Himba have an extrapair paternity rate of 48% with both men and women highly accurate at detecting cases of EPP.
Abstract
Among nonhuman species, social monogamy is rarely accompanied by complete fidelity. Evolutionary theory predicts that the rate of extrapair paternity (EPP) should vary according to socioecological conditions. In humans, however, geneticists contend that EPP is negligible and relatively invariable. This conclusion is based on a limited set of studies, almost all of which describe European-descent groups. Using a novel, double-blind method designed in collaboration with a community of Himba pastoralists, we find that the rate of EPP in this population is 48%, with 70% of couples having at least one EPP child. Both men and women were very accurate at detecting cases of EPP. These data suggest that the range of variation in EPP across human populations is substantially greater than previously thought. We further show that a high rate of EPP can be accompanied by high paternity confidence, which highlights the importance of disaggregating EPP from the notion of “cuckoldry.”
INTRODUCTION
The traditional view in animal behavior is that socially monogamous species should have relatively low rates of extrapair paternity (EPP) (1). However, the advent of DNA fingerprinting led to sweeping changes in the categorization of birds, with the vast majority of species previously categorized as socially monogamous now being labeled polyandrous (2). Among mammals with stable breeding bonds, paternity loss is generally lower than in birds but can be as high as 80% (3, 4). As these results accumulated, the focus of biologists shifted toward understanding the determinants of both inter- and intraspecies variation in EPP, leading to important insights into the operation of sexual selection and challenging assumptions about the relationships between paternity, monogamy, and paternal care (5).
In the human literature, anthropologists have long emphasized the limits of social monogamy. On one hand, the emergence of stable partnerships (either through monogamous or polygynous marriage) is believed to be an essential preadaptation for paternal investment, lineage and inheritance structures, and a myriad of other elements of human social dynamics (6, 7). On the other hand, despite the customary expectation of sexual exclusivity within marriage, extramarital relationships often occur. The frequency of extramarital partnerships across populations is quite variable, but 57% of societies in the Standard Cross-Cultural Sample are reported to have moderate to universal rates of female infidelity (8). In some societies, these concurrent partnerships occur in a formally or informally sanctioned manner (e.g., wife lending and partible paternity), while in others, extramarital relationships are strictly covert and carry severe punishments if detected. These data fit with widespread expectations that women can benefit from polyandry under certain circumstances (9–11).
High-quality genetic evidence of EPP in humans has only begun to accumulate in the last 5 to 10 years and is focused almost exclusively on populations of European descent (12). Unlike the picture of variable rates of concurrency emphasized by anthropologists, genetic data from both contemporary and historical populations have consistently revealed very low rates of EPP (~1 to 2%) (13–19). The only modern study of paternity in a non-Western population, among the Dogon of Mali, shows a similar rate of 1.8% (20). At first glance, these data might raise concerns about the reliability of anthropological accounts. However, the currently studied populations represent patriarchal socioeconomic systems with inheritance of property and increasing focus on the nuclear family, female mobility and autonomy that was historically (or currently in the case of Dogon) limited, and strong religious and cultural norms to promote and help to enforce marital fidelity (20, 21). From this perspective, it is expected that there is such uniformity in the existing data. However, to document the full extent of variation in EPP among humans, more studies are needed, particularly from populations where concurrency is common, and the aforementioned socioeconomic norms and conditions are less pervasive.
Studying EPP in humans raises a host of logistical and ethical challenges that are not present when assessing paternity in other species. These include the need to obtain clear and interpretable consent, a process for ensuring continued community engagement, and, most importantly, a way to ensure that the collection and dissemination of paternity results do not affect patterns of parental care. In some populations, these obstacles may be insurmountable, but in all cases, the methods used to ascertain EPP should be derived in collaboration with the community and tailored to meet the particular needs and constraints of that population. Here, through a 7-year collaborative process, we designed a double-blind protocol for the collection, analysis, and dissemination of genetic paternity data (see the Supplementary Materials for details). Only aggregate results were reported to the community and to the anthropologists working in the community, and no member of the research team is privy to both genetic data and individual identifiers. This prevents intentional or unintentional disclosure of results, while still allowing for the integration of demographic and genetic data. Before reporting aggregate rates, our results were compared with the community-reported perception of EPP to further protect against disruption of parenting norms and practice (fig. S1). Ethical approval for this study was granted by University of California, Los Angeles (UCLA) (IRB-10-000238), the State University of New York, Stony Brook (IRB-636415-12), Namibian Ministry of Home Affairs, and the University of Namibia Office of Academic Affairs and Research. Informed consent and, where necessary, parental assent were obtained from all participants, in addition to permission to work in the community from Chief Basekama Ngombe.
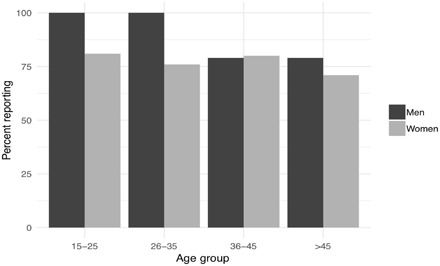
The decision to conduct this study among Himba pastoralists emerged from a series of demographic interviews conducted in 2010 and 2011, in which both men and women reported that a significant proportion of their children were fathered by someone other than the husband (22). Among Himba, the presence of both formal (marital) and informal (nonmarital) partners is common, and concurrent relationships are rarely stigmatized (23). A range of social norms and customs protect people’s ability to maintain informal partnerships, which can span decades and be equally or even more emotionally significant than a marital union (23, 24). That said, there are risks to these partnerships, ranging from sexual jealousy to physical harm. To assess the frequency of concurrent partnerships in this population, we conducted a survey of 171 married individuals and recorded their current number of informal partners. Seventy-seven percent of women and 85% of men had at least one nonmarital partner (Fig. 1), with the modal number of reported extramarital partners reported to be 1 and 2, respectively.
Fig. 1. Percentage of married men and women with at least one concurrent partner, grouped by age.
RESULTS
To determine the frequency of EPP in this population, we collected 718 saliva and buccal swab samples from the population for DNA analysis. From these, 177 husband-child pairs were derived from demographic records, and these were the basis of this study. This included 47 men who potentially fathered 257 children. While convenience sampling of the population was used, 35 of the 41 male-headed households included in a 2017 census (85%) were among those included. Of the six that were missing, two were sampled and had insufficient DNA to process. The raw rate of EPP in the sample, calculated as the percentage of time the husband was not the biological father, is 49%. To go from sample to population, we first estimated an error rate for paternity determination of 5% by calculating the percentage of genetic mismatches in recorded mother-offspring pairs due to sample swaps or other issues (see Materials and Methods). A statistical model accounting for this 5% false-positive rate estimates the rate as 46% (95% Bayesian credible interval, 38 to 53%). Because individual women and unique women-husband pairs may have different EPP rates, we also estimated EPP in a model that clusters by women and women-husband dyads (see the Supplementary Materials). After accounting for variation among mothers and mother-husband dyads, the mean EPP rate is 48% (95% Bayesian credible interval, 32 to 66%; Fig. 2). This closely matches the EPP rate that was presumed by the community, which was 47% (see Materials and Methods for details on how the community perceived EPP rate was derived). Among married couples, with polygynously married men included in multiple pairs, 70% had at least one child whose father was not the husband. This is a minimum threshold, as not all children within a pair were necessarily sampled.
Fig. 2. Posterior distribution of the EPP rate after clustering by mother and mother-husband dyad.
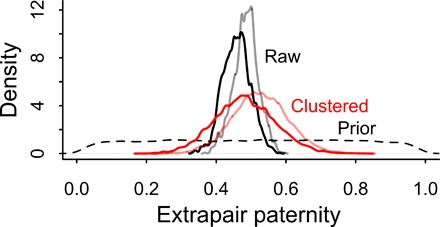
The posterior distribution EPP rate is plotted in red, against both the prior (dashed) and the naïve posterior rate from ignoring clustering by mothers and mother-father dyads (black). Faded curves are estimates without accounting for false-positive paternity.
Next, we compared genetic results with paternity assertions collected during demographic interviews to ascertain the level of paternity confidence for both men and women. For men, this represents their assessment of whether a particular child is their genetic offspring. For women, confidence represents their assessment of the likelihood that their child is the genetic offspring of their husband. High confidence represents accuracy in estimating paternity. In total, 151 maternal paternity assertions and 161 paternal paternity assertions were available. Correct assertions are where the parent accurately identifies the child’s biological father as either the husband or someone other than the husband. Incorrect assertions occur when a parent misattributes paternity either to the husband or to another man. Men were correct in their assertions 73% of the time, and women were correct 72% of the time. When these results were reported back to the community, Himba reported that they believed that this was an underestimate of their accuracy, stating that people likely reported to us that the husband had fathered a child, even when they did not believe he had. In line with this contention, in the vast majority of instances where Himba men and women were incorrect in their assertions, they erred in this direction (Fig. 3). Therefore, we believe that our results represent a conservative estimate of paternity confidence in this population.
Fig. 3. Percentage of correct paternity assertions by men and women.
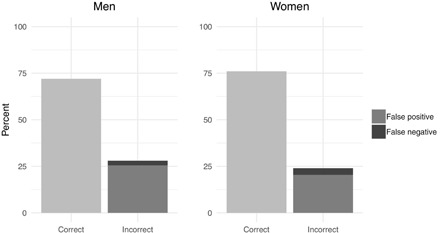
False positives are cases where the child was stated to be the biological offspring of the husband when he/she was not, and false negatives are where the child was claimed not to be the biological offspring of the husband when he/she was.
DISCUSSION
These data provide a stark contrast to the prevailing opinion in the genetics literature that EPP is negligible in humans. While Himba may be at the far end of the range of human variation on this trait, they are not alone in having frequent concurrent partnerships. To assess the true range of variation, more studies from a wider variety of economic and social settings are needed. In addition, we currently know very little about what factors might contribute to variation in EPP among humans. Higher rates of female concurrency have been linked to matrilineal inheritance, reliance on foraging and horticulture, a male-biased adult sex ratio, and prolonged periods of spousal absence (11, 25). However, causal links between these traits and the rate of EPP are opaque at best.
The high level of paternity confidence in this population is also notable. EPP is often presumed to amount to “cuckoldry,” where a male unwittingly invests in another male’s offspring. This threat has been linked to a suite of psychological mechanisms that are thought to exist to prevent and detect nonpaternity events and avoid such costly errors (26). While previous studies have shown general concordance between paternity confidence and actual paternity (27), ours is the first nonclinical study to assess paternity confidence using genetic data and individual assertions, and we show that both men and women are unexpectedly accurate at detecting cases of EPP. Men’s accuracy at detecting EPP events in this study far surpasses that seen previously. Whereas Anderson (27) showed that men with low paternity confidence were correct in their suspicions about 30% of the time, Himba men are more than 70% accurate. This has significant implications for our understanding of men’s motivations for providing care in high EPP contexts. The combination of high paternity confidence and high EPP means that Himba men are not, on the whole, being cuckolded. In contrast, in some contexts, men may be choosing to provide care for nonbiological children as part of the duties of social fatherhood in return for greater security for their other children or the benefits of strong male alliances, or because socioecological conditions such as a male-skewed sex ratio make polyandry the best choice for some males (11, 28–30).
We hope that these findings open the door to more paternity studies from a broader range of populations so that we can have greater clarity about the extent of EPP in humans. We further hope that future studies continue to disambiguate EPP from cuckoldry and instead concentrate on whether, how, and why EPP affects practices of parental care and partner choice in a particular population.
MATERIALS AND METHODS
Experimental design
Study population
Himba reside in northwest Namibia in an area called the Kaokoveld, in the Kunene region, as well as in southern Angola on opposite banks of the Kunene river. They are a Bantu group, closely related to Herero, with a population size of ~50,000 people. Himba remain seminomadic, with a largely pastoralist economy, but horticulture is common in areas close to the Kunene and neighboring tributaries. Our study was conducted in the community of Omuhonga, halfway between the regional capital of Opuwo and the town of Epupa, on the Angolan border.
Demographic interviews were conducted between 2010 and 2017 by B.A.S. and S.P.P. Ages and other dates were determined using a year-name system that is matched to Roman calendar years. See Scelza (31) for details on this method. Additional demographic information collected during these interviews, but not analyzed for this publication, included history of schooling and childhood illness, fosterage history, number of siblings, number of co-wives (for women), vital status of parents, and relation to head of household in current residence.
Marriage among Himba is arranged by parents, although “love match” marriages are common, particularly after the first marriage. Polygyny is common, but so is divorce, meaning most Himba adults have more than one spouse over their lifetime (either sequentially or concurrently). Age at marriage is later for men than women. Men marry for the first time in their late 20s, but this is typically to a child bride. Child marriages are not consummated, and coresidence typically does not begin until after the bride reaches menarche. Even then, a considerable portion of these marriages never come to fruition, although the husband is considered the “social father” of children born to her until she marries again. These children were considered out of wedlock for our purposes and not included in our paternity analysis (see the “Deriving the EPP and paternity confidence rates” section for details).
Double-blind protocol
To conduct analyses that incorporate both individualized demographic and genetic data, we designed, in collaboration with the community, a double-blind method for analyzing and reporting paternity results. This ensured that the anthropologists on our team would have the same access to information as participants and protects against the possibility that reporting of results could alter investment patterns or marital dynamics within the community. We further compared the community-derived aggregate rate (see below) with the actual aggregate rate, demonstrating that our data reflected existing perceptions of EPP in this community. To enact the double-blind procedure, each member of the team was blinded to certain kinds of data. Following this procedure, none of the parties had access to both individualized data and paternity results, yet analyses were able to maintain important links between individuals. The key is to ensure that the person who is collecting and maintaining identifiable data and returning to the community to report back results does not have access to individualized paternity results.
For our purposes, the three parties involved were the “anthropologists,” the “geneticists,” and the “statistician.” The anthropologists (B.A.S. and S.P.P.) were in charge of collecting both genetic samples and individualized data. It is these people, and these people only, who retain identifiable data linking an ID to attributes such as names, dates, and relationships to others in the study. Upon completion of fieldwork, B.A.S. sent samples (labeled with ID number only) to the geneticists (N.S., S.G., and B.M.H.), who extracted DNA and performed paternity analyses. The geneticists thus have access to coded genetic relationships, family pedigrees, and paternity results for particular dyads but did not have access to any individualized identifiers. Last, the statistician (R.M.) received paternity results from the geneticists and individual-level data (i.e., paternity assertions and an ID-coded pedigree) from the anthropologists. The statistician (R.M.) used IDs to link these results, calculate EPP rates, and perform other analyses. Last, the anthropologists were in charge of community reporting. Aggregate paternity and paternity confidence results were reported back to the community in July 2018. At that time, the chief and acting chief of the community provided an additional layer of post hoc consent to publish the results.
Deriving the EPP and paternity confidence rates
Paternity assertions were derived slightly differently for men and women. Using responses from semistructured interviews (see section SB), each birth was coded as either marital (taking place within a marriage) or nonmarital (taking place either before or between marriages). For each marital birth, women were then asked whether the biological father of that child was the husband or someone else. Women reported children fathered by a man other than the husband to be omoka. To derive men’s paternity assertions, male informants listed each of the children born to their wives and then stated whether he thought he was the father or whether the child was omoka. If they were unsure, then this was noted, but this occurred rarely and typically only if the child was still very young (<1 year old). An assertion of “unsure” was counted as omoka for these analyses.
In its simplest form, the rate of EPP is the proportion of children born to married couples, whose biological father is someone other than the husband. Here, we add just a couple of notes to clarify: (i) All children born to a man after he marries a woman, and in the case of divorce until she marries someone else, “count” as his. This means that children born between marriages have a social father, although the chance that this man is their biological father is close to zero. So as not to artificially inflate the rate of EPP in this population, we excluded children born between marriages from our analysis, categorizing these children as “out of wedlock.” (ii) Similarly, there are cases of child marriage, which are never consummated, but where the husband remains the social father of any children that woman has until she marries again (if she does). We also counted these children as out of wedlock and excluded them from our analysis.
Together, these exclusions mean that the proportion of cases where the social father is not the biological father exceeds what we report in our paper. This has important considerations for understanding the relationship between paternity and paternal care, which we will take up in future work. However, for now, we decided to provide the most conservative estimate of EPP for this group.
Samples
DNA via saliva or buccal swab (Oragene kits), age, sex, and ethnographic information were collected from Himba individuals with informed individual/parental consent. Samples were extracted using the DNA Genotek prepIT-L2P kit and protocol. One hundred eighty-eight samples were genotyped on the Illumina MEGAex array, and 530 samples were genotyped on the Illumina H3Africa array. DNA from 87 women, 47 social fathers, and 257 children were included in these analyses (Table 1).
Table 1. Sample characteristics.
| Women | Social father | |
| N in sample | 87 | 47 |
| Age: mean (min, max) | 49 (16, 83) | 58 (30, 99) |
| Parity*: mean (min, max) | 6.6 (1, 12) | 9.1 (1, 29) |
| Number of marriages: mean | 1.7 | 3.7 |
| % Currently married | 73 | 98 |
*Parity for men refers to the number of purported children, not actual paternity.
Community perception of EPP
To determine community-level perceptions of EPP in the study area, a short vignette was used. Participants (n = 21) were shown 10 small plastic babies and asked if a typical Himba man had 10 children with his wives, how many of them would be his, and how many would be from a boyfriend. Participants divided the babies into two piles, resulting in their perception of a typical distribution. While this does not mirror perfectly the way we obtained the population-level EPP rate using DNA, it was a clear and simple representation of people’s understanding of how paternity is distributed in this population. Figure S1 shows the range in responses by participants. There is significant variation in how participants responded (just as there is variation across dyads in the genetic data), while the average response mapped very well onto the actual average EPP rate.
Statistical analysis
Data processing
Raw intensity files from Illumina for both H3Africa and MEGAex arrays were clustered using Genome Studio’s Genotyping Module (v. 2.0). Array-specific cluster files and a GenCall threshold of 0.15 were also used. A total of 4 individuals from the MEGAex dataset and 10 individuals from the H3Africa dataset were removed from further analysis. Single nucleotide polymorphisms (SNPs) with a call frequency below 0.85, cluster separation values below 0.02, a heterozygote rate greater than or equal to 0.85, and/or with over two replicate errors were removed from subsequent analysis. Rare variants (having a minor allele frequency of less than 0.05) were flagged.
zCall
Rare variants from Genome Studio were recalled with zCall using all quality-controlled SNPs. We looked for but identified no samples with excess heterozygosity. We followed the standard zCall pipeline, using a Z threshold that produced the highest global concordance for each dataset (a Z threshold of 5 for the H3Africa platform and a Z threshold of 4 for the MEGA platform). Rare variants identified previously were then extracted from the zCall dataset and merged with common variants called by Genome Studio.
MEGA dataset
We used a custom file to update the locus information for several variants on the MEGA array that are reported to be on “chromosome 0” and “position 0.”
SNP array quality control
We converted each dataset from an A and B allele format to the Illumina top strand allele and updated all SNP names to those in dbSNP v. 144 using Python scripts. Loci duplicates were identified with an R script and then removed from each dataset using PLINK v1.90b3v (32). Next, we aligned each dataset to the 1000 Genomes reference hg19 using a Python script to identify indels, nonmatching SNPs, and flipped loci and then used PLINK to flip necessary loci and remove other SNP inconsistencies. We used a Python script to identify A/T and C/G loci and removed them in PLINK.
We used PLINK to filter out SNPs from each dataset that had a missing call rate greater than 5%, a minor allele frequency (MAF) less than or equal to 1%, or a Hardy-Weinberg equilibrium exact test with a P value below 0.0001. The two datasets were then merged in PLINK, and SNPs that had a missingness greater than 5% across platforms were excluded. The resulting merged dataset contained 377,995 autosomal and 8282 X chromosome SNPs left for analysis. We performed a sex check based on X chromosome heterozygosity using PLINK to check for possible sample swaps or contamination, resulting in the exclusion of one sample. Sample duplicates were then identified and removed using PLINK and R, resulting in a final set of 678 individuals (fig. S9).
Paternity analysis
We used KING 2.1.3 (33) to infer relatedness among individuals with the identical by descent (IBD) segment flag and generated a list of parent-offspring (PO) pairs. Similar data have shown negligible false positives with this method (34). We verified all recorded mother-offspring pairs first as a quality control measure and to assess a possible error rate. Samples from individuals and their identified social fathers were then queried against the list of PO pairs and recorded as either a positive or negative paternity match.
Estimating error rate
To assess the error rate, we calculated the percentage of mismatches for ethnographically recorded mother-offspring pairs. A mother-offspring mismatch was flagged when both the mother and child IDs appeared in the dataset but did not show a genetic PO relationship. This resulted in an error rate estimate of 5% (n = 257 pairs). These mismatches may be due to recording error in the lab or in the field, a sample swap, fosterage, or miscommunication of information between the investigator and participant. Using additional genetic and ethnographic information, we were able to rectify most of these mismatches.
Posterior distributions of EPP
Posterior distributions of EPP were assessed using a Bayesian multilevel model (Bernoulli outcome), which included varying effects for mother and mother-husband dyads. Weakly regularizing priors were used for all parameters. The model was implemented in Stan 2.18, using 8000 samples from four chains. All R-hat values were less than 1.01, and visual inspection of trace plots and rank histograms indicated no evidence of divergent transitions or biased posterior exploration. Posteriors from the model were used to generate an average posterior EPP rate with a mean of 0.53 (95% Bayesian credible interval from 0.37 to 0.68). Full model description and diagnostic plots are located in the “Multilevel models of EPP” section in the Supplementary Materials.
Supplementary Material
Acknowledgments
First and foremost, we thank the community of Omuhonga for continued support and for the trust they have instilled in us to undertake this project. J. Jakurama, C. Louis, and K. Ngombe acted as research assistants and translators in Namibia. Students in the Henn Lab (G. Bari, T. Rosenlicht, P. Romano, and J. Gaige) participated in DNA extraction. Members of the Behavior, Evolution, and Culture (BEC) group at UCLA provided valuable feedback on the manuscript. Funding: This work was funded by the NSF (BCS-1534682 to B.A.S.) and the NIH (IRACDA postdoctoral grant K12-GM102778 for E.G.A.). Author contributions: B.A.S. conceived the research. B.A.S., B.M.H., and J.S. designed the double-blind protocol and procedures for community dissemination along with community members in Namibia. B.A.S., S.P.P., and E.G.A. collected the genetic data, and B.A.S. and S.P.P. collected the demographic data. B.M.H., S.G., and N.S. extracted and analyzed the genetic data to determine paternity. S.P.P. and B.A.S. cleaned and coded the data, and R.M., B.M.H., and N.S. conducted the statistical analysis. B.A.S. and B.M.H. wrote the paper with feedback from all other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The Illumina genotype array data that support the findings of this study have been deposited in dbGaP under accession 37103 with controlled access. Paternity assertion data and paternity results are not available as linking assertions or other identifiable data with paternity data would compromise research participants’ privacy and could potentially bring harm to participants. This complies with the double-blind protocol that the investigators and the community designed and are subject to, as well as the approved IRB protocol for this study and our NSF-approved Data Management Plan. Data and code for figures in the main text and the Supplementary Materials not linked to paternity data are available at https://data.mendeley.com/datasets/xtgbg39jmv/draft?a=f60a3364-74b4-480e-8808-8878878bcc4d. Code for the multilevel models, together with the WAIC model comparison, and also data from the MEGA dataset can be found in the script at https://github.com/rmcelreath/Himba_EPP. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/8/eaay6195/DC1
Supplementary Materials and Methods
Fig. S1. Density plot of EPP Perception Task.
Fig. S2. Comparison of age for men in our paternity sample (n = 47) with those not living in Omuhonga but not included in our sample (n = 68).
Fig. S3. Comparison of age for women in our paternity sample (n = 59) with those not living in Omuhonga but not included in our sample (n = 129).
Fig. S4. Comparison of parity for women in our paternity sample (n = 59) with those not living in Omuhonga but not included in our sample (n = 129).
Fig. S5. Frequency distribution showing number of children in the sample, by mother.
Fig. S6. Frequency distribution showing number of children in the sample, by social father.
Fig. S7. Number of sampled children, by mother’s age.
Fig. S8. Trace plots (top) and rank histograms (bottom) for model clustering mother and mother-husband dyad.
Fig. S9. Flowchart of DNA sample quality control, beginning with total number of population samples and the final number of samples available for paternity analysis.
REFERENCES AND NOTES
- 1.T. H. Clutton-Brock, The Evolution of Parental Care (Princeton Univ. Press, 1991). [Google Scholar]
- 2.Griffith S. C., Owens I. P. F., Thuman K. A., Extra pair paternity in birds: A review of interspecific variation and adaptive function. Mol. Ecol. 11, 2195–2212 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Clutton-Brock T. H., Isvaran K., Paternity loss in contrasting mammalian societies. Biol. Lett. 2, 513–516 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohas A., Allainé D., Social structure influences extra-pair paternity in socially monogamous mammals. Biol. Lett. 5, 313–316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrie M., Kempenaers B., Extra-pair paternity in birds: Explaining variation between species and populations. Trends Ecol. Evol. 13, 52–58 (1998). [DOI] [PubMed] [Google Scholar]
- 6.B. Chapais, Primeval Kinship: How Pair-Bonding Gave Birth to Human Society (Harvard Univ. Press, 2009). [Google Scholar]
- 7.Gavrilets S., Human origins and the transition from promiscuity to pair-bonding. Proc. Natl. Acad. Sci. U.S.A. 109, 9923–9928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broude G. J., Greene S. J., Cross-cultural codes on twenty sexual attitudes and practices. Ethnology 15, 409–429 (1976). [Google Scholar]
- 9.Hrdy S. B., The optimal number of fathers: Evolution, demography, and history in the shaping of female mate preferences. Ann. N. Y. Acad. Sci. 907, 75–96 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Scelza B. A., Choosy but not chaste: Multiple mating in human females. Evol. Anthropol. 22, 259–269 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Starkweather K. E., Hames R., A survey of non-classical polyandry. Hum. Nat. 23, 149–172 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Larmuseau M. H., Matthijs K., Wenseleers T., Cuckolded fathers rare in human populations. Trends Ecol. Evol. 31, 327–329 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Greeff J. M., Erasmus J. C., Three hundred years of low non-paternity in a human population. Heredity 115, 396–404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larmuseau M. H. D., Vanoverbeke J., Van Geystelen A., Defraene G., Vanderheyden N., Matthys K., Wenseleers T., Decorte R., Low historical rates of cuckoldry in a Western European human population traced by Y-chromosome and genealogical data. Proc. Biol. Sci. 280, 20132400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larmuseau M. H., Claerhout S., Gruyters L., Nivelle K., Vandenbosch M., Peeters A., van den Berg P., Wenseleers T., Decorte R., Genetic-genealogy approach reveals low rate of extrapair paternity in historical Dutch populations. Am. J. Hum. Biol. 29, e23046 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Boattini A., Sarno S., Pedrini P., Medoro C., Carta M., Tucci S., Ferri G., Alù M., Luiselli D., Pettener D., Traces of medieval migrations in a socially stratified population from Northern Italy. Evidence from uniparental markers and deep-rooted pedigrees. Heredity 114, 155–162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solé-Morata N., Bertranpetit J., Comas D., Calafell F., Y-chromosome diversity in Catalan surname samples: Insights into surname origin and frequency. Eur. J. Hum. Genet. 23, 1549–1557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf M., Musch J., Enczmann J., Fischer J., Estimating the prevalence of nonpaternity in Germany. Hum. Nat. 23, 208–217 (2012). [DOI] [PubMed] [Google Scholar]
- 19.King T. E., Jobling M. A., What’s in a name? Y chromosomes, surnames and the genetic genealogy revolution. Trends Genet. 25, 351–360 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Strassmann B. I., Kurapati N. T., Hug B. F., Burke E. E., Gillespie B. W., Karafet T. M., Hammer M. F., Religion as a means to assure paternity. Proc. Natl. Acad. Sci. U.S.A. 109, 9781–9785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. Goody, The Development of the Family and Marriage in Europe (Cambridge Univ. Press, 1983). [Google Scholar]
- 22.Scelza B. A., Female choice and extra-pair paternity in a traditional human population. Biol. Lett. 7, 889–891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scelza B. A., Prall S. P., Partner preferences in the context of concurrency: What Himba want in formal and informal partners. Evol. Hum. Behav. 39, 212–219 (2018). [Google Scholar]
- 24.Van Wolputte S., Sex in troubled times: Moral panic, polyamory and freedom in north-west Namibia. Anthropol. South. Afr. 39, 31–45 (2016). [Google Scholar]
- 25.Hartung J., Paternity and inheritance of wealth. Nature 291, 652–654 (1981). [Google Scholar]
- 26.S. M. Platek, T. K. Shackelford, Female Infidelity and Paternal Uncertainty: Evolutionary Perspectives on Male Anti-Cuckoldry Tactics (Cambridge Univ. Press, 2006). [Google Scholar]
- 27.Anderson K., How well does paternity confidence match actual paternity? Curr. Anthropol. 47, 513–520 (2006). [Google Scholar]
- 28.K. R. Hill, A. M. Hurtado, Ache Life History: The Ecology and Demography of a Foraging People (Transaction Publishers, 1996). [Google Scholar]
- 29.S. Beckerman, P. Valentine, Cultures of Multiple Fathers: The Theory and Practice of Partible Paternity in Lowland South America (University Press of Florida, 2002). [Google Scholar]
- 30.Walker R. S., Flinn M. V., Hill K. R., Evolutionary history of partible paternity in lowland South America. Proc. Natl. Acad. Sci. U.S.A. 107, 19195–19200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scelza B. A., Female mobility and postmarital kin access in a patrilocal society. Hum. Nat. 22, 377–393 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., Lee J. J., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manichaikul A., Mychaleckyj J. C., Rich S. S., Daly K., Sale M., Chen W.-M., Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramstetter M. D., Dyer T. D., Lehman D. M., Curran J. E., Duggirala R., Blangero J., Mezey J. G., Williams A. L., Benchmarking relatedness inference methods with genome-wide data from thousands of relatives. Genetics 207, 75–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/8/eaay6195/DC1
Supplementary Materials and Methods
Fig. S1. Density plot of EPP Perception Task.
Fig. S2. Comparison of age for men in our paternity sample (n = 47) with those not living in Omuhonga but not included in our sample (n = 68).
Fig. S3. Comparison of age for women in our paternity sample (n = 59) with those not living in Omuhonga but not included in our sample (n = 129).
Fig. S4. Comparison of parity for women in our paternity sample (n = 59) with those not living in Omuhonga but not included in our sample (n = 129).
Fig. S5. Frequency distribution showing number of children in the sample, by mother.
Fig. S6. Frequency distribution showing number of children in the sample, by social father.
Fig. S7. Number of sampled children, by mother’s age.
Fig. S8. Trace plots (top) and rank histograms (bottom) for model clustering mother and mother-husband dyad.
Fig. S9. Flowchart of DNA sample quality control, beginning with total number of population samples and the final number of samples available for paternity analysis.


