MSN and NONO and their interaction-activated CREB signaling pathway may be new targets for the treatment and prognosis of TNBC.
Abstract
Triple-negative breast cancer (TNBC) is life-threatening because of limited therapies and lack of effective therapeutic targets. Here, we found that moesin (MSN) was significantly overexpressed in TNBC compared with other subtypes of breast cancer and was positively correlated with poor overall survival. However, little is known about the regulatory mechanisms of MSN in TNBC. We found that MSN significantly stimulated breast cancer cell proliferation and invasion in vitro and tumor growth in vivo, requiring the phosphorylation of MSN and a nucleoprotein NONO-assisted nuclear localization of phosphorylated MSN with protein kinase C (PKC) and then the phosphorylation activation of CREB signaling by PKC. Our study also demonstrated that targeting MSN, NONO, or CREB significantly inhibited breast tumor growth in vivo. These results introduce a new understanding of MSN function in breast cancer and provide favorable evidence that MSN or its downstream molecules might serve as new targets for TNBC treatment.
INTRODUCTION
Breast cancer is the most common malignant tumor in women (1). In recent years, the incidence of female breast cancer has increased rapidly, and the age of onset has gradually become younger (2). Among the multiple breast cancer subtypes, because of the lack of specific therapeutic targets, triple-negative breast cancer (TNBC) is the most malignant and lethal, making it extremely urgent to find novel and effective targets for TNBC treatment (3, 4).
Moesin (MSN) belongs to ERM (ezrin-radixin-MSN) family proteins (5, 6). Its protein structure includes N-terminal FERM domain, which can interact with transmembrane receptors, the α helix domain in the middle, and C-terminal F-actin binding domain, which can interact with cytoskeleton such as microfilaments. Such a protein structure causes MSN to be generally located in the cytoplasm or inside the bulge of the cell membrane (7, 8). As a connective protein between the cytoskeleton and the cell membrane, MSN regulates cell proliferation, movement, adhesion, and cell signal transduction (9).
MSN is associated with the occurrence and development of various cancers, such as cervical cancer, head and neck squamous cell carcinoma, and prostate cancer (10, 11). In our previous studies, we also found that miR-200c can play a crucial regulatory role in glioma by targeting MSN (6). These results showed that MSN is positively correlated with the malignancy of cancers. However, the role of MSN in breast cancer is still unclear. Moreover, the regulatory mechanisms of MSN on cancer progression remains poorly elucidated.
Our current studies have uncovered new regulatory mechanisms of MSN and biological processes that have never been noticed before. First, we discovered that MSN not only exists in the inner cell membrane and cytoplasm but also has a strong nuclear localization, which is associated with the phosphorylation of threonine residue at its 558 site. Second, the nuclear localization of the MSN protein is very important for its function in promoting breast cancer progression. Third, we demonstrated with solid results that the nucleoprotein NONO brings MSN and kinase PKC (protein kinase C) that interact with MSN into the nucleus, which leads to the phosphorylation of cyclic adenosine 3,5-monophosphate (cAMP) response element–binding protein (CREB) and activation of the downstream cAMP response signaling pathway to promote the development of breast cancer. These discoveries of the role of MSN enrich the basic understanding to MSN for researchers and provide strong evidence for its development as a new target for the treatment of TNBC.
RESULTS
MSN was highly expressed in TNBC and positively correlated with the breast cancer malignancy
To find the TNBC-enriched molecules and expect to discover new molecular targets for TNBC, we analyzed the RNA sequencing (RNA-seq) data of 43 TNBC and 38 non-TNBC breast cancer cell lines and clinical samples and found that MSN was highly expressed in TNBC samples (Fig. 1A and fig. S1A). More precisely, 85% of samples with high MSN expression were TNBC, while only 22% percent of samples with low MSN expression were TNBC. In addition, according to statistical analysis, the expression of MSN in TNBC was significantly higher than in non-TNBC. The average expression of MSN in TNBC was three times as much as that in non-TNBC (Fig. 1B). Looking through the MSN expression in 836 breast cancer samples with clear subtypes from The Cancer Genome Atlas (TCGA) Breast Cancer Illumina HiSeq percentile data, we found that the MSN level was higher in basal subtypes significantly (Fig. 1C). Then, we analyzed Breast Cancer Cell Lines (Heiser 2012) data with 48 different subtypes of breast cancer cell lines and also found that MSN showed nearly twice as high expression in basal-like as in luminal-like cell lines (Fig. 1D). To further confirm these results, we stained 22 TNBC and 28 non-TNBC tissue sections of patients with breast cancer with anti-MSN antibody and performed quantitative analysis. We have come to a similar conclusion that MSN is higher in TNBC (Fig. 1E). We analyzed the relationship between MSN expression and patient survival in the Nathan Kline Institute (NKI) database and found that patients with high MSN expression had poor prognosis (Fig. 1F). Quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot were used to access the expression of MSN in eight breast cancer cell lines, and results showed that the expression of MSN was higher in TNBC cell lines (Fig. 1G). These results reveal that MSN, highly expressed in TNBC, is closely related to the malignancy of breast cancer.
Fig. 1. MSN was highly expressed in TNBC and positively correlated with the breast cancer malignancy.
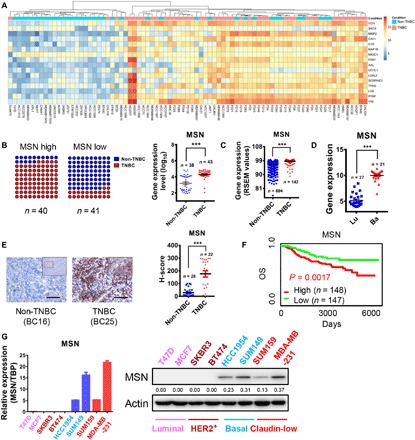
(A) We performed RNA-seq on 43 TNBC and 38 non-TNBC cell lines or clinical samples. The relative expression values of genes used in the heatmap are processed as follows: Relative expression value of gene = log2 (actual measured value +1). (B) The results of RNA-seq were statistically analyzed. The samples were divided into two groups by median of MSN expression, MSN high and MSN low. Then, the proportion of TNBC and non-TNBC in the two groups was shown as a dot plot (left). The results of RNA-seq were used to analyze the difference of MSN expression between TNBC and non-TNBC (right). (C) The expression of MSN was analyzed in different subtypes of breast cancer samples from the dataset (TCGA Breast Cancer Illumina HiSeq percentile), which was contained in the online University of California Santa Cruz (UCSC) Xena database. RSEM, RNA-seq by expectation-maximization. (D) The expression of MSN was analyzed in breast cancer cell lines of different subtypes from the dataset (Heiser 2012) contained in the online UCSC Xena database. Lu, luminal subtype; Ba, basal subtype. (E) Immunohistochemical staining was used to detect the expression of MSN in tumor sections from 22 cases of patients with TNBC and 28 cases of non-TNBC. Scale bars, 80 μm. The histo-score (H-score) statistics are shown on the right. (F) The overall survival (OS) of patients grouped by high and low MSN expression in breast cancer from the NKI dataset contained in an online database (PROGgeneV2). (G) MSN expression level measured by qRT-PCR (left) or Western blot (right) in eight breast cancer cell lines that contain four subtypes. TBP, TATA-box binding protein. ***P < 0.001 by unpaired t test of triplicates. Error bars, means ± SEM.
MSN positively regulated the progression of breast cancer
Since MSN expression is positively correlated with the malignancy of breast cancer, it might contribute to breast cancer progression. We established MSN-knockdown MDA-MB-231, SUM159, or overexpressing MDA-MB-231, T47D, and HCC1954 cell lines, which were confirmed by qRT-PCR and Western blot (Fig. 2A and fig. S1B). MSN knockdown significantly inhibited cell proliferation, invasion, and anchorage-independent growth, while MSN overexpression showed the opposite effects (Fig. 2, B to D, and fig. S1, C to E). Moreover, results of xenograft mouse models showed that MSN expression significantly affect the outgrowth of tumors in vivo (Fig. 2E, top and middle). After paraffin embedding and sectioning, we stained the tumor tissues with MSN and Ki67 antibodies. It was manifested that the positive rate of Ki67 was decreased in MSN knockdown and increased in MSN-overexpressing tumors significantly [Fig. 2E (bottom) and fig. S1F], which verified the impact of MSN on tumor cell proliferation in vitro. These results provide convincing evidences for the effect of MSN on breast tumor growth in vitro and in vivo.
Fig. 2. MSN positively regulated the progression of breast cancer.
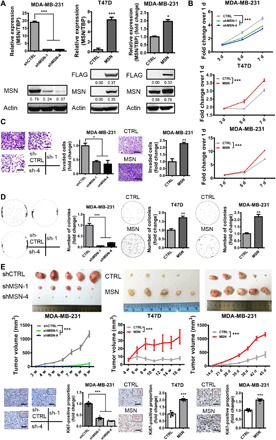
(A) qRT-PCR (top) and Western blot (bottom) was used to verify the knockdown or overexpression effect of MSN. (B) MTT assay was performed to determine the difference of cell proliferation ability after MSN knockdown or overexpression (n = 6). (C) Invasion assay was carried out with MSN knockdown (left) or MSN-overexpressing (right) MDA-MB-231 cells. Quantitative analysis of the total invasive cells of triplicates is shown as a bar graph. Scale bars, 200 μm (left) and 400 μm (right). CTRL, control. (D) Soft agar colony formation assay was performed using MSN knockdown MDA-MB-231 cells and MSN-overexpressing T47D or MDA-MB-231 cells. Colonies were counted in the whole field showed on the right (n = 3). (E) MDA-MB-231 shCTRL or shMSN cells were implanted into the fourth mammary fat pads at two flanks of nude mice, 1 million cells per site (n = 5). The tumor volume was measured once a week. T47D CTRL or MSN-overexpressing cells were implanted into the fourth mammary fat pads at two flanks of nude mice, 2 million cells per site (n = 5). The tumor volume was measured once every 2 weeks. MDA-MB-231 CTRL or MSN-overexpressing cells of 0.5 million were implanted into the fourth mammary fat pads at two flanks of nude mice (n = 5). The tumor volume was measured at indicated time. At the end of experiments, the tumors were taken out and the images are shown. Ki67 staining was performed by IHC (immunohistochemistry), and Ki67-positive proportions are shown on the right. Scale bars, 80 μm. Photo credit: Yuanyuan Qin, University of Science and Technology of China. **P < 0.01 and ***P < 0.001 by unpaired t test of triplicates or test of two-way ANOVA versus shCTRL or CTRL group. Error bars, means ± SEM.
Effects of MSN in regulating breast cancer are dependent on its T558 phosphorylation
It has been reported that MSN can be phosphorylated on threonine residue of its 558 site (T558) (12). In addition, we found that T558 phosphorylation level of MSN was positively associated with the total MSN level (Fig. 3A). Next, to explore whether T558 phosphorylation was critical in regulating breast cancer progression, we introduced the T558 site mutation with positive-charged amino acid, alanine, to simulate constitutive inactivation (T558A), while with negative-charged amino acid, glutamic acid, to simulate constitutive activation (T558E). Then, qRT-PCR and Western blot were used to verify the effect of overexpression in each group (Fig. 3B). Our results showed that MSNT558E-overexpressing cells had higher proliferation, invasion, and anchorage-independent growth ability in vitro compared with the wild-type group (Fig. 3, C to E, and fig. S2, A and B), which were also proved by a faster rate of xenograft tumor growth and a higher Ki67-positive rate in vivo (Fig. 3, F and G). Furthermore, these phenomena were reversed in MSNT558A-overexpressing cells compared with wild-type group in vitro and in vivo (Fig. 3, C to G, and fig. S2, A and B). Together, these results confirmed that MSN played the regulatory role in breast cancer depending on the phosphorylation at its T558 site.
Fig. 3. Effects of MSN in regulating breast cancer are dependent on its T558 phosphorylation.
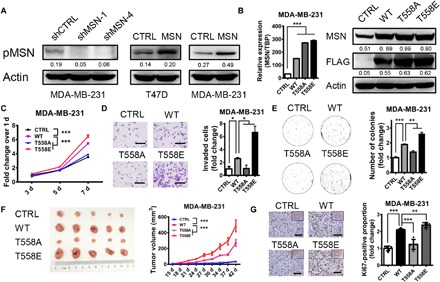
(A) Western blot was performed to determine the phosphorylation level of MSN T558 site after changing MSN expression. (B) qRT-PCR and Western blot were carried out to verify the overexpression of MSN with different status. WT, wild type; T558A, mutant of threonine 558 replaced by alanine. T558E, mutant of threonine 558 replaced by glutamic acid. (C) Cell proliferation ability was measured by MTT assay (n = 6). (D) Cell invasion ability was measured by the invasion assay (n = 3). The image (left) and quantitative analysis of the total invasive cells (right) are shown. Scale bars, 200 μm. (E) Anchorage-independent growth ability was measured by the soft agar colony formation assay (n = 3). The image (left) and quantitative analysis of colonies (right) are shown. (F) MDA-MB-231 cells of 0.5 million were implanted into the fourth mammary fat pads at two flanks of nude mice (n = 5). The figure shows the representative results of two independent repetitive experiments. The tumor volume was measured as indicated. At the end of experiments, the tumors were taken out and the images are shown. (G) Ki67 staining of tumor tissue was conducted by IHC, and positive proportion is shown on the right. Scale bars, 80 μm. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test of triplicates or test of two-way ANOVA. Error bars, means ± SEM.
Phosphorylated MSN enters nucleus to function for breast cancer progression with the assistance of a nuclear protein NONO
To investigate how MSN signals the cells to affect tumor progression, we used protein immunoprecipitation and mass spectrometry to find MSN-interacting proteins and unexpectedly detected a nucleoprotein NONO in anti–FLAG-MSN immunoprecipitated samples in MDA-MB-231 (Fig. 4A). The molecular weight size of NONO is very similar to the smaller one of the differential bands in the silver staining results of the protein samples obtained by immunoprecipitation (Fig. 4B). We also confirmed that exogenously overexpressed MSN protein could pull down endogenous NONO [Fig. 4C (top) and fig. S3A (left)], and exogenously overexpressed NONO protein could pull down endogenous MSN (fig. S3A, right). Similarly, endogenous MSN could pull down endogenous NONO protein (Fig. 4C, bottom). It was reported that MSN mainly distributes in the cytoplasm and the inner side of the cell bulge (13). By separating cell components (cytoplasm and nucleus) and observing the distribution of MSN protein in cells, we found that MSN was also located in the nucleus (Fig. 4D and fig. S3B), which is more obvious in the MSNT558E group compared to wild-type MSN-overexpressing cells, while not in the MSNT558A group (Fig. 4E and fig. S3C). Furthermore, immunofluorescence staining for endogenous or exogenous expression of MSN showed that MSN was distributed in both cytoplasm and nucleus, while NONO was mainly located in the nucleus. There is spatial accessibility of these two proteins in the nucleus (Fig. 4F and fig. S3D). At the same time, we also confirmed that MSNT558E has a stronger interaction with NONO compared to wild-type MSN, while MSNT558A has a weaker interaction (Fig. 4G and fig. S3E). These results provided strong evidences that MSN interacted with NONO.
Fig. 4. Phosphorylated MSN enters nucleus to function for breast cancer progression with the assistance of a nuclear protein NONO.
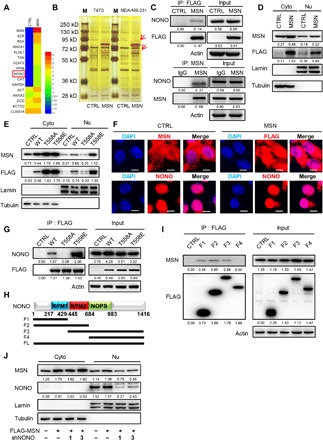
(A) We used anti-FLAG M2 magnetic beads to carry out immunoprecipitation experiments on MDA-MB-231–CTRL and FLAG-MSN–overexpressing cell lysates and identified the protein samples by mass spectrometry. The protein components and relative abundances are displayed by a heatmap. (B) Immunoprecipitated samples by anti-FLAG M2 Magnetic Beads of CTRL or FLAG-MSN–overexpressing T47D and MDA-MB-231 cells were separated by SDS–polyacrylamide gel electrophoresis and silver staining. The red box shows differential bands, which appear in both cell lines. M, marker. (C) CTRL or FLAG-MSN–overexpressing MDA-MB-231 cells were immunoprecipitated (IP) by anti-FLAG M2 Magnetic Beads and then immunoblotted (top). Anti-MSN antibody was incubated with Dynabeads protein A and Dynabeads protein G and then to immunoprecipitated samples of MDA-MB-231 cells and immunoblotted (bottom). IgG, immunoglobulin G. (D) Cytoplasmic (Cyto) and nuclear (Nu) proteins were separated according to instruction. Western blot was conducted to determine the distribution of MSN in CTRL or FLAG-MSN–overexpressing MDA-MB-231 cells. Tubulin, internal reference for cytoplasmic proteins and lamin for nuclear proteins. (E) Western blot was carried out to determine the distribution of wild-type MSN and its mutants in MDA-MB-231 cells. (F) Immunofluorescence assay was carried out, in which endogenous MSN was determined with anti-MSN antibody in CTRL cells, exogenous MSN was determined with anti-FLAG antibody in FLAG-MSN–overexpressing cells, and endogenous NONO was determined with anti-NONO antibody. Images were captured by confocal laser microscopy. Scale bars, 10 μm. (G) CTRL or different-status MSN-overexpressing MDA-MB-231 samples were immunoprecipitated with anti-FLAG M2 Magnetic Beads and immunoblotted. DAPI, 4′,6-diamidino-2-phenylindole. (H) Schematic diagram of NONO in different truncated forms. (I) CTRL or FLAG-tagged different NONO fragment-overexpressing MDA-MB-231 cells were immunoprecipitated with anti-FLAG M2 Magnetic Beads and immunoblotted. (J) Cytoplasmic and nuclear proteins were separated and immunoblotted after MSN overexpression and NONO knockdown in MDA-MB-231.
To explore the interacting domain of NONO with MSN, we established several truncated NONO-overexpressing cell lines (Fig. 4H). Our results suggested that only fragments containing RNA recognition motif 1 (RPM1) and/or RPM2 domain (F1, F2, or F3) can pull down MSN protein, which concluded that the critical region of NONO mediating interaction with MSN is RPM domains (Fig. 4I and fig. S3F). In addition, we found that MSN nuclear localization was decreased significantly after NONO knockdown (Fig. 4J and fig. S3, G and H), indicating that NONO has a vital contribution for MSN to enter the nucleus. These studies proved that MSN entered the nucleus by interacting with the nucleoprotein NONO.
The interaction between MSN and NONO is critical for MSN function on breast tumor progression
To know whether NONO-MSN interaction is important for MSN function on breast cancer progression, we knocked down NONO in MSN-overexpressing cell lines (fig. S3G) and found that NONO knockdown could abolish a series of cell functions caused by MSN overexpression (fig. S4, A to F). In vivo, the tumor growth and the positive rate of Ki67 in tumor sections were also abolished (fig. S4, G to I). These results state clearly that NONO is indispensable for MSN in promoting malignant progression of breast tumor.
MSN-NONO interaction activated CREB signaling to promote breast cancer progression by facilitating the nuclear localization of pPKC
NONO has a vital contribution for MSN to enter the nucleus. Therefore, exploring the details of NONO action might play a key role in elucidating the profound mechanisms of MSN in regulating breast cancer progression.
It has been reported that NONO is a component of cAMP response signaling pathway that activates transcription of downstream genes by stimulating interactions between CREB and coactivator (CRTC) (14, 15), and once phosphorylated PKA (pPKA) or pPKC phosphorylated serine 133 of CREB, the interaction between CREB and CRTC occurs (16, 17). In the cAMP response signal pathway, NONO promoted gene transcription by narrowing RNA polymerase and the promoter region of downstream genes with CREB response elements (18). Our result showed that NONO overexpression significantly increased the phosphorylation of CREB (Fig. 5A). We found that the phosphorylation level of CREB was decreased after knocking down MSN while it increased after overexpressing MSN (Fig. 5, B and C, and fig. S5, A and B), and phosphorylation of CREB was highly aggravated after overexpressing constitutively activated MSNT558E but alleviated by MSNT558A (Fig. 5D).
Fig. 5. MSN-NONO interaction activated CREB signaling to promote breast cancer progression by facilitating the nuclear localization of pPKC.
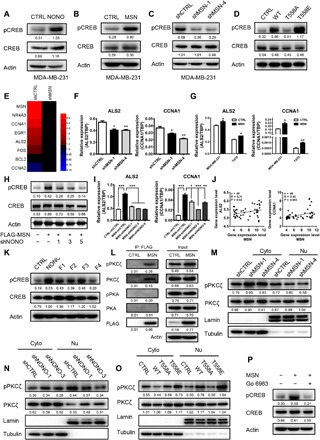
(A) CREB phosphorylation level was measured by Western blot after overexpressing NONO in MDA-MB-231 cells by Western blot. (B) CREB phosphorylation level was determined in MSN-overexpressing MDA-MB-231 cells by Western blot. (C) CREB phosphorylation level was determined in MSN knockdown MDA-MB-231 cells by Western blot. (D) Western blot was used to measure CREB phosphorylation level after overexpressing MSN with different mutation status in MDA-MB-231 cells. (E) The downstream genes of CREB signaling from the results of RNA-seq of MDA-MB-231–shCTRL and MDA-MB-231–shMSN cells were shown as heatmap according to relative expression level. (F) The expression of CREB downstream genes ALS2 and CCNA1 was measured by qRT-PCR in MSN knockdown MDA-MB-231 cells. (G) The expression of CREB downstream genes ALS2 and CCNA1 was measured by qRT-PCR in MSN-overexpressing MDA-MB-231 or T47D cells. (H) Phosphorylation level and total proteins of CREB were determined in MSN-overexpressing and NONO-knocking down MDA-MB-231 cells by Western blot. (I) The expression of CREB downstream genes ALS2 and CCNA1 was measured in MSN-overexpressing and NONO-knocking down MDA-MB-231 cells by qRT-PCR. (J) The expression relationship between MSN and CREB downstream genes ALS2 or CCNA1 was analyzed in breast cancer cell lines from the dataset (Heiser 2012) contained in the UCSC Xena database. (K) CREB phosphorylation level was detected in MDA-MB-231 cells overexpressing NONO in different truncated forms by Western blot. (L) The immunoprecipitated samples obtained with anti-FLAG M2 Magnetic Beads of CTRL or FLAG-MSN–overexpressing MDA-MB-231 cells were conducted with Western blot. (M) Nuclear protein level of pPKCζ was observed after MSN knockdown by Western blot. (N) Nuclear protein level of pPKCζ was observed after NONO knockdown by Western blot. (O) pPKCζ level in cytoplasm and nucleus was determined by Western blot after overexpressing MSN with different mutagenesis. (P) We examined the effect of PKC inhibitor on the phosphorylation of CREB when overexpressing MSN by Western blot. *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired t test of triplicates. Error bars, means ± SEM.
Furthermore, we analyzed the expression of CREB downstream genes in the RNA-seq data of MSN-knockdown MDA-MB-231 cells and confirmed that most downstream genes were down-regulated after knocking down MSN (Fig. 5E). We chose ALS2 and CCNA1 as the representative CREB downstream genes to carry out the following experiments. The expression level of ALS2 and CCNA1 was positively regulated by MSN expression (Fig. 5, F and G, and fig. S6A). NONO knockdown in MSN-overexpressing cells significantly blocked the increase of CREB phosphorylation and its downstream gene expression caused by MSN overexpression (Fig. 5, H and I, and fig. S6B). Given the interaction between MSN and NONO, these results indicated that MSN regulated the CREB downstream gene expression by interaction with NONO. In addition, we analyzed the expression of ALS2 and CCNA1 in different breast cancer cell lines in the database (Heiser 2012) and found that they were positively correlated with MSN level (Fig. 5J). The overexpression of NONO truncated forms containing RPM1 or RPM2 domain promoted the phosphorylation of CREB (Fig. 5K and fig. S6C), further confirming that the MSN-NONO interaction played an important role in activating CREB signaling. Accordingly, knocking down CREB or using a specific CREB activation inhibitor, 666-15, can significantly block the effects caused by MSN overexpression, including cell proliferation, invasion, soft agar colony formation ability, and the expression of CREB downstream genes (fig. S5, C to P).
As mentioned previously, NONO promoted gene transcription by narrowing RNA polymerase and the promoter region of downstream genes with CREB response elements (18, 19), which does not involve in the phosphorylation of CREB. Therefore, in our study, we need to define the mechanisms of CREB phosphorylation. CREB is a nuclear-localized protein, and its phosphorylation requires the existence of kinase in the nucleus (20, 21). Other studies reported that the upstream kinases of CREB, pPKA, or pPKC also can phosphorylate MSN (22–24). We detected both PKCζ and pPKCζ in the immunoprecipitation samples obtained by anti-FLAG magnetic beads using FLAG-tagged MSN-overexpressing MDA-MB-231, while PKA and pPKA were not detected (Fig. 5L). The nuclear localization of pPKCζ in MSN- or NONO-knockdown cells appeared to be significantly down-regulated (Fig. 5, M and N, and fig. S7, A and B). Because phosphorylated MSN has stronger interaction with NONO, the pPKC interacting with constitutively activated MSNT558E has a higher tendency to enter the nucleus but not in the constitutively inactivated MSNT558A group (Fig. 5O and fig. S7C). In addition, after treatment with the PKC inhibitor (Go 6983), the increase of CREB phosphorylation level caused by MSN overexpression was completely reversed (Fig. 5P). In addition, knocking down PKCζ (PRKCZ) resulted in an obvious rescue of the pCREB up-regulation caused by overexpression of MSN (fig. S7D). In conclusion, our results demonstrated that, while MSN interacted with NONO and entered the nucleus, the kinase PKCζ was also transported into the nucleus as a complex formed with MSN, which phosphorylated CREB in the nucleus, activated the CREB pathway, and promoted the progression of breast cancer.
MSN-NONO complex and downstream CREB signaling pathway could be targeted for TNBC
MSN promoted the phosphorylation of CREB by PKCζ and the expression of downstream genes by interacting with NONO. This whole signaling pathway provides multiple possible therapeutic targets for TNBC. In addition, MSN belongs to ERM family proteins. Only MSN has been screened out to be highly expressed in TNBC (Fig. 1, D and G, and fig. S8, A and B). The expression analysis of NONO, ALS2, and CCNA1 in breast cancer cell lines showed that these genes were highly expressed in basal subtype with higher malignancy (Fig. 6, A and B). Patients with higher expression of these genes had poorer prognosis (Fig. 6, C and D). We integrated data from multiple datasets (25–27) and found that patients with higher expression of MSN and NONO at the same time had poorer lung free metastasis survival than patients with higher expression of only one, not both, and those with lower expression of both had the best lung free metastasis survival rate (Fig. 6E). We divided the samples into different categories for further analysis. The lung free metastasis survival analysis of estrogen receptor (ER)–negative population showed similar results, that is, patients with high expression of MSN and NONO had the worst prognosis. However, because of the specific high expression of MSN in the TNBC, the lack of MSN expression in ER-positive patients may cause this phenomenon to be insignificant (fig. S9A). In addition, we used a TNBC patient-derived xenograft (PDX) model (#USTC11) to carry out the treatment experiment of targeting CREB. We found that the CREB inhibitor 666-15 alone can play a good role in inhibiting the growth of breast cancer, and the combination with docetaxel (DOC) showed a better effect (Fig. 6F). After the measurement and statistical analysis of the body weight of mice, it was found that 666-15 treatment had no obvious effect on mouse body weight (fig. S9B). During the treatment, mice were in good condition and had no obvious side effects.
Fig. 6. MSN-NONO complex and downstream CREB signaling pathway could be targeted for TNBC.
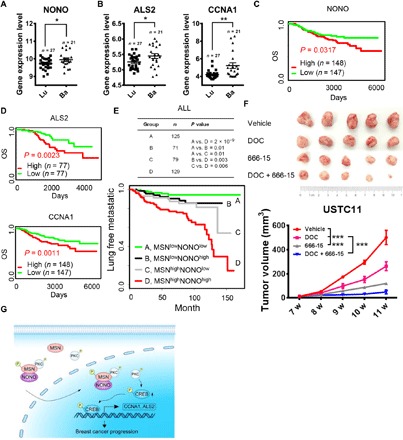
(A) The expression of NONO was analyzed in different subtype breast cancer cell lines from the dataset (Heiser 2012) contained in the UCSC Xena database. (B) The expression of CREB downstream genes ALS2 or CCNA1 was analyzed in different subtype breast cancer cell lines from the dataset (Heiser 2012) contained in the UCSC Xena database. (C) The OS influenced by NONO expression in breast cancer from the dataset (NKI) contained in an online database (PROGgeneV2). (D) The OS influenced by ALS2 expression in breast cancer from the dataset (GSE9893) and CCNA1 expression from the dataset (NKI) contained in an online database (PROGgeneV2). (E) The lung free metastasis survival influenced by different combinations of high and low expression of MSN and NONO from integrated datasets including GSE5327, GSE2603, and GSE2034. (F) One hundred thousand cells digested from PDX of TNBC (#USTC11 established by our group) were implanted into the fourth mammary fat pads at two flanks of nude mice. After tumor initiation, mice were divided into four groups and treated with vehicle, DOC (10 mg/kg), 666-15 (10 mg/kg), or both once a week intraperitoneally. Tumor size was monitored once a week, and tumor image was taken after mice were euthanized (n = 5). Photo credit: Yuanyuan Qin, University of Science and Technology of China. (G) Schematic diagram of the whole research results. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test. Error bars, means ± SEM.
In conclusion, our study illustrated that phosphorylated MSN and its kinase PKCζ entered the nucleus under the assistance of the nucleoprotein NONO. The nuclear localization of PKCζ subsequently phosphorylated CREB and activated the CREB signaling pathway via PKC and enhanced the expression of genes containing cAMP-related elements, such as ALS2 or CCNA1, thus promoting the progress of breast cancer (Fig. 6G).
DISCUSSION
TNBC refers to breast cancer lacking ER, progesterone receptor, and proto-oncogene HER2 (28, 29). Histological grade and cell proliferation ratio of this kind of breast cancer is higher (30). Because of the lack of effective therapeutic targets, the prognosis of TNBC is worse than in other subtypes of breast cancer (31). The clinical manifestation of TNBC is an aggressive course with a higher risk of distant metastasis, a higher chance of visceral such as lung metastasis, and a higher probability of brain metastasis (32, 33).
On the basis of our previous studies, we found that MSN was regulated by miR-200c in glioma and thus exhibited oncogenic characteristics (6). MSN belongs to ERM family proteins, which mainly include EZR, radixin (RDX), and MSN (34). They all have similar structures and functions and play a role as oncogenes in many cancers (35). By measuring the expression level of EZR, RDX, and MSN in different subtypes of breast cancer cell lines, we found that only MSN showed a very distinct subtype-dependent specific expression and was highly expressed in TNBC cells. These results confirm that MSN may play an important role and be a specific therapeutic target for TNBC. In addition, other researchers showed that MSN can control the function of regulatory T cells (Tregs) and the abundance and stability of transforming growth factor–β receptors on the cell surface. This study demonstrated that MSN inhibition can decrease Treg generation to restore antitumor immunity. Together, these findings indicate that targeting MSN might not only target TNBC but also become a potential therapeutic target for cancer immunotherapy or Treg-related immune diseases (36).
It has been reported that MSN can promote the proliferation and metastasis of glioma cells by interacting with CD44 and promoting WNT signaling pathway (37). Compound DX-52-1 can inhibit the invasion and metastasis of glioma by blocking the combination of MSN and CD44, thus playing an antitumor role in many aspects of tumor development (37, 38). However, phase 1 clinical results showed that patients have poor tolerance to DX-52-1, which limits the use of DX-52-1 as an antineoplastic drug (39). In addition to compound DX-52-1, the development of inhibitors targeting MSN is very limited, and less work has been done in breast cancer.
We focused on the high expression of MSN in highly malignant TNBC. Our results showed that MSN was an important gene for regulating the progression of breast cancer, and phosphorylation of threonine residue at its 558 site and NONO-mediated nuclear translocation were crucial for its function. NONO is involved in a variety of nuclear events such as gene transcription regulation, gene splicing, and DNA damage repair (40). In addition, NONO is involved in a variety of biological processes including regulation of circadian rhythm and influence of cancer progression (41). Some studies have shown that the survival and prognosis of patients with high expression of NONO are poor (42). In addition, there are significant differences in gene expression between tumors and normal tissues (43). However, the regulatory mechanism of NONO in tumors is still unclear. Our results indicated that MSN was transported into the nucleus by NONO and pPKCζ phosphorylating MSN was also transported into the nucleus together, where pPKCζ phosphorylated CREB and increased the expression of downstream oncogenes ALS2 and CCNA1, which have cAMP response elements. MSN, NONO, ALS2, and CCNA1 were all highly expressed in TNBC with high malignancy. In addition, according to our analysis of survival curve, patients with high expression of MSN and NONO had the worst survival, while patients with low expression of both had the best survival. These results suggest that these genes might serve as novel prognostic indicators for breast cancer. In addition, the signaling pathway that we studied provided several possible therapeutic targets, such as the interaction between MSN and NONO, the nuclear entering process of MSN and pPKCζ, or the kinase activity of pPKCζ and the phosphorylation of CREB. We hope to develop small-molecule inhibitors or antibodies based on these targets to develop therapeutic drugs for clinical application in TNBC.
Because the phosphorylation of MSN is regulated by its upstream kinases, overexpression of MSN does not guarantee its phosphorylation. Therefore, we introduced mutations at site 558 of MSN to simulate different phosphorylation states, making it persistently phosphorylated or phosphorylation inactivated. Although this mature approach has been used in several authoritative reports (44, 45), overexpression of mutants may affect the function of endogenous MSN by introducing artificial products, which might be solved in two ways. First, endogenous MSN can be knocked out with CRISPR and then introduce the mutants to the cells in future research. Second, overexpression of mutated MSN can be carried out in cell lines hardly expressing endogenous MSN, such as T47D cell line as we used in our study. Our results showed that overexpression of phosphorylated MSN could further promote the proliferation of tumor cells and had stronger interaction with NONO.
In conclusion, our studies demonstrated that ERM family protein MSN was highly expressed in TNBC. The T558-phosphorylated MSN interacted with the nuclear protein NONO and promoted the nuclear localization of pPKCζ interacting with MSN, which leads to CREB phosphorylation and the up-regulation of downstream gene expression and promotes the progression of breast cancer. Our current study provides the evidence that this signaling pathway can be used to develop novel therapeutic approaches for TNBC.
MATERIALS AND METHODS
Cell culture
Breast cancer cell lines SUM149 and SUM159 were from Asterland; MCF7, T47D, SKBR3, BT474, HCC1954, and MDA-MB-231 were purchased from the American Type Culture Collection. The culture medium for SUM149 and SUM159 was Ham’s F-12 (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA), insulin (5 μg/ml), hydrocortisone (1 μg/ml; both from Sigma-Aldrich, St. Louis, USA), and 1% penicillin-streptomycin (pen-strep) (Beyotime, China). The culture medium for MCF7 was Eagle’s minimum essential medium (Gibco, USA) supplemented with 10% FBS, insulin (10 μg/ml), and 1% pen-strep. The culture medium for T47D was RPMI 1640 (Gibco, USA) supplemented with 10% FBS, insulin (5 μg/ml), and 1% pen-strep. The culture medium for SKBR3, BT474, HCC1954, and MDA-MB-231 was RPMI 1640 supplemented with 10% FBS and 1% pen-strep. All the cell lines were tested and authenticated. These cell lines were maintained at 37°C in an atmosphere of 5% CO2.
Quantitative real-time polymerase chain reaction
Total RNA was extracted with TRIzol (9109; Takara Bio), and the RNA concentration was measured with NanoDrop (Thermo Scientific, USA). RNA (1 μg) was reverse- transcribed to complementary DNA (cDNA) using the HiScript II 1st Strand cDNA Synthesis Kit (R211-02, Vazyme) and the T100 Thermal Cycler (Bio-Rad). qRT-PCR was carried out to detect the expression level of related genes in this study using AceQ qPCR SYBR Green Master Mix (Q111-02, Vazyme), and it was performed using 7300 Plus Real-Time PCR System (Applied Biosystems). RT-PCR data for mRNA are shown relative to reference gene. The formula of relative expression value was as follows: 2(−ΔCt). The gene-specific primers used are listed below in Table 1.
Table 1. The gene-specific primers used in qRT-PCR.
| Sequence (5′→3′) | ||
| MSN | Forward: ATCACTCAGCGCCTGTTCTT | Reverse: CCCACTGGTCCTTGTTGAGT |
| TBP | Forward: CACGAACCACGGCACTGATT | Reverse: TTTTCTTGCTGCCAGTCTGGAC |
| ALS2 | Forward: AGATGGTGAGGTCTACAGCTT | Reverse: GAATGGGGCTACTTGGACAAAT |
| CCNA1 | Forward: GAGGTCCCGATGCTTGTCAG | Reverse: GTTAGCAGCCCTAGCACTGTC |
| NONO | Forward: GGCAGGCGAAGTCTTCATTCA | Reverse: TGGCAATCTCCGCTAGGGT |
| CREB | Forward: ATTCACAGGAGTCAGTGGATAGT | Reverse: CACCGTTACAGTGGTGATGG |
Western blot
For sample preparation, when the cell culture reaches to 80% confluence, the cells were collected and lysed for 40 min on ice. Extracted protein samples were denatured in 5× loading buffer containing SDS in boiling water for 10 min. Five percent nonfat dry milk for detection of conventional proteins or 5% bovine serum albumin for detection of phosphorylated proteins dissolved in tris-buffered saline containing 0.1% Tween 20 was used for blocking and antibody dilution. The following antibodies and dilutions were used: MSN (1:2000, ab52490, Abcam), actin (1:1000, HC201, TransGen), FLAG (1:2000, F7425, Sigma-Aldrich), phosphorylated MSN (1:1000, ab177943, Abcam), lamin (1:1000, ab133256, Abcam), tubulin (1:2000, HC101, TransGen), NONO (1:1000, 11058-1-AP, Proteintech), pCREB [1:1000, 87G3, Cell Signaling Technology (CST)], CREB (1:1000, 48H2, CST), pPKCζ (1:1000, 9378S, CST), PKCζ (1:500, sc-17781, Santa Cruz Biotechnology), pPKA (1:1000, 4781, CST), PKA (1:1000, 4782, CST), goat anti-mouse immunoglobulin G (IgG)–horseradish peroxidase (HRP) (1:5000, HS201-01, TransGen), and goat anti-rabbit IgG-HRP (1:5000, HS101-01, TransGen). Western HRP Substrate (WBKLS0500, Millipore) was used to detect HRP-conjugated secondary antibodies. Software (ImageJ) was used to quantify the relative expression of proteins, which was presented as the ratio of test protein integrated density to internal control integrated density. All uncropped images from Western blots displayed in the study are presented in supplementary data (fig. S10).
Separation of nuclear and cytoplasmic proteins
Nuclear and Cytoplasmic Protein Extraction Kit (P0028, Beyotime) was used to separate cytoplasm and nuclear protein according to the instruction. Under low osmotic pressure, the cells expand sufficiently, then the cell membrane was destroyed, and cytoplasmic proteins were released. The nucleus was then precipitated by centrifugation. Last, nuclear protein was extracted by high-salt nuclear protein extraction reagent. Phenylmethylsulfonyl fluoride (ST506, Beyotime) was used to inhibit protein degradation.
Plasmid/short hairpin RNA construction and virus infection
MSN and NONO were amplified from the reverse-transcribed cDNA from MDA-MB-231 cell line and cloned into pSIN-FLAG (puromycin or blasticidin resistant) vector (Addgene, USA), and the authenticity was verified by sequencing, respectively. Primer design was used to introduce site mutation to construct MSN-overexpressing plasmids with different phosphorylation patterns. Plasmid constructs expressing short hairpin RNAs (shRNAs) were designed by Sigma-Aldrich; the sense sequence was designed as follows in Table 2.
Table 2. The sense sequence of shRNAs.
| Sequence (5′→3′) | ||
| MSN | sh-1 | CCGGGCTAAATTGAAACCTGGAATTCTCGAGAATTCCAGGTTTCAATTTAGCTTTTTG |
| sh-4 | CCGGGCATTGACGAATTTGAGTCTACTCGAGTAGACTCAAATTCGTCAATGCTTTTTG | |
| NONO | sh-1 | CCGGGCCAGAATTCTACCCTGGAAACTCGAGTTTCCAGGGTAGAATTCTGGCTTTTTG |
| sh-3 | CCGGGCAGGCGAAGTCTTCATTCATCTCGAGATGAATGAAGACTTCGCCTGCTTTTTG | |
| sh-5 | CCGGCAGGCGAAGTCTTCATTCATACTCGAGTATGAATGAAGACTTCGCCTGTTTTTG | |
| CREB | sh-1 | CCGGGCTCGATAAATCTAACAGTTACTCGAGTAACTGTTAGATTTATCGAGCTTTTT |
| sh-4 | CCGGCAGTGGATAGTGTAACTGATTCTCGAGAATCAGTTACACTATCCACTGTTTTT | |
| PRKCZ | sh-1 | CCGGCAAGCTCACAGACTACGGCATCTCGAGATGCCGTAGTCTGTGAGCTTGTTTTT |
| sh-3 | CCGGACCTAATCAGAGTCATCGGGCCTCGAGGCCCGATGACTCTGATTAGGTTTTTT |
A highly efficient lentiviral system was used to generate MSN- or NONO-overexpressing plasmid DNA and the shRNA plasmid DNA. The cell lines were infected with the lentiviruses, and the stable cell lines were established. The lentiviral transfection efficiency was more than 90% in all cell lines.
MTT assay
Breast cancer cells (500 to 800 cells per well) were seeded in 96-well culture plates and cultured for 3, 5, or 7 days. MTT (M-2128, Sigma-Aldrich) was added to reach the final concentration of 0.5 mg/ml, and the cells were incubated at 37°C for 4 hours; then the supernatant was removed, and 100 μl of dimethyl sulfoxide was added. The optical density value was measured at 490 nm after shaking it for 10 min.
Invasion assay
Twenty thousand cells were seeded in the chambers with an 8-μm pore size (0216; BD Biosciences), coated with Matrigel (354234; Corning), and placed into 24-well culture plate. After 36 hours, cells in lower chambers were fixed and stained with 0.1% crystal violet, and the invaded cells were photographed by microscope for analysis.
Soft agar colony formation assay
Breast cancer cells (4000 to 20,000 cells per well) were seeded in 2× medium mixed with soft agar in six-well culture plates and cultured in the incubator for approximately 4 weeks. Colonies were stained with 0.005% crystal violet, and then the number of colonies was counted.
Tumorigenicity in nude mice
Tumorigenicity was determined by injecting MDA-MB-231 or T47D cells with Matrigel into fourth mammary fat pads of 1-month-old female nude mice. The number of tumors in each group was five. The animals were euthanized when the tumors were 1.0 to 1.5 cm in diameter. A portion of each tumor was fixed in formalin and embedded in paraffin for histological analysis. The tumor sizes were measured with a caliper and calculated as tumor volume = length × width2/2.
Immunohistochemistry
The tumor tissues of mice were fixed in formalin and processed for paraffin embedding. Sectioned samples were deparaffined in xylene and rehydrated in graded alcohol. Antigen retrieval was done according to the manufacturer’s protocol (MVS-0100, Maxvision), and then the endogenous peroxidase was inactivated with 3% hydrogen peroxide methanol solution, blocked with animal nonimmune serum (SP KIT-B, Maxvision), and incubated with primary antibodies overnight at 4°C and then with secondary antibodies for 15 min. Slides were stained using the detection kit (DAB-0031, Maxvision); cell nucleus was stained with hematoxylin (ZLI-9610, ZSGB-BIO). The following antibodies and dilutions were used for immunohistochemistry (IHC): MSN (1:100, ab52490, Abcam), Ki67 (1:100, ZM-0166, ZSGB-BIO), NONO (1:100, 11058-1-AP, Proteintech), and peroxidase-conjugated secondary antibody (KIT-5010, Maxvision). MSN expression in breast cancer tissue slices was scored semiquantitatively by a manual histo-score (H-score) methodology based on staining intensity and percentage of positive tumor cells. Strongly staining scored 3, moderately staining scored 2, weakly staining scored 1, and negatively staining scored 0. The H-score of MSN expression is obtained by the formula 3 × percentage of strongly staining + 2 × percentage of moderately staining + percentage of weakly staining, giving a range of 0 to 300.
Immune fluorescence staining and confocal imaging
Breast cancer cells (5000 cells per well) were seeded in chamber (154526, Thermo Scientific) and cultured for 2 days. Cells were fixed with cold methanol, and the membrane was perforated with 0.15% Triton X-100 (TB0198, Sangon Biotech), blocked with animal nonimmune serum (SP KIT-B, Maxvision), and incubated with primary antibodies overnight at 4°C and then with secondary antibodies for 1 hour. Cell nucleus was stained with 4′,6-diamidino-2-phenylindole (P36931, Life Technologies). Images were captured with a confocal microscope (LEICA SP5) with a 63× objective lens. The following antibodies were used for immune fluorescence: MSN (1:100, ab52490, Abcam), NONO (1:100, 11058-1-AP, Proteintech), FLAG (1:100, F7425, Sigma-Aldrich), pPKCζ (1:50, 9378S, CST), and goat anti-rabbit IgG secondary antibody Alexa Fluor 546 (1:200, A-11010, Life Technologies).
Immunoprecipitation assay
For sample preparation, when the cell culture reached 80% confluence, the cells were collected and lysed under the conditions of rotation in the mild lysis buffer for 1 hour. For the selection of magnetic beads, Dynabeads protein A (10001D, Life Technologies) and Dynabeads protein G (10003D, Life Technologies) were used for the experiment of endogenous protein, and the antibody was incubated with protein A/G in advance according to the manufacturer’s instructions. Anti-FLAG M2 Magnetic Beads (M8823, Sigma-Aldrich) were used for the experiment of exogenous protein. Except for a small fraction of input group, the remaining supernatant was incubated with the magnetic beads gently for the night at 4°C. After removing the supernatant, beads were washed five times and 10 min each. Proteins were eluted using competitive elution of 3× FLAG fusion proteins (0.4 mg/ml; F4799, Sigma-Aldrich) and were denatured in 5× loading buffer containing SDS in boiling water for 10 min. Boiled samples can be followed by silver staining and Western blot.
Mass spectrometry
Gel containing samples was decolorized and washed to make it transparent and then freeze-dried. Samples were reduced of disulfide bond by dithiothreitol and alkylation before enzymatic hydrolysis. Then, the peptide segment was extracted and dried in vacuum. Samples were desalted, and supernatant was added to the sample flask for mass spectrometry (Q Exactive) detection. The MaxQuant search database was retrieved.
RNA sequencing
After RNA extraction, the RNA integrality (RNA Quality Number) was first validated by Agilent 2100, and the qualified RNA was used to establish the RNA library, which was checked by quality control with Agilent 2100 before sequencing. When analyzing the expression levels of each gene in results of RNA-seq, the threshold of expression value was set to 0.1 greater than what was believed to be credible.
Chemical inhibitors
A selective CREB inhibitor, 666-15 (HY-101120, MedChemExpress), was purchased and used in our experimental design to treat MDA-MB-231 cells at a concentration of 73 nM. A pan-PKC inhibitor (Go 6983, Selleck) was purchased and used to treat MDA-MB-231 cells at a concentration of 5 μM.
Survival analysis and gene expression analysis with online database
NKI data for overall survival (OS) of MSN, NONO, and CCNA1 and GSE9893 for OS of ALS2 were from Gene Expression Omnibus of National Center for Biotechnology Information. Gene expression of these genes was bifurcated into high and low level at median. Available URL, www.ncbi.nlm.nih.gov/geo. Results were obtained with the online tool PROGgeneV2, for which the available URL is http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene". We integrated three datasets, GSE5327, GSE2603, and GSE2034, to analyze the lung free metastasis survival with different levels of MSN and NONO. Breast Cancer Cell Line (Heiser 2012) data for analyzing the expression of genes in different cell lines and TCGA Breast Cancer Illumina HiSeq percentile for analyzing the expression genes in clinical samples were from University of California Santa Cruz (UCSC) Genome Browser, for which the available URL is https://xenabrowser.net/datapages/.
Statement
Written informed consent was obtained from all patients before surgery, as advocated by the regional ethics committee. The study was approved by Fudan University Shanghai Cancer Center Institutional Review Board (050432-4-1212B).
All nude mice were bred and housed in Association for Assessment and Accreditation of Laboratory Animal Care International–accredited specific pathogen–free rodent facilities at University of Science and Technology of China or Fudan University Shanghai Medical College. Mice were housed in sterilized, ventilated microisolator cages and supplied with autoclaved commercial chow and sterile water. All mouse experiments were conducted in accordance with standard operating procedures approved by the Institutional Animal Care and Use Committee at University of Science and Technology of China or Fudan University Shanghai Medical College.
Statistics
Bar graphs were generated with GraphPad Prism 7, and all values are reported as the means ± SEM. A two-way analysis of variance (ANOVA) was used for multiple comparisons. Unless otherwise indicated, comparisons between two groups were performed using an unpaired, two-tailed t test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Thanks to S. Ethier for providing the breast cancer cell lines SUM149 and SUM159. Funding: This work was supported by the National Key Research and Development Program of China (Stem Cell and Translational Research 2016YFA0101202), the MOST grant (2015CB553800), NSFC grant (81772799, 81530075, and 81930075), Program for Outstanding Medical Academic Leader (2019LJ04), Fudan-SIMM Joint Research Fund FU-SIMM20172007, Fudan University Research Foundation IDH 1340042, and Research Foundation of the Fudan University Shanghai Cancer Center YJRC1603. Author contributions: Y.Q., W.C., and S.L. designed research studies. Y.Q. and W.C. conducted experiments. Y.Q., W.C., G.J., H.-l.W., Y.-b.Z., and S.H. analyzed data. L.Z., X.Y., X.H., and H.L. provided reagents and materials. Y.Q., W.C., and S.L.wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/8/eaaw9960/DC1
Fig. S1. MSN regulates breast cancer progression.
Fig. S2. Effects of MSN in regulating breast cancer are dependent on its T558 phosphorylation.
Fig. S3. MSN can interact with NONO and enter the nucleus with the assistance of NONO.
Fig. S4. The interaction between MSN and NONO is critical for MSN function on breast tumor progression.
Fig. S5. The function of MSN was mediated by phosphorylation of CREB.
Fig. S6. MSN-NONO interaction enhanced CREB signaling pathway.
Fig. S7. MSN-NONO interaction promoted CREB phosphorylation by facilitating the nuclear localization of pPKCζ.
Fig. S8. There was no significant difference in the expression of EZR and RDX in different subtypes of breast cancer cell lines.
Fig. S9. MSN-NONO complex and downstream CREB signaling pathway could be targeted for TNBC.
Fig. S10. Uncropped images from Western blots.
REFERENCES AND NOTES
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer Statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A., Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Denkert C., Liedtke C., Tutt A., von Minckwitz G., Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 389, 2430–2442 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Chen W., Qin Y., Wang D., Zhou L., Liu Y., Chen S., Yin L., Xiao Y., Yao X.-H., Yang X., Ma W., Chen W., He X., Zhang L., Yang Q., Bian X., Shao Z.-m., Liu S., CCL20 triggered by chemotherapy hinders the therapeutic efficacy of breast cancer. PLOS Biol. 16, e2005869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pines M., Levi O., Genin O., Lavy A., Angelini C., Allamand V., Halevy O., Elevated expression of moesin in muscular dystrophies. Am. J. Pathol. 187, 654–664 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Qin Y., Chen W., Liu B., Zhou L., Deng L., Niu W., Bao D., Cheng C., Li D., Liu S., Niu C., MiR-200c inhibits the tumor progression of glioma via targeting moesin. Theranostics 7, 1663–1673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagresle-Peyrou C., Luce S., Ouchani F., Soheili T. S., Sadek H., Chouteau M., Durand A., Pic I., Majewski J., Brouzes C., Lambert N., Bohineust A., Verhoeyen E., Cosset F. L., Magerus-Chatinet A., Rieux-Laucat F., Gandemer V., Monnier D., Heijmans C., van Gijn M., Dalm V. A., Mahlaoui N., Stephan J. L., Picard C., Durandy A., Kracker S., Hivroz C., Jabado N., de Saint Basile G., Fischer A., Cavazzana M., Andre-Schmutz I., X-linked primary immunodeficiency associated with hemizygous mutations in the moesin (MSN) gene. J. Allergy Clin. Immunol. 138, 1681–1689.e8 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty P. K., Zhang Y., Coomes A. S., Kim W.-J., Stupay R., Lynch L. D., Atkinson T., Kim J. I., Nie Z., Daaka Y., G protein–coupled receptor kinase GRK5 phosphorylates moesin and regulates metastasis in prostate cancer. Cancer Res. 74, 3489–3500 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Gautreau A., Louvard D., Arpin M., ERM proteins and NF2 tumor suppressor: The Yin and Yang of cortical actin organization and cell growth signaling. Curr. Opin. Cell Biol. 14, 104–109 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Yu H., Zhang Y., Ye L., Jiang W. G., The FERM family proteins in cancer invasion and metastasis. Front. Biosci. 16, 1536–1550 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita T., Nohata N., Fuse M., Hanazawa T., Kikkawa N., Fujimura L., Watanabe-Takano H., Yamada Y., Yoshino H., Enokida H., Nakagawa M., Okamoto Y., Seki N., Tumor suppressive microRNA-133a regulates novel targets: Moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem. Biophys. Res. Commun. 418, 378–383 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Vitorino P., Yeung S., Crow A., Bakke J., Smyczek T., West K., McNamara E., Eastham-Anderson J., Gould S., Harris S. F., Ndubaku C., Ye W., MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature 519, 425–430 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Slowing I. I., Trewyn B. G., Lin V. S.-Y., Mesoporous silica nanoparticles for intracellular delivery of membrane-impermeable proteins. J. Am. Chem. Soc. 129, 8845–8849 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Amelio A. L., Miraglia L. J., Conkright J. J., Mercer B. A., Batalov S., Cavett V., Orth A. P., Busby J., Hogenesch J. B., Conkright M. D., A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 20314–20319 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu T., Zhao Y., Hu Z., Li J., Chu D., Zhang J., Li Z., Chen B., Zhang X., Pan H., Li S., Lin H., Liu L., Yan M., He X., Yao M., MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 77, 5782–5794 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Gao H., Zhang L., Chen Z., Liu S., Zhang Q., Zhang B., Effects of intravenous anesthetics on the phosphorylation of cAMP response element-binding protein in hippocampal slices of adult mice. Mol. Med. Rep. 18, 627–633 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Ito J., Iijima M., Yoshimoto N., Niimi T., Kuroda S., Maturana A. D., Scaffold protein enigma homolog activates CREB whereas a short splice variant prevents CREB activation in cardiomyocytes. Cell. Signal. 27, 2425–2433 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Escoubas C. C., Silva-Garcia C. G., Mair W. B., Deregulation of CRTCs in aging and age-related disease risk. Trends Genet. 33, 303–321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altarejos J. Y., Montminy M., CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen S. M., Li B., Tsien R. W., Ma H., Evolutionary and functional perspectives on signaling from neuronal surface to nucleus. Biochem. Biophys. Res. Commun. 460, 88–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivares-Florez S., Czolbe M., Riediger F., Seidlmayer L., Williams T., Nordbeck P., Strasen J., Glocker C., Jänsch M., Eder-Negrin P., Arias-Loza P., Mühlfelder M., Plačkić J., Heinze K. G., Molkentin J. D., Engelhardt S., Kockskämper J., Ritter O., Nuclear calcineurin is a sensor for detecting Ca(2+) release from the nuclear envelope via IP3R. J. Mol. Med. 96, 1239–1249 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Zach S., Felk S., Gillardon F., Signal transduction protein array analysis links LRRK2 to Ste20 kinases and PKC zeta that modulate neuronal plasticity. PLOS ONE 5, e13191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boratkó A., Csortos C., PKC mediated phosphorylation of TIMAP regulates PP1c activity and endothelial barrier function. Biochim. Biophys. Acta Mol. Cell Res. 1864, 431–439 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi K., Yoshida S., Hatano R., Asano S., Pathophysiological Roles of Ezrin/Radixin/Moesin Proteins. Biol. Pharm. Bull. 40, 381–390 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., Jatkoe T., Berns E. M., Atkins D., Foekens J. A., Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massague J., Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minn A. J., Gupta G. P., Padua D., Bos P., Nguyen D. X., Nuyten D., Kreike B., Zhang Y., Wang Y., Ishwaran H., Foekens J. A., van de Vijver M., Massague J., Lung metastasis genes couple breast tumor size and metastatic spread. Proc. Natl. Acad. Sci. U.S.A. 104, 6740–6745 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loi S., Michiels S., Salgado R., Sirtaine N., Jose V., Fumagalli D., Kellokumpu-Lehtinen P. L., Bono P., Kataja V., Desmedt C., Piccart M. J., Loibl S., Denkert C., Smyth M. J., Joensuu H., Sotiriou C., Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 25, 1544–1550 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Penault-Llorca F., Viale G., Pathological and molecular diagnosis of triple-negative breast cancer: A clinical perspective. Ann. Oncol. 23, vi19–vi22 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Carey L., Winer E., Viale G., Cameron D., Gianni L., Triple-negative breast cancer: Disease entity or title of convenience? Nat. Rev. Clin. Oncol. 7, 683–692 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Balko J. M., Giltnane J. M., Wang K., Schwarz L. J., Young C. D., Cook R. S., Owens P., Sanders M. E., Kuba M. G., Sanchez V., Kurupi R., Moore P. D., Pinto J. A., Doimi F. D., Gomez H., Horiuchi D., Goga A., Lehmann B. D., Bauer J. A., Pietenpol J. A., Ross J. S., Palmer G. A., Yelensky R., Cronin M., Miller V. A., Stephens P. J., Arteaga C. L., Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 4, 232–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall C., Karhade M., Laubacher B., Anderson A., Kuerer H., DeSynder S., Lucci A., Circulating tumor cells after neoadjuvant chemotherapy in stage I–III triple-negative breast cancer. Ann. Surg. Oncol. 22 (Suppl. 3), S552–S558 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Tung N., Gaughan E., Hacker M. R., Lee L. J., Alexander B., Poles E., Schnitt S. J., Garber J. E., Outcome of triple negative breast cancer: Comparison of sporadic and BRCA1-associated cancers. Breast Cancer Res. Treat. 146, 175–182 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Yap T. A., Aerts J. G., Popat S., Fennell D. A., Novel insights into mesothelioma biology and implications for therapy. Nat. Rev. Cancer 17, 475–488 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Clucas J., Valderrama F., ERM proteins in cancer progression. J. Cell Sci. 127, 267–275 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Ansa-Addo E. A., Zhang Y., Yang Y., Hussey G. S., Howley B. V., Salem M., Riesenberg B., Sun S., Rockey D. C., Karvar S., Howe P. H., Liu B., Li Z., Membrane-organizing protein moesin controls Treg differentiation and antitumor immunity via TGF-β signaling. J. Clin. Invest. 127, 1321–1337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Morales F. C., Agarwal N. K., Dogruluk T., Gagea M., Georgescu M. M., Moesin is a glioma progression marker that induces proliferation and Wnt/beta-catenin pathway activation via interaction with CD44. Cancer Res. 73, 1142–1155 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Hong S.-H., Ren L., Mendoza A., Eleswarapu A., Khanna C., Apoptosis resistance and PKC signaling: Distinguishing features of high and low metastatic cells. Neoplasia 14, 249–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunnell C. A., Supko J. G., Eder J. P., Clark J. W., Lynch T. J., Kufe D. W., Shulman L. N., Phase I clinical trial of 7-cyanoquinocarcinol (DX-52-1) in adult patients with refractory solid malignancies. Cancer Chemother. Pharmacol. 48, 347–355 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Izumi H., McCloskey A., Shinmyozu K., Ohno M., p54nrb/NonO and PSF promote U snRNA nuclear export by accelerating its export complex assembly. Nucleic Acids Res. 42, 3998–4007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lahti T., Merikanto I., Partonen T., Circadian clock disruptions and the risk of cancer. Ann. Med. 44, 847–853 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Schwamborn K., Gaisa N. T., Henkel C., Tissue and serum proteomic profiling for diagnostic and prognostic bladder cancer biomarkers. Expert Rev. Proteomics 7, 897–906 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Ishiguro H., Uemura H., Fujinami K., Ikeda N., Ohta S., Kubota Y., 55 kDa nuclear matrix protein (nmt55) mRNA is expressed in human prostate cancer tissue and is associated with the androgen receptor. Int. J. Cancer 105, 26–32 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Esashi F., Christ N., Gannon J., Liu Y., Hunt T., Jasin M., West S. C., CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434, 598–604 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Nicoll R. A., Tomita S., Bredt D. S., Auxiliary subunits assist AMPA-type glutamate receptors. Science 311, 1253–1256 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/8/eaaw9960/DC1
Fig. S1. MSN regulates breast cancer progression.
Fig. S2. Effects of MSN in regulating breast cancer are dependent on its T558 phosphorylation.
Fig. S3. MSN can interact with NONO and enter the nucleus with the assistance of NONO.
Fig. S4. The interaction between MSN and NONO is critical for MSN function on breast tumor progression.
Fig. S5. The function of MSN was mediated by phosphorylation of CREB.
Fig. S6. MSN-NONO interaction enhanced CREB signaling pathway.
Fig. S7. MSN-NONO interaction promoted CREB phosphorylation by facilitating the nuclear localization of pPKCζ.
Fig. S8. There was no significant difference in the expression of EZR and RDX in different subtypes of breast cancer cell lines.
Fig. S9. MSN-NONO complex and downstream CREB signaling pathway could be targeted for TNBC.
Fig. S10. Uncropped images from Western blots.


