Abstract
Myelofibrosis (MF) is a myeloproliferative neoplasm characterized by cytopenia and extramedullary hematopoiesis, resulting in splenomegaly. Multiple pathological mechanisms (e.g., circulating cytokines and genetic alterations, such as JAKV617F mutation) have been implicated in the etiology of MF, but the molecular mechanism causing resistance to JAK2V617F inhibitor therapy remains unknown. Among MF patients who were treated with the JAK inhibitor ruxolitinib, we compared noncoding RNA profiles of ruxolitinib therapy responders versus nonresponders and found miR-543 was significantly upregulated in nonresponders. We validated these findings by reverse transcription–quantitative PCR. in this same cohort, in 2 additional independent MF patient cohorts from the United States and Romania, and in a JAK2V617F mouse model of MF. Both in vitro and in vivo models were used to determine the underlying molecular mechanism of miR-543 in MF. Here, we demonstrate that miR-543 targets the dioxygenases ten-eleven translocation 1 (TET1) and 2 (TET2) in patients and in vitro, causing increased levels of global 5-methylcytosine, while decreasing the acetylation of histone 3, STAT3, and tumor protein p53. Mechanistically, we found that activation of STAT3 by JAKs epigenetically controls miR-543 expression via binding the promoter region of miR-543. Furthermore, miR-543 upregulation promotes the expression of genes related to drug metabolism, including CYP3A4, which is involved in ruxolitinib metabolism. Our findings suggest miR-543 as a potentially novel biomarker for the prognosis of MF patients with a high risk of treatment resistance and as a potentially new target for the development of new treatment options.
Keywords: Hematology, Oncology
Keywords: Bone marrow, Molecular diagnosis, Noncoding RNAs
miR-543 is identified as a prognostic biomarker for myelofibrosis patients with high risk of treatment resistance.
Introduction
Myelofibrosis (MF) is a hematological malignancy characterized by the expansion of a myeloid neoplastic clone, leading to progressive bone marrow (BM) fibrosis with cytopenia and extramedullary hematopoiesis (1). The most prominent pathological feature in MF is the BM fibrosis. BM fibrosis leads to the deposition of reticulin or collagen fibers that create fibrous scar-like tissue (1), and the extent of BM fibrosis in MF patients has been correlated with clinical manifestations of advanced disease, together with a worse prognosis (1, 2). Inadequate production of blood cells (anemia, thrombocytopenia), bone pain, weakness and fatigue, a high risk of infection and bleeding, night sweats, and progressive spleen enlargement are all characteristics of advanced MF. Patients with MF have a median survival of 4–7 years (3, 4) and an overall median survival of 5.75 years after diagnosis (5).
Somatic mutations in genes such as JAK2 (specific mutation at JAK2V617F in 50%–60% of patients), MPL (thrombopoietin receptor; specific mutation at MPLW515L/K in 5%–10% of patients), and CALR (calreticulin; specific mutation at exon 9 in 20%–30% of patients), among others, have been identified in patients with MF (6–11). These mutations directly result in the constitutive activation of the JAK/STAT pathway. The JAK1/2 tyrosine kinase inhibitor ruxolitinib, the only FDA-approved therapy for MF, significantly improves splenomegaly and patients’ quality of life. However, ruxolitinib does not eliminate BM fibrosis, nor does it eradicate neoplastic clones (12), indicating that other pathogenic players need to be discovered and therapeutically exploited. Currently, allogeneic stem cell transplantation is the only curative therapy. However, fewer than 10% of patients are eligible and treatment-related mortality is common (13). Therefore, novel and effective therapies are needed posthaste.
MicroRNAs (miRNAs) are small, noncoding RNAs with regulatory functions that play a significant role in many human diseases, including cancer (14–20). Emerging studies show that miRNAs can act either as oncogenes or as tumor suppressor genes or sometimes as both (21, 22). Profiling of miRNA expression, both in tissues or circulating biofluids, has enabled advancements in diagnostics, disease staging, monitoring of disease progression, prognosis, and the evaluation of treatment response of various cancers and diseases (23–26). Moreover, miRNAs are proved to have important regulatory roles in hematopoiesis (27–32).
In this study, we hypothesized that response to ruxolitinib is related to deregulated miRNA expression. Therefore, miRNAs could represent one of the missing links for deciphering the mechanism of resistance to ruxolitinib therapy in MF (33). Here, we evaluated a cohort of 107 patients with MF who were treated with ruxolitinib at our center as a part of a phase I–II study and stratified them according to whether they had complete elimination of palpable splenomegaly (“responders”) or had less than 25% reduction in spleen size (“nonresponders”) (34, 35). By profiling the expression of miRNAs, we found the miR-543 cluster on chromosome 14q and the miR-182 cluster on chromosome 7q are deregulated. Notably, we found that miR-543 acts by targeting the epigenetic regulators ten-eleven translocation 1 (TET1) and 2 (TET2), which are members of a family of proteins that play a direct role in the DNA methylation process and gene activation and an indirect role in protein acetylation. We further identified that activation of STAT3 by the phosphorylation of JAK2 influences miR-543 transcription through its direct binding to miR-543 promoter.
Results
Genome-wide expression profiles of specific miRNAs stratify MF patients according to their responsiveness to ruxolitinib treatment.
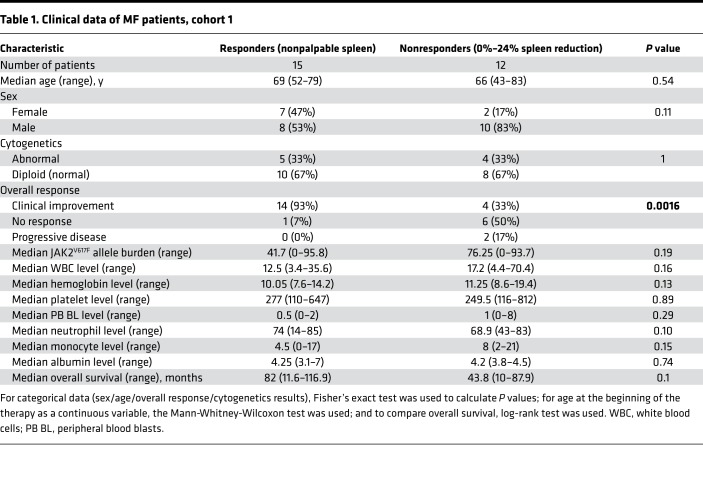
Because miRNAs have been widely explored to assess their clinical value as markers of cancer progression and treatment effectiveness (36), we analyzed the expression of miRNAs in patients with MF treated with ruxolitinib. From a study population of 107 patients with MF treated with ruxolitinib, we focused on the 2 extreme groups: the “responders” (n = 15, with complete elimination of palpable splenomegaly) and the “nonresponders” (n = 12, with 0%–24% spleen size reduction) to ruxolitinib treatment. We rationalized that by analyzing only these opposite groups and eliminating the patients with intermediate benefit, we would be able to identify the functionally most important miRNAs from the many differentially expressed ones in response to ruxolitinib. The disease progression of these patients was monitored for up to 4 years. The clinical features of the patients with MF in this study are described in Table 1. To identify the miRNAs differentially expressed between the 2 groups, we selected 7 patients from each group that had sufficient high-quality RNA available and performed Affymetrix miRNA microarrays for baseline (before the start of the treatment) BM samples. The analysis revealed that 227 miRNAs were significantly downregulated, while 164 were upregulated in the nonresponders (P < 0.05). Among these, we selected the miRNAs that had at least 2.5-fold difference relative to the responders and a P value less than 0.01 (8 downregulated miRNAs and 15 upregulated miRNAs; Figure 1A) for further validation with reverse transcription–quantitative PCR (RT-qPCR).
Table 1. Clinical data of MF patients, cohort 1.
Figure 1. miRNA expression profiles in BM of MF patients differ according to ruxolitinib treatment responsiveness, specifically for miR-543 levels.
(A) Heatmap displaying the microRNAs most differently expressed in nonresponders (0%–24% spleen size reduction after ruxolitinib treatment; n = 7) compared with responders (100% or complete elimination of palpable splenomegaly after ruxolitinib treatment; n = 7). Color key represents normalized expression values, where, for each row, red represents the maximum expression value and green the lowest. (B) Differential expression of miR-543 according to responsiveness to ruxolitinib treatment was validated by RT-qPCR in the same cohort of patients (cohort 1). (C) miRNA network analysis of the miRNA array profile from responders (n = 7) and nonresponders (n = 7). (D) Differential expression of miR-543 was further measured by RT-qPCR in an independent cohort of healthy controls (n = 9) and untreated patients with MF (n = 11) (cohort 2). (E) Differential expression of miR-543 was also measured by RT-qPCR in an independent cohort of healthy controls (n = 7) and untreated patients with MF (n = 16) (cohort 3). Nonresponder patients are identified with red triangles and responders with blue circles. Two-tailed t test was used to assess statistical significance for B, D, and E. Each dot represents 1 human sample. All experiments were repeated independently 3 times.
Differentially expressed miRNAs between patients with MF who responded or did not respond to ruxolitinib treatment.
The differential expression of miR-543, miR-382, miR-182, and miR-183 was then supported, by RT-qPCR, in the extended group (n = 27) of samples of responders and nonresponders, including the 14 tested by microarray. Our results showed that miR-543 (Figure 1B) and miR-382 (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121781DS1) were significantly upregulated in nonresponders (n = 12) versus responders (n = 15) (cohort 1), whereas miR-182 and miR-183 were significantly downregulated (Supplemental Figure 1, B and C). When analyzing miR-543 expression profiling for the available patients with MF developed from essential thrombocythemia (ET) or polycythemia vera (PV), we found no statistical difference in the number of responders (2 with previous PV and 4 with previous ET) versus nonresponders (4 with previous PV) (data not shown). Because miR-182 and miR-183 belong to the miR-96/182/183 cluster located within the 5-kb region on human chromosome 7q32.2, we tested the expression of miR-96 also and found it to be significantly downregulated in nonresponders also (Supplemental Figure 1D). miR-382 is part of the miRNA cluster B at 14q32, encompassing 42 miRNAs, out of which 15 were identified by the array as differentially expressed between the 2 groups. Overall, we identified miR-543 and miR-382 to be significantly overexpressed, while miR-96, miR-182, and miR-183 were significantly decreased in nonresponders when compared with ruxolitinib treatment responders.
Lower connectivity of the miRNA network is present in responders to ruxolitinib.
As a second approach for identifying the functionally most relevant miRNAs among the 23 found to be differentially expressed by microarray, we built an expression network for these miRNAs (37). This represents a high-level correlation between the miRNA expression in a given group of patients and is composed of nodes (the miRNAs) and of edges (high correlations between the expression of 2 miRNAs) (38, 39). The miRNA network of nonresponders is far more connected compared with responders (Figure 1C): it contains 104 edges, being a well-connected network with 14 nodes with over 10 edges each (nodes with high node connectivity, 0.4783–0.6087). On the other hand, responders’ miRNA network is a less connected network, with 57 edges and a lower node connectivity at 0.4783. This network contains 6 isolated nodes and 2 other nodes that are linked only to each other (miR-485 and miR-543). The hubs of responders’ network have a lower number of connections compared with the hubs of nonresponders’ network. The connectivity of the nodes of responders network was significantly lower compared with nonresponders network (P = 0.0137; Supplemental Figure 2). When analyzing the behavior of single miRNA nodes, we observed that a group of miRNAs with low connectivity in responders’ network (0 edges or 1 edge) are key elements of the nonresponders’ network, containing 14 edges each with an increase in the node connectivity. The miRNAs that showed the highest increase in the number of edges between the 2 groups are miR-100 and miR-134, with 14 edges in the nonresponders’ network, and miR-543 and miR-487a, with 13 edges. We can conclude that response to treatment implies a dramatic reorganization of the miRNA network, with the exclusion of 6 important nodes having a marked decrease in the number of edges. Very interestingly, a different research group also used miRNA microarray expression data with in silico target prediction data to build miRNA-gene networks for primary MF and identified miR-543 as the most connected node of the network (40).
Because our interest was specifically to establish why certain patients do not respond to the ruxolitinib therapy, and the only miRNA in common between the confirmed significantly differentially expressed genes and the network analysis studying the most network connected miRNAs independently of the expression was miR-543, as a potentially key element of the nonresponder’s miRNA network, we directed our focus toward the overexpression of miR-543.
miR-543 is overexpressed in MF samples from independent cohorts of patients.
We next assessed the expression of miR-543 in BM cells derived from patients with MF in comparison with BM cells from healthy individuals. We observed that this miRNA was significantly overexpressed in an independent MF patient cohort (cohort 2) when compared with healthy controls (Figure 1D and Supplemental Table 1). Similar results were obtained when measuring the expression levels of miR-543 in an independent cohort (cohort 3) (Figure 1E and Supplemental Tables 1 and 2). Furthermore, we analyzed the publicly available data sets (41) (microarray GSE53482) of CD34+ cells from primary MF (n = 42) and healthy donors (n = 31) and detected a significant overexpression of miR-543 in CD34+ cells from patients with MF (Supplemental Figure 3A). Survival analysis showed a trend of lower overall survival in MF patients with high miR-543 levels compared with those with low levels (Supplemental Figure 3B). Taken together, these data from multiple sets of patients from different institutions confirm that miR-543 is overexpressed in MF, with the highest expression in nonresponders to ruxolitinib.
Functional role of miR-543 in transcriptomic profile.
To determine the role of miR-543 in MF, we performed target prediction of the miRNA binding sites using multiple algorithms (9 algorithms as described in the Methods section) and selected those that were strongly predicted (at least in 6/9 algorithms) or experimentally validated (regardless of the target prediction results) using miRTarBase database. Pathway analyses of the miR-543 predicted targets are shown in Supplemental Table 3.
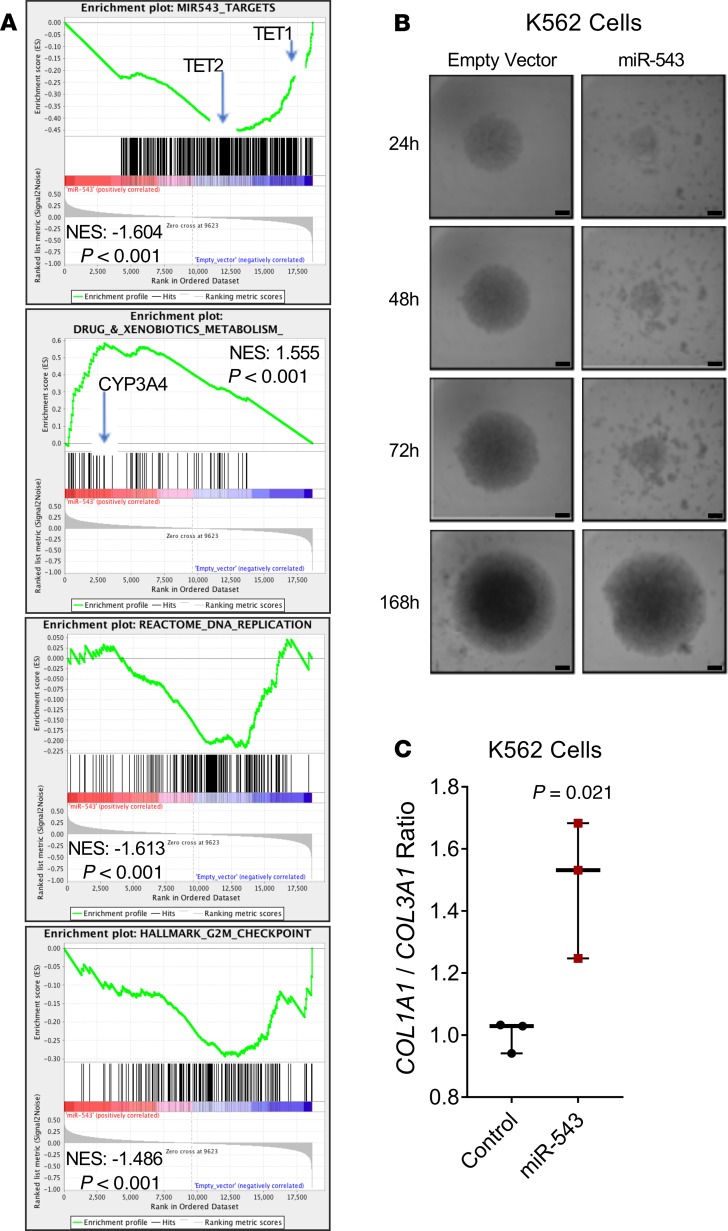
In the absence of established MF cell lines, we generated K562 (human chronic myelogenous leukemia) and OCI-AML3 (human acute myeloid leukemia) cells with stable overexpression of miR-543 (Supplemental Figure 4, A and B) to study the role of miR-543 in MF in vitro. Enrichment analysis of gene expression microarrays (GEMs) of K562 cells overexpressing miR-543 showed that the enrichment of selected miR-543 target genes is reduced as expected (Figure 2A, and Supplemental Table 4). Interestingly, overexpression of miR-543 promotes an increased enrichment of genes related to drug and xenobiotic metabolism and a decreased enrichment of genes related to DNA replication and to G2/M checkpoint (Figure 2A and Supplemental Tables 5–7). Using the available RNA samples from the patients analyzed for the miRNA array analysis, we performed transcriptomic profiling for GEM analyses. These functional analyses showed, in nonresponders, a decreased enrichment of about 53% of the selected miR-543 target genes used above and increased enrichment of genes related to drug and xenobiotic metabolism (Supplemental Figure 5). Collectively, these concordant data from stable clones and patients indicated that high miR-543 expression could be considered a prognosis marker and might be involved in MF progression.
Figure 2. miR-543 promotes drug metabolism–related gene enrichment and disrupts cell sphere formation.
(A) Gene enrichment analysis in K562 cells overexpressing miR-543. (B) The effect of miR-543 in formation and growth of 3D multicellular tumor spheroids (MCTS) was monitored at different time points for 7 days (24, 48, 72, and 168 hours) in K562 cells overexpressing miR-543 compared with those cells stably transduced with empty vector. Scale bars: 300 μm. (C) Expression ratio of COL1A1/COL3A1 transcripts in K562 cells overexpressing miR-543 compared with those cells stably transduced with empty vector. GAPDH and 18S levels were used as normalizers. Statistical differences were determined with 2-tailed t test. All experiments were repeated independently 3 times, and representative blots are shown. NES, normalized enrichment score.
Role of miR-543 in 3D multicellular spheroid formation.
The overexpression of miR-543 also markedly delayed the formation and growth of 3D multicellular tumor spheroids (MCTS) in K562 cells, and to a lesser extent in OCI-AML3 cells, on low adherent round-bottom plates (Figure 2B and Supplemental Figure 6, A and B), showing that this miRNA is involved in tissue structural formation of hematopoietic cells. Moreover, we found that overexpression of miR-543 in K562 cells caused increase of the COL1A1/COL3A1 mRNA ratio (Figure 2C), which is a key characteristic in lung (42) and liver (43) fibrosis. Both genes encode the collagen found in extensible connective tissues, being involved in tissue structural formation (44, 45).
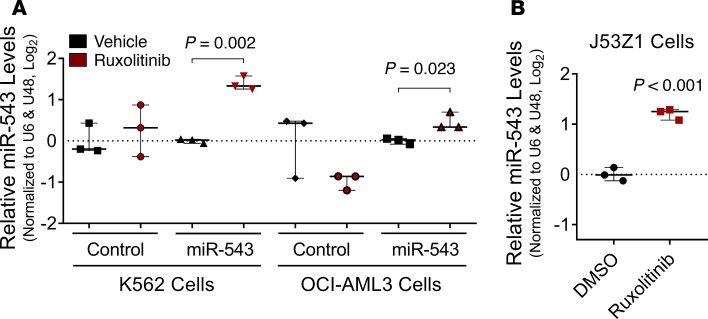
miR-543 levels are increased by ruxolitinib and by JAK2V617F mutation.
To evaluate whether ruxolitinib treatment affects the expression of miR-543, we stimulated with this drug the wild-type K562 and OCI-AML3 cells and the respective isogenic stable clones overexpressing miR-543, as well as the J53Z1 erythroid cells isolated from the JAK2V617F p53–/– mouse (46). We found that ruxolitinib treatment induced significantly higher expression levels of miR-543 in cells with already overexpressed miR-543 and in J53Z1 cells containing JAK2V617F mutation (Figure 3, A and B).
Figure 3. Ruxolitinib treatment induces increase in miR-543 levels when miR-543 and JAK2V617F mutation are present.
(A) Expression of miR-543 after treatment with ruxolitinib measured by RT-qPCR in K562 cells and OCI-AML3 cells stably transfected with empty vector (control) or p–miR-543 expression vector. U6 and RNU48 levels were used as normalizers. (B) Expression of miR-543 after treatment with ruxolitinib measured by RT-qPCR in J53Z1 cells containing the JAK2V617F mutation. U6 and RNU48 levels were used as normalizers. Comparisons were made using 2-tailed t test or Mann-Whitney-Wilcoxon nonparametric test. All experiments were repeated independently 3 times, and representative blots are shown.
To evaluate the role of miR-543 in MF in vivo and the possible role of the JAK2V617F mutation in miR-543 regulation, we evaluated the expression of miR-543 in JAK2V617F-transgenic (JAK2V617F-TG) mice that develop age-dependent MF (Figure 4A; mice were 60–70 weeks old with fully developed disease) (47, 48). We detected significantly higher expression levels of the miR-543 in BM cells of JAK2V617F-TG mice, compared to the wild-type mice of the same age (Figure 4B). In the JAK2V617F-TG mice of a younger age (13–22 weeks), suffering from PV, the expression of miR-543 was inconsistent and modest (data not shown), suggesting that miR-543 overexpression may be an MF-specific phenotype, as supported also by the patients’ data.
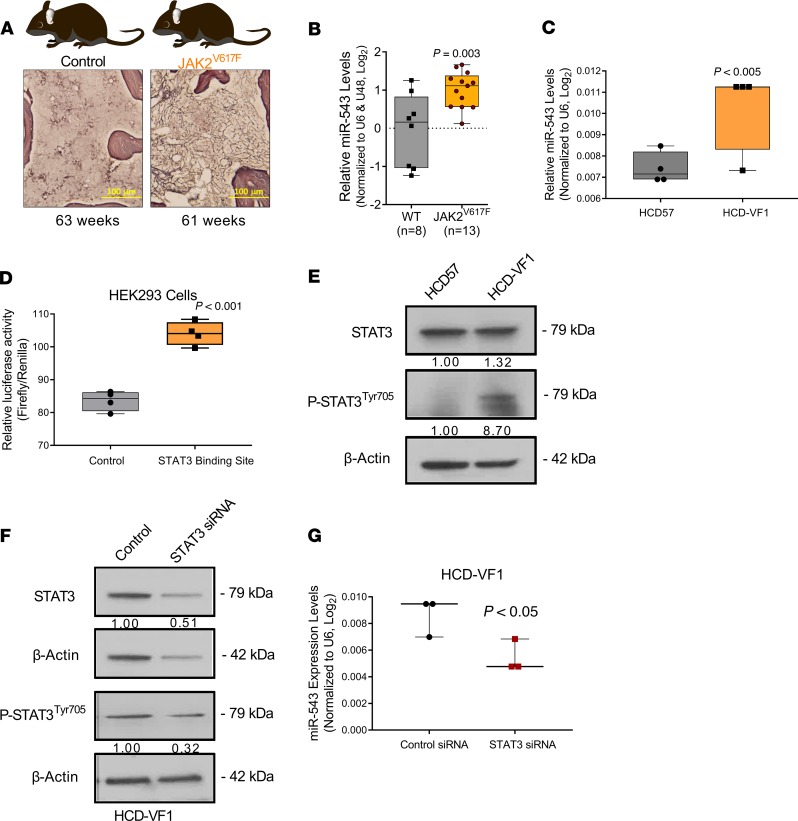
Figure 4. JAK2V617F mutation promotes increase in miR-543 levels.
(A) Representative diagram of the JAK2V617F-transgenic mouse model of MF and the representative histology slides of the BM tissue. Scale bars: 100 μm. (B) Differential expression of miR-543 measured by RT-qPCR in wild-type control mice and in JAK2V617F-transgenic mice. (C) Differential expression of miR-543 measured by RT-qPCR in the HCD57 parental control cells and HCD57 erythroleukemia cells containing the JAK2V617F mutation (HCD-VF1). U6 levels were used as normalizer. (D) Relative luciferase activity in HEK293 cells transiently cotransfected with the control vector and miR-543 promoter sequence with putative STAT3 binding site. (E) Expression levels of STAT3 and p-STAT3 Tyr705 were determined by Western blot analysis in HCD57 and HCD-VF1 cells. β-Actin was used as a loading control. (F) Expression levels of STAT3 and p-STAT3 Tyr705 were determined by Western blot analysis in HCD-VF1 cells in which STAT3 was silenced by siRNA. β-Actin was used as a loading control. (G) Silencing of STAT3 in HCD-VF1 erythroleukemia cells led to decreased miR-543 expression levels as assessed by RT-qPCR. Two-tailed t test was used to determine statistical significance for B and D. Mann-Whitney-Wilcoxon nonparametric test was used for C and E. Each dot represents 1 animal. All experiments were performed independently 3 times, and representative blots are shown.
To further determine whether the JAK2V617F mutation is involved in the regulation of miR-543 expression, we compared the expression level of miR-543 in HCD57 erythroleukemia cells containing the JAK2V617F mutation (HCD-VF1) with the parental HCD57 cells. We found that the HCD-VF1 cells had significantly higher expression of miR-543 in comparison with the HCD57 parental cells, supporting the connection between the presence of JAK2V617F mutation and the high expression of miR-543 (Figure 4C).
Activation of STAT3 induces miR-543 expression through promoter binding.
We decided to investigate whether JAK2V617F mutation may control miR-543 expression. Because STAT3 is a transcription factor that JAK2 activates (49, 50), we investigated the existence of STAT3 binding sites in the surrounding genomic regions of miR-543. miR-543 is located at chr14:101,031,987-101,032,064. This genomic region is enriched for several miRNAs; however, no host genes have been annotated for miR-543 (UCSC Genome Browser on Human Dec. 2013, GRCh38/hg38). In silico analysis of RNA-Seq reads from normal tissue and cell lines identified potential host gene transcriptional start sites within the genomic region chr14:101,012,000-101,021,000. In proximity to this region, 4 putative STAT3 binding sites were located (UCSC Genome Browser, ENCODE 3). Sequence analysis of these regions identified 1 STAT3 transcription factor consensus binding site (TTCCCGaAA, chr14:101,001,972-101,001,980) (51). From this region, we subcloned a 300-bp fragment containing the consensus site into the promoter region of a luciferase vector and performed luciferase-based reporter gene assay in HEK293 cells. Transient transfection of the vector containing the STAT3 putative binding site induced an increase in the luciferase activity when compared with the empty vector as control (Figure 4D). Thus, activation by phosphorylation of STAT3 through JAK2V617F mutation leads to stimulation of miR-543 expression. To confirm this regulation, we performed 2 sets of experiments. First, using the HCD-VF1 cells containing the JAK2V617F mutation, we found a higher STAT3 and p-STAT3Tyr705 protein expression compared with the parental nonmutated cell line (Figure 4E). Second, we examined whether STAT3 knockdown can affect expression of miR-543: in vitro siRNA transfection using STAT3 siRNA or control siRNA in HCD-VF1 cells (Figure 4F) significantly reduced the expression of miR-543 in HCD-VF1 cells when compared with siRNA control–transfected cells (Figure 4G). Overall, JAK pathway activation by JAK2V617F mutation initiates transcription of miR-543 through STAT3 transcription factor.
Role of miR-543 in targeting both TET1 and TET2.
Out of the thousands of predicted mRNA targets by multiple algorithms, we selected for further investigation TET1 and TET2 because both proteins have been previously reported to have functionally significant roles in the development of fibrosis (52–54). Our functional analysis of selected miR-543 target genes presented above showed TET1 and TET2 among the genes with significant decrease in gene enrichment (as identified with arrows in Figure 2A, and Supplemental Figure 5, left). Furthermore, loss-of-function mutations in Tet2 account for 5%–17% of myeloproliferative neoplasms (MPNs) (55, 56). Moreover, loss of Tet2 in combination with JAK2V617F expression in MPN mouse models appears to render a competitive advantage to hematopoietic stem cells, resulting in a more aggressive disease (57–59). Therefore, we evaluated the expression of these 2 proteins by Western blot analysis in BM samples from patients belonging to ruxolitinib treatment responder and nonresponder groups (Figure 5A). We also measured the expression of miR-543 in the PB samples from the same patients (Figure 5A) and observed an inverse correlation between the expression of miR-543 and the expression of the 2 proteins, TET1 and TET2 (88.8% of the cases; 8/9). All patients were positive for the JAK2V617F mutation, and from these 9 patients, 8 adhered to the characteristics of the group they belonged to (nonresponder, high miR-543; responder, low miR-543). This suggests that the majority of patients who do not respond to ruxolitinib therapy have loss of TET1 and TET2 expression potentially via miR-543 upregulation, in addition to having the JAK2V617F driver mutation.
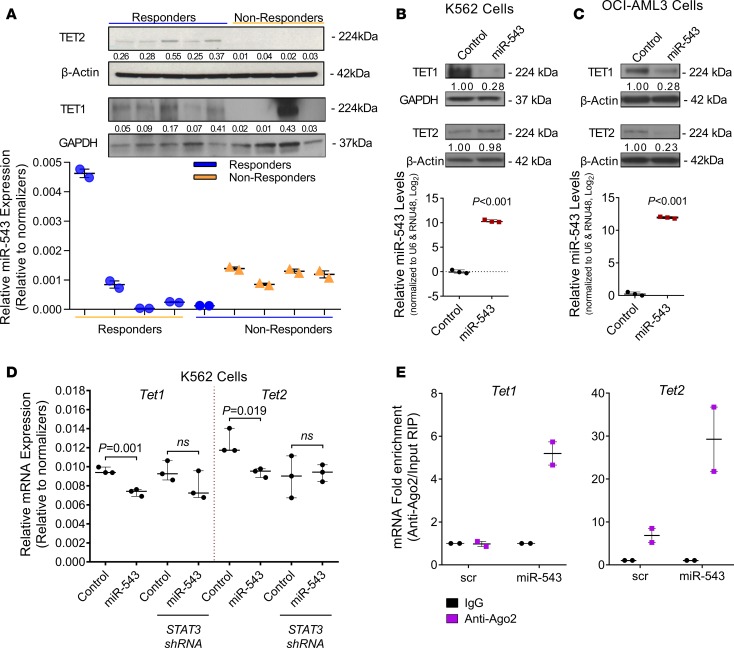
Figure 5. TET1 and TET2 are direct targets of miR-543.
(A) Top: Western blot analysis of TET1 and TET2 proteins in BM samples from patients with MF positive for the JAK2V617F mutation who responded to ruxolitinib treatment (complete elimination of palpable splenomegaly, low miR-543) or did not respond to ruxolitinib treatment (0%–24% spleen size reduction, high miR-543). GAPDH (for TET1) or β-actin (for TET2) protein levels were used as normalizers. Bottom: Expression levels of miR-543 in the peripheral blood (PB) samples derived from the same patients. Relative expression of miR-543 was measured by RT-qPCR and normalized to U6 and RNU48 levels. (B) Representative Western blot analysis of TET1 and TET2 proteins in K562 cells stably transduced with empty vector (control) or p–miR-543 expression vector. GAPDH or β-actin protein levels were used as normalizers. Relative expression of miR-543 measured by RT-qPCR and normalized to U6 and RNU48 levels (n = 3; bottom). (C) Representative Western blot analysis of TET1 and TET2 proteins in OCI-AML3 cells stably transduced with empty vector (control) or p–miR-543 expression vector. Vinculin or β-actin protein levels were used as normalizers. Relative expression of miR-543 measured by RT-qPCR and normalized to U6 and RNU48 levels (n = 3; bottom). (D) Expression of TET1 or TET2 in K562 cells stably transduced with STAT3 shRNA or control shRNA treated with ruxolitinib after being transiently cotransfected with the miR-543 mimic or control miRNA (scramble mimic). Results presented as relative mRNA levels relative to the geometric mean of the 2 normalizers: β-actin and GAPDH. (E) Levels of TET1 and TET2 mRNAs bound to the Ago2 complex were pulled down with IgG or anti-Ago2 antibody from HEK293 cells transiently transfected with empty vector (control) or with p–miR-543 expression vector. Two-tailed t test was used to assess statistical significance for B and C. Mann-Whitney-Wilcoxon nonparametric test was used for D. All experiments were performed independently 2 or 3 times, and representative blots are shown. Ago2, protein argonaute-2; RIP, RNA immunoprecipitation; NS, not significant.
To validate whether miR-543 indeed targets TET1 and TET2 mRNAs, we assessed their protein expression in K562 and OCI-AML3 cells overexpressing miR-543 (Figure 5, B and C, and Supplemental Figure 7, A and B) and after inhibition of miR-543 in SET2 cells (human ET cell line, carrying the JAK2V617F mutation with high miR-543 expression) by transient transfection with mirVana miR-543 inhibitor (Supplemental Figure 7C). We observed a clear downregulation of TET1 in cells overexpressing miR-543 compared with control cells for the K562 clones, but we could not confirm the changes in TET2 protein levels (Figure 5B and Supplemental Figure 7A). The downregulation of both TET1 and TET2 with miR-543 overexpression was confirmed in OCI-AML3 cells (Figure 5C and Supplemental Figure 7B). Additionally, the transient downregulation of miR-543 in SET2 cells showed inverse correlation between the expression of the miRNA and the 2 proteins, TET1 and TET2 (Supplemental Figure 7D). Because STAT3 can induce the transcription of miR-543 through binding to its promoter, we tested the role of STAT3 in the miR-543–promoted changes of TET1 and TET2 mRNA levels. We specifically assessed the TET1 and TET2 mRNA levels in K562 cells with STAT3 knockdown by shRNA with an miR-543 mimic versus control random sequence. We found that miR-543 mimic statistically significantly decreased both TET1 and TET2 mRNA levels in K562 cells, whereas after the knockdown of STAT3 by shRNA, the mimic could not significantly decrease TET2 or TET1 mRNA levels (Figure 5D).
To validate the direct interaction between miR-543 and TET1/TET2 mRNAs, we immunoprecipitated the RNA bound to Ago2 in transfected HEK293 cells overexpressing miR-543 and in control cells with low endogenous miR-543 expression. We detected 5.3- and 4.3-fold more TET1 and TET2 mRNA, respectively, bound to the Ago2 protein in the cells with high miR-543 expression, confirming the direct interaction of the miRNA with its targets (Figure 5E).
Additionally, we detected lower TET2/miR-543 ratios of expression (by ISH and IHC, respectively) in BM histological samples derived from the JAK2V617F-TG mice (40–70 weeks old) compared with the wild-type mice (Supplemental Figure 8), which translates into lower TET2 protein expression when correlated with high miR-543 expression. We did not observe statistically significant differences in TET1/miR-543 expression ratios, potentially because of the limited number of mouse specimens available for analysis. Therefore, the association between miR-543 and TET2 can be validated in an age-dependent JAK2V617F-induced MF mouse model, suggesting a role of the miR-543/TET2 axis in the development of MF in vivo.
Epigenetic effects of miR-543.
TET1 and TET2 are dioxygenases responsible for the conversion of 5-methyl cytosine (5-mC) into 5-hydroxymethyl cytosine (5-hmC), thus controlling DNA demethylation (60). Considering that the canonical function of TET1 and TET2 is to regulate DNA methylation, we analyzed the levels of 5-mC and 5-hmC DNA in cells with altered miR-543 expression. Overexpression of miR-543 in K562 cells led to an increased percentage of the global 5-mC (genome wide; Figure 6A) but no significant changes to the percentage of 5-hmC. Overexpression of miR-543 in K562 cells also led to an increased percentage of the long noncoding RNA H19 DMR of 5-mC (at the H19-S3 genomic locus), but knockdown of TET1 or TET2 did not completely inhibit this miR-543–induced effect (Figure 6A). Therefore, this indicates decreased activation of DNA demethylation pathways. These results suggest that miR-543 does not primarily affect the hydroxymethylation function of TET proteins, but their decreased levels are causing an enhanced global DNA methylation, which corresponds to reduced transcription of genes (61).
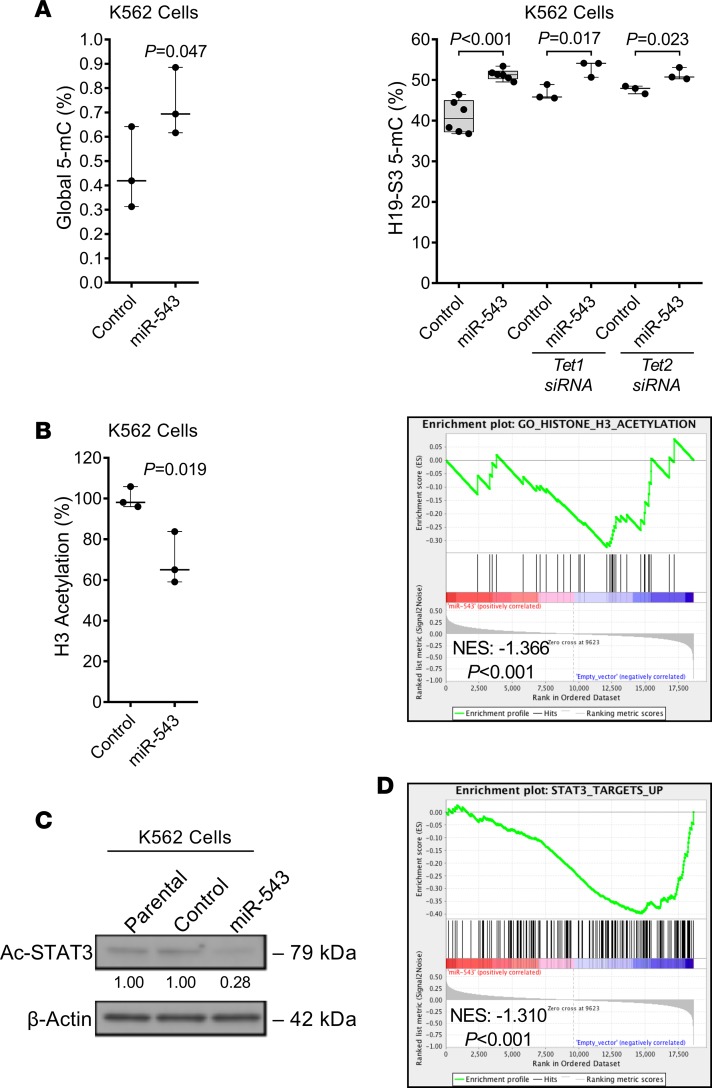
Figure 6. miR-543 leads to decreased protein acetylation.
(A) Percentage of overall 5-mC methylation levels in K562 cells stably transduced with empty vector (control) or p–miR-543 expression vector. Percentage of H19 differentially methylated region (DMR) 5-mC methylation levels in K562 cells stably transduced with empty vector (control) or p–miR-543 expression vector after being transiently transfected with TET1 or TET2 siRNA. (B) Percentage of overall histone 3 (H3) acetylation levels in K562 cells stably transduced with empty vector (control) or p–miR-543 expression vector. Gene enrichment analysis of genes related to H3 acetylation in K562 cells overexpressing miR-543. (C) Representative Western blot analysis of acetylated STAT3 protein in K562 parental cells and stably transduced with empty vector (control) or p–miR-543 expression vector. β-Actin protein levels were used as normalizer. (D) Gene enrichment analysis of genes upregulated by STAT3 pathway activation in K562 cells overexpressing miR-543. Two-tailed t test or Mann-Whitney-Wilcoxon nonparametric test was used to assess statistical significance for A and B. All experiments were repeated independently 3 times, and representative blots are shown.
Recently published data show the indirect role of TET1 on protein acetylation (e.g., H4K16 acetylation) (62). We identified that overexpression of miR-543 decreased global histone 3 acetylation, supported by a significant decrease in the enrichment of genes related to histone 3 acetylation in K562 cells (Figure 6B). Surprisingly, we also found that overexpression of miR-543 in K562 cells decreased STAT3 acetylation (Figure 6C and Supplemental Figure 9A), supported by a significant decrease in the enrichment of STAT3 target genes actively transcribed when STAT3 signaling is stimulated (Figure 6D). Accordingly, miR-543 inhibition in SET2 cells increased the acetylation of STAT3 (Supplemental Figure 9B). Notably, overexpression of miR-543 in the BM cells from patients with MF and in K562 cells showed a decrease in the levels of acetylated p53 protein (Supplemental Figure 9, C and D), supported by a significant decrease in the enrichment of genes related to p53 pathway activation (Supplemental Figure 9E). Interestingly, phosphatase and tensin homolog (PTEN) protein level, which is known to be decreased in primary MF (63), was decreased in representative ruxolitinib-resistant patients with high miRNA-543 levels (Supplemental Figure 10A), yet increased in SET2 cells upon miR-543 inhibition (Supplemental Figure 10B). These findings were corroborated with enrichment analysis of K562 cells overexpressing miR-543 that showed a significant increase in the enrichment of genes transcriptionally active when PTEN is downregulated (Supplemental Figure 10C). These data suggest that overexpressed miR-543 targets proteins (e.g., TET) that may have a global effect on protein acetylation (e.g., histone 3, STAT3, and p53) that leads to the development of MF and resistance to ruxolitinib.
Discussion
Ruxolitinib improves spleen size, symptoms, and overall prognosis in many patients with MF but to different extents and for different durations. In this study, we generated differential miRNA profiles, showing increased (e.g., miR-543, miR-382) and decreased (e.g., miR-182, miR-183) miRNAs in patients with MF who did not respond to ruxolitinib treatment. In parallel with our study, other groups showed that miR-543 is deregulated in MF granulocytes (64) and in PB CD34+ cells (40), validating the importance of miR-543 in the pathophysiology of MF. Collectively, our results demonstrate that increased levels of miR-543 correlate with ruxolitinib treatment “resistance” in patients with MF by promoting genes related to ruxolitinib metabolism (e.g., CYP3A4) and by directly targeting the TET1 and TET2 transcripts. This regulatory axis was found in the JAK2V617F-TG MF mouse model. Somatic mutations in TET2 have been described in about 19%, 12%, 24%, and 22% of patients with myelodysplastic syndromes, myeloproliferative disorders, secondary acute myeloid leukemia (AML), and chronic myelomonocytic leukemia, respectively (65). Here we show that patients with MF who do not respond well to ruxolitinib therapy have high levels of miR-543. This miR-543–mediated effect also correlates with the downstream effects that show decreases in protein levels of acetylated STAT3, PTEN, acetylated p53, and acetylated histone-3. PTEN is known to be decreased in MF (63).
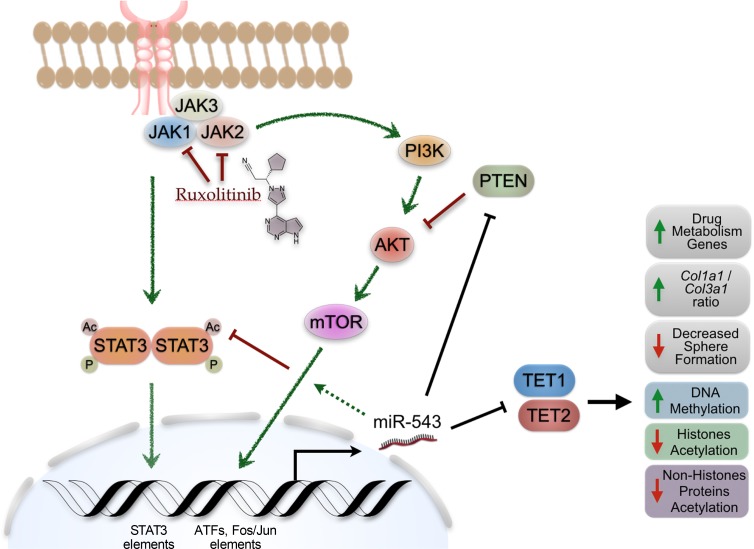
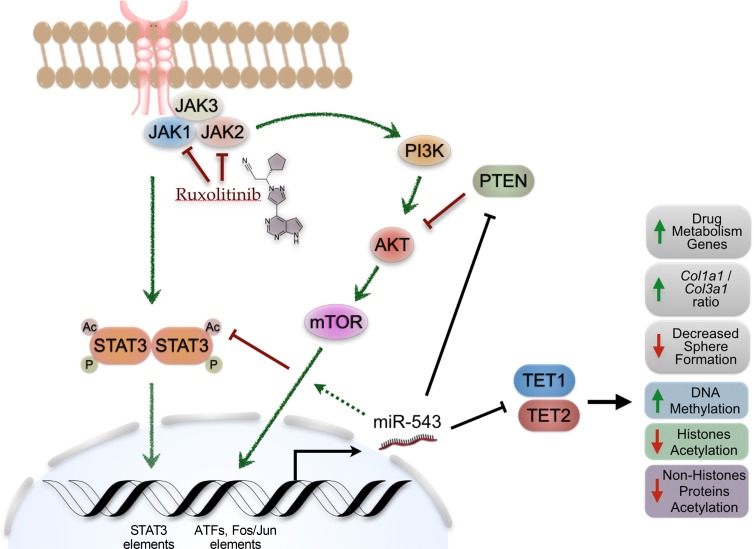
Interestingly, our results showed that constant activation of the JAK2/STAT3 signaling pathway by the JAK2V617F mutation induces an increase in miR-543 expression. Genomic screening of the DNA region upstream of the miR-543 host gene shows a STAT3 element, which suggests a direct role of STAT3 in regulating miR-543 expression. We found that phosphorylation of STAT3 by mutant JAK2 can control miR-543 expression epigenetically; however, inhibition of JAK2/STAT3 signaling by ruxolitinib can further induce miR-543 expression levels when miR-543 is already overexpressed. These observations, with unpublished data from our group showing that the upstream region of the miR-543 host gene also contains several other DNA elements for the binding of downstream effector transcription factors (e.g., ATFs, FOS/JUN) from other oncogenic signaling pathways (e.g., PI3K/AKT), strongly suggest that the regulation of miR-543 expression is targeted by multiple signaling pathways related to MF (Figure 7). Further research is needed to describe in detail the molecular regulation of miR-543 expression. However, the main scope of this work was to describe the role of miR-543 in the resistance to ruxolitinib treatment in MF.
Figure 7. Proposed model of the role of miR-543 in MF.
In the K562 cells overexpressing miR-543, we observed decreased STAT3 acetylation, which would decrease STAT3 signaling pathway activation. This decrease in STAT3 acetylation was corroborated with the decrease in the enrichment of target genes actively transcribed by STAT3. This suggests that the miR-543–promoted decrease in activation of the STAT3 downstream pathway may reduce the magnitude of the effect of ruxolitinib treatment in the inhibition of the signaling activation. In addition to the decrease in the magnitude of the intended molecular effect of ruxolitinib, the miR-543 can induce the enrichment of transcription of genes related to drug and xenobiotic metabolism. Therefore, miR-543 can decrease the ruxolitinib effect and half-life, resulting in an overall lack of the expected systemic responses (e.g., decrease in palpable splenomegaly).
Recently, it has been shown that TET1 can activate the transcription and expression of PTEN by direct binding to its promoter region, leading to demethylation of CpG islands (66). Moreover, in silico analysis demonstrated that miR-543 is predicted to target PTEN, and PTEN is known to be downregulated in primary MF (41, 63). There is a suggested crosstalk of the AKT signaling pathway linking PTEN to the hypoxia response and to the cAMP-related calcium-linked cellular processes (63). Additionally, it has been shown that TET1 has a role in protein acetylation (e.g., H4K16 acetylation) (62). Other groups have shown that miR-543 can suppress colorectal cancer growth by targeting KRAS and metastasis by targeting MTA1 and HMGA2 (67), and it can promote gastric cancer cell proliferation by targeting SIRT1 (68), which is a deacetylase of tumor protein p53 (TP53). Here we demonstrated that miR-543 induces decreased levels of TET1, correlating with decreased protein acetylation (e.g., acetylated histone 3, acetylated STAT3, and acetylated TP53 protein levels). Recently, Liu et al. investigated the involvement of miR-543 in 5-fluorouracil (5-FU) resistance in colon cancer cells and showed that downregulation of miR-543 significantly increased the sensitivity of 5-FU through suppressing the PTEN/PI3K/AKT signaling pathway in colorectal cancer cells (69). We also showed the negative correlation between miR-543 expression and TET1 and TET2 protein levels, demonstrating that TET1/2 are targeted directly by miR-543. However, the downregulation of both TET proteins in K563 and OCI-AML3 cells overexpressing miR-543 was not simultaneously observed. Our results suggested that this is most probably due to the presence of different mutations in epigenetic modifiers or cancer drivers between the OCI-AML3 and K562 cell lines (Supplemental Tables 8 and 9). OCI-AML3 cells are DNMT3A mutated (DNMT3AR882C) while K562 cells are wild-type. In vitro experiments indicate that the DNMT3AR882C mutation acts in a dominant negative manner to disrupt the de novo methyltransferase activity of wild-type homotetramers (70). AML patient BM harboring DNMT3AR882C mutations was similarly demonstrated to be hypomethylated when compared with patients with wild-type DNMT3A (71). These aforementioned studies also indicated that non-DNMT3AR882C mutations may act in a functionally distinct manner from DNMT3AR882C mutations. Independent prognostic outcomes and treatment protocols may need to be considered for these 2 classes of DNMT3A mutations.
The limitation of the research work presented here is the small number of patients treated with ruxolitinib. We only focused on 2 very different groups — that is, the complete responders (100% spleen size reduction) and the nonresponders (0%–24% spleen size reduction) to ruxolitinib treatment. We rationalized that by analyzing the “exceptional” responders and the worst responders, we would be able to identify the most important miRNAs from the many differentially expressed ones. Because of the limited amount of material and variable quality of RNA from clinical samples, we could ultimately use only 7 patient samples per group that preserved the RNA quality suitable for further miRNA array profile and protein analyses. However, using the data from patients, in vitro models, and an in vivo TG mouse model, our study provides a global overview showing the role of specific miRNAs, such as miR-543, involved in the transcriptional and posttranscriptional dysregulations of genes that are usually found mutated in patients with MF. In conclusion, we propose miR-543 as a potentially novel biomarker for MF patients with a high risk of resistance to ruxolitinib and as a potential specific target for RNA-based therapeutics.
Methods
Patients and sample collection.
BM samples used for miRNA profiling were collected from patients with MF recruited at the Department of Leukemia at The University of Texas MD Anderson Cancer Center (UT-MDACC) and at the Department of Leukemia at the University of Medicine and Pharmacy in Cluj-Napoca (UMF Cluj-Napoca, Romania). There were 107 MF patients recruited at UT-MDACC (34, 35, 72, 73). Before the treatment regimen, BM cells were collected and stored for research purposes (baseline samples). Spleen reduction was used as the main measure of therapy response. This study focused on 2 subgroups, those with 0%–24% spleen size reduction and no clinical improvement (representing the nonresponders to ruxolitinib treatment, n = 12) and those with 100% spleen size reduction (representing the responders, n = 15), accounting for 27 patients analyzed. PB samples for protein analysis were also collected from these patients. Control samples from healthy control individuals were obtained from external companies in the United States and Romania.
RNA extraction and miRNA and mRNA expression analyses.
Total RNA was extracted from BM aspirates and PB samples of patients with MF, as well as cell lines using TRIzol (Invitrogen, Thermo Fisher Scientific) by following the manufacturer’s protocol as has been previously described (74, 75).
RNA samples extracted from BM aspirates of patients with MF were analyzed using an Affymetrix GeneChip miRNA 3.0 Array. RNA samples extracted from cell lines’ stable clones were analyzed using an Affymetrix GeneChip Clariom S array, human. All microarray data are available at the National Center for Biotechnology Information’s Gene Expression Omnibus database under the accession number GSE142240. The CEL files generated from Affymetrix DNA microarray image analysis software were processed through Transcriptome Analysis Console 4.0, which normalizes (and applies the log2 function to) array signals using a robust multiarray averaging algorithm. Differential expression of miRNAs and mRNAs in comparative analysis was determined by a fold change in absolute value equal or greater to 1.1, a P value obtained from the moderated t-statistic from LIMMA package less than 0.05, and a false discovery rate less than 0.1. To support visual exploration of the miRNA data, a heatmap was generated using the heatmap.2 function of gplots library. Gene set enrichment analysis (GSEA) of differentially expressed genes was performed using GSEA software from Broad Institute.
For RT-qPCR validation of the miRNA expression analysis, 10 ng of total RNA was reverse-transcribed and amplified using the TaqMan miRNA Reverse Kit (Applied Biosystems, Thermo Fisher Scientific) with primers/probes specific for each miRNA. RT-qPCR was performed using the synthesized cDNA and SsoFast Probes SuperMix (Bio-Rad Laboratories) as previously described (76–78). Each amplification was performed in triplicate, where Ct values beyond the upper limit of the measuring system were input as 40 (78). miRNA expression was normalized by the ΔCt method, for samples with a Ct less than 40, using the small nuclear RNA U6 and the small nucleolar RNA U48 as reference genes (78). For mRNA analysis, cDNA was synthesized from 1 μg of isolated RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific) according to the manufacturer’s instructions. All gene expression primers were purchased commercially from Thermo Fisher Scientific. mRNA expression data were normalized to GAPDH and 18S.
Generating miRNA networks.
We used the correlation coefficient method to build miRNA networks (38, 39, 79). Briefly, we used the normalized microarray level of the 23 differentially expressed miRNAs to build a correlation matrix of 23 elements for each of the 2 patient groups. The existence of an edge between 2 miRNA nodes depends on the threshold value. As in our previous publication, we chose a high threshold value, a Pearson’s correlation r of ±0.8 (38), which corresponds to a statistically significant P value of 0.027. The value of this threshold determines whether an edge exists between 2 miRNA nodes. We selected the miRNA pairs from the matrix that correlated above the threshold and drew a line between the corresponding nodes of the network. We constructed a differential miRNA network for each patient cohort. To check whether there were any statistical differences between the numbers of edges of the 2 networks, we calculated the node connectivity of each node and used a paired Wilcoxon’s test to evaluate statistical difference between the 2 networks.
Animal model.
JAK2V617F-TG mice were generated by expressing human JAK2V617F in the hematopoietic system under the control of the vav-1 promoter (36). These mice have been crossed with wild-type C57BL/6 mice for over 10 generations. Homozygous line A JAK2V617F mice were used in this study, and wild-type C57BL/6 mice were used as controls as previously described (80, 81).
RIP.
For the TET1 and TET2 mRNA analysis, the RNA bound to the Ago2 protein was immunoprecipitated with the Magna RIP kit (MilliporeSigma) following the manufacturer’s protocol. Briefly, 15,000,000 cells overexpressing either scramble (scr) control or miR-543 were prepared in RIP lysis buffer, and the RNA-protein complexes were immunoprecipitated using anti-Ago2 and normal rabbit IgG (as control) antibodies. RNA was purified using the phenol-chloroform-isoamyl alcohol method and further used for cDNA synthesis using random hexamers, deoxynucleoside triphosphates, and SuperScript III Reverse Transcriptase (all from Invitrogen, Thermo Fisher Scientific). RT-qPCR was performed using the synthesized cDNA and primers specific for TET1 (forward: 5′-GCCCCAAGTCAGAATCCATG-3′; reverse: 5′ TCCACTTCTCCACCTCGATG-3′) and TET2 (forward: 5′-GAGCAGGTCCTAATGTGGCA-3′; reverse: 5′-TTCTGCGAACCACCCACTTA-3′). The fold enrichment was calculated using the 2–ΔCt method, normalized to the input and to the IgG control.
Identification of miR-543 targets.
To predict in silico hsa-miRNA-543 target genes and predicted binding sites, we retrieved experimentally validated miRNA-mRNA interactions from miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw), TarBase database version 8 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex), and miRecords database (http://c1.accurascience.com/miRecords/). We also retrieved miRNA-target interaction information from the miRWalk2.0 database (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk2) that hosts miRNA-target predictions from 12 programs. We selected targets that were predicted by at least 6 algorithms (half the total number of programs checked) or experimentally validated (regardless of the target prediction results). The selected target genes were used as a gene set for GSEA of transcriptomic data from K562 stable clones overexpressing miR-543 in comparison with those with empty vector (see Figure 2A).
Cell culture, transduction, and transfection.
The leukemia cancer cell lines K562 and OCI-AML3 (82) and derivative clones were cultured in RPMI cell culture medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 1% nonessential amino acids, and 1% HEPES buffer. SET2 cells were maintained in RPMI medium with 20% FBS. J53Z1, HCD-57, and HCD-VF1 cell lines were maintained as previously described (46, 83). HEK293 cells were grown in DMEM, supplemented with 10% FBS. All cell cultures were maintained in a humidified environment at 37°C and 5% CO2 and passaged no more than 10 times before the experiments. Before the start of the in vitro experiments, cells were authenticated by MDACC Characterized Cell Line Core Facility and were tested for mycoplasma contamination.
K562 and OCI-AML3 cells overexpressing hsa-miR-543 were generated using the pMIRNA1 lentiviral vector (System Bioscience) by following the manufacturer’s protocol (see Supplemental Methods). Briefly, the hsa-miR-543 precursor was subcloned into the vector, and 293FT cells were transduced with either the pMIRNA-543 vector or the pMIRNA empty vector (control). The virus in the supernatant was collected 48 hours later. K562 and OCI-AML3 cells were infected with the virus, and the GFP+ cells were sorted by FACS. Inhibition of miR-543 expression in SET2 cells was performed by cell transfection with 100 nM of anti–miR-543 mirVana miRNA inhibitor or with 100 nM scr negative control (all from Ambion, Life Technologies), using Lipofectamine 2000 reagent (Invitrogen, Thermo Fisher Scientific), according to the manufacturer’s protocol. Knockdown of STAT3 in HCD-VF1 cells was performed by cell transfection with 50 nM of STAT3 siRNA (MilliporeSigma) or with 50 nM negative control (MilliporeSigma).
3D MCTS formation.
K562 and OCI-AML3 cells were seeded in a 96-well low adherence round-bottom plate (Greiner Bio One) in 100 μL of medium at 10,000 cells per well. Images of the MCTS were captured using the IN Cell 6000 imager (GE Healthcare) at different time points (24, 48, 72, and 168 hours) to monitor their formation and growth. Medium was changed every 72 hours.
DNA isolation, DNA methylation, and histone 3 acetylation measurements.
After transfection, DNA from cell lines was extracted using the Quick-DNA Miniprep Kit (Zymo Research). For DNA methylation measurements, cell pellets were harvested 24 hours after transfection as described above. Then, the DNA samples were processed and DNA methylation or hydroxymethylation was determined using the MethylFlash Methylated Quantification Kit or MethylFlash Hydroxymethylated DNA Quantification Kit following the manufacturer’s recommendation (Epigentek). DNA methylation analysis of the genome was also assessed by pyrosequencing methylation analysis at UT-MDACC Epigenomics Profiling Core Facility. For global histone H3 acetylation measurements, cell pellets were harvested 24 hours after transfections as described above. Then, the DNA samples were processed and global histone H3 acetylation was determined using the EpiQuik Global Histone H3 Acetylation Assay Kit following the manufacturer’s recommendations (Epigentek).
Mutation screening of genes involved in epigenetic regulation (TET1, TET2, IDH1, IDH2, and DNMT3A) and JAK2 was performed in DNA from OCI-AML3, SET2, and K562 cell lines using a standard PCR amplification protocol followed by Sanger sequencing. Sequencing was performed on an ABI Sequencer 3730, and the data were analyzed using SeqMan Pro (DNASTAR, Lasergene). Primer sequences are shown in Supplemental Methods.
Protein analysis.
Protein analyses were performed using lysates from whole-cell pellet samples in radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitor cocktails (MilliporeSigma), as previously described (75). Protein concentration was quantified by the Bradford assay (Bio-Rad), and equal amounts were loaded on 4%–20% (or 4%–15% for TET2 hybridization) acrylamide Criterion TGX precast gels (Bio-Rad) for Western blot analysis. Resolved proteins were semidry-transferred to nitrocellulose membranes, which were then probed overnight with the following anti-human primary antibodies: TET1 (MilliporeSigma, SAB2700730; mouse monoclonal, 1:1,000), TET2 (Cell Signaling Technology, 4510S; rabbit polyclonal, 1:1,000), acetyl-TP53 (Cell Signaling Technology, 2570; rabbit polyclonal, 1:1,000), acetyl-STAT3 (Cell Signaling Technology, 2523; rabbit polyclonal, 1:1,000), STAT3 (Cell Signaling Technology, 9139; mouse monoclonal, 1:3,000), p-STAT3 (Tyr705) (Cell Signaling Technology, 9145; rabbit monoclonal, 1:1,000), GAPDH (Santa Cruz Biotechnology, SC51905; mouse monoclonal, 1:1,000), β-actin (MilliporeSigma, A2066; rabbit polyclonal, 1:10,000; or MilliporeSigma, A9357, mouse polyclonal, 1:4000), and vinculin (Santa Cruz Biotechnology, SC7649; goat polyclonal, 1:1,000). Membranes were then incubated with appropriate HRP-conjugated secondary antibody, followed by incubation with ECL detection substrate (GE Healthcare) or SuperSignal West Femto substrate (Thermo Fisher Scientific), and the chemoluminescent signal was detected with autoradiographic paper.
ISH and IHC analysis.
The formalin-fixed, paraffin-embedded tissue sections were dewaxed in xylene and rehydrated through an ethanol dilution series. Tissue sections were digested with 5 μg/mL proteinase K for 10 minutes at room temperature, then loaded onto Ventana Discovery Ultra for ISH analysis. The tissue slides were incubated with double digoxigenin–labeled mercury LNA microRNA probe (Exiqon) for 2 hours at 55°C. Slides were treated with 3% H2O2 to inactivate endogenous peroxidase activity and blocked with 5% bovine serum albumin in PBS (w/v), followed by primary and secondary antibody incubation in 1% bovine serum albumin (w/v) with 0.1% Tween-20 (v/v) in PBS) and washes in 0.01% Tween-20 (v/v) in PBS. Tyramine-conjugated fluorochrome was applied to the slide, and the tyramide signal amplification (TSA) reaction was allowed to proceed for 10–30 minutes. For the detection of protein, the tissue slides were incubated with primary antibody for 1 hour at 37°C after deparaffinization, and the secondary antibody (OmniMap anti-Rb HRP, Ventana, 760-4311) incubation and TSA reaction followed the same protocol. Fluorescent images were analyzed using LAS v4.3 software (Leica).
Statistics.
Statistical analysis and graphical representation were performed with GraphPad Prism (version 7.01). P values less than 0.05 were considered significant. The distributions of the data are displayed in box-and-whisker plots showing all points and outlying the minimum, first quartile, median, third quartile, and maximum values in duplicate or triplicate experiments. Shapiro-Wilk normality test was performed for each group to assess whether data were normally distributed. For normally distributed data, 2-tailed t test was used to compare mean values between different groups. When 1 of the groups did not pass the normality test, Mann-Whitney-Wilcoxon nonparametric test was used to assess statistical differences between the different groups.
Study approval.
All clinical data and human samples were obtained from participants who had given written informed consent, according to protocols approved by the Oncology Institute Ion Chiricuta Ethics Committee in accordance with the Declaration of Helsinki and UT-MDACC Institutional Review Board. All animal experiments were conducted in accordance with American Association for Laboratory Animal Science regulations and with the approval of Institutional Animal Care & Use Committees from UT-MDACC and the University of Oklahoma Health Sciences Center.
Author contributions
EFM, RSR, SV, and GAC conceived and designed the study. EFM, RB, RSR, AMS, TM, DG, CI, LF, EK, PN, NSDA, PM, CP, ETC, LG, MS, XZ, MRE, GB, ABP, CT, and IBN acquired data. EFM, RB, RSR, AMS, CI, MPD, MRE, IBN, PAZM, ZE, SV, and GAC analyzed and interpreted data. ZJZ directed the research related to JAK2V617F mice, WZ and WTH conducted experiments with JAK2V617F mice, and XZ performed the ISH and IHC analyses from mouse tissues. EFM, RSR, and GAC drafted the manuscript. All authors engaged in critical revision of the manuscript. SV and GAC acquired funding.
Supplementary Material
Acknowledgments
The work in GAC’s laboratory was partly supported by NIH/National Center for Advancing Translational Sciences grant UH3TR00943-01, the NIH/National Cancer Institute (NCI) grant 1 R01 CA182905-01, U54 grant UPR/MDACC Partnership for Excellence in Cancer Research 2016 Pilot Project, Team Department of Defense grant CA160445P1, a Ladies Leukemia League grant, a Chronic Lymphocytic Leukemia Moonshot Flagship project, a Sister Institution Network Fund 2017 grant, and the Estate of C. G. Johnson, Jr. EFM was supported in part by award number P50 CA140388 from the NCI and by the NIH Clinical Research Loan Repayment Program. AMS was supported by Fundação para a Ciência e a Tecnologia through fellowship SFRH/BD/85968/2012. ZJZ’s work was supported by grants from the MPN Foundation and the Oklahoma Center for the Advancement of Science and Technology. DG, MPD, and IBN were supported in part by a Programul Operațional Competitivitate grant nr35/01.09.2016, ID 37_796, titled “Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance” (CANTEMIR). NSDA was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo, BEPE 2016/09349-4.
Version 1. 01/16/2020
Electronic publication
Footnotes
EK’s current address is: Department of Medical Biology, Faculty of Health Sciences, Arctic University of Norway, Tromsø, Norway.
XZ’s current address is: Medical and Molecular Genetics Department, Indiana University, Indianapolis, Indiana, USA.
PAZM’s current address is: ImmunoGen, Inc., Boston, Massachusetts, USA.
RSR’s current address is: ProQR Therapeutics, Leiden, Netherlands.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(1):e121781.https://doi.org/10.1172/jci.insight.121781.
Contributor Information
Enrique Fuentes-Mattei, Email: efuentes1@mdanderson.org.
Recep Bayraktar, Email: RBayraktar@mdanderson.org.
Taghi Manshouri, Email: tmanshou@mdanderson.org.
Cristina Ivan, Email: CIvan@mdanderson.org.
Diana Gulei, Email: diana.c.gulei@gmail.com.
Linda Fabris, Email: fabrislinda@gmail.com.
Pilar Mur, Email: pmur@iconcologia.net.
Elizabeth Torres-Claudio, Email: elizabeth.torres9@upr.edu.
Mihnea P. Dragomir, Email: mihnea.p.dragomir@gmail.com.
Erik Knutsen, Email: erik.knutsen@uit.no.
Pranav Narayanan, Email: mail2pranav@yahoo.com.
Leonard Golfman, Email: lgolfman@yahoo.ca.
Masayoshi Shimizu, Email: mshimizu@mdanderson.org.
Xinna Zhang, Email: xz48@iu.edu.
Wanke Zhao, Email: wanke-zhao@ouhsc.edu.
Geoffrey Bartholomeusz, Email: gbarthol@mdanderson.org.
Ciprian Tomuleasa, Email: ciprian.tomuleasa@gmail.com.
Ioana Berindan-Neagoe, Email: ioananeagoe29@gmail.com.
Patrick A. Zweidler-McKay, Email: Patrick.Zweidler-McKay@ImmunoGen.com.
Zeev Estrov, Email: zestrov@mdanderson.org.
Srdan Verstovsek, Email: sverstov@mdanderson.org.
George A. Calin, Email: gcalin@mdanderson.org.
Roxana S. Redis, Email: roxana_redis@yahoo.com.
References
- 1.Reilly JT, et al. Guideline for the diagnosis and management of myelofibrosis. Br J Haematol. 2012;158(4):453–471. doi: 10.1111/j.1365-2141.2012.09179.x. [DOI] [PubMed] [Google Scholar]
- 2.Vener C, et al. Prognostic implications of the European consensus for grading of bone marrow fibrosis in chronic idiopathic myelofibrosis. Blood. 2008;111(4):1862–1865. doi: 10.1182/blood-2007-09-112953. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes F, et al. Identification of ‘short-lived’ and ‘long-lived’ patients at presentation of idiopathic myelofibrosis. Br J Haematol. 1997;97(3):635–640. doi: 10.1046/j.1365-2141.1997.782714.x. [DOI] [PubMed] [Google Scholar]
- 4.Tefferi A, et al. Validation and comparison of contemporary prognostic models in primary myelofibrosis: analysis based on 334 patients from a single institution. Cancer. 2007;109(10):2083–2088. doi: 10.1002/cncr.22630. [DOI] [PubMed] [Google Scholar]
- 5.Cervantes F, et al. New prognostic scoring system for primary myelofibrosis based on a study of the International Working Group for Myelofibrosis Research and Treatment. Blood. 2009;113(13):2895–2901. doi: 10.1182/blood-2008-07-170449. [DOI] [PubMed] [Google Scholar]
- 6.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 7.Campbell PJ, et al. V617F mutation in JAK2 is associated with poorer survival in idiopathic myelofibrosis. Blood. 2006;107(5):2098–2100. doi: 10.1182/blood-2005-08-3395. [DOI] [PubMed] [Google Scholar]
- 8.Pikman Y, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tefferi A, et al. One thousand patients with primary myelofibrosis: the mayo clinic experience. Mayo Clin Proc. 2012;87(1):25–33. doi: 10.1016/j.mayocp.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vannucchi AM, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27(9):1861–1869. doi: 10.1038/leu.2013.119. [DOI] [PubMed] [Google Scholar]
- 11.Oh ST, et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116(6):988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahr AA, et al. Bone marrow fibrosis in myelofibrosis: pathogenesis, prognosis and targeted strategies. Haematologica. 2016;101(6):660–671. doi: 10.3324/haematol.2015.141283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verstovsek S. Therapeutic potential of Janus-activated kinase-2 inhibitors for the management of myelofibrosis. Clin Cancer Res. 2010;16(7):1988–1996. doi: 10.1158/1078-0432.CCR-09-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 15.Bayraktar R, et al. Dual suppressive effect of miR-34a on the FOXM1/eEF2-kinase axis regulates triple-negative breast cancer growth and invasion. Clin Cancer Res. 2018;24(17):4225–4241. doi: 10.1158/1078-0432.CCR-17-1959. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 17.Fabbri M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305(1):59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci. 2011;121(4):141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 19.Sevignani C, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA. 2007;104(19):8017–8022. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Roosbroeck K, et al. The involvement of microRNA in the pathogenesis of Richter syndrome. Haematologica. 2019;104(5):1004–1015. doi: 10.3324/haematol.2018.203828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayraktar R, Bertilaccio MTS, Calin GA. The interaction between 2 worlds: microRNAs and Toll-like receptors. Front Immunol. 2019;10:1053. doi: 10.3389/fimmu.2019.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113(4):569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrajoli A, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122(11):1891–1899. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNA therapeutics in cancer - an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling H, Fabbri M, Calin GA. MicroRNAs and other noncoding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuentes-Mattei E, et al. Plasma Viral miRNAs indicate a high prevalence of occult viral infections. EBioMedicine. 2017;20:182–192. doi: 10.1016/j.ebiom.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Báez A, et al. Gene and miRNA expression profiles of hematopoietic progenitor cells vary depending on their origin. Biol Blood Marrow Transplant. 2014;20(5):630–639. doi: 10.1016/j.bbmt.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 28.Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37(1):33–44. doi: 10.1007/s10555-017-9724-7. [DOI] [PubMed] [Google Scholar]
- 29.Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Mol Oncol. 2017;11(12):1673–1686. doi: 10.1002/1878-0261.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghavachari N, et al. Integrated analysis of miRNA and mRNA during differentiation of human CD34+ cells delineates the regulatory roles of microRNA in hematopoiesis. Exp Hematol. 2014;42(1):14–27.e1. doi: 10.1016/j.exphem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan H, Cardozo C, Raza A. MicroRNAs in myeloproliferative neoplasms. Br J Haematol. 2013;161(4):471–483. doi: 10.1111/bjh.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Sankaran VG, Lodish HF. MicroRNAs in erythroid and megakaryocytic differentiation and megakaryocyte-erythroid progenitor lineage commitment. Leukemia. 2012;26(11):2310–2316. doi: 10.1038/leu.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008;22(6):1095–1105. doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- 34.Verstovsek S, et al. Long-term outcomes of 107 patients with myelofibrosis receiving JAK1/JAK2 inhibitor ruxolitinib: survival advantage in comparison to matched historical controls. Blood. 2012;120(6):1202–1209. doi: 10.1182/blood-2012-02-414631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verstovsek S, et al. Long-term survival in patients treated with ruxolitinib for myelofibrosis: COMFORT-I and -II pooled analyses. J Hematol Oncol. 2017;10(1):156. doi: 10.1186/s13045-017-0527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating noncoding RNAs - an update. Nat Rev Clin Oncol. 2018;15(9):541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 37.Dragomir M, Mafra ACP, Dias SMG, Vasilescu C, Calin GA. Using microRNA networks to understand cancer. Int J Mol Sci. 2018;19(7):E1871. doi: 10.3390/ijms19071871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasilescu C, et al. Circulating miRNAs in sepsis-A network under attack: an in-silico prediction of the potential existence of miRNA sponges in sepsis. PLoS ONE. 2017;12(8):e0183334. doi: 10.1371/journal.pone.0183334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batushansky A, Toubiana D, Fait A. Correlation-based network generation, visualization, and analysis as a powerful tool in biological studies: a case study in cancer cell metabolism. Biomed Res Int. 2016;2016:8313272. doi: 10.1155/2016/8313272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Identification of potential therapeutic target genes and miRNAs for primary myelofibrosis with microarray analysis. Exp Ther Med. 2017;14(4):2743–2750. doi: 10.3892/etm.2017.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norfo R, et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014;124(13):e21–e32. doi: 10.1182/blood-2013-12-544197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molina-Molina M, et al. Anti-fibrotic effects of pirfenidone and rapamycin in primary IPF fibroblasts and human alveolar epithelial cells. BMC Pulm Med. 2018;18(1):63. doi: 10.1186/s12890-018-0626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao R, et al. MicroRNA-29b-3p prevents Schistosoma japonicum-induced liver fibrosis by targeting COL1A1 and COL3A1. J Cell Biochem. 2018;119(4):3199–3209. doi: 10.1002/jcb.26475. [DOI] [PubMed] [Google Scholar]
- 44.Beridze N, Frishman WH. Vascular Ehlers-Danlos syndrome: pathophysiology, diagnosis, and prevention and treatment of its complications. Cardiol Rev. 2012;20(1):4–7. doi: 10.1097/CRD.0b013e3182342316. [DOI] [PubMed] [Google Scholar]
- 45.Marini JC, et al. Osteogenesis imperfecta. Nat Rev Dis Primers. 2017;3:17052. doi: 10.1038/nrdp.2017.52. [DOI] [PubMed] [Google Scholar]
- 46.Zhao W, Zou K, Farasyn T, Ho WT, Zhao ZJ. Generation and characterization of a JAK2V617F-containing erythroleukemia cell line. PLoS ONE. 2014;9(7):e99017. doi: 10.1371/journal.pone.0099017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing S, et al. Transgenic expression of JAK2V617F causes myeloproliferative disorders in mice. Blood. 2008;111(10):5109–5117. doi: 10.1182/blood-2007-05-091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao W, Ho WT, Zhao ZJ. Quantitative analyses of myelofibrosis by determining hydroxyproline. Stem Cell Investig. 2015;2:2. doi: 10.3978/j.issn.2306-9759.2015.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012;36(4):529–541. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan D, Jobe F, Hutchison RE, Mohi G. Deletion of Stat3 enhances myeloid cell expansion and increases the severity of myeloproliferative neoplasms in Jak2V617F knock-in mice. Leukemia. 2015;29(10):2050–2061. doi: 10.1038/leu.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tripathi SK, et al. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 2017;19(9):1888–1901. doi: 10.1016/j.celrep.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Rampal R, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123(22):e123–e133. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hattori M, et al. Global DNA hypomethylation and hypoxia-induced expression of the ten eleven translocation (TET) family, TET1, in scleroderma fibroblasts. Exp Dermatol. 2015;24(11):841–846. doi: 10.1111/exd.12767. [DOI] [PubMed] [Google Scholar]
- 54.Ha JS, Jeon DS, Kim JR, Ryoo NH, Suh JS. Analysis of the Ten-Eleven Translocation 2 (TET2) gene mutation in myeloproliferative neoplasms. Ann Clin Lab Sci. 2014;44(2):173–179. [PubMed] [Google Scholar]
- 55.Brecqueville M, et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012;51(8):743–755. doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- 56.Nangalia J, Green TR. The evolving genomic landscape of myeloproliferative neoplasms. Hematology Am Soc Hematol Educ Program. 2014;2014(1):287–296. doi: 10.1182/asheducation-2014.1.287. [DOI] [PubMed] [Google Scholar]
- 57.Chen E, et al. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood. 2015;125(2):327–335. doi: 10.1182/blood-2014-04-567024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kameda T, et al. Gene expression profiling of loss of TET2 and/or JAK2V617F mutant hematopoietic stem cells from mouse models of myeloproliferative neoplasms. Genom Data. 2015;4:102–108. doi: 10.1016/j.gdata.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kameda T, et al. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: disease sustainer and disease accelerator. Blood. 2015;125(2):304–315. doi: 10.1182/blood-2014-04-555508. [DOI] [PubMed] [Google Scholar]
- 60.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13(1):28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma N, et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat Genet. 2018;50(1):83–95. doi: 10.1038/s41588-017-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong J, et al. TET1 modulates H4K16 acetylation by controlling auto-acetylation of hMOF to affect gene regulation and DNA repair function. Nucleic Acids Res. 2017;45(2):672–684. doi: 10.1093/nar/gkw919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calura E, et al. A data-driven network model of primary myelofibrosis: transcriptional and post-transcriptional alterations in CD34+ cells. Blood Cancer J. 2016;6(6):e439. doi: 10.1038/bcj.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guglielmelli P, et al. Small RNA sequencing uncovers new miRNAs and moRNAs differentially expressed in normal and primary myelofibrosis CD34+ cells. PLoS ONE. 2015;10(10):e0140445. doi: 10.1371/journal.pone.0140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 66.Pei YF, et al. TET1 inhibits gastric cancer growth and metastasis by PTEN demethylation and re-expression. Oncotarget. 2016;7(21):31322–31335. doi: 10.18632/oncotarget.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan C, et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget. 2016;7(16):21825–21839. doi: 10.18632/oncotarget.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Dong G, Wang B, Gao W, Yang Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun. 2016;469(1):15–21. doi: 10.1016/j.bbrc.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 69.Liu G, Zhou J, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019;39(3):BSR20190249. doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russler-Germain DA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122(25):4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verstovsek S, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363(12):1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verstovsek S, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366(9):799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phan LM, et al. Hepatocyte growth factor/cMET pathway activation enhances cancer hallmarks in adrenocortical carcinoma. Cancer Res. 2015;75(19):4131–4142. doi: 10.1158/0008-5472.CAN-14-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuentes-Mattei E, et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer. J Natl Cancer Inst. 2014;106(7):dju158. doi: 10.1093/jnci/dju158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferrajoli A, et al. Epstein-Barr virus microRNAs are expressed in patients with chronic lymphocytic leukemia and correlate with overall survival. EBioMedicine. 2015;2(6):572–582. doi: 10.1016/j.ebiom.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tudor S, et al. Cellular and Kaposi’s sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma. Cell Death Dis. 2014;5:e1559. doi: 10.1038/cddis.2014.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 79.Kose F, Weckwerth W, Linke T, Fiehn O. Visualizing plant metabolomic correlation networks using clique-metabolite matrices. Bioinformatics. 2001;17(12):1198–1208. doi: 10.1093/bioinformatics/17.12.1198. [DOI] [PubMed] [Google Scholar]
- 80.Shi K, Zhao W, Chen Y, Ho WT, Yang P, Zhao ZJ. Cardiac hypertrophy associated with myeloproliferative neoplasms in JAK2V617F transgenic mice. J Hematol Oncol. 2014;7:25. doi: 10.1186/1756-8722-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin X, et al. Elevated levels of mast cells are involved in pruritus associated with polycythemia vera in JAK2V617F transgenic mice. J Immunol. 2014;193(2):477–484. doi: 10.4049/jimmunol.1301946. [DOI] [PubMed] [Google Scholar]
- 82.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17(3):637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 83.Chen Y, Guo Y, Zhao W, Tina Ho WT, Fu X, Zhao ZJ. Identification of an orally available compound with potent and broad FLT3 inhibition activity. Oncogene. 2016;35(23):2971–2978. doi: 10.1038/onc.2015.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.