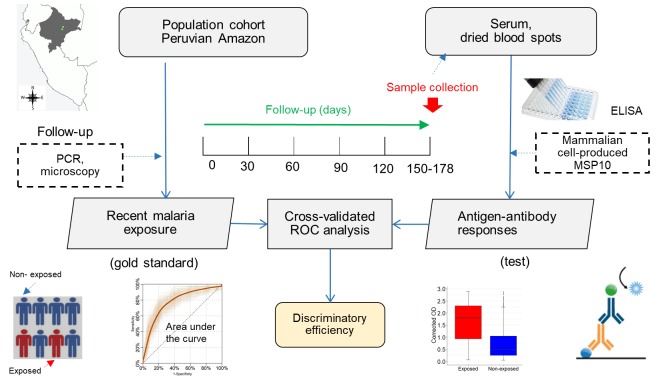
Abstract
BACKGROUND
Serological tools for the accurate detection of recent malaria exposure are needed to guide and monitor malaria control efforts. IgG responses against Plasmodium vivax and P. falciparum merozoite surface protein-10 (MSP10) were measured as a potential way to identify recent malaria exposure in the Peruvian Amazon.
METHODS
A field-based study included 470 participants in a longitudinal cohort who completed a comprehensive evaluation: light microscopy and PCR on enrollment, at least 1 monthly follow-up by light microscopy, a second PCR, and serum and dried blood spots for serological analysis at the end of the follow-up. IgG titers against novel mammalian cell–produced recombinant PvMSP10 and PfMSP10 were determined by ELISA.
RESULTS
During the follow-up period, 205 participants were infected, including 171 with P. vivax, 26 with P. falciparum, 6 with infections by both species but at different times, and 2 with mixed infections. Exposure to P. vivax was more accurately identified when serological responses to PvMSP10 were obtained from serum (sensitivity, 58.1%; specificity, 81.8%; AUC: 0.76) than from dried blood spots (sensitivity, 35.2; specificity, 83.5%; AUC: 0.64) (PAUC < 0.001). Sensitivity was highest (serum, 82.9%; dried blood spot, 45.7%) with confirmed P. vivax infections occurring 7–30 days before sample collection; sensitivity decreased significantly in relation to time since last documented infection. PvMSP10 serological data did not show evidence of interspecies cross-reactivity. Anti-PfMSP10 responses poorly discriminated between P. falciparum–exposed and nonexposed individuals (AUC = 0.59; P > 0.05).
CONCLUSION
Anti-PvMSP10 IgG indicates recent exposure to P. vivax at the population level in the Amazon region. Serum, not dried blood spots, should be used for such serological tests.
FUNDING
Cooperative agreement U19AI089681 from the United States Public Health Service, NIH/National Institute of Allergy and Infectious Diseases, as the Amazonian International Center of Excellence in Malaria Research.
Keywords: Immunology, Infectious disease
Keywords: Diagnostics, Epidemiology, Malaria
In a prospective, longitudinal cohort study, anti-Merozoite Surface Protein 10 antibodies indicated recent exposure to Plasmodium vivax in the Peruvian Amazon.
Introduction
Malaria remains a serious public health threat in the Americas with more than 120 million people living at risk for infection in 21 countries, within the context of hypo- and co-endemic Plasmodium vivax and Plasmodium falciparum transmission (1). In 2015, Peru reported 66,609 cases of malaria, accounting for about 19% of total reported malaria cases in the Americas (1). The vast majority (95%) of cases occur in northeastern Peru, the Loreto Department of the Amazon region. Here, malaria due to P. vivax is more common than P. falciparum malaria (Pv/Pf ratio of 4/1 in 2015), and transmission is highly heterogeneous (2). More than 80% of malaria cases reported by Loreto Ministry of Health–operated health posts (passive case detection; PCD) are concentrated among 10%–20% of communities (3). Cross-sectional studies using light microscopy (LM) for malaria diagnosis show that malaria parasite prevalence varies considerably among communities around Iquitos (the capital of Loreto) (4, 5). The detection of submicroscopic infections using PCR further indicates a higher level of heterogeneous transmission than would otherwise be recognized using LM alone (5, 6). In regions where malaria elimination continues to be an important public health goal, the logistical burden of using microscopic or molecular techniques to identify ongoing malaria transmission in heterogenous and focal transmission settings might be overcome by using simple serological techniques, such as antibody detection.
Focal malaria transmission has increasingly been reported in several areas with low or declining transmission and continues to pose a major challenge to National Malaria Control Programs (NMCPs) (7, 8) because reemergence is always a risk. Historically, NMCPs have used the number of reported malaria cases (e.g., microscopically confirmed symptomatic infections in Peru) to stratify malaria risk and identify hotspots, prioritize intervention areas, and monitor the impact of control interventions (9). As malaria transmission decreases, measuring the incidence of clinical malaria becomes increasingly difficult and insensitive (10), mainly because of the substantial number of asymptomatic and submicroscopic infections not identified by routine PCD (11). Accurately measuring malaria transmission is not straightforward, and different factors (i.e., sensitivity/specificity of diagnostic tests, costs, feasibility, acceptability, seasonal variations in transmission) must be taken into account before choosing adequate metrics (12, 13).
Serological tools are increasingly proposed as useful alternatives for estimating malaria transmission and monitoring its changes over time for both P. falciparum and P. vivax (3, 14–18). Previous studies have shown that seroconversion rate (SCR) (i.e., the rate at which seronegative individuals became seropositive) estimated from age-stratified seroprevalence data provide key information on malaria transmission patterns, especially when parasite rates are low (14, 17, 19). Seroprevalence rates provide a reliable tool for assessing malaria transmission in low endemic areas with seasonal patterns because antibodies, reflecting exposure to parasite antigens, remain in the blood longer than malaria parasites and are thus easier to detect and less subject to seasonal variations (20). However, a number of knowledge and technical gaps need to be addressed to develop or optimize serological tests according to the needs of the NMCPs’ surveillance systems (21). Few proteins have been investigated as potential antigens for serosurveillance, and most of them are primarily vaccine candidates with conserved orthologous genes between P. vivax and P. falciparum species likely to generate cross-reactivity in co-endemic areas (e.g., merozoite surface protein-1, 19-kDa C-terminal region, MSP119; apical membrane antigen-1; and circumsporozoite protein) (3, 21, 22).
Two recent studies using an Escherichia coli–based cell-free expression system to generate genome-level protein microarrays of P. vivax and P. falciparum proteins identified P. vivax merozoite surface protein-10 (PvMSP10) and P. falciparum (PfMSP10) as top hits, suggesting their potential as biomarkers of malaria infection (23, 24). The present study was designed to test the hypothesis that IgG responses to properly folded, conformationally correct, mammalian cell–produced recombinant Plasmodium MSP10 proteins would have improved performance characteristics (sensitivity, specificity, and interspecies cross-reactivity) for determining recent malaria exposure in individuals residing in malaria-endemic areas of the Peruvian Amazon. The study was rigorous in that follow-up of longitudinal, population-based cohorts was carried out, which enabled comprehensive identification and registration of most malaria infections (both asymptomatic and symptomatic) over the study period.
Results
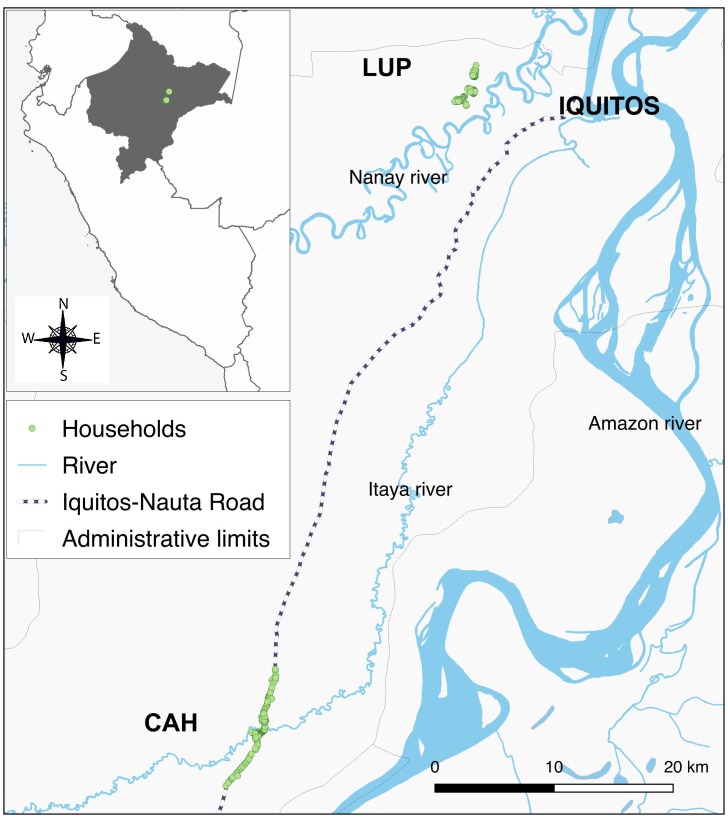
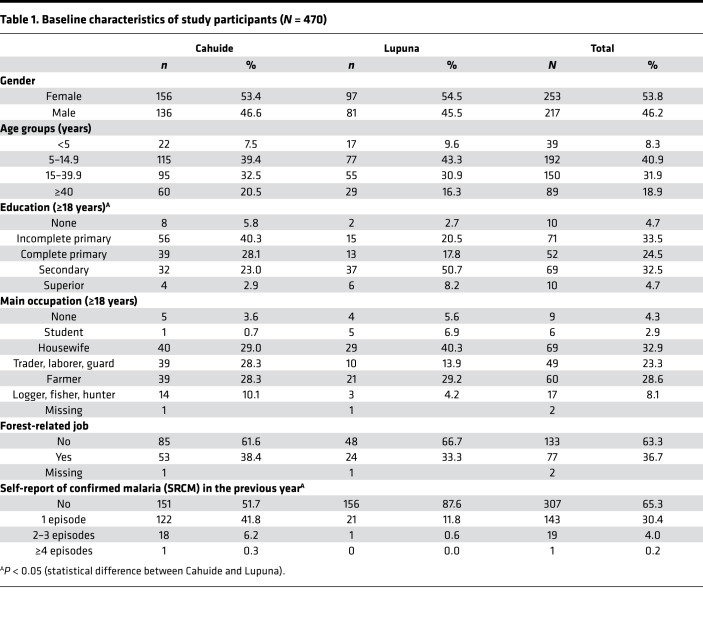
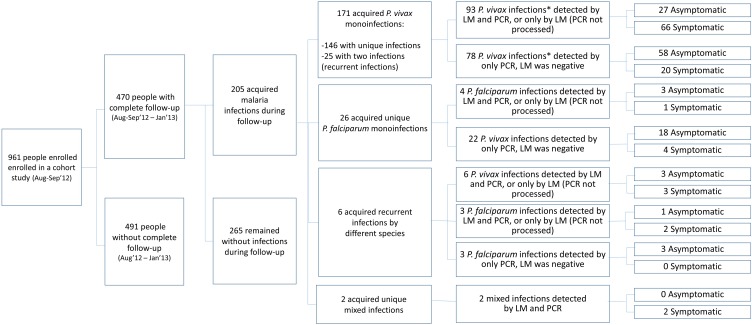
Of the 961 enrolled individuals in August through September 2012 in the study area (Figure 1), 470 (292 in Cahuide and 178 in Lupuna) completed follow-up through January 2013 and met the study selection criteria (baseline characteristics, Table 1). With a mean period of 127 ± 23 days (range: 90–178 days), the follow-up allowed for the identification of 146 (31.1%) individuals with single monoinfections by P. vivax, 25 (5.3%) with recurrent monoinfections by P. vivax (i.e., a first P. vivax infection detected by LM, and 1 month or more later, a second P. vivax infection detected by either LM or PCR), and 26 (5.5%) individuals with single monoinfections by P. falciparum. In addition, 6 (1.3%) individuals acquired recurrent monoinfections by different species (P. falciparum or P. vivax), and 2 (0.4%) had mixed infections by both species. Median times between infection and end of follow-up were 45 (range: 0–169 days) and 101 days (range: 21–165 days) for P. vivax and P. falciparum, respectively (Figure 2). At the time of sample collection for serological testing, cohort data showed that 26 individuals had P. vivax parasite confirmation in the previous 6 days, 35 between 7 and 30 days before, 67 between 31 and 60 days before, and 51 individuals between 61 and 169 days before, respectively. Regarding P. falciparum infection, 2 individuals had parasite confirmation in the past 6 days, 6 between 7 and 30 days before, 8 between 31 and 60 days before, and 18 individuals between 61 and 169 days before, respectively.
Figure 1. Study area.
CAH, Cahuide; LUP, Lupuna.
Table 1. Baseline characteristics of study participants (N = 470).
Figure 2. Flow diagram of study participants and characteristics of malaria infections detected during their follow-up.
*Only last infections were considered in individuals with recurrent infections during the follow-up.
Two types of samples, 470 serum and 466 dried blood spot (DBS) samples, were available for ELISA analysis. Antibody responses to PvMSP10 from serum samples (median corrected OD = 0.19) were significantly higher than those from blood spots (median corrected OD = 0.11) (P < 0.001), with both responses showing a significant but weak positive correlation (Spearman’s r = 0.31, and P < 0.001). Seropositivity to PvMSP10 was found in 33.4% (157/470) and 23.6% (110/466) of serum and DBS samples, respectively. Fair agreement in the P. vivax serological status (74.7%, 348/466) was observed between serological tests performed on serum and DBS samples (P < 0.001). Regarding antibody responses to PfMSP10 on serum samples, the median corrected OD was 0.06, and seropositivity was found in 36.8% (173/470) of individuals.
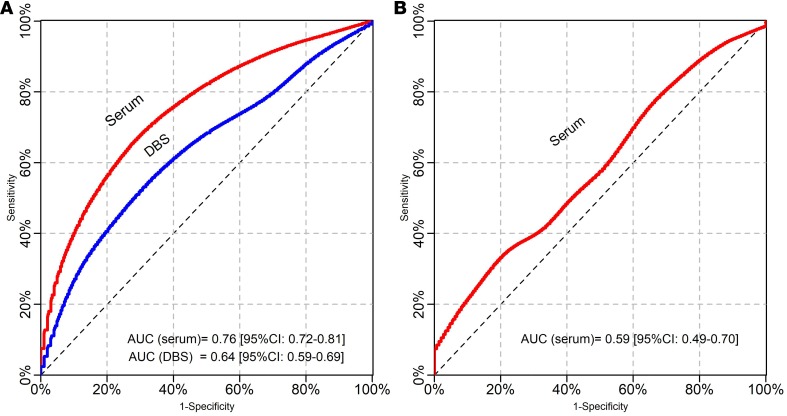
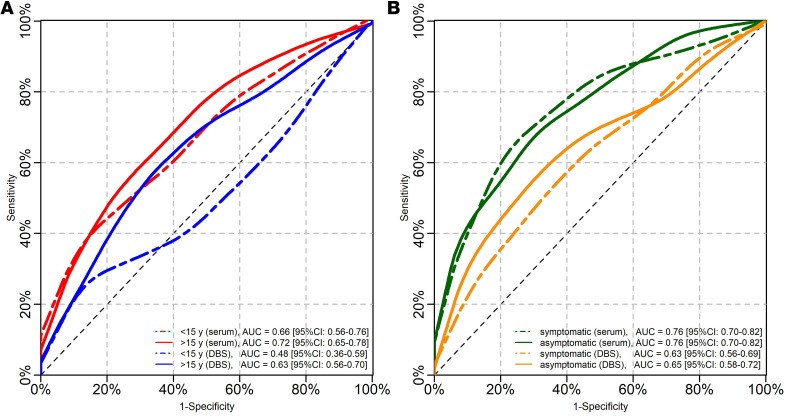
The cross-validated receiver operating characteristic (ROC) analysis (Figure 3) confirmed that the discriminatory ability for P. vivax malaria exposure was significantly more efficient when serological tests were performed with serum samples (AUC = 0.76, 95% CI: 0.72–0.81) than with DBS samples (AUC = 0.64, 95% CI: 0.59–0.69) (P < 0.001). Although this discriminatory ability was higher in individuals aged 15 years or more than in younger ones with both types of samples (Figure 4A), significant differences in the AUC between both age groups were found only with DBS samples (AUC in ≥15 years: 0.63, 95% [95% CI: 0.56–0.70]; AUC in <15 years: 0.48, 95% [95% CI: 0.36–0.59]; P = 0.02).
Figure 3. ROC curves and AUCs of serological responses for detecting malaria exposure.
(A) using PvMSP10 antigen, (B) using PfMSP10 antigen.
Figure 4. ROC curves and AUCs of serological responses for detecting P.
vivaxmalaria exposure. (A) Discrimination assessment in individuals aged less than 15 years and in those aged at least 15 years. (B) Discrimination between individuals with asymptomatic infections and those without infections and between individuals with symptomatic infections and those without infections.
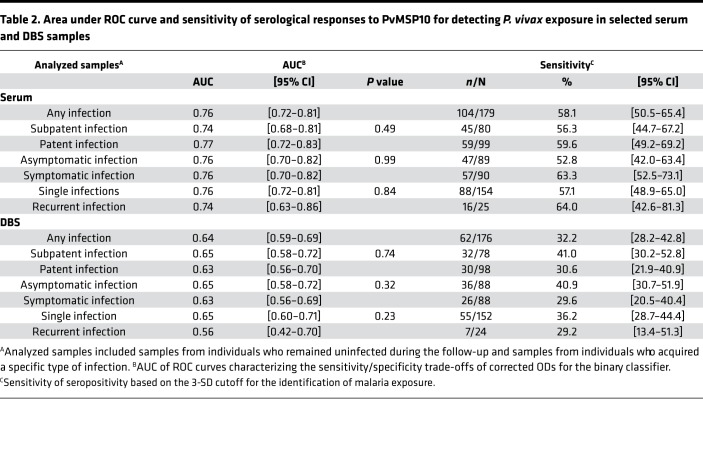
Seropositivity to PvMSP10 on serum samples (defined by the cutoff) identified individuals who acquired P. vivax infections at any time during the follow-up with sensitivity (SN) of 58.1% (95% CI: 50.5–65.4), specificity (SP) of 81.8% (95% CI: 76.9–86.1), positive predictive value (PPV) of 66.2% (95% CI: 59.9–72.1), and negative predictive value (NPV) of 76.0% (95% CI: 72.6–79.2). Comparatively, seropositivity to PvMSP10 on DBS showed an overall lower performance for the identification of P. vivax exposure: SN 35.2% (28.2–42.8), SP 83.5% (78.7–87.5), PPV 56.4% (48.2–64.2), and NPV 68.0% (65.3–70.5). AUCs and SN of serological tools for P. vivax exposure on both serum and DBS did not significantly differ between sample subgroups that included only patent or subpatent infections (P > 0.05), only symptomatic or asymptomatic infections (P > 0.05) (Figure 4B), and only single or recurrent infections (P > 0.05) (Table 2).
Table 2. Area under ROC curve and sensitivity of serological responses to PvMSP10 for detecting P. vivax exposure in selected serum and DBS samples.
In contrast to PvMSP10, seropositivity to PfMSP10 on serum samples showed a suboptimal performance to identify individuals who acquired P. falciparum infections during the follow-up: SN 47.1% (30.2–64.6), SP 64.0% (59.3–68.5), PPV 9.9% (6.5–13.0), and NPV 92.2% (91.8–95.5). This poor performance for discriminating between P. falciparum–exposed and nonexposed individuals was also supported by ROC curves (AUC = 0.59, 95% CI: 0.49–0.70) (Figure 3) and led to the decision not to process the DBS samples.
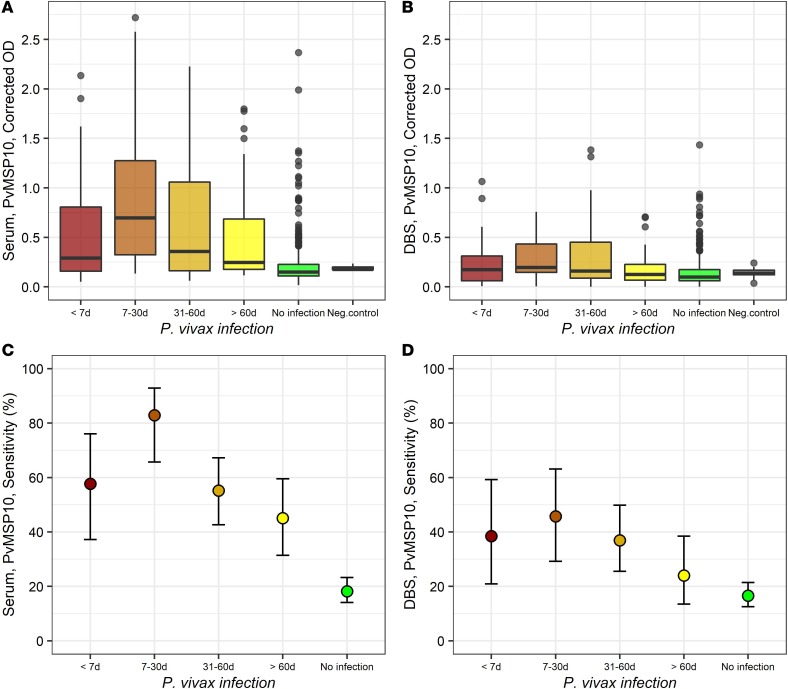
Median corrected ODs on serum samples were the highest in individuals with infections between 7 and 30 days (median: 0.70, and IQR: 0.951) and then decreased with the time since last infection 31–60 days (median: 0.36, and IQR: 0.90) and more than 60 days (median: 0.25, and IQR: 0.51) (P < 0.001, Kruskal-Wallis test) (Figure 5A). Pairwise comparison found that all individuals with detected infections (regardless of their time) had higher ODs than individuals without infections (P < 0.001, post hoc Dunn’s test) and that the ODs of this latter group (0.15, 0.12) did not differ from ODs of negative controls (0.18, 0.28) (P > 0.05). Corrected ODs on DBS seemed to follow a similar trend as those on serum samples (Figure 5B); however, ODs only in individuals with infections between 7 and 30 days (median: 0.20, and IQR: 0.28) and between 31 and 60 days (median: 0.16, and IQR: 0.37) were significantly higher than those in individuals without infections (median: 0.10, and IQR: 0.11) (P < 0.001).
Figure 5. Serological responses against PvMSP10 with the time since last P.
vivaxinfection. (A and B) Box plots (25th–75th percentiles) of corrected OD values on, respectively, serum and DBS samples. Horizontal bars inside the boxes represent the median of OD values in each group. (C and D) Dot plots of the sensitivity of dichotomized serological results in detecting malaria exposure using respectively serum and DBS samples, showing mean and 95% confidence limits in each group estimated by the Wilson score method.
The SN of dichotomized serological results for detecting P. vivax exposure was 57.7% (95% CI: 37.2–76.0) on serum samples from individuals with P. vivax infections of less than 7 days and increased to 82.9% (95% CI: 65.7–92.8) with infections between 7 and 30 days, but then SN decreased significantly with the time since last infection (trend P = 0.01) 31–60 days (55.2%, 95% CI: 42.6–67.2) and more than 60 days (45.1%, 95% CI: 31.4–59.5) (Figure 5C). A similar variation in the SN of serological responses to PvMSP10 with the time since last infection was observed with DBS samples but at lower levels: less than 7 days (38.5%, 20.9–59.3), 7–30 days (45.7%, 29.2–63.1), 30–60 days (36.9%, 25.5–49.8), and more than 60 days (24.0%, 13.5–38.5) (Figure 5D). Interestingly, 3 individuals from Lupuna village who received treatment at the time of the study enrollment after being diagnosed with P. vivax malaria, and did not experience any other infection during the cohort period, were found to have positive serological results on their serum samples at the end of the follow-up. Two of them were also positive when DBS samples were analyzed.
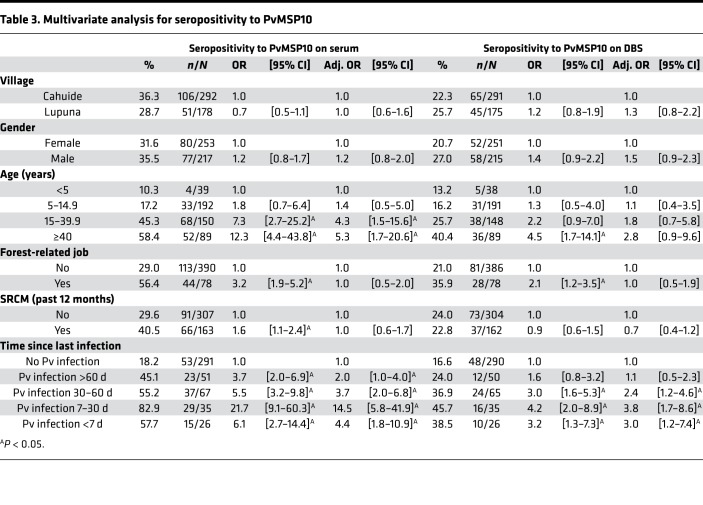
The multivariate logistic model found that only age and time since last infection were independently associated with seropositivity to P. vivax determined on serum samples (Table 3). Although individuals aged 15–39 years (adj. OR = 4.3 [1.5–15.6]) and those at least 40 years (adj. OR = 5.3 [1.7–20.6]) were, respectively, 4.3 times and 5.3 times more likely to be seropositive than those less than 5 years old, individuals with P. vivax infections of 7–30 days had the highest odds (adj. OR = 14.5 [5.8–41.9]) of being seropositive (among other individuals with P. vivax infections) in comparison with those who remained uninfected during the follow-up. Considering the analysis of DBS samples, individuals with P. vivax infections of 7–30 days also had the highest odds (adj. OR = 3.8 [1.7–8.6]) of being seropositive when compared with individuals who did not present with infection.
Table 3. Multivariate analysis for seropositivity to PvMSP10.
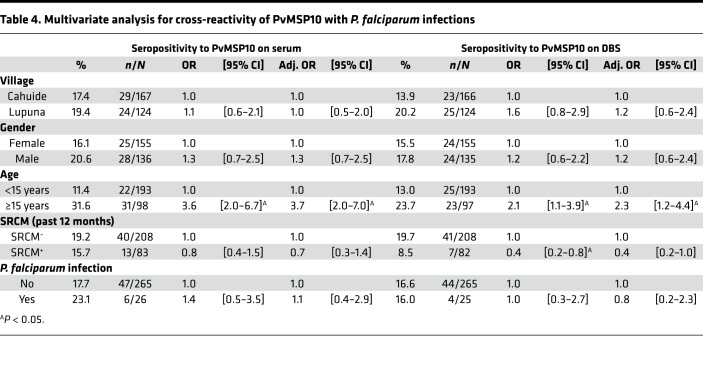
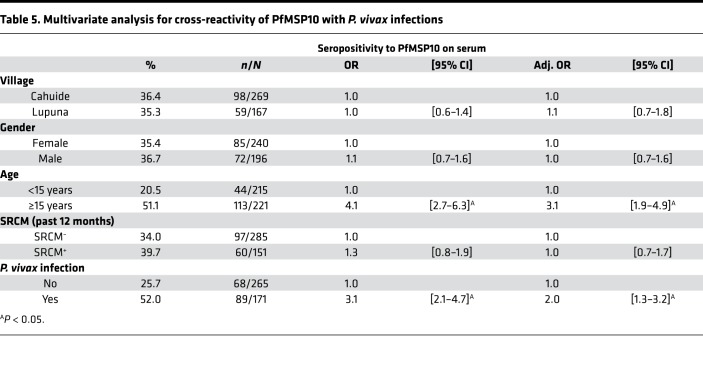
Cross-reactivity analyses showed no differences in seropositivity levels to PvMSP10 between individuals who acquired P. falciparum infections and those without any infection during the follow-up, with both serum (23.1% vs. 17.7%, P = 0.50) and DBS samples (16.0% vs. 16.6%, P = 0.94). The lack of differences remained even after adjusting for village, sex, age, and malaria history in the multivariate analysis for P. vivax seropositivity (Table 4). Conversely, individuals acquiring P. vivax infections during the follow-up (52.0%, 89/171) were 2 times (adj. OR = 2.0 [1.3–3.2]) more likely to be seropositive to PfMSP10 than those without any detected infection (25.7%, 68/265) (Table 5). ELISA analysis of plasma samples from Senegalese adults with documented recent P. falciparum infection showed IgG reactivity to recombinant PfMSP10 but not to recombinant PvMSP10, indicating lack of cross-reactivity of antibodies between these antigens (data not shown).
Table 4. Multivariate analysis for cross-reactivity of PvMSP10 with P. falciparum infections.
Table 5. Multivariate analysis for cross-reactivity of PfMSP10 with P. vivax infections.
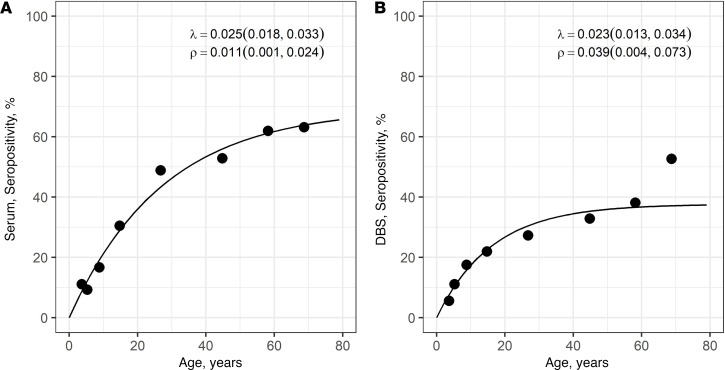
Age-seroprevalence plots for PvMSP10 on both serum and DBS samples confirmed the increasing P. vivax seropositivity with age (Figure 6). Despite the lower seroprevalence with DBS mainly in individuals older than 15 years, SCRs (λ) estimated from catalytic models for the study participants were not different according to the type of samples: serum (λ = 0.025 [0.018–0.033]) and DBS (λ = 0.023 [0.013–0.034]).
Figure 6. P. vivax age-seroprevalence curves according to sample type (serum or dried blood spot) for study participants.
(A) Serum and (B) DBS samples. Dots represent observed prevalence, while lines represent fitted curves derived from catalytic models. SCRs (λ) and seroreversion rates (SRRs, ρ) estimated by catalytic models and corresponding 95% CIs estimated by Bootstrap methods are shown on the top right of each plot.
Discussion
Here we show that human IgG responses to Plasmodium species–specific, mammalian cell–produced recombinant MSP10 were a useful and effective biomarker to identify recent P. vivax infection in the Peruvian Amazon. Such a biomarker is key to the identification of populations affected by ongoing malaria transmission, even at low intensity, which is fundamental to malaria control and elimination efforts.
Complete and well-characterized P. vivax and P. falciparum infections were identified in cohort participants during a 5-month follow-up, which enabled longitudinal assessment of specific IgG responses to MSP10 as an indication of P. vivax and P. falciparum malaria exposure. The ability to trigger specific antibody responses with acceptable sensitivity in serum samples for detecting recent malaria infection of less than 30 days, the relatively rapid decay of serological responses in infected individuals, and the lack of immunological cross-reactivity with P. falciparum suggest the potential use of IgG antibodies against PvMSP10 as a serosurveillance marker for recent P. vivax exposure in the co-endemic Peruvian Amazon. Conversely, antibody responses against PfMSP10 showing poor discrimination between individuals recently exposed and not exposed to P. falciparum may not be suitable as markers of P. falciparum exposure in co-endemic areas.
Several cross-sectional sero-epidemiological studies in Amazonian countries have reported the association of antibody responses with previous malaria infections by P. vivax and P. falciparum (3, 18, 25, 26). However, because these previous studies relied on individual recall of past malarial infection, they could not reliably date or confirm past infections. The few studies that have prospectively assessed the relationship between antigen-specific seropositivity and the occurrence of malarial infection focused on individuals with premunition (clinical immunity to malaria) using asexual blood stage vaccine candidates as antigens (27–30). In Brazil, immune responses to MSP-1 (N-terminus) were found to be significantly associated with time elapsed until next infection, as well as with a reduction of the risk of clinical infection after 1 year of follow-up (27, 28). In Peru, the assessment of the humoral dynamics before, during, and after P. falciparum infections showed that responses against MSP119 can rapidly develop in both children and adults and be maintained through a 4-month follow-up period in hypoendemic Amazonian areas, in contrast to shorter responses previously reported in high-transmission areas (29). MSP119 for both P. falciparum and P. vivax are currently proposed as markers of protection against malaria, as well as surrogate markers for malaria exposure, following the accumulation of positive evidence from different populations around the world (31, 32). Longitudinal population cohorts with rigorous active follow-up, able to detect all or most malaria infections, both asymptomatic and symptomatic, offer the most powerful study design to assess the role of potential serosurveillance markers for malaria (33, 34). To our knowledge, this study is the first to report the association between levels of naturally acquired IgG antibodies against recombinant PvMSP10 antigen and time since last infection, using this advantageous study design.
Our group previously reported that PvMSP10 was the P. vivax antigen with the highest reactivity after probing sera from symptomatic subjects with P. vivax infections living around Iquitos, using a protein microarray with more than 2000 P. vivax proteins expressed by an E. coli–based cell-free expression system (23). Thus, the significant differences in seropositivity levels between individuals who presented with P. vivax infections and those who did not confirm the excellent antigenicity of the PvMSP10 proteins expressed in mammalian cell expression systems. Modest sensitivity (around 45%) and high specificity (95%) of IgG responses against PvMSP10 on serum samples for the diagnosis of malaria in symptomatic infected P. vivax patients have been reported in a Korean study that assessed the role of this protein as a vaccine candidate for P. vivax (35). However, the performance of serological tests for malaria diagnosis is always affected by the antibody kinetics, and this can have wide variability among individuals and fluctuate in levels over time (36, 37), affecting the sensitivity of antigen-antibody responses. Different antibody kinetics of IgG subclasses (not quantified in our study) composing the IgG total may also influence this performance (21). Because antibodies can take several days to develop and persist for weeks or months after the infection has resolved (38), serological tools may not be good for the direct diagnosis of actively infected individuals (21). Indeed, the sensitivity of PvMSP10 responses for detecting recent exposure from serum samples was 57.7% in individuals with microscopically confirmed diagnosis of P. vivax around the end of the cohort study (0–6 days since last infection). Sensitivity was highest (82.9%) within 7–30 days since last infection, but following this initial phase, a significant decay of antibodies occurred, causing a decline in the sensitivity of antibody responses (45.1% in infections >60 days, Figure 5C). Interestingly, seropositive levels were also observed in 3 individuals with confirmed and treated P. vivax infections only at the beginning of the cohort, suggesting that PvMSP10 antibodies could persist for more than 5 months in the absence of a new infection.
Because potential cross-reactivity of antibodies to antigens from different malaria species could affect test specificity in areas where multiple malaria species are present (39, 40), further analyses were conducted to explore the possibility of interspecies cross-reactivity and its influence in the specificity of antigen-antibody responses. Although previous studies have reported large conserved regions of MSP10 across P. vivax and P. falciparum (e.g., sequence similarity in the C-terminal EGF-like domain) (41), we did not find evidence of cross-reactivity between PvMSP10 antibodies and P. falciparum in our study samples or in the Senegalese samples used as control because of their origin in a non–P. vivax-endemic region (data not shown). A possible explanation for not obtaining a better specificity for detecting recent P. vivax exposure may be an overclassification of some individuals as “not infected” by the gold standard. Those individuals could have acquired P. vivax infections and been cured before the onset of follow-up, and their PvMSP10 antibody levels could have remained in the absence of a new infection. The evidence of persisting antibody responses longer than the follow-up period further supports this hypothesis. In contrast to PvMSP10, the analysis with antibodies against PfMSP10 suggested high cross-reactivity with P. vivax in analyzed samples, likely affecting the test specificity and performance for discriminating between exposed and nonexposed individuals by P. falciparum. Cross-reactivity would make the PfMSP10 antibody responses not suitable for detecting recent P. falciparum exposure in a co-endemic region (21), particularly in areas where P. vivax infections are markedly more common than P. falciparum infections; however, further studies with enough samples from malaria and other endemic infections will be required to confirm these preliminary findings.
DBSs are thought to be a potentially useful and convenient source of antibodies for sero-epidemiological surveys, especially those in remote locations (42). However, we found that the efficiency of PvMSP10 antibody responses for identifying recent exposure to P. vivax was significantly lower on DBSs than on serum samples. This low efficiency may be explained by the inherent limitations of the sample and the short time that DBSs spent at tropical conditions in the field, despite the efforts to comply with all recommendations for their transport, conservation, storage, and lab processing (42). For serosurveillance purposes, capillary blood obtained by finger stick as a source of serum could be evaluated given the operational limitations of the serum collection by venipuncture and the problems in antibody recovery from DBSs.
Despite the potential limitation of only a 5-month follow-up period, the data presented here suggest that PvMSP10 antibody responses are short duration (<6 months) and therefore have the ability to identify recent infections (with modest sensitivity), irrespective of the presence of symptoms (symptomatic or not) and parasite density levels (patent or subpatent). These antibody characteristics and the good fit of age-specific seroconversion models to the serological data support the specific role and potential application of PvMSP10 IgG responses as a surveillance tool for monitoring malaria transmission at a population level. In combination with spatial analysis techniques, serological responses to PvMSP10 could be used to predict clusters with high malaria risk, across the spectrum of P. vivax infection phenotypes (i.e., symptomatic, asymptomatic, and subpatent infections, as well as hypnozoite carriers) (43). Recent-exposure tools would be more suitable in settings where malaria transmission is highly heterogeneous at the geographical level and over time and where hotspot locations vary from year to year (unstable hotspots) (44). Antibody responses to PvMSP10 could also be used to assess short-term changes (within months) in malaria transmission following control interventions (33), by calculating pre-intervention and post-intervention age-seroconversion rates mainly in younger age groups. Indeed, the lack of differences observed in P. vivax SCRs obtained from serum samples and DBSs (despite big differences in overall seropositivity levels between them) are explained mainly by the similarity in seropositivity levels in younger individuals.
Sensitive and specific serological tools for the accurate detection of malaria exposure are urgently needed by NMCPs to overcome the limitations of currently used surveillance tools in low and moderate endemic areas where transmission is seasonal, is increasingly focal, and is sustained mainly by asymptomatic and subpatent infections. The high immunogenicity of PvMSP10, the relatively short-lived antigen-antibody responses, and the limited cross-reactivity with P. falciparum make specific PvMSP10 antibodies candidates for markers of recent exposure to P. vivax malaria in co-endemic areas, such as the Amazon region. Future research should include kinetics assessment of IgG subclasses against PvMSP10, identification of prevalent allelic variants in the antigen (i.e., polymorphisms), performance assessment of antibody responses for detecting malaria exposure in other P. vivax–endemic regions in South America and Southeast Asia, and their subsequent operative evaluation in identifying hotspots of P. vivax infections and monitoring short-term changes in P. vivax transmission in different epidemiological settings. The combination with other good markers of malaria exposure (recent or remote) may improve the sensitivity of serological tools to extend the scope of surveillance activities. An innovative use of sensitive serological tools for recent P. vivax exposure would be for the identification of individuals with a high probability to carry hypnozoites. Because any individual carrying P. vivax antibodies is likely to harbor hypnozoites, irrespective of active infection (presence of blood forms), this serological tool could be used to guide the administration of radical cure treatment during large-scale population screenings. This operational strategy may be preferred over mass drug administration as a malaria elimination strategy to avoid unnecessary treatments and the related risk of severe hemolysis in glucose-6-phosphate dehydrogenase–deficient individuals.
Methods
Study design and area
A test validation design was used to assess the performance of serological tests for detecting recent malaria exposure by P. vivax and P. falciparum, taking advantage of biological samples and malaria infection data previously collected within a short-term malaria population cohort study in 2 rural villages of the Peruvian Amazon: Cahuide and Lupuna (Figure 1). Cahuide is situated along Iquitos-Nauta Road 60 km from the city of Iquitos, on the bank of the Itaya River (latitude 04° 13.785’S longitude 73° 276’ W), while Lupuna is located in the forest 10 km from Iquitos, crossing the Nanay River (latitude 03° 44.591’S longitude 73° 19.615’W). The climate is tropical, warm and humid with a rainy season from November to May and a dry season from June to October (3). The annual average temperature is around 27°C, the relative humidity above 80%, and average annual rainfall 4 m. Both P. vivax and P. falciparum cases are reported in both villages throughout the entire year (45), with a peak between February and July. The highly anthropophilic Anopheles darlingi is the main malaria vector, and a recent report showed that this vector was now biting mainly outdoors whereas it was previously known to bite equally in- and outdoors (45).
Longitudinal malaria cohort and follow-up
From August to September 2012, 961 inhabitants in Cahuide and Lupuna were enrolled in a longitudinal cohort study aimed at estimating the prevalence and incidence of asymptomatic malaria parasite parasitemia. Data on sociodemographic variables (age, sex, education level, occupation) and SRCM episodes in the previous year (i.e., the report of malaria infections in the past 12 months, which were confirmed by LM in health facilities) were recorded for each participant and finger prick blood samples taken for immediate LM and for storage on filter paper (Whatman grade 3, Whatman) for subsequent PCR. After enrollment, participants were followed up using 3 detection methods: routine PCD (i.e., health post–based case detection for symptomatic malaria), weekly active case detection (ACD) of symptomatic individuals (i.e., door-to-door canvassing of malaria-related symptoms and blood sampling if such symptoms were present), and monthly (October 2012–January 2013) population screening (PS) for blood sampling, regardless of symptoms. In PCD, individuals presenting with fever or any other malaria symptoms at health posts of Cahuide and Lupuna were tested by LM, and microscopically confirmed infections were treated and registered. During ACD through weekly household visits, axillary temperature was taken from enrolled participants, and history of fever or any other malaria symptoms was registered. If the participant had fever (temperature ≥ 37.5°C), or reported fever, headache, chills, or concern for malaria in the past 7 days, a finger prick blood sample was taken for immediate diagnosis by LM, and microscopically confirmed infections were treated. Monthly PS coincided once a month with weekly household visits and consisted of systematic finger prick blood sampling of all enrolled participants and the treatment of all microscopically positive infections, irrespective of the presence of symptoms. Following Peruvian national treatment guidelines (46), all P. falciparum infections confirmed by LM were treated with mefloquine-artesunate and P. vivax infections with chloroquine and primaquine at health posts. In December 2012, the scheduled PS allowed for another blood spot on filter paper from each participant for a second PCR. Two weeks later, a blood spot on filter paper and a whole-blood sample were collected for serological tests, in addition to their use for diagnosis by a trained pathologist.
After drying for a minimum of 2 hours, DBS samples were inserted into a closed bag with desiccant, transported the same day of collection to the field lab in Iquitos, and then stored at –20°C until processing. Whole blood was collected by venipuncture in tubes with EDTA (BD Vacutainer, BD) and transported at 4°C to the field lab. There, blood samples were centrifuged at 2205 g to obtain plasma, which was later stored at –70°C until processing. Serum samples were only thawed 1 time for time inventory and a second time for ELISA processing.
Study selection criteria
Because performance assessment of serological tests for detecting recent malaria exposure required accurate information about the occurrence of malaria infections throughout the cohort period (33, 34), only individuals with complete follow-up through January 2013 were included in the analysis. Individuals who met the selection criteria had the following: malaria diagnoses by LM and PCR at enrollment (August–September 2012); at least 1 monthly follow-up and diagnosis by LM (August 2012–January 2013), during either weekly ACD or PS; a second malaria diagnosis by PCR (December 2012); and available serum and DBS for serological analyses 2 weeks later (January 2013).
Laboratory procedures
Microscopy.
Thick and thin smears were stained for 10 minutes with a 10% Giemsa solution, and parasite density was computed after counting the number of asexual parasites for 200 white blood cells (WBCs) in the thick smear and assuming a concentration of 8000 WBCs/μL. Slides were read on-site and then again the following day by a microscopy expert at the study’s reference laboratory in Iquitos. A slide was declared negative if no malaria parasite was found after examining 100 fields (47). Quality control was done blindly on all positive slides and 10% of randomly chosen negative slides by a senior technician at the Institute of Tropical Medicine-Alexander von Humboldt at Universidad Peruana Cayetano Heredia in Lima. Discordant results were reassessed by a second senior technician until agreement.
Real-time PCR.
DBSs on filter paper were cut into pieces of approximately 6 mm2 for DNA extraction using QIAamp DNA Micro Kit from QIAGEN, following the manufacturer’s instructions. Subsequent amplification was performed by a quantitative real-time PCR method targeting the 18s rRNA gene region. Following the protocol reported by Mangold et al. (48), 5′-TAACGAACGAGATCTTAA-3′ and 5′-GTTCCTCTAAGAAGCTTT-3′ were used as primers, and PCR conditions consisted of an initial denaturation at 95°C for 2 minutes, followed by amplification for 45 cycles of 20 seconds at 95°C, 20 seconds at 52°C, and 30 seconds at 68°C. Amplification was immediately followed by a melt program consisting of 5 seconds at 65°C and a stepwise temperature increase of 0.5°C/s up to 95°C. The analysis of the differences in melting curves provided an accurate differentiation between Plasmodium species.
Serology.
MSP10 is an immunogenic protein encoded by a single copy gene (in P. vivax, PVX_114145; in P. falciparum, GenBank accession PF3D7_0620400) expressed in the asexual blood stages of P. vivax and P. falciparum (49). For this study, PvMSP10 and PfMSP10 were expressed in a human embryonic kidney (HEK293) mammalian cell line (Supplemental Appendix; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130769DS1) using procedures as described elsewhere (50, 51). See complete unedited blots in the supplemental material. Of note, N-linked glycosylation sequons were mutated to prevent glycosylation when proteins were expressed in human cells. Total IgG against both proteins was detected on serum and DBS samples using an ELISA protocol previously described (3, 52) with slight modifications. Both serum and DBS (5 mm diameter disc/sample) were eluted overnight at 4°C in PBS–nonfat milk–0.05% Tween 20 to obtain eluates at 1:500. For each sample type, 100 μL of the eluate was added in duplicate to blocked ELISA plates coated separately with each antigen (1 μL/mL) in carbonate buffer at pH 9.6. Pooled sera from infected patients (either by P. vivax or P. falciparum) as positive controls, and from 8 individuals with no previous exposure to malaria (living in nonendemic areas and with no reported trips to endemic areas, and with confirmed negative PCR) as negative controls were diluted at 1:500 in PBS–nonfat milk–0.05% Tween 20 and included in, respectively, 4 and 8 wells of each 96-well plate. Seventy-eight wells containing 39 serum/DBS samples in duplicate, 4 antigen-uncoated blank wells, and 2 zero concentration control wells completed the plate. Affinity-purified, horseradish peroxidase–labeled goat anti-human IgG (H+L) (KPL; catalog 62-8420) diluted to 1:10,000 in PBS-Tween was used as a secondary antibody (100 μL/well) and incubated for 1 hour before development of the ELISA using 100 μL TMB peroxidase substrate (KPL). OD values in antigen-coated wells were read at 450 nm (VersaMax ELISA Microplate Reader, Molecular Devices, Thermo Fisher Scientific) and corrected by subtracting from them the mean of OD values in antigen-uncoated blank wells. The number of SDs from the corrected OD mean of negative controls to the corrected OD of a given study sample was calculated in each plate, allowing for the dichotomized classification of each sample as either seropositive (greater than or equal to cutoff of 3 SDs) or seronegative (less than cutoff of 3 SDs).
Data management and statistics
Data were obtained using paper forms and double (independently) entered and cross-checked in Excel (Microsoft). Analysis was done using R v.2.15 software (R Development Core Team, R Foundation for Statistical Computing).
Five groups of individuals were identified in the malaria cohort: those with no infections during the follow-up, those with single (unique) monoinfections, those with recurrent monoinfections by same parasite species, those with recurrent monoinfections by different species, and those with mixed infections. The malaria cohort data were used as a gold standard reference to define malaria exposure in study participants. An individual with malaria exposure to a particular species was an individual with the confirmed presence of species-specific parasites in the blood by LM and/or PCR (monoinfection or mixed infection) at any time during the follow-up. For recurrent infections by the same parasite species, only data of the last infection were considered in the analysis. Individuals with no infections during the follow-up and those who acquired monoinfections due to a species other than that assessed by serological tools were considered not exposed.
Cross-validation logistic models assessed the relationship between antigen-antibody responses (i.e., corrected ODs) on biological samples (serum or DBS) and malaria exposure using a resampling method (100 times) whereby two-thirds of participants were randomly selected to be in a training data set and the remaining one-third in the testing data set. Average cross-validated ROC curves characterized the sensitivity/specificity trade-offs for the binary classifier, and their AUCs with corresponding DeLong 95% CI (53) assessed the model discriminatory efficiency for malaria exposure. Performance indicators of dichotomized serological results (seropositive or seronegative) for the identification of malaria exposure (i.e., SN, SP, PPV, and NPV) were also calculated and their 2-sided 95% CIs estimated using the Wilson score method.
Locally weighted smoothing generating a smoothed fit curve with a confidence region described the relationship between corrected ODs obtained from serum and DBS samples and the time since last infection (Supplemental Figure 1) and supported the classification of participants into 5 groups: infection within less than 7 days, 7–30 days, 31–60 days, or more than 60 days or no infection during follow-up. Corrected ODs between those groups were further compared using the Kruskal-Wallis test with Dunn’s multiple-comparisons post hoc tests. Moreover, the χ2 test for trend was used to evaluate changes in the SN of serological results for detecting exposure with the time since last malaria infection, while univariate and multivariate logistic models assessed the association of the latter time-dependent variable with seropositivity, adjusting for potential confounders, such as village, sex, age groups, main economic activity, and SRCM in the past year. All statistical analyses considered a P value less than 0.05 as statistically significant.
Interspecies cross-reactivity of antibodies was also evaluated using multivariate logistic models that compared serological responses against PvMSP10 or PfMSP10 between individuals who had only monoinfections by, respectively, P. falciparum or P. vivax and individuals who did not present infection during the cohort period, with adjustment for village, sex, age groups, and SRCM in the past year. In addition, plasma samples from Senegalese adult subjects with documented P. falciparum infection in the past 2–3 months were used to examine cross-reactivity between P. falciparum sera and recombinant PvMSP10.
Seropositivities in both serum and DBS samples were stratified into yearly age decile groups and then analyzed using a reverse catalytic conversion model (14). The model allows for the generation of age-seroprevalence curves and the estimation of 2 parameters: (a) an SCR (λ) or force of infection, defined as the annual rate at which individuals change from seronegative to seropositive; and (b) an SRR (ρ), defined as the annual rate at which seropositive individuals revert to a seronegative state (14). Bootstrapped 95% CIs for SCR and SRR were estimated using the bias-corrected and accelerated bootstrap resampling method (54).
Study approval
Ethical clearance for the cohort study was obtained from the Ethics Review Board of the Universidad Peruana Cayetano Heredia, Lima, Peru (SIDISI code 57395). Permission was obtained from health and local authorities after they received an explanation of the purpose and procedures of the study. Written informed consent was obtained before participation and blood sampling from all adults and the parents of all participating children less than 18 years. Along with their parents’ consent, children older than 7 years provided written informed consent.
Author contributions
ARA, DG, ALC, RHG, and JMV worked on the clinical/field study design and execution. KPP, MC, KT, DG, EA, EM, KG, CF, YA, and JMV worked on the laboratory study design and execution. GT designed and carried out ELISA assays; contributed to manuscript writing and editing. Data analysis was carried out by ARA, KPP, MC, KT, DG, ALC, RHG, and JMV. Statistical analysis was done by ARA, DG, and ALC. The manuscript was drafted by ARA, KPP, DG, ALC, RHG, and JMV and circulated to all coauthors for corrections, additions, modifications, and final approval.
Supplementary Material
Acknowledgments
We thank all residents and local authorities from the Loreto Villages of Cahuide and Lupuna for their enthusiastic participation in the study, as well as all field workers for their dedication during the fieldwork. We also thank Amy Bei of the Yale School of Public Health and her team and human volunteers in Senegal for the use of blood samples in this study. This study was funded by cooperative agreement U19AI089681 from the United States Public Health Service, NIH/National Institute of Allergy and Infectious Diseases, as the Amazonian International Center of Excellence in Malaria Research.
Version 1. 11/26/2019
In-Press Preview
Version 2. 01/16/2020
Electronic publication
Footnotes
KPP’s present address is: Cell and Developmental Biology, Division of Biology; and Tata Institute for Genetics and Society, University of California San Diego School of Medicine, La Jolla, California.
GT’s present address is: Jacobs School of Medicine and Biomedical Sciences, University at Buffalo, Buffalo, New York, USA.
YA’s present address is: Pediatrics, BC Children’s Hospital, University of British Columbia, Vancouver, British Columbia, Canada.
JMV’s present address is: Section of Infectious Diseases, Department of Internal Medicine, Yale School of Medicine, New Haven, Connecticut, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Role of funding source: The funding agency (NIH) had no role in the design of this study nor any role in its execution, analyses, and interpretation of data or the decision to submit results.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(1):e130769.https://doi.org/10.1172/jci.insight.130769.
Contributor Information
Angel Rosas-Aguirre, Email: angelrosasa@gmail.com.
Kailash P. Patra, Email: kpatra@ucsd.edu.
Maritza Calderón, Email: mmcalderons@yahoo.es.
Katherine Torres, Email: kathjess2000@yahoo.com.
Dionicia Gamboa, Email: dionigamboa@yahoo.com.
Edith Arocutipa, Email: thedi.see@gmail.com.
Edith Málaga, Email: edith.malaga@gmail.com.
Katherine Garro, Email: katherine.garro@upch.pe.
Carlos Fernández, Email: carlos.fernandez.m@upch.pe.
Grace Trompeter, Email: grace.trompeter@gmail.com.
Yossef Alnasser, Email: yossefalnasser@gmail.com.
Alejandro Llanos-Cuentas, Email: elmer.llanos@upch.pe.
Robert H. Gilman, Email: gilmanbob@gmail.com.
Joseph M. Vinetz, Email: Joseph.vinetz@yale.edu.
References
- 1. World Health Organization. World Malaria Report 2016. World Health Organization website. https://www.who.int/malaria/publications/world-malaria-report-2016/report/en/ Accessed December 2, 2019.
- 2.Rosas-Aguirre A, et al. Epidemiology of Plasmodium vivax malaria in Peru. Am J Trop Med Hyg. 2016;95(6 Suppl):133–144. doi: 10.4269/ajtmh.16-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosas-Aguirre A, et al. Hotspots of malaria transmission in the Peruvian Amazon: rapid assessment through a parasitological and serological survey. PLoS One. 2015;10(9):e0137458. doi: 10.1371/journal.pone.0137458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roshanravan B, et al. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg. 2003;69(1):45–52. doi: 10.4269/ajtmh.2003.69.45. [DOI] [PubMed] [Google Scholar]
- 5.Branch O, et al. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J. 2005;4:27. doi: 10.1186/1475-2875-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosas-Aguirre A, et al. Micro-heterogeneity of malaria transmission in the Peruvian Amazon: a baseline assessment underlying a population-based cohort study. Malar J. 2017;16(1):312. doi: 10.1186/s12936-017-1957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousema T, et al. Identification of hot spots of malaria transmission for targeted malaria control. J Infect Dis. 2010;201(11):1764–1774. doi: 10.1086/652456. [DOI] [PubMed] [Google Scholar]
- 8.Bousema T, et al. Hitting hotspots: spatial targeting of malaria for control and elimination. PLoS Med. 2012;9(1):e1001165. doi: 10.1371/journal.pmed.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roll Back Malaria Partnership. Framework for Monitoring Progress and Evaluating Outcomes and Impact. World Health Organization. https://apps.who.int/iris/bitstream/handle/10665/66745/WHO_CDS_RBM_2000.25.pdf;jsessionid=7258E62509BE6FD9C58ED2619297D3E5?sequence=1 Accessed December 2, 2019.
- 10.malERA Consultative Group on Monitoring. Evaluation, Surveillance A research agenda for malaria eradication: monitoring, evaluation, and surveillance. PLoS Med. 2011;8(1):e1000400. doi: 10.1371/journal.pmed.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yekutiel P. Problems of epidemiology in malaria eradication. Bull World Health Organ. 1960;22:669–683. [PMC free article] [PubMed] [Google Scholar]
- 12.Hay SI, Smith DL, Snow RW. Measuring malaria endemicity from intense to interrupted transmission. Lancet Infect Dis. 2008;8(6):369–378. doi: 10.1016/S1473-3099(08)70069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tusting LS, Bousema T, Smith DL, Drakeley C. Measuring changes in Plasmodium falciparum transmission: precision, accuracy and costs of metrics. Adv Parasitol. 2014;84:151–208. doi: 10.1016/B978-0-12-800099-1.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drakeley CJ, et al. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci USA. 2005;102(14):5108–5113. doi: 10.1073/pnas.0408725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart L, et al. Rapid assessment of malaria transmission using age-specific sero-conversion rates. PLoS One. 2009;4(6):e6083. doi: 10.1371/journal.pone.0006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corran P, Coleman P, Riley E, Drakeley C. Serology: a robust indicator of malaria transmission intensity? Trends Parasitol. 2007;23(12):575–582. doi: 10.1016/j.pt.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Cook J, et al. Serological markers suggest heterogeneity of effectiveness of malaria control interventions on Bioko Island, equatorial Guinea. PLoS One. 2011;6(9):e25137. doi: 10.1371/journal.pone.0025137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunha MG, et al. Serologically defined variations in malaria endemicity in Pará state, Brazil. PLoS One. 2014;9(11):e113357. doi: 10.1371/journal.pone.0113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiang MS, et al. Surveillance for malaria elimination in Swaziland: a national cross-sectional study using pooled PCR and serology. PLoS One. 2012;7(1):e29550. doi: 10.1371/journal.pone.0029550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bousema T, et al. Serologic markers for detecting malaria in areas of low endemicity, Somalia, 2008. Emerging Infect Dis. 2010;16(3):392–399. doi: 10.3201/eid1603.090732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott SR, Fowkes FJ, Richards JS, Reiling L, Drew DR, Beeson JG. Research priorities for the development and implementation of serological tools for malaria surveillance. F1000Prime Rep. 2014;6:100. doi: 10.12703/P6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim CS, et al. Seroprevalence to the circumsporozoite protein peptide antigen of Plasmodium vivax in Korean children. Microbiol Immunol. 2005;49(6):521–527. doi: 10.1111/j.1348-0421.2005.tb03757.x. [DOI] [PubMed] [Google Scholar]
- 23.Chuquiyauri R, et al. Genome-scale protein microarray comparison of human antibody responses in Plasmodium vivax relapse and reinfection. Am J Trop Med Hyg. 2015;93(4):801–809. doi: 10.4269/ajtmh.15-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres KJ, et al. Genome-level determination of Plasmodium falciparum blood-stage targets of malarial clinical immunity in the Peruvian Amazon. J Infect Dis. 2015;211(8):1342–1351. doi: 10.1093/infdis/jiu614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosas-Aguirre A, et al. Assessing malaria transmission in a low endemicity area of north-western Peru. Malar J. 2013;12:339. doi: 10.1186/1475-2875-12-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ladeia-Andrade S, et al. Naturally acquired antibodies to merozoite surface protein (MSP)-1(19) and cumulative exposure to Plasmodium falciparum and Plasmodium vivax in remote populations of the Amazon Basin of Brazil. Mem Inst Oswaldo Cruz. 2007;102(8):943–951. doi: 10.1590/S0074-02762007000800009. [DOI] [PubMed] [Google Scholar]
- 27.Nogueira PA, et al. A reduced risk of infection with Plasmodium vivax and clinical protection against malaria are associated with antibodies against the N terminus but not the C terminus of merozoite surface protein 1. Infect Immun. 2006;74(5):2726–2733. doi: 10.1128/IAI.74.5.2726-2733.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Versiani FG, et al. High levels of IgG3 anti ICB2-5 in Plasmodium vivax-infected individuals who did not develop symptoms. Malar J. 2013;12:294. doi: 10.1186/1475-2875-12-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres KJ, Clark EH, Hernandez JN, Soto-Cornejo KE, Gamboa D, Branch OH. Antibody response dynamics to the Plasmodium falciparum conserved vaccine candidate antigen, merozoite surface protein-1 C-terminal 19kD (MSP1-19kD), in Peruvians exposed to hypoendemic malaria transmission. Malar J. 2008;7:173. doi: 10.1186/1475-2875-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark EH, et al. Plasmodium falciparum malaria in the Peruvian Amazon, a region of low transmission, is associated with immunologic memory. Infect Immun. 2012;80(4):1583–1592. doi: 10.1128/IAI.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cutts JC, Powell R, Agius PA, Beeson JG, Simpson JA, Fowkes FJ. Immunological markers of Plasmodium vivax exposure and immunity: a systematic review and meta-analysis. BMC Med. 2014;12:150. doi: 10.1186/s12916-014-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowkes FJ, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: A systematic review and meta-analysis. PLoS Med. 2010;7(1):e1000218. doi: 10.1371/journal.pmed.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King CL, et al. Biosignatures of exposure/transmission and immunity. Am J Trop Med Hyg. 2015;93(3 Suppl):16–27. doi: 10.4269/ajtmh.15-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helb DA, et al. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci USA. 2015;112(32):E4438–E4447. doi: 10.1073/pnas.1501705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y, Wang B, Sattabongkot J, Lim CS, Tsuboi T, Han ET. Immunogenicity and antigenicity of Plasmodium vivax merozoite surface protein 10. Parasitol Res. 2014;113(7):2559–2568. doi: 10.1007/s00436-014-3907-8. [DOI] [PubMed] [Google Scholar]
- 36.Fowkes FJ, et al. New insights into acquisition, boosting, and longevity of immunity to malaria in pregnant women. J Infect Dis. 2012;206(10):1612–1621. doi: 10.1093/infdis/jis566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White MT, et al. Dynamics of the antibody response to Plasmodium falciparum infection in African children. J Infect Dis. 2014;210(7):1115–1122. doi: 10.1093/infdis/jiu219. [DOI] [PubMed] [Google Scholar]
- 38.Drakeley C, Cook J. Chapter 5. Potential contribution of sero-epidemiological analysis for monitoring malaria control and elimination: historical and current perspectives. Adv Parasitol. 2009;69:299–352. doi: 10.1016/S0065-308X(09)69005-9. [DOI] [PubMed] [Google Scholar]
- 39.Woodberry T, et al. Antibodies to Plasmodium falciparum and Plasmodium vivax merozoite surface protein 5 in Indonesia: species-specific and cross-reactive responses. J Infect Dis. 2008;198(1):134–142. doi: 10.1086/588711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Igonet S, et al. Cross-reactivity studies of an anti-Plasmodium vivax apical membrane antigen 1 monoclonal antibody: binding and structural characterisation. J Mol Biol. 2007;366(5):1523–1537. doi: 10.1016/j.jmb.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Leal O, et al. Identifying and characterising the Plasmodium falciparum merozoite surface protein 10 Plasmodium vivax homologue. Biochem Biophys Res Commun. 2005;331(4):1178–1184. doi: 10.1016/j.bbrc.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Corran PH, et al. Dried blood spots as a source of anti-malarial antibodies for epidemiological studies. Malar J. 2008;7:195. doi: 10.1186/1475-2875-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturrock HJ, et al. Targeting asymptomatic malaria infections: active surveillance in control and elimination. PLoS Med. 2013;10(6):e1001467. doi: 10.1371/journal.pmed.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bejon P, et al. Stable and unstable malaria hotspots in longitudinal cohort studies in Kenya. PLoS Med. 2010;7(7):e1000304. doi: 10.1371/journal.pmed.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno M, et al. Implications for changes in Anopheles darlingi biting behaviour in three communities in the peri-Iquitos region of Amazonian Peru. Malar J. 2015;14:290. doi: 10.1186/s12936-015-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ministerio de Salud del Perú. Norma técnica para la atención de la malaria y malaria severa en el Perú. MINSA. http://bvs.minsa.gob.pe/local/MINSA/4378.pdf Accessed December 2, 2019.
- 47. Ministerio de Salud del Perú. Norma técnica de salud para el control de calidad del diagnóstico microscópico de malaria. MINSA. http://www.ins.gob.pe/insvirtual/images/otrpubs/pdf/FINAL%20MALARIA%2028.12.10%5B1%5D.pdf Accessed December 2, 2019.
- 48.Mangold KA, et al. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005;43(5):2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pacheco MA, et al. Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp. Infect Genet Evol. 2012;12(5):978–986. doi: 10.1016/j.meegid.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bushell KM, Söllner C, Schuster-Boeckler B, Bateman A, Wright GJ. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 2008;18(4):622–630. doi: 10.1101/gr.7187808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hostetler JB, Sharma S, Bartholdson SJ, Wright GJ, Fairhurst RM, Rayner JC. A library of Plasmodium vivax recombinant merozoite proteins reveals new vaccine candidates and protein-protein interactions. PLoS Negl Trop Dis. 2015;9(12):e0004264. doi: 10.1371/journal.pntd.0004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook J, et al. Sero-epidemiological evaluation of changes in Plasmodium falciparum and Plasmodium vivax transmission patterns over the rainy season in Cambodia. Malar J. 2012;11:86. doi: 10.1186/1475-2875-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under 2 or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 54.DiCiccio TJ, Efron B. Bootstrap confidence intervals. Stat Sci. 1996;11(3):189–212. doi: 10.1214/ss/1032280214. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.