Abstract
Colitis, an inflammatory bowel disease, is caused by a variety of factors, but luminal microbiota are thought to play crucial roles in disease development and progression. Indole is produced by gut microbiota and is believed to protect the colon from inflammatory damage. In the current study, we investigated whether indole-3-carbinol (I3C), a naturally occurring plant product found in numerous cruciferous vegetables, can prevent colitis-associated microbial dysbiosis and attempted to identify the mechanisms. Treatment with I3C led to repressed colonic inflammation and prevention of microbial dysbiosis caused by colitis, increasing a subset of gram-positive bacteria known to produce butyrate. I3C was shown to increase production of butyrate, and when mice with colitis were treated with butyrate, there was reduced colonic inflammation accompanied by suppression of Th17 and induction of Tregs, protection of the mucus layer, and upregulation in Pparg expression. Additionally, IL-22 was increased only after I3C but not butyrate administration, and neutralization of IL-22 prevented the beneficial effects of I3C against colitis, as well as blocked I3C-mediated dysbiosis and butyrate induction. This study suggests that I3C attenuates colitis primarily through induction of IL-22, which leads to modulation of gut microbiota that promote antiinflammatory butyrate.
Keywords: Immunology, Inflammation
Keywords: Inflammatory bowel disease, T cells
Indole-3-carbinol attenuates colitis primarily through induction of IL-22, which leads to modulation of gut microbiota that promote antiinflammatory butyrate.
Introduction
Colitis is a disorder characterized by chronic inflammation and tissue destruction within the colon and intestinal tract. Ulcerative colitis and Crohn’s disease are 2 forms of colitis defined by the affected areas of inflammation and depth of destruction in the gastrointestinal tissue (1). In ulcerative colitis, the inflammation associated with the disease is regulated to the colon and rectum, with damage occurring at the superficial epithelial surface, whereas in Crohn’s disease, the inflammation can be found throughout the gastrointestinal tract and often extends deeper into the colonic tissue layer (2). The etiology of colitis, while not fully understood, can be attributed to dietary, genetic, and environmental factors, as well as dysregulation of the immune response and the luminal microbiota (3).
Research has shown that disruption in the normal distribution of gut microbiome, known as microbial dysbiosis, and metabolomic activity of the gut microbiota, such as certain short chain fatty acid (SCFA) production, promotes inflammation and colitis development (4). SCFAs have been shown to play a crucial role in colitis, in terms of both the promotion and regulation of this disease. For example, acetic acid is known to induce colitis in animal models (5, 6), whereas butyrate, the conjugate base form of butyric acid, was shown to reduce symptoms in ulcerative colitis patients by acting as an energy source for colonic epithelial cells for healing (7). Interestingly, research revealed that patients with Crohn’s disease have decreased expression of aryl hydrocarbon receptor (AhR) in lamina propria (LP) mononuclear cells (8). Also, mice deficient in AhR were more susceptible to developing colitis, having more severe symptoms compared with WT controls (9). AhR has gained recognition as a potential target in immunological processes by way of affecting the development of Tregs, associated with maintenance of intestinal homeostasis, and Th17 cells, which are often associated with proinflammatory responses leading to colitis (10, 11). Thus, AhR is a potential therapeutic target against colitis, and known ligands for this receptor could serve as critical preventive and therapeutic agents.
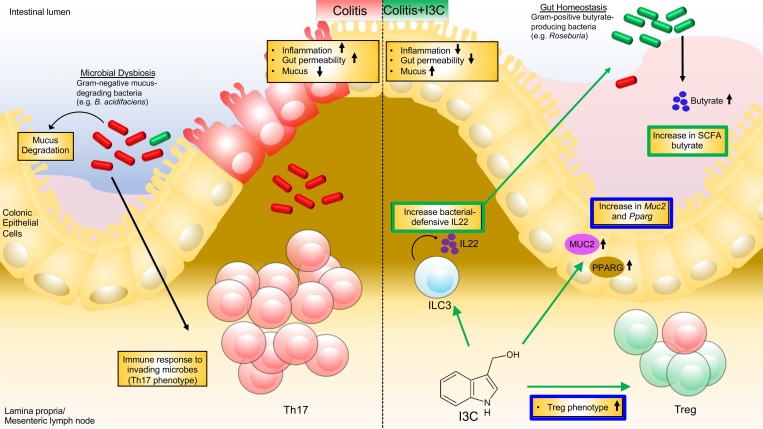
Indole-3-carbinol (I3C), a natural plant product found in many cruciferous vegetables, is a promising candidate for the treatment of colitis because it is both a known ligand for AhR (12, 13) and was shown to affect bacteria and bacterial byproducts, key components of the gut microbiome (14, 15). Some early reports showed that I3C was able to reduce the severity of dextran sodium sulfate–induced (DSS-induced) colitis, and this effect was lost in AhR–/– mice, resulting in a more severe disease state (16). However, currently, there are no studies reported that focus on how I3C treatment of colitis affects the gut microbial environment and, consequently, the inflammation in the gut. In the current study, therefore, we investigated the effect of I3C on 2,4,6-trinitrobenzenesulfonic acid–induced (TNBS-induced) colitis with concurrent analysis of the gut microbiome and metabolites. We demonstrate that I3C can modulate the gut microbiome during colitis in such a way that it prevents the overgrowth of potential pathogenic bacteria while selectively allowing antiinflammatory butyrate production, thereby inducing an antiinflammatory state. However, induction of IL-22, which has been shown to promote host defense against microbes (17), was found to occur after I3C administration but not with butyrate supplementation. Neutralizing IL-22 negated I3C-mediated protective effects against colitis, particularly by preventing alterations in the gut microbiome and increases in butyrate levels, suggesting that this cytokine was essential in regulating colitis-associated microbial dysbiosis.
Results
I3C treatment reduces colitis symptoms and alters CD4+ Th17 and Treg subsets in the mesenteric lymph node.
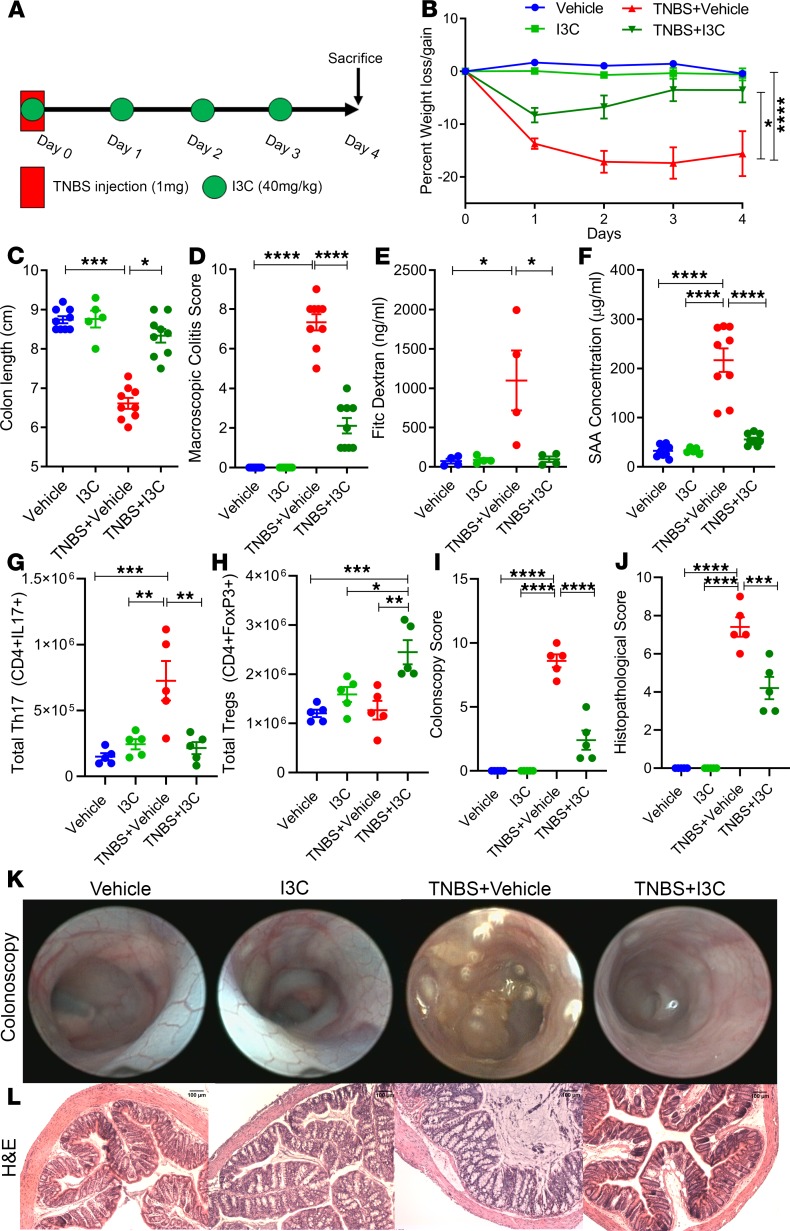
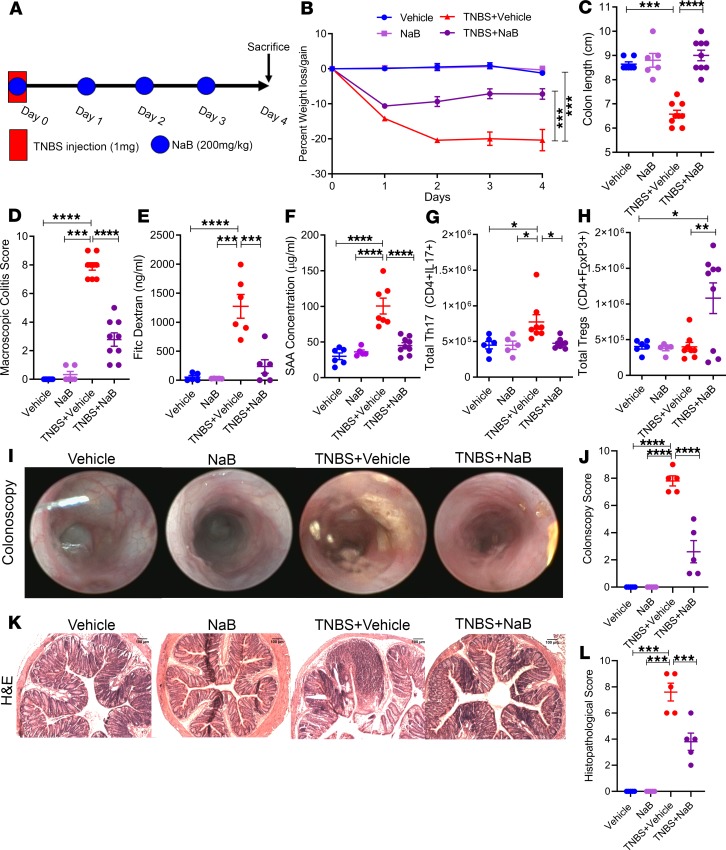
In order to test the efficacy of I3C treatment in the TNBS colitis model, mice with colitis were treated with an I3C regime previously reported by us to be effective in other types of inflammatory disorders (14, 18, 19), as illustrated in Figure 1A. We used 4 groups of mice: Vehicle alone, I3C alone, TNBS + Vehicle, and TNBS + I3C. We observed a 15%–20% reduction in weight of TNBS + Vehicle–treated mice (Figure 1B). However, in the TNBS + I3C group, this percentage of weight loss was significantly reduced. Mice induced with TNBS colitis also had significant reduction (~2 cm) in the overall length of the colon compared with control groups (Figure 1C). However, colitis mice treated with I3C prevented this colon-shortening effect. For the macroscopic colitis score, TNBS + Vehicle mice had significantly higher scores compared with controls, which were reduced in theTNBS + I3C group (Figure 1D). FITC-dextran gut permeability assay was also performed, and TNBS + Vehicle mice showed increased levels of detectable fluorescein in the circulating serum compared with controls, indicative of increased intestinal permeability (Figure 1E). In contrast, TNBS + I3C mice had much lower levels of FITC-dextran circulating in the blood, comparable with the baseline levels of controls. Next, we measured circulating levels of serum amyloid A (SAA), a common inflammatory biomarker found to be increased in patients with Crohn’s disease and ulcerative colitis (20). TNBS + Vehicle mice had significantly higher concentrations of SAA when compared with control mice, but these levels were reduced upon treatment with I3C (Figure 1F). To test if I3C was also effective against another model of colitis, we used DSS-induced colitis and found that I3C was able to significantly reduce disease parameters in the DSS model as it did in TNBS model, to include effects in weight loss, colon length, macroscopic colitis score, and SAA levels (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127551DS1).
Figure 1. Treatment with I3C resulted in amelioration of TNBS-induced colitis.
(A) TNBS colitis was induced in mice as described in Methods, and the efficacy of treatment with I3C was tested with the following experimental groups unless otherwise stated: Vehicle (n = 9), I3C (n = 5), TNBS + Vehicle (n = 9), TNBS + I3C (n = 9). (B–D) Disease parameters assessed included percent weight loss (B), colon length (C), and macroscopic score (D). (E) FITC-dextran was given to mice by oral gavage on day 3 of the TNBS model, and serum was collected 4 hours later to test gut permeability for Vehicle (n = 4), I3C (n = 4), TNBS + Vehicle (n = 4), and TNBS + I3C (n = 4), which are combined data from 2 independent experiments. (F) On day 3, serum was also collected to determine the levels of circulating SAA. (G and H) Bar plots depicting total cell numbers of Th17 (G) and Tregs (H) from MLN of experimental mice (n = 5). (I) Bar graph depicting colonoscopy scores from experimental mice (n = 5). (J) Bar graph depicting histopathological scores of H&E-stained colons from experimental mice (n = 5) (K) Representative colonoscopy images taken during day 3 of the TNBS model (n = 5). (L) Representative H&E stains of colons from experimental mice (n = 5). Scale bars: 100 μm (original magnification, ×100). Data are shown as mean ± SEM of combined data from at least 3 independent experiments unless otherwise stated. Significance was determined using 1-way ANOVA and Tukey’s multiple comparisons test; *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Previous reports — including results from our laboratory — have shown that, in the TNBS and DSS models of colitis, there is a significant increase in the inflammatory Th17 subset in the colon and colonic-associated mesenteric lymph node (MLN) (21–24), which plays a role in pathogenesis. To that end, we studied Th17/Treg subsets in the MLN and found that, in the TNBS + Vehicle group, there was a significant increase in the number of Th17 cells (Figure 1G). However, treatment with I3C prevented Th17 expansion and significantly increased Tregs in the MLN (Figure 1H).
Colonoscopy images revealed that the TNBS + Vehicle group had evidence of ulcerations and tissue sloughing in the lining of the colon, whereas these observations were far less in colonic linings of controls or colitis mice treated with I3C, resulting in a less severe colonoscopy score (Figure 1I). In support of these endoscopic observations, histological evaluation revealed overall tissue destruction in mucosa, submucosa, and LP layers; loss of crypts; and increased evidence of cellular infiltration in diseased mice when compared with controls (Figure 1J). Colons from TNBS-induced colitis mice treated with I3C, however, maintained crypt formation and normal colonic tissue architecture, and they showed reduced signs of cellular infiltration. Representative colonoscopy and H&E colon stains are depicted in Figure 1, K and L. Collectively, these data effectively demonstrate that I3C was able to protect mice from TNBS-mediated colitis, which was associated with a decrease in Th17 and increase in Tregs.
Treatment with I3C prevents TNBS colitis–associated microbial dysbiosis.
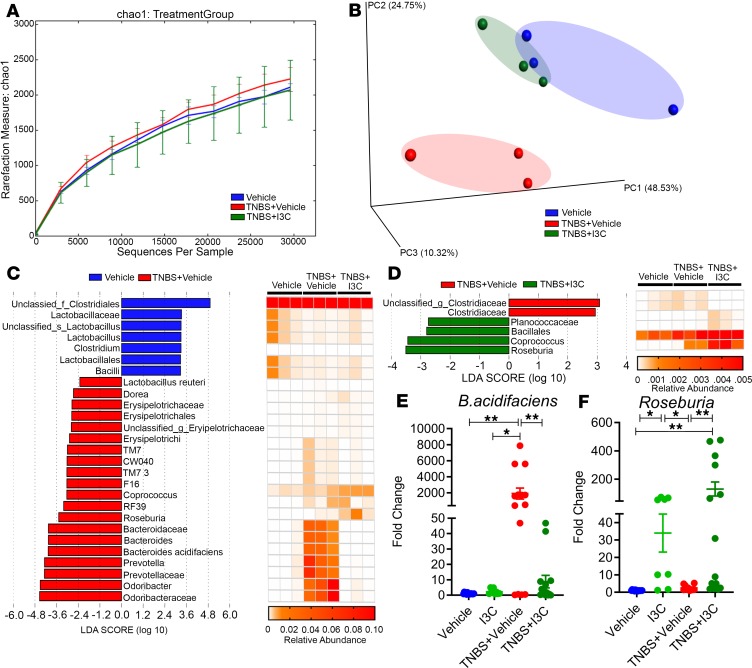
Next, we studied microbial dysbiosis induced by TNBS and the potential protective effect of I3C. We performed bacterial 16S rRNA gene sequencing studies using the MiSeq platform. Analysis of the sequenced data obtained from the colonic flushes was performed using the standardized online NIH-based analysis tool Nephele (25) — specifically, the 16S paired-end QIIME option — in order to determine differences in gut microbiome diversity, phylogeny, and function. In terms of the α diversity, or overall species richness within each sample, there were no significant differences detected, as illustrated by the chao1 rarefaction measurement (Figure 2A). However, in terms of the β diversity, which measures the similarity or dissimilarity between the experimental groups, there was evidence of clear separation of the disease group (TNBS + Vehicle) from the control (Vehicle) and treatment (TNBS + I3C) groups (Figure 2B). While the colitis disease samples clustered and separated from the other 2 groups, the control and I3C treatment samples clustered together much closer to one another. This suggested that the microbiome composition in the TNBS + I3C group resembled more closely the controls than the TNBS + Vehicle mice.
Figure 2. I3C alters gut microbiome composition in the TNBS colitis model.
(A and B) 16S rRNA sequencing from the colonic flushes was performed on Vehicle (n = 3), TNBS + Vehicle (n = 3), and TNBS + I3C (n = 3) experimental mice and represent 1 independent experiment. Sequenced reads were analyzed using Nephele to determine chao1 α diversity (A) and β diversity by PCA plot (B). (C) LDA scores (left) from LefSe analysis was performed on relative OTU abundances, depicted in corresponding heatmaps (right), between Vehicle vs. TNBS + Vehicle. (D) LDA scores (left) from LefSe analysis was performed on relative OTU abundances, depicted in corresponding heatmaps (right), between TNBS + Vehicle vs. TNBS + I3C. For LefSe data, the α factorial Kruskal-Wallis test among classes was set to 0.05, and the threshold on the logarithmic LDA score for discriminative features was set at 2. Sequencing data was validated by qPCR using primers for the various bacteria at the genus or species level for Vehicle (n = 14), I3C (n = 8), TNBS + Vehicle (n = 14), and TNBS + I3C (n = 14). Validation, expressed as fold change compared with Vehicle, was performed for B. acidifaciens (E) and Roseburia (F). Data are shown as ± SEM of data combined from 3 independent experiments. Significance was determined using 1-way ANOVA and Tukey’s multiple comparisons test; *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Phylogenetic analysis of the gut microbiome from the phylum to the species level was performed on the samples using standard operational taxonomic unit (OTU) classification (Supplemental Figures 2–7). A distinct bacterial signature was found in the TNBS + Vehicle mice that differed from the I3C-treated and controls — particularly in the enrichment of mostly gram-negative bacteria in the Bacteriodetes phylum. At the phylum level, mice with colitis had increased abundance in both Proteobacteria and the more recently discovered TM7 bacteria compared with control or I3C-treated mice (Supplemental Figure 3). A similar lineage pattern was seen at the class level, as Gammaproteobacteria and TM7-3 were enriched in TNBS + Vehicle mice, as well as at the order level with increased abundance of Pseudomonadales, a subset of Gammaproteobacteria, and CW040, a subset of TM7-3 (Supplemental Figures 4 and 5). At the family to genus level, TNBS + Vehicle mice were enriched in gram-negative bacteria (Bacteroides, Odoribacter, Prevotella, and Pseudomonas) compared with the other groups (Supplemental Figures 6 and 7). Of those disease-enriched bacteria, only Bacteroides and Prevotella were found to be significantly increased in the TNBS + Vehicle group when compared with controls or TNBS + I3C (Supplemental Figure 8, A and B). At the species level, Bacteroides acidifaciens (B. acidifaciens) was shown to be significantly more abundant in TNBS + Vehicle mice but was effectively reduced with I3C treatment (Supplemental Figure 8C). In almost all the bacterial taxa, the TNBS + I3C group had very similar microbial gut composition as the controls, except for 2 gram-positive Firmicutes enriched only in the TNBS + I3C treatment group (Planococcaceae and Roseburia) (Supplemental Figure 7). Among these 2, Roseburia was the only one found to be significantly enriched in the TNBS + I3C–treated mice compared with TNBS + Vehicle and control groups (Supplemental Figure 8D). Linear discriminant analysis effect size (LefSe) (26) was performed to identify any key microbial biomarkers that might differentiate both the disease and I3C-treated states. In comparisons of the Vehicle versus TNBS + Vehicle groups, linear discriminant analysis (LDA) scores (Figure 2C) and the corresponding cladogram (Supplemental Figure 9A) showed that colitis mice were most enriched in gram-negative Odoribacter, Prevotella, and Bacteroides, including B. acidifaciens. However, the Vehicle control mice had higher enrichment of gram-positive Firmicutes to include Bacilli (e.g., Lactobacillus) and Clostridium microbes. When comparing the TNBS + Vehicle with TNBS + I3C groups, Roseburia was found to have the highest LDA score and enrichment in the I3C-treated mice (Figure 2D and Supplemental Figure 9B). Collectively, these data show that a combination of enriched gram-negative bacteria (Odoribacter, Prevotella, and Bacteroides) could serve as potential biomarkers of colitis, whereas effective treatment with I3C was characterized by an enrichment in gram-positive bacteria in the genus Roseburia.
In order to further confirm results from the Nephele and LefSe data outputs, PCR was performed with primers specific for the species B. acidifaciens (Figure 2E) and genus Roseburia (Figure 2F). Results from the PCR validation studies confirmed observations from the sequencing data. Specifically, B. acidifaciens was found to be significantly increased in the TNBS + Vehicle mice (Figure 2E), while Roseburia was significantly higher in the TNBS + I3C group (Figure 2F). Taken together, the phylogenetic data reveal that I3C treatment effectively prevented TNBS-induced microbial dysbiosis, which was characterized by increases in gram-negative bacteria, but I3C also selectively increased gram-positive Roseburia, which are known to increase production of SCFA butyrate.
Treatment of TNBS colitis with I3C results in alterations in microbial function and SCFA production.
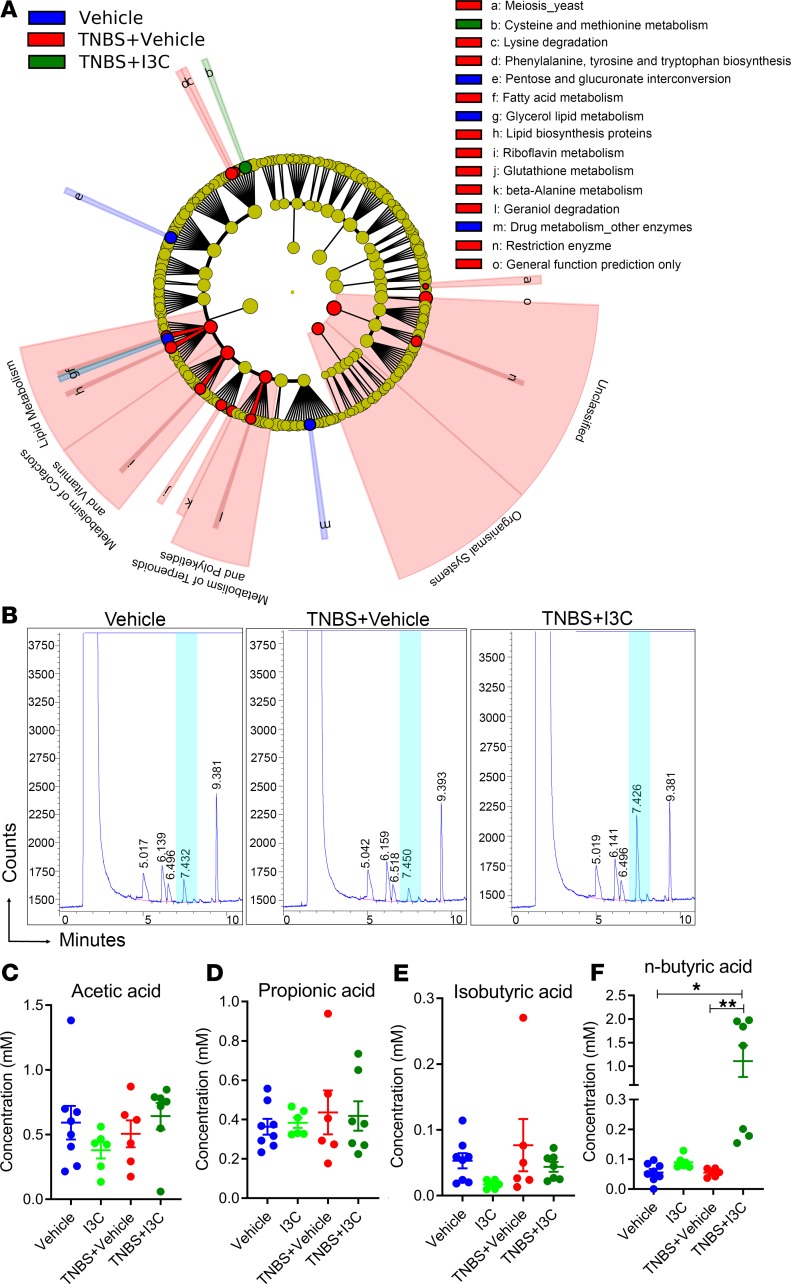
In addition to the phylogenetic results acquired from the 16S rRNA gene sequencing data, the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PiCRUSt) option in the Nephele analysis pipeline was used to determine how changes in the gut microbiome were affecting Level 2 (L2) and L3 KEGG pathways, a theoretical method used to determine potential changes in microbial functions based on bacterial OTU composition. At L2 pathways, TNBS mice had significant increases in percent abundances in the digestive system and amino acid metabolism, but these were reduced with I3C treatment (Supplemental Figure 10A). Of the 328 identified L3 pathways, 43 were found to be significantly altered either between the control and TNBS + Vehicle groups or between the TNBS + Vehicle and TNBS + I3C groups (Supplemental Figure 10B). Among those 43, 4 were found to be significantly enriched in the colitis samples compared with controls but were reduced after treatment with I3C. These pathways included riboflavin metabolism, tropane, piperidine and pyridine alkaloid biosynthesis, α-linolenic acid (ALA) metabolism, and ubiquitin system. Pentose and glucoronate interconversions and electron transfer carriers were the only 2 L3 pathways found to be significantly lower in abundance in the TNBS + Vehicle mice compared with controls, but they were restored in the TNBS + I3C–treated groups. Among all aforementioned significantly altered pathways, LefSe analysis showed that riboflavin metabolism and the pentose and glucoronate interconversion pathways could serve as biomarkers for the TNBS + Vehicle groups (Figure 3A and Supplemental Figure 11).
Figure 3. I3C alters gut microbial function and SCFA production in the TNBS colitis model.
16S rRNA sequencing from the colonic flushes from 1 independent experiment was performed on Vehicle (n = 3), TNBS + Vehicle (n = 3), and TNBS + I3C (n = 3) experimental mice. Sequenced reads were analyzed using the Nephele platform to determine Level 2 and Level 3 functions. (A) Cladogram generated from LefSe analysis of Level 2 and Level 3 KEGG pathways. For LefSe data, the α factorial Kruskal-Wallis test among classes was set to 0.05, and the threshold on the logarithmic LDA score for discriminative features was set at 2. (B) SCFA analysis was performed on Vehicle (n = 8), I3C (n = 6), TNBS + Vehicle (n = 6), and TNBS + I3C (n = 7) experimental mice from colonic flushes that were acidified using metaphosphoric acid and analyzed by GC-FID, with represented GC plots depicted. Data depicted are combined from 2 independent experiments. Highlighted in blue is the significantly altered peak corresponding to n-butyric acid. (C–F) Detected SCFAs included acetic acid (C), propionic acid (D), isobuytric acid (E), and n-butyric acid (F). Data are shown as mean ± SEM, with significance determined by 1-way ANOVA and Tukey’s multiple comparisons test; *P < 0.05; **P < 0.01.
Next, we measured SCFA concentrations in colonic flushes, as shown in representative GC plots in Figure 3B. Acetic acid (Figure 3C), propionic acid (Figure 3D), and i-butyric/isobutyric (Figure 3E) were not significantly altered among control, TNBS + Vehicle, and TNBS + I3C treatment samples. However, I3C treatment was found to significantly increase the concentration of n-butyric acid (Figure 3F) compared with control; disease studies showed that alterations in microbial composition and function caused by TNBS-induced colitis can be effectively prevented with I3C treatment and that I3C is able to alter butyrate production, perhaps through the ability of this compound, to selectively increase bacterial species in the genus Roseburia, which are known to produce high levels of butyrate.
Treatment with sodium butyrate prevents TNBS-induced colitis–associated symptoms.
Collectively, the results thus far showed that I3C was able to increase colonic butyrate levels in the TNBS model of colitis, which correlated with the observation that I3C treatment leads to an increase in butyrate-producing Roseburia spp. With this information, butyrate supplementation experiments were performed in lieu of standard microbiome transfer experiments to determine what I3C effects, if any, could be mediated through increased butyrate production. To that end, TNBS colitis mice were treated with sodium butyrate (NaB) as detailed in Figure 4A. The dose and route of NaB mimicked previous reports in other mouse models (27, 28) and correlated with similar colonic butyric acid concentrations seen in the I3C treatment experiments (29). In the experiments replacing I3C with NaB, striking similarities in results and outcomes were observed. NaB treatment was able to significantly reduce weight loss (Figure 4B), colon shortening (Figure 4C), and macroscopic colitis scores (Figure 4D) compared with the disease controls, similar to I3C. Treatment of TNBS colitis mice with NaB also reduced colitis-associated gut permeability (Figure 4E) and SAA levels (Figure 4F). T cell subtyping in the MLNs showed that NaB treatment was also able to effectively reduce the increases in disease-associated Th17 cells (Figure 4G) and significantly induce antiinflammatory Tregs (Figure 4H). Colonoscopy images on day 3 revealed that the diseased mice had ulcerations and tissue sloughing, which were not present in the TNBS mice treated with NaB (Figure 4I), reflected in an increased colonoscopy score (Figure 4J). Also, in the TNBS-induced colitis mice, the colon histology preparations showed significant amounts of cellular infiltration and complete destruction of the colonic tissue normal architecture, whereas in diseased mice treated with NaB, there were minimal signs of damage, in the form of elongated cellular crypt formation indicative of tissue healing processes (Figure 4K), reflected in the histopathological scores (Figure 4L). All of these observations indicated that NaB was able to ameliorate colitis disease parameters, but more importantly, the fact that these results closely resembled the outcomes in the I3C experiments strongly suggested that butyrate-induction by I3C is a key mechanistic factor in the natural compound’s effectiveness in counteracting the detrimental effects caused by colitis induction.
Figure 4. Treatment with NaB resulted in amelioration of TNBS-induced colitis.
(A) TNBS colitis was induced in mice to test the efficacy of treatment with NaB with the following experimental groups: Vehicle (n = 7), NaB (n = 6), TNBS + Vehicle (n = 9), and TNBS + NaB (n = 9), with data combined from 3 independent experiments unless otherwise stated. (B–D) Disease parameters accessed included percent weight loss (B), colon length (C), and macroscopic score (D). (E) FITC-dextran was given to experimental mice by oral gavage on day 3 of the TNBS model, and serum was collected 4 hours later to test gut permeability for Vehicle (n = 5), NaB (n = 5), TNBS + Vehicle (n = 6), and TNBS + NaB (n = 6). Data depicted are combined from 2 independent experiments. (F) On day 3, serum was also collected to determine the levels of circulating SAA for Vehicle (n = 6), NaB (n = 5), TNBS + Vehicle (n = 7), and TNBS + NaB (n = 9). Data are combined from 2 independent experiments. (G and H) Total cell numbers of T cell subsets in the MLN were determined for Th17 (G) and Tregs (H) for Vehicle (n = 6), NaB (n = 5), TNBS + Vehicle (n = 9), and TNBS + I3C (n = 9). (I and J) Representative colonoscopy images (I) taken during day 3 of the TNBS model and scored (J; n = 5). (K and L) Representative H&E stains (K) were scored appropriately (L; n = 5). Scale bars: 100 μm (original magnification, ×100). Images are combined representative samples from 3 independent experiments. Data are shown as mean ± SEM, and significance was determined using 1-way ANOVA and Tukey’s multiple comparisons test; *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
I3C and NaB prevent bacterial localization into colonic crypts, maintain protective mucus production, and increase Pparg expression during TNBS-induced colitis, but only I3C increases IL-22 production.
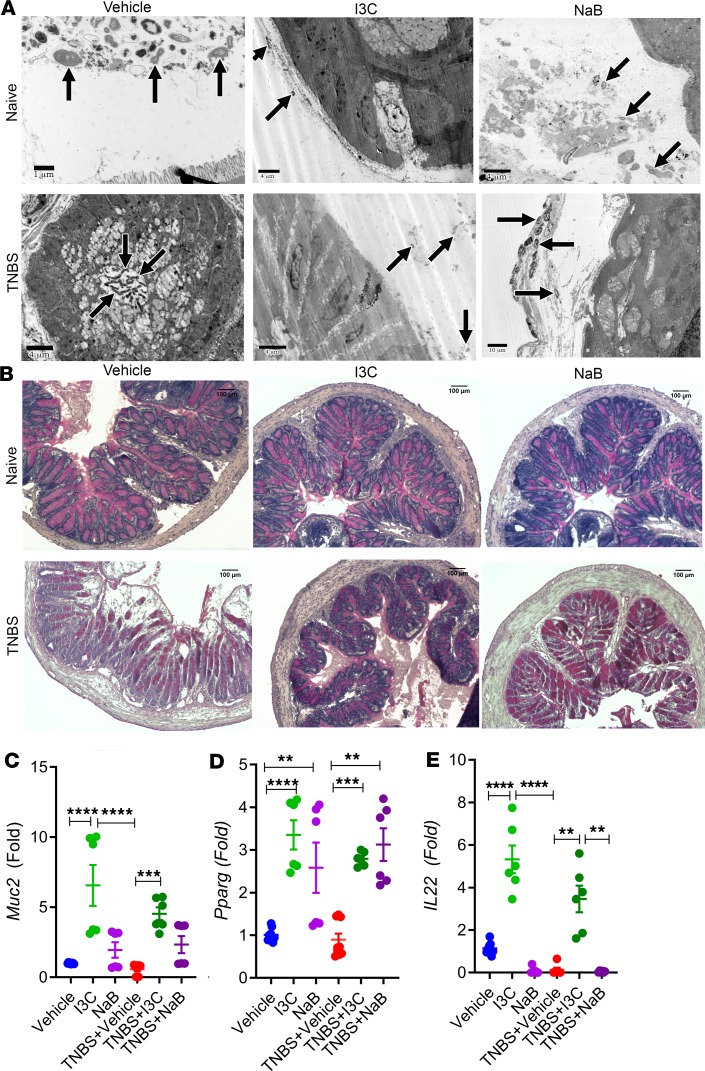
Because a continuous layer of mucus in the colon prevents lumen-bound bacteria from coming into close contact with the epithelial surface, and the fact that increases in mucus-degrading bacteria play a role in ulcerative colitis (30), we investigated the effect of I3C on mucus production in the colon. To examine bacterial localization, transmitting electron microscopy (TEM) was performed on colon tissues extracted from all experimental groups. Collected TEM images show the relative proximity of bacteria in the mouse colon (Figure 5A). In the Vehicle, I3C, and NaB groups, clear separation of intestinal microbes away from the intestinal epithelium surface was maintained. In the TNBS + Vehicle control, the image is a colonic crypt in cross-section with bacteria entering the crypt lumen, which was not present in any of the colon sections collected. Treatment of TNBS-induced colitis by I3C or NaB treatment reestablished the distance between intestinal epithelium and gut microbes that was disrupted by TNBS-induced colitis. Such localization of the bacteria in the colonic crypt region is indicative of breakdown in the protective mucus layer. To examine this further, PAS staining was performed on colon tissue sections. Control tissues (Vehicle, I3C, NaB) showed normal staining with bright magenta highlighting goblet cells and mucus secretion in the crypt lumen (Figure 5B). However, in the disease tissue of TNBS + Vehicle mice, the presence of goblet cells and mucus production was greatly reduced. Treatment of colitis mice with I3C or NaB restored the presence of crypt-associated goblet cells and mucus, indicating that these treatments were maintaining the protective mucus layer of the colon, which could be preventing bacteria from coming into close contact to the epithelial surface. To further substantiate this, colonic expression of Muc2 was determined, as Muc2 serves as a key component of the mucus layer in the colon (31). Results of colonic Muc2 expression showed that treatment with I3C significantly increased expression of mRNA for this protein when compared with TNBS + Vehicle or normal control groups (Figure 5C), thus reinforcing results from the PAS stains. While NaB was shown to trend toward an increase in Muc2 expression, especially compared with the disease state, the results were not found to be significant.
Figure 5. Treatment with I3C or NaB prevents bacterial localization into the colonic crypts, maintains mucus production, and leads to upregulation in Pparg, but it only I3C increases IL-22 in the TNBS model.
(A) Colonic tissue sections were fixed and imaged with TEM. Representative images are shown for Vehicle (n = 6), I3C (n = 6), NaB (n = 6), TNBS + Vehicle (n = 6), TNBS + I3C (n = 6), and TNBS + NaB (n = 6). Black arrows indicate bacteria present in proximity to the colonic luminal surface microvilli, with TNBS + Vehicle image being a cross-section showing bacteria in the colonic crypt area. Scale bars indicated within images. (B) Colonic tissue sections were fixed and subjected to PAS staining. Representative images are shown for Vehicle (n = 6), I3C (n = 6), NaB (n = 6), TNBS + Vehicle (n = 6), TNBS + I3C (n = 6), and TNBS + NaB (n = 6). Scale bars: 100 μm (original magnification, ×100). Images are representative from 1 experiment out of 2 independent experiments performed. (C–E) Real-time PCR detecting colonic mRNA expression for Muc2 (C), Pparg (D), and IL-22 (E) was performed on colonic tissue samples (Vehicle n = 9, I3C n = 6, NaB n = 6, TNBS + Vehicle n = 9, TNBS + I3C n = 6, and TNBS + NaB n = 6) run in triplicate for each experimental group, and data are combined from 2 independent experiments. Data are shown as mean ± SEM, and significance was determined using 1-way ANOVA and Tukey’s multiple comparisons test; **P < 0.01; ***P < 0.005; ****P < 0.001.
More recent reports have shown that receptor PPARG and cell signaling IL-22 play significant roles in intestinal health, where both are implicated in regulating the immune response in the colon epithelial, particularly to gut microbes (17, 32, 33). There is also evidence that butyrate can act as an agonist to the PPARG (34). With this knowledge, expression of mouse Pparg mRNA was determined. Colitis mice treated with either I3C or NaB had significant increases in Pparg compared with either naive controls or TNBS + Vehicle mice (Figure 5D). Interestingly, when examining the expression of colonic IL-22, only I3C, and not NaB, was able to significantly increase this cytokine (Figure 5E). Taken together, these studies highlight how I3C, perhaps through butyrate-dependent and butyrate-independent manners, can affect important key regulators of host-microbe interactions in the colon.
IL-22 neutralization negates protective effects of I3C against TNBS-induced colitis.
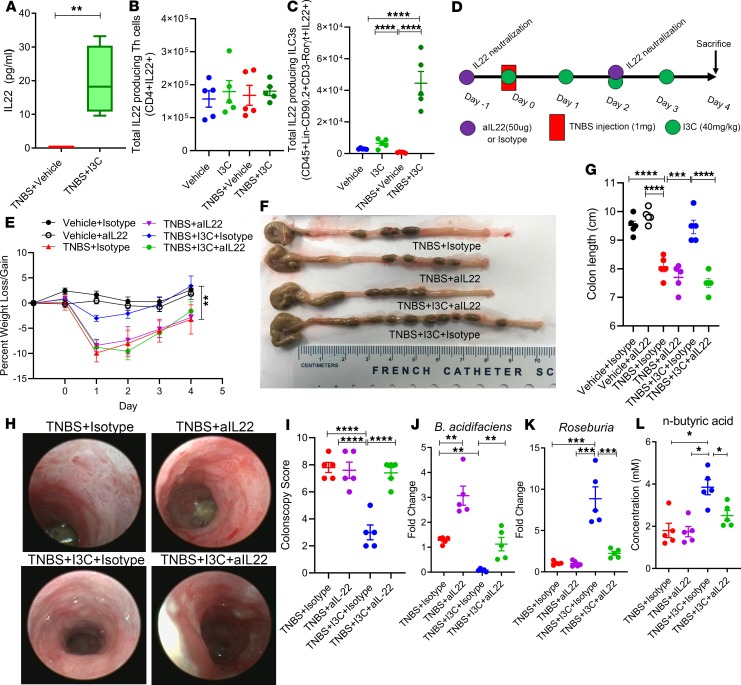
Because IL-22 expression levels were shown to increase after I3C treatment in the colon, secretion of this cytokine was evaluated during disease (TNBS + Vehicle) and treated (TNBS + I3C) states. Colon explants from experimental mice were excised and cultured overnight before detecting IL-22 secretion levels. As shown in Figure 6A, IL-22 was found to be secreted at higher levels after treatment with I3C compared with disease controls. With this in mind, the source of increased IL-22 secretion was determined by phenotyping the colonic LP fraction using flow cytometry. T helper cells that produce IL-22 were not found to be significantly altered among the experimental groups (Figure 6B). However, IL-22 in innate lymphoid Type 3 cells (ILC3s) was found to be significantly higher after treating with I3C (Figure 6C).
Figure 6. Neutralization of IL-22 prevents I3C-mediated protection against colitis and ability to increase butyric acid.
(A) Washed colonic explanted tissue (1 cm from cecum) were cultured for 24 hours (n = 5 per group). Supernatants were collected for analysis of IL-22 secretion by ELISA. (B) Total number of IL-22–expressing Th cells (n = 5 per group). (C) Total number of ILC3s expressing IL-22 (n = 5 per group). (D) TNBS colitis was induced in mice as described in Methods, and the efficacy of treatment with I3C in the presence or absence of neutralizing IL-22 antibody (aIL-22) was tested with the following experimental groups: TNBS + Isotype (n = 5), TNBS + IL-22 (n = 5), TNBS + I3C + Isotype (n = 5), and TNBS + I3C + aIL-22 (n = 5). Vehicle + Isotype (n = 5) and Vehicle + aIL-22 (n = 5) were included to assess weight and colon length compared with disease controls. (E–G) Weight loss (E) and colon length (F and G) were assessed. (H) Representative colonoscopy images taken during day 3 of the TNBS model are depicted, along with colonoscopy scores (I, n = 5). (J and K) Fold change of bacteria from colonic flushes using real-time PCR were determined for B. acidifaciens (J) and Roseburia (K). (L) SCFA n-butyric concentration was determined using GC-FID. Data are representative of 2 independent experiments. Data within dot plots and box-and-whisker plots equal the mean ± SEM, and significance was determined using 1-way ANOVA and Tukey’s multiple comparison test; *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
In order to determine if IL-22 secretion was necessary for I3C-mediated protection during colitis, I3C treatment of TNBS colitis experiments was repeated to include a neutralizing antibody against IL-22 (aIL-22), as detailed in Figure 6D. Interestingly upon aIL-22 treatment, the ability of I3C to prevent colitis-associated weight loss was reversed when compared with the isotype controls (Figure 6E). In addition, I3C treatment with the neutralizing antibody was not able to prevent colitis-associated shortening of the colon (Figure 6, F and G) nor damage to the colon (Figure 6, H and I). Most interesting was the loss in the ability of I3C to alter the gut microbiome after IL-22 neutralization. B. acidifaciens, which was decreased after I3C treatment with the isotype control (TNBS + I3C + Isotype), was no longer decreased after neutralizing IL-22 (TNBS + I3C + aIL-22) (Figure 6J). Even in the untreated disease state with neutralization of IL-22 (TNBS + Vehicle + aIL-22), there was a significant increase in B. acidfaciens when compared with the isotype control (TNBS + Vehicle + Isotype), suggesting that IL-22 plays an important role in regulating this colitis-associated bacterial species. Additionally, butyrate-producing Roseburia, which increased after I3C treatment (TNBS + I3C + Isotype), was no longer increased upon I3C + aIL-22 treatment (Figure 6K). In correlation with this observation of Roseburia regulation, while butyrate levels increased in I3C isotype controls (TNBS + I3C + Isotype), it was decreased significantly after aIL-22 treatment (Figure 6L). Collectively, these data demonstrated that IL-22 is important in I3C-mediated protective effects during colitis, particularly in regulating the gut microbiome and butyrate levels.
Discussion
Research and development of novel and safe therapeutics to combat this colitis are of prominent importance. Studies show that intestinal inflammation is linked to the loss of balance maintained by regulatory mechanisms occurring at the complex interaction of the host intestinal barrier and the lumen-bound microbes. There is a growing consensus that alterations in the gut microbiota are directly linked to the disease; whether this alteration is causative of the disease or a product of the disease state is still an area of research that is being explored. However, there appears to be some consistency in a microbial profile that occurs during colitis, which is further reinforced by this study.
In TNBS-induced colitis in rats, microbiome analysis revealed that the disease groups had increases in overall Bacteriodetes and a decrease in Firmicutes at the phylum level (35). In a murine DSS-induced colitis model, researchers found a significant increase in Bacteroidales, which correlated with intestinal mucin degradation, whereas Clostridiales decreased during inflammation, correlating with a reduction in butyrate (36, 37). These microbial profiles are not limited to animal models, as similar results have been found in the human populations diagnosed with colitis. In human inflammatory bowel disease (IBD) patients, certain bacteria seem to become more prominent when compared with healthy controls, such as members of the family Enterobacteriaceae, Bacteriodaceae, and Bacteriodes (38–42). 16S rRNA gene profiles of mucosal tissue samples from ulcerative colitis patients found that this patient population had higher abundances of Prevotella and Bacteriodes (43). Reports also indicated that an expansion in Proteobacteria is consistently found in IBD patients (44). TM7, a recently defined and cultured bacterium, often resides almost exclusively in the oral cavity, with limited abundance in the gut microflora. However, TM7 was found to be significantly higher in mucosal samples in Crohn’s disease and ulcerative colitis patients when compared with normal controls (45). Interestingly, 16S rRNA microbiome profiling in the current study identified these alterations in the TNBS-induced mouse model, as well, which demonstrates that the current colitis model and microbiota study closely mimics what is seen in the human disease. In view of this, highlighted in this report is the increase in B. acidifaciens during TNBS-induced colitis, which was also observed in a mouse DSS model (37). This bacterial species displays key characteristics that could very well classify this as a colitis-specific biomarker — or, at the very least, a species that promotes colitis development and progression. B. acidifaciens was recently shown to act as a mucin degrader in vivo (46). Mucin serves as a natural barrier in the normal intestinal environment, and the degradation of this layer would help promote microbial dysbiosis and intestinal inflammation. In addition, B. acidifaciens is known to produce SCFA end products acetate, propionate, and succinate (47). These SCFAs — particularly acetate — when absorbed and transported to the liver, can lead to a decrease in butyrate, which serves as an energy source for colonic epithelial cells (48, 49). However, it should be noted that propionate supplementation in the DSS-induced colitis model was found to be protective (50), suggesting that the combination and concentration of these SCFAs might be playing a role in overall disease severity or regulation. Interestingly, in a recent study, B. acidifaciens was inoculated into mice to prevent obesity and resulted in increased weight loss and reduction in fat mass (51) — an outcome seen in colitis, as well.
Phylogenetic bacterial profiles were not the only aspects of microbial dysbiosis that were shared between the current study and the human disease condition. Bacterial function in the disease state was altered similarly, which is more than likely due to the changes in microbial diversity and abundance. For example, the current report highlights that microbial functions such as riboflavin metabolism, ALA metabolism, and ubiquitin system were all increased in the TNBS-induced disease state. Riboflavin metabolism, which is indicative of an increase in gram-negative bacteria, is a hallmark of an inflammatory environment characterized by increases in oxidative stress, and increased metabolism of riboflavin was observed in colitis patients (40, 52, 53). ALA is not produced by the human host and has be consumed as part of the host diet. This omega fatty acid is known to reduce the inflammatory response by reducing NF-κB activation and oxidative stress, and ALA supplementation was shown to reduce colitis severity (54). As this process is increased in the disease state, it can be reasoned that the microbes are metabolizing ALA and competing with the host, therefore depleting a potential antiinflammatory source. Lastly, an increase in ubiquitin system is well documented as an exploitative mechanism by human pathogenic bacteria on the host (55, 56), and this process was found to be altered in colitis (57). The fact that these alterations in microbial composition and function in the current study matched reports in human colitis populations is encouraging, but even more important is the fact that treatment with I3C prevented TNBS colitis–induced microbial dysbiosis, which could be attributed to the antimicrobial activities of I3C presented in earlier reports (58).
Of particularly note was the fact that I3C was shown to almost selectively lead to an increase in abundance of gram-positive bacteria known to produce butyrate (Roseburia spp.). This is a very significant finding, especially considering that supplementation of microbiota from Crohn’s disease patients with Roseburia spp. led to increased butyrate production and enhancement in the intestinal epithelial barrier integrity (59). In addition, a recent report shows how R. intestinalis is able to prevent IL-17 secretion and increase Treg differentiation in TNBS-induced colitis (60). There are many reports showing that butyrate is effective at alleviating the burdens inflicted by colitis, particularly through mechanisms that include increases in antimicrobial peptides (AMPs) and mucus production (61–64). More recently, the PPARG signaling pathways (e.g., NF-κB and MAPK) have become a target for amelioration of colitis, and findings suggest that this receptor activation negatively regulates key inflammatory mediators (65–67). In the current study, I3C and NaB were able to significantly increase transcript expression of this receptor in the colon. It will be interesting to determine in future studies if these mechanisms regulated by I3C, to include increases in mucus and Pparg, are strictly dependent on butyrate or if this natural indole compound implores butyrate-independent mechanisms to elicit these responses. Regardless, the data in the current study is extremely encouraging, as it adds to the currently scarce amount of research studies providing evidence that I3C is a potential therapeutic agent in combating the effects of colitis. It is also exciting to show that I3C, along with its other beneficial properties, can modulate the gut microbiome during colitis in such a way that it prevents the overgrowth of potential pathogenic bacteria while selectively allowing an antiinflammatory butyrate-producer to thrive in the microenvironment.
As noted previously, I3C is a well-known ligand of AhR. In several different mouse models of colitis, activation of AhR by various known ligands resulted in protection against colitis, most notably through alterations in Treg and Th17 generation (68–71). More recently, it was shown that AhR ligands can stimulate AhR-expressing ILC3s, intraepithelial lymphocytes of the mucosa that are known to strengthen the intestinal epithelial cell barrier through an IL-22–dependent epithelial cell regeneration mechanism and produce AMPs (8, 72–75). In this regard, I3C may be able to prevent the expansion of pathogenic gram-negative bacteria in colitis through activating AhR-expressing cells (T cells, ILCs, colonic epithelial cells) to produce IL-22, promote the release of host AMPs, or alter the immune response in other ways. As butyrate treatment alone did not result in an increase in IL-22 mRNA expression, perhaps due to the fact that this SCFA generally acts through GPRs and not AhR signaling (76), these findings suggested that I3C is able to incorporate mechanisms independently of increasing butyrate production to prevent the negative effects associated with colitis. In this particular report, IL-22 production by ILC3s seemed to be altered after I3C treatment. This would suggest that an increase in butyrate-producers by I3C is a beneficial consequence, particularly in shifting Th17 to Treg populations, but the host defense to microbial dysbiosis by way of an increase in IL-22 is dependent solely on I3C, an activator of AhR. IL-22, via a CARD9-dependent mechanism, was shown to play an important role in attenuating intestinal inflammation during colitis, and mice deficient in CARD9 and IL-22 had altered microbiomes in addition to being susceptible to colitis development (77). Future studies investigating the regulation of IL-22 by I3C, and whether or not it is dependent on AhR activation, will have to be conducted. In addition, while male and female mouse microbiomes have been shown to be similar (78), sex differences have been observed in both susceptibility to TNBS-colitis induction (79, 80) and response to AhR ligands (81–83). Inasmuch as the current report focuses on findings in female mice only, disease outcomes and key findings (e.g., increased IL-22 production by ILC3s) will have to be investigated in male mice, as well. In summary, the current study has identified IL-22 as the key molecule through which I3C mediates its beneficial effects against colitis, including reversal of colitis-mediated dysbiosis, increased butyrate production, and antiinflammatory response.
Methods
Animals.
Female BALB/cJ and C57BL/6 mice were purchased from the Jackson Laboratory, and experimental mice used were between 8 and 10 weeks of age. All mice were housed at the Association for Assessment and Accreditation of Laboratory Animal Care–accredited (AAALAC-accredited) animal facility at the University of South Carolina School of Medicine. Mice were housed in specific pathogen-free conditions, under 12-hour light/12-hour dark cycles.
Induction of colitis and treatment with I3C, NaB, or neutralization of IL-22.
TNBS colitis was induced in susceptible BALB/cJ mice (84). Mice were given a total volume of 50 μL intrarectal injections of 1 mg of TNBS (Sigma-Aldrich) in 50% ethanol under light isoflurane anesthesia, as previously reported (84). For treatment groups, mice were given either 100 μL i.p. injections of I3C (40 mg/kg in 0.05% DMSO/corn oil) or oral gavage of NaB (200 mg/kg in water), both purchased from Sigma-Aldrich, 2 hours after injection of TNBS. Subsequent treatments were given daily throughout the entire experiment. The dose and route of I3C is consistent with our previous reports (14, 18), and the NaB dosage and route were determined based on other published reports (27–29, 85). All experimental groups in the TNBS model were given 50% ethanol intrarectal injections to ensure changes observed were not due to administration of the ethanol diluent. Vehicle controls in these experiments were also given i.p. injections of 0.05% DMSO/corn oil. For the DSS model, DSS was purchased from Chem-Impex International and introduced into drinking water (ab libitum) at a 3% concentration for 1 week, followed by a week of regular drinking water for female C57BL/6 mice, as previously described (86). For treatment groups in the DSS model, I3C treatment (i.p. injections of 40 mg/kg in 0.05% DMSO/corn oil) begin 1 hour after introduction of DSS into the drinking water and was given every other day until completion of the experiment. Vehicle groups in the DSS model were also given i.p. injections of 0.05% DMSO/corn oil. For neutralization of IL-22 experiments, functional grade IL-22 monoclonal antibody (IL-22JOP; 16-7222-85) was purchased from Thermo Fisher Scientific, along with rat IgG2a κ isotype control (eBR2a; 16-4321-85). To neutralize IL-22, the aIL-22 was given i.p. 24 hours prior to administration of TNBS or treatment with I3C at a 50-μg dose in 100 μL of PBS (Sigma-Aldrich), as well as during the middle of the colitis experiments (day 2). Dose and route of IL-22 neutralization were based on a previous report (87).
Assessment of colitis disease parameters.
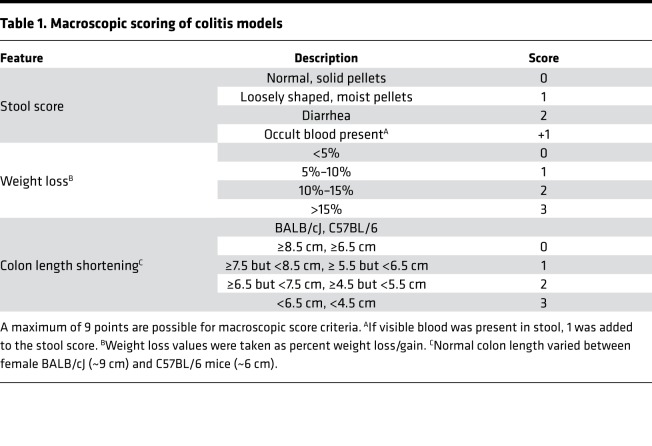
For both colitis models, mice were weighed daily, and colon lengths were measured at experimental end-points. To determine macroscopic colitis scores in the experimental groups, a slightly modified scoring system (Table 1) was used based on previous reports (88). To test in vivo gut permeability, the FITC-dextran assay was used as reported elsewhere (89). Briefly, on day 3, mice were given 600 mg/kg of 4kD FITC-dextran (Sigma-Aldrich) dissolved in 100 μL of PBS by oral gavage. Four hours later, blood was collected from mice by retroorbital bleeding, and FITC-dextran concentrations were determined using a PerkinElmer Life Sciences spectrophotometer with excitation wavelength at 480 nm. SAA levels, which have been used in the clinic to determine the severity of colitis in patients (90), were assessed using a SAA mouse ELISA purchased by Abcam. SAA levels were determined from serum collected at day 3 of the TNBS model and day 10 of the DSS model. Colonoscopy images were taken on day 3 of the TNBS model using a Karl Storz Tele Pack Vet X LED endoscope designed for small animals. The colonoscopy scoring parameters were based on a methodology previously reported (91).
Table 1. Macroscopic scoring of colitis models.
Microscopic evaluation of the colon tissues.
Colon histology was obtained by harvesting proximal colon samples on day 4 of the TNBS model. Tissues were fixed in methanol-Carnoy’s fixative solution for 24 hours and then embedded in paraffin before cutting into 9-μm sections. For H&E staining, sections were stained as previously described (92). Histopathological scoring parameters were based on previously published reports (93). For periodic acid-schiff (PAS) staining, a staining kit purchased from Sigma-Aldrich was used following manufacturer’s instructions. Images of stained sections were taken using a Leica DM2500 with LASX image capture suite (Leica Microsystems Inc.). For TEM images, mouse colons were harvested and prepared as previously described (92). Images were captured on a JEOL 1400S TEM located in the Instrumentation Resource Facility at the University of South Carolina School of Medicine.
Analysis of Treg and Th17 subsets in MLN during colitis.
MLNs were excised from mice during experimental end-points and prepared for staining as previously described (14). To assess and quantify Treg/Th17 subsets, samples were stained with fluorescently labeled antibodies purchased from BioLegend. To quantify Treg subsets, True-Nuclear Transcription Factor Buffer Set (BioLegend) was used as per the instructions from the manufacturer to label cells with antibodies for CD3-FITC (clone 17A2), CD4-APC (clone GK1.5), and FoxP3-PE (clone MF-14). For Th17 subsets, BD Cytofix/Cytoperm from BD Biosciences was used to fix and permeabilize the cells for staining using CD4-APCand IL-17-PE (clone TC11-18H10.1) antibodies. BD FACS Celesta and FlowJo analysis software (BD Biosciences) were used to determine percentage of these T cell subsets, and total Treg/Th17 numbers were determined by multiplying this percentage by total number of cells isolated from the MLNs. Flow cytometry gating strategy and representative plots from experimental groups for Th17/Treg subsets are depicted in Supplemental Figure 12. Gates were positioned according to unstained (negative control) and single-color antibody controls (positive controls).
Examination of IL-22 secretion from colonic explants.
Explants of colon tissues were prepared as previously described (94). Briefly, colon tissue (length of 1 cm from the cecum) was excised from experimental mice (TNBS + Vehicle and TNBS + I3C) and washed 3 times with PBS. Tissues were placed in 24-well cell culture plates in complete RPMI 1640 media (Corning) (10% FBS, 1% penicillin/streptomycin, 1% HEPES; Sigma-Aldrich) and incubated at 37°C at 5% CO2 for 24 hours. Supernatants were collected following centrifugation at 8000 g at 4°C for 10 minutes and stored at –80°C for later analysis by ELISA. Mouse IL-22 ELISA kit was purchased from BioLegend for detection of IL-22 in the collected supernatants.
LP isolation and IL-22–positive cell phenotyping.
Cells from the LP fraction were isolated as previously described (24). IL-22 production by immune cells in the LP were phenotyped using flow cytometry (BD FACs Celesta) based on a previous reports (95). The gating strategy is summarized in Supplemental Figure 13. Gating and analysis of flow plots was performed using FlowJo version 10.5.3 (BD Biosciences). For phenotyping, the following antibodies from BioLegend were used: BV421-Lin (BioLegend, 133311), APC/Cyanine 7-CD45 (clone 30-F11), BV785-CD3 (clone:145-2C11), CD90.2-FITC (clone 53-21), Alexa-Fluor 647 IL-22 (clone Poly5164), and Rorγt-PE (clone Q31-378).
Microbial 16S rRNA gene analysis and PiCRUSt functional analysis.
To perform bacterial phylogenetic and functional analysis, genomic DNA was isolated from colonic flushes on day 4 of TNBS model and sequenced as previously described (96). For all downstream analysis after sequencing, this study used the Nephele platform from the National Institute of Allergy and Infectious Diseases (NIAID) Office of Cyber Infrastructure and Computational Biology (OCICB) in Bethesda, Maryland, USA (25). The LefSe Galaxy web application tool developed by the Huttenhower group was performed using default parameters as previously published (26).
SCFA analysis from colonic flushes.
Quantification of SCFA from colonic flushes collected from experimental groups was performed as previously described (96, 97). Briefly, colon contents (100 mg) were extracted using 25% metaphosphoric acid, centrifuged (12,000 g for 15 minutes at 4°C), and filtered with Ultra-free MC columns from Thermo Fisher Scientific. HP 5890 gas chromatograph configured with flame-ionization detectors (GC-FID) was used for SCFA analysis, using 2-ethylbutyric acid as an internal standard (IS) for all samples analyzed.
Quantitative PCR (qPCR).
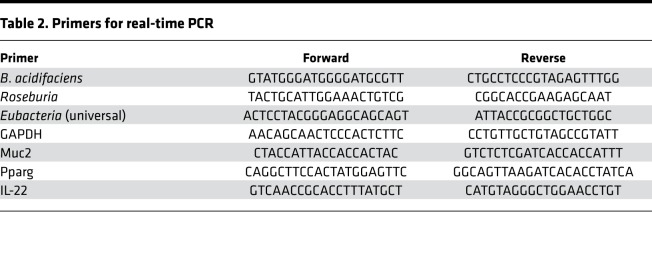
Real-time PCR was performed using bacterial primers designed to recognize the 16S rRNA gene subunit of bacteria (Table 2). 16S rRNA sequences were obtained from the National Center for Biotechnology Information (NCBI) database, and primers were designed and ordered by Intergrated DNA Technologies (IDT). Genomic DNA was isolated as described above. PCR reactions were performed on a CFX96 qPCR system from Bio-Rad. For gene transcript analysis in colonic tissue, RNA from colon tissues (25 mg of proximal colon) were ground up using Kontes RNase-free pellet pestle grinders from VWR International. RNA from colonic tissues was isolated using an RNAeasy Plus Mini kit from Qiagen following manufacturer’s instructions, and cDNA was synthesized as previously described (19). Primers for Muc2, Pparg, and IL-22 (Table 2) were designed using IDT website primer design interface, and PCR reaction and analysis was performed using Bio-Rad CFX96 PCR platform. For bacteria validation studies, levels were normalized to universal eubacterial primer. For genes, expression levels were normalized to GAPDH. Fold changes from PCR analysis were obtained by using the ΔΔCT method with comparison to the control groups, as previously described (98).
Table 2. Primers for real-time PCR.
Availability of data and material.
The authors declare that all the data supporting the findings of this study are provided within the manuscript, additional information file, and provided links. Raw sequenced reads generated from the Illumina platform have been deposited into the NCBI Sequence Read Archive public database, with SRA accession number PRJNA497446.
Statistics.
GraphPad Prism software was used for all statistical analysis unless otherwise indicated. In order to determine statistical significance, 1-way ANOVA and Tukey’s post hoc multiple comparisons tests were used when comparing 3 or more groups. For comparisons between only 2 groups, an unpaired, 2-tailed standard Student’s t test was used. A P value less than 0.05 was considered significant.
Study approval.
All procedures involving animals were performed according to NIH guidelines under protocols approved by the IACUC at the University of South Carolina.
Author contributions
PBB, MN, and PSN contributed to the overall research design and manuscript preparation. PBB performed the majority of experiments and the analysis of the data presented in the manuscript. MN and PSN were responsible for obtaining all the funding. LM contributed by preparing and providing TEM images of colon tissue from experimental groups. HA, ND, WB, KM, and US assisted with collecting data for the TNBS-induced colitis experiments to include performing flow cytometry, ELISA explant, and preparation of samples for SCFA analysis. CT and SC assisted in statistical analysis and interpretation of the data. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
This study used the Nephele platform from the NIAID OCICB. Special thanks to Mike Walla, Director of Mass Spectrometry Services at the University of South Carolina, for his assistance in SCFA quantification presented in these studies. The studies were supported, in part, by NIH grants P01AT003961, R01AI123947, R01AI129788, and P20GM103641. The funding agency had no role in the experimental design, data collection and analysis, decision to publish, or preparation of the manuscript.
Version 1. 01/16/2020
Electronic publication
Version 2. 01/16/2020
Udai P. Singh's name was corrected.
Version 3. 01/21/2020
Name correction for Haider Rasheed Alrafas
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(1):e127551.https://doi.org/10.1172/jci.insight.127551.
Contributor Information
Philip B. Busbee, Email: Brandon.Busbee@uscmed.sc.edu.
Nicholas Dopkins, Email: nicholas.dopkins@uscmed.sc.edu.
William Becker, Email: William.Becker@uscmed.sc.edu.
Kathryn Miranda, Email: Kathryn.Miranda@uscmed.sc.edu.
Saurabh Chatterjee, Email: schatt@mailbox.sc.edu.
Mitzi Nagarkatti, Email: mitzi.nagarkatti@uscmed.sc.edu.
References
- 1.Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res. 2017;10:63–73. doi: 10.2147/JIR.S116088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bang B, Lichtenberger LM. Methods of Inducing Inflammatory Bowel Disease in Mice. Curr Protoc Pharmacol. 2016;72:5.58.1–5.5842. doi: 10.1002/0471141755.ph0558s72. [DOI] [PubMed] [Google Scholar]
- 3.Taghipour N, et al. An experimental model of colitis induced by dextran sulfate sodium from acute progresses to chronicity in C57BL/6: correlation between conditions of mice and the environment. Gastroenterol Hepatol Bed Bench. 2016;9(1):45–52. [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed I, Roy BC, Khan SA, Septer S, Umar S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms. 2016;4(2):E20. doi: 10.3390/microorganisms4020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascolo N, Izzo AA, Autore G, Maiello FM, Di Carlo G, Capasso F. Acetic acid-induced colitis in normal and essential fatty acid deficient rats. J Pharmacol Exp Ther. 1995;272(1):469–475. [PubMed] [Google Scholar]
- 6.Pastrelo MM, et al. Effect of Concentrated Apple Extract on Experimental Colitis Induced by Acetic Acid. Int J Mol Cell Med. 2017;6(1):38–49. [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel W, et al. Butyrate increases colonocyte protein synthesis in ulcerative colitis. J Surg Res. 1994;57(1):210–214. doi: 10.1006/jsre.1994.1133. [DOI] [PubMed] [Google Scholar]
- 8.Monteleone I, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141(1):237–248, 248.e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Furumatsu K, et al. A role of the aryl hydrocarbon receptor in attenuation of colitis. Dig Dis Sci. 2011;56(9):2532–2544. doi: 10.1007/s10620-011-1643-9. [DOI] [PubMed] [Google Scholar]
- 10.Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 2009;127(3):299–311. doi: 10.1111/j.1365-2567.2009.03054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206(1):43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ociepa-Zawal M, Rubiś B, Laciński M, Trzeciak WH. The effect of indole-3-carbinol on the expression of CYP1A1, CYP1B1 and AhR genes and proliferation of MCF-7 cells. Acta Biochim Pol. 2007;54(1):113–117. doi: 10.18388/abp.2007_3276. [DOI] [PubMed] [Google Scholar]
- 13.Marconett CN, Sundar SN, Poindexter KM, Stueve TR, Bjeldanes LF, Firestone GL. Indole-3-carbinol triggers aryl hydrocarbon receptor-dependent estrogen receptor (ER)alpha protein degradation in breast cancer cells disrupting an ERalpha-GATA3 transcriptional cross-regulatory loop. Mol Biol Cell. 2010;21(7):1166–1177. doi: 10.1091/mbc.e09-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busbee PB, Nagarkatti M, Nagarkatti PS. Natural indoles, indole-3-carbinol and 3,3’-diindolymethane, inhibit T cell activation by staphylococcal enterotoxin B through epigenetic regulation involving HDAC expression. Toxicol Appl Pharmacol. 2014;274(1):7–16. doi: 10.1016/j.taap.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung WS, Lee DG. Mechanism of decreased susceptibility for Gram-negative bacteria and synergistic effect with ampicillin of indole-3-carbinol. Biol Pharm Bull. 2008;31(9):1798–1801. doi: 10.1248/bpb.31.1798. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147(3):629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Mizoguchi A, Yano A, Himuro H, Ezaki Y, Sadanaga T, Mizoguchi E. Clinical importance of IL-22 cascade in IBD. J Gastroenterol. 2018;53(4):465–474. doi: 10.1007/s00535-017-1401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouse M, Singh NP, Nagarkatti PS, Nagarkatti M. Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br J Pharmacol. 2013;169(6):1305–1321. doi: 10.1111/bph.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busbee PB, Nagarkatti M, Nagarkatti PS. Natural indoles, indole-3-carbinol (I3C) and 3,3’-diindolylmethane (DIM), attenuate staphylococcal enterotoxin B-mediated liver injury by downregulating miR-31 expression and promoting caspase-2-mediated apoptosis. PLoS ONE. 2015;10(2):e0118506. doi: 10.1371/journal.pone.0118506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plevy S, et al. Combined serological, genetic, and inflammatory markers differentiate non-IBD, Crohn’s disease, and ulcerative colitis patients. Inflamm Bowel Dis. 2013;19(6):1139–1148. doi: 10.1097/MIB.0b013e318280b19e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endharti AT, Permana S. Extract from mango mistletoes Dendrophthoe pentandra ameliorates TNBS-induced colitis by regulating CD4+ T cells in mesenteric lymph nodes. BMC Complement Altern Med. 2017;17(1):468. doi: 10.1186/s12906-017-1973-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin H, Zhang W, Jiang X, Chen R, Huang X, Huang Z. Total glucosides of paeony ameliorates TNBS‑induced colitis by modulating differentiation of Th17/Treg cells and the secretion of cytokines. Mol Med Rep. 2017;16(6):8265–8276. doi: 10.3892/mmr.2017.7598. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, et al. Th1/Th2 Balance and Th17/Treg-Mediated Immunity in relation to Murine Resistance to Dextran Sulfate-Induced Colitis. J Immunol Res. 2017;2017:7047201. doi: 10.1155/2017/7047201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh UP, et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143(3):478–489. doi: 10.1111/imm.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber N, et al. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics. 2018;34(8):1411–1413. doi: 10.1093/bioinformatics/btx617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D, et al. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J Gastroenterol. 2017;23(1):60–75. doi: 10.3748/wjg.v23.i1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathewson ND, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17(5):505–513. doi: 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C, Kim BG, Kim JH, Chun J, Im JP, Kim JS. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int Immunopharmacol. 2017;51:47–56. doi: 10.1016/j.intimp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Lennon G, et al. Influences of the colonic microbiome on the mucous gel layer in ulcerative colitis. Gut Microbes. 2014;5(3):277–285. doi: 10.4161/gmic.28793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawashima H. Roles of the gel-forming MUC2 mucin and its O-glycosylation in the protection against colitis and colorectal cancer. Biol Pharm Bull. 2012;35(10):1637–1641. doi: 10.1248/bpb.b12-00412. [DOI] [PubMed] [Google Scholar]
- 32.Speca S, Dubuquoy L, Desreumaux P. Peroxisome proliferator-activated receptor gamma in the colon: inflammation and innate antimicrobial immunity. J Clin Gastroenterol. 2014;48 Suppl 1:S23–S27. doi: 10.1097/MCG.0000000000000253. [DOI] [PubMed] [Google Scholar]
- 33.Bertin B, Dubuquoy L, Colombel JF, Desreumaux P. PPAR-gamma in ulcerative colitis: a novel target for intervention. Curr Drug Targets. 2013;14(12):1501–1507. doi: 10.2174/13894501113149990162. [DOI] [PubMed] [Google Scholar]
- 34.Russo R, et al. Sodium butyrate and its synthetic amide derivative modulate nociceptive behaviors in mice. Pharmacol Res. 2016;103:279–291. doi: 10.1016/j.phrs.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Zheng H, et al. Modulation of Gut Microbiome Composition and Function in Experimental Colitis Treated with Sulfasalazine. Front Microbiol. 2017;8:1703. doi: 10.3389/fmicb.2017.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munyaka PM, Rabbi MF, Khafipour E, Ghia JE. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J Basic Microbiol. 2016;56(9):986–998. doi: 10.1002/jobm.201500726. [DOI] [PubMed] [Google Scholar]
- 37.Schwab C, et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 2014;8(5):1101–1114. doi: 10.1038/ismej.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace KL, Zheng LB, Kanazawa Y, Shih DQ. Immunopathology of inflammatory bowel disease. World J Gastroenterol. 2014;20(1):6–21. doi: 10.3748/wjg.v20.i1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erickson AR, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS ONE. 2012;7(11):e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wills ES, Jonkers DM, Savelkoul PH, Masclee AA, Pierik MJ, Penders J. Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation. PLoS ONE. 2014;9(3):e90981. doi: 10.1371/journal.pone.0090981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagalingam NA, Kao JY, Young VB. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2011;17(4):917–926. doi: 10.1002/ibd.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55(Pt 5):617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 44.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9(4):219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 45.Kuehbacher T, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57(Pt 12):1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 46.Berry D, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci USA. 2013;110(12):4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyamoto Y, Itoh K. Bacteroides acidifaciens sp. nov., isolated from the caecum of mice. Int J Syst Evol Microbiol. 2000;50 Pt 1:145–148. doi: 10.1099/00207713-50-1-145. [DOI] [PubMed] [Google Scholar]
- 48.Plöger S, et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- 49.Leonel AJ, Alvarez-Leite JI. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care. 2012;15(5):474–479. doi: 10.1097/MCO.0b013e32835665fa. [DOI] [PubMed] [Google Scholar]
- 50.Tong LC, et al. Propionate Ameliorates Dextran Sodium Sulfate-Induced Colitis by Improving Intestinal Barrier Function and Reducing Inflammation and Oxidative Stress. Front Pharmacol. 2016;7:253. doi: 10.3389/fphar.2016.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang JY, et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017;10(1):104–116. doi: 10.1038/mi.2016.42. [DOI] [PubMed] [Google Scholar]
- 52.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reifen R, Karlinsky A, Stark AH, Berkovich Z, Nyska A. α-Linolenic acid (ALA) is an anti-inflammatory agent in inflammatory bowel disease. J Nutr Biochem. 2015;26(12):1632–1640. doi: 10.1016/j.jnutbio.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Ashida H, Kim M, Sasakawa C. Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol. 2014;12(6):399–413. doi: 10.1038/nrmicro3259. [DOI] [PubMed] [Google Scholar]
- 56.Steele-Mortimer O. Exploitation of the ubiquitin system by invading bacteria. Traffic. 2011;12(2):162–169. doi: 10.1111/j.1600-0854.2010.01137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cleynen I, et al. Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut. 2014;63(8):1265–1274. doi: 10.1136/gutjnl-2012-303205. [DOI] [PubMed] [Google Scholar]
- 58.Sung WS, Lee DG. In vitro antimicrobial activity and the mode of action of indole-3-carbinol against human pathogenic microorganisms. Biol Pharm Bull. 2007;30(10):1865–1869. doi: 10.1248/bpb.30.1865. [DOI] [PubMed] [Google Scholar]
- 59.Geirnaert A, et al. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep. 2017;7(1):11450. doi: 10.1038/s41598-017-11734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu C, et al. Roseburia intestinalis inhibits interleukin‑17 excretion and promotes regulatory T cells differentiation in colitis. Mol Med Rep. 2018;17(6):7567–7574. doi: 10.3892/mmr.2018.8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J, et al. Therapeutic Potential to Modify the Mucus Barrier in Inflammatory Bowel Disease. Nutrients. 2016;8(1):E44. doi: 10.3390/nu8010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cresci G, Nagy LE, Ganapathy V. Lactobacillus GG and tributyrin supplementation reduce antibiotic-induced intestinal injury. JPEN J Parenter Enteral Nutr. 2013;37(6):763–774. doi: 10.1177/0148607113486809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52(10):1442–1447. doi: 10.1136/gut.52.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song M, Xia B, Li J. Effects of topical treatment of sodium butyrate and 5-aminosalicylic acid on expression of trefoil factor 3, interleukin 1beta, and nuclear factor kappaB in trinitrobenzene sulphonic acid induced colitis in rats. Postgrad Med J. 2006;82(964):130–135. doi: 10.1136/pgmj.2005.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y, et al. A novel pyrazole-containing indolizine derivative suppresses NF-κB activation and protects against TNBS-induced colitis via a PPAR-γ-dependent pathway. Biochem Pharmacol. 2017;135:126–138. doi: 10.1016/j.bcp.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 66.Yang Y, et al. Andrographolide derivative AL-1 ameliorates TNBS-induced colitis in mice: involvement of NF-кB and PPAR-γ signaling pathways. Sci Rep. 2016;6:29716. doi: 10.1038/srep29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soubh AA, Abdallah DM, El-Abhar HS. Geraniol ameliorates TNBS-induced colitis: Involvement of Wnt/β-catenin, p38MAPK, NFκB, and PPARγ signaling pathways. Life Sci. 2015;136:142–150. doi: 10.1016/j.lfs.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 68.Goettel JA, et al. AHR Activation Is Protective against Colitis Driven by T Cells in Humanized Mice. Cell Rep. 2016;17(5):1318–1329. doi: 10.1016/j.celrep.2016.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh NP, Singh UP, Singh B, Price RL, Nagarkatti M, Nagarkatti PS. Activation of aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of experimental colitis. PLoS ONE. 2011;6(8):e23522. doi: 10.1371/journal.pone.0023522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benson JM, Shepherd DM. Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn’s disease. Toxicol Sci. 2011;120(1):68–78. doi: 10.1093/toxsci/kfq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Díaz-Díaz CJ, et al. The Aryl Hydrocarbon Receptor is a Repressor of Inflammation-associated Colorectal Tumorigenesis in Mouse. Ann Surg. 2016;264(3):429–436. doi: 10.1097/SLA.0000000000001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12(1):21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 73.Goldberg R, Prescott N, Lord GM, MacDonald TT, Powell N. The unusual suspects--innate lymphoid cells as novel therapeutic targets in IBD. Nat Rev Gastroenterol Hepatol. 2015;12(5):271–283. doi: 10.1038/nrgastro.2015.52. [DOI] [PubMed] [Google Scholar]
- 74.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334(6062):1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 75.Qiu J, Zhou L. Aryl hydrocarbon receptor promotes RORγt+ group 3 ILCs and controls intestinal immunity and inflammation. Semin Immunopathol. 2013;35(6):657–670. doi: 10.1007/s00281-013-0393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sivaprakasam S, Bhutia YD, Ramachandran S, Ganapathy V. Cell-Surface and Nuclear Receptors in the Colon as Targets for Bacterial Metabolites and Its Relevance to Colon Health. Nutrients. 2017;9(8):E856. doi: 10.3390/nu9080856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campbell JH, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6(11):2033–2044. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bábíčková J, et al. Sex Differences in Experimentally Induced Colitis in Mice: a Role for Estrogens. Inflammation. 2015;38(5):1996–2006. doi: 10.1007/s10753-015-0180-7. [DOI] [PubMed] [Google Scholar]
- 80.Kozik AJ, Nakatsu CH, Chun H, Jones-Hall YL. Age, sex, and TNF associated differences in the gut microbiota of mice and their impact on acute TNBS colitis. Exp Mol Pathol. 2017;103(3):311–319. doi: 10.1016/j.yexmp.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 81.Lee J, Prokopec SD, Watson JD, Sun RX, Pohjanvirta R, Boutros PC. Male and female mice show significant differences in hepatic transcriptomic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. BMC Genomics. 2015;16:625. doi: 10.1186/s12864-015-1840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu H, Lei X, Klaassen C. Gender differences in renal nuclear receptors and aryl hydrocarbon receptor in 5/6 nephrectomized rats. Kidney Int. 2006;70(11):1920–1928. doi: 10.1038/sj.ki.5001880. [DOI] [PubMed] [Google Scholar]
- 83.van Esterik JC, et al. Compound- and sex-specific effects on programming of energy and immune homeostasis in adult C57BL/6JxFVB mice after perinatal TCDD and PCB 153. Toxicol Appl Pharmacol. 2015;289(2):262–275. doi: 10.1016/j.taap.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 84.Elson CO, et al. Hapten-induced model of murine inflammatory bowel disease: mucosa immune responses and protection by tolerance. J Immunol. 1996;157(5):2174–2185. [PubMed] [Google Scholar]
- 85.Zhang L, et al. Sodium Butyrate Protects -Against High Fat Diet-Induced Cardiac Dysfunction and Metabolic Disorders in Type II Diabetic Mice. J Cell Biochem. 2017;118(8):2395–2408. doi: 10.1002/jcb.25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh UP, et al. Resveratrol (trans-3,5,4’-trihydroxystilbene) induces silent mating type information regulation-1 and down-regulates nuclear transcription factor-kappaB activation to abrogate dextran sulfate sodium-induced colitis. J Pharmacol Exp Ther. 2010;332(3):829–839. doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeaman MR, et al. Mechanisms of NDV-3 vaccine efficacy in MRSA skin versus invasive infection. Proc Natl Acad Sci USA. 2014;111(51):E5555–E5563. doi: 10.1073/pnas.1415610111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Engel MA, Kellermann CA, Burnat G, Hahn EG, Rau T, Konturek PC. Mice lacking cannabinoid CB1-, CB2-receptors or both receptors show increased susceptibility to trinitrobenzene sulfonic acid (TNBS)-induced colitis. J Physiol Pharmacol. 2010;61(1):89–97. [PubMed] [Google Scholar]
- 89.An G, et al. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204(6):1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Niederau C, Backmerhoff F, Schumacher B, Niederau C. Inflammatory mediators and acute phase proteins in patients with Crohn’s disease and ulcerative colitis. Hepatogastroenterology. 1997;44(13):90–107. [PubMed] [Google Scholar]
- 91.Kodani T, et al. Flexible colonoscopy in mice to evaluate the severity of colitis and colorectal tumors using a validated endoscopic scoring system. J Vis Exp. 2013;(80):e50843. doi: 10.3791/50843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saeed AI, Rieder SA, Price RL, Barker J, Nagarkatti P, Nagarkatti M. Acute lung injury induced by Staphylococcal enterotoxin B: disruption of terminal vessels as a mechanism of induction of vascular leak. Microsc Microanal. 2012;18(3):445–452. doi: 10.1017/S1431927612000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akgun E, Caliskan C, Celik HA, Ozutemiz AO, Tuncyurek M, Aydin HH. Effects of N-acetylcysteine treatment on oxidative stress in acetic acid-induced experimental colitis in rats. J Int Med Res. 2005;33(2):196–206. doi: 10.1177/147323000503300207. [DOI] [PubMed] [Google Scholar]
- 94.Perera AP, et al. MCC950, a specific small molecule inhibitor of NLRP3 inflammasome attenuates colonic inflammation in spontaneous colitis mice. Sci Rep. 2018;8(1):8618. doi: 10.1038/s41598-018-26775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bauché D, et al. LAG3+ Regulatory T Cells Restrain Interleukin-23-Producing CX3CR1+ Gut-Resident Macrophages during Group 3 Innate Lymphoid Cell-Driven Colitis. Immunity. 2018;49(2):342–352.e5. doi: 10.1016/j.immuni.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 96.Chitrala KN, et al. CD44 deletion leading to attenuation of experimental autoimmune encephalomyelitis results from alterations in gut microbiome in mice. Eur J Immunol. 2017;47(7):1188–1199. doi: 10.1002/eji.201646792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao G, Nyman M, Jönsson JA. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed Chromatogr. 2006;20(8):674–682. doi: 10.1002/bmc.580. [DOI] [PubMed] [Google Scholar]
- 98.Alrafas HR, Busbee PB, Nagarkatti M, Nagarkatti PS. Resveratrol modulates the gut microbiota to prevent murine colitis development through induction of Tregs and suppression of Th17 cells. J Leukoc Biol. 2019;106(2):467–480. doi: 10.1002/JLB.3A1218-476RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.