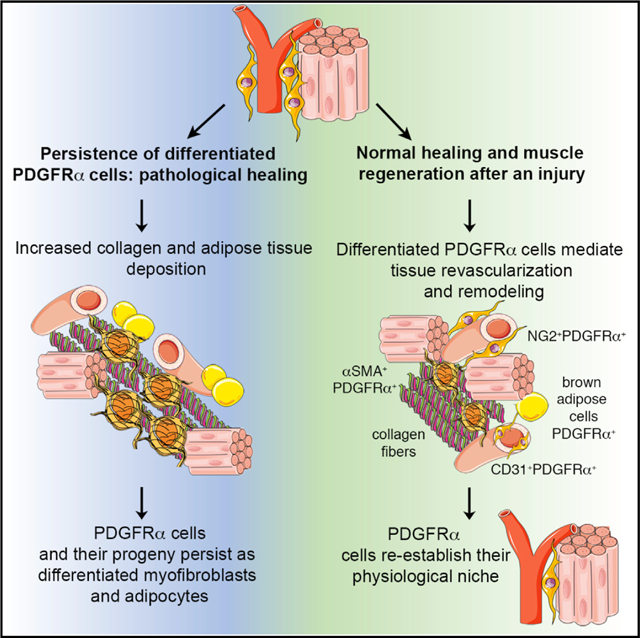
SUMMARY
PDGFRα+ mesenchymal progenitor cells are associated with pathological fibro-adipogenic processes. Conversely, a beneficial role for these cells during homeostasis or in response to revascularization and regeneration stimuli is suggested, but remains to be defined. We studied the molecular profile and function of PDGFRα+ cells in order to understand the mechanisms underlying their role in fibrosis versus regeneration. We show that PDGFRx+ cells are essential for tissue revascularization and restructuring through injury-stimulated remodeling of stromal and vascular components, context-dependent clonal expansion, and ultimate removal of pro-fibrotic PDGFRα+-derived cells. Tissue ischemia modulates the PDGFRα+ phenotype toward cells capable of remodeling the extracellular matrix and inducing cell-cell and cell-matrix adhesion, likely favoring tissue repair. Conversely, pathological healing occurs if PDGFRα+-derived cells persist as terminally differentiated mesenchymal cells. These studies support a context-dependent “yin-yang” biology of tissue-resident mesenchymal progenitor cells, which possess an innate ability to limit injury expansion while also promoting fibrosis in an unfavorable environment.
Graphical Abstract
In Brief
Santini et al. show that progenitor PDGFRα+ cells residing in skeletal muscle are mesenchymal stromal cells with a dual function, which on the one hand can stabilize newly formed blood vessels and limit injury expansion after ischemia, but on the other hand are also capable of promoting fibrosis in an unfavorable environment.
INTRODUCTION
Stromal tissues support parenchymal functioning by providing extracellular matrix (ECM), paracrine signaling cues, nutrients, and oxygen (Farahani and Xaymardan, 2015). Mesenchymal cells resident within the stroma are heterogeneous. However, the population of cells expressing platelet-derived growth factor receptor α (PDGFRα) exhibits in vitro and in vivo features of mesenchymal progenitor cells (Farahani and Xaymardan, 2015; Santini et al., 2016).
In adult tissues, cells expressing PDGFRα typically reside in an interstitial/perivascular niche (Chong et al., 2011, 2013; Pannérec et al., 2013; Santini et al., 2016; Uezumi et al., 2014a) and may play a role in various disease pathologies, including fibrosis (Olson and Soriano, 2009), with other roles, including formation of a small percentage of gastrointestinal stromal tumors (Heinrich et al., 2003; Hirota et al., 2003) and scleroderma-related pathologies (Gabrielli et al., 2007; Lozano et al., 2006; Okamoto, 2006; Tan, 2006). For example, a subset of perivascular PDGFRα+ cells expressing ADAM12 (a disintegrin and metalloprotease 12) are a major source of pro-fibrotic cells after injury (Dulauroy et al., 2012). Similarly, perivascular PDGFRα+ cells that co-express Gli1 generate myofibroblasts after injury of the heart, kidney, lung, and liver (Kramann et al., 2015). In the aorta, PDGFRα+ and Sca1+ cells potentially contribute to vascular calcification by differentiating into osteoblasts (Chong et al., 2013), whereas resident cardiac PDGFRα+ cells likely contribute to fibro-fatty infiltration in arrhythmogenic cardiomyopathy (Lombardi et al., 2016; Paylor et al., 2013) and PDGFRα+/PDGFRβ+ co-positive cells participate in cardiac and skeletal muscle fibrosis (Murray et al., 2017). In murine skeletal muscle and skeletal muscle from Duchenne muscular dystrophy patients, PDGFRα+ cells also exhibit adipogenic and fibrogenic potential (Uezumi et al., 2010, 2014a, 2014b).
These studies are counterbalanced by other reports suggesting beneficial functions for PDGFRα+ cells. For example, PDGFRα+ Sca1+ cell injection after myocardial infarction increased cardiac function by augmenting angiogenesis (Noseda et al., 2015). Furthermore, Sca1+PDGFRα+ fibro-adipogenic progenitors enhance the differentiation of primary myogenic progenitors in co-cultivation experiments (Joe et al., 2010), while recent studies have shown that PDGFRα+ fibro-adipogenic progenitors support muscle stem cell expansion and muscle regeneration after injury (Wosczyna et al., 2019). In addition, neural crest-derived PDGFRα+ mesenchymal cells can differentiate into bone and dermal cells during digit tip regeneration and wound healing (Carr et al., 2019).
Based on these data, a general hypothesis has arisen that differing subsets of resident mesenchymal cells are responsible for pro-fibrotic effects after injury, versus homeostatic and repair roles (Di Carlo and Peduto, 2018). However, it remains possible that a single mesenchymal stromal population could perform these dual functions and have both pro- and anti-fibrotic functionality. We elected to address this possibility, and using various approaches, we disclosed the dual “yin-yang” functionality of PDGFRα+ mesenchymal cells. On the one hand, these cells were associated with vascular stabilization, reduced vascular leakiness, and a more mature vascular architecture in regenerating tissues. On the other hand, by subtly manipulating these cells or their environment, PDGFRα+ cells enhanced fibrosis and vessel leakage.
RESULTS
PDGFRα+ Cell Characterization in Murine Skeletal Muscle
We characterized PDGFRα+ cells in mouse skeletal muscle. Consistent with prior studies (Chong et al., 2011, 2013; Uezumi et al., 2010, 2014a, 2014b), we observed using flow cytometry that PDGFRα+ cells are a rather rare population (4.0% ± 0.8% of total cells) that expresses a broad range of mesenchymal markers (Sca1, CD105, CD73, and CD29) (Figures 1A and 1B), including the pericyte and perivascular mesenchymal markers NG2 and PDGFR b (PDGFRβ) (Figure 1B). Only a minor proportion of PDGFRα+ cells expressed CD44 or CD31. Matching isotype control antibody data are presented as overlay histograms (Figure 1B), and fluorescence minus one (FMO) controls are shown in Figures S1A and S1B. PDGFRα+ cells were generally located in the interstitial region between skeletal muscle fibers and in contact with capillaries (Figures 1C, 1D, and S1C), and they often co-expressed NG2 (Figures 1E, 1F, and S1D). We also used PDGFRαH2B-eGfp transgenic mice to confirm these results. These mice express the H2B-eGfp fusion gene from the endogenous PDGFRα locus, and fluorescence patterns mimic endogenous PDGFRα expression (Hamilton et al., 2003). We confirmed that GFP+ cells in skeletal muscle express NG2 and are located in the proximity of CD31+ cells (Figure S1E).
Figure 1. PDGFRα+ Cells Are Tissue-Resident Mesenchymal Cells.
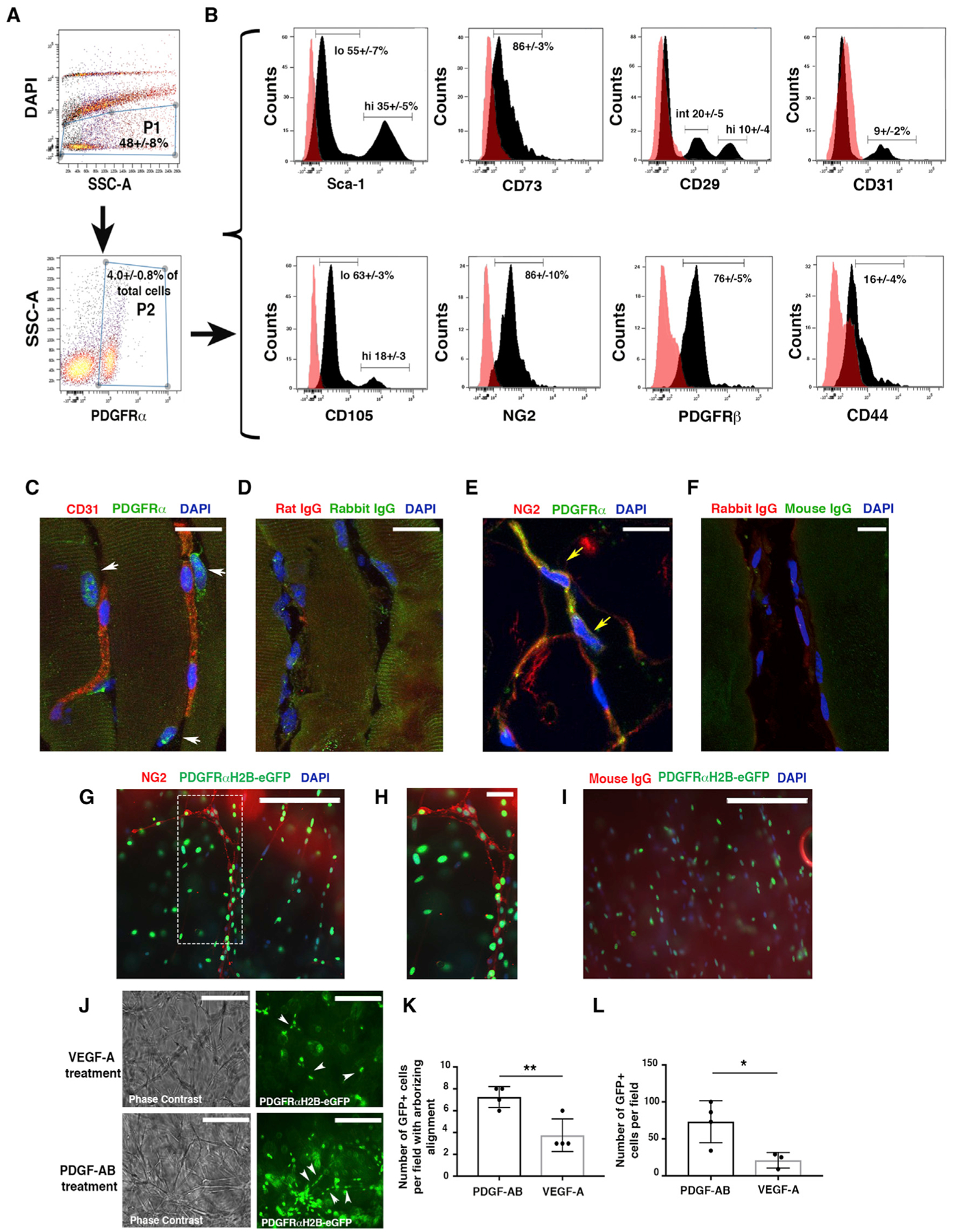
(A) Flow cytometric analysis of PDGFRα+ cells from murine skeletal muscle obtained from wild-type (WT) mice at 10–12 weeks of age. Cells were labeled with allophycocyanin (APC)- and phycoerythrin (PE)-conjugated antibodies, then gated as the DAPI− population (P1) and then for PDGFRα+-APC antibody (P2).
(B) PDGFRα+ cells were analyzed for the expression of endothelial (CD31), mesenchymal (Sca1, CD73, CD105, CD29, CD44), and pericyte markers (NG2 and PDGFRβ) from n = 3 independent experiments. PDGFRα+Sca1+ and PDGFRα+CD105+ cells were present as high (hi)- and low (lo)-expression populations. An intermediate (int) population was detected for PDGFRα+CD29+ cells. As a control, PDGFRα+ cells were labeled with immunoglobulin G (IgG) isotype antibodies. Representative histograms (dark peaks) are overlaid with IgG isotype controls (pink peaks).
(C–F) Immunofluorescence staining of PDGFRα+ cells with anti-PDGFRα antibody and anti-CD31 (C) or anti-NG2 (E) antibodies in the skeletal muscle of 4-month-old uninjured mice. White arrows show PDGFRα+CD31− cells (C); yellow arrows show PDGFRα+NG2+ cells (E). (D) and (F) represent control IgG immunofluorescence images. Images acquired with a Leica SP5 DM confocal as merged z stacks. Scale bars, 20 μm.
(G) Thoracic aortas from 4-month-old PDGFRαH2B-eGfp mice were isolated, dissected to 1-mm slices (“rings”), and plated in Matrigel. GFP+ cell migration was monitored daily for 5 days with an EVOS AMG imaging system (Thermo Fisher Scientific). Vessel-like formation of PDGFRαH2B-eGfp+ cells was assessed after 5 days by staining with anti-NG2 antibody (red) and DAPI (blue). Scale bar, 200 μm.
(H) High-power magnification of the dashed rectangle in (G). Scale bar, 30 μm. Images acquired with an EVOS AMG imaging system (Thermo Fisher Scientific). Images are representative of n = 3 independent experiments.
(I) Control immunofluorescence analysis with rabbit IgG (DAKO) as primary antibody. Scale bar, 200 μm.
(J) Aortic sections were treated with serum free-EBM media (Lonza) supplemented with VEGF-A or PDGF-AB for 5 days. Images were acquired with an EVOS AMG cell imaging system and are representative of n = 3 independent experiments. White arrows show scattered (VEGF treatment) versus linear arborizing (PDGF-AB treatment) localization of PDGFRα+ cells. Scale bars, 200 μm.
(K) Quantification of PDGFRαH2B-eGfp+ cells aligned as linear arborizing tubule-like structures after aortic explant and treatment with PDGF-AB or VEGF-A for 5 days as detected in a field of 0.2 mm2. The graph shows the average of n = 4 independent experiments for both groups. Data represent means ± SDs and were calculated by Student’s t test. **p < 0.01.
(L) Quantification of PDGFRαH2B-eGfp+ cells migrating outward from explanted aortas. Cells were counted in a field of 0.2 mm2 in n = 4 independent experiments for PDGF-AB and n = 3 for VEGF-A. *p < 0.05. Data represent means ± SDs.
To begin to understand whether PDGFRα+ cells contribute to regenerative processes, we performed ex vivo studies mimicking revascularization using aortic tissues from PDGFRαH2B-eGfp mice. Notably, in the uninjured murine aorta, PDGFRα+ cells are widely present in the adventitial compartment, but they do not generally express NG2 (Figures S1F and S1G). By plating dissected aortic samples from PDGFRαH2B-eGfp mice in Matrigel, we observed that PDGFRα+ cells aligned along newly formed tubules (Figures 1G–1I). We observed occasional areas of PDGFRα+ cells that co-expressed NG2 (Figures 1G and 1H), suggesting that under specific conditions, aortic mesenchymal PDGFRα+cells migrate and act as pericyte-like cells. The addition of PDGF-AB ligand to these explants was associated with increased vascular maturation (Figures 1J and 1K) and PDGFRα+ cell content (Figure 1L), compared to vascular endothelial growth factor (VEGF)-treated explants. These data suggest a potential role for PDGFRα+ mesenchymal cells in regulating vessel formation.
PDGFRα+ Cells Promote Revascularization and Modulate Fibrosis
To understand whether PDGFRα+ cells modulate revascularization and regeneration in vivo, we induced hindlimb ischemia (HLI) in 7-month-old inbred athymic nude mice (NU/J) and performed local adoptive transfer of purified PDGFRαH2B-eGfp+ cells, or PBS (control), into the ischemic zone. Although skeletal muscle may regenerate after injury, in older mice the regenerative program is impaired (Conboy et al., 2003; Grounds, 1998; Welle, 2002). Cells for adoptive transfer were obtained by fluorescence-activated cell sorting (FACS) for GFP+PDGFRα+ co-positive cells from the adductor thigh muscles of 3-month-old PDGFRαH2B-eGfp mice (Figures S2A and S2B). Before adoptive transfer, sorted GFP+PDGFRα+ cells were cultured for 7–10 days in hypoxic conditions (5% O2), without passaging, to allow recovery after tissue digestion while avoiding spontaneous cell differentiation (Atkuri et al., 2007; Drela et al., 2014; Fehrer et al., 2007; Panchision, 2009; Parrinello et al., 2003). Flow cytometry assessment of sorted GFP+PDGFRα+ cells showed that after 7 days in culture, 0.31% ± 0.38% of cells expressed CD31, 0.25% ± 0.29% expressed NG2, and 81.08% ± 9.98% expressed PDGFRα (n = 4 experiments). After HLI, blood flow analysis showed the same extent of recovery in control and cell-injected mice (Figures 2A and S2C). Consistent with this, there was no difference between groups in capillary density in ischemic tissues (Figures 2B and S2D). However, adoptive transfer of GFP+PDGFRα+ cells was associated with a 2.5-fold decrease in vessel leakage (Figure 2C). In addition, GFP+PDGFRα+ cell-injected mice exhibited reduced fibrosis (Figures 2D–2G), which correlated with a significant decrease in the mean diameter of regenerating vessels (Figures 2H and 2I). In accordance with these data, GFP+PDGFRα+ cell-injected mice exhibited a more normal vessel structure, with NG2+ cells tightly interacting with CD31+ cells, compared to PBS-injected mice in which the relation between NG2+ and CD31+ cells was more dispersed (Figures 2J and 2K).
Figure 2. Undifferentiated PDGFRα+ Cells Promote Favorable In Vivo Tissue Revascularization and Healing.
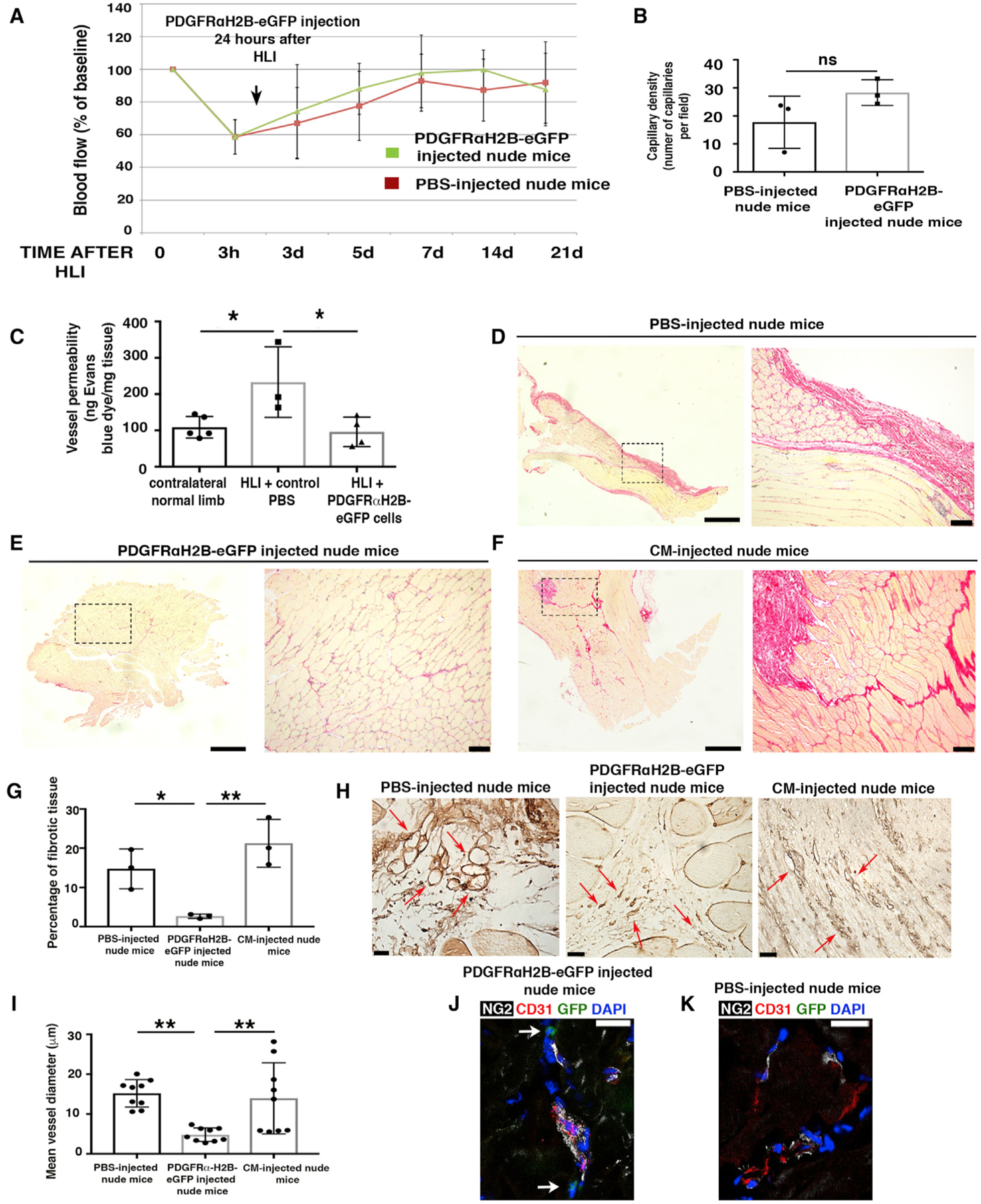
(A) HLI was induced in 7-month-old female nude mice (athymic). Adoptive transfer by local injection of 2 × 105 undifferentiated GFP+PDGFRα+ cells from 3-month-old male PDGFRαH2B-eGfp mice was performed 24 h after HLI induction. Blood flow was assessed as indicated in STAR Methods. Sixteen mice were analyzed at time 0 and after HLIs were divided randomly as control (PBS injected, n = 8) or cell-injected (n = 8) mice. Data represent means ± SDs and were analyzed by 1-way ANOVA and Tukey’s multiple comparison test. There were no differences between groups.
(B) Hindlimbs were harvested 21 days after HLI and sections were stained for isolectin B4 as described (Wen et al., 2005) and capillaries were counted in an area of 0.1 mm2 (capillary density). Groups were compared using Student’s t test (n = 3 independent experiments, p = 0.15). Data represent means ± SDs.
(C) Vessel permeability was assessed 21 days after HLI as reported (Radu and Chernoff, 2013). Values were normalized per milligram of tissue isolated. Groups were compared using 1-way ANOVA and Tukey’s multiple comparison test (n = 5 for contralateral normal limb, n = 3 for control PBS, and n = 4 for GFP+PDGFRα+ cells). Overall ANOVA p = 0.019. Data represent means ± SDs. Tukey’s post-test, *p < 0.05.
(D–F) Picrosirius red staining of skeletal muscle from PBS- (D), cell- (E), and conditioned media (CM)-injected (F) nude mice 21 days after HLI. Left panels are low magnification and dashed squares show where higher magnification (right panels) was acquired. Scale bars, 1 mm for left panels and 100 μm for right panels. Images representative of n = 3 independent experiments and were acquired with a Leica DMi8, Application Suite X, and a DCM2900 camera.
(G) Percentage of fibrotic tissue was calculated using ImageJ (NIH) and compared using 1-way ANOVA and Tukey’s multiple comparison test. *p < 0.05, **p < 0.01. Data represent means ± SDs.
(H) Isolectin B4 staining of PBS-, CM- and cell-injected ischemic adductor muscles 3 weeks after HLI. Red arrows show small and defined capillaries in mice injected with GFP+PDGFRα+ cells, whereas in control samples they were enlarged and appeared disorganized. Scale bars, 25 μm. Images acquired with a Leica DMi8, Application Suite X, and a DCM2900 camera.
(I) Average diameter of ~9 vessels measured in PBS-, CM- and cell-injected ischemic muscles. Quantification performed with ImageJ (n = 3 mice). **p < 0.01 and groups were compared using 1-way ANOVA and Tukey’s multiple comparison test. Data represent means ± SDs.
(J and K) Immunofluorescence analysis of GFP+PDGFRα+ (J) and PBS-injected (K) adductor muscles with antibodies against CD31 (red), NG2 (white), GFP+PDGFRα+ cells (green), and DAPI (blue). Scale bars, 20 μm. Images representative of n = 3 independent experiments. Images obtained with a confocal Leica SP5 DM as z stacks.
We also evaluated paracrine signaling in this model by assessing the effect of conditioned media from GFP+PDGFRα+ cells. Compared to GFP+PDGFRα+ cell-injected mice, conditioned media-injected mice showed impaired skeletal muscle regeneration 21 days after HLI (Figures 2D–2G). Furthermore, vessel diameters in the regenerating regions after HLI were similar between PBS- and conditioned media-injected nude mice, but were reduced in GFP+PDGFRα+ cell-injected mice (Figures 2H and 2I). These data suggest that paracrine signaling from PDGFRα+ cells plays only a minor role in augmenting regeneration, while cell-specific effects are the predominant factor driving our observations.
We then studied the fate of GFP+PDGFRα+ cells in our HLI model. On close examination, 7 days after HLI induction, GFP+ cells were found to be associated with what appeared to be newly forming vascular structures (Figures 3A and S2F). The expression of GFP decreased in certain cells, likely due to their increased endothelial lineage commitment and reduced PDGFRα expression, resulting in reduced GFP expression (Figure 3B). GFP+ cells were also observed co-expressing NG2 (Figures 3C, 3D, and S2F), in which NG2 is present on the cell membrane (and GFP is a nuclear protein). Many GFP+ cells were present in scar tissue and co-expressed a smooth muscle actin (αSMA) (Figures 3E, 3F, and S2F). Overall, GFP+CD31+ and GFP+NG2+ cells were less prevalent than GFP+αSMA+ cells, but the numbers of these subsets were insufficient for reliable quantitation.
Figure 3. Differentiation Potential of PDGFRα+ Cells after Ischemic Insult.
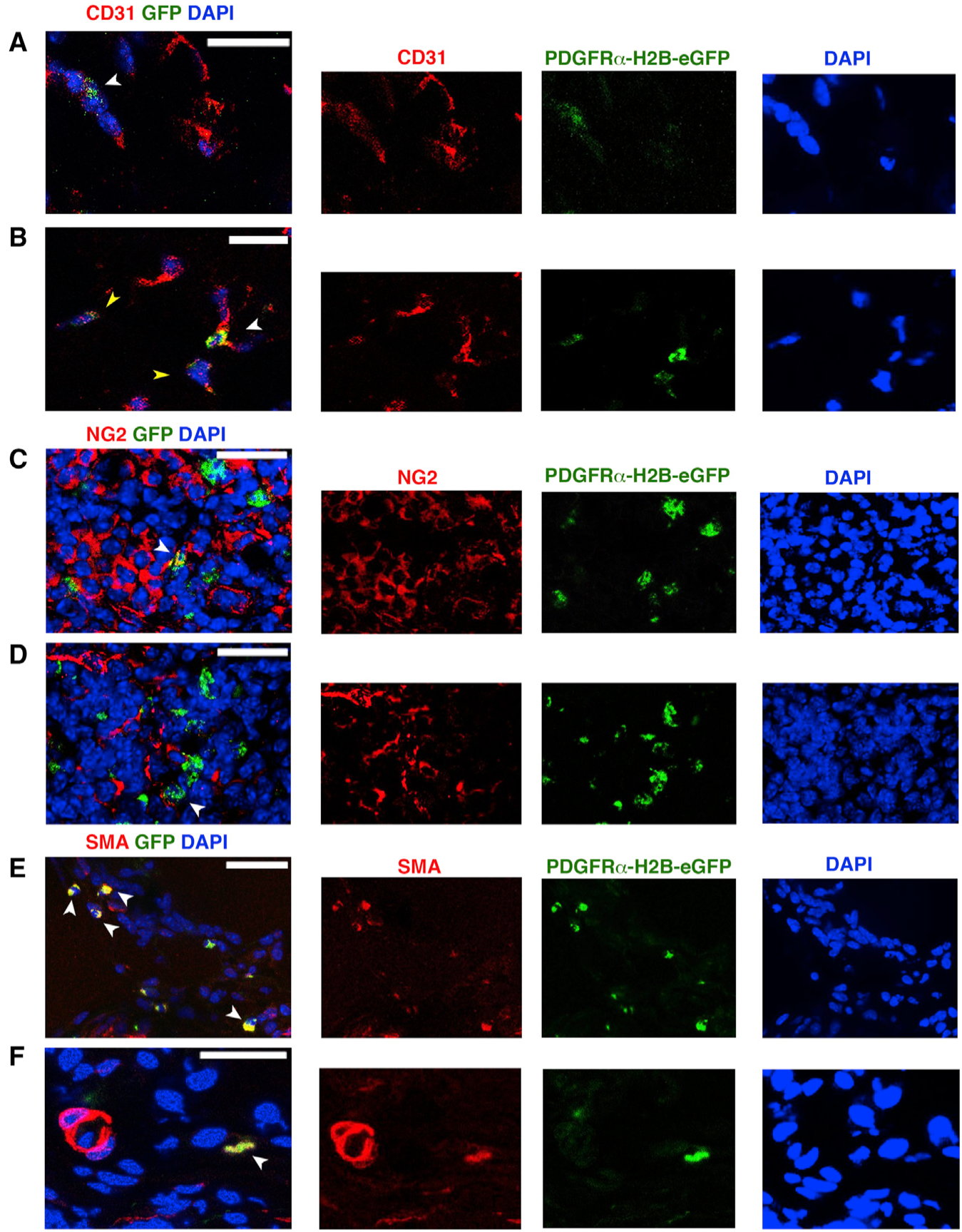
(A–F) Seven days after HLI induction and adoptive transfer of undifferentiated GFP+PDGFRα+ cells, we tracked injected cells by nuclear GFP expression and co-staining with anti-CD31 (A and B), anti-NG2 (C and D) and Cy3-conjugated anti-αSMA antibody (E and F). White arrowheads indicate GFP+ cells; yellow arrowheads in (B) show decreased expression of GFP. Images representative of n = 3 independent experiments and were obtained with a confocal Leica SP5 DM as z stacks.
In contrast, at 21 days after HLI, there was a dramatic decrease in the number of GFP+ cells compared to the 7-day time point (Figure S2E). The remaining GFP+ cells were adjacent to vascular components in direct contact with NG2+ cells (Figure S2G) or as GFP+CD31+ cells (Figure S2H), but they were not αSMA+ (Figure S2I), suggesting that the remaining GFP+ cells were integrated in the vasculature, but were not further involved in fibrotic tissue remodeling. Nevertheless, it is important to consider that GFP expression by GFP+PDGFRα+ cells obtained from PDGFRαH2B-eGfp mice may be reduced or lost as these cells undergo lineage commitment (e.g., into endothelial cells or myofibroblasts) (Chong et al., 2011; Pelekanos et al., 2012).
These data indicate that while PDGFRα+ cells do not contribute to the extent of neovascularization after HLI, they enhance the quality of vessel formation by increasing vessel stability while also reducing fibrosis. In addition, these data suggest that PDGFRα+ cells can transdifferentiate into stromal components that likely favor regeneration.
PDGFRα+ Cell Ablation Impairs Revascularization and Increases Tissue Damage after Ischemia
Next, we analyzed the effect of HLI in mice in which PDGFRα+ cells were ablated (MCM+/iDTR+/− mice). To generate MCM+/iDTR+/− mice, we crossed mice with an inducible human diphtheria toxin (DTX) receptor allele (iDTR) with PDGFRαMerCreMer (PDGFRαMCM) mice bearing a tamoxifen-inducible Cre recombinase in one allele of the PDGFRα gene (Figure 4A) (Ding et al., 2013). Mice received tamoxifen for 7 days and then DTX for 3 days before HLI surgery, followed by DTX every other day until tissue harvesting 5 days after HLI induction (Figure 4B). The model ablated ~85% of PDGFRα+ cells in hindlimb adductor muscles (Figure 4C), and ablation efficiency was monitored by flow cytometric analysis and immunofluorescence staining of a small fragment of adductor muscle from every experimental mouse (Figures S3A–S3D).
Figure 4. PDGFRα+ Cell Ablation Is Associated with Impaired Tissue Healing.
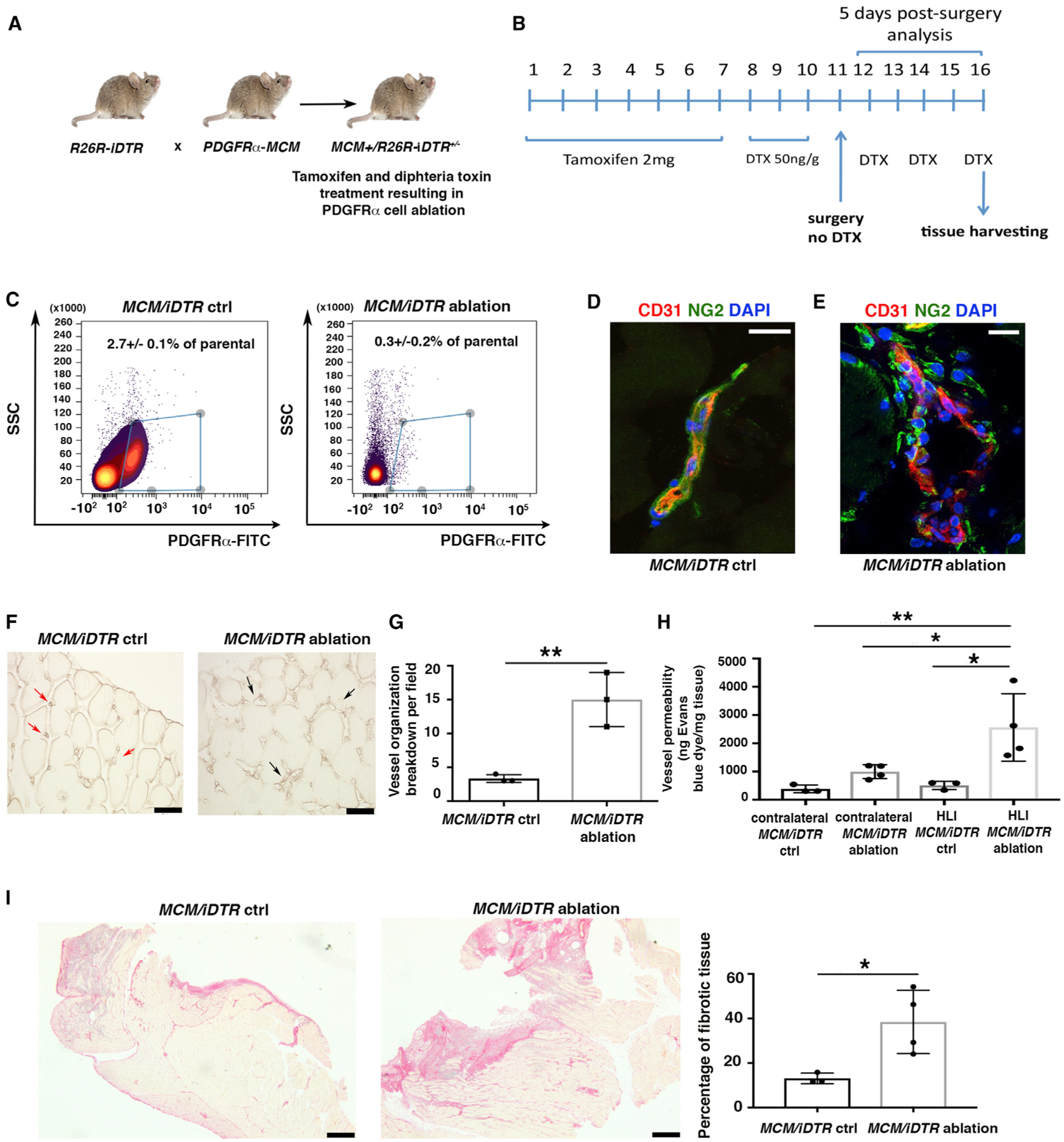
(A) Breeding strategy to generate R26R/iDTR mice harboring a knockin for CreER in the PDGFRα locus.
(B) To induce ablation, 3-month-old MCM+/iDTR+/− mice received tamoxifen and diphtheria toxin (DTX) as indicated.
(C) Flow cytometric analysis of whole hindlimb muscles to confirm cell ablation in MCM+/iDTR+/− mice treated with tamoxifen and DTX (MCM/iDTR ablation) versus peanut oil and DTX (MCM/iDTR control, ctrl) 5 days after HLI induction. PDGFRα+ cells were labeled with fluorescein isothiocyanate (FITC)-conjugated anti-PDGFRα antibody (1 μg/million cells). Data were analyzed using the Cytobank web application. Subsequently, each MCM/iDTR mouse was validated with flow cytometric analysis using a small piece of tissue, and only those with successful PDGFRα+ cell ablation were used for further experiments (see Figure S3A).
(D and E) Immunofluorescence analysis of MCM/iDTR ctrl (D) and ablated (E) adductor muscles with anti-CD31 (red) and anti-NG2 (green). Scale bars, 20 μm. Images representative of n = 3 independent experiments. Images obtained with a confocal Leica SP5 DM as z stacks.
(F) Isolectin B4 staining of MCM/iDTR ctrl and ablated adductor muscles. Red arrows show small and defined capillaries in the control sample, whereas black arrows show enlarged and disorganized capillaries in PDGFRα+ cell-ablated muscles. Scale bars, 50 μm. Images acquired with a Leica CTR 5500 microscope and DFC340FX camera.
(G) Vessel breakdown was counted in an area of 0.1 mm2, and the graph represents the average of n = 3 independent experiments. **p < 0.01. Data represent means ± SDs.
(H) Vessel permeability was assessed 5 days after HLI, as reported (Radu and Chernoff, 2013). Values were normalized per milligram of tissue isolated. Groups compared using 1-way ANOVA and Tukey’s multiple comparison test (n = 3 for control and n = 4 for ablated samples). Overall ANOVA p = 0.0051. Data represent means ± SDs. *p < 0.05, **p < 0.01.
(I) Picrosirius red staining of MCM/iDTR ctrl and ablated adductor muscles. Images representative of n = 3 independent experiments for MCM/iDTR ctrl and n = 4 for MCM/iDTR ablation. Right panel shows the quantification of the injured area, *p < 0.05. Data represent means ± SDs. Scale bar, 1 mm. Images acquired with a Leica DMi8, Application Suite X, and a DCM2900 camera.
We observed that PDGFRα+ cell ablation was associated with a macroscopically visible increase in tissue bleeding (Figure S3E). At the microscopic level, there was an increase in abnormally enlarged blood vessels (Figure 4F), with the majority of vessels presenting a dishomogeneous distribution of NG2+ and CD31+ cells compared to control vessels (Figures 4D, 4E, and S3F). Quantification showed the increased breakdown of vessel organization in ablated compared to control samples (Figure 4G). Furthermore, vessel permeability was significantly increased in ablated mice with HLI compared to control mice (Figure 4H). This phenotype correlated with an increase in fibrotic tissue after PDGFRα+ cell ablation (Figure 4I). No differences were observed in blood flow 5 days after HLI (Figure S3G). Furthermore, PDGFRα+ cell ablation alone did not induce tissue disruption or other changes before ischemia induction (Figures S3H–3J). These data are concordant with our adoptive transfer experiments and suggest that skeletal muscle PDGFRα+ cells are necessary for vessel stabilization and organization and to promote tissue healing after ischemic injury.
PDGFRα+ Cells Upregulate Stromal Remodeling Pathways during Skeletal Muscle Regeneration
To understand the mechanisms underlying PDGFRα+ cell function, we used PDGFRαH2B-eGfp mice and studied FACS-purified GFP+PDGFRα+ cells from the adductor muscle of uninjured (skeletal muscle [SM]) animals and also 7 days after HLI (skeletal muscle injury [SMI]). Consistent with prior reports (Chong et al., 2011; Kramann et al., 2015), we observed that GFP+PDGFRα+ cells harvested 7 days after HLI do not express the CD45 pan-hematopoietic marker (Figure S4A). PDGFRα+ cells harvested 7 days after HLI (SMI) showed altered mesenchymal marker expression (Figures S4B–S4D) compared to the same cells isolated from uninjured animals (Figure 1B), including a reduction in Sca1+PDGFRα+ cells but an increase in CD29+PDGFRα+ and CD44+PDGFRα+ cells. In addition, 7 days after HLI, there was an increase in CD31+PDGFRα+ cells (Figures S4C and S4D) compared to uninjured tissues (Figure 1B). Isotype antibodies are shown as pink histograms in Figure S4C and FMO controls are shown in Figures S5A and S5B.
We also undertook transcriptomic profiling by RNA sequencing (RNA-seq) of GFP+PDGFRα+ cells from SM and SMI tissues. Principal-component analysis (Figure 5A) and a block diagonal heatmap (Figure 5B) showed that GFP+PDGFRα+ cells harvested from ischemic (SMI) hindlimbs cluster separately from cells isolated from uninjured hindlimbs (SM), and they exhibit differential expression of 300 genes (Figures 5C and 5D; Table S1). As analyzed by the molecular signature database (mSigDB database), differentially expressed transcripts between the SM and SMI populations were represented in pathways related to matri-some, ECM, extracellular glycoproteins, and stem cell biology (Table S2).
Figure 5. HLI Induces Pro-fibrotic and Pro-inflammatory PDGFRα+ Cell Activation.
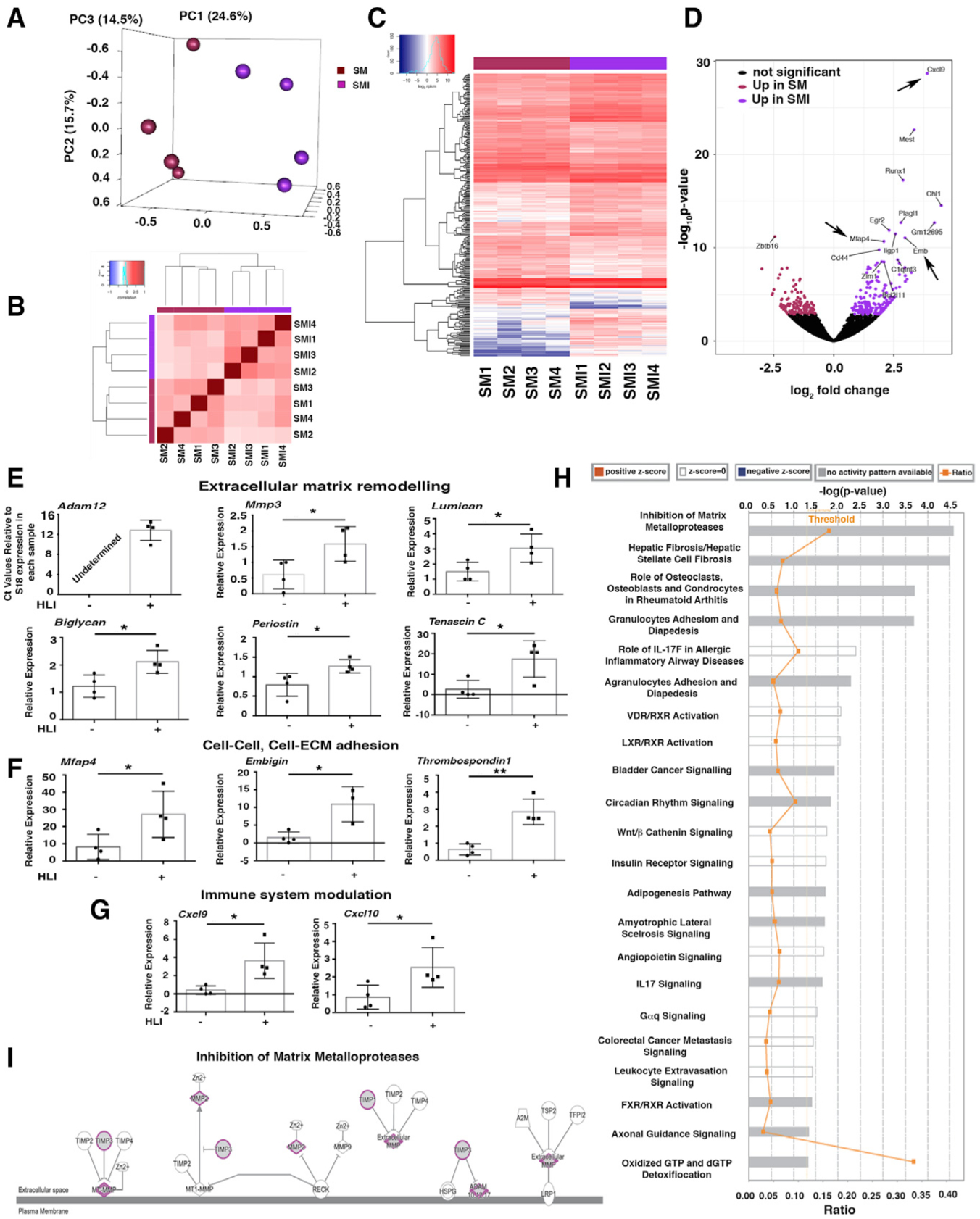
(A) Principal-component analysis (PCA) using RNA-seq data from PDGFRα+ cells from uninjured skeletal muscle (SM) and 7 days after HLI induction (skeletal muscle injury [SMI]); n = 4 different samples for both SM and SMI.
(B) Block diagonal heatmap of PDGFRα+ cells from SM and SMI showing clustering among the analyzed populations.
(C) Heatmap analysis and dendrogram of differential gene expression comparing PDGFRα+ cells from SM and SMI mice.
(D) Volcano plot showing differentially expressed genes in PDGFRα+ cells comparing SM versus SMI (300 genes had significantly different expressions). Arrows indicate upregulated genes in the inflammatory response and cell-cell, cell-ECM adhesion group.
(E–G) Quantitative real-time PCR validation of key differentially expressed transcripts for SM versus SMI PDGFRα+ cell populations, as identified by RNA-seq, using Taqman probes (Thermo Fisher Scientific) that are specific for the indicated genes. Transcripts are grouped according to their role in ECM remodeling (E), in cell-ECM and cell-cell adhesion (F), and immune modulation (G). Samples were normalized against 18S RNA levels and Student’s t test was performed. *p < 0.05, **p < 0.01. In detail: panels in (E): n = 4 for mmp3, p = 0.035; lumican, p = 0.032; biglycan, p = 0.02; periostin, p = 0.03; tenascin C, p = 0.02. Adam12 was not detected in SM PDGFRα+ cells. In (F), panels show n = 4 for mfap4, p = 0.04; embigin, p = 0.014; thrombospondin 1, p = 0.0018. In (G), panels show n = 4 for cxcl9, p = 0.017; cxcl10, p = 0.04. Data represent means ± SDs.
(H) Canonical pathways identified by Ingenuity Pathway Analysis (IPA; Ingenuity Systems, https://www.ingenuity.com) for SM versus SMI PDGFRα+ cells. Significance is expressed as p value. Bars correspond to the top 22 canonical pathways that surpassed the IPA statistical threshold. Orange squares represent the ratio value for each canonical pathway.
(I) The top IPA pathway of SM versus SMI PDGFRα+ cells (by p value) was “inhibition of matrix metalloproteases.” Genes/proteins are illustrated as nodes and molecular relations as connecting lines between nodes (direct relations as normal lines; indirect relations as dashed lines), with those highlighted (purple outline, gray fill) being differentially expressed between SM and SMI PDGFRα+ cell populations.
We performed quantitative real-time PCR to validate key differentially expressed genes identified by RNA-seq (Figures 5E–5G). Among ECM remodeling genes, HLI induced an upregulation of transcripts for the metalloproteinases Adam12 and Mmp3 and of lumican (Nikitovic et al., 2008) and biglycan (Pogány et al., 1994; Schönherr et al., 1995), which are implicated in collagen fibril assembly and organization (Figure 5E). Furthermore, after HLI, GFP+PDGFRα+ cells had increased transcript levels of the ECM components tenascin C and periostin (Figure 5E). We also observed increased levels of cell-cell and cell-ECM adhesion transcripts such as microfibrillar-associated glycoprotein 4 (Mfap4) (Milićević et al., 2016), embigin (Guenette et al., 1997), and thrombospondin (Chen et al., 2000; Lawler, 2000) (Figure 5F). After HLI, GFP+PDGFRα+ cells also produced high levels of transcripts associated with monocyte chemoattraction such as Cxcl9 and Cxcl10 (Figure 5G). We did not observe any change in the expression of the Gli1 transcript (not shown) (Kramann et al., 2015, 2016).
We used Ingenuity Pathway Analysis (IPA; Ingenuity Systems, https://www.ingenuity.com) to investigate signaling networks in the differentially expressed genes in GFP+PDGFRα+ cells after HLI. As shown in Figure 5H, pathways showing differential regulation included those governing matrix metalloproteases (“inhibition of metalloproteases”) (Figure 5I), conversion of pericytes into myofibroblasts (“hepatic fibrosis/hepatic stellate cell fibrosis”), regulation of inflammatory response pathways (“granulocyte adhesion and diapedesis,” “role of IL17F in allergic inflammatory airway disease,” “leukocyte extravasation signaling”), and networks implicated in the transport of lipids outside cells (“FXR/RXR activation” and “LXR/RXR activation”) (Figure S5C). Genes involved in lipid transport such as Abcg1 and Atp11c were also upregulated in GFP+PDGFRα+ cells after HLI (Figures S5D and S5E). In contrast, genes involved in the hydrolysis of Acyl-coenzyme A (CoA) were downregulated in GFP+PDGFRα+ cells after HLI (Acot8; Figure S5F).
These data indicate that ischemia induces a modulation in the PDGFRα+ cell phenotype and signaling pathways toward cells capable of remodeling ECM, inducing cell-cell and cell-matrix adhesion, and regulating adipocytic and inflammatory responses, likely favoring tissue repair and vessel stability.
Terminally Differentiated PDGFRα+ Cells Lose Beneficial Regenerative Properties
The activation of PDGFRα signaling may lead to fibrosis (Olson and Soriano, 2009) or vascular calcification (Cho et al., 2013; Hayes et al., 2014; Olson and Soriano, 2009), while PDGFRα signaling inhibition using imatinib reduces tissue degeneration after spinal cord injury (Abrams et al., 2012). To understand how PDGFRα+ cells could adversely affect tissue regeneration, we reproduced cell differentiation and activation conditions in vitro (Drela et al., 2014; Fehrer et al., 2007; Parrinello et al., 2003) by culturing and passaging sorted GFP+PDGFRα+ cells from PDGFRαH2B-eGfp mice in high oxygen conditions (20% O2). After only 1 passage in high oxygen, PDGFRα+ cells acquired a hypertrophied morphology with increased cell area and fine αSMA cytoskeletal structure resembling myofibroblast-like cells (Figures S6A–S6C). There were no significant levels of senescence under either culture condition (Figure S6D). Quantitative real-time PCR comparing undifferentiated (unpassaged, 5% O2) and differentiated (passaged, 20% O2) PDGFRα+ cells showed the upregulation of genes involved in cell matrix remodeling and deposition (Adam12 and collagen1a1) and fibrosis (Tgfβ1 and fibroblast activation protein [FAP]) (Figure S6E). These data indicate that with activation, as may be expected with tissue injury, PDGFRα+ cells differentiate into pro-fibrotic myofibroblast-like cells.
To investigate whether these PDGFRα+ cell-derived differentiated myofibroblast-like cells could contribute to fibrosis and impair vessel maturation, we performed further adoptive transfer experiments and injected differentiated myofibroblast-like GFP+PDGFRα+ cells from PDGFRαH2B-eGfp mice into young 3-month-old nude mice 24 h after HLI induction. Adoptive transfer of differentiated myofibroblast-like GFP+PDGFRα+ cells did not affect blood flow after HLI (Figure 6A). However, adoptive transfer of differentiated GFP+PDGFRα+ cells increased vessel leakage (Figure 6B) without changing capillary density (Figure 6C). This phenotype was associated with impaired regeneration and an increase in fibrotic area in cell-injected mice (Figure 6D). Injected GFP+ cells were identified at 7 and 21 days (Figure 6E). At 21 days after HLI, GFP+ cells were associated with areas of ECM deposition (collagen1 staining) in the proximity of muscle fibers and in more extended collagenous patches (Figures 6F–6H and S6F).
Figure 6. Differentiated PDGFRα+-Derived Cells Impair Healing after HLI.
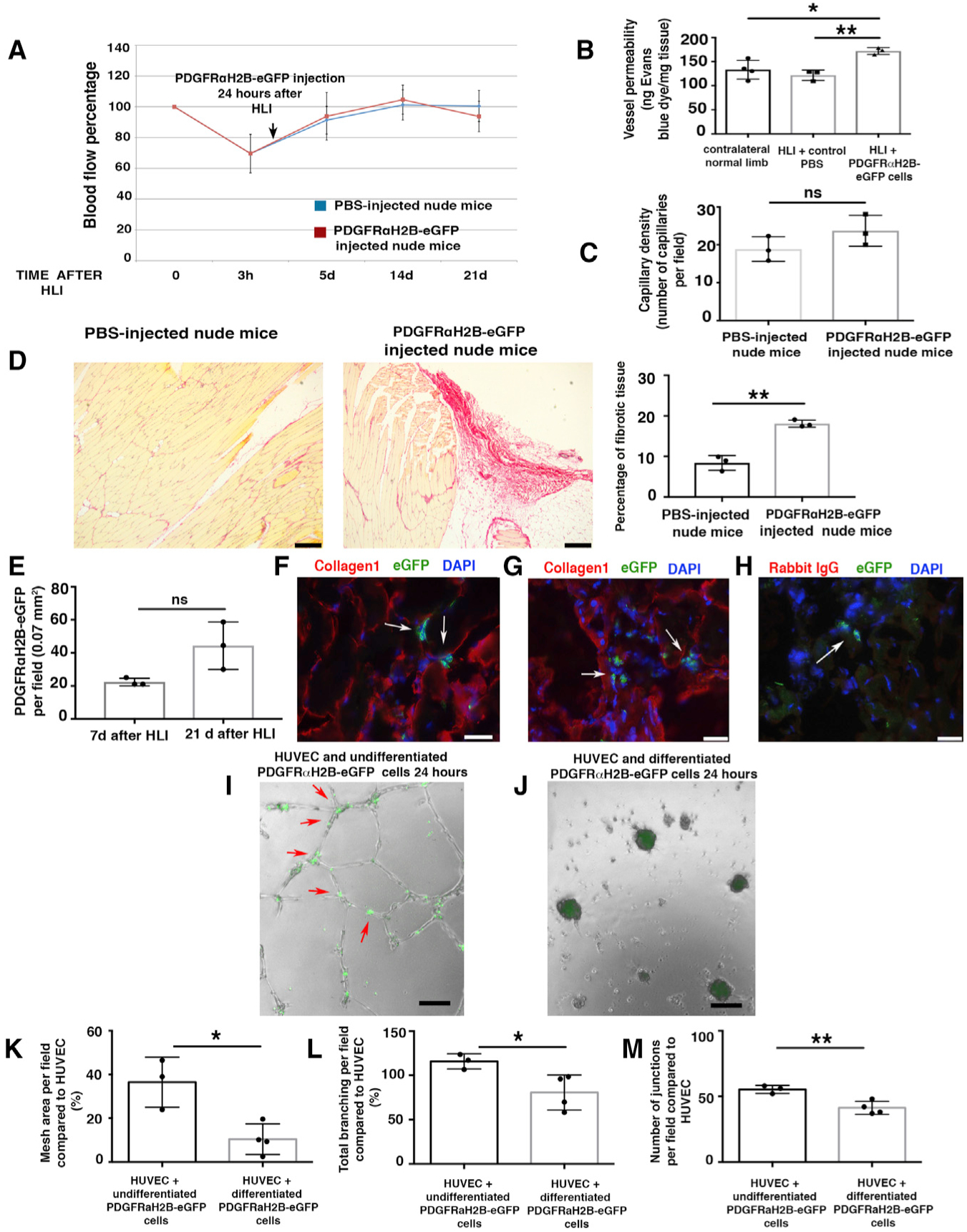
(A) HLI was induced in 3-month-old female nude mice. Local injection of 7 × 105 differentiated GFP+PDGFRα+ cells from 3-month-old male PDGFRαH2B-eGfp mice, or PBS, was performed 24 h after HLI induction. Blood flow was assessed as indicated in STAR Methods. Nineteen mice were analyzed at time 0 and randomized to control (PBS injected, n = 10) and cell-injected (n = 9) groups. Statistical analysis performed using 1-way ANOVA and Tukey’s multiple comparison test. There were no differences between the groups.
(B) Vessel permeability 21 days after HLI was determined as reported (Radu and Chernoff, 2013). Values were normalized per milligram of tissue isolated. The graph represents the average of n = 4 independent experiments for normal contralateral limb, n = 3 for both PBS-injected and differentiated PDGFRαH2B-eGfp+ cell-injected limbs. Groups compared using 1-way ANOVA and Tukey’s multiple comparison test. Data represent means ± SDs. *p < 0.05, **p < 0.01.
(C) Isolectin B4 staining performed as described (Wen et al., 2005) from cell- and PBS-injected mice. Capillaries were counted in an area of 0.1 mm2 (capillary density). The graph shows the average of n = 3 independent experiments as calculated by Student’s t test. ns represents p = 0.18. Data represent means ± SDs.
(D) Picrosirius red staining of skeletal muscle from PBS- and cell-injected nude mice 21 days after HLI. Scale bars, 250 Pro-inflammatory m. Images representative of n = 3 independent experiments. Scar area was calculated using ImageJ and analyzed by Student’s t test. **p < 0.01. Data represent means ± SDs.
(E) Injected cells were tracked by nuclear GFP expression. Quantification performed using ImageJ. GFP+PDGFRα+ co-positive cells were counted in each field, where the presence of the cells was maximal. Data represent means ± SDs and differences were not significant (ns) among groups.
(F–H) Adductor muscle sections were labeled with anti-GFP FITC-conjugated antibody and anti-collagen1 antibody. Images are representative overlays of n = 3 independent experiments and show the presence of PDGFRαH2B-eGfp+ cells (white arrows) in an area of fibrotic tissue in proximity to skeletal muscle fibers (F) or in areas with extended collagen deposition (G). Images acquired with a Leica CTR 5500 microscope and DFC340FX camera. Scale bar, 20 μm. (H) Control for collagen1 staining using rabbit IgG as the primary antibody. Anti-GFP FITC-conjugated antibody was used to detect injected cells. Individual color panels from (F), (G), and (H) are presented in Figure S6F.
(I–M) Modulation of in vitro tubulogenesis by PDGFRα+ cells. (I and J) HUVECs were plated and co-cultured with undifferentiated freshly sorted GFP+PDGFRα+ cells (I) or differentiated myofibroblast-like GFP+PDGFRα+ cells from PDGFRαH2B-eGfp mice (J). Cells were co-cultured and GFP+PDGFRα+ cells were tracked by GFP expression. Analysis was performed 24 h after plating and recorded with an EVOS AMG imaging system (Thermo Fisher Scientific). In (I), arrows show aligned GFP+PDGFRα+ cells associated with in vitro tubules. Scale bar, 100 μm. (K–M) Quantification of mesh area (total tube area) (K), total branching (amount of branches expanding from nodes) (L), and number of junctions (extent of vessel branching) (M) from cultures of HUVEC alone and HUVEC co-cultures with differentiated and undifferentiated PDGFRα+ cells after 7 h of co-culture. ImageJ angiogenesis software was used for quantification. Data represent n = 4 independent experiments for HUVEC+differentiated GFP+PDGFRα+ cells and n = 3 for HUVEC+undifferentiated GFP+PDGFRα+ cells. *p < 0.05 and **p < 0.01. Data calculated by Student’s t test and represent means ± SDs.
To understand how the adoptive transfer of differentiated GFP+PDGFRα+ cells increased vessel permeability in vivo after HLI (Figure 6B), while adoptive transfer of undifferentiated GFP+PDGFRα+ cells decreased vessel permeability (Figure 2C), human umbilical cord endothelial cells (HUVECs) were co-cultured in Matrigel with undifferentiated (Figure 6I) or differentiated GFP+PDGFRα+ cells (Figure 6J). Undifferentiated GFP+PDGFRα+ cells organized along the tubular-like structures and at junctions (Figure 6I, red arrows), whereas differentiated GFP+PDGFRα+ cells induced tubule breakage and collapse (Figure 6J). Differentiated GFP+PDGFRα+ cells impaired in vitro tubule formation by HUVECs, with a reduction in mesh area (Figure 6K), total branching (Figure 6L), and number of junctions (Figure 6M) compared to undifferentiated GFP+PDGFRα+ cells. In contrast and as distinct from co-culture, conditioned media from differentiated versus undifferentiated GFP+PDGFRα+ cells did not affect in vitro tubulogenesis (Figure S6G), suggesting that the mechanisms associated with altered vessel permeability are not related to paracrine factor release. Corroborating the importance of direct rather than paracrine effects of PDGFRα+ cells, angiogenin and VEGF-A transcript levels were unaltered in vivo in GFP+PDGFRα+ cells after HLI (SMI) compared to GFP+PDGFRα+ cells in uninjured tissue (SM) (Figure S6H). In addition, we did not observe the altered activation of IPA pathways associated with vessel formation when comparing GFP+PDGFRα+ cells from SM and SMI tissues in our RNA-seq analyses (Figure 5H), collectively indicating that cell-cell interactions rather than paracrine factors are likely to modulate vessel stability and leakage after ischemic insult.
In sum, these data show that the persistence of differentiated myofibroblast-like PDGFRα+ cells in ischemic skeletal muscles impairs the normal program of tissue regeneration and induces vessel leakage, abnormal ECM deposition, and fibrosis.
Single and Multicolor Lineage Tracking Show Clonal Responsiveness of PDGFRα+ Cells
To better understand the in vivo differentiation potential of PDGFRα+ cells, we generated MCM+/R26R-YFP+/− mice by crossing PDGFRαMCM mice (Ding et al., 2013) with R26R-eYfp mice (Figure S7A). Three-month-old MCM+/R26R-YFP+/− mice were treated with tamoxifen (1 mg/mL) for 7 days and HLI was induced 28 days after the final tamoxifen injection (Figure S7B) to allow complete metabolism of tamoxifen before HLI (Vaughan et al., 2015). A group of tamoxifen-induced control mice did not receive HLI (Figure S7). In uninjured (control) mice, consistent with the above data, we noted a relatively low number of YFP+ cells (Figure S7C), which often appeared in close proximity to CD31+ cells, NG2+ cells, and αSMA+ cells, but which rarely co-expressed these markers (Figures S7D–S7F). In contrast, 7 days after HLI, we noted an increase in the number of YFP+ cells, which were present mostly in the scar area (Figures S7C and S8). We further identified that after HLI, YFP+ cells are present in ischemic tissues as endothelial cells (Figures S8A and S8B), pericytes (Figures S8C and S8D), or in close proximity to small vessels and were co-positive for αSMA (Figures S8E and S8F). However, the integration of these YFP+ cells into newly forming vessels appeared to be an infrequent event.
To better define PDGFRα+ cell fate, we also tracked these cells using the Brainbow 2.1 line. By crossing R26R-Brainbow 2.1 (Livet et al., 2007; Snippert et al., 2010) with PDGFRαMCM mice to generate MCM+/R26R-Brainbow+/− mice, we clonally marked distinct populations of PDGFRα+-derived cells (Figures 7 and S9A). Mice were treated with tamoxifen before HLI, with the time window between tamoxifen and injury suggested in prior studies (Kanisicak et al., 2016). Samples were harvested and assessed at 7 and 21 days after HLI (Figure S9B).
Figure 7. In Vivo Expansion and Differentiation of PDGFRα+ Cells.
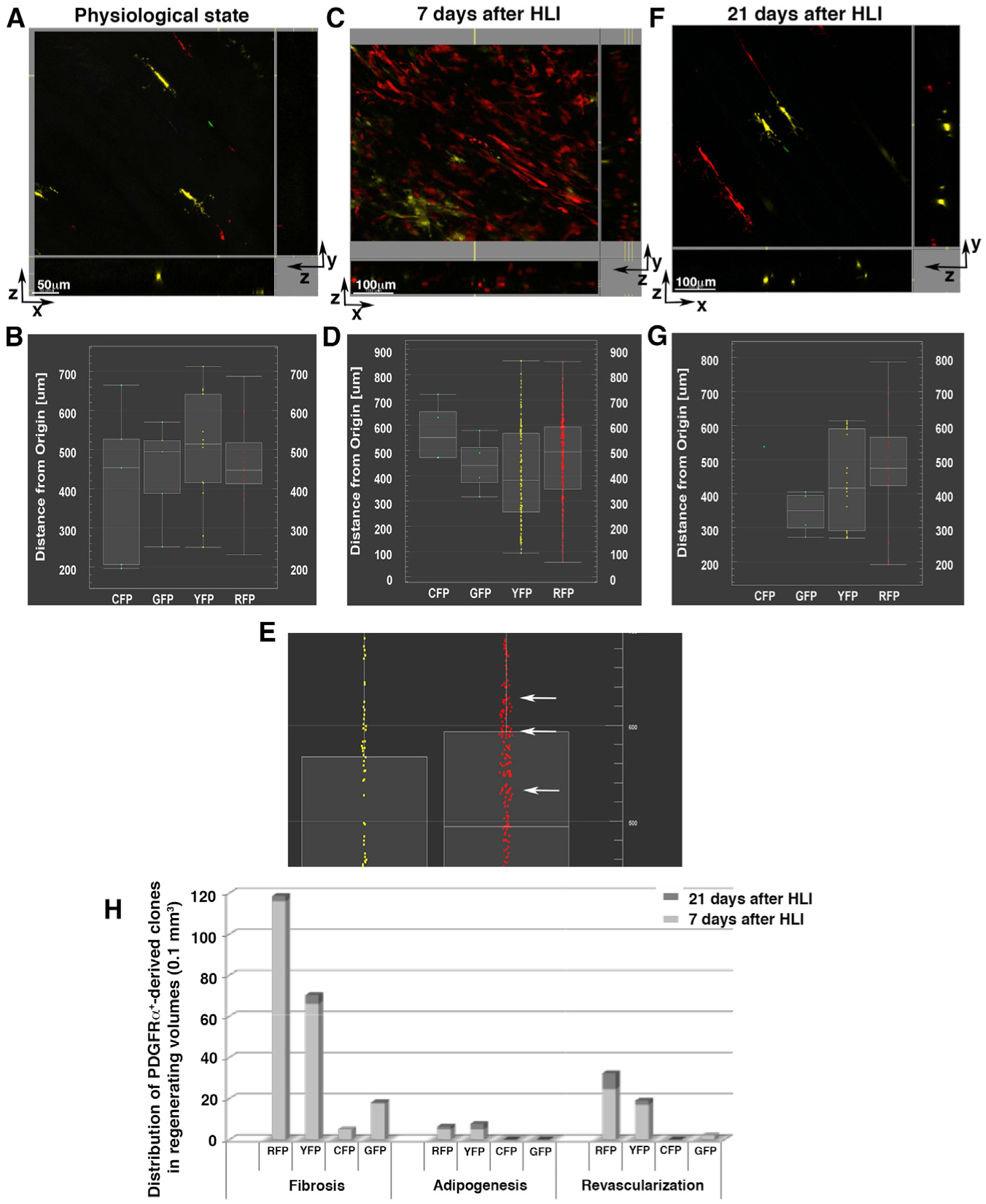
Using MCM+/R26R-Brainbow+/− mice, we studied the expansion and differentiation of PDGFRα+ cells and their progeny after HLI.
(A, C, and F) 3D microscopy images presented as x-y, x-z, and y-z views. Fluorescent channels (pseudo-colored cyan for CFP, green for GFP, yellow for YFP, red for RFP) are displayed in a physiological (uninjured) state (A), 7 days after HLI induction (C), and 21 days after HLI induction (F) and were used to analyze 1D-view plots. The images are representative of analyses performed in different locations in 3 independent mice for each condition. Scale bars as indicated.
(B, D, E, and G) 1D-view plots of cell distributions based on their distances from the origin (x-y axis) showing CFP+, GFP+, YFP+, and RFP+ cells in a physiological state (B), 7 days after HLI induction (D and E), and 21 days after HLI (G). (E) Enlarged view of (D) highlighting YFP+ and RFP+ cells and clones. At 7 days after HLI, PDGFRα+ YFP+ and RFP+ clones increased in number (see Figures S9 and S10). In addition, RFP+ cells formed small clusters throughout the 3D volume (E, white arrows), compared to scattered distributions at other time points, suggesting that at this time point, PDGFRα+-derived RFP+ cell clones were retained in proximity to their original niche.
(H) The average number of PDGFRα+-derived RFP+, YFP+, CFP+, and GFP+ clones was quantified in 3 independent mice for each time point after HLI induction (7 and 21 days) using second harmonic generation (SHG) images to distinguish fibrotic, adipocytic, and revascularized areas. Quantification was accomplished by counting PDGFRα+-derived cells present in z stacked images per 0.1 mm3 volume.
Assessment of freshly isolated skeletal muscle samples by whole-mount three-dimensional (3D) confocal/2-photon microscopy (Malide, 2016; Malide et al., 2012, 2014) showed that in uninjured conditions, PDGFRα+ cells reside along skeletal muscle fibers (Figures S9C and S9D) and around capillaries and small vessels (Figures S10A–S10C). PDGFRα+ cells were labeled with all 4 Brainbow fluorescent proteins, and consistent with prior reports (Chappell et al., 2016), YFP+ and red fluorescent protein-positive (RFP+) PDGFRα+ cells were more abundant (48.52% and 27.20%, respectively) than CFP+ (9.47%) and GFP+ (14.8%) cells (Figures 7 and S9I; Table S3).
At 7 days after HLI, there was an increased number of total PDGFRα+ and PDGFRα+-derived cells compared to physiological conditions (8.4-fold; Figure S9I). Labeled cells in the ischemic areas were associated with vessel-like structures (Figures S9E and S9F; Video S1), with fibrotic tissue (Figures S10D and S10E; Video S2) and as brown adipose cells (Figures S10F and S10G). The majority of PDGFRα+-derived cells were associated with vessels and fibrotic tissues, while a smaller proportion were identified in adipogenic regions (Figure S9J).
Three weeks after HLI, PDGFRα+ cell numbers (Figure S9I; Table S3) and their fluorescence distribution pattern (Figures S9G and S9H) generally returned to the physiological (uninjured) state (Figures 7 and S9; Table S3). Specifically, we observed that areas of tissue revascularization and adipogenesis were still present (Figures S9J and S10H–S10M; Video S3), but the number of labeled cells in these regions was reduced compared to 7 days after HLI (Figure S9J). A marked reduction in the number of PDGFRα+-derived cells was also observed in fibrotic regions (Figures 7H, S9J, S10L, and S10M). These results corroborate our prior analyses, in which PDGFRα+αSMA+ cells were cleared from the regenerated tissue 21 days after HLI induction (Figures 3 and S2).
Using the ImarisXT module, we measured the distribution frequency of Brainbow recombination and the clustering of PDGFRα+ cells and their progeny. We confirmed that 7 days after HLI, cells stochastically labeled by RFP or YFP were increased in number compared to both the uninjured state and also 21 days after HLI (Figures 7A–7G). After injury, PDGFRα+-derived cells maintained the same distribution in 3D space compared to uninjured tissues (Figures 7B, 7D, and 7G). Moreover, 7 days after HLI, RFP+ and to a lesser extent YFP+ cells were frequently observed as distinct, small clusters indicative of clonal PDGFRα+ cell expansion (Figures 7D and 7E, white arrows). This clustering indicates that early after ischemic injury, certain PDGFRα+-derived cell clones proliferate and are retained in proximity to their original niche. In addition, we detected cells marked by each of the four Brainbow fluorescent proteins within fibrotic regions, while only RFP+ and YFP+ clones were detected in adipogenic and revascularization areas (Figure 7H). We interpret these data as suggesting that after an ischemic injury, different PDGFRα+ cell clones coexist, with the majority destined to modulate the fibrotic response, while a limited number of PDGFRα+ cell clones are directed toward remodeling of the vasculature and adipose tissue. Furthermore, we did not detect any combinations of the four colors (i.e., cyan, red, green, and yellow, carried by the Brainbow cassette) occurring in wave-lengths that could suggest fusion events, indicating that PDGFRα+-derived cell fusion was unlikely to be a major factor in our model.
Our analyses revealed the phenotypic plasticity of PDGFRα+ cells, with clonal capacity to participate in vessel and adipose formation, and in the remodeling of injured and fibrotic tissues. These data indicate that tissue-resident PDGFRα+ cells are a mesenchymal progenitor population with the ability to synchronously modulate different programs and pathways in response to ischemia. We identified that skeletal muscle regeneration depends on PDGFRα+ cell-regulated reconstitution of the tissue parenchyma and stroma, with subsequent removal of differentiated PDGFRα+-derived stromal progeny.
DISCUSSION
We analyzed the role of mesenchymal PDGFRα+ cells in tissue homeostasis and healing following an ischemic event. The important in vivo results were as follows: (1) adoptive transfer of undifferentiated PDGFRα+ cells into aged nude mice improved healing after HLI (Figures 2 and 3); (2) the physiologic presence of PDGFRα+ cells was associated with favorable healing (Figures 7, S7, S8, S9, and S10); (3) PDGFRα+ cell ablation resulted in vessel disorganization and increased fibrosis (Figure 4); and (4) adoptive transfer of differentiated PDGFRα+ cells into nude mice resulted in increased fibrosis (Figure 6). Together with other data presented here, these findings permit us to draw several conclusions. First, PDGFRα+ cells and their progeny have the potential to heal an ischemic insult and to promote vessel stabilization. Second, after HLI, PDGFRα+ cells increased their activation state with the upregulation of pathways relevant to stromal tissue remodeling (Figure 5). Third, while undifferentiated PDGFRα+ cells promote favorable healing (Figures 2 and 3), the persistence of their differentiated progeny has adverse effects and promotes fibrosis after HLI (Figure 6). Fourth, PDGFRα+-derived progeny showed an initial injury-stimulated expansion, with the majority of PDGFRα+-derived clones programmed to modulate extracellular matrix and revascularization (Figures 7, S7, S8, S9, and S10). Later, there was a marked decrease in the number of PDGFRα+-derived cells in the previously injured region. Furthermore, while 7 days after HLI we detected PDGFRα+-derived cells that were co-positive for either CD31 (Figures S8A and S8B), NG2 (Figures S8C and S8D), or αSMA (Figures S8E and S8F), the substantial reduction in the number of PDGFRα+ and PDGFRα+-derived cells from 7 to 21 days after HLI (Figure S9I) suggests that PDGFRα+-derived cells do not make a substantial, direct, integrated contribution to durable new vessel formation in our model.
The switch of PDGFRα+ cells from quiescence to an activated state may be an important event after injury that re-establishes tissue stability and avoids disarray. Thus, early after HLI, PDGFRα+ cell ablation led to an increased injury area, blood vessel disorganization, and hemorrhage (Figure 4). This is consistent with studies showing impaired regeneration in the damaged muscle of animals treated with PDGFRα antagonist (Fiore et al., 2016). At present, the cell populations responsible for this increased fibrosis and tissue injury in the absence of PDGFRα+ cells remain to be determined, but may include macrophages and endothelial cells (Leach et al., 2013; Zhou et al., 2019) or cells not expressing PDGFRα but still participating in stromal tissue remodeling such as PDGFRα−NG2+, PDGFRα−Sca1+, or PDGFRα−CD44+ cells. Furthermore, and as a practical limitation in our studies, it is unknown whether PDGFRα+ cell ablation leads to vascular defects directly or whether this arises due to a more generalized failure of regeneration. However, highlighting their regenerative capacity and in contrast to ablation, adoptive transfer of undifferentiated PDGFRα+ cells into the ischemic hindlimbs of older mice favored vessel stabilization and mitigated fibrosis (Figures 2 and 3). Consistent with this, Noseda et al. (2015) showed amelioration of cardiac tissue ischemia after the injection of undifferentiated PDGFRα+Sca1+ cells. Furthermore, Fretto et al. (1993) observed that the delivery of the PDGF-BB ligand decreased cardiomyo-cyte death and preserved systolic function after myocardial infarction (Hsieh et al., 2006). In addition, the systemic administration of PDGF-BB improved vascular reactivity and hemodynamics in hemorrhagic shock (Liu et al., 2014) and induced healing in lower-extremity diabetic ulcers (Smiell et al., 1999). Our data are consistent with these studies and highlight the possibility of using undifferentiated PDGFRα+ cells as a therapeutic strategy to promote vessel stabilization and reduce fibrosis.
Although our study and the above analyses appear to contradict prior observations indicating a pro-fibrotic and adipogenic role of PDGFRα+ cells (Lombardi et al., 2016; Pannérec et al., 2013; Paylor et al., 2013; Uezumi et al., 2010, 2014a, 2014b), these earlier studies were performed in the setting of chronic diseases such as skeletal muscle dystrophy (Uezumi et al., 2014a, 2014b) and arrhythmogenic cardiomyopathy (Lombardi et al., 2016; Paylor et al., 2013), and in mdx mutant mice (Ieronimakis et al., 2016). It is possible that rather than being a primary cause of these diseases, PDGFRα+ cells may merely exacerbate these ongoing pathologies. Thus, depending on the inflammatory milieu, fibro-adipogenic progenitors residing in tissues for longer periods may cause fibrotic degeneration and chronic injury (Lemos et al., 2015). Accordingly, we found that differentiated PDGFRα+-derived cells were associated with poor healing and lack of regeneration when implanted into ischemic murine skeletal muscle, with reduced vessel stability and increased tissue fibrosis (Figure 6). These differentiated PDGFRα+ cells, unlike undifferentiated cells, appeared to escape cell clearance (Lemos et al., 2015) and remained abundant 3 weeks after injury (Figures S2E and 6E). As a whole, it appears that the timely removal or quiescence of specific differentiated PDGFRα+ cells is important for optimal tissue healing and/or preventing chronic pathological diseases. Consistent with this, cardiac periostin+ myofibroblasts revert to a quiescent state once the post-myocardial infarction fibrotic response has resolved (Kanisicak et al., 2016), while ADAM12+ myofibroblasts are eliminated as tissues recover following cardiotoxin injection (Dulauroy et al., 2012).
In summary, we demonstrated a dual (“yin-yang”) role for mesenchymal PDGFRα+ cells during tissue regeneration as supporting cells orchestrating vessel formation that concurrently limit tissue injury. Our data suggest that a reappraisal of PDGFRα+ cells in non-regenerating organs such as the heart and kidney may be appropriate, as their role in promoting fibrosis and a lack of healing may be related to injury chronicity rather than being any intrinsic property of PDGFRα+ cells them-selves. From a cell therapy perspective, there are also important implications for this yin-yang functionality of PDGFRα+ cells. For the last decade, there has been concern that the delivery of various mesenchymal progenitor populations as a cell therapy is associated with initial beneficial paracrine effects but with a failure of cell engraftment and eventual removal and/or death of the transferred cells (Golpanian et al., 2016). We suggest that, depending on the chronicity of the disease and the degree of lineage differentiation, this “failure of engraftment” may be essential for tissue recovery, and that if “successful” engraftment of specific lineage-committed mesenchymal cells were achieved, this may be pathologic. Fully dissecting the cues that direct these cells toward regeneration rather than fibro-adipogenic differentiation is clearly a task that is of major scientific and clinical importance.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents may be directed to and will be fulfilled by the Lead Contact, Dr. Jason Kovacic (jason.kovacic@mountsinai.org). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Murine transgenic models
All animal studies were performed in compliance with the regulations of the National Institutes of Health and the local institutional animal care and use committee of the Icahn School of Medicine at Mount Sinai. In all experimental procedures mice were anaesthetised with inhaled Isoflurane (1.5–2.5%) and 1.5 ml/min O2. Adequacy of anesthesia was monitored by foot pinch before any incision. For tissue extraction and primary cell isolation, mice were euthanized by cervical dislocation after being anesthetised with Isoflurane. After surgery, animals were allowed to recover with free access to food and water. Injection of analgesia (e.g., buprenorphine) was performed as required post-operatively.
Several strains were used in this study. 1) PDGFRαH2B-eGfp mice express the H2B-eGfp fusion gene from the endogenous PDGFRα locus (Jackson Laboratory; strain B6.129S4-PDGFRαtm11(EGFP)Sor/J; stock #007669). Fluorescence patterns mimic the expression pattern of the endogenous gene. Mice were kept in hemizygous breeding conditions. 2) PDGFRαMerCreMer mice, defined as MCM in this paper, were obtained from RIKEN (Japan), accession number CDB0674K, http://www2.clst.riken.jp/arg/mutant%20mice%20list.html. Mice were generated by knocking the tamoxifen inducible Cre-ER cDNA into the PDGFRα locus (Ding et al., 2013). MCM mice were kept in hemizygous breeding and crossed with iDTR, R26R-eYfp and R26R-Brainbow mice. 3) iDTR mice (Jackson Laboratories; strain C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J; stock #007900) have Cre-inducible expression of the human DTR that renders cells susceptible to ablation following DTX administration. 4) MCM+/iDTR+/− mice were generated by crossbreeding MCM and iDTR mice. 5) Nude mice (Jackson Laboratories, athymic nude, nu/j, inbred for 100 generations) were used for cell transplantation experiments. Mice were purchased from Jackson Laboratories Inc (stock #002019) and kept in SPF conditions. Animals kept in these conditions show premature aging as reported previously (Jutila, 1977). 6) The R26R-Brainbow (also called R26R-Confetti) line was purchased from Jackson Laboratories (stock #017492) and used to clonally track PDGFRα+ cells, including their expansion and differentiation. The R26R-Confetti conditional allele has a CAG promoter followed by a floxed-STOP cassette and the Brainbow 2.1 construct all targeted into the Gt(ROSA)26Sor locus. The R26R-Confetti allele functions as a stochastic multicolor Cre recombinase reporter of multiple fluorescent proteins from a single genomic locus. R26R-Confetti mice permit cell labeling and the distinguishing of individual/adjacent cells with nuclear localized, membrane-targeted, or cytoplasmic fluorescent proteins in Cre recombined cells. 7) R26R-Brainbow mice were bred with MCM mice to generate MCM+/R26R-Brainbow+/− mice that express one of the random fluorescent proteins specifically in PDGFRα+ cells upon tamoxifen injection. 8) R26R-eYfp mice were purchased from Jackson Laboratories (stock #006148) and used to lineage track PDGFRα+ cells in physiological conditions and in response to ischemia. R26R-eYfp mice have a loxP-flanked STOP sequence followed by the Enhanced Yellow Fluorescent Protein gene (eYfp) inserted into the Gt(ROSA)26Sor locus. When bred to mice expressing Cre recombinase, the STOP sequence is deleted and eYfp expression is observed in the Cre-expressing tissue(s). 9) R26R-eYfp mice were bred with MCM mice to generate MCM+/R26R-YFP+/− mice expressing specifically YFP in PDGFRα+ cells after tamoxifen injection. 10) Wild-type (WT) C57BL/6J mice (Jackson Laboratories stock #000664) were also used as indicated.
Cell culture
We used human umbilical cord endothelial cells (HUVECs) and GFP+PDGFRα+ cells from the hindlimbs of PDGFRαH2B-eGfp mice.
HUVECs were used in co-culture experiments in vitro with differentiated and undifferentiated GFP+PDGFRα+ cells. Briefly, 20,000 HUVECs were seated on 150 μL matrigel (Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, Corning) alone or in combination with 10,000 differentiated or undifferentiated GFP+PDGFRα+ cells. Co-culture with lower (5000 cells) or higher numbers (20,000 cells) of GFP+PDGFRα+ cells produced similar results (not presented). Matrigel was prepared 1h before and left at 37°C until plating of cells. The experiment was performed in 48 multiwells. Cells were monitored every 3h for 24h. Measurements were performed at 7h. Pictures were obtained with an EVOS AMG cell imaging system (Thermo Fisher Scientific) and tubulogenesis (mesh area, total branching, and number of junctions) were quantified with ImageJ angiogenesis analyzer software (NIH).
FACS-sorted GFP+PDGFRα+ cells from the hindlimbs of PDGFRαH2B-eGfp mice were used to obtain undifferentiated and differentiated PDGFRα+ cells. For undifferentiated PDGFRα+ cells, after tissue digestion and FACS for GFP and PDGFRα co-positive cells (see below), GFP+PDGFRα+ cells were plated in hypoxic conditions (5% O2) in DMEM (high glucose, 4500 g/L, Corning) supplemented with 1% antibiotic/antimycotic (Invitrogen) and 10% FBS (Invitrogen). When near-confluent, undifferentiated GFP+PDGFRα+ cells were used directly for experiments without passaging. For differentiated GFP+PDGFRα+ cells, GFP+PDGFRα+ cells were plated in identical media (DMEM with 10% FBS and antibiotic/antimycotic) but they were cultured in 20% O2 and passaged once, and then allowed to reach near-confluence prior to experiments. Both cell types were stained with Cy3-conjugated anti-αSMA antibody diluted in PBS 1x/0.3% Triton X-100 1:500 for 1h at room temperature. Nuclei were stained with DAPI for 10 min at room temperature. Images acquired using an EVOS AMG cell imaging system (Thermo Fisher Scientific) and cell size was measured with ImageJ (NIH). Cell-injection with differentiated and undifferentiated GFP+PDGFRα+ cells was performed in nude mice after HLI induction as described below.
METHOD DETAILS
Antibodies
For immunofluorescence we used the following antibodies against: NG2 (Abcam, ab83178, ab50009 or ab83508), PDGFRα (Santa Cruz, #sc-338 or sc-398206), αSMA-Cy3 (Sigma #C6198), GFP-FITC (Abcam, ab6662), CD31 (BD Bioscience, 550274) and Collagen1 (Abcam, ab34710). Secondary antibodies were purchased from Life Technologies and used at 1:100 or 1:200 dilution for 2h at room temperature. As control, rabbit, rat or mouse IgG were used at an identical dilution to the corresponding primary antibody.
For flow cytometric analysis and FACS we used the following antibodies: APC-conjugated PDGFRα (eBioscience, #17-1401-81), APC-conjugated CD31 (BioLegend, #102409), APC-conjugated NG2 (R&D, #FAB2585A), PE-conjugated Sca1 (Becton Dickinson, #553108), PE-conjugated NG2 (Becton Dickinson #562415), APC-Cy7-conjugated CD45 (Becton Dickinson, #557659), PE-conjugated CD44 (eBioscience, 12-0441-82), PE-conjugated CD31 (Biolegend, #102408), PE-conjugated CD73 (Biolegend, #127205), PE-conjugated CD29 (Biolegend, #102207), PE-conjugated CD105 (R&D systems, #FAB1320P) and PE-conjugated PDGFRα (eBioscience 12-1402-81). For isotype controls the following antibodies were used for each mesenchymal marker: PE rat IgG2a, kappa (control for Sca1 BD Bioscience, #553930), APC rat IgG2a kappa (control for PDGFRα eBioscience, #17-4321-81), PE mouse IgG2a, kappa (control for NG2, BD Bioscience #555574), PE rat IgG2A (control for CD105 R&D systems, IC006P), PE Armenian hamster IgG (control for CD29 Biolegend, #400907), PE rat IgG1, kappa (control for CD73 Biolegend, #400407), PE rat IgG2a, kappa (control for CD31 Biolegend, #400507), PE rat IgG2a, kappa (control for PDGFRα eBioscience, #12-4321-80), PE rat IgG2b, kappa (control for CD44 eBioscience, #12-4031-82).
Histology and immunofluorescence analysis
The adductor muscle was harvested from control and injured mice, washed in PBS and fixed in 4% paraformaldehyde (PFA) to further process for paraffin sectioning or for embedding in OCT to prepare samples for frozen sections.
For immunohistochemistry analysis, samples embedded in paraffin were sectioned at 8μm thickness and processed for staining of the microvasculature and to analyze scar and fibrosis formation. Microvessels were stained with endothelial cell marker isolectin B4 (Biotinylated, L2140, Sigma) as described (Wen et al., 2005). Microvessels were counted in 4–6 defined microscope fields in 3 different samples per condition (PBS and cell injected). Connective tissue was visualized using Masson’s Trichrome stain as described by the manufacturer (Sigma) or Picrosirius Red staining as described (Junqueira et al., 1979).
For immunofluorescence, 8–10μm sections of the adductor muscle were stained with the antibodies listed previously. PDGFRα and NG2 were used at 1:50 dilution, overnight at 4°C. GFP-FITC and αSMA-Cy3 were used at 1:500 dilution for 1h at room temperature. GFP-FITC and collagen1 were used at 1:500 and 1:100 dilution respectively overnight at 4°C. GFP-FITC and CD31 were used at 1:500 and 1:50 dilution respectively overnight at 4°C. For collagen1 anti-rabbit A546 (Life Technologies) was used at 1:200 and for PDGFRα anti-rabbit A488 (Life Technologies) was used at 1:100 dilution for 2h at RT. For NG2 anti-rabbit or anti-mouse A546 (Life Technologies) was used at 1:100 for 2h RT. For CD31 anti-rat A546 (Life Technologies) was used at 1:100 for 2h RT. Controls were performed with rabbit (DAKO, XD936), rat IgG (abcam ab18450) and mouse IgG (Life Technologies, #02–6200) at concentrations similar to or greater than the primary antibodies.
Images were acquired either with a Leica CTR 5500 microscope and DFC425 camera for brightfield or DFC340FX camera for immunofluorescence, an EVOS AMG cell imaging system (Thermo Fisher Scientific), or Leica DMi8 with DCF365FX camera for immunofluorescence or DMV2900 for brightfield, as indicated in the Figure Legends. Confocal microscopy was performed with a Leica SP5 DM and images acquired as z stacks on 10 μm frozen sections.
Aortic ring assay
An aortic ring assay was performed as described (Baker et al., 2011). Briefly, freshly isolated aortas from PDGFRαH2B-eGfp mice were cut in cross-section into 1μm ring segments and distributed on 50 μL of low growth factor matrigel (Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, Corning). Aortic rings and matrigel were left at 37°C for 30 min to allow the matrigel to solidify prior adding EBM media supplemented with VEGF-AA (30ng/ml, Peprotech) or PDGF-AB (30ng/ml, R&D). Sprouting of cells was observed for 5d and recorded on an EVOS AMG cell imaging system (Thermo Fisher Scientific). After 5d, the rings were fixed with 4% PFA, permeabilized with PBS/Triton 0.25% and stained with NG2 as described above.
Hindlimb ischemia model and cell injection
Unilateral high femoral artery ligation and excision was performed on male MCM+/iDTR+/− mice or nude female mice (old mice were 7 months of age; young mice were 3 months as indicated). Anesthesia was administered using Isoflurane as described above. Mice were placed in a dorsal position with their hindlimbs externally rotated. A skin incision was made over the femoral artery beginning at the inguinal ligament and continued caudally to the popliteal position. The femoral artery and vein were isolated from the neurovascular bundle along the entire limb, and ligated using a 6–0 prolene suture at the distal and the proximal ends. Major branch points were dissected free and the femoral artery and vein were excised in their entirety.
Nude mice were injected with undifferentiated (n = 8, 2×105) or differentiated (n = 9, 7×105) GFP+PDGFRα+ cells (originally from PDGFRαH2B-eGfp mice) 24h after induction of HLI. Control mice (n = 8 for undifferentiated control experiment and n = 10 for differentiated control experiment) were injected with an equal amount of 1× PBS. Undifferentiated cells were grown in 5% O2. Differentiated cells were grown in 20% O2 and passaged once as described above. In preparation for injection cells were trypsinized and prepared in 1× PBS to a total volume of 100 μL per mouse. Cells were then injected in two different areas of the adductor muscle in divided aliquots. Blood flow analysis and/or final experimental procedures were performed as described in the results section.
Tamoxifen and diptheria toxin treatment
MCM+/iDTR+/−, MCM+/R26R-YFP+/− and MCM+/R26R-Brainbow+/− mice were treated with 2mg tamoxifen (Sigma Aldrich) diluted in peanut oil (Sigma Aldrich) or with peanut oil alone as control for seven consecutive days. For MCM+/R26R-Brainbow+/− mice, HLI was induced 4–5d after completing tamoxifen, and confocal microscopy assessment was performed at 7 and 21d after HLI as described below. For MCM+/R26R-YFP+/− mice, after completing treatment with tamoxifen, these mice were left for 28d (to allow exogenous tamoxifen to dissipate) before inducing HLI. Immunofluoresence staining and confocal microscopy were performed 7d after HLI. For MCM+/iDTR+/− mice, after completing treatment with tamoxifen, these mice were further treated with DTX (Sigma Aldrich) by I.P. injection at 50ng/g for 3d prior to HLI induction, and then every other day for an additional 5d.
Conditioned Media preparation and injection
Conditioned media from GFP+PDGFRα+ cells was obtained by culturing cells as above for 24h in serum free media. Media were collected and gently centrifuged and proteins were concentrated using an Amicon Ultra-0.5 centrifugal filter (Millipore) as described by the Manufacturer. Equal amounts of proteins (approximately 5μg) were injected in 6 mice 24h after HLI induction and the regenerative outcomes were compared to the adoptive transfer of the cells prepared as described above.
Flow cytometric analysis and FACS
For FACS, adductor muscles from PDGFRαH2B-eGfp mice were excised and washed in PBS. Samples were minced in 1ml of serum free DMEM containing 0.8 Wunsch units/ml of Liberase DL (Roche Diagnostic) and left for 40 min at 37°C on a gently shaking thermo-block at 1200rpm. As indicated in the specific experiments, cells were incubated for 1h with anti-PDGFRα APC-conjugated antibody at a concentration of 1 μg per million cells and/or anti-CD45-APC-Cy7 (1μg/million cells) in DMEM containing 2% FBS. Dead cells were excluded by labeling with 1 μg/ml of DAPI 30 min prior sorting. Cells were sorted on an Influx FACS at the Flow Cytometry Core Facility at ISMMS.
For flow cytometric analysis using WT mice, cells from the adductor muscle were isolated as described above and labeled with 1 μg per million cells using anti-PDGFRα-APC antibody, and PE-conjugated or PE-conjugated isotype control for Sca1, CD44, NG2, CD31, CD73, CD105, CD29 and PDGFRα antibodies. In experiments where PDGFRα+GFP+ cells were isolated from PDGFRαH2B-eGfp mice, APC-conjugated antibodies for CD31 and NG2 were used at 1μg per million of cells. Cells were visualized with a LSRII flow cytometric analyzer and the analysis performed with the publicly available analytical platform Cytobank.
Confocal and two-photon microscopy, and image analysis
For the analysis of MCM+/R26R-Brainbow+/− mice, samples were processed fresh for microscopy within 20 min of harvesting. Skeletal muscles including the adductor, hamstring, and gastrocnemius muscles were analyzed at all time points. To maintain the integrity of the cellular components and the fluorescent signals from the Cre-recombined Brainbow cassette, the tissue was not fixed or permeabilized but processed immediately for confocal analysis. Microscopy imaging was performed using a Leica SP5 five channel confocal and multiphoton system (Leica microsystems, Mannheim, Germany) equipped with multi-line Argon, diode 561nm, HeNe 594nm, and HeNe 633nm visible lasers. For the 2-photon mode we used a pulsed femtosecond Ti:Sapphire (Ti-Sa) laser, tunable for excitation from 680 to 1,080nm with dispersion correction. Freshly excised skeletal muscles were placed onto 35mm number 0 coverglass culture dishes in 1× PBS and imaged using a HCX-IRAPO-L 25×/0.95 NA water dipping objective (WD = 2.5mm). Previously recorded fluorescence emission spectra were used to set four sequential confocal imaging channels as follows: CFP (458/468–482nm), eGFP (488/496–514 nm), YFP (514/522–558nm) and RFP (561/580–650nm). In addition, structural information was obtained by two-photon SHG microscopy, revealing signal from skeletal fibers and fibrillar collagen (using the Ti-Sa laser at 920nm collecting back-scattered signal) as we previously described (Malide et al., 2012). For 3D volume rendering and analysis, a series of x-y-z images (typically 1×1×4 μm3 voxel size) along the z axis at 5 μm intervals over a range of depths (150–200 μm) were collected throughout the skeletal muscles. Reconstruction and analysis of 3D volumes was performed with Imaris ×64 software version 8.4 to transform the stacks of raw images into volume-rendered (3D) data of fluorescent cells inside skeletal muscle fibers and to export them as 3D-rotation movies.
For skeletal muscle tissue preparations from nude mice after adoptive cell transfer, MCM+/R26R-YFP+/− and MCM+/iDTR+/− mice, a conventional laser-scanning confocal microscope was used (Leica SP5 DM). Images of PDGFRα+ cells labeled with YFP, GFP or with the rabbit antibody SC-20 (Santa Cruz) and associated fluorescence (CD31, NG2 and αSMA) were acquired using an Argon/2 ion laser for excitation at 488nm, a HeNe laser for excitation at 543nm. High-magnification image stacks were captured using a 63× oil immersion lens. Orthogonal views were created using Fiji and the respective movies by 3D Viewer Plugin in Fiji. The image stacks were acquired at a 1024×1024 spatial resolution.
RNA extraction, cDNA amplification and real-time PCR
RNA was extracted from freshly sorted GFP+PDGFRα+ cells using a RNAeasy mini kit (QIAGEN) as described by the manufacturer. For in vitro culture of undifferentiated and differentiated GFP+PDGFRα+ cells, RNA was extracted using a RNAeasy mini kit (QIAGEN) and cDNA was synthesized as described below. RNA was measured using a NanoDrop 2000c (Thermo Scientific). For cDNA amplification RNA was diluted at 100pg/μL for all cells and 500pg were used from all samples and amplified using the Ovation RNASeq System V2 32 reactions (NuGEN Technologies, Inc.). Amplification was performed according to manufacturer’s instructions. Briefly, total RNA was reverse-transcribed using a combination of random hexamers and a poly-T chimeric primer, followed by second strand synthesis using DNA polymerase. The cDNA was then amplified using single primer isothermal amplification technology (SPIA). A proprietary combination of enzymes and primers allows for preferential priming of non-rRNA sequences.
cDNA was synthesized using a Taqman reverse transcription kit (Applied Biosystems). 10ng/μl of RNA was reverse transcribed using random hexamers with the following conditions: 25°C 10 min, 48°C for 30 min and 95°C for 5 min. An aliquot of the synthesized cDNA was diluted 1:5 to obtain a concentration of 2ng/μl.
For quantitative real-time PCR, 6ng of Ovation RNaseq system-amplified cDNA or cDNA from undifferentiated and differentiated GFP+PDGFRα+ cells was used. Samples were normalized against 18sRNA transcript levels and relative expression was calculated using the double delta Ct method. Probes were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.).
RNA sequencing
RNaseq and bioinformatic analysis were performed at the Institute for Genomics and Multiscale Biology at the ISMMS. As an initial step, Qubit QC values were evaluated and a cut-off of above 500ng total was applied. Libraries from the SPIA amplified cDNA as described above were made using the Ultralow DR library kit (NuGEN) according to manufacturer’s instructions. High-throughput RNaseq was performed using a HiSeq 2500 (Illumina) with reads between 7–20 million among the different samples.
Bioinformatic analysis
RNaseq reads were aligned to GRCm38/mm10with STAR v2.4.0g1 (Dobin et al., 2013). Uniquely mapping reads overlapping genes were counted with FeatureCounts v1.4.4 (Liao et al., 2014) using annotations from ENSEMBL GRCm38.75. All analysis used log2 counts per million (CPM) or log2 RPKM (Mortazavi et al., 2008) following TMM normalization (Robinson and Oshlack, 2010) implemented in edgeR v3.14.0 (Robinson and Oshlack, 2010). Genes with over 1 counts per million in at least 4 samples were retained. Differential expression analysis was performed using DESeq2 (Love et al., 2014). Downstream analysis including principal components analysis and heatmaps was performed in R (R Development Core Team, 2015). Hierarchical clustering was performed with the ward.D method in R. Gene set enrichment tests were performed in R with a hypergeomtric test using MSigDB (Subramanian et al., 2005). Additional enrichment and pathway tests were performed with Ingenuity Pathway Analysis and MetaCore.
Blood flow measurement
Blood flow was measured in mice anesthetized under constant-flow isoflurane, both in the ischemic leg and the control leg, preoperatively at time 0, postoperatively (between 2–3h after surgery), and at different time points as indicated after HLI, using a moorFLPI-2 unit (Moor Instruments, Ltd.). Recording was done at a processing mode set at low resolution/high speed, with a frequency of 25Hz.
Evans blue dye injection and vessel permeability analysis
Vessel permeability was assessed as described (Radu and Chernoff, 2013). Briefly, 200 μL of 1% Evans blue dye (Sigma Aldrich, Inc.) diluted in 1× PBS was injected via tail vein. The adductor muscle was isolated 30 min after injection and weighed. Dissected tissues were left for 36h in 500 μL of formamide at 55°C prior to spectrophotometric measurements (Abs = 610nm).
Senescence Assay
Cellular senescence was measured by a senescence histochemical staining kit (Sigma, CS0030). PDGFRα+ cells from 4-month old PDGFRαH2B-eGfp mice were sorted as described above and plated under hypoxic (5% O2) or normoxic conditions (20% O2). To induce differentiation, cells in 20% O2 were passaged once. Analysis was performed as indicated by the Manufacturer. Briefly, senescent cells were detected using a rapid staining procedure based on β-galactosidase activity at pH 6. Under these conditions β-galactosidase activity is easily detectable in senescent cells but not in quiescent, immortal or tumor cells. Quantification of senescent cells was performed by counting the number of β-galactosidase positive cells in 10 to 20 fields in n = 3 independent experiments for each condition, and expressing this as a percentage of the total number of cells in each field analyzed. Comparison among different groups was performed by analysis of variance (one-way ANOVA) with Tukey’s post-testing.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Values are expressed as mean ± standard deviation of the mean (SD). Statistical significance was assumed when p < 0.05. GraphPad Prism V.7® (GraphPad Software, La Jolla, USA) was used for statistical analyses. Comparison among different samples (as indicated in each Figure legend) was performed by analysis of variance (one-way ANOVA) with Tukey’s post-testing or by Student’s t test (two-tailed) as indicated.
Imaging and quantification in tissues
Imaging for Masson’s trichrome assay, biotinylated isolectin B4 staining, and for PDGFRα+ and NG2+ cells in skeletal muscle was acquired with a Leica CTR 5500 microscope and DFC425 camera for bright-field images, a DFC340FX camera for immunofluorescence pictures or with an EVOS AMG cell imaging system (Thermo Fisher Scientific). For the aortic ring experiments and fluorescence imaging of differentiated and undifferentiated GFP+PDGFRα+ cells, imaging was performed with an EVOS AMG cell imaging system (Thermo Fisher Scientific).
Quantification of microvessel density and vessel breakdown in MCM+/iDTR+/− mice was performed by counting in 4 defined microscope fields in 3 different samples per condition. Measurements were performed on an identically sized area for all samples (0.1mm2).
Maximal fibrotic area was measured with ImageJ. Slides for each condition after HLI (n = 3) were analyzed under the microscope to find the minimal and maximal fibrotic area covering the entire extension of injury. The maximal fibrotic area for the adoptive transfer and the ablation studies was quantified using values of maximal threshold depending on the level of injured tissue in the animals analyzed.
Quantification of vessel permeability
Evans blue dye leakage from vessels was measured by reading the prepared samples (n = 3) at 610nm and normalizing these values for mg of tissue. A standard curve of known Evans blue dye amounts was used to calculate the extravasated ng of Evans blue dye (Radu and Chernoff, 2013).
Quantification of blood flow recovery
Blood flow percentage recovery was measured using a moorFLPI-2 unit by dividing the average flow values of the ischemic limb by the healthy limb at the indicated time points. Values are expressed in percentage. The number of mice for each condition is described in the Figure Legends and Results section.
Quantification of tubulogenesis
In vitro tubulogenesis was analyzed in HUVECs in co-culture with undifferentiated and differentiated GFP+PDGFRα+ cells. Quantification of mesh area, total branching, and number of junctions was performed to determine the effects of PDGFRα+ cells on HUVEC function and analyzed by ImageJ angiogenesis analyzer software (NIH). Regulation of tubulogenesis was calculated by dividing the mesh area or the total branching values of each independent experiment (n = 4 for differentiated PDGFRα+ cell co-culture and n = 3 for undifferentiated PDGFRα+ cell co-culture) in co-culture set ups with the average mesh area or total branching values in HUVECs alone (n = 3). The values are expressed in percentage.
Quantification of injected GFP+PDGFRα+ cells in nude mice
Approximately 7×105 undifferentiated and differentiated GFP+PDGFRα+ cells were injected in both young and old nude mice respectively as previously described (hind limb ischemia model and cell injection sections). Analysis was performed at 7 and 21d after HLI. Three animals were used in each condition. Harvested skeletal muscles were frozen in OCT and 10mm sections were analyzed for the presence of GFP+PDGFRα+ cells. Ten slides for each mouse and each condition were analyzed and quantification was performed using the slides where cells were visibly present in higher number. Images acquired with a Leica DMi8, Application Suite X and a 40X objective. Cell amount was quantified using ImageJ and reported as number of cells in 0.07mm2.
Quantification of Brainbow studies
Analyses were performed on 3 mice for each condition (physiological uninjured state, 7 and 21d after HLI induction). For every mouse between 4 and 16 images were collected in different locations in the hindlimb skeletal muscle. Three of these images for each mouse in each condition were used for quantification and one or two for graphical representation in this manuscript.
For quantitative assessment of clone frequency and distribution, 3D z stack fluorescent images were further processed using user-guided software segmentation tools in the Imaris Surpass module (Malide et al., 2012; Takaku et al., 2010). Individual cells were segmented as surface objects in their individual fluorescent channels. Their numbers were exported in Excel software and displayed as relative average number in the z stack analyses. A total of 169 cells in uninjured mice, 1421 cells 7d after HLI and 287 cells 21d after HLI were counted. Percentages of cells were quantified by relating the relative average number of individual fluorescent cells to the total in the 3D datasets (z stacks). The collected images were used to quantify the average number and fluorescence distribution of PDGFRα+-derived clones in injured areas (fibrosis, adipose tissue and revascularization). Measurements were performed on three mice in each condition (7 and 21d post-HLI) for a total of 3–4 images where adipose, fibrotic, and vascular tissue could be distinguished by second harmonic generation (SHG). The total volume for these analyses was 0.1mm3. For quantitative assessment of 3D cell and clone frequency, positions, and distribution, z stacks were further processed using the ImarisXT module, which integrates MATLAB applications (MathWorks) by distance measurements (using a distance transformation algorithm) and cluster analysis (Malide et al., 2012). Cell coordinate data were exported in the Vantage module (Imaris software) and used to generate 1D view plots of the cells based on distance from the origin (intersection of x-y axis). These were analyzed for each clone at different time points for cell distribution and distances (clustering) between cells.
DATA AND CODE AVAILABILITY
Data Resources
The accession number for the bulk RNaseq data reported in this manuscript is GEO: GSE101930 and the datasets are available at the Genome Expression Omnibus (GEO) and in Tables S1 and S2 of this manuscript. The raw data including the analysis of vessel permeability in the adoptive transfer experiments (Figures 2C and 6B), the flow cytometry fcs files for sorting the cells for RNA sequence and adoptive transfer (Figures S2A, S2B, and S4A) and the majority of the flow cytometry data to characterize PDGFRα+ cells as stromal cells (Figures 1A, 1B, S1A, S1B, S4B, S4C, S5A, and S5B) are uploaded at Mendeley Data with the following https://doi.org/10.17632/wkg6844nwv.1.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NG2 | Abcam | Cat#ab83508; RRID:AB_2087616. ab50009; RRID:AB_881569. ab83178; RRID:AB_10672215 |
| PDGFRα | Santa Cruz | Cat#sc-338; RRID:AB_631064. sc-398206; RRID:N/A |
| αSMA-Cy3 | Sigma | Cat#C6198; RRID:AB_476856 |
| GFP-FITC | Abcam | Cat#ab6662; RRID:AB_305635 |
| PDGFRα-APC | eBioscience | Cat#17-1401-81; RRID:AB_529482 |
| NG2-APC | R&D | Cat#FAB2585A; RRID:AB_622049 |
| CD31-APC | BioLegend | Cat#102409; RRID:AB_312904 |
| Sca1-PE | Becton Dickinson | Cat#553108; RRID:AB_394629 |
| NG2-PE | BD PharMingen | Cat# 562415; RRID:AB_11154031 |
| CD45-APC-Cy7 | Becton Dickinson | Cat#557659; RRID:AB_396774 |
| CD44-PE | eBioscience | Cat#12-0441-82; RRID:AB_465664 |
| CD31-PE | Abcam | Cat#ab25644; RRID:AB_470721 |
| Collagen1 | Abcam | Cat#ab34710; RRID:AB_731684 |
| PE rat IgG2a, kappa | BD-Bioscience | Cat#553930; RRID:AB_479719 |
| APC rat IgG2a kappa | eBioscience | Cat#17-4321-81; RRID:AB_470181 |
| PE mouse IgG2a, kappa | BD-Bioscience | Cat#555574; RRID:AB_395953 |
| PE rat IgG2A | R&D systems | Cat# IC006P; RRID:AB_357256 |
| PE Armenian hamster IgG | Biolegend | Cat#400907; RRID:AB_326593 |
| PE rat IgG1, kappa | Biolegend | Cat#400407; RRID:AB_326513 |
| PE rat IgG2a, kappa | Biolegend | Cat#400507; RRID:N/A |
| PE rat IgG2a, kappa | eBioscience | Cat#12-4321-80; RRID:AB_1834380 |
| PE rat IgG2b, kappa | eBioscience | Cat#12-4031-82; RRID:AB_470042 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Evans blue dye | Sigma | Cat#E2129 |
| Paraformaldehyde | EMS | Cat#15710 |
| OCT | Tissue-Tek | Cat#4583 |
| Vascular endothelial growth factor A, VEGF-A | Peprotech | Cat#450–32 |
| Platelet derived-growth factor AB, PDGF-AB | R&D | Cat#1115-AB-010 |
| Biotinylated isolectin B4 | Sigma | Cat#L2140 |
| Tamoxifen (> 99%) | Sigma | Cat#T-5648 |
| Diphteria Toxin, DTX | Sigma | Cat#D0564 |
| Growth Factor Reduced BD Matrigel Membrane | BD Bioscience | Cat#356230 |
| DMEM, High glucose/L-glutamine | Life technologies | Cat#11965–096 |
| EGM-2 SingleQuot Kit Suppl. & Growth Factors | Lonza | Cat#CC4176 |
| EBM2 basal media | Lonza | Cat#CC3156 |
| Fetal Bovine Serum | Life Technologies | Cat#16000044 |
| Strepdavidin conjugated HRP | Abcam | Cat#ab64269 |
| DAB | Sigma | Cat#D3939–1SET |
| Critical Commercial Assays | ||
| RNAeasy mini kit | QIAGEN | Cat#74104 |
| Direct Red 80 | Sigma | Cat# 365548 |
| Ovation RNA sequence V2 | NuGen Technologies | Cat#7102–32 |
| Senescence Cells Histochemical Staining Kit | Sigma | Cat#CS0030 |
| Deposited Data | ||
| RNaseq data | This paper | GEO: GSE101930 |
| Experimental Models: Cell Lines | ||
| HUVEC | Lonza | Cat#C2517A |
| PDGFRαH2B-eGFP cells (from PdgfrαH2B-eGfp mice) | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Nude, Strain NU/J, inbred for 100 generations | Jackson Laboratories | Cat#002019; RRID:IMSR_JAX:002019 |
| Mouse: PdgfrαH2B-eGfp, Strain B6.129S4-Pdgfratm11(EGFP)Sor/J | Jackson Laboratories | Cat#007669; RRID:IMSR_JAX:007669 |
| Mouse: IDTR, Strain C57BL/6-Gt(ROSA)26Sortm1 (HBEGF)Awai/J | Jackson Laboratories | Cat#007900; RRID:IMSR_JAX:007900 |
| Mouse: MCM, Strain PdgfrαMerCreMer | Riken | Accession# CDB0674K, http://www2.clst.riken.jp/arg/mutant%20mice%20list.html; RRID:N/A |
| Mouse: Brainbow 2.1, Strain B6.129P2-Gt(ROSA) 26Sortm1(CAG-Brainbow2.1)Cle/J. | Jackson Laboratories | Cat#017492; RRID:IMSR_JAX:017492; |
| Mouse: R26R-eYfp, Strain B6.129X1-Gt (ROSA)26Sortm1(eYfp)Cos/J | Jackson Laboratories | Cat#006148; RRID:IMSR_JAX:006148 |
| Sequence-Based Reagents | ||
| Ovation RNA sequence V2 | NuGen Technologies | Cat#7102–32 |
| Software and Algorithms | ||
| GraphPad Prism V.6® | GraphPad Software, La Jolla, USA | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| ImageJ | NIH | https://imagej.nih.gov/ij/; RRID:SCR_003070 |
| Fiji | NIH | http://fiji.sc; RRID:SCR_002285 |
| Cytobank Community | Cytobank Inc. | https://www.cytobank.org/index.html; RRID:SCR_014043 |
| Imaris V8.4 | Bitplane | https://imaris.oxinst.com/packages; RRID:SCR_007370 |
Highlights.
Skeletal muscle PDGFRα+ cells have dual pro-regenerative and pro-fibrotic functions
PDGFRα+ cells are necessary for the restoration of tissue integrity after ischemia
PDGFRα+ cells remodel extracellular matrix and support revascularization after ischemia
PDGFRα+ cells induce pathological outcomes if they persist as differentiated cells
ACKNOWLEDGMENTS
We thank Fondation Leducq (Transatlantic Network of Excellence Awards) for funding these analyses. We acknowledge the assistance of the Microscopy, Genomics and Multiscale Biology, Histopathology, and Flow Cytometry Core Facilities at Icahn School of Medicine at Mount Sinai (ISMMS). M.P.S. acknowledges support from the National Institutes of Health (NIH) (R01HL135093). G.P. acknowledges support from the NIH (R01GM114434). V.D. and A.N.-K. were supported by NIH grant T32HL007824. J.C.K. acknowledges support from Fondation Leducq and the NIH (R01HL135093 and R01HL130423). R.P.H. acknowledges support from the National Health and Medical Research Council of Australia (NHMRC) (APP1118576 and 1074386), the Australian Research Counsel Special Initiative in Stem Cell Science (SR110001002), Foundation Leducq (15 CVD 03, 13 CVD 01), and the New South Wales Government Department of Health. T.F. is supported by the Fondation Leducq and the Progeria Research Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.12.045.
DECLARATION OF INTERESTS
R.P.H. declares a patent filed in the name of University of Sydney, Victor Chang Cardiac Research Institute, covering the use of the PDGF-AB ligand as a therapy for cardiovascular events, including myocardial infarction. Status: Provisional patent and PCT submitted, priority data 14.06.18. The other authors declare no competing interests.
REFERENCES
- Abrams MB, Nilsson I, Lewandowski SA, Kjell J, Codeluppi S, Olson L, and Eriksson U (2012). Imatinib enhances functional outcome after spinal cord injury. PLoS One 7, e38760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkuri KR, Herzenberg LA, Niemi AK, Cowan T, and Herzenberg LA (2007). Importance of culturing primary lymphocytes at physiological oxygen levels. Proc. Natl. Acad. Sci. USA 104, 4547–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Robinson SD, Lechertier T, Barber PR, Tavora B, D’Amico G, Jones DT, Vojnovic B, and Hodivala-Dilke K (2011). Use of the mouse aortic ring assay to study angiogenesis. Nat. Protoc 7, 89–104. [DOI] [PubMed] [Google Scholar]
- Carr MJ, Toma JS, Johnston APW, Steadman PE, Yuzwa SA, Mahmud N, Frankland PW, Kaplan DR, and Miller FD (2019). Mesenchymal Precursor Cells in Adult Nerves Contribute to Mammalian Tissue Repair and Regeneration. Cell Stem Cell 24, 240–256.e9. [DOI] [PubMed] [Google Scholar]
- Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, and Jørgensen HF (2016). Extensive Proliferation of a Subset of Differentiated, yet Plastic, Medial Vascular Smooth Muscle Cells Contributes to Neointimal Formation in Mouse Injury and Atherosclerosis Models. Circ. Res 119, 1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Herndon ME, and Lawler J (2000). The cell biology of thrombospondin-1. Matrix Biol. 19, 597–614. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Cho HJ, Lee HJ, Song MK, Seo JY, Bae YH, Kim JY, Lee HY, Lee W, Koo BK, et al. (2013). Vascular calcifying progenitor cells possess bidirectional differentiation potentials. PLoS Biol. 11, e1001534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, Heffernan C, Menon MK, Scarlett CJ, Rashidianfar A, et al. (2011). Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 9, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJ, Reinecke H, Iwata M, Torok-Storb B, Stempien-Otero A, and Murry CE (2013). Progenitor cells identified by PDGFR-alpha expression in the developing and diseased human heart. Stem Cells Dev. 22, 1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, and Rando TA (2003). Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577. [DOI] [PubMed] [Google Scholar]
- Di Carlo SE, and Peduto L (2018). The perivascular origin of pathological fibroblasts. J. Clin. Invest 128, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Tanaka Y, Hayashi M, Nishikawa S, and Kataoka H (2013). PDGF receptor alpha+ mesoderm contributes to endothelial and hematopoietic cells in mice. Dev. Dyn 242, 254–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drela K, Sarnowska A, Siedlecka P, Szablowska-Gadomska I, Wielgos M, Jurga M, Lukomska B, and Domanska-Janik K (2014). Low oxygen atmosphere facilitates proliferation and maintains undifferentiated state of umbilical cord mesenchymal stem cells in an hypoxia inducible factor-dependent manner. Cytotherapy 16, 881–892. [DOI] [PubMed] [Google Scholar]
- Dulauroy S, Di Carlo SE, Langa F, Eberl G, and Peduto L (2012). Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat. Med 18, 1262–1270. [DOI] [PubMed] [Google Scholar]
- Farahani RM, and Xaymardan M (2015). Platelet-Derived Growth Factor Receptor Alpha as a Marker of Mesenchymal Stem Cells in Development and Stem Cell Biology. Stem Cells Int. 2015, 362753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrer C, Brunauer R, Laschober G, Unterluggauer H, Reitinger S, Kloss F, Gülly C, Gassner R, and Lepperdinger G (2007). Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell 6, 745–757. [DOI] [PubMed] [Google Scholar]
- Fiore D, Judson RN, Low M, Lee S, Zhang E, Hopkins C, Xu P, Lenzi A, Rossi FM, and Lemos DR (2016). Pharmacological blockage of fibro/adipogenic progenitor expansion and suppression of regenerative fibrogenesis is associated with impaired skeletal muscle regeneration. Stem Cell Res. (Amst.) 17, 161–169. [DOI] [PubMed] [Google Scholar]
- Fretto LJ, Snape AJ, Tomlinson JE, Seroogy JJ, Wolf DL, LaRochelle WJ, and Giese NA (1993). Mechanism of platelet-derived growth factor (PDGF) AA, AB, and BB binding to alpha and beta PDGF receptor. J. Biol. Chem 268, 3625–3631. [PubMed] [Google Scholar]
- Gabrielli A, Svegliati S, Moroncini G, Luchetti M, Tonnini C, and Avvedimento EV (2007). Stimulatory autoantibodies to the PDGF receptor: a link to fibrosis in scleroderma and a pathway for novel therapeutic targets. Autoimmun. Rev 7, 121–126. [DOI] [PubMed] [Google Scholar]
- Golpanian S, Wolf A, Hatzistergos KE, and Hare JM (2016). Rebuilding the Damaged Heart: Mesenchymal Stem Cells, Cell-Based Therapy, and Engineered Heart Tissue. Physiol. Rev 96, 1127–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds MD (1998). Age-associated changes in the response of skeletal muscle cells to exercise and regeneration. Ann. NY Acad. Sci 854, 78–91. [DOI] [PubMed] [Google Scholar]
- Guenette RS, Sridhar S, Herley M, Mooibroek M, Wong P, and Tennis-wood M (1997). Embigin, a developmentally expressed member of the immunoglobulin super family, is also expressed during regression of prostate and mammary gland. Dev. Genet 21, 268–278. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, and Soriano P (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol 23, 4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BJ, Riehle KJ, Shimizu-Albergine M, Bauer RL, Hudkins KL, Johansson F, Yeh MM, Mahoney WM Jr., Yeung RS, and Campbell JS (2014). Activation of platelet-derived growth factor receptor alpha contributes to liver fibrosis. PLoS One 9, e92925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. (2003). PDGFRA activating mutations in gastrointestinal stromal tumors. Science 299, 708–710. [DOI] [PubMed] [Google Scholar]
- Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, and Kitamura Y (2003). Gain-of-function mutations of platelet-derived growth factor receptor alpha gene in gastrointestinal stromal tumors. Gastroenterology 125, 660–667. [DOI] [PubMed] [Google Scholar]
- Hsieh PC, Davis ME, Gannon J, MacGillivray C, and Lee RT (2006). Controlled delivery of PDGF-BB for myocardial protection using injectable self-assembling peptide nanofibers. J. Clin. Invest 116, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieronimakis N, Hays A, Prasad A, Janebodin K, Duffield JS, and Reyes M (2016). PDGFRα signalling promotes fibrogenic responses in collagen-producing cells in Duchenne muscular dystrophy. J. Pathol 240, 410–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, and Rossi FM (2010). Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 12, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira LC, Bignolas G, and Brentani RR (1979). Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem. J 11, 447–455. [DOI] [PubMed] [Google Scholar]
- Jutila J (1977). Congenitally athymic (nude) mice and their application to the study of immunity and ageing In Immunology and Ageing, Makinoden Tand Yienis E, eds. (Springer; ), pp. 177–182. [Google Scholar]
- Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, J Lin SC, Aronow BJ, Tallquist MD, and Molkentin JD (2016). Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun 7, 12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, and Humphreys BD (2015). Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann R, Goettsch C, Wongboonsin J, Iwata H, Schneider RK, Kuppe C, Kaesler N, Chang-Panesso M, Machado FG, Gratwohl S, et al. (2016). Adventitial MSC-like Cells Are Progenitors of Vascular Smooth Muscle Cells and Drive Vascular Calcification in Chronic Kidney Disease. Cell Stem Cell 19, 628–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J (2000). The functions of thrombospondin-1 and-2. Curr. Opin. Cell Biol. 12, 634–640. [DOI] [PubMed] [Google Scholar]
- Leach HG, Chrobak I, Han R, and Trojanowska M (2013). Endothelial cells recruit macrophages and contribute to a fibrotic milieu in bleomycin lung injury. Am. J. Respir. Cell Mol. Biol 49, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos DR, Babaeijandaghi F, Low M, Chang CK, Lee ST, Fiore D, Zhang RH, Natarajan A, Nedospasov SA, and Rossi FM (2015). Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med 21, 786–794. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang J, Zhu Y, Xiao X, Peng X, Yang G, Zang J, Liu S, and Li T (2014). Beneficial effects of platelet-derived growth factor on hemorrhagic shock in rats and the underlying mechanisms. Am. J. Physiol. Heart Circ. Physiol 307, H1277–H1287. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, and Lichtman JW (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62. [DOI] [PubMed] [Google Scholar]
- Lombardi R, Chen SN, Ruggiero A, Gurha P, Czernuszewicz GZ, Willerson JT, and Marian AJ (2016). Cardiac Fibro-Adipocyte Progenitors Express Desmosome Proteins and Preferentially Differentiate to Adipocytes Upon Deletion of the Desmoplakin Gene. Circ. Res 119, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E, Segarra M, and Cid MC (2006). Stimulatory autoantibodies to the PDGF receptor in scleroderma. N. Engl. J. Med 355, 1278–1279. [DOI] [PubMed] [Google Scholar]
- Malide D (2016). In Vivo Cell Tracking Using Two-Photon Microscopy. Methods Mol. Biol 1444, 109–122. [DOI] [PubMed] [Google Scholar]
- Malide D, Métais JY, and Dunbar CE (2012). Dynamic clonal analysis of murine hematopoietic stem and progenitor cells marked by 5 fluorescent proteins using confocal and multiphoton microscopy. Blood 120, e105–e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malide D, Métais JY, and Dunbar CE (2014). In vivo clonal tracking of hematopoietic stem and progenitor cells marked by five fluorescent proteins using confocal and multiphoton microscopy. J. Vis. Exp (90), e51669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milićević NM, Schmidt F, Kunz N, Kalies K, Milićević Z, Schlosser A, Holmskov U, Sorensen GL, and Westermann J (2016). The role of microfibrillar-associated protein 4 (MFAP4) in the formation and function of splenic compartments during embryonic and adult life. Cell Tissue Res. 365, 135–145. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, and Wold B (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Murray IR, Gonzalez ZN, Baily J, Dobie R, Wallace RJ, Mackinnon AC, Smith JR, Greenhalgh SN, Thompson AI, Conroy KP, et al. (2017). av integrins on mesenchymal cells regulate skeletal and cardiac muscle fibrosis. Nat. Commun 8, 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitovic D, Katonis P, Tsatsakis A, Karamanos NK, and Tzanakakis GN (2008). Lumican, a small leucine-rich proteoglycan. IUBMB Life 60, 818–823. [DOI] [PubMed] [Google Scholar]
- Noseda M, Harada M, McSweeney S, Leja T, Belian E, Stuckey DJ, Abreu Paiva MS, Habib J, Macaulay I, de Smith AJ, et al. (2015). PDGFRα demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat. Commun 6, 6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H (2006). Stimulatory autoantibodies to the PDGF receptor in scleroderma. N. Engl. J. Med 355, 1278. [PubMed] [Google Scholar]
- Olson LE, and Soriano P (2009). Increased PDGFRαlpha activation disrupts connective tissue development and drives systemic fibrosis. Dev. Cell 16, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchision DM (2009). The role of oxygen in regulating neural stem cells in development and disease. J. Cell. Physiol 220, 562–568. [DOI] [PubMed] [Google Scholar]
- Pannérec A, Formicola L, Besson V, Marazzi G, and Sassoon DA (2013). Defining skeletal muscle resident progenitors and their cell fate potentials. Development 140, 2879–2891. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, and Campisi J (2003). Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor B, Fernandes J, McManus B, and Rossi F (2013). Tissue-resident Sca1+ PDGFRα+ mesenchymal progenitors are the cellular source of fibrofatty infiltration in arrhythmogenic cardiomyopathy. F1000Res. 2, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelekanos RA, Li J, Gongora M, Chandrakanthan V, Scown J, Suhaimi N, Brooke G, Christensen ME, Doan T, Rice AM, et al. (2012). Comprehensive transcriptome and immunophenotype analysis of renal and cardiac MSC-like populations supports strong congruence with bone marrow MSC despite maintenance of distinct identities. Stem Cell Res. (Amst.) 8, 58–73. [DOI] [PubMed] [Google Scholar]
- Pogány G, Hernandez DJ, and Vogel KG (1994). The in vitro interaction of proteoglycans with type I collagen is modulated by phosphate. Arch. Biochem. Biophys 313, 102–111. [DOI] [PubMed] [Google Scholar]
- Radu M, and Chernoff J (2013). An in vivo assay to test blood vessel permeability. J. Vis. Exp (73), e50062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2015). R: A language and environment for statistical computing (R Foundation for Statistical Computing).
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini MP, Forte E, Harvey RP, and Kovacic JC (2016). Developmental origin and lineage plasticity of endogenous cardiac stem cells. Development 143, 1242–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr E, Witsch-Prehm P, Harrach B, Robenek H, Rauterberg J, and Kresse H (1995). Interaction of biglycan with type I collagen. J. Biol. Chem 270, 2776–2783. [DOI] [PubMed] [Google Scholar]
- Smiell JM, Wieman TJ, Steed DL, Perry BH, Sampson AR, and Schwab BH (1999). Efficacy and safety of becaplermin (recombinant human platelet-derived growth factor-BB) in patients with nonhealing, lower extremity diabetic ulcers: a combined analysis of four randomized studies. Wound Repair Regen. 7, 335–346. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, and Clevers H (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku T, Malide D, Chen J, Calado RT, Kajigaya S, and Young NS (2010). Hematopoiesis in 3 dimensions: human and murine bone marrow architecture visualized by confocal microscopy. Blood 116, e41–e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FK (2006). Autoantibodies against PDGF receptor in scleroderma. N. Engl. J. Med 354, 2709–2711. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, and Tsuchida K (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 12, 143–152. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Ikemoto-Uezumi M, Nakatani M, Morita M, Yamaguchi A, Yamada H, Nishino I, Hamada Y, and Tsuchida K (2014a). Identification and characterization of PDGFRα+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis. 5, e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi A, Ikemoto-Uezumi M, and Tsuchida K (2014b). Roles of nonmyogenic mesenchymal progenitors in pathogenesis and regeneration of skeletal muscle. Front. Physiol 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A, Brumwell A, and Chapman H (2015). Tamoxifen Administration for Lineage Tracing Using CreERT2 Mice. Protocol Exchange. 10.1038/protex.2015.018. [DOI] [Google Scholar]
- Welle S (2002). Cellular and molecular basis of age-related sarcopenia. Can. J. Appl. Physiol 27, 19–41. [DOI] [PubMed] [Google Scholar]
- Wen PH, De Gasperi R, Sosa MA, Rocher AB, Friedrich VL Jr., Hof PR, and Elder GA (2005). Selective expression of presenilin 1 in neural progenitor cells rescues the cerebral hemorrhages and cortical lamination defects in presenilin 1-null mutant mice. Development 132, 3873–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosczyna MN, Konishi CT, Perez Carbajal EE, Wang TT, Walsh RA, Gan Q, Wagner MW, and Rando TA (2019). Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep. 27, 2029–2035.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zheng Y, Sun L, Badea SR, Jin Y, Liu Y, Rolfe AJ, Sun H, Wang X, Cheng Z, et al. (2019). Microvascular endothelial cells engulf myelin debris and promote macrophage recruitment and fibrosis after neural injury. Nat. Neurosci 22, 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Resources
The accession number for the bulk RNaseq data reported in this manuscript is GEO: GSE101930 and the datasets are available at the Genome Expression Omnibus (GEO) and in Tables S1 and S2 of this manuscript. The raw data including the analysis of vessel permeability in the adoptive transfer experiments (Figures 2C and 6B), the flow cytometry fcs files for sorting the cells for RNA sequence and adoptive transfer (Figures S2A, S2B, and S4A) and the majority of the flow cytometry data to characterize PDGFRα+ cells as stromal cells (Figures 1A, 1B, S1A, S1B, S4B, S4C, S5A, and S5B) are uploaded at Mendeley Data with the following https://doi.org/10.17632/wkg6844nwv.1.


