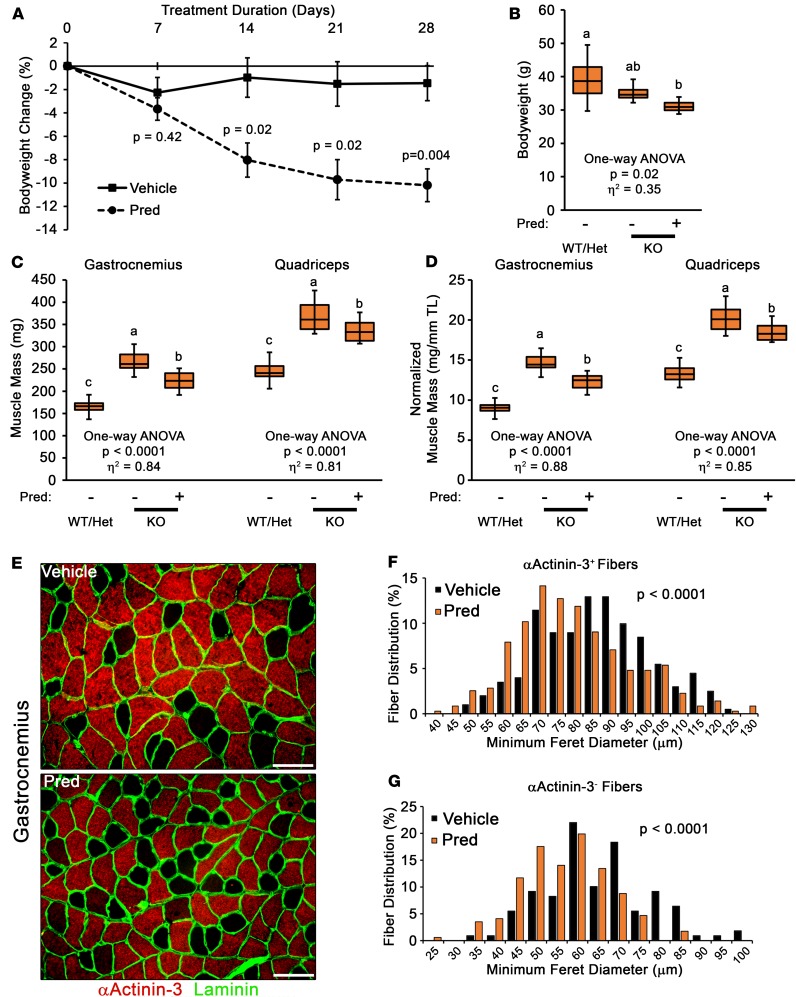
Previous reports have suggested that Mstn mediates dexamethasone-induced skeletal muscle atrophy in otherwise healthy muscle (42, 43). Intriguingly, our immunoblotting (Figure 4F) and transcriptomic data (Supplemental Figure 1D) suggest that Mstn gene expression is not increased with daily Pred treatment in D2.mdx mice; therefore, we directly tested the requirement of Mstn in Pred-induced muscle atrophy using the Mstn-KO mouse line. Adult male Mstn-KO mice received daily treatments with either vehicle or 5 mg/kg Pred for 28 days. Compared with vehicle, Pred-treated Mstn-KO mice progressively decreased in body weight during the 4-week treatment (Figure 9, A and B) and lost significant muscle mass by the end of the experiment (Figure 9, C and D). Analysis of muscle fiber types affected by Pred treatment revealed that gastrocnemius myofibers labeled by αActinin-3 (αActinin-3+ fibers), a protein primarily found in fast-glycolytic type IIX and IIB muscle fibers, as well as those not labeled by αActinin-3 (αActinin-3– fibers; consist of slow type I and fast-oxidative IIA fibers) both atrophy in response to Pred treatment (Figure 9, E–G). Overall, these data demonstrate that Pred can induce significant muscle atrophy in the absence of Mstn and further supports that glucocorticoid-induced muscle atrophy overpowers the hypertrophic program driven by Mstn loss of function.
Figure 9. Myostatin ablation does not prevent Pred-induced muscle atrophy.
Adult myostatin KO mice received daily oral treatments of vehicle or 5 mg/kg prednisolone (Pred) for 28 days (n = 4–5). (A) Mouse body weight change was evaluated during the course of the experiment. Data were analyzed using Welch’s t test (α = 0.05) and are displayed as mean ± SEM. (B–D) Body weight (B), absolute muscle masses (C), and tibia length–normalized muscle masses (D) were measured at terminal endpoint. WT and heterozygous (Het) littermate values are included for mass difference perspective. Data were analyzed using 1-way ANOVA (effect size reported as η2), followed by Tukey’s post hoc tests (α = 0.05). Data are presented as box-and-whisker plots, with minimum and maximum values indicated by error bars; data are shown as mean ± SEM. Groups that are significantly different from each other are indicated by nonoverlapping letter designations (P ≤ 0.05). (E–G) Immunofluorescent staining of vehicle- and Pred-treated KO gastrocnemius muscles for αActinin-3, a marker of fast-glycolytic muscle fibers, and laminin (E) allowed fiber size distribution analysis of αActinin-3+ (positive) fibers (F) and αActinin-3– (negative) fibers (G) of vehicle and Pred groups. Data were analyzed using Welch’s t test (α = 0.05) and are depicted as histograms of the entire data set. Scale bars: 100 μm.
Discussion
DMD is the most prevalent and severe of the muscular dystrophies, affecting ~1:5000 males (54) with a life expectancy into the mid-20s (55). The devastating and multifaceted pathological process of DMD suggests that the development of combinatorial therapies is needed to prolong the quality of life and life expectancy of patients. Therefore, it is important to understand how potential therapeutics interact with each other and established DMD treatment regimens, in addition to their efficacy as monotherapies. Identification of these potential interactive effects at preclinical stages can facilitate clinical trial design. Key findings of the current report include substantial muscle atrophy caused by long-term glucocorticoid treatment in mdx mice, inhibition of muscle protein synthesis by daily glucocorticoid administration, and prevention of Mstn inhibition–stimulated muscle hypertrophy by glucocorticoid use. We identify unique gene expression signatures elicited by chronic glucocorticoid treatment, and we demonstrate that Mstn is not required for Pred-induced muscle atrophy. The data of this study, therefore, suggest that targeting Mstn in DMD patients on chronic glucocorticoid treatment regimens is unlikely to show efficacy in terms of skeletal muscle hypertrophy and is a clear example of such a combinational therapy interaction that can substantially impact clinical development.
Off-label Pred use is frequent among DMD patients due to its ability to prolong patient ambulation (9), and recently, deflazacort became the first drug approved for all DMD patients, in part for further increased functional improvements in patients (10). In agreement with the potentially beneficial effect of glucocorticoids on dystrophic muscle function, we observe improved SPo in the Dp of D2.mdx mice treated with Pred for both 12 and 20 weeks, when initiated at 4 weeks of age. This, however, comes at the expense of atrophy and force decrements of the EDL, suggesting that the effects of Pred on muscle contractility are not uniform across all muscles.
Although Pred has long been known to induce skeletal muscle atrophy in patients (56, 57), the mechanism responsible for the observed wasting has been ambiguous. Previous reports have suggested that glucocorticoid-induced muscle atrophy is driven by the ubiquitin-proteasome system and/or Mstn activation (35, 42, 43). In agreement with the involvement of the ubiquitin-proteasome system, we find trends toward higher poly-ubiquitin levels and increased Trim63 expression following Pred treatment; however, we identify larger decrements in rates of protein synthesis, as determined by puromycin incorporation into elongating polypeptide chains, an indicator of active translation (58). This is in agreement with findings of reduced insulin-stimulated protein synthesis by Pred in healthy individuals, as measured by radio-labeled amino acid incorporation (37). Interestingly, classical investigations report decreased amino acid transport into cells as a result of glucocorticoid administration (59, 60), suggesting that reduced amino acid availability for protein translation is a possible mechanism contributing to reduced protein synthesis and skeletal muscle atrophy caused by chronic Pred treatment. Such a mechanism makes physiological sense, as endogenous glucocorticoid (cortisol) levels are increased during periods of organismal stress to promote the mobilization of glucose and amino acids from skeletal muscle to tissues vital for organism survival.
We additionally demonstrate that Mstn is not required for Pred-induced atrophy, as D2.mdx mice do not upregulate Mstn in response to chronic Pred treatment and Mstn-KO mice display robust muscle atrophy upon Pred treatment. In fact, the Pred-induced reduction of phospho-SMAD3 we observe in D2.mdx mice may even impair Mstn-induced signaling in a chronic Pred-treatment setting, particularly because expression of Smad7, a SMAD3 inhibitor molecule, is increased by Pred (Supplemental Figure 1D). Via this mechanism, it is possible that Pred induces a state of Mstn resistance, as binding to ActRIIB-ALK4/5 is uncoupled from SMAD2/3 activation. While previous reports have suggested that Mstn-null mice do not exhibit glucocorticoid-induced skeletal muscle atrophy (42), the conflicting findings of the current work could be due to differences in glucocorticoids used, duration of glucocorticoid administration, and/or background of mouse strain used for the study.
The mechanism of action for glucocorticoid-based therapeutics is traditionally considered to be the suppression of inflammatory gene expression (61). Paradoxically, we find that chronic Pred treatment in D2.mdx mice, in fact, increases gene signatures associated with heightened inflammation, including Spp1, a gene that encodes a negative modifier of DMD disease progression, osteopontin (62–64). Additional notable genes strongly activated by Pred include Lcn2, a marker of increased systemic inflammation (65); Alox5, an enzyme that catalyzes inflammatory lipid mediator formation (66); and Mmp9, a matrix metalloproteinase associated with more severe pathology within dystrophic muscle (67).
The major finding of the current work is the demonstration that Mstn inhibition does not elicit muscle mass gains when combined with chronic Pred treatment. This is clinically significant, as Mstn-targeting therapeutics are in clinical or preclinical development (refs. 24–27; NCT02515669, NCT01524224, NCT01958970, NCT02310763, and NCT02145234) for DMD and other muscle diseases. Our findings in D2.mdx mice suggest that trials for Mstn inhibition using skeletal muscle mass increase, as a primary outcome measure will ultimately fail if patients are on glucocorticoids. This phenomenon could underlie the negative data from the recently completed phase 2 clinical trial for the Mstn neutralizing antibody, domagrozumab, in DMD patients (NCT02310763) and additionally could have major implications for the ongoing phase 1/2 trial for another Mstn targeting antibody, RO7239361, in ambulatory DMD patients (NCT02515669).
As the identification of effective small molecule therapeutics to combat inflammation, fibrosis, and muscle wasting in DMD and BMD will continue to remain an unmet medical need, our findings highlight the importance of evaluating candidate therapeutic efficacy in combination with standard-of-care therapies. Additionally, these data further support the rationale to explore alternative standard-of-care regimens for dystrophinopathy patients other than chronic glucocorticoids, such as intermittent glucocorticoid dosing (12) and/or nonsteroidal NF-κB inhibitors (68, 69), both of which demonstrate efficacy and safety without promoting skeletal muscle atrophy.
Methods
Animals.
This study used male mdx mice of the C57BL/10 (B10.mdx; Jax 001801) and DBA/2J (D2.mdx; Jax 013141) backgrounds, as well as WT DBA/2J mice (Jax 000671), originally obtained from Jackson Laboratory. Mstn-KO mice used in this study were originally obtained from Se-Jin Lee (the Jackson Laboratory; ref. 13) and are congenic on the C57BL/6J background. Mice were housed 3–5 mice per cage; randomly assigned into treatment groups; provided ad libitum access to food, water, and enrichment; and maintained on a 12-hour light/dark system. Pred suspensions were prepared in cherry syrup vehicle for daily per oral (PO) treatment of 0 (vehicle control), 1, or 5 mg/kg doses. AAV-mediated Mstn inhibition was achieved by i.p. injection of 0 (PBS vehicle control) or 1 × 1012 gc of AAV8.LSP.dnMstn vector consisting of dnMstn driven by a liver-specific promoter, as previously described (17, 19, 70), at 6 weeks of age.
Ex vivo muscle function assessment.
Maximal tetanic tension assessments of the EDL and Dp muscles were evaluated as previously described (19) by the University of Florida Physiological Assessment Core. Briefly, the muscles of anesthetized mice were dissected and placed in physiological Ringer’s solution gas equilibrated with 95% O2 /5% CO2. After determining optimum length, muscles were subjected to 3 isometric contractions (stimulated at 120 Hz for 500 ms) to determine maximum Po. Following experimental procedures, muscles were weighed, either frozen embedded in OCT or snap frozen, and stored at –80°C until further use. Force measurements and dissections were performed by investigators blinded to treatment groups.
Protein balance investigation.
To assess protein ubiquitination resulting from daily glucocorticoid treatment, male DBA/2J mice were treated with vehicle (n = 6) or 5 mg/kg Pred (n = 5) for 10 days, with the final Pred dose being administered 2 hours prior to terminal endpoint. Twenty-four hours prior to terminal endpoint, vehicle- and Pred-treated mice received a single s.c. injection with 20 mg/kg of the proteasome inhibitor, MG-132, to allow accumulation of poly-ubiquitinated proteins (38). Assessment of protein synthesis changes caused by daily glucocorticoid treatment was performed using the surface sensing of translation (SUnSET) methodology (39, 58), with male DBA/2J mice treated with vehicle or 5 mg/kg Pred (n = 6) for 10 days and with the final Pred dose being administered 2 hours prior to terminal endpoint. At exactly 30 minutes prior to terminal endpoint, mice of both treatment groups received single i.p. injections of 20 mg/kg puromycin dissolved in 100 μL PBS to label actively elongating peptide chains. At terminal endpoint of each experiment, mice were euthanized by CO2 inhalation, and quadriceps muscles were quickly dissected, snap-frozen in liquid nitrogen, and stored at –80°C until biochemical analysis.
Immunoblotting.
Snap-frozen muscle samples were finely crushed and homogenized in T-PER buffer (Thermo Fisher Scientific) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific). Protein concentration of resulting supernatant was determined using Bio-Rad Protein Assay. Protein samples were boiled in 4× sample buffer, subjected to SDS-PAGE using 4%–12% SDS-polyacrylamide gels (Invitrogen), and transferred to nitrocellulose membranes using the iBlot system (Invitrogen). Membranes were blocked in 5% BSA-TBST and incubated with primary antibody overnight at 4°C. Following TBST washes, membranes were incubated in the appropriate HRP-conjugated secondary antibody for 1 hour at room temperature, washed, incubated for 5 minutes in ECL reagent (Thermo Fisher Scientific), and imaged using the LI-COR C-DiGit (LI-COR Biosciences) imaging system. Primary antibodies used in this study include: anti-polyubiquitin (K-48 linkage specific; Cell Signaling; 8081; 1:1000), anti-puromycin (clone 12D10; MilliporeSigma; MABE343; 1:4000), anti–Mstn N-terminus (1:1000; ref. 17), anti–phospho-SMAD3 (S425/427; Cell Signaling Technologies; 9520; 1:1000), anti-SMAD2/3 (Cell Signaling Technologies; 5678; 1:500), and anti-GAPDH (Santa Cruz Biotechnology Inc.; sc-25778). Membranes were stained with Ponceau red and imaged for loading normalization, which demonstrates linear staining relationships for the protein loading range used in this study (20–40 μg; ref. 71). Band signal intensities were measured using Image Studio Lite software (LI-COR Biosciences), normalized to sample loading, and reported relative to respective control samples.
Immunofluorescence.
Gastrocnemius muscles embedded in OCT were cryosectioned at 10 μm, fixed in ice-cold acetone, and stained for immunofluorescent analysis using anti–αActinin-3 (Thermo Fisher Scientific; PA5-36233; 1:250) and anti-laminin (Novus; NBP2-44751; 1:800) primary antibodies, followed by donkey anti–rabbit Alexa 568 (Invitrogen; A10042; 1:500) and donkey anti–rat Alexa 488 (Invitrogen; A21208; 1:500) secondary antibodies. Slides were visualized with a Leica DMR microscope, and images were acquired using a Leica DFC310FX camera interfaced with Leica LAS X software. Immunofluorescent images were processed and analyzed for muscle fiber size distribution by a blinded investigator using ImageJ (NIH) software.
Real-time PCR.
RNA was isolated from finely crushed snap-frozen mouse quadriceps samples using Trizol Reagent (Invitrogen), treated with DNAse (Promega), and reverse transcribed using the SuperScript III kit (Invitrogen). Resulting cDNA was subjected to real-time PCR using RQG SYBR Green supermix (Qiagen) in a Rotor Gene Q real-time PCR machine (Qiagen). The following mouse-specific primers were used: Trim63 (forward) 5′-AGG GCT CCC CAC CAC TGT GT-3′ and (reverse) 5′-TTG CCC CTC TCT AGG CCA CCG-3′; Gapdh (forward) 5′-AGC AGG CAT CTG AGG GCC CA-3′ and (reverse) 5′-TGT TGG GGG CCG AGT TGG GA-3′. Relative gene expression quantification was performed using the ΔΔCt method with Gapdh as the normalization gene.
Transcriptomic analysis.
The RNA-seq cDNA library was generated from 100–400 ng total RNA using TruSeq RNA Sample Preparation Kit (Illumina) according to the manufacturer’s protocol. Briefly poly-A tailed RNA molecules were pulled down with poly-T oligonucleotide attached magnetic beads. Following purification, mRNA was fragmented with divalent cations at 85°C, and cDNA was generated by random primers using SuperScript II enzyme (Invitrogen). Second-strand synthesis was performed followed by end repair, single A-base addition and ligation of barcode indexed adaptors to the DNA fragments. Adapter-specific PCRs were performed to generate sequencing libraries. Libraries were size selected with E-Gel EX 2% agarose gels (Invitrogen) and purified by QIAquick Gel Extraction Kit (Qiagen). Libraries were sequenced on an HiSeq 2500 instrument. Four biological replicates were sequenced for each population.
TopHat2 (72) was used to align the reads to the mm10 mouse assembly. Further downstream analysis of the aligned reads was performed using StrandNGS software (Version 2.8, Build 230243; Strand Life Sciences). Normalization of the raw read counts was performed using the DeSeq method (73). One-way ANOVA and Tukey’s post hoc test was performed for the normalized counts. Changes in genes with fold change > 1.5 and P < 0.05 were considered to be statistically significant, and the genes were differentially expressed. Heatmaps were drawn using the R package pheatmap. The accession numbers for the RNA-seq reported in this paper are SRP215469 and have been assigned to Bioproject: PRJNA555085 (http://dataview.ncbi.nlm.nih.gov/object/PRJNA555085?reviewer=4msm4tro4p7he2rluvapmnh0m9).
Lists of differentially expressed genes (P < 0.05 and fold change [FC] > 2) were analyzed using the REACTOME Pathway Knowledgebase (51) and WikiPathways (52) databases to identify cellular pathways most affected by each treatment.
Statistics.
Statistical analysis was performed using unpaired, 2-tailed Welch’s t test (effect size reported as Cohen’s d [d]) and 1- or 2-way ANOVA (effect size reported as eta-squared [η2]), followed by Tukey’s HSD post hoc tests, where appropriate. A P value less than 0.05 was considered significant. Values are displayed as box-and-whisker plots, mean ± SEM, or as histograms.
Study approval.
All procedures and experiments were approved by and conducted in accordance of the University of Florida IACUC.
Author contributions
Research and study design were contributed by DWH, CCH, LN, and HLS. Experimental procedures and data acquisition were conducted by DWH, CCH, AP, MKM, and LAW. All authors were involved in data analysis, interpretation, and manuscript writing. Order of co–first authors was determined alphabetically.
Supplementary Material
Acknowledgments
This work was funded by a Wellstone Muscular Dystrophy Cooperative Center grant (U54-AR-052646) from the NIH to HLS, a Parent Project Muscular Dystrophy grant to HLS, Leducq Foundation funding (13CVD04) to HLS, and NIH funding (R01DK115924) to LN. DWH was supported by a career development grant from the Muscular Dystrophy Association (MDA549004). The authors thank Jason Puglise and Zachary Wakefield for technical support during this study.
Version 1. 12/12/2019
In-Press Preview
Version 2. 01/16/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(1):e133276.https://doi.org/10.1172/jci.insight.133276.
Contributor Information
David W. Hammers, Email: dhammers@ufl.edu.
Cora C. Hart, Email: coracricket@ufl.edu.
Andreas Patsalos, Email: apatsalos@jhmi.edu.
Michael K. Matheny, Email: mathemk@ufl.edu.
Lillian A. Wright, Email: l.wright@ufl.edu.
Laszlo Nagy, Email: lnagy@jhmi.edu.
H. Lee Sweeney, Email: lsweeney@ufl.edu.
References
- 1.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90(8):3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch EM, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447(7140):87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 4.Mendell JR, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74(5):637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 5.Mendell JR, et al. Gene therapy for muscular dystrophy: lessons learned and path forward. Neurosci Lett. 2012;527(2):90–99. doi: 10.1016/j.neulet.2012.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amoasii L, et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med. 2017;9(418):eaan8081. doi: 10.1126/scitranslmed.aan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäckman E, Henriksson KG. Low-dose prednisolone treatment in Duchenne and Becker muscular dystrophy. Neuromuscul Disord. 1995;5(3):233–241. doi: 10.1016/0960-8966(94)00048-E. [DOI] [PubMed] [Google Scholar]
- 9.Mendell JR, et al. Randomized, double-blind six-month trial of prednisone in Duchenne’s muscular dystrophy. N Engl J Med. 1989;320(24):1592–1597. doi: 10.1056/NEJM198906153202405. [DOI] [PubMed] [Google Scholar]
- 10.Griggs RC, et al. Efficacy and safety of deflazacort vs prednisone and placebo for Duchenne muscular dystrophy. Neurology. 2016;87(20):2123–2131. doi: 10.1212/WNL.0000000000003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barp A, et al. Genetic Modifiers of Duchenne Muscular Dystrophy and Dilated Cardiomyopathy. PLoS ONE. 2015;10(10):e0141240. doi: 10.1371/journal.pone.0141240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quattrocelli M, et al. Intermittent glucocorticoid steroid dosing enhances muscle repair without eliciting muscle atrophy. J Clin Invest. 2017;127(6):2418–2432. doi: 10.1172/JCI91445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 14.Zimmers TA, et al. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296(5572):1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]
- 15.Wolfman NM, et al. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci USA. 2003;100(26):15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 17.Liu M, Hammers DW, Barton ER, Sweeney HL. Activin Receptor Type IIB Inhibition Improves Muscle Phenotype and Function in a Mouse Model of Spinal Muscular Atrophy. PLoS ONE. 2016;11(11):e0166803. doi: 10.1371/journal.pone.0166803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bish LT, et al. Long-term systemic myostatin inhibition via liver-targeted gene transfer in golden retriever muscular dystrophy. Hum Gene Ther. 2011;22(12):1499–1509. doi: 10.1089/hum.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morine KJ, Bish LT, Pendrak K, Sleeper MM, Barton ER, Sweeney HL. Systemic myostatin inhibition via liver-targeted gene transfer in normal and dystrophic mice. PLoS ONE. 2010;5(2):e9176. doi: 10.1371/journal.pone.0009176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morine KJ, et al. Activin IIB receptor blockade attenuates dystrophic pathology in a mouse model of Duchenne muscular dystrophy. Muscle Nerve. 2010;42(5):722–730. doi: 10.1002/mus.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornegay JN, et al. Dystrophin-deficient dogs with reduced myostatin have unequal muscle growth and greater joint contractures. Skelet Muscle. 2016;6:14. doi: 10.1186/s13395-016-0085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner KR, McPherron AC, Winik N, Lee SJ. Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Ann Neurol. 2002;52(6):832–836. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- 23.Bogdanovich S, et al. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420(6914):418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- 24.St Andre M, et al. A mouse anti-myostatin antibody increases muscle mass and improves muscle strength and contractility in the mdx mouse model of Duchenne muscular dystrophy and its humanized equivalent, domagrozumab (PF-06252616), increases muscle volume in cynomolgus monkeys. Skelet Muscle. 2017;7(1):25. doi: 10.1186/s13395-017-0141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell C, et al. Myostatin inhibitor ACE-031 treatment of ambulatory boys with Duchenne muscular dystrophy: Results of a randomized, placebo-controlled clinical trial. Muscle Nerve. 2017;55(4):458–464. doi: 10.1002/mus.25268. [DOI] [PubMed] [Google Scholar]
- 26.Pirruccello-Straub M, et al. Blocking extracellular activation of myostatin as a strategy for treating muscle wasting. Sci Rep. 2018;8(1):2292. doi: 10.1038/s41598-018-20524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latres E, et al. Myostatin blockade with a fully human monoclonal antibody induces muscle hypertrophy and reverses muscle atrophy in young and aged mice. Skelet Muscle. 2015;5:34. doi: 10.1186/s13395-015-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker C, et al. Myostatin antibody (LY2495655) in older weak fallers: a proof-of-concept, randomised, phase 2 trial. Lancet Diabetes Endocrinol. 2015;3(12):948–957. doi: 10.1016/S2213-8587(15)00298-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y, et al. LC-MS/MS multiplexed assay for the quantitation of a therapeutic protein BMS-986089 and the target protein Myostatin. Bioanalysis. 2016;8(3):193–204. doi: 10.4155/bio.15.238. [DOI] [PubMed] [Google Scholar]
- 30.Coley WD, et al. Effect of genetic background on the dystrophic phenotype in mdx mice. Hum Mol Genet. 2016;25(1):130–145. doi: 10.1093/hmg/ddv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukada S, et al. Genetic background affects properties of satellite cells and mdx phenotypes. Am J Pathol. 2010;176(5):2414–2424. doi: 10.2353/ajpath.2010.090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heydemann A, et al. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J Clin Invest. 2009;119(12):3703–3712. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison-Nozik A, et al. Glucocorticoids enhance muscle endurance and ameliorate Duchenne muscular dystrophy through a defined metabolic program. Proc Natl Acad Sci USA. 2015;112(49):E6780–E6789. doi: 10.1073/pnas.1512968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baehr LM, Furlow JD, Bodine SC. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J Physiol (Lond) 2011;589(Pt 19):4759–4776. doi: 10.1113/jphysiol.2011.212845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey JL, Wang X, Price SR. The balance between glucocorticoids and insulin regulates muscle proteolysis via the ubiquitin-proteasome pathway. Miner Electrolyte Metab. 1999;25(4-6):220–223. doi: 10.1159/000057451. [DOI] [PubMed] [Google Scholar]
- 37.Short KR, Bigelow ML, Nair KS. Short-term prednisone use antagonizes insulin’s anabolic effect on muscle protein and glucose metabolism in young healthy people. Am J Physiol Endocrinol Metab. 2009;297(6):E1260–E1268. doi: 10.1152/ajpendo.00345.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tawa NE, Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest. 1997;100(1):197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman CA, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25(3):1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 41.Cohen S, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185(6):1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilson H, et al. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology. 2007;148(1):452–460. doi: 10.1210/en.2006-0539. [DOI] [PubMed] [Google Scholar]
- 43.Ma K, et al. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metab. 2003;285(2):E363–E371. doi: 10.1152/ajpendo.00487.2002. [DOI] [PubMed] [Google Scholar]
- 44.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol, Cell Physiol. 2009;296(6):C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 45.Chen JL, et al. Elevated expression of activins promotes muscle wasting and cachexia. FASEB J. 2014;28(4):1711–1723. doi: 10.1096/fj.13-245894. [DOI] [PubMed] [Google Scholar]
- 46.McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22(3):260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- 47.Carlson CJ, Booth FW, Gordon SE. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am J Physiol. 1999;277(2 Pt 2):R601–R606. doi: 10.1152/ajpregu.1999.277.2.r601. [DOI] [PubMed] [Google Scholar]
- 48.McDonald CM, et al. Longitudinal pulmonary function testing outcome measures in Duchenne muscular dystrophy: Long-term natural history with and without glucocorticoids. Neuromuscul Disord. 2018;28(11):897–909. doi: 10.1016/j.nmd.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Willcocks RJ, et al. Multicenter prospective longitudinal study of magnetic resonance biomarkers in a large duchenne muscular dystrophy cohort. Ann Neurol. 2016;79(4):535–547. doi: 10.1002/ana.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baltgalvis KA, Call JA, Nikas JB, Lowe DA. Effects of prednisolone on skeletal muscle contractility in mdx mice. Muscle Nerve. 2009;40(3):443–454. doi: 10.1002/mus.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. doi: 10.1093/nar/gkz1031. Jassal B, et al. The reactome pathway knowledgebase [published online ahead of print November 6, 2019]. Nucleic Acids Res. [DOI] [Google Scholar]
- 52.Slenter DN, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46(D1):D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49(1):59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendell JR, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71(3):304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 55.Passamano L, et al. Improvement of survival in Duchenne Muscular Dystrophy: retrospective analysis of 835 patients. Acta Myol. 2012;31(2):121–125. [PMC free article] [PubMed] [Google Scholar]
- 56.Mastaglia FL. Adverse effects of drugs on muscle. Drugs. 1982;24(4):304–321. doi: 10.2165/00003495-198224040-00003. [DOI] [PubMed] [Google Scholar]
- 57.Horber FF, Scheidegger JR, Grünig BE, Frey FJ. Thigh muscle mass and function in patients treated with glucocorticoids. Eur J Clin Invest. 1985;15(6):302–307. doi: 10.1111/j.1365-2362.1985.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 59.Gelehrter TD, McDonald RA. Steroid specificity of the glucocorticoid inhibition of amino acid transport in rat hepatoma cells. Endocrinology. 1981;109(2):476–482. doi: 10.1210/endo-109-2-476. [DOI] [PubMed] [Google Scholar]
- 60.Le Cam A, Freychet P. Effect of glucocorticoids on amino acid transport in isolated rat hepatocytes. Mol Cell Endocrinol. 1977;9(2):205–214. doi: 10.1016/0303-7207(77)90121-6. [DOI] [PubMed] [Google Scholar]
- 61.Desmet SJ, De Bosscher K. Glucocorticoid receptors: finding the middle ground. J Clin Invest. 2017;127(4):1136–1145. doi: 10.1172/JCI88886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Capote J, et al. Osteopontin ablation ameliorates muscular dystrophy by shifting macrophages to a pro-regenerative phenotype. J Cell Biol. 2016;213(2):275–288. doi: 10.1083/jcb.201510086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vetrone SA, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J Clin Invest. 2009;119(6):1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pegoraro E, et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76(3):219–226. doi: 10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moschen AR, Adolph TE, Gerner RR, Wieser V, Tilg H. Lipocalin-2: A Master Mediator of Intestinal and Metabolic Inflammation. Trends Endocrinol Metab. 2017;28(5):388–397. doi: 10.1016/j.tem.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 66.Chen XS, Sheller JR, Johnson EN, Funk CD. Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372(6502):179–182. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Mittal A, Makonchuk DY, Bhatnagar S, Kumar A. Matrix metalloproteinase-9 inhibition ameliorates pathogenesis and improves skeletal muscle regeneration in muscular dystrophy. Hum Mol Genet. 2009;18(14):2584–2598. doi: 10.1093/hmg/ddp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammers DW, et al. Disease-modifying effects of orally bioavailable NF-κB inhibitors in dystrophin-deficient muscle. JCI Insight. 2016;1(21):e90341. doi: 10.1172/jci.insight.90341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finanger E, et al. Phase 1 Study of Edasalonexent (CAT-1004), an Oral NF-κB Inhibitor, in Pediatric Patients with Duchenne Muscular Dystrophy. J Neuromuscul Dis. 2019;6(1):43–54. doi: 10.3233/JND-180341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hammers DW, Merscham-Banda M, Hsiao JY, Engst S, Hartman JJ, Sweeney HL. Supraphysiological levels of GDF11 induce striated muscle atrophy. EMBO Mol Med. 2017;9(4):531–544. doi: 10.15252/emmm.201607231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilda JE, Gomes AV. Stain-Free total protein staining is a superior loading control to β-actin for Western blots. Anal Biochem. 2013;440(2):186–188. doi: 10.1016/j.ab.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.