Abstract
The T helper 2 (Th2) inflammatory cytokine interleukin-13 (IL-13) has been associated with both obstructive and fibrotic lung diseases; however, its specific effect on the epithelial stem cells in the gas exchange compartment of the lung (alveolar space) has not been explored. Here, we used in vivo lung models of homeostasis and repair, ex vivo organoid platforms, and potentially novel quantitative proteomic techniques to show that IL-13 disrupts the self-renewal and differentiation of both murine and human type 2 alveolar epithelial cells (AEC2s). Significantly, we find that IL-13 promotes ectopic expression of markers typically associated with bronchiolar airway cells and commonly seen in the alveolar region of lung tissue from patients with idiopathic pulmonary fibrosis. Furthermore, we identify a number of proteins that are differentially secreted by AEC2s in response to IL-13 and may provide biomarkers to identify subsets of patients with pulmonary disease driven by “Th2-high” biology.
Keywords: Pulmonology, Stem cells
Keywords: Adult stem cells, Proteomics
Evidence that IL-13 directly impairs differentiation of alveolar epithelial cells provides insight into the cell biology of chronic lung disease.
Introduction
Chronic lung diseases are associated with significant morbidity and mortality and, collectively, constitute the fourth leading cause of death in the United States. Chronic obstructive pulmonary disease (COPD) and pulmonary fibrosis are 2 such diseases with increasing prevalence. Both conditions involve pathological changes in the gas exchange (or alveolar) region of the lung, which normally consists of a delicate arrangement of thin-walled air sacs with type 1 and type 2 alveolar epithelial cells (AEC1s and AEC2s), stromal cells, and blood vessels. COPD can involve airspace enlargement and loss of this lung tissue, while fibrosis is associated with alveolar scarring and overproduction of extracellular matrix. In addition, lung fibrosis is typically associated with the appearance in the alveoli of tracts of epithelial cells expressing ectopic markers. Such tissue includes hyperplastic type 2 pneumocytes lining alveolar septae (1, 2) and the “bronchiolarized” epithelium typically present in areas of honeycomb cysts (3), the variably sized cystic structures found in scarred areas of lung in advanced pulmonary fibrosis.
Understanding the etiology of COPD and pulmonary fibrosis and devising new treatments for both diseases require knowledge of the cellular and molecular mechanisms that regulate the growth and differentiation of cells in the alveolar region. We hypothesize that the development and progression of both diseases involves a failure in the reciprocal interactions that normally sustain the AEC2s, which function as the epithelial stem cells in the alveolar space, and the fibroblasts that inhabit the stem cell niche (4). Over the past few years, considerable progress has been made in identifying some of the critical trophic factors in the stem cell niche using technologies such as lineage tracing, transcriptomics, and in vitro organoid assays (in which isolated AEC2s are cultured with niche fibroblasts) (5). This has led to the recognition that FGF, EGF, Wnt, and bone morphogenetic protein (BMP) signaling pathways are all involved in alveolar growth and differentiation, both in vivo and in organoid cultures (6–9). Less attention, however, has been paid to a potential effect of inflammatory cytokines on the proliferation and differentiation of alveolar cells (10). To address this question, we conducted a targeted screen of cytokines implicated in respiratory disease. Unexpectedly, we made the potentially novel observation that interleukin-13 (IL-13) has a profound effect on AEC2 stem cell self-renewal and differentiation.
IL-13 is a pleiotropic T helper type 2 (Th2) cytokine that has a variety of functions in pulmonary biology. Typically, these have been associated with allergy and allergic airway disease. In this context, IL-13 promotes mucus synthesis (11) from airway epithelial cells, production of IgE by B cells (12), and regulation of cytokine production by macrophages (12). Significantly, however, a polymorphism in the IL13 gene promoter that enhances gene transcription has been associated with both COPD (13) and worse pulmonary function in patients with pulmonary fibrosis (14). In addition, increased IL13 gene and protein expression have been observed in lung tissue (15) and bronchoalveolar lavage fluid (16, 17), from a subset of patients with idiopathic pulmonary fibrosis (IPF), notably in patients with rapidly progressive disease (15). In animal models, inducible pulmonary overexpression of IL-13 causes alveolar airspace enlargement, increased lung compliance, and mucus metaplasia (18, 19), characteristics usually associated with an emphysema phenotype. IL-13 signaling has also been shown to be involved in tissue fibrosis (20), where it appears to activate fibroblast proliferation and extracellular matrix deposition through transforming growth factor–β (TGF-β) production (20–22). Surprisingly, given the strong evidence supporting a role for IL-13 in alveolar biology and disease, the effect of IL-13 on alveolar epithelial stem cell function and response to injury has not been previously studied.
Here, we exploit a combination of in vivo lung models of both homeostasis and repair, ex vivo organoid platforms, and potentially novel quantitative proteomic techniques to show that IL-13 disrupts the normal differentiation of murine and human AEC2s. Specifically, we find that IL-13 promotes ectopic expression in AEC2s of markers typically associated with bronchiolar cells and with a phenotype similar to that of the “hyperplastic AEC2s” seen in IPF lungs. We also identify a number of factors AEC2s secrete in response to IL-13 that could be used as clinical biomarkers to distinguish subsets of patients with chronic and heterogeneous lung disease who have a “high Th2 phenotype.” Collectively, these data support a role for IL-13 in lung biology that moves beyond IL-13–mediated chemokine and inflammation-driven responses. Our data not only demonstrate that IL-13 has specific and direct interactions with alveolar epithelial cells but also suggest how dysregulated or unchecked IL-13 expression can impair alveolar regeneration and contribute to persistence and progression of chronic lung diseases.
Results
IL-13 overexpression in vivo leads to airspace enlargement and an altered ratio of AEC2s to AEC1s.
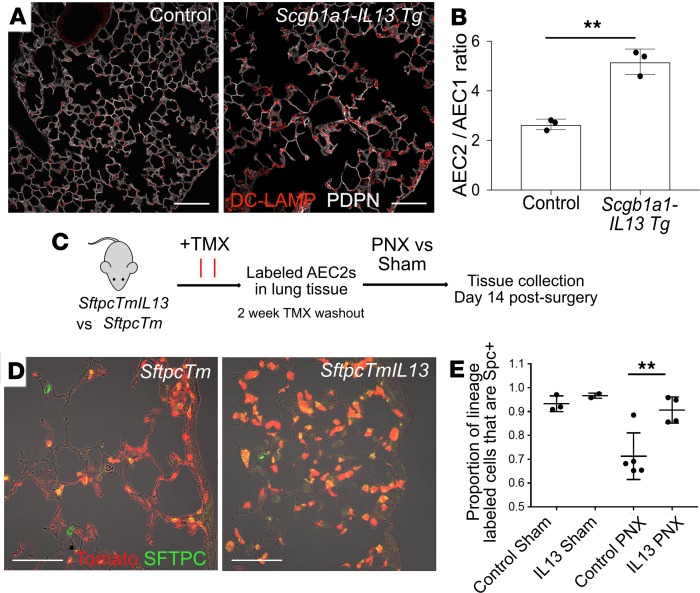
Models have been established previously to explore the role of IL-13 overexpression in the murine lung. Constitutive overexpression of IL-13 under the control of the uteroglobin (Scgb1a1/Cc10) promoter (23) in Scgb1a1-Il-13-Tg mice results in numerous airway changes, including tissue inflammation, mucus hyperproduction, goblet cell hyperplasia, and subepithelial airway fibrosis as well as alveolar airspace enlargement. Further studies using a doxycycline-inducible IL-13 transgene (19) revealed that this IL-13–induced alveolar enlargement is not a developmentally driven phenotype but rather can be due to destruction of previously normal alveoli in adult tissue. Although these studies highlighted the contributions of matrix metalloproteinases and cysteine proteases to the IL-13 phenotype, they did not address a potential direct effect of IL-13 on AEC2s. Here, we demonstrated the presence of airspace enlargement in the Scgb1a1-Il-13-Tg mice by histology (Figure 1A) and found a trend toward increased proliferation of AEC2s at steady state in transgenic lungs compared with controls (P = 0.052; Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131232DS1). Moreover, the significantly higher ratio of AEC2s to AEC1s in the transgenic lungs raises the possibility that IL-13, directly or indirectly, blocks the generation of AEC1s from AEC2s (Figure 1B).
Figure 1. IL-13 overexpression leads to an increase in the proportion of AEC2s to AEC1s.
(A) Constitutive overexpression of IL-13 from airway epithelial cells in the Scgb1a1-Il-13–transgenic mouse leads to airspace enlargement and a proportional increase in the number of AEC2s (labeled with DC-LAMP [LAMP3]) compared with AEC1s (marked by podoplanin [PDPN] expression). (B) The proportion of AEC2s to AEC1s was assessed by counting AEC2s (DC-LAMP+ cells) and AEC1s (cells with nuclear expression of homeobox only protein X, HOPX) and expressing the results as a proportion (AEC2/AEC1). There are more AEC2s relative to AEC1s in the IL-13–overexpressing mice. Unpaired t test; error bars indicate mean ± SD. (C) Schematic for lineage-labeling AEC2s in adult mice with (SftpcTmIL13) and without (SftpcTm) constitutive overexpression of IL-13 and subsequent pneumonectomy procedure (PNX). (D) Control lungs 14 days after PNX contain many lineage-labeled AEC1s. (E) IL-13–overexpressing lungs (IL-13 PNX) contain fewer lineage-labeled AEC1s after PNX when compared with controls (control PNX) despite higher levels of post-PNX AEC2 proliferation (data not shown and Supplemental Figure 1). Unpaired t test; error bars indicate mean ± SD. Scale bars: 100 μm (A), 75 μm (D). **P < 0.005.
To test the hypothesis that IL-13 directly affects differentiation of AEC2s in vivo, it was necessary to use an experimental model in which AEC2s are induced to both robustly proliferate and differentiate into AEC1s because their turnover in the normal lung is very slow. The injury model we chose was left lobe pneumonectomy (PNX). This manipulation prompts regrowth of the remaining lung lobes to restore the gas exchange area of the lung and involves both proliferation of AEC2s and their differentiation into AEC1s. We carried out this procedure in both WT mice and mice overexpressing IL-13 and compared the outcome.
To accomplish these experiments, we generated mice with the genotypes Sftpc-CreERT2/+ R26R-tdTomato/+ Scgb1a1-Il-13-Tg/+ (experimental) and Sftpc-CreERT2/+ R26R-tdTomato/+ (control) (hereafter abbreviated SftpcTmIl-13 and SftpcTm) and dosed them with tamoxifen (TMX) at least 2 weeks before PNX to induce lineage label expression in AEC2s. PNX and sham control surgeries were performed, and after 14 days (Figure 1C), the right accessory lobe was analyzed with immunofluorescence and quantitative confocal microscopy to determine the ratio of lineage-labeled AEC2s (defined by the presence of surfactant associated protein C [SFTPC] staining) and lineage-labeled AEC1s (lineage-labeled cell body with no SFTPC staining). As shown in Figure 1D, we found fewer lineage-labeled AEC1s in the SftpcTmIl-13 mice compared with controls after PNX (Figure 1, D and E). By contrast, the proliferation rates of lineage-labeled AEC2s in the SftpcTmIl-13 mice were higher than in controls (Supplemental Figure 1, C–E). We did not detect any qualitative change in the fibroblast population in the alveolar space of these mice (Supplemental Figure 1F). These data suggest that overexpression of IL-13 in the murine lung promotes AEC2 proliferation but at the same time inhibits their normal differentiation into AEC1s that occurs in the setting of PNX.
IL-13 inhibits differentiation of AEC2s in organoid culture.
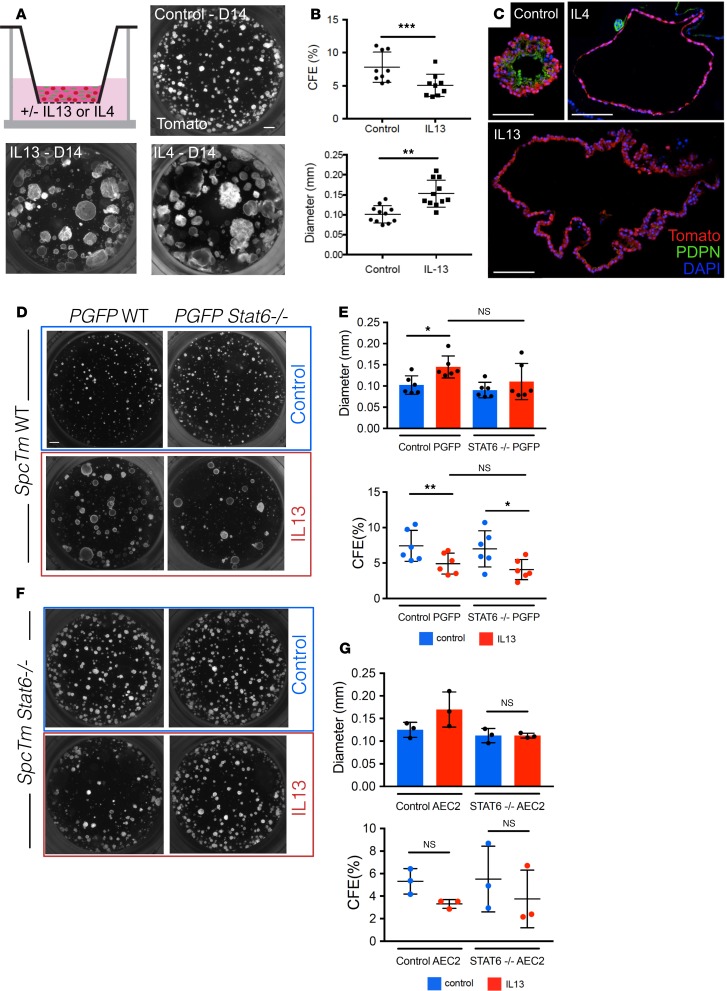
We next examined the effect of IL-13 on AEC2s cultured in our ex vivo organoid “alveolosphere” culture system (5). To accomplish this, we isolated lineage-labeled cells from SftpcTm mice. As we have previously shown (5), the vast majority of cells labeled in these mice are AEC2s; less than 1% of labeled cells are dual-positive SFTPC+SCGB1A1+ cuboidal cells in the terminal bronchioles (bronchoalveolar stem cells, or BASCs; ref. 24) and alveoli. Isolated AEC2s were cocultured in Matrigel with alveolar fibroblasts expressing platelet-derived growth factor receptor–α (Pdgfra), with or without IL-13 (or IL-4) (Figure 2A). After 14 days, the morphology of the cytokine-treated alveolospheres was dramatically different from that of controls; the IL-13–treated organoids were much larger although the colony-forming efficiency was lower (Figure 2B). These results were recapitulated with IL-4, another Th2 type cytokine that shares receptor subunits and signaling molecules with IL-13 (ref. 25 and Figure 2C). Approximately 55% of the IL-13–treated organoids had a “simple” (monolayer) morphology, distinct from the multilayer morphology displayed by nearly all the control spheres (Supplemental Figure 2, A and B). Reduced colony-forming efficiency (CFE) and increased sphere diameter occurred in a dose-dependent manner with IL-13 (Supplemental Figure 2C). The increase in average sphere diameter is a result of IL-13 treatment promoting a statistically significant increase in organoids that are more than 200 μm in diameter (Supplemental Figure 2D). The IL-13–induced phenotype occurred if IL-13 was added to the culture system at any time up to day 6. Addition beyond this time did not generate as exaggerated a phenotype (Supplemental Figure 3). Interestingly, IL-13 does not appear to permanently alter the ability of AEC2s to differentiate because the normal day 14 alveolosphere phenotype can be rescued if the IL-13 is withdrawn as late as day 10 (Supplemental Figure 4). Of note, although 45% of the IL-13–treated spheres displayed a multilayer morphology, none of these IL-13–treated organoids was phenotypically normal when analyzed for cellular composition and polarity (see below; Supplemental Figure 5). Also of note, it does not appear that the change in architecture of the organoids is related to changes in fluid and electrolyte transport. We treated control and IL-13–treated organoids with forskolin, a diterpenoid that is known to induce swelling of organoids through modulation of ion channels via activation of adenylate cyclase and increase in cyclic adenosine monophosphate levels. Although we saw an increase in diameter of organoids derived from tracheal basal cells, we saw no change in the diameter of AEC2-derived organoids treated with forskolin either in the presence or absence of IL-13 (Supplemental Figure 6).
Figure 2. AEC2-derived organoids grown with IL-13 display altered morphology.
(A) 3D organoid cultures containing lineage-labeled AEC2s and PdgfraGFP+ stromal cells were grown with and without IL-13. Organoids grown with IL-13 appear larger and less dense. This morphology is observed with treatment with another Th2 cytokine, IL-4, as well. (B) When compared with control organoids, IL-13–treated organoids display lower colony-forming efficiency (CFE) and higher average diameter. Paired t tests; error bars indicate mean ± SD. (C) IL-13–treated spheres lack AEC1 staining (PDPN). Organoids treated with IL-4 display similar histological morphology. (D and F) Lineage-labeled AEC2s were isolated from WT SftpcTm mice (SpcTm WT) or from Stat6–/– SftpcTm mice (SpcTm Stat6–/–). PdgfraGFP+ stromal cells were isolated from WT PdgfraGFP mice or from Stat6–/– PdgfraGFP mice (PGFP Stat6–/–). These cells were plated in organoid culture in different combinations and grown to day 14. (D and E) Organoids derived from WT lineage-labeled AEC2s grow normally in control conditions, even if STAT6 signaling is abolished in the stroma. When IL-13 is added to WT organoids supported by either WT or Stat6–/– stromal cells, the typical IL-13 effect is observed. (F and G) Stat6–/– organoids grown with IL-13 do not display an increase in average sphere diameter, and the morphology appears grossly normal. These data suggest that the IL-13 effect is due to STAT6 signaling in the epithelium. One-way ANOVA; error bars indicate mean ± SD. Scale bars: 500 μm (A), 100 μm (C), and 500 μm (D). *P < 0.05; **P < 0.005.
IL-13–induced epithelial phenotype is STAT6 dependent.
Our initial organoid studies could not distinguish whether the effects of IL-13 are due to direct actions on AEC2s or via paracrine signaling by IL-13–stimulated fibroblasts. Because it has been well described that IL-13 uses STAT6 as a downstream effector in both cell types, we generated mice of the genotypes Sftpc-CreERT2/+ R26R-tdTomato/+ Stat6–/– (hereafter SftpcTm Stat6) and Pdgfra:H2B-GFP Stat6–/– (hereafter PGFP Stat6). Using FACS, we isolated lineage-labeled AEC2s from TMX-dosed SftpcTm Stat6 and SftpcTm mice and GFP+ stromal cells from PGFP Stat6 and PGFP mice. We then established organoid cultures with mutant and WT cells in different combinations, with and without IL-13 (Figure 2, D and F). When STAT6 signaling was abolished in the Pdgfra+ stromal cells, IL-13 treatment induced the typical phenotype (i.e., larger, simpler spheres) (Figure 2, D and E). In contrast, when STAT6 signaling was abolished in the SFTPC+ AEC2s, the IL-13–induced alveolosphere phenotype no longer developed (Figure 2, F and G). This suggests that the effect of IL-13 on AEC2 stem cell behavior is mediated directly by STAT6 in the epithelium and is not contingent upon a STAT6-dependent, IL-13–induced change in the supporting stromal cells.
IL-13 promotes an immature phenotype in AEC2s in organoid culture.
We performed detailed histological analysis of alveolospheres from both control and IL-13 conditions and scored sections for markers of alveolar epithelial cells. As described previously, control spheres had a multilayered architecture, with AEC2-like cells on the outside and AEC1-like cells on the inside, typically positive for HOPX, PDPN (also known as T1-α) (Figure 3A), or advanced glycosylation end-product specific receptor (RAGE) (Supplemental Figure 5A). By contrast, IL-13–treated spheres lacked this organization, and none of these spheres (neither simple nor multilayer) contained AEC1s positioned with a clear inside preference (Supplemental Figure 5B). The expression of alveolar epithelial markers was noticeably attenuated in the IL-13–treated spheres (Figure 3A). Of note, a significantly higher proportion of cells in IL-13–treated spheres were negative for typical AEC2 and AEC1 markers (Figure 3B and Supplemental Figure 5B).
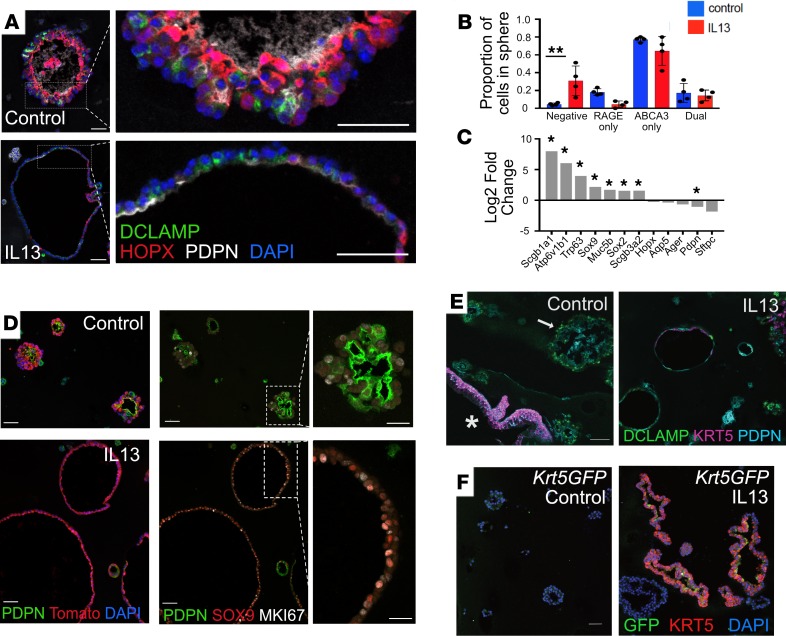
Figure 3. IL-13 disrupts AEC2 differentiation in vitro.
(A) Control organoids express both AEC2 (DC-LAMP) and AEC1 (HOPX, PDPN) markers. The morphology of most IL-13–treated spheres is abnormal, with only a single layer of epithelial cells. There are AEC2 and AEC1 markers present, but these occur in an apparently random distribution not seen in control spheres. (B) Compared with control organoids, IL-13–treated organoids contain a higher proportion of cells that lack both AEC2 and AEC1 markers. One-way ANOVA; error bars indicate mean ± SD. **P < 0.005. (C) RNA-Seq of day 16 organoids grown in the presence of IL-13 (compared with control conditions) reveals increased expression of bronchiolar markers (n = 3 biological replicates per condition; asterisk indicates genes significantly differentially expressed). (D) Lineage-labeled (Tomato+) organoids grown in the presence of IL-13 express more SOX9 and display more proliferation (MKI67) than lineage-labeled organoids grown in control conditions. (E) IL-13 induces ectopic expression of KRT5 in AEC2s. An AEC2-derived sphere (arrow) grown in control conditions expresses AEC2 (DC-LAMP) and AEC1 (PDPN) markers but not KRT5. A rare contaminating basal cell–derived organoid (asterisk) serves as a positive control, demonstrating expected KRT5 staining. In contrast, a proportion of IL-13–treated spheres express KRT5. (F) KRT5 expression in IL-13–treated spheres is supported with use of a Krt5GFP reporter mouse line. Scale bars: 50 μm (A); 50 μm, insets 25 μm (D); 75 μm (E); and 50 μm (F).
Bulk RNA-Seq of lineage-labeled epithelial cells isolated from organoids at day 16 showed that the IL-13–treated cells expressed lower levels of both AEC2 and AEC1 markers (Hopx, Ager, Pdpn, Sftpc) (Figure 3C, Supplemental Figure 7A, and ref. 26) and genes associated with lipid processing (Supplemental Figure 7B) versus controls. Of particular interest is that fact that IL-13 induced significant upregulation of transcripts for genes such as Scgb1a1 (+8-fold), Atp6v1b1 (+6-fold), Trp63, and Muc5b (Figure 3C) that are normally expressed in airway epithelium. In addition, immunofluorescence staining showed that IL-13–treated spheres express higher levels of SOX9 (Figure 3D), a transcription factor expressed in the developing lung that is involved in the balance of proliferation and differentiation of epithelial tip progenitor cells (27). We did not observe increased expression of airway-related protein markers (e.g., SCGB1A1, SOX2, TRP63) or specific mucins (e.g., MUC5B) with conventional immunofluorescence staining (data not shown). Taken together, these results suggest that IL-13 treatment promotes dedifferentiation of AEC2s and a reprogramming of these cells toward a bronchiolar phenotype. These effects on the AEC2 transcriptome were also observed when cultures were treated with IL-4 (Supplemental Figure 7C).
IL-13 induces expression of keratin 5 in a subset of AEC2-derived organoids.
In our analysis of immunofluorescence staining for bronchiolar marker proteins in IL-13–treated organoids, we found expression of keratin 5 (KRT5) in murine AEC2-derived, IL-13–treated spheres. KRT5 is a cytoskeletal protein that is characteristic of basal cells in the mouse trachea and intralobar airways of normal human lung. Significantly, KRT5 is expressed ectopically in many SFTPC+ “hyperplastic AEC2s” found in the alveolar space of IPF lungs (Supplemental Figure 8, A and B, and refs. 1, 2). Immunofluorescence analysis of day 14 organoids revealed the presence of KRT5 in 7.9% of lineage-labeled IL-13–treated, AEC2-derived alveolospheres; there was no KRT5 expressed in control spheres (Figure 3E). This finding was supported by using a KRT5GFP reporter line (Figure 3F). These data support the hypothesis that AEC2s are induced to express KRT5 in the presence of IL-13.
Interestingly, our data show that when AEC2s are supported by Stat6–/– fibroblasts and grown in the presence of IL-13, there is an increase in the proportion of organoids that contain KRT5+ cells. In attempting to determine the mechanism behind this, we assessed by ELISA levels of a known IL-13 decoy receptor, IL-13 receptor subunit α 2 (IL-13Rα2) (28) in conditioned medium collected at different times during organoid growth. IL-13Rα2 levels went up significantly by day 10 in the presence of IL-13 when organoids were supported by normal fibroblasts, but IL-13Rα2 levels remained near or at baseline when organoids were supported by Stat6–/– fibroblasts (Supplemental Figure 9).
Alveolar epithelial cells coexpressing SFTPC and SCGB1A1 may preferentially respond to IL-13 treatment.
Because our data suggest that the AEC2s that respond to IL-13 take on a bronchiolar-like phenotype (with upregulation of bronchiolar-related gene transcripts), we asked whether a subset of AEC2s that normally express low levels of the bronchiolar marker SCGB1A1 would be primed to respond to IL-13. Alveolar epithelial cells that are positive for SFTPC and SCGB1A1 have been shown to be located at and close to the BADJ (BASCs) (24) or scattered in low numbers throughout the alveoli (29); these cells have also been shown to respond preferentially in a model of injury with Streptococcal pneumoniae (30). To test our hypothesis that SCGB1A1-expressing AECs respond preferentially to IL-13, we isolated from Scgb1a1-CreER/+ R26R-tdTomato/+ mice (hereafter Scgb1a1Tm) lineage-labeled cells that were also “high” for LysoTracker (LT), a validated marker of differentiated AEC2s (ref. 31 and Figure 4A) and compared their growth with that of “bulk” AEC2s from the same mice (i.e., non–lineage-labeled, LT+ cells). In this experiment, the CFE of IL-13–treated, lineage-labeled epithelial cells was higher than that of control cells (Figure 4, B and C). This suggests that the subset of AEC2s that express SCGB1A1 are capable of more robust proliferation in the presence of IL-13 than non–SCGB1A1-expressing AEC2s. A similar proportion of organoids derived from SCGB1A1-expressing AEC2s grown in the presence of IL-13 express KRT5 (Figure 4D; quantitative data not shown).
Figure 4. AEC2s that coexpress SCGB1A1 are more capable of growth in the presence of IL-13 than AEC2s that do not express SCGB1A1.
(A) Lungs were isolated from Scgb1a1CreER/+ Tm/+ (Scgb1a1Tm) mice following TMX administration. Airway cells and scattered alveolar epithelial cells (white arrowheads) are labeled in these mice; rare BASCs (SFTPC+SCGB1A1+ cells located in the terminal bronchiolar region; yellow arrowhead) are labeled. Lungs were dissociated and a single-cell suspension was stained for LysoTracker (LT) in order to mark AEC2s. Both Tomato+ (Sgcb1a1–lineage-labeled) LT+ cells (AEC2s that coexpress SCGB1A1; gray box) and Tomato– (non–lineage-labeled) LT+ cells (AEC2s that do not coexpress SCGB1A1; magenta box) were sorted via FACS and cultured in equal numbers with PGFP+ stromal support cells for 14 days with and without IL-13. (B and C) The CFE of Tomato+ Scgb1a1–lineage-labeled AEC2s increases with IL-13 exposure (calculated based on fold change in CFE from control). The baseline CFE of Scgb1a1Tm LT+ control cultures is 3.58%. This CFE increases to 5.25% in the presence of IL-13. The baseline CFE of non–lineage-labeled AEC2s that are LT+ is 14.42% in control conditions. This CFE drops to 11.68% in the presence of IL-13. The average sphere diameter of IL-13–treated organoids derived from Tomato+ Scgb1a1-expressing AEC2s is higher than IL-13–treated organoids derived from Tomato– Scgb1a1 lineage– AEC2s. (D) There is notable KRT5 staining in organoids derived from Tomato+ Scgb1a1–lineage-labeled, LT+ AEC2s compared with control spheres. One-way ANOVA with multiple comparisons; error bars represent mean ± SD. Scale bars: 100 μm (A), 500 μm (B), and 100 μm (D). *P < 0.05; **P < 0.005.
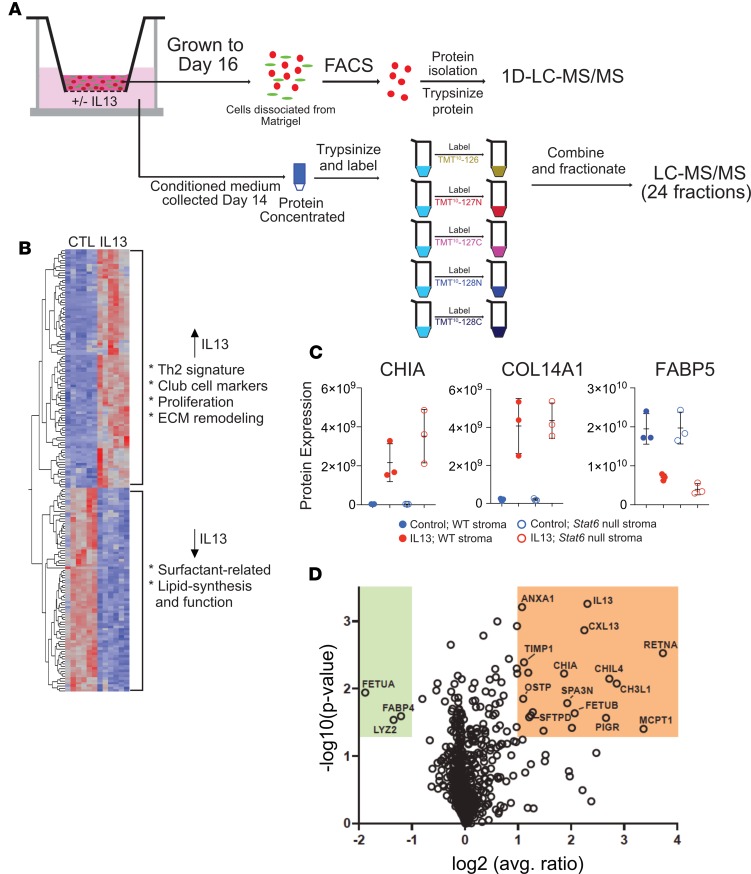
Potentially novel quantitative proteomics approach reveals IPF-related proteins in IL-13–treated AEC2s.
To better understand the phenotype of AEC2s exposed to IL-13 treatment, we isolated lineage-labeled (Tomato+) epithelial cells from day 16 organoids derived from lineage-labeled AEC2s grown with and without IL-13 after removing supporting stromal cells by FACS. We then analyzed protein expression in the Tomato+ cells by mass spectrometry–based proteomics (Figure 5) and compared the results to bulk RNA-Seq data from the same cells. The upregulation of a number of transcripts and their corresponding gene products (Figure 5, B and C) — including chitinase-like proteins (CHIA, CHIL3, CHIL4, and CHI3L1) and resistin-like–α (FIZZ1; Supplemental Table 1) — mirrored a Th2 inflammatory phenotype that has been well described in murine lungs (32). In addition, the differential expression of these proteins (and all other proteins subsequently detected) was maintained in AEC2 cells cultured with Stat6–/– fibroblasts, demonstrating an effect of IL-13 on the AEC2s that is not dependent on an IL-13–driven, STAT6-dependent response by supporting fibroblasts (Figure 5C).
Figure 5. Proteomic analysis.
(A) Schematic. For cell-based proteomic analysis, lineage-labeled cells were dissociated from Matrigel, and protein isolated from them was trypsinized and peptides analyzed by nanoflow liquid chromatography–tandem mass spectrometry (nano–LC-MS/MS). For secretomic analysis, conditioned medium was collected at day 14 and protein trypsinized, labeled, fractionated, and analyzed by LC-MS/MS. (B) Heatmap showing differential expression of proteins in lineage-labeled epithelial cells from control and IL-13–exposed cultures. Proteins depicted are significant at P < 0.05 and fold change > ±1.5 in IL-13 vs. control. (C) Three representative differentially expressed proteins from cell-based proteomic analysis in B. Chitinase-like protein A (CHIA) and collagen α 1(XIV) chain (COL14A1) were significantly upregulated in AEC2-derived cells in IL-13–treated cultures compared with controls. Conversely, there was less expression of fatty acid binding protein 5 (FABP5) in AEC2-derived cells from IL-13–treated cultures compared with controls. CHIA WT, IL-13 vs. control, P = 0.001965; CHIA STAT6, IL-13 vs. control, P = 0.005662; COL14A1 WT, IL-13 vs. control, P = 0.011875; COL14A1 STAT6, IL-13 vs. control, P = 0.0.00319; FABP5 WT, IL-13 vs. control, P = 0.015991; FABP5 STAT6, IL-13 vs. control, P = 0.0.004382. *P < 0.05; **P < 0.005; paired, 2-tailed t test. (D) Volcano plot depicting proteins that are differentially expressed in the secretome of cultures treated with IL-13 versus control. Error bars represent mean ± SD. Analysis done with 3 biological replicates for each condition.
We mapped differentially expressed genes/proteins to numerous other relevant pathways, including proliferation, surfactant function, bronchiolar cell markers, and extracellular matrix remodeling (Figure 5B and Supplemental Table 1). An IL-13–dependent hyperproliferative phenotype was suggested by an increase in expression of proliferation marker protein Ki-67 (MKI67) as well as condensin subunits SMC2 and SMC4, all of which localize to mitotic chromosomes (33). Also consistent with this phenotype was increased expression of keratin 18 (34). IL-13–dependent AEC2 metabolic dysfunction was suggested by lower levels of fructose-1,6-bisphosphatase 1 (FBP1), which is expressed in human AEC2s but not AEC1s (35), as well as other key enzymes in surfactant homeostasis (e.g., fatty acid synthase [FASN], acetyl-coA carboxylase [ACACA], and choline-phosphate citidylyltransferase A) (36, 37) and the surfactant-associated lysozyme C-2 (LYZ2) (38–40). In addition, FABP5, a transporter of fatty acids that is believed to protect against oxidative damage to lipids and to regulate inflammatory functions of macrophages (41, 42), was downregulated in IL-13–treated AEC2s. Of note, the levels of intracellular SFTPA, SFTPB, SFTPC, and SFTPD were not significantly altered by IL-13 treatment; secreted surfactant was not measured.
IL-13–induced extracellular matrix modification was suggested by increased expression in AEC2s of COL14A1, a fibril-associated collagen that has been shown to be increased in IPF lung (43, 44) and procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (PLOD2), which catalyzes a critical step in collagen cross-linking (45). Although expression of surfactant-related genes is inhibited by IL-13, levels of the club cell marker cytochrome P450 isoform 2F2 (CP2F2) was higher in IL-13–treated organoids, consistent with our findings that IL-13–responsive AEC2s take on bronchiolar characteristics in the setting of IL-13 treatment. Furthermore, because club cells can produce surfactant proteins but not surfactant lipid, these data suggest that IL-13 promotes a secretory phenotype that may be defective in normal surfactant function.
In addition to the cell proteome, we used a potentially novel proteomic approach to quantify the secreted proteins from conditioned medium (Figure 5A). Specifically, we used isobaric labeling, fractionation, and microflow liquid chromatography–tandem mass spectrometry (LC-MS/MS) to identify and quantify proteins secreted by cells into the organoid culture medium. Analysis of the conditioned medium versus sham allowed us to differentiate these secreted factors from the background of FBS and Matrigel, a basement membrane preparation from mouse sarcoma cells that contains extracellular matrix proteins. This analysis identified a small subset of proteins differentially expressed in medium from IL-13–treated organoids and present at much higher levels than in the medium-only control (Figure 5D). As in the cell proteome/transcriptome, the most highly upregulated proteins included the STAT6-inducible chitinases, FIZZ1, and polymeric Ig receptor. Likewise, levels of lysozyme C2 (LYZ2), a potentially important component of surfactant, were significantly lower (Figure 5D).
Impaired differentiation of human alveolar epithelial cells in the presence of IL-13.
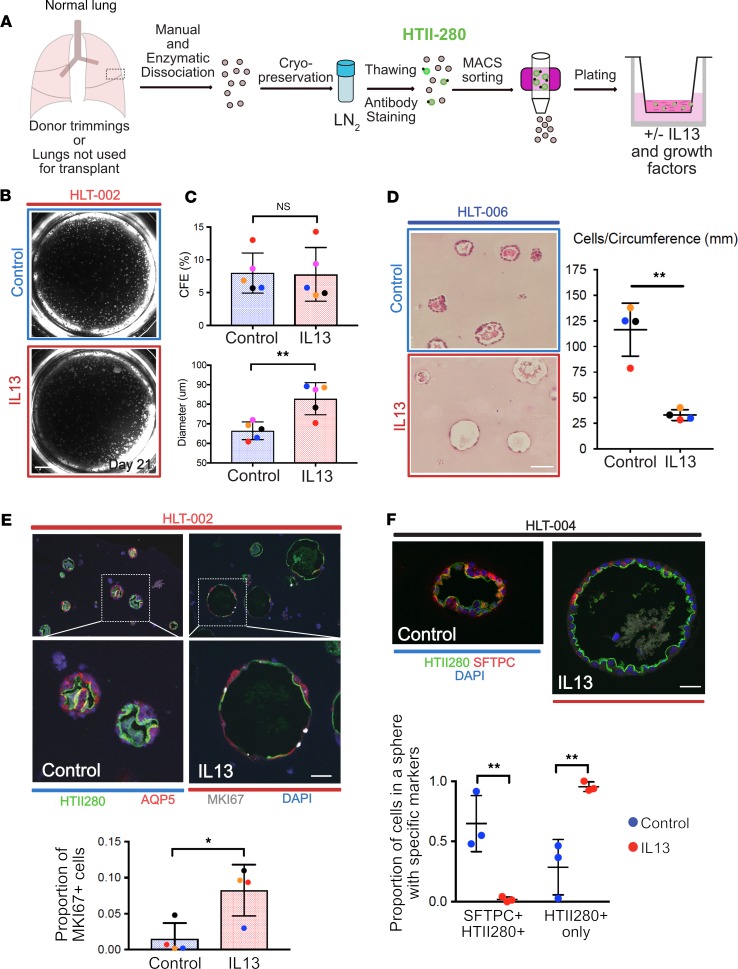
We next asked whether IL-13 also impairs the proliferation and differentiation of human alveolar epithelial cells (hAEC2s) in organoid culture. We adapted our previously reported protocol (5) and isolated HTII-280+ hAEC2s (46) from normal human lung tissue (Figure 6A). Cells were grown in Matrigel with air-liquid interface (ALI) medium (47) for 21 days with and without IL-13 (Figure 6, A and B). There was no difference in CFE between control and IL-13–treated hAEC2-derived organoids, but there was a statistically significant increase in average sphere diameter in IL-13–treated organoids (Figure 6C). IL-13 promotes an abnormal morphology in human alveolospheres. Strikingly, cells in IL-13–treated spheres have a more flattened appearance and are less cuboidal. We confirmed this finding by quantifying the number of cells per unit of circumference distance in each sphere (Figure 6D). Immunohistochemistry showed that HTII-280 was present on the luminal (apical) surface of hAEC2-derived control organoids (Figure 6E), suggesting AEC2 identity. Some cells in the spheres also expressed the AEC1 marker aquaporin 5 (AQP5) (Figure 6E), although none of the AQP5+ cells resembled the highly attenuated AEC1s that we saw in murine alveolospheres. This is consistent with reports from other groups suggesting that there is a paucity of AEC1s in control hAEC2-derived organoids (6, 48), regardless of treatment condition. Assessment of proliferation using MKI67 revealed an increased rate of epithelial cell proliferation in IL-13–treated hAEC2s at day 21 compared with controls (Figure 6E), a finding consistent with the murine alveolosphere proteomic data. Analysis of immunofluorescence markers revealed that there were fewer HTII-280+SFTPC+ dual-positive AEC2s in IL-13–treated spheres. A significant proportion of cells in IL-13–treated spheres were HTII-280+ only, lacking normal coexpression of SFTPC (Figure 6F).
Figure 6. Human lung cell culture.
(A) Schematic for processing of normal human lung for culture of hAEC2s. (B) Representative example of human alveolospheres grown with and without IL-13. (C) CFE is similar with and without IL-13, but average sphere diameter is higher in IL-13–treated cultures. (D) Quantification of cell number per unit circumference. The cells in IL-13–treated spheres cover more surface area and appear elongated. Two-tailed t test; error bars represent mean ± SD. (E) Representative immunohistochemistry of AEC2s (HTII-280+) and AEC1s (AQP5+) in human alveolospheres grown with and without IL-13. Note the morphology of IL-13–treated spheres is different from controls and there is less staining for both AEC2 and AEC1 markers. There are more proliferating (MKI67+) cells in the IL-13–treated organoids. Two-tailed t test; error bars represent mean ± SD. (F) Representative spheres from a separate donor. Note larger diameter of IL-13–treated sphere and less expression of the AEC2 marker, SFTPC. One-way ANOVA; error bars represent mean ± SD. Dot colors correspond to separate donors. Donors are listed as HLT-xxx. Scale bars: 500 μm (B), 60 μm (D), 50 μm (E), 30 μm (F). *P < 0.05; **P < 0.005.
Discussion
In this work, we used a combination of in vivo lineage tracing in an injury/repair model, ex vivo organoid culture, and quantitative proteomics to reveal previously unappreciated STAT6-dependent effects of the Th2 cytokine, IL-13, on alveolar epithelial stem cells. Our observations have important implications for AEC2 stem cell biology and potentially for the diagnosis and treatment of patients with chronic lung disease.
Our in vivo lineage-tracing experiments clearly showed that IL-13 causes impaired differentiation of AEC2s to AEC1s in the PNX model of compensatory lung regrowth, suggesting that IL-13 can affect the stem cell responses of the alveolar epithelium. We did not observe any obvious phenotypic changes in the alveolar fibroblasts in histological sections, although we did not perform transcriptomic or proteomic profiling on this population to evaluate their response to IL-13 more completely. We are able to discern, however, from our in vitro organoid culture models, that the effect of IL-13 on AEC2s is mediated by STAT6 signaling in the epithelium and that this effect is not mediated by a STAT6-dependent response of the supporting stromal cells. This is important because many prior studies have focused on the effect of IL-13 on fibroblasts (49), with the dogma being that lung fibroblasts act as effector cells in the lung after activation by IL-13 or IL-4. In contrast, our study suggests that IL-13–exposed AEC2s can serve as effector cells to perpetuate an IL-13–induced inflammatory response through production of profibrotic and proinflammatory proteins (such as FIZZ1 [ref. 50] and periostin, POSTN [ref. 51]) and through expression of increased levels of structural and ECM-relevant proteins, such as COL14A1 and PLOD2.
To be clear, this work does not rule out a contribution of the stromal cells that could be contributing to the epithelial phenotype that we observe. In fact, our findings that IL-13Rα2 is present in conditioned medium (CM) when organoids are supported by normal fibroblasts (but is not present in CM at similar levels when organoids are supported by Stat6–/– fibroblasts) and that the presence of IL-13Rα2 attenuates the level of KRT5 expression in IL-13–treated organoids suggests that an IL-13–induced response of the stromal cells can affect the epithelial phenotype. Further, our experiments have not ruled out that IL-13 signals in fibroblasts in a STAT6-independent manner to promote the production of a secreted factor that, in turn, acts on AEC2s in a STAT6-dependent fashion. It will be important to investigate this possibility in future work so that the specific IL-13–driven pathway(s) that lead(s) to AEC2 stem cell dysfunction can be elucidated further.
One of the most interesting findings from this work is the suggestion that IL-13 can promote a bronchiolar phenotype in AEC2s. Our transcriptomic data show that IL-13 impairs the stem cell function of AEC2s and induces ectopic expression of genes normally associated with bronchiolar and more proximal airway cell types. Specifically, IL-13 promotes increased transcript levels of Scgb1a1, Scgb3a2, Atp6v1b1, and Trp63 and increased protein expression of transformation related protein 63 (TRP63) and CP2F2 (among others). One possible explanation for these changes is that they are orchestrated by ectopic expression of SOX9, a transcription factor normally involved in distal epithelial progenitor fate decisions in the developing lung (27), holding AEC2s in an immature state. Our work does not reveal, however, whether all AEC2s are capable of developing this bronchiolar phenotype or whether only a subset of AEC2s can do so. Our data show that Scgb1a1–lineage-traced AEC2s have a competitive growth advantage in culture with IL-13 compared with bulk AEC2s, and this suggests that alveolar epithelial cells that already express low levels of bronchiolar markers such as SCGB1A1 are primed and more capable of turning on a bronchiolar program in the presence of IL-13. To determine whether there is heterogeneity in the response of AEC2s to IL-13, it would be reasonable to perform single-cell RNA-Seq analysis of AEC2s grown with and without IL-13 in culture to explore further the heterogeneity that exists in the alveolar response to IL-13.
As IL-13–exposed AEC2s become more bronchiolarized, it is perhaps not surprising to see that IL-13 exposure also causes AEC2s to express reduced levels of typical AEC2 genes, including Sftpc and Lyz2, as well as proteins that are involved with fatty acid metabolism and processing, such as FBP1, FABP5, FASN, and ACACA. How these fatty acid–based transcriptomic and proteomic changes affect the reparative capacity of alveolar epithelial stem cells is currently unknown and is worthy of future study.
In the context of our ex vivo human experiments, we have shown that IL-13 affects the growth of hAEC2s in organoid culture. At this time, we cannot determine whether the effect is direct (as it appears to be in the mouse) or indirect (mediated through the supporting stromal cells). However, IL-13 clearly has an effect on the self-renewal capacity of AEC2s and on the morphology of hAEC2-derived organoids. This supports our hypothesis that IL-13 influences human lung remodeling in vivo. Taken together, our murine and human data suggest that IL-13 exerts pleiotropic effects on alveolar epithelial stem cells and that the AEC2 response to IL-13 has the potential to influence lung remodeling, either directly or indirectly.
As outlined in the introduction, IL-13 has been implicated in the pathogenesis of IPF (14–17). Most of the data supporting the role of IL-13 in IPF are correlative: IL-13 mRNA levels are increased in lung tissue from patients with IPF (15), and IL-13 protein levels are increased in bronchoalveolar lavage fluid (BALF) from a subset of patients with IPF (16); however, the biology behind this association has not been elucidated. Studies have shown that alveolar macrophages can produce IL-13 in fibrotic lungs (17) and that fibrotic lung tissue has increased levels of expression of IL-13 receptors (52). Nevertheless, a clear cellular source of IL-13 production in IPF lungs and biological data suggesting downstream target(s) of IL-13 signaling in the context of disease have not been shown. Our study, therefore, provides a foundation from which to consider how the AEC2-specific effects of IL-13 may relate to the development, pathogenesis, and progression of IPF.
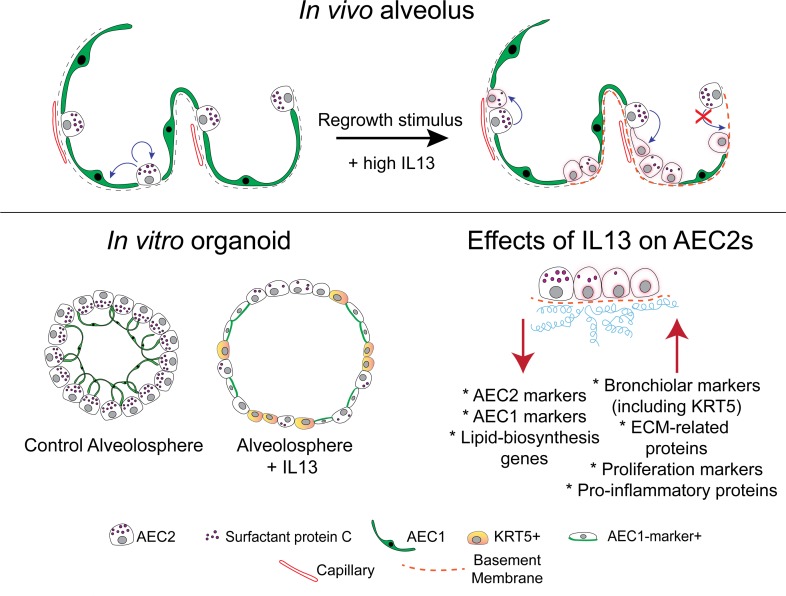
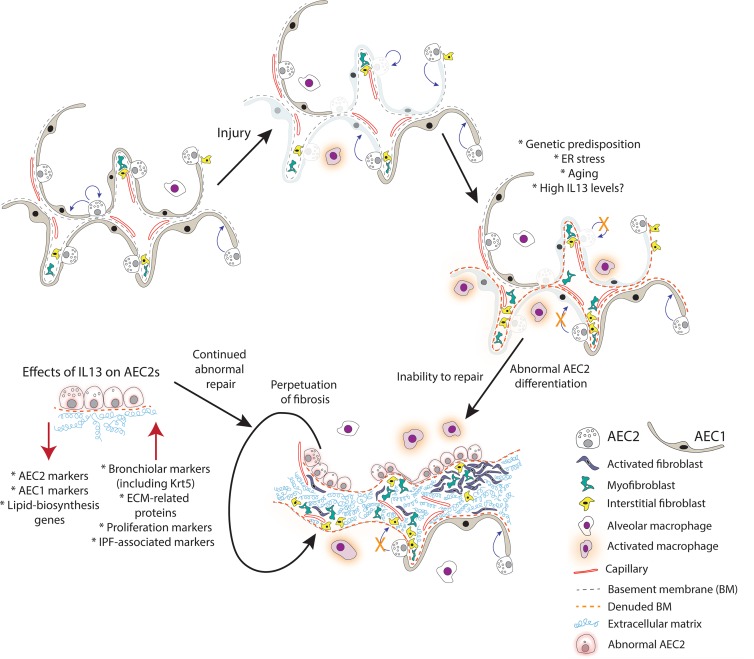
In beginning to consider how AEC2-specific effects of IL-13 can promote disease pathogenesis, we have considered the IL-13–induced phenotypic changes to AEC2s in the context of published IPF single-cell sequencing data. Interestingly, the “intermediate” epithelial cell type in IPF lungs identified by Xu et al. (53) shares many of the characteristics of our AEC2-derived IL-13–treated cells (increase in KRT5, SOX9, MKI67; decrease in SFTPC and FABP5) (54). We hypothesize that these “intermediate” epithelial cells may in fact be similar to the “hyperplastic AEC2s” found in regions of fibrotic alveolar septae lacking typical AEC1s. Currently, the origin of these cells is not known, but our data suggest that at least some of these cells could arise from AEC2s responding to high levels of IL-13 in the alveolar regions. According to our model, once the phenotypic switch has occurred and high IL-13 levels persist, the “hyperplastic cells,” having lost a normal AEC2 phenotype, are unable to differentiate to AEC1s and normal repair is disrupted. In turn, incomplete epithelial repair leads to secretion of profibrotic and ECM remodeling–related proteins, which sets up a negative feedback loop (see schematic, Figure 7). We hypothesize that this biological scenario could be relevant in a subset of patients with IPF who have high levels of Th2 cytokines in the BALF.
Figure 7. Model for the potential role of IL-13 in the pathogenesis of IPF.
Normal lung is capable of epithelial repair, characterized by self-renewal and differentiation of AEC2s. In normal circumstances, injury can be repaired through these mechanisms. In the setting of genetic predisposition, endoplasmic reticulum stress, aging, or, as we propose, high IL-13 levels, AEC2s proliferate but are unable to differentiate normally, leading to denuded basement membrane, proliferation, and differentiation of mesenchymal cell types, and deposition of extracellular matrix. AEC2s are “hyperplastic” in appearance, have reduced expression of lipid biosynthesis genes, and acquire a more bronchiolar phenotype. As such, they are not capable of normal repair and a fibrotic response is perpetuated.
Given our experimental findings and what has been previously reported regarding IL-13 and the pathogenesis of fibrosis, we hypothesized that anti–IL-13 treatments might improve the clinical course in patients with IPF. Three recent phase II trials (55–58), however, did not demonstrate evidence of benefit associated with anti–IL-13 therapy. It is important to consider these clinical trial results in light of our experimental data and the limited clinical data that exist about lung IL-13 levels in patients with IPF. First of all, we contend that only a subset of patients included in these studies had a “high” IL-13 phenotype, and as a result, the clinical trials were not powered adequately to observe a benefit. The limited published data demonstrating IL-13 and IL-4 elevations in IPF BALF reveal that there is substantial heterogeneity in the levels of these Th2 cytokines in individual patients (59, 60). Although 1 trial attempted to address this concern (with analysis of POSTN-dichotomized groups), we contend that POSTN may not be a reliable surrogate marker for IL-13 activity in IPF (61) given that it can be induced not only by IL-13, but also by other factors, such as TGF-β1, -β2, and -β3; BMPs 2 and 4; VEGF; and other interleukins (briefly reviewed in ref. 62). POSTN has been identified as a biomarker that is associated with IPF prognosis (63, 64), but to the best of our knowledge, POSTN levels have not been directly correlated with IL-13 levels in IPF. Next, it has been proposed by others that direct targeting of IL-13 may enhance the expression of profibrotic/inflammatory transcripts (65), and alternative indirect IL-13–targeting strategies should be considered. Moreover, our data suggest that IL-4 and IL-13 have redundant effects on AEC2 stem cell biology, and targeting only 1 of these cytokines with a specific monoclonal antibody may be insufficient. This redundancy was addressed in the recent trial with SAR156597, a drug that neutralizes both IL-13 and IL-4, and notably, a positive trend on acute exacerbations was observed (56). Finally, we argue that targeting IL-13 (or IL-13 and IL-4) beyond the early stages of IPF is probably too late because remodeling of the ECM will have occurred and the lung is stiff. The resulting biomechanical alterations in the niche may prevent alveolar epithelial repair from proceeding normally (66).
Moving forward, we envision a number of scenarios in which it would be beneficial to phenotype patients with chronic lung disease based on Th2 cytokine levels so that clinical care and clinical trial design can become more personalized. Because IL-13 is present in the serum at very low levels (making detection difficult) (59), alternative biomarkers must be explored. Our data show that IL-13 induces the expression of a number of other Th2-related proteins from AEC2s. This raises the possibility that a biomarker panel incorporating these proteins to phenotype patients would have great clinical utility, allowing the clinical research community to characterize disease trajectory or treatment response in Th2-high versus -low patients and allowing for more nuanced clinical trial design in the future. Using this approach, inhibition of IL-13 in IPF and other respiratory diseases may represent a promising path for treatment (60).
Methods
Mice.
Mice used were Sftpc-CreERT2 (67) and Rosa26R-CAG-tdTm (Rosa-Tm) (previously described in ref. 67); B6.129S4-Pdgfratm11(EGFP)sor/J (Pdgfra-H2B:GFP) from The Jackson Laboratory (strain 007669); Stat6-null mice from The Jackson Laboratory (strain 005977, ref. 68); Krt5GFP-Tg mouse line (previously described in ref. 69); and Scgb1a1-CreERT2 (previously described in ref. 29); and Scgb1a1-Il-13-Tg mice, a gift from Jack Elias (previously described in ref. 23). All mice were on a C57BL/6 background and were older than 8 weeks at the outset of experiments detailed herein. Males and females were used in roughly equal numbers, 3–11 per group.
TMX administration.
TMX (T5648; MilliporeSigma) was a 20 mg/mL stock solution in corn oil and given via oral gavage. Mice received 2–4 doses of 0.2 mg/g every other day to induce cell labeling.
PNX and sham surgery.
Mice were anesthetized with isoflurane for 3 minutes until breathing slowed. The left chest and neck were shaved and cleaned with betadine and 70% ethanol. The neck was dissected to reveal the trachea, and the trachea was intubated for mechanical ventilation. Buprenorphine SR (1 mg/mL) 1 μL/g was injected subcutaneously at the outset of the procedure for pain control. The chest wall was dissected by layers, and the thorax was opened to expose the left lung. For mice undergoing PNX, the main bronchus and blood vessels were exposed, isolated, and clamped. The left lung was removed. Sham-operated mice had the thorax opened similarly but no lung removal. The rib cage was sutured back together, and chest wall layers were re-approximated with staples after aspiration of 0.5 mL remaining air inside the hemithorax. Additional anesthetic (50 μL bupivacaine) was injected into each wound. Anesthetic gas was weaned and mice remained on a ventilator until able to breathe without support. Mice were sacrificed 2 weeks from surgery date, and lungs were embedded for immunohistochemical staining.
Lung dissociation and FACS.
Murine lungs were dissociated as described previously (67). Sorting was performed on FACSVantage SE, and data were analyzed with FACSDiva (BD Biosciences). LT staining was performed per manufacturer instructions (LysoTracker Deep Red; Invitrogen, Thermo Fisher Scientific L12492). FACS-sorted cells were used in cell culture, as described below. Human tissue was dissociated in a similar manner with protease solution; important differences in processing and sorting for culture are described below.
Murine cell culture.
Sorted cells were plated as described previously (5). For secretomic analysis, medium was collected for each condition separately before each feeding starting at day 6. Collected medium was stored at –80°C.
Histology and immunohistochemistry.
Lungs were inflated to 20 cm H2O pressure with 4% paraformaldehyde for 20 minutes. After inflation, lungs and airways were removed together and fixed for 3 hours in 4% PFA at 4°C with the trachea tied off, followed by PBS wash overnight. Cell culture inserts were fixed with 4% paraformaldehyde for 30 minutes at 4°C, followed by a wash in PBS overnight. Tissue/inserts were embedded in paraffin or OCT. Paraffin sections (7 μm) and cryosections (12 μm) were prepared as described previously (67). Paraffin sections underwent 10 mM sodium citrate antigen retrieval. Antibodies were as follows: rat, DC-LAMP/CD208 (1:250; DDX0191; Dendritics); rabbit, SFTPC (1:1000; ab3786; Abcam); rabbit, HOPX (1:250; SC-30216; Santa Cruz Biotechnology); rabbit, keratin 5 (1:1000; PRB-160P; Covance); hamster, PDPN (T1-α) (1:1000; clone 8.1.1; DSHB); rabbit, RFP (1:250; 600–401379; Rockland); goat, SCGB1A1 (CC10) (1:200; gift from Barry Stripp, Cedars Sinai, Los Angeles, California, USA); guinea pig, LPCAT (1:2000, gift from John Shannon, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA); rat, KI-67 (1:250; 14-5698-82; Invitrogen, Thermo Fisher Scientific); chicken, GFP (1:500; GFP1020; Aves Lab); rabbit, SOX9 (1:1000, ab53121; Abcam); mouse, p63 (1:200; 58033; One World Lab); rat, RAGE (1:200, MAB1179, R&D Systems); rabbit, AQP5 (1:200, ab78486, Abcam); rabbit, ABCA3 (1:250, WRAB-ABCA3, Seven Hills); mouse, MUC-5ac (1:200, ab77995, Abcam); mouse, α-SMA (1:250, A2547, MilliporeSigma); and human, HTII-280 (1:60, gift from Leland Dobbs, UCSF, San Francisco, California, USA). Alexa Fluor–coupled secondary antibodies (Invitrogen, Thermo Fisher Scientific) were used at 1:500: 488–anti-rat (A21208), 488–anti-rabbit (A21206), 488–anti-goat (A11055), 488–anti-mouse (A21202), 488–anti-hamster (A21110), 555–anti-mouse (A31570), 555–anti-rabbit (A31572), 555–anti-rat (A21434), 647–anti-goat (A21447), 647–anti-hamster (A21451), 647–anti-mouse (A31571), 647–anti-rabbit (A31573), and 647–anti-rat (A21247).
Confocal microscopy.
All images for scoring cells and immunohistochemical staining analysis consisted of a Z stack of multiple optical sections and were captured using either an Olympus Fluoview FV3000 confocal microscope (associated software system used was Fluoview FV31S-SW) or a Zeiss 710 Inverted Confocal. Image analysis and counting were performed using ImageJ/Fiji (NIH).
Cell harvest from Matrigel for FACS sorting and downstream RNA-Seq, proteomic analysis, and Western blot.
Matrigel inserts were dissociated with Dispase (60 μL per well; 30 minutes at 37°C; catalog 354235, 5 U/mL; BD Biosciences) and organoids were retrieved. Organoids were trypsinized (0.1% trypsin-EDTA) to generate a single-cell suspension, and this was subjected to FACS for separation and collection of lineage-labeled epithelial cells. Cells were pelleted and frozen at –80°C before protein isolation.
ELISA for IL-13Rα2 levels in conditioned medium.
Conditioned medium was collected at defined time points. ELISA was performed with the Mouse IL-13 R alpha 2 Quantikine ELISA Kit (M13RA2; R&D Systems) per manufacturer instructions.
RNA-Seq.
RNA was isolated from FACS-isolated, lineage-labeled epithelial cells using RNAqueous-Micro Total RNA Isolation Kit (AM1931, Invitrogen, Thermo Fisher Scientific). cDNA synthesis and sequencing were performed by IGSP Genome Sequencing and Analysis Core Resource (Duke University). Data quality control was performed to remove adapter and low-quality bases using Cutadapt. Reads were aligned to reference using TopHat2 with strand-specific and multiple-hit options. Differential expression analysis was performed using Cuffdiff (upper quartile normalization, multiple-hit corrections, and strand-specific options) (Supplemental Table 2). The data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO accession number GSE142629 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142629).
Proteomic analysis of sorted cells.
Cell pellets in residual sorting buffer were adjusted to 0.2% acid-labile surfactant synthesized as described previously (70) in 50 mM ammonium bicarbonate, pH 8, followed by probe sonication (Misonix 1/8″ microprobe). After centrifugation at 15,000 g for 10 minutes, protein concentrations were determined by Bradford assay (Pierce Coomassie Plus), and 10 μg of each sample was reduced with 10 mM DTT at 80°C for 5 minutes and alkylated with 25 mM iodoacetamide (IAM) at room temperature for 30 minutes. Excess IAM was quenched with an additional 10 mM DTT, and proteins were digested overnight with 1:25 (w/w) trypsin/protein as previously described (71).
Protein digests were analyzed by nanoflow LC-MS/MS using a nanoACQUITY UPLC system (Waters) coupled to a Fusion Lumos high-resolution accurate mass tandem mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ionization source. Briefly, 800 ng of peptide was trapped on a Symmetry C18 180 μm × 20 mm trapping column (5 μL/min at 99.9/0.1 v/v H2O/MeCN) followed by an analytical separation using a 1.7-μm ACQUITY UPLC HSS T3 C18 75 μm × 250 mm column (Waters) with a 90-minute gradient of 5%–30% acetonitrile (MeCN) with 0.1% formic acid at a flow rate of 400 nL/min and column temperature of 55°C. Data collection on the Fusion Lumos MS was performed in data-dependent acquisition (DDA) mode with a 240,000-resolution (at m/z 200) full MS scan from m/z 375 to m/z 1600 with a target automatic gain control (AGC) value of 2e5 ions and 50-ms maximum injection time (IT) and advanced precursor determination and internal calibration enabled. Peptides were selected for MS/MS using charge state filtering (2–5), monoisotopic precursor selection, and a dynamic exclusion of 20 seconds. MS/MS was performed using higher energy C-Trap dissociation with a collision energy of 30% ± 5% with detection in the ion trap using rapid scanning, an AGC target of 1e4, and maximum IT of 30 ms. A time-dependent (1 second) method was used.
Proteomic analysis of conditioned media.
Five hundred microliters of conditioned media was concentrated using Millipore Amicon Ultra-0.5 centrifugal filters (10 kDa) followed by buffer exchange with 50 mM triethammonium bicarbonate (TEAB) at pH 8.5 and Bradford assay. One hundred micrograms of each sample was adjusted to 5% (w/v) deoxycholate and reduced and alkylated as above, then digested with 1:10 (w/w) N-tosyl-l-phenylalanine chloromethyl ketone–trypsin for 4 hours. Samples were acidified with a final 1% (v/v) trifluoroacetic acid (TFA) and 2% (v/v) MeCN followed by centrifugation 15,000 g for 5 minutes, and supernatants were lyophilized. Peptides were reconstituted in 200 mM TEAB followed by labeling with Tandem Mass Tag (TMT) reagents (Thermo Fisher Scientific) according to the manufacturer’s protocol. Labeling was as follows: 127N, control rep 1; 127C, control rep 2; 128N, control rep 3; 129N, IL-13 rep 1; 129C, IL-13 rep 2; 130N, IL-13 rep 3; 130C, control media rep 1; 131N, control media rep 2; and 131C, control media rep 3. After quenching with hydroxylamine, TMT reactions were combined to a single sample and lyophilized.
Labeled peptides were reconstituted in 0.1% (v/v) formic acid, and 550 μg of total peptide was fractionated using high pH reversed phase chromatography. Briefly, peptides were fractionated using a 2.1 mm × 5 cm ethylene bridged hybrid C18 column (Waters) and Agilent 1100 HPLC. Separations utilized a flow rate of 0.4 mL/min and column temperature of 55°C, and mobile phases consisted of 20 mM ammonium formate, pH 10 (MPA), and neat MeCN (MPB). Separations used a gradient of 7%–35% MPB over 50 minutes and 35%–90% MPB over 10 minutes. Ninety-six equal fractions were collected over 60 minutes (~0.25 mL each fraction) and reconcatenated into 24 fractions (e.g., fraction 1 was pooled from wells 1, 25, 49, and 73) followed by lyophilization and reconstitution in 1% TFA/2% MeCN.
Individual fractions were analyzed using microflow LC-MS/MS using a Waters ACQUITY LC interfaced to a Thermo Fisher Scientific Q-Exactive HF-X MS. Briefly, approximately 20 μg of fractionated peptide was separated on a 1 mm × 15 mm 1.7-μm charged surface hybrid C18 column using a flow rate of 100 μL/min, a column temperature of 55°C, and a gradient of 3%–28% (v/v) MeCN/H2O containing 0.1% formic acid over 60 minutes and interfaced to the HF-X via a heated electrospray ionization source with default tune parameters for the 100 μL/min flow rate. DDA used the TMT method (with advanced peak detection disabled) with 120,000-resolution precursor ion (MS1) scan from 375–1500 m/z, AGC target of 3E6, and maximum IT of 50 ms. MS/MS used a top10 method with 45,000 resolution, AGC target of 1e5 and minimum AGC target of 1e4 with 96-ms IT, an normalized collision energy of 32, isolation width of 0.7 m/z, and dynamic exclusion of 20 seconds.
Proteomics data analysis.
Data were analyzed using Proteome Discoverer v. 2.2 (Thermo Fisher Scientific). For label-free quantitation of the AEC2 proteome, MS/MS data were searched against a UniProt database with Mus musculus taxonomy (downloaded on September 5, 2017), which contained additional entries for EGFP, tdTomato, BSA, and yeast alcohol dehydrogenase 1 (16,898 total entries). Search parameters included trypsin specificity, up to 2 missed cleavage sites, 5-ppm precursor and 0.6-Da product ion tolerances, fixed modification on Cys (carbamidomethyl) and variable deamidation (NQ), Gln → pyroGlu, and acetyl protein N-terminus. Percolator was used for estimation of false discovery rate (FDR), and data were annotated at a 1% peptide and protein FDR. Label-free quantitation used the Minora Feature Detection and Feature Mapper nodes with chromatographic alignment using fine parameter tuning, a max rt shift of 5 minutes and tolerance of 5 ppm, and feature linking using a 2-minute retention time (rt) tolerance, a 5-ppm mass tolerance, and min signal-to-noise (s/n) threshold of 1. Protein quantitation used unique and razor peptides. Data were normalized to total peptide quantity across, with the exclusion of serum albumin peptides, and imputation used low-abundance resampling. Fold changes were calculated based on the average of each of the individual paired ratios, and P values were calculated from log2-transformed data using a paired, 2-tailed t test in Excel.
For TMT quantitation of the organoid secretome, database searching used Sequest HF as described above with semitryptic specificity, a 0.02-Da product ion tolerance, fixed Cys carbamidomethylation, Lys and peptide N-terminal TMT label, and variable deamidation. The Reporter Ion Quantifier processing node used a 10-ppm integration tolerance, and the Reporter Ion Quantifier consensus node used unique and razor peptides, a coisolation threshold of 50%, and an average reporter s/n threshold of 5. Quan values were corrected for TMT reagent isotype impurities. Normalization, imputation, and statistical analyses were as described for the cell proteome.
Proteomic data availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRoteomics IDEntifications partner repository with the data set identifier PDX013006.
Human cell culture.
Dissociated human lung tissue was resuspended in 10 mL of DMEM/F12 + 10% at 37°C. Cell suspension was centrifuged for 5 minutes at 300 g. Supernatant was aspirated before resuspending in blocking solution (2% BSA, 2% FBS in PBS with 1:100 Human True Stain FcX block; BioLegend) for 15 minutes at 4°C. Primary antibodies (human, HTII-280 1:60) were added directly to blocking solution for 1 hour away from light. Samples were rinsed with 5 mL FACS buffer, centrifuged, and resuspended with secondary antibody (40 μL magnetic beads and 160 μL FACS buffer) for 30 minutes at 4°C. Samples were rinsed with 5 mL FACS buffer, centrifuged, and resuspended in 1 mL FACS buffer before being sorted via magnetic-activated cell sorting (MACS). All cells expressing HTII-280 were collected after samples passed through the MACS column, then were centrifuged and resuspended in FACS buffer at a concentration of 1000 cells/μL, confirmed by manual counting. MRC5s (fetal human fibroblast cell line obtained from ATCC and verified mycoplasma clean by distributor) were cocultured with human AEC2s: 10,000 AEC2s and 50,000 MRC5s were plated in each 24-well 0.4-μm Transwell insert (Falcon) combined in a 1:1 ratio with growth factor–reduced Matrigel (BD Biosciences). Each well was fed every other day with 500 μL of ALI medium (47) in the lower chamber. Rho-associated protein kinase inhibitor (10 μM, Y0503; MilliporeSigma) was included in the medium for the first 4 days of growth. Cultures were incubated at 37°C in 5% CO2/air. Plates were imaged every 7 days.
Statistics.
Data are expressed as the mean ± SD in the main text and figure legends. All experiments were repeated 3 or more times, with the exception of the IL-13 dose-response experiment (Supplemental Figure 2) and the experiment to test the addition/removal of IL-13 at different times from culture (Supplemental Figures 3 and 4). No animals were excluded from analysis. Biological replicates were sex and age matched whenever possible. Variable differences between experimental and control groups were assessed using 2-tailed t test. One-way ANOVA was used for multiple comparisons. Statistical significance is indicated by the following: *P < 0.05; **P < 0.005; and ***P < 0.0005.
Study approval.
Mouse experiments were performed under IACUC guidelines and approved protocols at Duke University. Human lung epithelial cells were isolated under protocols approved by the Institutional Review Boards at both Duke University and University of North Carolina at Chapel Hill.
Author contributions
KMG performed experiments, interpreted data, and wrote the manuscript. AJS performed experiments and interpreted data. SVP performed experiments and interpreted data. AB performed experiments. MAM supervised proteomics work. MWF performed proteomics work, interpreted data, and wrote the manuscript. CEB designed experiments, performed experiments, interpreted data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Brigid Hogan, Rob Tighe, Claude Piantadosi, and Lindsay Marjoram for critical reading of the manuscript. We thank our patients and their families for generous donation of tissue. This work was supported by K08 HL122521 (CEB), Burroughs Wellcome Fund Career Award for Medical Scientists (CEB), Duke Health Scholars Award (CEB), and NIH 2T32 HL007-36 (Institutional Training Grant, KMG).
Version 1. 01/16/2020
Electronic publication
Version 2. 02/24/2020
Corrected publication date in pdf file.
Funding Statement
Awarded to CEB
Awarded to CEB
Institutional Training Grant, KMG
Awarded to CEB
Footnotes
SVP’s current address is: Department of Molecular Biology, Princeton University, Princeton, New Jersey, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(1):e131232.https://doi.org/10.1172/jci.insight.131232.
Contributor Information
Kristen M. Glisinski, Email: kristen.glisinski@duke.edu.
Adam J. Schlobohm, Email: adam.schlobohm@duke.edu.
Sarah V. Paramore, Email: paramore@princeton.edu.
Anastasiya Birukova, Email: anastasiya.birukova@duke.edu.
M. Arthur Moseley, Email: arthur.moseley@duke.edu.
Matthew W. Foster, Email: mwfoster@duke.edu.
Christina E. Barkauskas, Email: christina.barkauskas@duke.edu.
References
- 1.Iyonaga K, Miyajima M, Suga M, Saita N, Ando M. Alterations in cytokeratin expression by the alveolar lining epithelial cells in lung tissues from patients with idiopathic pulmonary fibrosis. J Pathol. 1997;182(2):217–224. doi: 10.1002/(SICI)1096-9896(199706)182:2<217::AID-PATH833>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Smirnova NF, Schamberger AC, Nayakanti S, Hatz R, Behr J, Eickelberg O. Detection and quantification of epithelial progenitor cell populations in human healthy and IPF lungs. Respir Res. 2016;17(1):83. doi: 10.1186/s12931-016-0404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seibold MA, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS ONE. 2013;8(3):e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkauskas CE, Noble PW. Cellular mechanisms of tissue fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am J Physiol, Cell Physiol. 2014;306(11):C987–C996. doi: 10.1152/ajpcell.00321.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zacharias WJ, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555(7695):251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zepp JA, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170(6):1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BLM. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145(9):dev163014. doi: 10.1242/dev.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiraishi K, et al. Mesenchymal-epithelial interactome analysis reveals essential factors required for fibroblast-free alveolosphere formation. iScience. 2019;11:318–333. doi: 10.1016/j.isci.2018.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsura H, Kobayashi Y, Tata PR, Hogan BLM. IL-1 and TNFα Contribute to the Inflammatory Niche to Enhance Alveolar Regeneration. Stem Cell Reports. 2019;12(4):657–666. doi: 10.1016/j.stemcr.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuperman DA, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med. 2002;8(8):885–889. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 12.de Vries JE. The role of IL-13 and its receptor in allergy and inflammatory responses. J Allergy Clin Immunol. 1998;102(2):165–169. doi: 10.1016/S0091-6749(98)70080-6. [DOI] [PubMed] [Google Scholar]
- 13.Liao N, Zhao H, Chen ML, Xie ZF. Association of the IL-13 polymorphisms rs1800925 and rs20541 with chronic obstructive pulmonary disease risk: An updated meta-analysis. Medicine (Baltimore) 2017;96(47):e8556. doi: 10.1097/MD.0000000000008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding M, et al. Association of the SNP rs1800925(C/T) in the interleukin-13 gene promoter with pulmonary function in Chinese Han patients with idiopathic pulmonary fibrosis. Cell Biochem Biophys. 2013;67(3):905–909. doi: 10.1007/s12013-013-9580-1. [DOI] [PubMed] [Google Scholar]
- 15.Murray LA, et al. Targeting interleukin-13 with tralokinumab attenuates lung fibrosis and epithelial damage in a humanized SCID idiopathic pulmonary fibrosis model. Am J Respir Cell Mol Biol. 2014;50(5):985–994. doi: 10.1165/rcmb.2013-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SW, et al. Interleukin-13 and its receptors in idiopathic interstitial pneumonia: clinical implications for lung function. J Korean Med Sci. 2009;24(4):614–620. doi: 10.3346/jkms.2009.24.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock A, Armstrong L, Gama R, Millar A. Production of interleukin 13 by alveolar macrophages from normal and fibrotic lung. Am J Respir Cell Mol Biol. 1998;18(1):60–65. doi: 10.1165/ajrcmb.18.1.2627. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z, et al. IL-13-induced chemokine responses in the lung: role of CCR2 in the pathogenesis of IL-13-induced inflammation and remodeling. J Immunol. 2002;168(6):2953–2962. doi: 10.4049/jimmunol.168.6.2953. [DOI] [PubMed] [Google Scholar]
- 19.Zheng T, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000;106(9):1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med. 2006;12(1):99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 21.Lee CG, et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1) J Exp Med. 2001;194(6):809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passalacqua G, et al. IL-13 and idiopathic pulmonary fibrosis: Possible links and new therapeutic strategies. Pulm Pharmacol Ther. 2017;45:95–100. doi: 10.1016/j.pupt.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103(6):779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 25.McCormick SM, Heller NM. Commentary: IL-4 and IL-13 receptors and signaling. Cytokine. 2015;75(1):38–50. doi: 10.1016/j.cyto.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rockich BE, et al. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci USA. 2013;110(47):E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiaramonte MG, et al. Regulation and function of the interleukin 13 receptor alpha 2 during a T helper cell type 2-dominant immune response. J Exp Med. 2003;197(6):687–701. doi: 10.1084/jem.20020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlins EL, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4(6):525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khatri A, Kraft BD, Tata PR, Randell SH, Piantadosi CA, Pendergast AM. ABL kinase inhibition promotes lung regeneration through expansion of an SCGB1A1+ SPC+ cell population following bacterial pneumonia. Proc Natl Acad Sci USA. 2019;116(5):1603–1612. doi: 10.1073/pnas.1816030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Velden JL, Bertoncello I, McQualter JL. LysoTracker is a marker of differentiated alveolar type II cells. Respir Res. 2013;14:123. doi: 10.1186/1465-9921-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills-Karp M, Finkelman FD. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci Signal. 2008;1(51):pe55. doi: 10.1126/scisignal.1.51.pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takagi M, et al. Ki-67 and condensins support the integrity of mitotic chromosomes through distinct mechanisms. J Cell Sci. 2018;131(6):jcs212092. doi: 10.1242/jcs.212092. [DOI] [PubMed] [Google Scholar]
- 34.Woodcock-Mitchell J, Mitchell JJ, Reynolds SE, Leslie KO, Low RB. Alveolar epithelial cell keratin expression during lung development. Am J Respir Cell Mol Biol. 1990;2(6):503–514. doi: 10.1165/ajrcmb/2.6.503. [DOI] [PubMed] [Google Scholar]
- 35.Gizak A, Rakus D, Kolodziej J, Zabel M, Ogorzalek A, Dzugaj A. Human lung fructose-1,6-bisphosphatase is localized in pneumocytes II. Histol Histopathol. 2001;16(1):53–55. doi: 10.14670/HH-16.53. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y, Zhou R, Rehg JE, Jackowski S. Role of phosphocholine cytidylyltransferase alpha in lung development. Mol Cell Biol. 2007;27(3):975–982. doi: 10.1128/MCB.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniscalco WM, Finkelstein JN, Parkhurst AB. De novo fatty acid synthesis by freshly isolated alveolar type II epithelial cells. Biochim Biophys Acta. 1983;751(3):462–469. doi: 10.1016/0005-2760(83)90307-7. [DOI] [PubMed] [Google Scholar]
- 38.Beers MF, Kim CY, Dodia C, Fisher AB. Synthesis of type II cell lamellar body lysozyme-15 kD protein (lbl-15) by perfused rat lung. Am J Respir Cell Mol Biol. 1994;11(2):240–248. doi: 10.1165/ajrcmb.11.2.8049085. [DOI] [PubMed] [Google Scholar]
- 39.Gibson KF, Phadke S. Intracellular distribution of lysozyme in rat alveolar type II epithelial cells. Exp Lung Res. 1994;20(6):595–611. doi: 10.3109/01902149409031739. [DOI] [PubMed] [Google Scholar]
- 40.Yogalingam G, Doyle IR, Power JH. Expression and distribution of surfactant proteins and lysozyme after prolonged hyperpnea. Am J Physiol. 1996;270(3 Pt 1):L320–L330. doi: 10.1152/ajplung.1996.270.3.L320. [DOI] [PubMed] [Google Scholar]
- 41.Gally F, Kosmider B, Weaver MR, Pate KM, Hartshorn KL, Oberley-Deegan RE. FABP5 deficiency enhances susceptibility to H1N1 influenza A virus-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2013;305(1):L64–L72. doi: 10.1152/ajplung.00276.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makowski L, Hotamisligil GS. The role of fatty acid binding proteins in metabolic syndrome and atherosclerosis. Curr Opin Lipidol. 2005;16(5):543–548. doi: 10.1097/01.mol.0000180166.08196.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzortzaki EG, et al. Active remodeling in idiopathic interstitial pneumonias: evaluation of collagen types XII and XIV. J Histochem Cytochem. 2006;54(6):693–700. doi: 10.1369/jhc.5A6835.2006. [DOI] [PubMed] [Google Scholar]
- 44.McDonough JE, et al. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight. 2019;4(22):131597. doi: 10.1172/jci.insight.131597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Slot AJ, et al. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278(42):40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez RF, Allen L, Gonzales L, Ballard PL, Dobbs LG. HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J Histochem Cytochem. 2010;58(10):891–901. doi: 10.1369/jhc.2010.956433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 48.Jacob A, et al. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell. 2017;21(4):472–488.e10. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doucet C, Brouty-Boyé D, Pottin-Clémenceau C, Canonica GW, Jasmin C, Azzarone B. Interleukin (IL) 4 and IL-13 act on human lung fibroblasts. Implication in asthma. J Clin Invest. 1998;101(10):2129–2139. doi: 10.1172/JCI741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T, et al. Regulation of found in inflammatory zone 1 expression in bleomycin-induced lung fibrosis: role of IL-4/IL-13 and mediation via STAT-6. J Immunol. 2004;173(5):3425–3431. doi: 10.4049/jimmunol.173.5.3425. [DOI] [PubMed] [Google Scholar]
- 51.Ito Y, et al. IL-13 induces periostin and eotaxin expression in human primary alveolar epithelial cells: comparison with paired airway epithelial cells. PLoS ONE. 2018;13(4):e0196256. doi: 10.1371/journal.pone.0196256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakubzick C, et al. Augmented pulmonary IL-4 and IL-13 receptor subunit expression in idiopathic interstitial pneumonia. J Clin Pathol. 2004;57(5):477–486. doi: 10.1136/jcp.2003.012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1(20):e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du Y, et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax. 2017;72(5):481–484. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker JM, et al. A phase 2 randomized controlled study of tralokinumab in subjects with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2018;197(1):94–103. doi: 10.1164/rccm.201704-0784OC. [DOI] [PubMed] [Google Scholar]
- 56.Raghu G, et al. SAR156597 in idiopathic pulmonary fibrosis: a phase 2 placebo-controlled study (DRI11772) Eur Respir J. 2018;52(6):1801130. doi: 10.1183/13993003.01130-2018. [DOI] [PubMed] [Google Scholar]
- 57.Maher TM, et al. The RIFF Study (Cohort B): a phase II, randomized, double-blind, placebo-controlled trial of lebrikizumab in combination with pirfenidone in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2018;197:A6168 [Google Scholar]
- 58.Swigris JJ, et al. The RIFF Study (Cohort A): a phase II, randomized, double-blind, placebo-controlled trial of lebrikizumab as monotherapy in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2018;197:A6167 [Google Scholar]
- 59.Arron JR, Choy DF, Scheerens H, Matthews JG. Noninvasive biomarkers that predict treatment benefit from biologic therapies in asthma. Ann Am Thorac Soc. 2013;10 Suppl:S206–S213. doi: 10.1513/AnnalsATS.201303-047AW. [DOI] [PubMed] [Google Scholar]
- 60.Wijsenbeek MS, Kool M, Cottin V. Targeting interleukin-13 in idiopathic pulmonary fibrosis: from promising path to dead end. Eur Respir J. 2018;52(6):1802111. doi: 10.1183/13993003.02111-2018. [DOI] [PubMed] [Google Scholar]
- 61.Izuhara K, et al. Roles of periostin in respiratory disorders. Am J Respir Crit Care Med. 2016;193(9):949–956. doi: 10.1164/rccm.201510-2032PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Norris RA, Moreno-Rodriguez R, Hoffman S, Markwald RR. The many facets of the matricelluar protein periostin during cardiac development, remodeling, and pathophysiology. J Cell Commun Signal. 2009;3(3-4):275–286. doi: 10.1007/s12079-009-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neighbors M, et al. Prognostic and predictive biomarkers for patients with idiopathic pulmonary fibrosis treated with pirfenidone: post-hoc assessment of the CAPACITY and ASCEND trials. Lancet Respir Med. 2018;6(8):615–626. doi: 10.1016/S2213-2600(18)30185-1. [DOI] [PubMed] [Google Scholar]
- 64.Okamoto M, et al. Periostin, a matrix protein, is a novel biomarker for idiopathic interstitial pneumonias. Eur Respir J. 2011;37(5):1119–1127. doi: 10.1183/09031936.00059810. [DOI] [PubMed] [Google Scholar]
- 65.Habiel DM, Espindola MS, Coelho AL, Hogaboam CM. Modeling idiopathic pulmonary fibrosis in humanized severe combined immunodeficient mice. Am J Pathol. 2018;188(4):891–903. doi: 10.1016/j.ajpath.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burgess JK, Mauad T, Tjin G, Karlsson JC, Westergren-Thorsson G. The extracellular matrix - the under-recognized element in lung disease? J Pathol. 2016;240(4):397–409. doi: 10.1002/path.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011;108(52):E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4(3):313–319. doi: 10.1016/S1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 69.Schoch KG, Lori A, Burns KA, Eldred T, Olsen JC, Randell SH. A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol. 2004;286(4):L631–L642. doi: 10.1152/ajplung.00112.2003. [DOI] [PubMed] [Google Scholar]
- 70.Yu YQ, Gilar M, Lee PJ, Bouvier ESP, Gebler JC. Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem. 2003;75(21):6023–6028. doi: 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]
- 71.Foster MW, et al. Proteomic analysis of primary human airway epithelial cells exposed to the respiratory toxicant diacetyl. J Proteome Res. 2017;16(2):538–549. doi: 10.1021/acs.jproteome.6b00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.