Abstract
Backgrounds
p53 is a tumor suppressor that prevents cancer onset and progression, and mutations in the p53 gene cause loss of the tumor suppressor function of the protein. The mutant p53 protein in tumor cells can form aggregates which contribute to the dominant-negative effect over the wild-type p53 protein, causing loss of p53 tumor suppression or gain of novel oncogenic functions. Mutations in p53 have been implicated in the pathogenesis of primary prostate cancer (PCa), and are often detected in recurrent and metastatic disease. Thus, targeting mutant p53 may constitute an alternative therapeutic strategy for advanced PCa for which there are no other viable options.
Methods
In this study, we used immunoprecipitation, immunoflurorenscence, clonogic survival and cell proliferation assays, flow cytometric analysis and in vivo xenograft to investigate the biological effects of ReACp53, a cell-permeable peptide inhibitor of p53 aggregation, on mutant p53-carrying PCa cells.
Results
Our results show that ReACp53 targets amyloid aggregates of mutant p53 protein and restores the p53 nuclear function as transcriptional factor, induces mitochondrial cell death and reduces DNA synthesis of mutant p53-carrying PCa cells; ReACp53 also inhibits xenograft tumor growth in vivo.
Conclusions
The data presented here suggest a therapeutic potential of targeting mutant p53 protein in advanced PCa setting, which has a clinical impact for aggressive PCa with transforming how such tumors are managed.
Keywords: Castration-resistant Prostate Cancer, Tp53, amyloid aggregates, mitochondrial cell death, cell cycling
Introduction
Prostate Cancer (PCa) is the most commonly diagnosed non-cutaneous malignancy in men and the second leading cause of cancer death in the United States [1]. When the tumor is low grade and organ-confined, surgery or radiation therapy is curative. For advanced and metastatic tumors as well as for recurrent tumors after local therapy, the treatment of choice is hormonal therapy by inhibition of androgen production or block of androgen receptor (AR) function. Although initially hormonal therapy relieves symptons and disease progression in nearly all patients, the tumor eventually recurs and progresses to become castration-resistant PCa (CRPC). CRPC is characterized by continued AR activation, PSA (prostate-specific antigen) production and tumor growth in an androgen-deprived environment. Although p53 mutations are relatively uncommon in untreated prostatic adenocarcinoma, they are present in about 40% of CRPC and in 70% of small cell neuroendocrine prostate cancer (SCNC), a more aggressive type of CRPC, suggesting a possible involvement of p53 mutation with disease progression [2, 3].
The p53 protein plays essential roles in cells by integrating and regulating multiple signals and ensuring adequate temporal and spatial responses to stresses. Mutation of p53 is one of the common genetic events detected in half of all human tumors. Studies have revealed that p53 mutation significantly correlates with disease progression, resistance to cancer treatments and worse clinical outcomes of cancer patients [4–7].
p53 can interact with various binding partners, this flexible nature of p53 allows p53 to regulate different cellular processes such as apoptosis, DNA repair, cell cycle arrest and metabolism[8–10]. Mutant p53 typically loses its wild-type (WT) functions and may exert a dominant-negative effect over the remaining WT p53 allele[11]. Functional loss of p53 may result from the inability of mutant p53 to bind DNA, while gain of function can arise from an altered mutant p53 interactome[12]. More recently, mutations in p53 were shown to favor the protein aggregation, which in turn result in protein sequestration and/or inactivation [9, 13, 14]. Studies further suggest that the dominant-negative effect and gain-of-functions of mutant p53 can occur via oligomerization and co-aggregation of mutant p53 with WT p53 and other proteins in the p53 pathway, such as p63 and p73[15, 16].
Protein aggregation is implicated in many human diseases[14, 17, 18]. Accumulating evidence have demonstrated that WT or mutant p53 could form amyloid-like aggregates in cancer cells, and that formation of p53 protein aggregates may participate in carcinogenesis[9, 19–21]. Several portions of the p53 protein have been predicted in silico or shown experimentally to be involved in aggregate formation, including the N-terminal domain, the DNA-binding domain and the C-terminal tetramerization domain [13, 22–24]. While full-length p53 is likely to aggregate in vitro using any number of its aggregation-prone regions, in vivo it is conceivable that, due to partial unfolding and interaction with binding partners, only a subset of these segments is available and solvent-exposed at any given time and thus promotes aggregation[13]. Among these, amino acids 251–258 within the DNA binding domain has been pinpointed as important for p53 aggregation in cells and in vivo[15]. Of interest, a recent study showed that ReACp53, a therapeutic cell-penetrating peptide designed to interfere with the aggregation of the 252–258 segment of mutant p53 protein, has anti-cancer properties in vitro and in vivo in ovarian cancer models[25].
In this study, we tested the potential therapeutic effect of ReACp53 in CRPC cells expressing mutant p53 protein. Our results showed that treatment with ReACp53 can change the aggregation status of mutant p53, resulting in promoted p53-Bax interaction with consequent mitochondrial cell death in CRPC cells. ReACp53 treatment also inhibits xenograft tumor growth of CWRR1, a CRPC cell line that express mutant p53 protein, but has less effect on xenograft tumor growth of C4–2 cells that harbor WT p53. The results suggest a potential of targeting mutant p53 in advanced PCa as a treatment strategy that can transform the treatment landscape for PCa, which may benefit many PCa patients who have no other options.
Materials and Methods
Reagents and Cell line
Mutant p53 aggregation inhibitor, ReACp53, and a sequence-scrambled control peptide (SCR) were kindly provided by Dr. Soragni (UCLA)[25]. The human prostate cancer cell lines DU145 and PC-3 were from American Type Tissue Collection (Manassas, VA), which has provided certifications of analysis for karyotyping and short tandem repeat (STR) profiling. CWRR1 and LNCaP-derived C4–2 were kindly provided by Drs. Tindall and E. Wilson[26]. Engineered PC-3 cells and C4–2 cells with expressions of wild-type p53 or mutant p53 (p53R175His and p53P223Leu) were generated as reported in our previous study[27]. CWRR1, DU145 and C4–2 cells were grown in RPMI-1640 medium supplemented with 10% FBS; PC-3 cells were grown in Ham’s F12K medium with 2 mM L-glutamine, 1.5 g/L sodium bicarbonate and 10% FBS. All cell lines were tested negative for mycoplasma contamination using a Cell Culture Contamination Detection Kit (ThermoFisher Scientific).
p53 stability/aggregation experiment
For SDS-resistance assay, log phase growing cells were pelleted and resuspended in cold Pierce IP Lysis Buffer (Thermo Fisher) freshly supplemented with 2× HALT protease and phosphatase inhibitor (Thermo Fisher). After incubation on ice for 30 minutes and cleared by centrifugation, 200 μg of native lysates were brought to a 500 μl volume in IP buffer and heated at 42°C in the presence of vehicle, SCR or 20 μM ReACp53 for 10 minutes, followed by a 30 minute incubation on ice. The protein samples were used for Immunoprecipitations with 5 μg of PAb240 antibody (Santa Cruz Biotech) at 4°C overnight, and analyzed with 15% SDS-PAGE gel electrophoresis and immunoblotting with anti-p53/DO-1 antibody[25].
For p53 stability, 200 μg of cell lysate in RIPA buffer was immunoprecipitated with 2 μg of anti-p53/DO-1 antibody. The immunoprecipitated complexes were purified with Pierce Direct Magnetic IP/Co-IP kit (Pierce) and analyzed with western blot.
Western blot analysis
Cells were lysed in HALT inhibitor-containing RIPA buffer with mild sonication for indicated western blot assays. The mitochondrial fraction was prepared from cell pellets utilizing a reagent-based method per manufacturer’s instruction (Mitochondria Isolation Kit; Pierce), and lysed in RIPA buffer and analyzed with Western blot as previously reported[27]. Antibodies were from Santa Cruz Biotech (p53/DO-1, Bax/N-20, PCNA/PC10 and p21/C-19), Calbiochem (Mdm2/Ab3), abcam (ubiquitin/ab7780, HSP90/AC88 and cyclin E/ab33911), Dako-Agilent (AR/AR441) and Sigma-Aldrich (Cyclin A/C4710). Anti-β-actin (abcam, ab8227) was included for equivalent protein loading.
Assay of mitochondrial membrane potential (MMP)
Cells were treated for 72 hours with 10 μM ReACp53 or 10 μM SCR, or DMSO as control, and 1 × 106 cells were collected for MMP analysis by JC-1 staining according to manufacturer instruction (MitoProbe JC-1 Assay kit, Life technologies).
Cell proliferation and clonogenic survival assays
Cell proliferation and viability assay was performed with using MTS Cell Proliferation Colorimetric Assay Kit (Biovision) per manufacturer’s instruction. Briefly, 1000 cells/well were plated into 96-well plate and incubated overnight. Cells were then treated with indicated chemicals for 72 hours. After incubation with MTS Reagent for one hour, the soluble formazan was measured as the absorbance at 490 nm using a SpectraMax M3 reader, and data acquisition and analysis were performed with SoftMax Pro 6 software (Molecular Devices).
For clonogenic survival assay, 500 cells were plated in 60 mm dish, and were exposure to 10 μM of ReACp53 or SCR, or PBS as control, for 72 hours. Cells were then maintained in fresh culture medium for 7–10 days. Formed cell colonies were stained with crystal violet, and colonies consisting of >50 cells were considered as surviving colonies. Average numbers for surviving colonies were plotted versus control to determine clonogenic survival fractions for each treatment. In the experiments for combination treatment of ReACp53 and Androgen deprivation (ADT), cells were pretreated with 10 μM ReACp53 for 72 hours, and 500 cells were then plated in 60 mm dish and cultured in ReACp53-containing RPMI-1640 medium supplied with 10% of Dextran-stripped FBS (Denville).
BrdU (bromodeoxyuridine) incorporation assay
8×104 log phase cells were plated in 6-well plate overnight. Cells were then treated with 10 μM ReACp53 or 10 μM SCR, or DMSO as control, for 72 hours. 10 μmol/L BrdU (BD Biosciences) was added 2 hours before collection. The percentage of BrdU incorporation was analyzed with flow cytometric analysis per the manufacturer’s instruction (BD FITC BrdU Flow Kit).
Immunofluorescence analysis
Cells grown on glass slide were labelled with 100 nM of MitoTracker® probes (Mitotracker Green/Red FM, Invitrogen) for thirty minutes before cell collection, and were then fixed with 2% paraformaldehyde (Fisher Scientific). Immunofluorescence staining were performed with anti-p53 (DO-1) or PAb240 antibodies (EMD Millipore). Images were acquired with LSM 510 confocal microscope (Zeiss) with 40× objective (oil) and processed by Photoshop (Adobe).
RNA extraction and quantitative real-time PCR
Total RNA was extracted using RNeasy mini kits (Qiagen). The PrimeScript RT Master Mix (Takara) was used for cDNA synthesis. Quantitative real-time PCR (qRT-PCR) was performed on ABI 7500 (Applied Biosystems) using SYBR Green PCR Master Mix (Quanta Biosciences). The primers used in the assays are listed in supplementary table.
Xenograft Tumor Growth Assay and Immunohistochemistry
All animal work was conducted in accordance with the NIH Guidelines of Care and Use of Laboratory Animals and approved by Duke Institutional Animal Care and Use Committee (IACUC/A092-16-04). Immunocompromised NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice were from The Jackson Laboratories. 2×106 CWRR1 or C4–2 cells in 0.1ml 1× HBSS with 50% Matrigel (Corning) were inoculated subcutaneously into the right thigh of 6–8 weeks old male NSG mouse. When tumor volumes reached a size of 50–100 mm3 (approx. day 7 after inoculation), mice were randomly grouped into 3 groups (n=4) that received treatments of PBS (0.2ml), ReACp53 (15 mg/kg) or SCR (15 mg/kg) with intraperitoneal (IP) injections every 48 hours for two weeks. Tumors were measured every three days and tumor volumes were determined from caliper measurements of tumor length (L) and width (W) according to the formula (L×W2)/2.
Xenograft tumor were collected and paraffin-embedded for immunohistochemistry analysis.
Statistical Analyses
Statistical analyses were performed using Student’s t-test. A p-value <0.05 was considered as significant.
Results
ReACp53 acts on stability and aggregates of mutant p53 protein in PCa cells
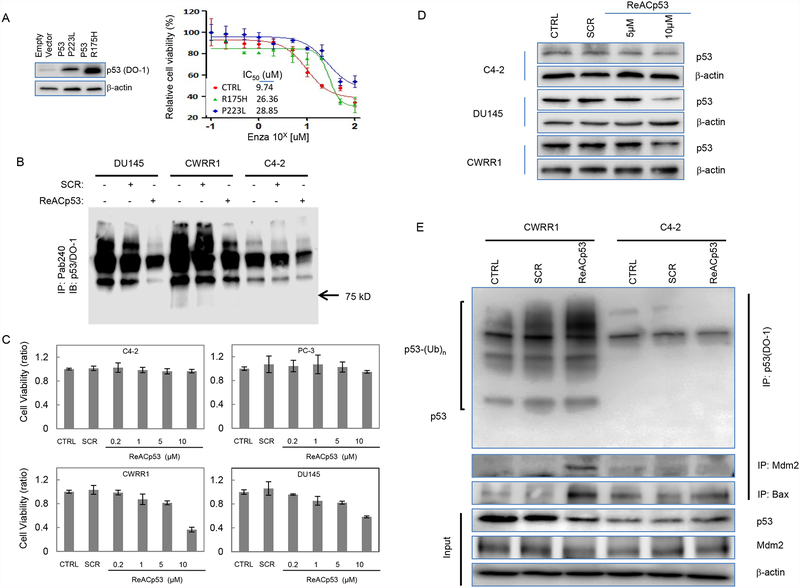
We first tested the potential effect of mutant p53 expression on the response of CRPC cells to the treatment of Enzalutamide (ENZA), a newer agent targeting AR signaling approved by FDA as a standard therapeutic regimen for the treatment of castration-resistant PCa (CRPC) that is no longer responding to the first line androgen deprivation therapy (ADT). In this study, we engineeringly expressed mutant p53 of p53 P223L and p53 R175H, two hot spot mutations of p53 occuring in nearly 2% of cancer cells [28] in C4–2 cells that carry wild-type p53 [29], and then treated these cells with ENZA. Our results showed that expression of mutant p53 in C4–2 cells dramatically increased IC50 of ENZA (from 9.74 μM for control C4–2 cells to 26.36 μM for C4–2/p53R175H and 28.85 μM for C4–2/p53P223L cells, respectively) (figure 1A), suggesting that expression of mutant p53 protein may cause treatment resistance in PCa cells. Of interest, while no changes were observed in AR expression, we noticed that expression of p53 R175H in C4–2 cells increased PSA production while p53 P223L showed no such effect on PSA level (supplemental figure A), suggesting that different mutants of p53 may have diversity on regulating AR signaling and thus differently affect the PCa progression.
Figure 1:
Effects of ReACp53 on p53 aggregates and stability in PCa. A. Engineered expression of mutant p53 induces ENZA-resistance in PCa C4–2 cells. Left: Western blot showing expression of p53R175H and p53P223L in C4–2 cells. Right: Graphs showing the change of IC50 for ENZA in engineered C4–2 cells; B. SDS-resistance assay showing ReACp53 reduces the aggregate formation of mutant p53 protein in PCa cells; C. Graphs showing the ReACp53 treatment decreases cell vialility of mutant p53-bearing PCa cells; D. Western blot analysis showing treatment of ReACp53 downregulates p53 protein levels in DU145 and CWRR1 cells, but not in C4–2 cells; E. Immunoprecipitation assays showing the exposure to ReACp53 induces ubiquintination of mutant p53 and increases protein interactions of mutant p53 with Mdm2 and Bax in CWRR1 cells. Western blot analysis for input cell lysates showing the negative regulation effect of ReACp53 treatment on protein expression of Mdm2 in PCa cells. Data represent the average of three independent experiments. Error bars indicate standard deviation. * indicates statistical significance (p<0.05).
We next tested the biological effects of ReACp53 on PCa cells expressing WT p53 or mutant p53. In these experiments, we used DU145 cells which contains two mutant p53 alleles (P223L and V274F) [30, 31], PCa CWR22-derived castration-resistant CWRR1 cells bearing mutations of p53 in exon 8 (R273H) and exon 9 (Q331R)[31], and PCa LNCaP-derived androgen-independent C4–2 cells carrying wild-type p53 [29].
We used a SDS-resistance assay[25] to determine the effect of ReACp53 on protein aggregation formed with mutant p53. As shown in figure 1B, we found that the presence of 20 μM ReACp53 in native cell lysates during heating at 42°C could remarkably reduce SDS-resistant p53 aggregate formation in DU145 and CWRR1 cells. However, no such effect was observed for C4–2 cells, although slight change was also observed in C4–2 cells which indicates the nature of WT p53 can also form aggregates and the drug could also impact WT p53 funtion in live cancer cells. These results indicate ReACp53 can target the mutant p53 aggregates in PCa cells.
In dose-dependence analyses using trypan blue exclusion assay, we detected dramatically decreased cell viabilities of CWRR1 and DU145 cells when cells were treated with 10 μM ReACp53 for 72 hours. However, ReACp53 treatment did not show similar effects for C4–2 cells or PC-3 cells, a more aggressive CRPC cell model but expressing no p53 [32] (figure 1C).
To test the potential effects of ReACp53 on p53 protein expression and stability, we determined the change in protein level and the potential ubiquintination of p53 in cells after exposure to ReACp53. Our results showed that ReACp53 treatment resulted in decreased p53 protein level in DU145 and CWRR1 cells, but not in C4–2 cells. As controls, treatment with scrambled (SCR) peptide (10 μM) or PBS showed no obvious effects on change of p53 protein level in all three cell lines (figure 1D). Immunoprecipitation (IP) assay further showed that exposure to 10 μM ReACp53 for 72 hours induced p53 ubiquintination in CWRR1 cells (figure 1E). These results thus indicate that, consistent with previous observations in ovarian cancer cells[25], ReACp53 acts on the protein aggregation and stability of mutant p53 in PCa cells.
In normal unstressed cells, rapid degradation of WT p53 is tightly associated with the ubiquitin ligase Mdm2. Mdm2 can also drive the degradation of mutant p53, which indicates that the mutant p53 proteins are not intrinsically resistant to Mdm2-associated degradation, although in tumor cells this regulation is disrupted because p53-dependent transcription of Mdm2 is inhibited by the loss-of-function of mutant p53 or by the dominant-negative effect of mutant p53 over WT p53[12]. Of interest, we detected Mdm2 protein in p53-immunoprecipitated cell lysates prepared from ReACp53-treated CWRR1 cells; however, we also noticed that ReACp53 treatment did not change the protein expression of Mdm2 (figure 1E). These results suggest that ReACp53 could affect the physical interaction of mutant p53 protein with Mdm2 when cells are exposed to ReACp53.
The binding studies using p53 truncation mutants have shown that the minimum p53 binding site for Mdm2 can be mapped to the first 50 amino acids of p53 or specifically, a segment between amino acids 18 and 23 [33]. The necessity for an intact p53 tetramerization domain for rapid binding to Mdm2 was also identified[34]. In addition, studies have revealed that several posttranslational modifications of mutant p53 can promote Mdm2-mediated degradation of p53 protein [35]. It is thus suggested that the interaction of Mdm2 and p53 protein is conformation-sensitive, and amyloid aggregation of mutant p53 protein may block the proper docking sites of Mdm2-p53 complex. To this setting, we hypothesized that the conformational change of amyloid aggregated mutant p53 protein induced by ReACp53 treatment could also affect the interaction of p53 with other molecules. Indeed, IP assay showed that ReACp53 treatment also increased the protein association of mutant p53 with Bax in CWRR1 cells. Interestingly, ReACp53 treatment had no obvious effect on the association of WT p53 with Bax (figure 1E).
ReACp53 induces mitochondrial cell death in PCa cells carrying mutant p53 protein
p53 protein can promote an apoptotic cell death of cancer cells not only through target gene activation in the nucleus but also through direct physical interaction of p53 with antiapoptotic Bcl-2 family proteins[12, 36]. For example, activation of Bax by p53 can initiate mitochondrial apoptosis[27, 36]. The evidence showing that treatment with ReACp53 increases the protein-protein association of Bax and mutant p53 thus leads to a hypothesis that exposure to ReACp53 can initiate the mitochondrial proapoptotic signaling in cancer cells bearing mutant p53 protein.
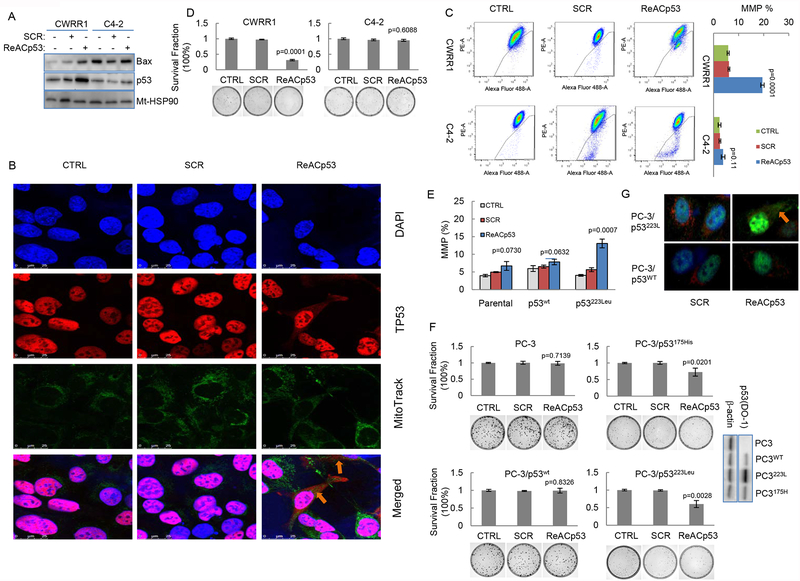
To test this, we determined the potential effects of ReACp53 on mitochondrial accumulations of p53 and Bax proteins in PCa cells. For this, we isolated the mitochonrial fractions (supplemental figure B) from cells treated with or without ReACp53 (10 μM for 48 hours). Western blot results showed that ReACp53 treatment induced mitochondrial accumulations of p53 and Bax in CWRR1 cells (figure 2A). Immunoflurorescence staining with DO-1 antibody, which recognizes both WT and mutant p53, also revealed that, while p53 was detected mainly in the nucleus of untreated cells, ReACp53 was able to induce obvious p53 mitochondrial staining in a cell population of near 17% in CWRR1 cells (figure 2B). However, in C4–2 cells, p53 staining with DO-1 antibody was barely detectable, and no mitochondrial staining was found when cells were treated with ReACp53 (supplemental figure C).
Figure 2:
ReACp53 induces p53-mediated mitochondrial cell death of PCa cells carrying mutant p53 protein. A. Western blot analysis showing ReACp53 treatment induces mitochondrial accumulation of p53 and Bax in CWRR1 cells. Mt-HSP90 was included as a loading control for mitochondrial fraction protein; B. Immunofluorence analysis showing the accumulation of mutant p53 protein in mitochondria in CWRR1 cells treated with ReACp53. Mitochondria was labeled with Mito-Green FM, p53 (DO-1) was labeled with Alexa Fluro 594; C. ReACp53 induces MMP in CWRR1 cells, but not in C4–2 cells. Left: representative results of flow cytometry for MMP detection; Right: Graphs showing the effects of ReACp53 treatment on changing of MMP in CWRR1 and C4–2 cells; D. Clonogenic survival assay showing ReACp53 treatment reduced the clonogenic survival of CWRR1 cells; E. Graphs showing the effects of ReACp53 on MMP in PC-3 cells with engineered expressions of WT or mutant p53; F. ReACp53 inhibits clonogenic cell survival of PC-3 cells with expression of mutant p53 (p53223L or p53175H), but had no effect on MMP in parental PC-3 cells or PC-3 cells expressing WTp53. Western blot showing the engineered expressions of different types of p53 in PC-3 cells Data represent the average of three independent experiments. Error bar indicates standard deviation. Arrows indicate p53 mitochondrial staining; G. Representative images showing ReACp53 induces mitochondrial accumulation of mutant p53223L in PC-3 cells. Mitochondria was labeled with Mito-Red FM, p53 (DO-1) was labeled with Alexa Fluro 488. Cells were treated with 10 μM of ReACp53 for 48 hours.
We next examined the mitochondrial membrane potential (MMP) in these PCa cells. The flow cytometry results showed that exposure to 10 μM ReACp53 for 48 hours caused an increase of MMP from 5.74±0.47 to 19.20±1.40 (%, p=0.0001) in CWRR1 cells (figure 2C). Although ReACp53 treatment also increased MMP percentage from 0.82±0.48 to 3.22±0.36 in C4–2 cells, the difference was not statistically significant (figure 2C). Clonogenic survival analysis further revealed that ReACp53 treatment reduced clonogenic survival of CWRR1 (to percentage of 31.2±2.19, p=0.0001) and DU145 cells (to percentage of 64.2±6.32, p=0.0011) cells. However, no such cell killing effect was observed in clonogenic survival analysis for C4–2 cells (figure 2D and supplemental figure D).
In p53-null PC-3 cells with engineered p53 expression, we also detected that treatment with ReACp53 increased MMP % (from 4.03±0.26 to 13.08±0.25, p=0.0007) in cells that express mutant p53 (p53223Leu). However, no such changes were observed in parental PC-3 or PC-3 cells expressing WT p53 (figure 2E). Consistent with these MMP changes, similar regulatory effects of ReACp53 on clonogenic survival were also observed in engineered PC-3 cells: ReACp53 treatment decreased clonogenic survival of PC-3/p53223Leu and PC-3/p53175His cells, but did not change the clonogenic survival of PC-3/WT p53 or parental PC-3 cells (figure 2F). In addition, IF staining with DO-1 antibody also showed that exposure to ReACp53 induced mitochondrial translocation of p53 protein only in PC-3 cells that express engineered p53223Leu, but not in PC-3 cells expressing WT p53 (figure 2G). These results thus suggest that ReACp53 treatment can cause conformational change of aggregated mutant p53 protein and increase the association of p53 protein with Bcl-2 family proteins which leads to mitochondrial apoptosis in PCa cells carrying mutant p53 protein.
ReACp53 treatment reduces DNA synthesis and inhibits cell proliferation of PCa cells expressing WT or mutant p53 protein
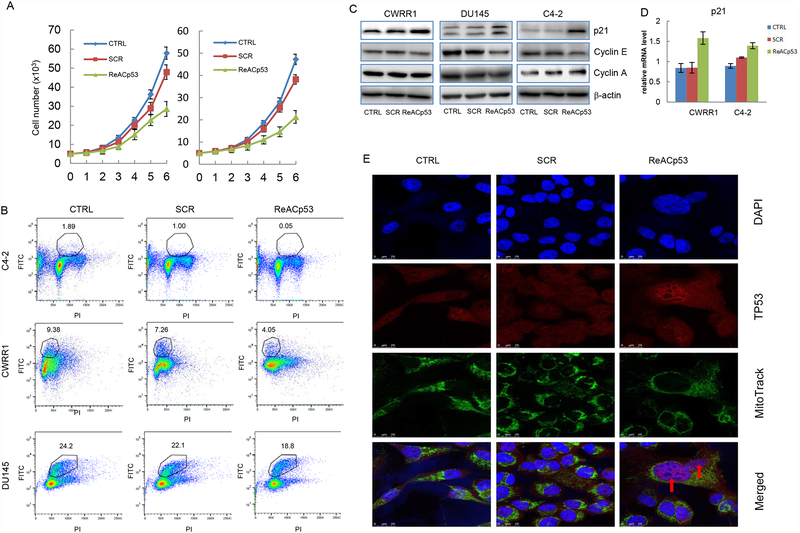
The data shown above demonstrated the inhibitory effect of ReACp53 on cell survival of mutant p53-carrying PCa cells. Of interest, however, we also observed that continuous exposure to 10 μM ReACp53 could inhibit cell proliferation for both C4–2 and CWRR1 cells (figure 3A). In addition, flow cytometry analysis revealed that, while exposure to SCR for 48 hour slightly reduced the BrdU incorporation rates, treatment with 10 μM ReACp53 caused remarkable decreases of BrdU incorporation in C4–2, CWRR1 and DU145 cells (figure 3B and supplemental figure E). These results indicate that treatment with ReACp53 may act on DNA synthesis of PCa cells, regardless of p53 mutation status.
Figure 3.
ReACp53 restores nuclear function of mutant p53 and inhibits DNA synthesis of PCa cells. A. Cell proliferation assays showing the effects of continuous exposure to 10 μM of ReACp53 on cancer cell proliferations of C4–2 (left) and CWRR1 (right) cells; B. BrdU incorporation assay showing ReACp53 treatment decreases DNA synthesis of C4–2, CWRR1 and DU145 cells; C. Western blot analyses showing the ReACp53 treatment increases protein expression of p21 and reduces Cyclin E expression in PCa cells; D. Graphs showing ReACp53 treatment increases p21 transcripts in C4–2 and CWRR1 cells; E. Immunofluorence analysis showing the nuclear accumulation of mutant p53 protein in CWRR1 cells induced by ReACp53 treatment. Nuclear was stained with DAPI, mitochondria was labeled with Mito-Green FM, mutant p53 (PAb240) was labeled with Alexa Fluro 594. Data represent the average of three independent experiments. Error bars represent standard deviation. Arrows indicate p53 nuclear staining.
Dysregulated cell cycle control, including unchecked process of DNA synthesis, is a fundamental feature of cancer cells. In eukaryotic cells, the transition through the G1 to S phase or the transition of the S to G2 phase are tightly controlled by several cell cycle checkpoints, and the kinase activity of Cdk/cyclin complexes and stability of p21, a transcriptional target of p53[37], play vital roles in these processes [38]. We found in our study that exposure to ReACp53 elevated p21 protein level significantly in CWRR1 cells and slightly in DU145 and C4–2 cells (figure 3C). ReACp53 treatment also slightly reduced protein level of cyclin E in these cells, but had no effect on cyclin A expression (figure 3C). We also noticed that exposure to ReACp53 increased p21 transcript level in both C4–2 and CWRR1 cells (figure 3D). These results suggest that ReACp53 treatment may restore the nuclear function of p53 in CWRR1 and DU145 cells, or enhances p53 transcriptional activity in C4–2 cells, resulting in reduced transit of cell cycling from G1 to S. Indeed, the IF immunostaining showed that when cells were stained with PAb240, a conformation-specific antibody that binds only to inactive mutant-like p53, mutant p53 localized to the cytosol and was barely detectable in nuclear in untreated or SCR-treated CWRR1 cells, but in ReACp53-treated cells, a portion of cells (around 30%) were observed with p53 nucleus staining, indicating that ReACp53 can restore p53 to an active conformation and induce nulear localization (figure 3E).
Therapeutic potential of ReACp53 on PCa
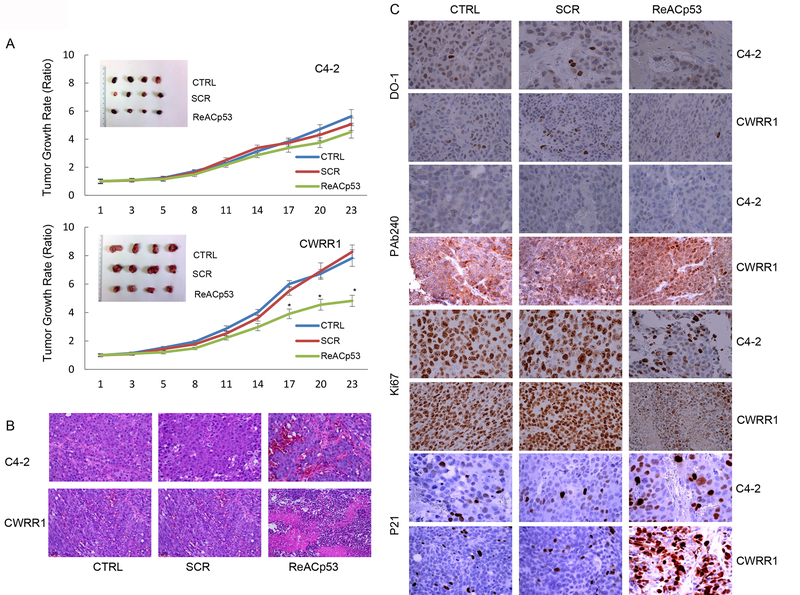
We further tested the therapeutic potential of ReACp53 on PCa. We performed in vivo tumor growth assay with established mouse xenografts of CWRR1 and C4–2 cells. Our results showed that ReACp53 treatment remarkably suppressed the tumor growth of CWRR1-derived, not C4–2-derived xenograft tumors (figure 4A and B, and supplemental figure F). IHC staining showed that: DO-1 antibody-stained p53 protein located in nucleus of xenograft tumor cells, and ReACp53 treatment led to decreased p53 staining only in CWRR1-derived xenograft tumor tissues; Although PAb240 detects both WT and mutant p53 in fixed tissues under denaturation conditions, we barely detected PAb240-stained p53 in C4–2-derived xenograft tumor cells. p53 staining could be detected as puncta in the cytosol of CWRR1-derived xenograft tumor cells, and, in ReACp53-treated xenograft tumor, we observed a portion of tumor cells showing nuclear p53 staining with reduced number of cells with cytosol puncta (figure 4C). We also noticed that ReACp53 treatment reduced Ki67 labeling index, a proliferative biomarker, in tumor cells of xenografts derived from C4–2 and CWRR1 cells. Of interest, we detected increased nuclear staining of p21 in ReACp53-treated xenograft tumors for both C4–2 and CWRR1(figure 4C).
Figure 4.
ReACp53 inhibits tumor growth of PCa xenografts. A. ReACp53 treatment inhibits tumor growth of mouse xenograft derived from CWRR1 cells, but not from C4–2 cells. The growth curves represent the average values for 4 xenograft tumors in each group. Error bars represent standard deviation. * indicates statistical significance (p<0.05); B. H&E staining of xenograft tumors; C. IHC staining showing the effect of ReACp53 treatment on expressions and cellular localization of p53 protein (DO-1), mutant p53 protein (PAb240), p21 protein and Ki67 protein in xenografts established from C4–2 or CWRR1 cells.
ReACp53 increases sensitivity of PCa cells to hormonal therapy and AR-signaling targeting therapy
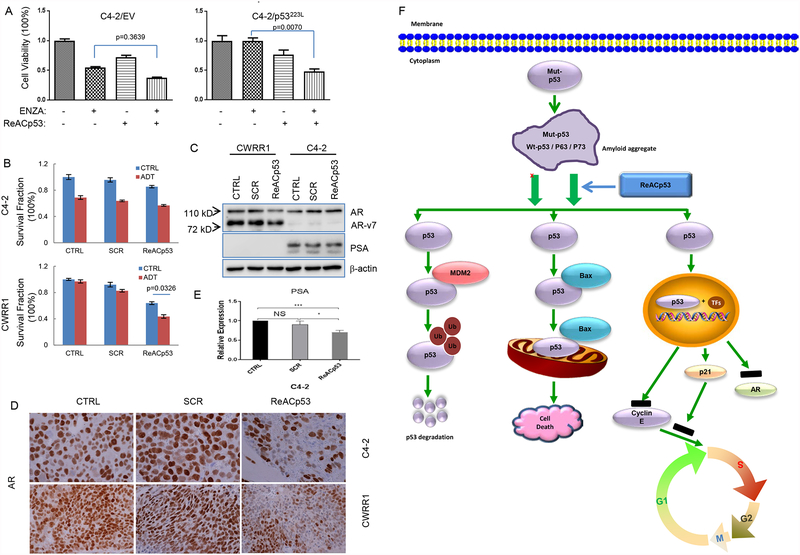
To test whether the observed effects of ReACp53 targeting mutant p53 could also affect the responses of PCa cells to the commonly used treatments for PCa, we used clonogenic survival assay to determine the responses of engineered C4–2 cells to combination treatment of ReACp53 and ENZA. In this study, we used C4–2 cells with overexpression of p53P223L because our results showed p53P223L expression caused more significant resistance to ENZA treatment in C4–2 cells (figure 1A). We found in this experiment that exposure to ReACp53 increased the sensitivity of C4–2 cells with engineered expression of p53P223L to the treatment of ENZA, however, no such effect was observed for control C4–2 cells (figure 5A). We also tested the inhibitory effects of ReACp53 on cell clonogenic survival of CWRR1 and C4–2 cells, and found that ReACp53 pretreatment decreased clonogenic cell survial of CWRR1 cells but not C4–2 cells when cells were undergoing ADT (figure 5B). These results suggest a potential of ReACp53 sensitizing the mutant p53-bearing PCa cells to antiandrogen therapy. Indeed, we detected a slight decrease of AR protein level in cultured CWRR1 cells and in CWRR1-derived xenograft tumor cells after ReACp53 treatment, although no change of PSA was observed because CWRR1 cells lack PSA protein expression (figure 5C and 5D). In C4–2 cells, however, we found ReACp53 treatment decreased transcript level of PSA (figure 5E). These observations indicate that ReACp53 may also affect the AR expression through restored p53 function of the mutant p53 [39], or through the “off target” pathway, in PCa cells.
Figure 5.
ReACp53 increases sensitivity of PCa cells receiving treatments of ADT or ENZA. A. Graphs showing the effects of combination treatment of ReACp53 and ENZA on clonogenic survival of C4–2 cells with engineering expression of mutant p53; B. Graphs showing the effects of ReACp53 on cell vialbility of PCa cells receiving ADT; C. Western blot results showing the effect of ReACp53 on expressions of AR and PSA in PCa cells; D. Representative images of IHC staining showing the effect of ReACp53 treatment on expressions of AR protein in xenografts established from CWRR1 or C4–2 cells; E. Graphs showing the changes of PSA transcripts in C4–2 cells treated with 10 μM of ReACp53 for 48 hours. Data represent the average of three independent experiments. Error bars represent standard deviation. * indicates p<0.05; *** indicates p<0.001; NS represents no statistic significance; F. Model for the action of ReACp53 on cell death and cell proliferation of PCa cells carrying mutant p53 protein. ReACp53 treatment segregates the amyloid aggregation of mutant p53 protein complex and induces binding of Mdm2 and Bax to p53 protein, resulting in the degradation of p53 protein and the translocation of p53-Bax to mitochondria to induce mitochondrial cell death in PCa cells, or restores p53 nuclear function as a transcription factor.
Discussion
Germline or somatic mutations occur constantly in the human body and contribute to not only cancer development but also disease progression[40, 41]. Numerous genetic alterations, such as mutation in TP53, SPOP and FOXA1, gene copy number alteration of MYC, RB1 and PTEN, and DNA structural rearreangment of E26 transformation-specific fusions, have been identified in primary PCa[2, 3]. Clinical studies further revealed that some genetic alterations in primary PCa can predict recurrence/relapse of the disease [42]. Recently, a multi-institutional clinical sequencing study [3]conducted prospective whole-exome and transcriptome sequencing of tumor biopsies from advanced PCa patients (mCRPC), and the results showed 90% of mCRPC patients harbor clinically actionable pathogenic germline or somatic gene alterations. The genetic landscape discovered in this study included gene signatures previsously identified in PCa and new genomic alterasions such as PIK3CA/B, APC, R-spondin and ZBTB16/PLZF. Of interest, TP53 aberration is not only one of the genetic events with highest frequency in mCRPC, but also appears to be enriched in mCRPC compared to primary PCa. Similar results were also reported in other studies [43, 44].
p53 alterations are less common in PCa than in other cancers [5]. However, p53 nuclear overexpression, a surrogate marker of p53 mutation, as measured by IHC staining, has been found to correlate with poor prognosis in primary PCa[45, 46]. Our previous studies have demonstrated that pathogenesis of SCNC involves the inactivation of the p53 pathway as a fundamental molecular change [47]. In addition, studies have also revealed that AR gene amplification or AR regulatory effects on its target genes might associate with p53 mutation, or inactivation, in a subset of CRPC, or hormone-refractory PCa[39, 48]. These findings indicate the importance of p53 mutation in the development of therapeutic failure for PCa, and may also suggest a novel strategy of targeting this molecular alteration for the treatment of aggressive PCa.
ReACp53 is a peptide designed to cap the specific exposed aggregation-prone sequence, centered on Ile254, fused with a cell-penetrating polyarginine tag[25]. Studies have revealed that ReACp53 can readily affect the stabilization and the nuclear localization of mutant p53 protein, and re-activate normal p53 transcriptional activity in p53 mutant ovarian cancer cells, resulting in regulation of protein expression of p53-target genes and interaction of mutant p53 with p53 homologs of p63/p73, increasing apoptotic cell death and reducing cell viability, and inhibiting xenograft tumor growth of p53-mutant tumors; targeting mutant p53 could sensitize lung cancer cells carring mutant p53 to cisplatin treatment[25, 49]. Of interest, ReACp53 has also been reported to affect cancer cells survival of cells expressing WT p53[50].
In this study, we present evidence showing that ReACp53 treatment can act on mutant p53 aggregation, leading to a conformational change of p53 protein which promotes the association of mutant p53 with protein factors that are important for p53 function and stability. As the results of such change, we found that treatment with ReACp53 enhanced the association of mutant p53 with Bax protein and induced mitochondrial cell death; or restored the nuclear function of p53 as a tumor suppressor to regulate gene expressions of cell cycle-related proteins such as p21 and cyclin E with consequences on the transit of cell cycling from G1 to S; exposure to ReACp53 could also downregulate AR expression and change the response of cancer cells to the ADT treatment in PCa cells bearing mutant p53 (figure 5E). Of interest, we noticed that ReACp53 showed slight effects on the expression of cell cycle-related genes and on DNA synthesis of CRPC cells that express WT p53, suggesting a potential of ReACp53 may target the misfolding or amyloid assembly of WT p53 that prevent its normal function [19]. Our results further revealed the inhibitory effect of ReACp53 on tumor growth of PCa xenografts. These observations indicate a potential of ReACp53 to act as a single agent or combination with antiandrogen drugs for the clinical management of advanced PCa, and this novel therapeutic strategy may benefit the patients who are undergoing or failed conventional hormonal therapies.
In conclusion, our data demonstrated that ReACp53 targets amyloid aggregation of and restores the nuclear function of mutant p53, leading to inhibition of cancer cell proliferation. Of interest, our results revealed that ReACp53 treatment also changed response of CRPC cells bearing mutant p53 to AR-targeting therapy. We thus presume that ReACp53 treatment may also have “off target” effects on cancer cell survial, and this needs to be further studied. Nevertherless, the results presented here indicate a potential of ReACp53 as a single therapeutic regimen that could be applied to the treatment of advanced PCa. Additional research along this direction may eventually establish novel, p53 mutation-targeting treatment strategies that can benefit a large portion of advanced PCa (CRPC and SCNC) patients.
Supplementary Material
Acknowledgments
The authors thank Dr. David Eisenberg and Dr. Alice Soragni from David Geffen School of Medicine, UCLA for providing ReACp53 and its scramble control, and for their helpful comments and advice on experimental design.
Fundings
This work is supported by Stand Up to Cancer–Prostate Cancer Foundation–Prostate Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0812). This research grant was made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Jiaoti Huang is supported by grants from the National Institutes of Health (1R01CA181242, 1R01CA172603, 1R01CA205001, 1U54CA217297, 1R01CA212403, 1R01CA200853), Department of Defense Prostate Cancer Research Program (PC150382), and Prostate Cancer Foundation (Joyce and Larry Stupski Prostate Cancer Precision Oncology Special Challenge award).
Lingfan Xu is supported by grants from Youth Culturing Plan of National Natural Science Foundation (2018Kj16) and Anhui Natural Science Foundation (1908085QH337).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Reference
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;162(2):454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009;137(3):413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Leroy B, Fournier JL, Ishioka C, Monti P, Inga A, Fronza G, et al. The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res 2013;41(Database issue):D962–9. doi: 10.1093/nar/gks1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2010;2(3):a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He C, Li L, Guan X, Xiong L, Miao X. Mutant p53 Gain of Function and Chemoresistance: The Role of Mutant p53 in Response to Clinical Chemotherapy. Chemotherapy 2017;62(1):43–53. doi: 10.1159/000446361. [DOI] [PubMed] [Google Scholar]
- 8.Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, et al. The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J 2006;25(21):5191–200. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva JL, De Moura Gallo CV, Costa DC, Rangel LP. Prion-like aggregation of mutant p53 in cancer. Trends Biochem Sci 2014;39(6):260–7. doi: 10.1016/j.tibs.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem 2008;77:557–82. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 11.Sabapathy K The Contrived Mutant p53 Oncogene - Beyond Loss of Functions. Front Oncol 2015;5:276. doi: 10.3389/fonc.2015.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 13.Lei J, Qi R, Wei G, Nussinov R, Ma B. Self-aggregation and coaggregation of the p53 core fragment with its aggregation gatekeeper variant. Phys Chem Chem Phys 2016;18(11):8098–107. doi: 10.1039/c5cp06538k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa DC, de Oliveira GA, Cino EA, Soares IN, Rangel LP, Silva JL. Aggregation and Prion-Like Properties of Misfolded Tumor Suppressors: Is Cancer a Prion Disease? Cold Spring Harb Perspect Biol 2016;8(10). doi: 10.1101/cshperspect.a023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stindt MH, Muller PA, Ludwig RL, Kehrloesser S, Dotsch V, Vousden KH. Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene. 2015;34(33):4300–10. Epub 2014/11/25. doi: 10.1038/onc.2014.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rangel LP, Costa DC, Vieira TC, Silva JL. The aggregation of mutant p53 produces prion-like properties in cancer. Prion 2014;8(1):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefani M Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta 2004;1739(1):5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol 2010;11(4):301–7. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva JL, Vieira TC, Gomes MP, Bom AP, Lima LM, Freitas MS, et al. Ligand binding and hydration in protein misfolding: insights from studies of prion and p53 tumor suppressor proteins. Acc Chem Res 2010;43(2):271–9. doi: 10.1021/ar900179t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J Biol Chem 2012;287(33):28152–62. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy CB, Stumbo AC, Ano Bom AP, Portari EA, Cordeiro Y, Silva JL, et al. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int J Biochem Cell Biol 2011;43(1):60–4. doi: 10.1016/j.biocel.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 22.Higashimoto Y, Asanomi Y, Takakusagi S, Lewis MS, Uosaki K, Durell SR, et al. Unfolding, aggregation, and amyloid formation by the tetramerization domain from mutant p53 associated with lung cancer. Biochemistry. 2006;45(6):1608–19. doi: 10.1021/bi051192j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigacci S, Bucciantini M, Relini A, Pesce A, Gliozzi A, Berti A, et al. The (1–63) region of the p53 transactivation domain aggregates in vitro into cytotoxic amyloid assemblies. Biophys J 2008;94(9):3635–46. doi: 10.1529/biophysj.107.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishimaru D, Andrade LR, Teixeira LS, Quesado PA, Maiolino LM, Lopez PM, et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42(30):9022–7. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- 25.Soragni A, Janzen DM, Johnson LM, Lindgren AG, Thai-Quynh Nguyen A, Tiourin E, et al. A Designed Inhibitor of p53 Aggregation Rescues p53 Tumor Suppression in Ovarian Carcinomas. Cancer cell 2016;29(1):90–103. Epub 2016/01/11. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory CW, Johnson RT Jr., Mohler JL, French FS, Wilson EM Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer research. 2001;61(7):2892–8. Epub 2001/04/18. [PubMed] [Google Scholar]
- 27.Chen X, Wong JY, Wong P, Radany EH. Low-dose valproic acid enhances radiosensitivity of prostate cancer through acetylated p53-dependent modulation of mitochondrial membrane potential and apoptosis. Molecular cancer research : MCR. 2011;9(4):448–61. Epub 2011/02/10. doi: 10.1158/1541-7786.MCR-10-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. http://p53.free.fr/Database/Cancer_cell_lines/cell_lines_1.0.pdf.
- 29.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer research. 1994;54(10):2577–81. [PubMed] [Google Scholar]
- 30.Gurova KV, Rokhlin OW, Budanov AV, Burdelya LG, Chumakov PM, Cohen MB, et al. Cooperation of two mutant p53 alleles contributes to Fas resistance of prostate carcinoma cells. Cancer research. 2003;63(11):2905–12. [PubMed] [Google Scholar]
- 31.van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57(3):205–25. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 32.Tai S, Sun Y, Squires JM, Zhang H, Oh WK, Liang CZ, et al. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate. 2011;71(15):1668–79. doi: 10.1002/pros.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2;--fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9(9):2523–9. [PubMed] [Google Scholar]
- 34.Burch LR, Midgley CA, Currie RA, Lane DP, Hupp TR. Mdm2 binding to a conformationally sensitive domain on p53 can be modulated by RNA. FEBS Lett 2000;472(1):93–8. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto M, Furihata M, Ohtsuki Y. Posttranslational phosphorylation of mutant p53 protein in tumor development. Med Mol Morphol. 2006;39(2):79–87. doi: 10.1007/s00795-006-0320-0. [DOI] [PubMed] [Google Scholar]
- 36.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303(5660):1010–4. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 37.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2010;2(8):a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–93. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 39.Alimirah F, Panchanathan R, Chen J, Zhang X, Ho SM, Choubey D. Expression of androgen receptor is negatively regulated by p53. Neoplasia. 2007;9(12):1152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A 2006;103(48):18238–42. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014;111(30):11139–44. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soundararajan R, Aparicio AM, Logothetis CJ, Mani SA, Maity SN. Function of Tumor Suppressors in Resistance to Antiandrogen Therapy and Luminal Epithelial Plasticity of Aggressive Variant Neuroendocrine Prostate Cancers. Front Oncol 2018;8:69. doi: 10.3389/fonc.2018.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai HK, Lehrer J, Alshalalfa M, Erho N, Davicioni E, Lotan TL. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer. 2017;17(1):759. doi: 10.1186/s12885-017-3729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khemlina G, Ikeda S, Kurzrock R. Molecular landscape of prostate cancer: implications for current clinical trials. Cancer Treat Rev 2015;41(9):761–6. doi: 10.1016/j.ctrv.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer. 2014;135(6):1369–80. doi: 10.1002/ijc.28784. [DOI] [PubMed] [Google Scholar]
- 47.Chen H, Sun Y, Wu C, Magyar CE, Li X, Cheng L, et al. Pathogenesis of prostatic small cell carcinoma involves the inactivation of the P53 pathway. Endocr Relat Cancer. 2012;19(3):321–31. doi: 10.1530/ERC-11-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guseva NV, Rokhlin OW, Bair TB, Glover RB, Cohen MB. Inhibition of p53 expression modifies the specificity of chromatin binding by the androgen receptor. Oncotarget. 2012;3(2):183–94. doi: 10.18632/oncotarget.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Hu Y, Wang JL, Yao H, Wang H, Liang L, et al. Proteomic identification of ERP29 as a key chemoresistant factor activated by the aggregating p53 mutant Arg282Trp. Oncogene. 2017;36(39):5473–83. doi: 10.1038/onc.2017.152. [DOI] [PubMed] [Google Scholar]
- 50.Wang G, Fersht AR. Multisite aggregation of p53 and implications for drug rescue. Proc Natl Acad Sci U S A 2017;114(13):E2634–E43. doi: 10.1073/pnas.1700308114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.