A comparison of different CRISPR systems and promoters using Agrobacterium-mediated transformation in wheat shows the SpCas9 system to be the most efficient for genome editing, and a high haploid induction rate is achieved by editing TaMTL.
Keywords: Agrobacterium-mediated transformation, genome editing, haploid induction, TaMTL, TaWaxy, wheat
Abstract
The use of CRISPR/LbCpf1 and CRISPR/xCas9 systems in wheat have not yet been reported. In this study, we compared the efficiencies of three CRISPR editing systems (SpCas9, LbCpf1, and xCas9), and three different promoters (OsU6a, TaU3, and TaU6) that drive single-guide (sg)RNA, which were introduced into wheat via Agrobacterium-mediated transformation. The results indicated that TaU3 was a better choice than OsU6a or TaU6. The editing efficiency was higher using two sgRNAs than one sgRNA, and mutants with a large fragment deletion between the two sgRNAs were produced. The LbCpf1 and xCas9 systems could both be used successfully. Two endogenous genes, TaWaxy and TaMTL, were edited with high efficiency by the optimized SpCas9 system, with the highest efficiency (80.5%) being achieved when using TaU3 and two sgRNAs to target TaWaxy. Rates of seed set in the TaMTL-edited T0 transgenic plants were much lower than that of the wild-type. A haploid induction rate of 18.9% was found in the TaMTL-edited T1 plants using the CRISPR/SpCas9 system. Mutants with reverse insertion of the deleted sequences of TaMTL and TaWaxy between the two sgRNAs were identified in the edited T0 plants. In addition, wheat grains lacking embryos or endosperms were observed in the TaMTL-edited T1 generation.
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) is a type of bacterial defense system that degrades alien DNA, and it functions with various CRISPR-associated proteins (Cas9). Since its discovery, this system has been widely used in animals and plants for precise gene modification, especially the type-II CRISPR/SpCas9 system from Streptococcus pyogenes. The CRISPR/SpCas9 system is characterized by its efficiency and simplicity, and can recognize the protospacer-adjacent motif (PAM) site NGG with the assistance of trans-activating crRNA (tracrRNA). To date, genes of many crops have been edited using this technique, including maize, rice, wheat, soybean, barley, sorghum, potato, tomato, flax, and cotton (Ricroch et al., 2017; Zhang et al., 2018a; Abe et al., 2019). The most frequent applications of the CRISPR/SpCas9 system are to produce gene-knockouts or null alleles, which are mainly achieved by the introduction of small indels that lead to frame-shift mutations. However, the CRISPR/SpCas9 system can only recognize DNA sequences upstream of the appropriate 5´-NGG-3´ PAMs, which limits the number of potential target sites. SpCas9 variants are therefore needed to overcome this restriction.
The recently identified type-II system, Cpf1 (CRISPR from Prevotella and Francisella 1), has distinct features compared to SpCas9. It is a single RNA-guided endonuclease that recognizes the thymidine-rich PAM and generates cohesive ends with four or five nucleotide overhangs rather than blunt-end breaks. Cpf1 is a dual nuclease that not only cleaves target DNA but also processes its own CRISPR RNA (crRNA) (Fonfara et al., 2016; Zetsche et al., 2017). Moreover, the maturation of crRNA by Cpf1 does not require the assistance of tracrRNA. The CRISPR/Cpf1 system also considerably expands the characteristics of SpCas9. It not only has genome-editing activity in mammalian cells, rice, Arabidopsis, soybean, and tobacco, but also has multiple gene-editing activity in mammalian cells and rice, where up to four genes can be simultaneously edited by Cpf1 using a single crRNA array spaced by mature direct repeats (Tang et al., 2017; Wang et al., 2017b; Zetsche et al., 2017).
The SpCas9 variant xCas9, generated using phage-assisted continuous evolution, is reported to recognize a broad range of PAM sequences in mammals and rice, including NG, GAA, and GAT (Hu et al., 2018; Wang et al., 2018). The xCas9 3.7 system can recognize GAT, GAA, and NG PAM sites in mammalian cells and rice, and performs better than other Cas9 variants (Hu et al., 2018; Wang et al., 2018). Moreover, xCas9 can efficiently induce mutations at target sites with NG and GAT PAM sequences in rice. Hua et al. (2019) reported comparable editing efficiencies between xCas9 3.6 and xCas9 3.7 at the CGG, TGA, and CGT PAM sites, while xCas9 3.6 is 7.4 and 3.4 times as efficient as xCas9 3.7 at the AGC and GAT PAM sites, respectively. The most recent study found that xCas9 3.7 exhibits nearly equivalent editing efficiency to SpCas9 at most canonical NGG PAM sites, whereas it shows limited activity at non-canonical NGH (H=A, C, T) PAM sites (Zhong et al., 2019a).
The editing efficiencies of CRISPR/Cas9 and CRISPR/Cpf1 have recently been compared in maize (Lee et al., 2019), but a similar comparison has not yet been carried out in wheat. Although there have been some studies on the application of the CRISPR/SpCas9 system for genome editing in wheat (Wang et al., 2014; Zhang et al., 2018b, 2019b), the use of the CRISPR/Cpf1 and CRISPR/xCas9 has not yet been reported. This may be due to the complicated genome of common wheat: many genes exist in at least three copies, and thus targeted genome editing is more difficult to achieve. In addition, wheat transformation is still a difficult task in many laboratories, and gene editing using CRISPR/Cpf1 and CRISPR/xCas9 requires a highly efficient transformation system as its basis.
Doubled-haploid technology substantially accelerates the breeding process for many crop species. In the past four decades, anther culture and microspore culture have been widely used to produce wheat haploid plants (Machii et al., 1998; Liu et al., 2002). However, strong genotype dependency exists in these two culture techniques as well as low plant regeneration frequency, and they involve complicated manipulation steps. Chromosome elimination techniques through wide crossing between wheat and maize or barley can generate wheat haploids, but well-controlled environmental are required conditions for growing the plants and for the rescue culture for the immature haploid embryos (Barclay 1975; Laurie et al., 1986; Zhang et al., 2014b). In maize, haploid plants can easily be obtained in vivo using an inbred haploid-inducer line, and many good inbred lines have been developed for efficient and fast breeding using this technique (Ishii et al., 2016; Yao et al., 2018).
Despite the problems, the CRISPR mutation system has great potential for editing economically important endogenous genes of wheat, such as MATRILINEAL (MTL) and Waxy. MTL is a pollen-specific phospholipase and can trigger haploid induction in maize by a frame-shift mutation (Kelliher et al., 2017; Liu et al., 2017). Knockout of ZmDMP can increase the haploid induction rate (HIR) by 5–6-fold in the presence of MTL/ZmPLA/NLD (Zhong et al., 2019b). The knockout of OsMATL can reduce seed-set and lead to a 2–6% haploid induction rate in rice (Yao et al., 2018). In wheat, three orthologs of TaMTL, namely TraesCS4A02G018100, TraesCS4B02G286000, and TraesCS4D02G284700, have been located on chromosomes 4A, 4B, and 4D, respectively. Waxy in wheat encodes granule-bound starch synthase I, which is required for the synthesis of amylose (Yamamori et al., 1994) and influences starch composition and flour quality. In wheat, three orthologs of TaWaxy, namely TraesCS4A02G418200, TraesCS7A02G070100, and TraesCS7D02G064300, are located on chromosomes 4A, 7A, and 7D, respectively.
The objectives of this study were to develop a well-performing editing system for wheat, and to implement it to edit selected important example genes. First, we compared the efficiencies of the three editing systems, CRISPR/SpCas9, CRISPR/LbCpf1, and CRISPR/xCas9 using Agrobacterium-mediated transformation of a marker-free transgenic wheat line H29, which only carries a single copy of the GUS gene as the target (Wang et al., 2017a; Liu et al., 2020b). We then applied the optimized system to edit TaMTL and TaWaxy to develop the induction of haploids and to improve starch quality.
Materials and methods
Plant materials, plasmids, and bacteria strains
A marker-free transgenic wheat (Triticum aestivum) line H29 and the two varieties Fielder and Ningchun4 were used for transformation in this study. Line H29 was obtained by our laboratory from a transformation experiment using the cultivar Xinchun9 as a receptor, and it carries a single copy of the GUS gene and lacks the bar gene. In addition, GUS was inserted into the distal region of a pair of wheat chromosomes in H29 (Liu et al., 2020b). Fielder and Ningchun4 were acquired from the Crop Germplasm Bank of China. All the plants were grown in pots (20×30 cm) in a growth chamber maintained at 24 °C, 16/8 h light/dark with 300 μmol m–2 s–1 light intensity at 45% humidity. For full details of cultivation see Wang et al., (2017a).
The plasmid pWMB110 containing the bar gene as a selection marker for generating transgenic plants and the maize (Zea mays) ubi promoter for driving the expression of a target gene or DNA sequence within a T-DNA region had previously been constructed by our laboratory (Supplementary Fig. S1 at JXB online). Vectors containing SpCas9 and LbCpf1 (Lachnospiraceae bacterium ND2006 Cpf1) were kindly provided by Prof. Yaoguang Liu (Southern China Agricultural University) and Prof. Chuanxiao Xie (Institute of Crop Sciences of Chinese Academy of Agricultural Sciences), respectively. Agrobacterium strain C58C1 and an E. coli strain containing the helper plasmid pRK2013 were kindly provided by Dr Tom Clemente (University of Nebraska-Lincoln, USA).
Construction of vectors for gene editing
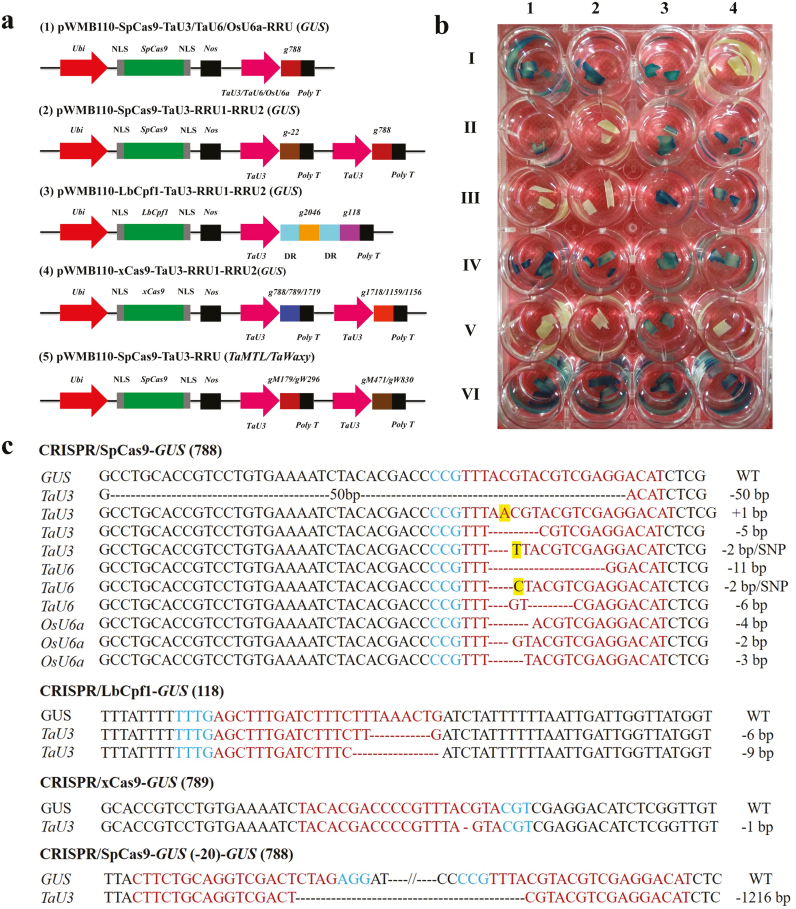
The recombination vectors for the CRISPR/SpCas9, CRISPR/LbCpf1, and CRISPR/xCas9 systems were constructed based on methods described previously (Ma et al., 2015; Wang et al., 2017b; Hu et al., 2018), and in addition xCas9 was synthesized using the sequence described by Hu et al. (2018). The promoters of TaU3 (GenBank accession number X63065.1), TaU6 (X63066.1), and rice (Oryza sativa) OsU6a (KR029106.1) were synthesized and cloned into the plasmid pWMBX110-SpCas9 vector to generate the plasmids pWMB110-SpCas9-TaU3, pWMB110-SpCas9-TaU6, and pWMB110-SpCas9-OsU6a (Supplementary Fig. S1; see also Results). The TaU3 promoter was also cloned into the vectors pWMB110-LbCpf1 and pWMB110-xCas9 to generate the plasmids pWMB110-LbCpf1-TaU3 and pWMB110-xCas9-TaU3, respectively. The single-guide (sg)RNAs of the GUS gene (JN593326.1) were driven by the TaU3, TaU6 and OsU6a promoters for the CRISPR/SpCas9 system and by the TaU3 promoter for both the CRISPR/LbCpf1 and CRISPR/xCas9 systems. The sgRNAs of TaMTL and TaWaxy were all driven by the TaU3 promoter. To target GUS in line H29, we designed both a single sgRNA (g788) and two sgRNAs (g-22 and g788) for the CRISPR/SpCas9 system, two crRNAs (g118 and g2046) for the CRISPR/LbCpf1 system, and three pairs of sgRNAs (g788 and g1718, g789 and g1159, g1719 and g1156) for the CRISPR/xCas9 system (Supplementary Table S1). Two pairs of sgRNAs, one to target TaMTL (TaMTL-179 and TaMTL-471) and the other to target TaWaxy (TaWaxy-296 and TaWaxy-830) were designed for the CRISPR/SpCas9 system.
The detailed single-guide (sg)RNA sequences of GUS were designed based on the respective PAM sites of the CRISPR/SpCas9, CRISPR/LbCpf1, and CRISPR/xCas9 systems and the restriction enzyme sites. The gRNA sequences of the three copies each of TaMTL and TaWaxy were designed by aligning their conserved sequences and further selecting 20 nucleotides upstream of a PAM motif (5´-NGG-3´) and restriction enzyme sites for the CRISPR/SpCas9 system (see Results). The primers for plasmid construction and detection of transgenic plants are listed in Supplementary Table S2. All the expression vectors were introduced into Agrobacterium strain C58C1 by triparental mating (Ditta et al., 1980).
Agrobacterium-mediated transformation
Immature wheat grains were collected 2 weeks after anthesis, sterilized with 75% ethanol for 1 min and 5% sodium hypochlorite for 15 min, and then washed five times with sterile water in aseptic conditions. Immature wheat embryos were isolated and Agrobacterium-mediated transformation was used to obtain transgenic plants following the protocol described by Wang et al. (2017a). Transgenic plants were transplanted into pots and cultivated in a growth chamber under the same conditions as described above.
Detection analysis of edited mutations
Genomic DNA was extracted from candidate T0 transgenic mutant plants using a FastPure Plant DNA Isolation Mini Kit (Vazyme Biotech Co., Ltd) for digestion and deep sequencing. The targeted genes GUS, TaMTL and TaWaxy were amplified using their respective specific primers (Supplementary Table S2). A PCR-restriction enzyme (PCR-RE) assay was performed for these genes, where the reactions consisted of the corresponding restriction enzymes (1 U each) in 20 μl reaction buffer including 10 μl PCR product and were digested for 2 h at 37 °C. The resultant products were separated in a 2% agarose gel and visualized using a GelDoc XR System (BioRad). To distinguish different mutant types, the PCR products were subcloned into the pMD18-T vector (TaKaRa) and sequenced. For each mutant sample, at least five positive colonies were randomly selected and sequenced. The mutations were identified by aligning the reference sequences.
GUS-edited T0 transgenic plants that were detected by enzyme digestion or sequencing were confirmed by histochemical staining of fresh young leaves for GUS expression, as described by Jefferson et al. (1987).
Cytological and phenotypic detection of TaMTL-edited mutations
Immature embryos at 2 weeks post-anthesis (from sterilized immature grains) of the TaMTL-edited T0 transgenic plants (as detected by enzyme digestion or Sanger sequencing) were cultured on MS medium. After 1 week of growth, the root tips from the germinated embryos were collected for chromosome observation as described by Han et al. (2004). The spikes and grains of the TaMTL-edited T0 transgenic plants at near-maturity stage were visually examined and rates for seed set were recorded.
Pieces of flag leaf of 2 cm length were sampled from the TaMTL-edited plants at the booting stage, and placed on glass slides with the abaxial epidermis uppermost. The mesophyll tissues of the samples were carefully scraped off using a sharp knife. The stomatal guard cells on the epidermis were observed and measured under an optical microscope incorporating a mirror micrometer (Zhang et al., 2014b).
Quantitative reverse-transcriptase PCR
Anther was sampled from the TaMTL-edited plants QD33-3, QD33-14, and QD33-26 and the wild-type Fielder plants in order to determine the transcript abundances of TaMTL-4A, TaMTL-4B, and TaMTL-4D by quantitative reverse-transcriptase (qRT-)PCR. Total RNA was extracted using Trizol reagent (Invitrogen). cDNA synthesis was then carried out using HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech Co., Ltd), and 1 μg of the total RNA was reverse-transcribed using 5×HiScript III qRT SuperMix at 37 °C for 15 min and at 85 °C for 5 s. Finally, qRT-PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd) in a 7500 Fast Real-Time PCR system (Applied Biosystems). The specific primers for TaMTL-4A, TaMTL-4B, and TaMTL-4D are listed in Supplementary Table S2. Transcript abundance was expressed relative to that of TaADP using the 2–2ΔΔCT method (Livak and Scmittgen, 2001).
Results
Optimization of promoters for sgRNA regulation in the CRISPR/SpCas9 system
Editing efficiencies were determined using three promoters for controlling sgRNA expression: the rice U6 promoter (OsU6a), and the wheat U3 (TaU3) and U6 promoters (TaU6). The widely used SpCas9 was driven by the maize ubiquitin promoter, and the same sgRNA targeting GUS was controlled by the OsU6a, TaU3, or TaU6 promoters (Fig. 1a). The three different vectors were transformed using A. tumefaciens into the marker-free transgenic wheat line H29, which only has a single copy of GUS. PCR-RE assays and Sanger sequencing showed that the editing efficiencies of the different promoters in plants of the T0 generation were very different (Table 1, Supplementary Fig. S2). The editing efficiency of the OsU6a promoter was only 21.6% and most of the mutant plants were heterozygous. The editing efficiency of the TaU6 promoter was 36.0%, and only 4.9% of the mutants were bi-allelic (3 out of 61 plants) while 31.1% were heterozygous (19 out of 61 plants). Interestingly, the mutation efficiency was 61.4% for the TaU3 promoter; moreover, 20.0% and 41.4% of the mutants were bi-allelic and heterozygous, respectively (Table 1). These results clearly demonstrated that of the three promoters tested, TaU3 was the best choice for driving sgRNA expression in Agrobacterium-mediated genome editing in wheat.
Fig. 1.
Genome editing in wheat using the CRISPR/SpCas9, CRISPR/LbCpf1, and CRISPR/xCas9 systems. (a) Linearized CRISPR/SpCas9, CRISPR/LbCpf1, and CRISPR/xCas9 constructs. ubi, Zea mays ubi promoter; NLS, nuclear localization signal; Nos, Nos terminator; TaU3/TaU6/OsU6a, different sgRNA promoters; Poly T, a 7-bp poly T sequence; RRU, sgRNA (GUS, TaWaxy, TaMTL) or crRNA (GUS) ribozyme units; DR, direct repeat. (b) The expression of GUS protein in edited T0 transgenic plants. The heterozygous mutants are I1–2, II3–4, III3–4, IV1–4; the bi-allelic mutants are II2, III1–2, V1–2, 4; non-mutant plants are V3, VI1–4., VI2, VI3 and VI4; H29 is I3; and wild-type Xinchun9 is I4. (c) InDel mutations in GUS from edited T0 transgenic plants. Blue letters indicate the PAM sequences, red letters indicate the sgRNA or crRNA sequences, and the dashed lines represent nucleotide deletions. Insertions and SNPs are shaded in yellow, and the size of the deletion or insertion is shown on the right. SNP, single-nucleotide polymorphism.
Table 1.
Summary of the target sequences and mutations in wheat T0 plants obtained using the CRISPR/SpCas9, /LbCpf1, and /xCas9 editing systems
| Target loci | Promoter | PAM-guide sequence (5´–3´) | No. of transgenic plants | No. of mutant plants | Mutation rate % | Genotypes obtained |
|---|---|---|---|---|---|---|
| SpCas9-S (one sgRNA) | ||||||
| GUS-788 | TaU3 | CCGTTTACGTACGTCGAGGACAT | 70 | 43 | 61.4 | 14Bi + 29He + 27WT |
| GUS-788 | TaU6 | CCGTTTACGTACGTCGAGGACAT | 61 | 22 | 36.0 | 3Bi + 19He + 39WT |
| GUS-788 | OsU6a | CCGTTTACGTACGTCGAGGACAT | 74 | 16 | 21.6 | 2Bi + 14He + 58WT |
| SpCas9-D (two sgRNAs) | ||||||
| GUS--22 | TaU3 | CTTCTGCAGGTCGACTCTAGAGG | 164 | 75 | 45.7 | 24Bi + 51He + 89WT |
| GUS-788 | TaU3 | CCGTTTACGTACGTCGAGGACAT | 164 | 106 | 64.6 | 34Bi + 72He + 58WT |
| LbCpf1 | ||||||
| GUS-118 | TaU3 | TTTGAGCTTTGATCTTTCTTTAAACTG | 65 | 2 | 3.1 | 2He + 63WT |
| GUS-2046 | TaU3 | TTTCGGCTACAAGAACGCTAGCCATCAC | 65 | 0 | 0 | 65WT |
| xCas9 3.7 | ||||||
| GUS-788 | TaU3 | CCGTTTACGTACGTCGAGGACAT | 71 | 32 | 45.1 | 32He + 39WT |
| GUS-789 | TaU3 | TACACGACCCCGTTTACGTACGT | 67 | 1 | 1.5 | 1He + 66WT |
PAM, protospacer-adjacent motif, with the target sequences highlighted in bold. SpCas9-S, containing a single sgRNA; SpCas9-D, containing two sgRNAs; Bi, bi-allele; He, heterozygote; WT, wild-type.
Comparison of the editing efficiencies of the three CRISPR systems
Two sgRNAs (g-22 and g788) together targeting GUS were designed to be driven by the TaU3 promoter and their total editing efficiencies were compared with the single sgRNA g788 targeting the GUS gene (Fig. 1a). Among the 164 transgenic plants obtained, PCR-RE assays and Sanger sequencing identified 75 and 106 plants with mutations at the g-22 and g788 sites, respectively, and 70 plants with simultaneous mutations at both sites. Their editing efficiencies were 45.7%, 64.6%, and 42.7%, respectively (Table 1, Supplementary Table S3). The total editing efficiency was up to 70.1% for plants in which a mutation occurred in one or two loci (Supplementary Table S3). Simultaneous mutations at the two sites, which can lead to a large fragment deletion, had an efficiency of 37.2% (Table 2). The results indicated that the combination of the two sgRNAs could result in a higher editing efficiency compared to a single sgRNA. Furthermore, the combination of sgRNAs could lead to a large fragment deletion.
Table 2.
Summary of the target sequences and mutations of GUS, and different TaMTL and TaWaxy homologous genes in T0 wheat plants edited using the CRISPR/SpCas9-D editing system containing two sgRNAs
| Target loci | PAM-guide sequence (5´–3´) | No. of transgenic plants | No. of mutant plants | Mutation rate % | Genotypes obtained | LFD | LFD rate (%) |
|---|---|---|---|---|---|---|---|
| GUS-22 | CTTCTGCAGGTCGACTCTAGAGG | 164 | 75 | 45.7 | 24Bi + 51He + 89WT | 61 | 37.2 |
| GUS-788 | CCGTTTACGTACGTCGAGGACAT | 106 | 64.6 | 34Bi + 72He + 58WT | |||
| TaMTL4A-179 | CCAAGCTGCAGGAGCTGGACGGC | 101 | 35 | 34.7 | 11Bi + 24He + 66WT | 9 | 8.9 |
| TaMTL4A-471 | CCGCGGTGACCGCATCGCTGAGG | 36 | 35.6 | 13Bi + 23He + 65WT | |||
| TaMTL4B-179 | CCAAGCTGCAGGAGCTGGACGGG | 101 | 19 | 18.8 | 6Bi + 13He + 82WT | 6 | 5.9 |
| TaMTL4B-471 | CCGCGGTGACCGCGTCGCTGAGG | 9 | 8.9 | 7Bi + 2He + 92WT | |||
| TaMTL4D-179 | CCAAGCTGCAGGAGCTGGACGGG | 101 | 24 | 23.8 | 13Bi + 11He + 77WT | 8 | 7.9 |
| TaMTL4D-471 | CCGCGGTGACCGCGTCGCTGAGG | 16 | 15.8 | 8Bi + 8He + 85WT | |||
| TaWaxy4A-296 | GGCGGCCTCGGCGACGTCCTCGG | 87 | 35 | 40.2 | 25Bi + 10He + 52WT | 10 | 11.5 |
| TaWaxy4A-830 | AAGACCAAGGAGAAGATCTACGG | 36 | 41.4 | 29Bi + 7He + 51WT | |||
| TaWaxy7A-296 | GGCGGCCTCGGCGACGTCCTCGG | 87 | 39 | 44.8 | 19Bi + 20Bi + 48WT | 11 | 12.6 |
| TaWaxy7A-830 | AAGACCAAGGAGAAGATCTATGG | 47 | 54.0 | 25Bi + 22He + 40WT | |||
| TaWaxy7D-296 | GGCGGCCTCGGCGACGTCCTCGG | 87 | 23 | 26.4 | 10Bi + 13He + 64WT | 14 | 16.1 |
| TaWaxy7D-830 | AAGACCAAGGAGAAGATCTACGG | 31 | 35.6 | 18Bi + 13He + 56WT |
PAM, protospacer-adjacent motif, with the target sequences highlighted in bold. Bi, bi-allele; He, heterozygote; WT, wild-type; LFD, large fragment deletion.
The TaU3 promoters were also used to drive the expression of two sgRNAs targeting GUS in order to compare the editing efficiencies of CRISPR/SpCas9, CRISPR/xCas9, and CRISPR/LbCpf1 in wheat. The PAM site for LbCpf1 is TTTN, and two sgRNAs (g118 and g2046) with direct repeats (DRs) were designed to target this site (Fig. 1a). PCR-RE assays and Sanger sequencing identified only two of the 65 transgenic plants as having mutations in the g118 target, and the editing efficiency was only 3.1% (Table 1). The two plants were heterozygous mutants; one had a 6-bp deletion and the other had a 9-bp deletion in the target sequence (Fig. 1c). In contrast, no mutations were detected within the g2046 target (Table 1).
The xCas9 3.7 system was employed to edit GUS at different PAMs, namely NGG (788), NGA (1718), NGT (789), NGC (1159), GAA (1719), and GAT (1156), and the target sequences of NGG and NGA, NGT and NGC, and GAA and GAT were designed as pairs in the CRISPR/xCas9 vector (Fig. 1a). Out of 71 transgenic plants obtained, PCR-RE assays and Sanger sequencing identified 32 plants with mutations at the NGG PAM site, with an editing efficiency of 45.1%. All the mutant plants were heterozygous. With regards to the NGT PAM site, only one mutant was identified out of 67 transgenic plants obtained (Table 1); this plant was heterozygous with a 1-bp deletion (Fig. 1c). There were no mutations detected at the NGA, NGC, GAA, or GAT PAM sites in the 71, 67, 79, and 79 transgenic plants obtained, respectively. Even though the target sequence was the same in the two systems xCas9 edited the NGG PAM site with lower efficiency than SpCas9.
Inheritance of the mutation sites in the edited plants
The segregation of the bi-allelic and heterozygous mutants derived from the CRISPR/SpCas9 system were examined in the T1 generation, in which the expression of the sgRNA for GUS was driven by the TaU3 promoter. All the original bi-allelic mutants were homozygous or bi-allelic in the T1 generation and 21 transgene-free mutants were obtained from a total of 87 plants. Of the T1 progeny from the mutants that were heterozygous in the T0 generation, 19 were homozygous, 56 were heterozygous, and 20 had no mutations, as determined by PCR-RE assays. The T1 segregation ratio was 1:2:1 (P>0.05) and in accordance with Mendelian heritance patterns. GUS was silenced in the homozygous and bi-allelic mutants in which the deleted base pairs could not be divided by 3 (Fig. 1b), implying that the GUS gene-silencing mutations were induced by the editing system. Sanger sequencing revealed that all the mutations in GUS in the T1 generation were consistent with those in the T0 generation and no additional mutations were observed. These results demonstrated that CRISPR/SpCas9 induced-mutations were heritable in wheat.
Confirmation of the optimized CRISPR system by editing the endogenous genes TaMTL and TaWaxy
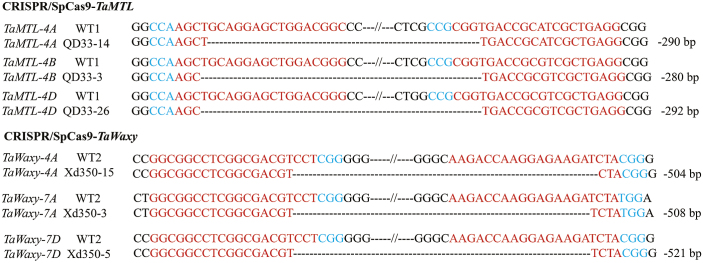
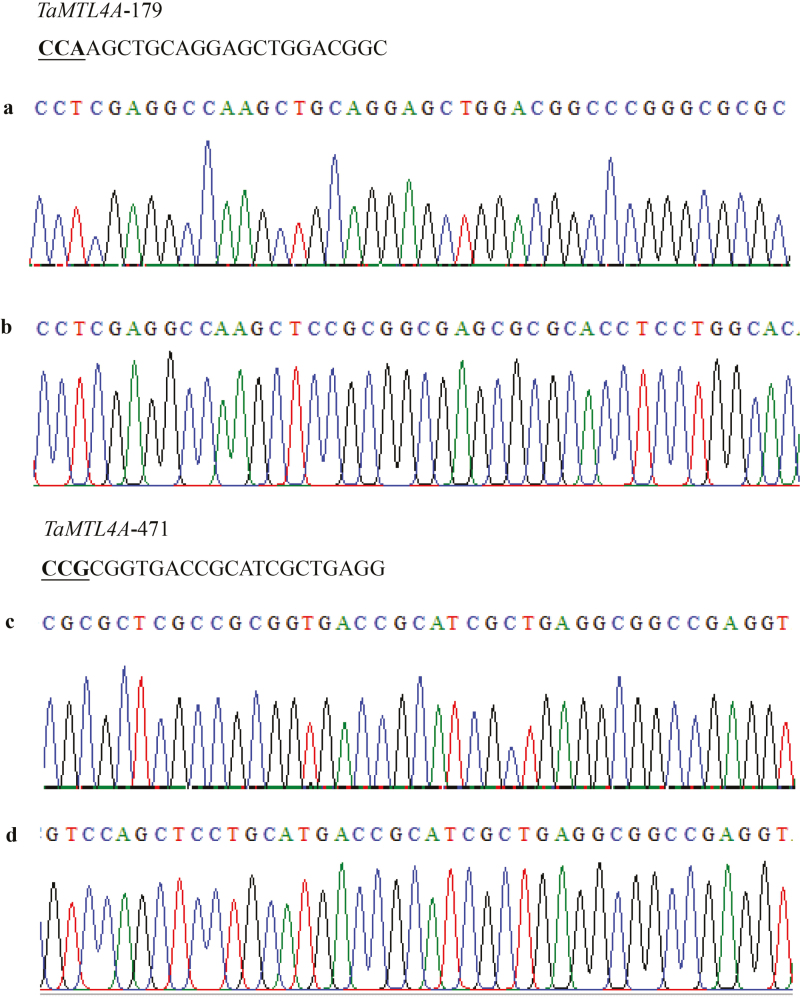
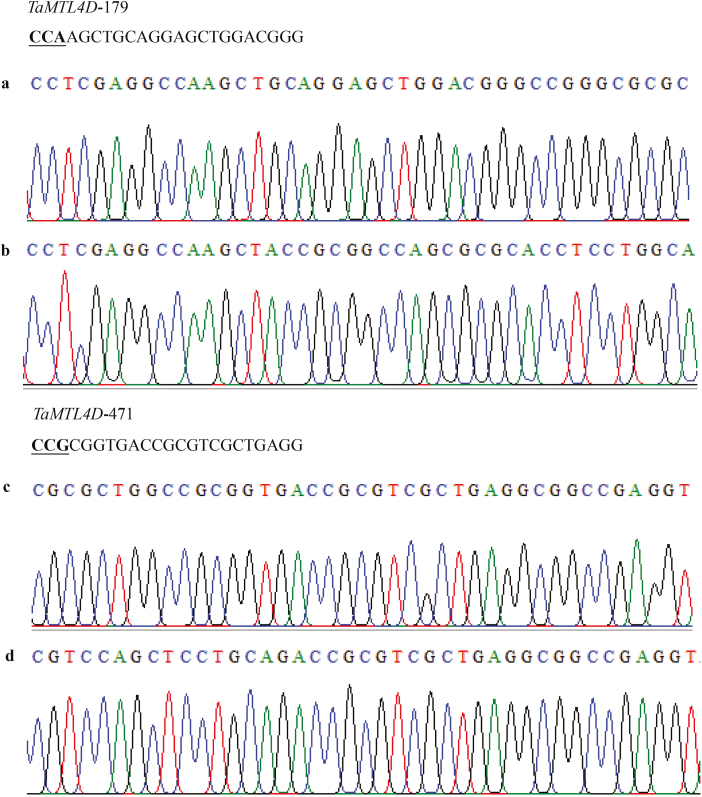
The CRISPR/SpCas9 system was further used to edit two wheat endogenous genes, TaMTL and TaWaxy. The expression of the sgRNAs of the two genes was driven by the TaU3 promoter, and the constructs were introduced into the varieties Fielder and Ningchun4 via Agrobacterium-mediated transformation (Fig. 1a). For TaMTL, two sgRNAs (TaMTL-179 and TaMTL-471) were designed to target the first and second exons of the three homologous genes on chromosomes 4A, 4B, and 4D genomes in Fielder. The editing efficiencies of this gene were up to 46.5% and 38.6% at the targets of TaMTL-179 and TaMTL-471, respectively (Table 2, Supplementary Fig. S2). The total editing efficiency was up to 57.5% for plants in which a mutation occurred at one or two loci (Table 3). At the target TaMTL-179, the editing efficiencies for the homologous genes on chromosomes 4A, 4B, and 4D were 34.7%, 18.8%, and 23.8%, respectively (Table 2; Supplementary Figs S2, S3), and at the target TaMTL-471, the efficiencies for the homologous genes on the three chromosomes were 35.6%, 8.9%, and 15.8%, respectively (Table 2; Supplementary Figs S2, S3). The editing efficiency was 12.9% for simultaneous mutations at the three loci and 13.9% for simultaneous mutations at two loci (Table 3). In addition, a large fragment deletion was also identified when the two targets were simultaneously edited on any one of chromosomes 4A, 4B, or 4D, with efficiencies of 8.9%, 5.9%, and 7.9% for the three homologous genes, respectively (Fig. 2, Supplementary Fig. S5a, Table 2). Interestingly, we found a mutant, QD33-3, with reverse insertion of the deleted sequences within the TaMTL homologous genes on chromosomes 4A and 4D in the T0 generation, which lays a foundation for target gene replacement in wheat (Fig 3, Fig. 4, Supplementary Figs S6, S7).
Table 3.
Frequency distribution of the chromosomal locus mutations for TaMTL and TaWaxy in T0 wheat plants using the CRISPR/SpCas9 system
| Number of loci | ||||
|---|---|---|---|---|
| Mutation | 3 | 2 | 1 | 0 |
| TaMTL | 13 (12.9%) | 14 (13.9%) | 31 (30.7%) | 43 (42.5%) |
| TaWaxy | 28 (32.2%) | 20 (23.0%) | 22 (25.3%) | 17 (19.5%) |
Fig. 2.
Sequences of the large fragment deletions in TaMTL and TaWaxy in different loci of chromosomes of wheat plants edited using the CRISPR/SpCas9 system. WT1, Fielder variety (wild-type); WT2, Ningchun4 variety (wild-type). TaMTL-edited plants are named as QD33, and TaWaxy-edited plants are named as Xd350. Blue letters indicate the PAM sequences, red letters indicate the sgRNA sequences, and the dashed lines represent nucleotide deletions.
Fig. 3.
Detailed reverse sequence insertion analysis in wheat TaMTL at the 4A chromosomal loci. (a, c) TaMTL-4A sequence in the wild-type Fielder variety, and (b, d) TaMTL-4A sequence in QD33-3, which was edited using the CRISPR/SpCas9 system.
Fig. 4.
Detailed reverse sequence insertion analysis in wheat TaMTL at the 4D chromosomal loci. (a, c) TaMTL-4D sequence in the wild-type Fielder variety, and (b, d) TaMTL-4D sequence in QD33-3, which was edited using the CRISPR/SpCas9 system.
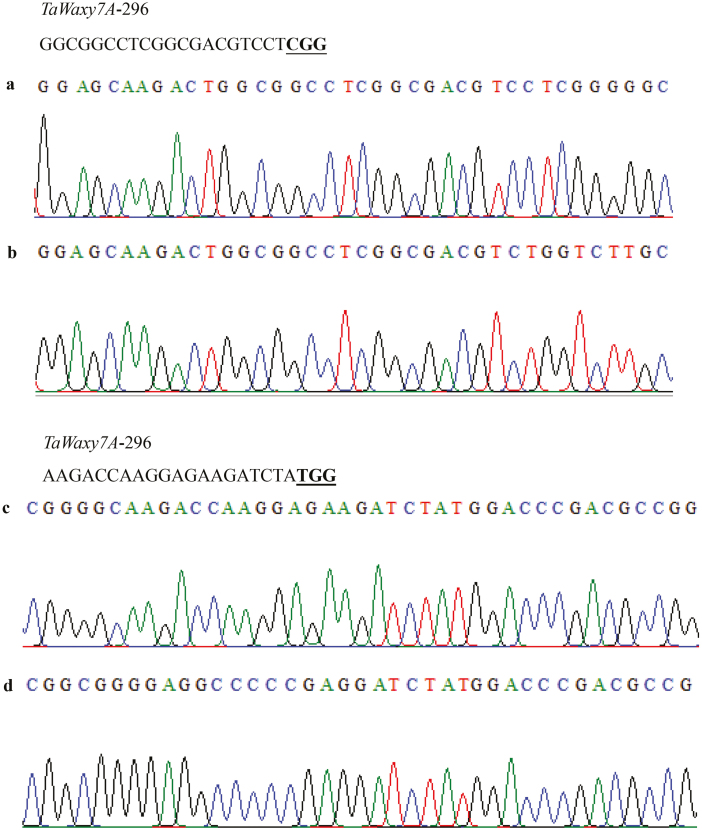
The editing efficiencies of the sgRNAs targeting TaWaxy-296 and TaWaxy-830 on the first and fourth exons of chromosomes 4A, 7A, and 7D in the widely cultivated commercial hexaploid variety Ningchun4 were as high as 47.1% and 71.3%, respectively (Table 2; Supplementary Fig. S2). The total editing efficiency was up to 80.5% for mutations occurring at one or two loci (Table 3). At the target TaWaxy-296, the editing efficiencies for the three homologous genes on chromosomes 4A, 7A, and 7D were 40.2%, 44.8%, and 26.4%, respectively (Table 2, Supplementary Fig. S8), and at the target TaWaxy-830, the efficiencies were 41.4%, 54.0%, and 35.6%, respectively (Table 2, Supplementary Fig. S9). The efficiency was 32.2% for simultaneous mutations at the three loci and 23.0% for simultaneous mutations at two loci (Table 3). In addition, a large fragment deletion was also identified where the two targets were simultaneously edited on chromosomes 4A, 7A, and 7D, and the efficiencies were 11.5%, 12.6%, and 16.1%, respectively (Fig. 2, Supplementary Fig. S5b, Table 2). We also identified a mutant, Xd350-15, with reverse insertion of the deleted sequences within the TaWaxy homologous genes on chromosome 7A in the T0 generation (Fig. 5, Supplementary Fig. S10). Four selected TaWaxy-edited lines (Xd350-3, Xd350-5, Xd350-8, and Xd350-15) were found to be simultaneous mutations in the three homologous genes TaWaxy-4A, TaWaxy-7A, and TaWaxy-7D (Fig. 2, Supplementary Figs S8, S9).
Fig. 5.
Detailed reverse sequence insertion analysis in wheat TaWaxy at the 7A chromosomal loci. (a, c) TaWaxy-7A sequence in the wild-type Ningchun4 variety, and (b, d) TaWaxy-7A sequence in Xd350-15, which was edited using the CRISPR/SpCas9 system.
Detection of off-target mutations using Sanger sequencing
We next assessed the potential off-target effects using GUS, TaMTL, and TaWaxy in the CRISPR/SpCas9, CRISPR/LbCpf1, and CRISPR/xCas9 systems. The potential off-targets of the homologous positions of the sgRNAs for these genes were searched in the wheat reference genome (EnsemblPlants: http://plants.ensembl.org/Triticum_aestivum/Tools/Blast), and six potential off-target sites were found for GUS, six for TaMTL, and four for TaWaxy. Compared with the target sequences they had 2–4 bp mismatches (Supplementary Table S4). To detect the off-target events, we designed specific primers and amplified possible off-target areas in the T0 transgenic plants that contained the guide RNA (Supplementary Table S4) and then the PCR amplicons were detected by Sanger sequencing. No mutation was found to occur at all the 16 possible off-target sites. These results indicated that the possible off-targets could be ignored, and that the CRISPR system could highly specifically target the selected sites for mutation in the edited plants obtained in this study.
Editing of TaMTL induces a reduction in seed set and an increase in haploid production
Of the 55 TaMTL-edited plants, seven lines were examined further: QD33-3, QD33-14, QD33-20, QD33-26, QD33-43, QD33-46, and QD33-52 (Figs 2–4, Supplementary Figs S6, S7 at JXB online). There was a large fragment deletion (290 bp) between the two targets in the homologous genes on chromosome 4A in QD33-14, QD33-20, and QD33-26. Lines QD33-3 and QD33-43 had a large fragment deletion (280 bp) on chromosome 4B, and there was a reverse insertion between the two targets in the homologous genes on chromosomes 4A and 4D in QD33-3. Another large fragment deletion (292 bp) was found in QD33-26 and QD33-46 between the two targets in the homologous genes on chromosome 4D. Seed-set rates of the TaMTL-edited T0 transgenic plants were much lower than that of the wild-type Fielder (Fig. 6a, Table 4). Among them, TaMTL-4A, TaMTL-4B, and TaMTL-4D were simultaneously edited in QD33-3, QD33-14, QD33-20, QD33-26, QD33-43, and QD33-46, and TaMTL-4A and TaMTL-4D were simultaneously edited in QD33-52 (Table 4). qPCR analysis showed that the post-transcriptional expression levels of TaMTL-4A, TaMTL-4B, and TaMTL-4D in the anthers of plants QD33-3, QD33-14, and QD33-26 were significantly reduced in comparison with the wild-type (Fig. 6b).
Fig. 6.
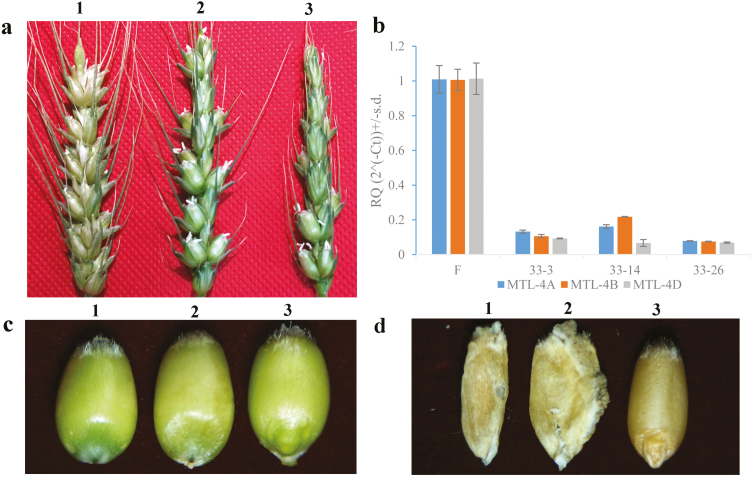
Phenotypes of TaMTL-edited wheat lines using the CRISPR/SpCas9 system. (a) Spikes of the wild-type Fielder variety (1) and the TaMTL-knockout plants (2, 3). (b) Relative amounts of TaMTL-4A, TaMTL-4B, and TaMTL-4D RNA in wild-type Fielder (F) and TaMTL-edited plants (QD33-3, QD33-14, and QD33-26). The expression of the wild-type was set to 1 for each gene. (c) Seeds of TaMTL-edited plants with no embryo (1, 2) compared with the wild-type (3). (d) Seeds of TaMTL-edited plants with no endosperm (1, 2) compared with the wild-type (3).
Table 4.
Seed-set and haploid induction rates of TaMTL-edited plants of wheat using the CRISPR/SpCas9 system
| Plant ID | Genotype of TaMTL-4A | Genotype of TaMTL-4B | Genotype of TaMTL-4D | TSSN | RSSN | No. of seeds with no embryo | SSR (%) | No. of haploid plants | HIR (%) |
|---|---|---|---|---|---|---|---|---|---|
| QD33-3 | Bi | L | Bi | 86 | 19 | 2 | 22.1 | 3 | 26.3 |
| QD33-14 | L | He | He | 86 | 17 | 2 | 19.8 | 0 | 11.8 |
| QD33-20 | L | He | Bi | 89 | 21 | 3 | 23.6 | 1 | 19.0 |
| QD33-26 | L | Bi | L | 85 | 19 | 2 | 22.4 | 4 | 31.6 |
| QD33-43 | Bi | L | Bi | 84 | 17 | 0 | 20.2 | 2 | 11.8 |
| QD33-46 | He | He | L | 87 | 19 | 1 | 21.8 | 3 | 21.1 |
| QD33-52 | Bi | WT | Bi | 90 | 20 | 1 | 22.2 | 1 | 10.0 |
| Fielder | WT | WT | WT | 92 | 83 | 0 | 90.2 | 0 | 0 |
Bi, bi-allele; He, heterozygote; L, large fragment deletion; WT, wild-type; TSSN, theoretical seed-set number; RSSN, real seed-set number; SSR, seed-set rate; HIR, haploid induction rate.
Interestingly, some grains lacking an embryo were found in QD33-3, QD33-14, QD33-20, QD33-26, QD33-46, and QD33-52 (Fig. 6c), and two shriveled grains without endosperm and embryo were found in QD33-3 (Fig. 6d). The ploidy of the immature seeds in the TaMTL-edited plants was determined through chromosome counting, and 25 haploids were identified from a total of 132 seeds: the haploid seeds had 21 chromosomes while the diploid seeds had 42. The haploids had shorter plant height, fewer tillers, narrower leaves, and shorter guard-cell length compared with the diploids (Fig. 7). A haploid induction rate of 10.0–31.6% was observed in the gene-edited plants (Table 4).
Fig. 7.
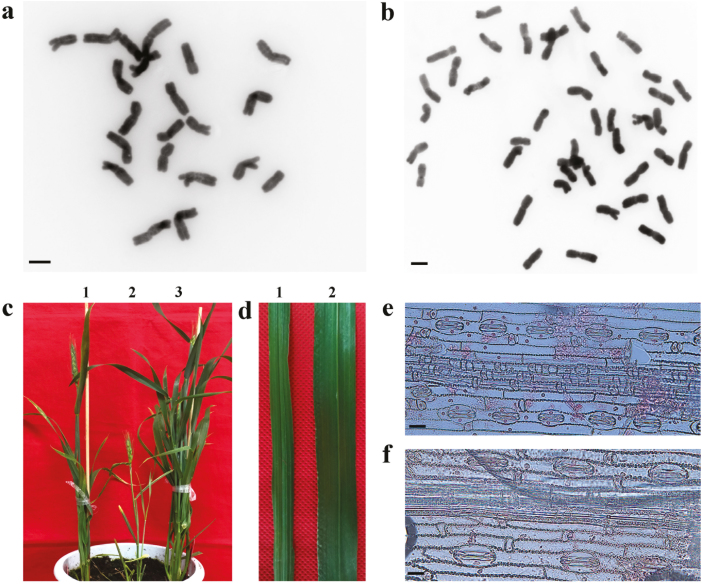
Characteristics of the wheat haploid plants in the TaMTL-edited mutants using the CRISPR/SpCas9 system. (a, b) Chromosomes of haploids (a) and diploids (b) in TaMTL-edited T1 plants. The haploid plants had 21 chromosomes and the diploid plants had 42. (c) Plant height in the haploid (2) and the diploid (1, 3). (d) Leaf width in the haploid (1) and the diploid (2). Guard cell length in the haploid (e) and the diploid (f). Scale bars in (a, b, e, f) are 10 μm.
Discussion
The CRISPR/Cas9 system has been widely used for plant genome editing since its first development and, as an example, a number of wheat genes have been edited with this system over the last year by Agrobacterium-mediated transformation (Abe et al., 2019; Liu et al., 2020a; Okada et al., 2019; Zhang et al., 2019b). However, the editing efficiency is lower in wheat than in other plants (Ma et al., 2015; Feng et al., 2018). In the genome editing of other plant species including maize and rice, the promoters used for both the sgRNAs and Cas9 are thought to significantly influence the targeting efficiency (Xu et al., 2014; Zhang et al., 2014a; Ma et al., 2015; Xie et al., 2015; Zhu et al., 2016; Feng et al., 2018). The ubi promoter works better than the CaMV35S promoter for regulating Cas9 in rice, Arabidopsis, and maize (Feng et al, 2014; Xing et al., 2014). The sgRNA sites are also very important for the generation of target mutations in the gene of interest and greatly influence the gene-editing efficiency (Zhu et al., 2016; Feng et al., 2018). In the present study, the maize ubi promoter was used to control SpCas9, and the wheat TaU3 and TaU6 and rice OsU6a promoters were used to control sgRNAs to edit GUS by Agrobacterium-mediated transformation. The editing efficiencies of TaU3, TaU6, and OsU6a were 61.4%, 36.0%, and 21.6%, respectively (Table 1). The TaU3 promoter thus appeared to be the best choice for regulating sgRNA expression when performing Agrobacterium-mediated CRISPR gene editing in wheat. When two sgRNAs were designed to target GUS, the editing efficiency was up to 70.1% (Supplementary Table S3), and large fragment deletions were also detected between the two targets with efficiencies up to 37.2% (Table 2). The editing efficiency using two sgRNAs was therefore higher than by using a single sgRNA to target the gene. Interestingly, although editing mutations including large fragment deletions were detected in TaMTL-edited, TaWaxy-edited, and GUS-edited plants, the efficiencies for TaMTL-4A, TaMTL-4B, and TaMTL-4D, and TaWaxy-4A, TaWaxy-7A, and TaWaxy-7D were lower than that for GUS (Table 2). GUS was an exogenous gene that was inserted onto the distal region in a pair of wheat chromosomes with a single copy in the H29 line used in this study (Liu et al., 2020b), while the endogenous TaMTL and TaWaxy genes were all present in the wheat genome with three copies. We therefore conclude that the copy number and chromosomal locations of the target genes might influence the editing efficiencies.
Compared with CRISPR/SpCas9, CRISPR/xCas9 edited the NGG PAM site with lower efficiency even though the target sequence was the same in the two systems, which was consistent with a previous report (Wang et al., 2018). In the xCas9 system, the target sequence in the NGT PAM site was almost similar to the NGG PAM site, but its editing efficiency was quite low. In addition, although the target sequences at the NGA, NGC, GAA, and GAT PAM sites in xCas9 were different from the sequence at the NGG PAM site in the SpCas9 system, no mutant plant was detected at all four sites. The editing efficiencies at the two target sites in CRISPR/LbCpf1 (g118 and g2046) and the six target sites in xCas9 (g788, g1718, g789, g1159, g1719, and g1156) were clearly much lower than those at the two target sites in SpCas9 (g-22 and g788) (Table 1, Supplementary Table S1). A recent study found that xCas9 possesses limited activity at non-canonical NGH (H=A, C, T) PAM sites in rice protoplasts (Zhong et al., 2019a), and other studies have also reported that the editing efficiency of Cpf1 in maize and Arabidopsis is significantly lower than that in rice (Tang et al., 2017; Wang et al., 2017b; Lee et al., 2019; Malzahn et al., 2019). We therefore conclude that species diversity may be the reason for the lower editing efficiencies of the CRISPR/LbCpf1 and CRISPR/xCas9 systems in wheat than in other plants.
The genetic behavior of the edited GUS gene indicated that its segregation was accordance with Mendelian heritance patterns, and all the mutations of GUS in the T1 generation were consistent with those in the T0 generation with no additional mutations being observed. This was consistent with the report by Howells et al. (2018) in which no additional editing was observed in the T1 generation. In a recently published study, it was found that some new mutations happened and editing efficiencies increased in the next a few generations (Zhang et al., 2019a). Our results were different because we selected edited plants for detailed investigation at target sites that might not have harbored integration of SpCas9 and sgRNA. Generally, the transgenic plants might have been chimeric in T0, and some new edited plants might have missed detection in this generation but gone on to be detected in the next generation. Moreover, wheat genome editing might be closely associated with the design of target sites, application of promoters for sgRNA, and location of target genes (Howells et al., 2018).
By combining gene-editing technology with haploid induction technology important genes can be edited, and in addition homozygous edited plants can be quickly obtained. For example, Kelliher et al. (2019) transformed a vector expressing Cas9 and a gRNA targeting the putative wheat GRASSY TILLER1 orthologs TaGT1-4A, TaGT1-4B, and TaGT1-4D into a maize inbred line NP2222 and then this was pollenated to wheat, resulting in haploid and doubled-haploid wheat plants with the edited GRASSY TILLER1. The key gene controlling the haploid production trait in maize, ZmPLA, has recently been cloned, and knockout of this gene by CRISPR/Cas9 leads to the production of haploid seeds at a rate of 6–10% (Dong et al., 2018). Editing the homologous gene of ZmPLA to silence it in rice by using CRISPR/Cas9 resulted in the formation of haploid grains at a rate of 2–6% (Yao et al., 2018). In our study, we used an optimized CRISPR/SpCas9 system and edited wheat TaMTL, which is the homolog of ZmPLA for haploid induction. Our results showed that a double-knockout mutation of TaMTL-4A and TaMTL-4D resulted in haploid induction at a frequency of 10%, and triple-knockout of TaMTL-4A, TaMTL-4B, and TaMTL-4D resulted in haploid induction at a frequency of 11.8–31.6% (Table 4). A new study has recently reported that TaPLA-A and TaPLA-D knockout lines trigger haploid induction at a rate of 2–3% (Liu et al., 2020a). In our CRISPR/SpCas9 system, we used the TaU3 promoter to control two sgRNAs to target TaMTL-4A, TaMTL-4B, and TaMTL-4D simultaneously, whereas Liu et al. (2020a) used the TaU6 promoter to regulate one sgRNA to target TaPLA. Clearly, our haploid induction rate was the highest. In addition, we not only identified haploid plants in the T1 generation from the lines QD33-3, QD33-14, QD33-20, QD33-26, QD33-43, QD33-46, and QD33-52, but also found some grains lacking an embryo in the lines QD33-3, QD33-14, QD33-20, QD33-26, QD33-46, and QD33-52. Interestingly, two shriveled grains without endosperm and embryo were found in the line QD33-3 (Fig. 6c, d). Further research is needed to determine the mechanism of endosperm and embryo abortion.
The starch composition of wheat grains has an important influence on flour quality. Amylose is encoded by the Waxy gene, which is synthesized by granule-bound starch synthase I (Sano, 1984; Wang et al., 1990). In common wheat, Waxy is located on chromosomes 7AS, 4AL (translocated from the original locus on 7BS), and 7DS (Yamamori et al., 1994). The variants of Waxy (Wx-A1, Wx-B1, and Wx-D1) have been used to determine the effect of deficiencies on amylase content and starch pasting properties (Yamamori, 2009). However, the frequencies of variation at the Waxy locus are relatively low in modern wheat cultivars. In our system TaWaxy-4A, TaWaxy-7A, and TaWaxy-7D were successfully edited with high efficiency by CRISPR/SpCas9, and further research is planned to obtain homozygous strains and to test the starch with a view to enhancing the flour quality.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Schematic map of the vector pWMB110-SpCas9/LbCpf1/xCas9.
Fig. S2. Detection of mutations in GUS, TaMTL-179, TaMTL-471, TaWaxy-296, and TaWaxy-830.
Fig. S3. InDel mutations of TaMTL at the TaMTL-179 site from edited T0 transgenic plants.
Fig. S4. InDel mutations of TaMTL at the TaMTL-471 site from edited T0 transgenic plants.
Fig. S5. Detection of large fragment deletions from different TaMTL and TaWaxy homologous genes.
Fig. S6. Sequences of TaMTL at loci on the 4A chromosome in the wild-type and the edited plant QD33-3.
Fig. S7. Sequences of TaMTL at loci on the 4D chromosome in the wild-type and the edited plant QD33-3.
Fig. S8. InDel mutations of TaWaxy at the TaWaxy-296 site from edited T0 transgenic plants.
Fig. S9. InDel mutations of TaWaxy at the TaWaxy-830 site from edited T0 transgenic plants.
Fig. S10. Sequences of TaWaxy at loci on the 7A chromosome loci in the wild-type and the edited plant Xd350-15.
Table S1. Summary of the target sequences and mutations of GUS in T0 wheat plants obtained using the CRISPR/xCas9 editing system.
Table S2. PCR primers used for vector construction and editing identification.
Table S3. Frequency distribution of the mutations for GUS in T0 wheat plants using the CRISPR/SpCas9-D system.
Table S4. Off-target detection using designed sgRNAs for GUS, TaMTL, and TaWaxy.
Acknowledgements
This research was financially supported in part by grants from the Ministry of Agriculture and Rural Affairs of China (2016ZX08010004, 2016ZX08009001), the Science and Technology Department of Ningxia in China (2019BBF02020), and the Chinese Academy of Agricultural Sciences in China (2060302-2-19).
Author contributions
KW and XY conceived the research and designed the experiments; HL and KW conducted most of the experiments; ZJ, QG, and ZL performed the vector construction and observations of cytogenetic; LD and XP investigated the agronomic traits; HL, KW, and XY drafted and revised the manuscript.
References
- Abe F, Haque E, Hisano H, et al. . 2019. Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Reports 28, 1362–1369.e4. [DOI] [PubMed] [Google Scholar]
- Barclay IR. 1975. High frequencies of haploid production in wheat (Triticum aestivum) by chromosome elimination. Nature 256, 410. [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proceedings of the National Academy of Sciences, USA 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Li L, Liu C, Liu C, Geng S, Li X, Huang C, Mao L, Chen S, Xie C. 2018. Genome editing and double-fluorescence proteins enable robust maternal haploid induction and identification in maize. Molecular Plant 11, 1214–1217. [DOI] [PubMed] [Google Scholar]
- Feng C, Su H, Bai H, et al. . 2018. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnology Journal 16, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, et al. . 2014. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. [DOI] [PubMed] [Google Scholar]
- Han F, Liu B, Fedak G, Liu Z. 2004. Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theoretical and applied genetics 109, 1070–1076. [DOI] [PubMed] [Google Scholar]
- Howells RM, Craze M, Bowden S, Wallington EJ. 2018. Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biology 18, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, et al. . 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K, Tao XP, Han PJ, Wang R, Zhu JK. 2019. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Molecular Plant 12, 1003–1014. [DOI] [PubMed] [Google Scholar]
- Ishii T, Karimi-Ashtiyani R, Houben A. 2016. Haploidization via chromosome elimination: means and mechanisms. Annual Review of Plant Biology 67, 421–438. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Richbourg L, et al. . 2017. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542, 105–109. [DOI] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Su X, et al. . 2019. One-step genome editing of elite crop germplasm during haploid induction. Nature Biotechnology 37, 287–292. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. 1986. Wheat × maize hybridization. Canadian Journal of Genetics and Cytology 28, 313–316. [Google Scholar]
- Lee K, Zhang Y, Kleinstiver BP, et al. . 2019. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnology Journal 17, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li X, Meng D, et al. . 2017. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase a generates haploid induction in maize. Molecular Plant 10, 520–522. [DOI] [PubMed] [Google Scholar]
- Liu CX, Zhong Y, Qi XL, et al. . 2020. a Extension of the in vivo haploid induction system from maize to wheat. Plant Biotechnology Journal. In press. doi: 10.1111/pbi.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Wang K, Wang J, Du LP, Pei XW, Ye XG. 2020b Genetic and agronomic traits stability of marker-free transgenic wheat plants generated from Agrobacterium mediated co-transformation in T2 and T3 generation. Journal of Integrative Agriculture 19, 23–32. [Google Scholar]
- Liu W, Zheng MY, Polle EA, Konzak CF. 2002. Highly efficient doubled-haploid production in wheat (Triticum aestivum L.) via induced microspore embryogenesis. Crop Science 42, 686–692. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. . 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Machii H, Mizuno H, Hirabayashi T, Li H, Hagio T. 1998. Screening wheat genotypes for high callus induction and regeneration capability from anther and immature embryo cultures. Plant Cell Tissue and Organ 53, 67–74. [Google Scholar]
- Malzahn AA, Tang X, Lee K, et al. . 2019. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biology 17, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Arndell T, Borisjuk N, Sharma N, Watson-Haigh NS, Tucker EJ, Baumann U, Langridge P, Whitford R. 2019. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnology Journal 17, 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricroch A, Clairand P, Harwood W. 2017. Use of CRISPR systems in plant genome editing: toward new opportunities in agriculture. Emerging Topics in Life Sciences 1, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y. 1984. Differential regulation of waxy gene expression in rice endosperm. Theoretical and applied genetics 68, 467–473. [DOI] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, et al. . 2017. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nature Plants 3, 17103. [DOI] [PubMed] [Google Scholar]
- Wang JJ, Meng XB, Hu XX, Sun TT, Li JY, Wang KJ, Yu H. 2018. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnology Journal 17, 709–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liu HY, Du LP, Ye XG. 2017a Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnology Journal 15, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MG, Mao YF, Lu YM, Tao XP, Zhu JK. 2017b Multiplex gene editing in rice using the CRISPR-Cpf1 system. Molecular Plant 10, 1011–1013. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. 2014. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, Hong MM. 1990. Nucleotide sequence of rice waxy gene. Nucleic Acids Research 18, 5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences, USA 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori M. 2009. Amylose content and starch properties generated by five variant Wx alleles for granule-bound starch synthase in common wheat (Triticum aestivum L.). Euphytica 165, 607–714. [Google Scholar]
- Yamamori M, Nakamura T, Endo TR, Nagamine T. 1994. Waxy protein deficiency and chromosomal location of coding genes in common wheat. Theoretical and applied genetics 89, 179–184. [DOI] [PubMed] [Google Scholar]
- Yao L, Zhang Y, Liu C, Liu Y, Wang Y, Liang D, Liu J, Sahoo G, Kelliher T. 2018. OsMATL mutation induces haploid seed formation in indica rice. Nature Plants 4, 530–533. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, et al. . 2017. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nature Biotechnology 35, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, et al. . 2014. a The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnology Journal 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zhang RZ, Gao J, Gu TT, Song GQ, Li W, Li DD, Li YL, Li GY. 2019a Highly efficient and heritable targeted mutagenesis in wheat via the Agrobacterium tumefaciens-mediated CRISPR/Cas9 system. International Journal of Molecular Sciences 20, 4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang K, Lin ZS, Du LP, Ma HL, Xiao LL, Ye XG. 2014b Production and identification of haploid dwarf male sterile wheat plants induced by corn inducer. Botanical Studies 55, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Massel K, Godwin ID, Gao C. 2018a Applications and potential of genome editing in crop improvement. Genome Biology 19, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang R, Song G, Gao J, Li W, Han X, Chen M, Li Y, Li G. 2018b Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biology 18, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li WL. 2019b Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnology Journal 17, 1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Sretenovic S, Ren Q, et al. . 2019. a Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Molecular Plant 12, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Liu C, Qi X, et al. . 2019. b Mutation of ZmDMP enhances haploid induction in maize. Nature Plants 5, 575–580. [DOI] [PubMed] [Google Scholar]
- Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J. 2016. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. Journal of Genetics and Genomics 43, 25–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.