Arabidopsis flowering regulators, KHZ1 and KHZ2, are novel members of the autonomous pathway that promote flowering through repression of the splicing efficiency of FLC pre-mRNA.
Keywords: Arabidopsis, autonomous, flowering, FLOWERING LOCUS C, KHZ1, KHZ2
Abstract
As one of the most important events during the life cycle of flowering plants, the floral transition is of crucial importance for plant propagation and requires the precise coordination of multiple endogenous and external signals. There have been at least four flowering pathways (i.e. photoperiod, vernalization, gibberellin, and autonomous) identified in Arabidopsis. We previously reported that two Arabidopsis RNA-binding proteins, KHZ1 and KHZ2, redundantly promote flowering. However, the underlying mechanism was unclear. Here, we found that the double mutant khz1 khz2 flowered late under both long-day and short-day conditions, but responded to vernalization and gibberellin treatments. The late-flowering phenotype was almost completely rescued by mutating FLOWERING LOCUS C (FLC) and fully rescued by overexpressing FLOWERING LOCUS T (FT). Additional experiments demonstrated that the KHZs could form homodimers or interact to form heterodimers, localized to nuclear dots, and repressed the splicing efficiency of FLC pre-mRNA. Together, these data indicate that the KHZs could promote flowering via the autonomous pathway by repressing the splicing efficiency of FLC pre-mRNA.
Introduction
The transition from vegetative to reproductive development is a vital process in the life cycle of a flowering plant, and this process is coordinately regulated by various endogenous and exogenous factors (Mouradov et al., 2002; Yuan et al., 2016). Numerous molecular genetic studies on the model dicotyledonous plant Arabidopsis have identified at least four major flowering pathways: photoperiod, vernalization, gibberellin (GA), and autonomous (Srikanth and Schmid, 2011). Studies have demonstrated that these pathways are interconnected and ultimately converge to a set of floral integrators, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1). The floral integrators in turn regulate the expression of floral meristem identity (FMI) genes, such as APETALA1 (AP1) and FRUITFULL(FUL), to promote the floral transition (Yamaguchi et al., 2005; Corbesier et al., 2007; Jang et al., 2009; Yu et al., 2016).
Plants can perceive environmental light signals and use the circadian clock as a time-keeping mechanism to monitor day length and ensure flowering at the proper time (Samach and Coupland, 2000; Hayama and Coupland, 2003; Yanovsky and Kay, 2003). As a model plant, Arabidopsis flowers early under long-day conditions and flowers late under short-day conditions (Yuan et al., 2016). CONSTANS (CO) and FT are vital regulators in response to photoperiod in Arabidopsis. The circadian clock regulates the expression of CO, which further activates the expression of FT (Wigge et al., 2005; Yuan et al., 2016). The FT protein, which is synthesized by specialized companion cells in Arabidopsis, induces the expression of the FMI genes and initiates flowering (Abe et al., 2005; Corbesier et al., 2007; Chen et al., 2018).
Many plants, such as winter cereals (Sharma et al., 2017), require a prolonged period of cold treatment before flowering, which is a process known as vernalization (Kim et al., 2009). Genetic analyses of the winter-annual varieties of Arabidopsis revealed that FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) are two key genes that respond to vernalization (Michaels and Amasino, 1999; Johanson et al., 2000). FRI functions by activating the expression of FLC (Geraldo et al., 2009), which encodes a MADS-box transcription factor and acts as a repressor of flowering by suppressing the expression of flower-promoting genes (e.g. FT and SOC1) (Searle et al., 2006; Michaels, 2009). Vernalization promotes flowering primarily via the epigenetic repression of FLC (Michaels, 2009). Studies have revealed three regulators, VERNALIZATION INSENSITIVE 3 (VIN3), VERNALIZATION 1 (VRN1), and VERNALIZATION 2 (VRN2), that are needed for initiation and maintenance of FLC silencing after vernalization (Gendall et al., 2001; Levy et al., 2002; Sung and Amasino, 2004; Srikanth and Schmid, 2011). The inhibition of FLC is mainly achieved by the modification of FLC chromatin by histone deactetylation and methylation (He, 2009).
GA is a class of plant hormone that promotes flowering in Arabidopsis. Mutations in the GA biosynthesis and signaling pathways delay the flowering time, whereas exogenous application of GA accelerates flowering (Mouradov et al., 2002; Srikanth and Schmid, 2011). The GA1 gene, which encodes an ent-kaurene synthase, is needed for the first step of GA biosynthesis (Sun et al., 1992). Flowering was delayed for the ga1-3 mutant under continuous light and could not be induced under short-day conditions (Wilson et al., 1992). SPINDLY (SPY), which encodes an O-linked N-acetylglucosamine transferase, negatively regulates GA signaling (Jacobsen and Olszewski, 1993). GA signaling was constitutively active in the spy mutant. Mutation of SPY could overcome the late flowering of ga1 as demonstrated by the finding that the flowering time of the ga1 spy double mutant was similar to that of the wild-type (WT) plant (Jacobsen et al., 1996; Swain et al., 2001). Previous studies have demonstrated that there are several points of convergence between the GA pathway and other pathways. For instance, the promoter activity of the floral integrator gene LEAFY (LFY) was increased by GA treatment, but LFY expression was reduced 10-fold in the ga1-3 mutant (Blázquez et al., 1997, 1998). GA also plays an important role in the regulation of another floral integrator gene SOC1. SOC1 expression is increased by exogenous treatment with GA but is nearly undetectable in the ga1-3 mutant under short-day conditions (Moon et al., 2003).
In the absence of exogenous inducers, plants will flower after a particular stage of vegetative growth. This flowering pathway is known as the autonomous pathway. Many proteins have been identified in the autonomous pathway, such as FLOWERING LOCUS CA (FCA), FLOWERING LOCUS PA (FPA), FLOWERING LOCUS VE (FVE), FLOWERING LOCUS D (FLD), LUMINIDEPENDENS (LD), FLOWERING LOCUS Y (FY), RELATIVE OF EARLY FLOWERING 6 (REF6), FLOWERING LOCUS KH DOMAIN (FLK) and TBP-ASSOCIATED FACTOR 15b (TAF15b); these factors accelerate flowering by inhibiting FLC (Lim et al., 2004; Mockler et al., 2004; Simpson, 2004; Srikanth and Schmid, 2011; Eom et al., 2018). Mutants of the autonomous pathway have a late-flowering phenotype under both long-day and short-day conditions; however, the late-flowering phenotype of these mutants can be overcome by vernalization or exogenous GA (Mouradov et al., 2002; Moon et al., 2003). Generally, the factors involved in the autonomous pathway are divided into three functional categories: RNA processing, and transcriptional and epigenetic regulation (Eom et al., 2018). In the RNA processing category, FCA, FY, FPA, cleavage stimulation factor 64 (CstF64), CstF77, and Pcf11p-similar protein 4 (PCFS4) are likely to participate in the cleavage and polyadenylation of mRNA (Macknight et al., 1997; Schomburg et al., 2001; Simpson et al., 2003; Xing et al., 2008; Liu et al., 2010), while glycine-rich RNA-binding protein 7 (GRP7), GRP8, pre-mRNA processing protein 8 (PRP8), PRP39-1, and serine/arginine-rich 45 (SR45) are involved in RNA splicing (Ali et al., 2007; Wang et al., 2007; Streitner et al., 2008; Marquardt et al., 2014). For instance, FCA can interact with FY to regulate RNA 3' end-processing (Simpson et al., 2003). LD is part of the transcriptional regulation category. It is localized to the nucleus and contains a glutamine-rich region at its C-terminus that is similar to the glutamine-rich domains found in several other transcription factors (Lee et al., 1994; Aukerman et al., 1999). The last category is involved in the epigenetic process. One of the repressors of FLC (FLD) has an important role in the histone deacetylation of FLC chromatin. Mutation of FLD results in the hyperacetylation of histone in the FLC chromatin (He et al., 2003).
We previously reported that two RNA-binding proteins, CCCH zinc-finger and K homology (KH) domain proteins 1 and 2 (KHZ1 and KHZ2), are involved in the regulation of flowering time in Arabidopsis (Yan et al., 2017). In this study, we investigated the molecular mechanisms by which these two proteins regulated flowering. Our results indicate that KHZ1 and KHZ2 are novel members of the autonomous pathway that may function as homodimers or heterodimers to repress FLC expression primarily through the inhibition of its splicing efficiency. Taken together, our study demonstrates that KHZ1 and KHZ2 play important roles in regulating flowering via the autonomous pathway.
Materials and methods
Plant materials and growth conditions
All Arabidopsis (Arabidopsis thaliana) used in this study were of ecotype Columbia-0. The transgenic and mutants KHZ1 and KHZ2 lines were described previously (Yan et al., 2017). Plants were cultivated at 22 °C with a relative humidity of 60% under long-day (16 h light/8 h dark) or short-day (8 h light/16 h dark) conditions.
Flowering time measurements
Flowering time was measured by recording the number of rosettes and cauline leaves, and the days when the first flower became visible. The plants were cultivated under long-day or short-day conditions. For GA treatment, 20 μM GA was administered by spraying plants growing under long days twice a week until the first flower appeared. For vernalization treatment, the Arabidopsis seeds were held at 4 °C for 22 d or 6 weeks in darkness and subsequently transferred to long-day conditions (Koornneef et al., 1991). At least 20 plants were measured and averaged for each measurement and statistical analysis. One-way ANOVA was used for statistical analysis of flowering time (*P<0.05; **P<0.01).
Generation of flc single mutants and khz1 khz2 flc triple mutants
Two short guide (sgRNA) targets (Target13683, TCAAGATCCTTGAT CGATA-TGG; and Target24055, AATATATGCAGCAAGCTTG-TGG) in the FLC gene were selected and cloned into the pHEE2A-TRI vector (Wang et al., 2015). The CRISPR (clustered regularly interspaced short palindromic repeat) constructs were transformed into the WT and khz1 khz2 mutants to generate the flc mutants and khz1 khz2 flc triple mutants, respectively. To obtain the homozygous mutants, we amplified the fragment surrounding the target by PCR using gene-specific primers (FLC-cri-LP/RP), and the product was analyzed by restriction enzyme digestion (ClaI for Target1 and HindIII for Target2). Fragments that could not be digested were sequenced. The specific primer pair zCas9-IDF3-2/rbcS_E9t-IDR was used to identify non-transgenic lines. The primers used in this study are presented in Supplementary Table S1 at JXB online.
Plasmid constructions
To generate the FT overexpression plasmid, the full-length coding sequence (CDS) was cloned into the binary vector pSuper 1300, which contains the Cauliflower mosaic virus (CaMV) 35S promoter. The resulting construct is referred to as 35S::FT. For the yeast two-hybrid assay, the full-length CDSs for KHZ1 and KHZ2 were cloned into the pGBKT7 and pGADT7 vectors (Clontech) to generate the KHZ1-BD, KHZ2-BD, KHZ1-AD, and KHZ2-AD constructs. For the bimolecular fluorescence complementation (BiFC) assay in Arabidopsis protoplasts, the full-length CDSs for KHZ1 and KHZ2 were cloned into the pSPYNE173 and pSPYCE(M) vectors to produce YN-KHZ1, YN-KHZ2, YC-KHZ1, and YC-KHZ2 constructs (Waadt et al., 2008). PCR amplification, enzyme digestion, and plasmid transformation were performed using standard protocols. Primers used for cloning are presented in Supplementary Table S2.
RNA isolation and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was isolated as previously reported (Oñate-Sánchez and Vicente-Carbajosa, 2008). First-strand cDNA was synthesized from 1 μg of total RNA using the M-MLV reverse transcription system (Takara, Beijing, China) according to the manufacturer’s instructions (Promega, China). qRT-PCR was performed in a volume of 20 μl using the ABI7500 Fast Real-time PCR system (Applied Biosystems, USA) and SYBR Green I Master Mix (Takara). AT4G34270 served as the internal control. All qRT-PCR primers are listed in Supplementary Table S3.
Subcellular localization assay
Transgenic plants roots of 7-day-old seedlings grown on Murashige and Skoog medium were used for subcellular localization assay. According to the method previously described (Kim and Somers, 2010), the plasmids were transformed into Arabidopsis protoplasts to observe the transient expression of the green fluorescent protein (GFP) fusion proteins with a Zeiss 710 Meta laser scanning confocal microscope (Zeiss LSM710, Germany).
Splicing efficiency analysis of FLC
Splicing efficiency was measured as previously described (Marquardt et al., 2014; Wu et al., 2016). Unspliced primers were designed to specifically span the intron–exon junction, while the spliced primers were designed to cross the exon–exon junction. Total RNA (2.5 μg) was reverse transcribed using specific primers and oligo d(T)18 to generate the spliced and unspliced forms of FLC. qRT-PCR was performed to determine the levels of spliced and unspliced FLC. AT4G34270 was used as an internal control. The spliced/unspliced ratio was calculated as the splicing efficiency. Data are presented as the means of three biological replicates. All the specific primers used for this analysis are presented in Supplementary Table S3.
Yeast two-hybrid assay
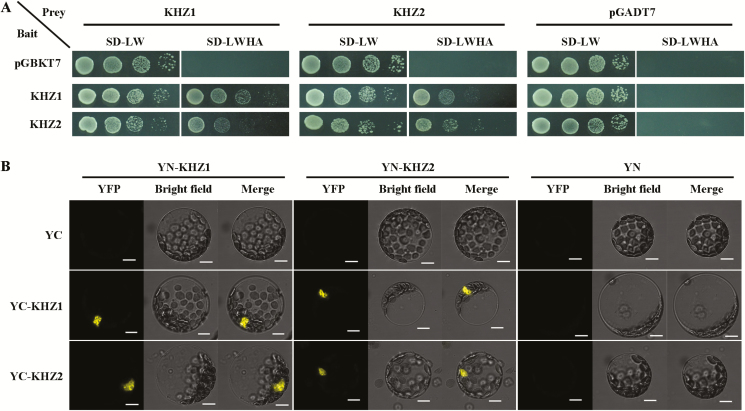
Yeast two-hybrid analysis was performed as previously described (Zhu et al., 2011). KHZ1-BD and KHZ2-BD served as the bait constructs, and KHZ1-AD and KHZ2-AD were the prey constructs. The empty vectors (pGBKT7 and pGADT7) were used as negative controls. Bait and prey constructs were co-transformed into yeast strain AH109. Transformants were picked from SD-Leu-Trp (SD-LW) medium, gradient-diluted, and plated on SD-Leu-Trp-His-Ade (SD-LWHA) medium. Pictures were taken after 4 d of growth at 30 °C. The experiment was performed with three independent biological replicates.
BiFC assay
BiFC assay was performed as described previously (Ho et al., 2009). Arabidopsis protoplasts were isolated from 4-week-old WT plants and transformed with different construct combinations. The protoplasts were incubated at 22 °C for 16–18 h, and then the BiFC signal was imaged using the channel for yellow fluorescent protein (YFP) fluorescence (Zeiss LSM710). Three independent biological replicates were performed.
Results
KHZ1 and KHZ2 are not components of the photoperiod pathway
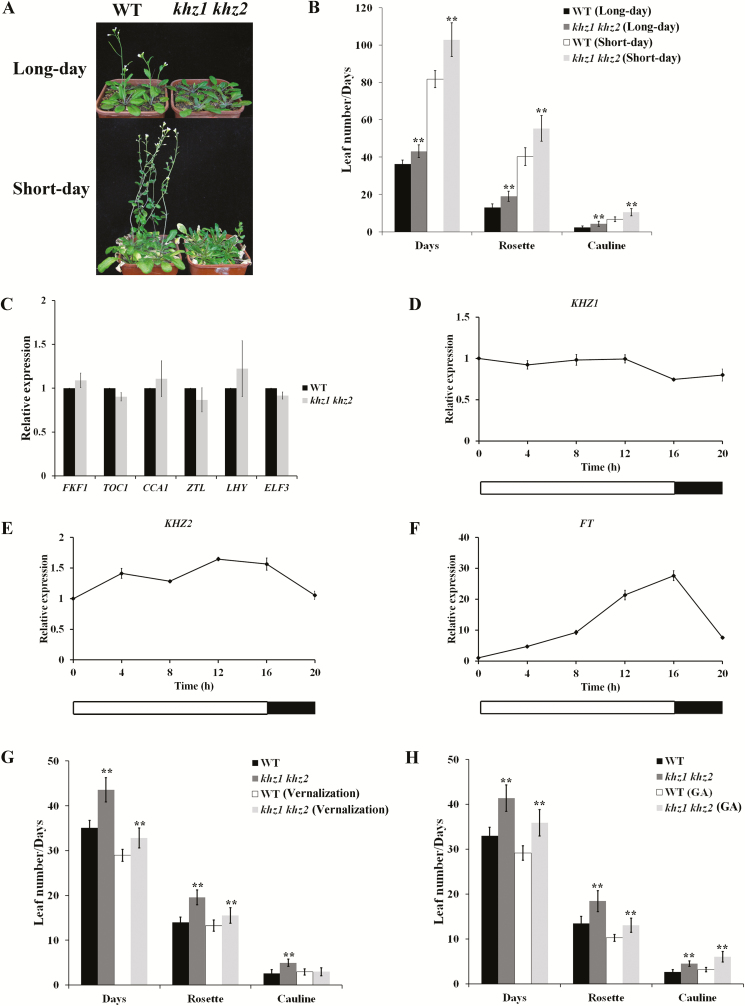
Our previous study revealed that KHZ1 (AT3G12130) and KHZ2 (AT5G06770) positively regulate flowering time redundantly in Arabidopsis (Yan et al., 2017). To explore the underlying mechanisms of the KHZ proteins in regulating flowering, we analyzed the flowering phenotype of khz1 khz2 double mutant plants under different photoperiod conditions. The flowering times of WT and khz1 khz2 plants were 80 d and 102 d, respectively, under short-day conditions, and 35 d and 43 d, respectively, under long-day conditions. Thus, the double mutant flowered later than the WT plants under both conditions. The late-flowering phenotype of the khz1 khz2 plants was confirmed by analyzing the number of leaves (Fig. 1A, B). These results demonstrated that the khz1 khz2 plants could respond to the photoperiod, suggesting that the KHZs might not be components of the photoperiod pathway.
Fig. 1.
khz1 khz2 flowering time analysis in response to different conditions. (A) The flowering phenotype of khz1 khz2 plants under long-day and short-day conditions. The pictures were taken on days 35 and 80 under long-day and short-day conditions, respectively. (B) The flowering time and leaf number of WT and khz1 khz2 plants grown under long-day and short-day conditions. (C) FKF1, TOC1, CCA1, ZTL, LHY, and ELF3 transcript levels in 10-day-old WT and khz1 khz2 plants. (D–F) Relative gene expression of KHZ1, KHZ2, and FT. Ten-day-old WT seedlings under long-day conditions were harvested every 4 h. Data are presented as the mean ±SD (n=3). (G) The flowering time and leaf number of WT and khz1 khz2 plants after vernalization treatment for 22 d. (H) The flowering time and leaf number of WT and khz1 khz2 plants following GA treatment. The flowering time and leaf number were recorded for at least 20 individual plants and are presented as the mean ±SD (one-way ANOVA, **P<0.01). (This figure is available in color at JXB online.)
Expression of the key genes of the photoperiod pathway were also examined, including FLAVINBINDING, KELCHREPEAT, F-BOX1 (FKF1), TIMING OF CAB EXPRESSION1 (TOC1), CIRCADIAN CLOCK ASSOCIATED1 (CCA1), ZEITLUPE (ZTL), LATE ELONGATED HYPOCOTYL (LHY), and EARLY FLOWERING3 (ELF3). As shown in Fig. 1C, expression of the tested photoperiod genes did not significantly differ between the khz1 khz2 and WT plants. The expression patterns of KHZ1, KHZ2, and FT were also assessed in WT plants under long-day conditions using qRT-PCR. The results showed that the expression of KHZ1 and KHZ2 was maintained at a constant level, whereas FT was rhythmic with a peak expression level in the late afternoon (Fig. 1D–F). These results further indicated that KHZ1 and KHZ2 are not components of the photoperiod pathway.
khz1 khz2 is responsive to vernalization and GA treatments
To examine the response of the khz1 khz2 plants to vernalization treatment, the seeds were incubated in a cold room at 4 °C for 22 d in darkness, and then the seedlings were transferred to long-day conditions. Vernalization-treated khz1 khz2 plants flowered earlier than untreated plants (32 d versus 43 d, respectively), albeit slightly later than the vernalization-treated WT plants (29 d) (Fig. 1G). The leaf numbers were also decreased by vernalization treatment, although the khz1 khz2 plants had more leaves than the WT plants (Fig. 1G). Vernalization treatment for 6 weeks produced similar results (Supplementary Fig. S1A, C). Consistent with the late-flowering phenotype of the khz1 khz2 double mutant, the FLC mRNA level was higher in untreated khz1 khz2 compared with WT plants, but the FLC mRNA level was drastically reduced after vernalization (Supplementary Fig. S1B). These results showed that the khz1 khz2 plants were sensitive to vernalization.
To analyze the effects of GA, a solution containing 20 μM GA3 was sprayed twice a week on the growing plants until they flowered. Similar to vernalization treatment, GA significantly promoted the flowering of khz1 khz2 plants. Flowering of GA-treated khz1 khz2 plants was initiated at 35 d with a rosette leaf number of ~13, while the untreated plants flowered at 41 d with a rosette leaf number of ~18. However, the GA-treated khz1 khz2 plants still flowered later than the GA-treated WT plants (Fig. 1H). These results demonstrated that the khz1 khz2 plants could respond to GA.
Loss of FLC function eliminates the late-flowering phenotype of khz1 khz2 plants
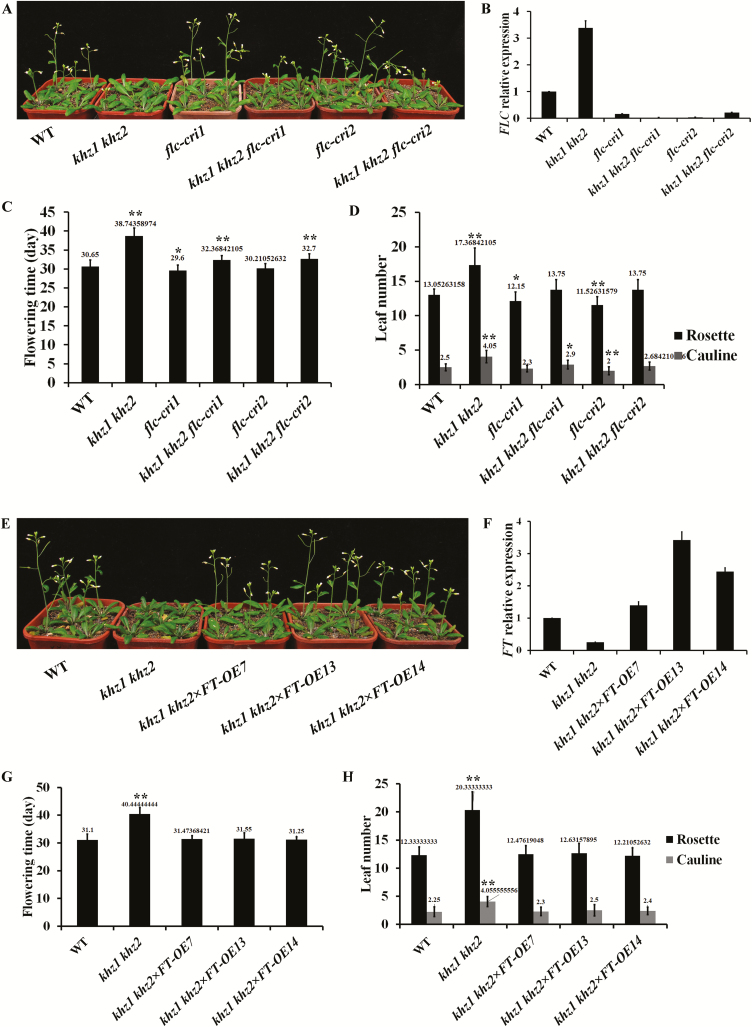
It is known that the mutants of the autonomous pathway genes flower late under both long-day and short-day conditions and are highly sensitive to vernalization and GA (Martinez-Zapater and Somerville, 1990; Koornneef et al., 1991; Mouradov et al., 2002; Moon et al., 2003; Lim et al., 2004). Because KHZ1 and KHZ2 share all these properties, we speculate that they may function in the autonomous pathway. The autonomous pathway genes promote flowering by repressing the central flowering repressor gene FLC (Cheng et al., 2017). Mutations in autonomous pathway genes combined with flc can rescue the late-flowering phenotype (Rouse et al., 2002; Eom et al., 2018). Thus, we investigated whether flowering time could be restored in khz1 khz2 mutants after combination with a mutation in FLC. We generated khz1 khz2 flc-cri triple mutants and flc-cri single mutants using CRISPR/Cas9 technology. Two sgRNA targets for FLC were selected and cloned into the pHEE2A-TRI vector and then transformed into khz1 khz2 and WT plants. We identified homozygous khz1 khz2 flc-cri and flc-cri lines among T2 progeny by sequencing. From this process, we obtained two khz1 khz2 flc-cri triple mutants and two flc-cri single mutants, of which khz1 khz2 flc-cri1 and flc-cri1 resulted from sgRNA Target1, khz1 khz2 flc-cri2 and flc-cri2 resulted from sgRNA Target2 (Supplementary Fig. S2A–D). Among these mutants, flc-cri1, flc-cri2, and khz1 khz2 flc-cri2 were non-transgene mutants (Supplementary Fig. S2E, F). The detection of FLC expression showed that the abundance of FLC mRNA was increased in khz1 khz2, and decreased in flc-cri and khz1 khz2 flc-cri mutants compared with the WT (Fig. 2B). Evaluation of the flowering phenotype of the mutants under long-day conditions revealed that the khz1 khz2 flc-cri triple mutants flowered much earlier than the khz1 khz2 double mutants, albeit slightly later than WT and flc-cri plants (Fig. 2A, C). We also examined the effect of flc mutation on the number of rosette leaves. The khz1 khz2 flc-cri triple mutants had a few more rosette leaves than WT and flc-cri plants, but far fewer rosette leaves than khz1 khz2 plants (Fig. 2D). The flowering phenotype of khz1 khz2 flc-cri triple mutants resembled the phenotype of flc plants, which flowered slightly earlier than WT plants. These results demonstrated that FLC loss of function largely rescued the late-flowering phenotype displayed by khz1 khz2 plants, which revealed that KHZ1 and KHZ2 are novel members of the autonomous pathway. Because mutation of FLC could not fully rescue the late-flowering phenotype of khz1 khz2, indicating that FLC is not the sole target of KHZs, there might be other factors involved in the pathway.
Fig. 2.
Mutation of FLC or overexpression of FT rescued the khz1 khz2 late-flowering phenotype. (A) The flowering phenotype of WT, khz1 khz2, flc-cri1, khz1 khz2 flc-cri1, flc-cri2, and khz1 khz2 flc-cri2 plants under long-day conditions. Pictures were taken on day 32. (B) FLC transcript levels in 10-day-old WT, khz1 khz2, flc-cri1, khz1 khz2 flc-cri1, flc-cri2, and khz1 khz2 flc-cri2 seedlings. (C) The flowering time and (D) leaf number of plants grown under long-day conditions. (E) The flowering phenotype of WT and khz1 khz2×FT-OE plants under long-day conditions. Pictures were taken on day 32. (F) FT transcript levels in 10-day-old WT, khz1 khz2, and khz1 khz2×FT-OE seedlings. (G) The flowering time and (H) leaf number of WT and khz1 khz2×FT-OE plants grown under long-day conditions. In (B) and (F), data are presented as the mean ±SD (n=3). The flowering time and leaf number were recorded for at least 20 individual plants and are presented as the mean ±SD (one-way ANOVA, *P<0.05, **P<0.01). (This figure is available in color at JXB online.)
FT can mediate signals from the photoperiod, vernalization, and autonomous pathways (Han et al., 2008; Blümel et al., 2015), and FT expression is repressed by FLC (Lee et al., 2000; Michaels et al., 2005; Helliwell et al., 2006). In addition, FT expression is significantly decreased in khz1 khz2 mutant plants (Yan et al., 2017). Therefore, we investigated whether FT overexpression could rescue the late-flowering phenotype observed for the khz1 khz2 plants. We introduced the 35S::FT plasmid into WT and khz1 khz2 plants to obtain the transgenic lines FT-OE and khz1 khz2×FT-OE, respectively. The FT-OE lines had two types of expression, low (1.5- to 3-fold) and high (1500- to 30 000-fold) (Supplementary Fig. S3A–D). FT expression in the khz1 khz2×FT-OE lines was similar to that in the low expression FT-OE lines (Fig. 2F; Supplementary Fig. S3B). The FT-OE high expression lines flowered much earlier than the WT (19 d versus 34 d), while the flowering time and rosette leaves of the FT-OE low expression lines and khz1 khz2×FT-OE lines were similar to those of the WT plants (Fig. 2E, G, H; Supplementary Fig. S3E, F).
The splicing of FLC pre-mRNA is altered in khz1 khz2 mutants
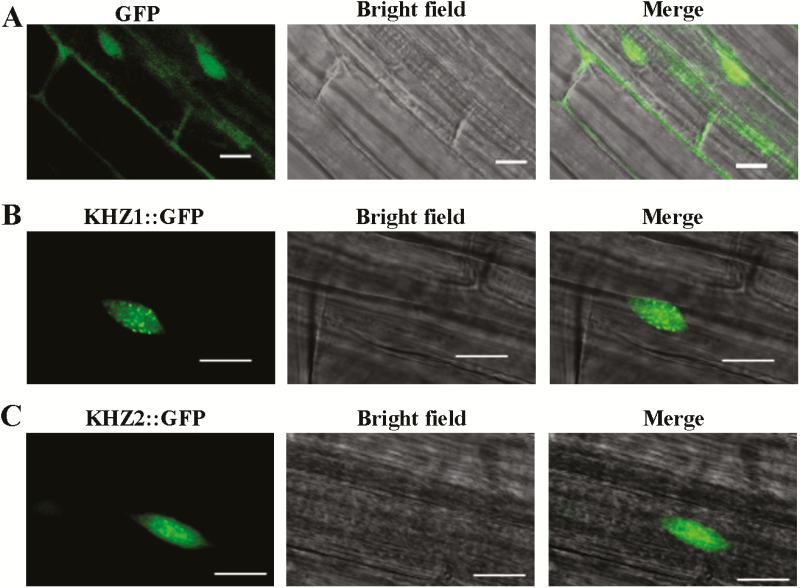
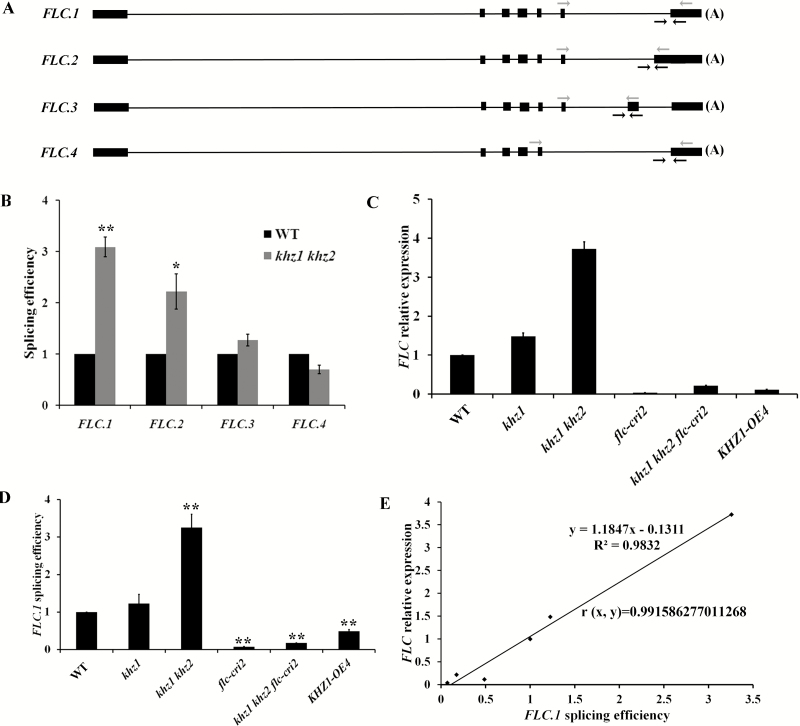
The subcellular localization of an RNA-binding protein and its biochemical function are closely related (Heintzen et al., 1997; Lorkovic and Barta, 2002). If the protein localized to the cytoplasm, it may regulate mRNA stability or translation. Alternatively, if the protein localized to the nucleus, it is more likely to modulate the pre-mRNA processing (e.g. pre-mRNA splicing) or mRNA transport (Lim et al., 2004). Here, we found that the GFP protein was evenly distributed in the cytoplasm and nucleus (Fig. 3A), while KHZ1::GFP and KHZ2::GFP were localized to the nucleus (Yan et al., 2017). In particular, KHZ1::GFP and KHZ2::GFP were localized to nuclear dots (Fig. 3B, C) that look like the nuclear speckles where many pre-mRNA splicing factors are localized (Wu et al., 2016; Xin et al., 2017). The nuclear dots of KHZ1::GFP were much brighter than those of KHZ2::GFP (Fig. 3B, C). Because KHZ1 and KHZ2 promote flowering mainly through the repression of FLC expression, we examined whether the splicing of FLC pre-mRNA was altered in khz1 khz2 mutants. We evaluated the splicing efficiency of specific introns of four different FLC isoforms (Fig. 4A). The splicing efficiency of intron 6 in isoform 1 (FLC.1) was significantly increased in the double mutant plants (Fig. 4B, D), while the splicing efficiency of FLC.1 in the KHZ1 overexpression lines was significantly decreased (Fig. 4D). The splicing efficiency of intron 6 in isoform 2 (FLC.2) was also significantly increased in the double mutant plants (Fig. 4B).
Fig. 3.
KHZ1::GFP and KHZ2::GFP are localized to nuclear dots in 7-day-old Arabidopsis root tips. (A) The GFP protein was evenly distributed in the cytoplasm and nucleus. The KHZ1::GFP (B) and KHZ2::GFP (C) proteins were distributed to nuclear dots. Scale bar=10 μm. (This figure is available in color at JXB online.)
Fig. 4.
Analysis of FLC splicing efficiency. (A) Schematic of FLC isoforms. The locations of the primers are indicated by arrows. The primers with gray arrows detected the spliced pre-mRNA, while the primers with black arrows detected the unspliced pre-mRNA. The spliced/unspliced ratio was calculated as the splicing efficiency. (B) Examination of splicing efficiency of select introns associated with the different FLC isoforms in WT and khz1 khz2 mutant plants. (C) FLC transcript levels in 10-day-old WT, khz1, khz1 khz2, flc-cri2, khz1 khz2 flc-cri2, and KHZ1-OE4 seedlings. (D) The splicing efficiency of FLC.1 in 10-day-old WT, khz1, khz1 khz2, flc-cri2, khz1 khz2 flc-cri2, and KHZ1-OE4 seedlings. (E) The relationship between FLC relative expression and FLC.1 splicing efficiency. The correlation coefficient was calculated by the Excel formula: =CORREL (array-x, array-y). *P<0.05, **P<0.01, data are presented as the mean ±SD for three replicates.
A previous report demonstrated that FLC.1 is the most abundantly expressed FLC isoform that encodes a functional FLC protein, while the other isoforms are expressed at much lower levels (Wu et al., 2016). Therefore, we examined the expression of FLC and the splicing efficiency of FLC.1 in WT, khz1, khz1 khz2, flc-cri2, khz1 khz2 flc-cri2, and KHZ1-OE4 plants. We found that the expression of FLC and the splicing efficiency of FLC.1 had a linear correlation (Fig. 4C–E) with a correlation coefficient as high as 0.99. These results further suggested that KHZ1 and KHZ2 down-regulated the FLC transcript level primarily by repressing the splicing efficiency of FLC.1.
KHZ1 and KHZ2 interact with each other or themselves in vitro and in vivo
The autonomous pathway accelerates flowering primarily by inhibiting the central flowering repressor FLC (Sang et al., 2017). Several autonomous pathway genes, such as FCA, FLD, FLK, FPA, FVE, FY, and LD, have been widely studied in this pathway (Simpson, 2004; Marquardt et al., 2006; Srikanth and Schmid, 2011; Cheng et al., 2017). However, none of these factors regulated one another at the mRNA level (Simpson, 2004). In the present study, we examined the relationship between the KHZ genes and other autonomous pathway genes in khz1 khz2 plants. No significant differences in the expression of autonomous pathway genes were observed between the khz1 khz2 and WT plants (Supplementary Fig. S4), indicating that KHZ1 and KHZ2 are not the regulators of the other autonomous pathway genes.
Previous studies had reported that some autonomous pathway proteins (e.g. FCA/FY) can physically interact and regulate RNA processing (Simpson et al., 2003). Thus, we determined whether KHZ1 and KHZ2 could interact with other autonomous pathway proteins using the yeast two-hybrid assay. The results showed that none of the tested proteins could interact with KHZ1 or KHZ2 (Supplementary Fig. S5). Interestingly, KHZ1 could interact with itself and KHZ2 in yeast (Fig. 5A). Using the BiFC assay, we also demonstrated that KHZ1 could interact with KHZ2 and itself in Arabidopsis protoplasts (Fig. 5B). In addition, KHZ2 could also interact with itself and KHZ1 in yeast and Arabidopsis protoplasts (Fig. 5). These results suggested that KHZ proteins could interact with each other to form heterodimers or with themselves to form homodimers.
Fig. 5.
Interactions of KHZ1 and KHZ2. (A) The interactions of KHZ1 and KHZ2 in yeast. Three independent experiments were performed with similar results. (B) KHZ1 and KHZ2 interactions in Arabidopsis protoplasts detected by BiFC assay. Scale bar=10 μm. Three independent experiments were performed with similar results. (This figure is available in color at JXB online.)
Discussion
We previously reported that the RNA-binding proteins KHZ1 and KHZ2 regulate flowering (Yan et al., 2017). However, to date, the underlying mechanisms have not been clear. Here, we provide an in-depth characterization of the mechanisms by which the KHZ proteins modulate flowering.
KHZ1 and KHZ2 are autonomous pathway factors
Autonomous pathway genes (e.g. FCA, FLD, FLK, FPA, FVE, FY, and LD) are thought to function largely in parallel with the vernalization pathway upstream of FLC and the photoperiod pathway (Boss et al., 2004; Simpson, 2004; Quesada et al., 2005). They repress FLC and thus act as promoters of the floral transition (Simpson, 2004). Mutations in these genes are generally recessive and delay flowering under both long-day and short-day conditions, and the inhibitory effect of the mutations can be overcome by vernalization or exogenous GA treatments (Abou-Elwafa et al., 2011). The regulation of flowering time by the autonomous pathway is mediated by FLC and the floral integrators downstream of FLC, such as FT. Mutations of FLC can eliminate the late-flowering phenotype caused by mutations in autonomous pathway genes (Koornneef et al., 1994; Lee et al., 1994; Sanda and Amasino, 1996; Michaels and Amasino, 2001; Abou-Elwafa et al., 2011). Similar to these autonomous pathway factors, the khz1 khz2 double mutant plants displayed a late-flowering phenotype under both long-day and short-day conditions when compared with the WT plants, and they were sensitive to vernalization and GA treatment (Fig. 1). Mutation of FLC in the khz1 khz2 double mutants significantly accelerated flowering. Moreover, the combination of khz1 khz2 with FT-OE lines fully rescued the late-flowering phenotype of khz1 khz2 double mutants (Fig. 2). Taken together, it can be concluded that Arabidopsis KHZ1 and KHZ2 are novel factors of the autonomous pathway.
Although vernalization promotes the flowering time significantly, it cannot fully rescue the late-flowering phenotype of khz1 khz2 mutants. Consistent with this effect, the khz1 khz2 flc-cri triple mutants flowered slightly later than the WT and the flc-cri mutants. We hypothesized that FLC is the main, but not sole, target of the KHZs; there may be other factors that participate in KHZ-mediated flowering regulation. This situation is similar to that of other autonomous pathway genes, such as FLK (Lim et al., 2004), LD (Lee et al., 1994), FLD (Sanda and Amasino, 1996), and PCFS4 (Xing et al., 2008). Furthermore, the autonomous pathway is known to mediate the effects of environmental cues (e.g. ambient growth temperature) (Blázquez et al., 2003; Halliday et al., 2003) and internal signals by ultimately modulating FT expression (Lim et al., 2004). Thus, we speculated that KHZs might also affect these factors to promote flowering.
KHZ1 and KHZ2 regulate the splicing efficiency of FLC
RNA-binding proteins play key roles in RNA metabolism (i.e. splicing, polyadenylation, mRNA stability, mRNA localization, and translation) (Lee and Schedl, 2006). The CCCH domain and the KH domain are two critical RNA-binding elements in RNA-binding proteins. Proteins that contain these domains have multiple functions in regulating plant development and hormone and stress responses. For instance, HUA1, which contains six CCCH-type zinc finger motifs, is a regulator of stamen and carpel identities (Li et al., 2001). The KH domain RNA-binding protein, HOS5, is important for pre-mRNA splicing in Arabidopsis and contributes to the tolerance against various stresses and the responses to the hormones abscisic acid (ABA) and jasmonic acid (JA) (Xiong et al., 1999; Chen et al., 2013; Thatcher et al., 2015).
RNA-binding proteins are also involved in the autonomous pathway. FLC levels are primarily regulated by autonomous pathway proteins through RNA processing, post-transcriptional regulation, and chromatin modification (Cheng et al., 2017; Eom et al., 2018). For example, FCA and FPA, two RNA recognition motif-containing RNA-binding proteins, regulate alternative polyadenylation of antisense RNAs and 3' end formation of FLC (Macknight et al., 1997; Schomburg et al., 2001; Liu et al., 2007; Hornyik et al., 2010; Liu and Mara, 2010). FLK, an RNA-binding protein with three KH domains, is part of the autonomous pathway and represses FLC at the transcriptional level or through RNA-mediated chromatin regulation (Veley and Michaels, 2008; Ripoll et al., 2009). To date, the precise molecular mechanisms by which the majority of autonomous pathway proteins regulate FLC expression are unknown.
KHZ1 and KHZ2 are two RNA-binding proteins that contain both the CCCH and KH domains, and they are both localized to the nucleus (Yan et al., 2017). In the current study, we found that KHZ1 and KHZ2 are specifically localized to nuclear dots (Fig. 3), which are very like nuclear speckles, where pre-mRNA splicing factors are localized (Spector and Lamond, 2011). This result indicated that the KHZ proteins might be involved in the regulation of the pre-mRNA process, similar to other autonomous pathway proteins.
In Arabidopsis, FLC is the key repressor of flowering and the integrator of the autonomous pathway. It has four isoforms, and the most abundantly expressed isoform is FLC.1, which encodes the functional FLC protein (Wu et al., 2016; Cheng et al., 2017). Our study revealed that the KHZ proteins repressed the splicing efficiency of FLC pre-mRNA, especially FLC.1, and there was a positive linear correlation between splicing efficiency and FLC levels (Fig. 4). These results suggest that the KHZ proteins promote flowering mainly by suppressing the production of mature FLC mRNA.
KHZ proteins may function as dimers
The flowering time of plants is affected by well-delineated factors that are involved in complex genetic networks in A. thaliana. Eom et al (2018) demonstrated that the factors in the autonomous pathway were independent of each other at the transcriptional level. For example, mutation in FLK does not affect the expression of FCA or FPA, and the expression of FLK is normal in fca or fpa mutants (Lim et al., 2004). These findings are consistent with our own results that the khz1 khz2 double mutants did not alter the expression of other autonomous pathway genes. We believed that KHZ proteins might act in parallel to other autonomous pathway proteins to promote FLC expression.
At the translation level, Simpson et al (2003) revealed that a physical interaction between FCA and FY is important for RNA processing. In this study, KHZ1 and KHZ2 did not physically interact with any of the other autonomous pathway proteins tested (Supplementary Fig. S5). They did, however, interact with each other and themselves (Fig. 5), suggesting that KHZ1 and KHZ2 form hetero- or homodimers. In our previous work, we showed that KHZ1 and KHZ2 are redundant for promoting flowering, but KHZ1 activity is more predominant than that of KHZ2 (Yan et al., 2017). Therefore, we proposed that KHZ1–KHZ1 is the primary dimer, with dimers utilizing KHZ2 used as an alternative.
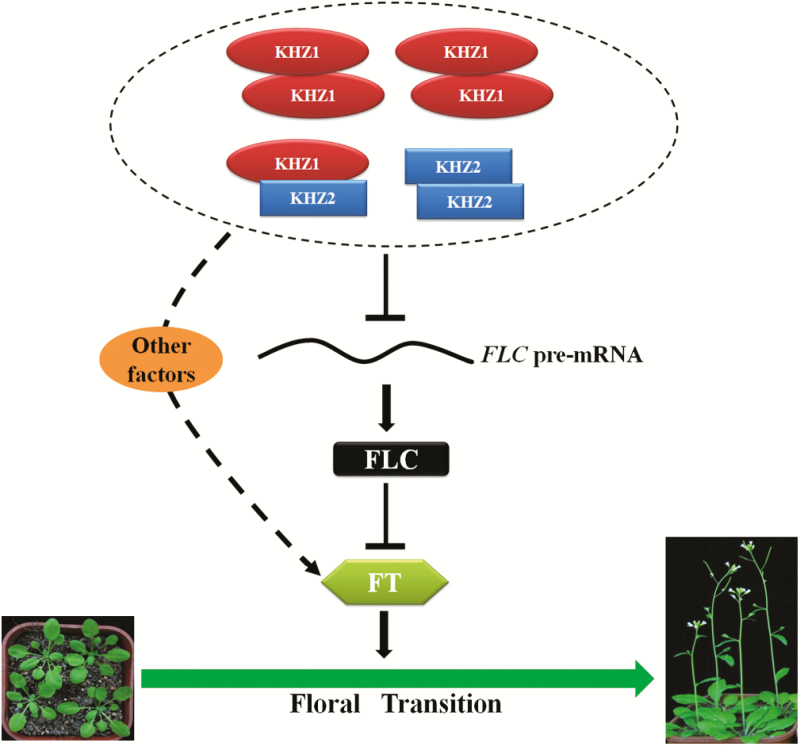
In our work, we identified KHZ1 and KHZ2 as novel members of the autonomous pathway. They may act as homo- or heterodimers to repress FLC mRNA levels through the suppression of the splicing efficiency of FLC.1, which subsequently leads to the up-regulation of FT expression (Fig. 6). Based on the fact that the late-flowering phenotype of khz1 khz2 was almost completely rescued by mutation of FLC or completely rescued by FT overexpression, the KHZ proteins might also promote flowering by modulating FT through other factors in parallel. Our results suggest that KHZ1 and KHZ2 are involved in the regulation of the floral transition through the autonomous pathway. This study sheds more light on the molecular mechanisms of the autonomous pathway in Arabidopsis and provides a better understanding of KH and CCCH domain-containing proteins. However, further investigation is still needed to fully understand the exact mechanism.
Fig. 6.
A working model of KHZ proteins in the regulation of the floral transition. KHZ proteins may function as homodimers or heterodimers (most prominently KHZ1–KHZ1 homodimers) to inhibit the splicing efficiency of FLC, which in turn relieves the inhibition of FT. These events promote the transition from vegetative to reproductive growth. Mutation of FLC cannot fully rescue the late-flowering phenotype of khz1 khz2, but FT overexpression can completely rescue the late-flowering phenotype of khz1 khz2, which indicates that KHZ proteins may also regulate other factors which then modulate FT to promote flowering. (This figure is available in color at JXB online.)
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Flowering time analysis after vernalization treatment for 6 weeks.
Fig. S2. The generation of flc-cri and khz1 khz2 flc-cri mutants by CRISPR/Cas9.
Fig. S3. The analysis of FT-overexpressing lines.
Fig. S4. Expression levels of autonomous pathway genes in 10-day-old WT and khz1 khz2 seedlings.
Fig. S5. Interaction analysis of KHZ1 and KHZ2 with autonomous pathway proteins in yeast.
Table S1. Primers used for genotyping.
Table S2. Primers used for plasmid construction.
Table S3. Primers used for qRT-PCR.
Acknowledgement
The study was supported by a grant from the National Natural Science Foundation of China (no. 31570254). The authors declare that they have no conflict of interest.
Author contributions
YH conceived the research, and supervised the experiments; ZY designed the experiments and prepared the figures; ZY and HS performed the experiments; YL and MJ provided technical assistance; and ZY, HS, and YH wrote the manuscript.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Abou-Elwafa SF, Büttner B, Chia T, Schulze-Buxloh G, Hohmann U, Mutasa-Göttgens E, Jung C, Müller AE. 2011. Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. Journal of Experimental Botany 62, 3359–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali GS, Palusa SG, Golovkin M, Prasad J, Manley JL, Reddy AS. 2007. Regulation of plant developmental processes by a novel splicing factor. PLoS One 2, e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Lee I, Weigel D, Amasino RM. 1999. The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. The Plant Journal 18, 195–203. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D. 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Green R, Nilsson O, Sussman MR, Weigel D. 1998. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. The Plant Cell 10, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. 1997. LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844. [DOI] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C. 2015. Flowering time regulation in crops—what did we learn from Arabidopsis? Current Opinion in Biotechnology 32, 121–129. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. 2004. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell 16(Suppl), S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Payyavula RS, Chen L, Zhang J, Zhang C, Turgeon R. 2018. FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proceedings of the National Academy of Sciences, USA 115, 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Cui P, Chen H, Ali S, Zhang S, Xiong L. 2013. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genetics 9, e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JZ, Zhou YP, Lv TX, Xie CP, Tian CE. 2017. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiology and Molecular Biology of Plants 23, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. . 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Eom H, Park SJ, Kim MK, Kim H, Kang H, Lee I. 2018. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. The Plant Journal 93, 79–91. [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C. 2001. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Geraldo N, Bäurle I, Kidou S, Hu X, Dean C. 2009. FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiology 150, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC. 2003. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. The Plant Journal 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H. 2008. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. The Plant Journal 55, 253–265. [DOI] [PubMed] [Google Scholar]
- Hayama R, Coupland G. 2003. Shedding light on the circadian clock and the photoperiodic control of flowering. Current Opinion in Plant Biology 6, 13–19. [DOI] [PubMed] [Google Scholar]
- He Y. 2009. Control of the transition to flowering by chromatin modifications. Molecular Plant 2, 554–564. [DOI] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM. 2003. Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. 1997. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 94, 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. The Plant Journal 46, 183–192. [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG. 2010. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Developmental Cell 18, 203–213. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proceedings of the National Academy of Sciences, USA 93, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. 1993. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. The Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G. 2009. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. The Plant Journal 60, 614–625. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM. 2009. Vernalization: winter and the timing of flowering in plants. Annual Review of Cell and Developmental Biology 25, 277–299. [DOI] [PubMed] [Google Scholar]
- Kim J, Somers DE. 2010. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiology 154, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Blankestijndevries H, Hanhart C, Soppe W, Peeters T. 1994. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. The Plant Journal 6, 911–919. [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular & General Genetics 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes & Development 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. 1994. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. The Plant Cell 6, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T. 2006. RNA-binding proteins. Wormbook: the online review of C elegans biology, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. 2002. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. [DOI] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X. 2001. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. The Plant Cell 13, 2269–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM. 2004. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. The Plant Cell 16, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. 2010. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97. [DOI] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C. 2007. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Molecular Cell 28, 398–407. [DOI] [PubMed] [Google Scholar]
- Liu Z, Mara C. 2010. Regulatory mechanisms for floral homeotic gene expression. Seminars in Cell & Developmental Biology 21, 80–86. [DOI] [PubMed] [Google Scholar]
- Lorković ZJ, Barta A. 2002. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Research 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, et al. . 1997. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Marquardt S, Boss PK, Hadfield J, Dean C. 2006. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. Journal of Experimental Botany 57, 3379–3386. [DOI] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. 2014. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Molecular Cell 54, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Somerville CR. 1990. Effect of light quality and vernalization on late-flowering mutants of Arabidopsis thaliana. Plant Physiology 92, 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. 2009. Flowering time regulation produces much fruit. Current Opinion in Plant Biology 12, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. The Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. 2005. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiology 137, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Yu X, Shalitin D, et al. . 2004. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proceedings of the National Academy of Sciences, USA 101, 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. 2003. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. The Plant Journal 35, 613–623. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. 2002. Control of flowering time: interacting pathways as a basis for diversity. The Plant Cell 14(Suppl), S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Dean C, Simpson GG. 2005. Regulated RNA processing in the control of Arabidopsis flowering. International Journal of Developmental Biology 49, 773–780. [DOI] [PubMed] [Google Scholar]
- Ripoll JJ, Rodríguez-Cazorla E, González-Reig S, Andújar A, Alonso-Cantabrana H, Perez-Amador MA, Carbonell J, Martínez-Laborda A, Vera A. 2009. Antagonistic interactions between Arabidopsis K-homology domain genes uncover PEPPER as a positive regulator of the central floral repressor FLOWERING LOCUS C. Developmental Biology 333, 251–262. [DOI] [PubMed] [Google Scholar]
- Rouse DT, Sheldon CC, Bagnall DJ, Peacock WJ, Dennis ES. 2002. FLC, a repressor of flowering, is regulated by genes in different inductive pathways. The Plant Journal 29, 183–191. [DOI] [PubMed] [Google Scholar]
- Samach A, Coupland G. 2000. Time measurement and the control of flowering in plants. Bioessays 22, 38–47. [DOI] [PubMed] [Google Scholar]
- Sanda SL, Amasino RM. 1996. Ecotype-specific expression of a flowering mutant phenotype in Arabidopsis thaliana. Plant Physiology 111, 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang S, Chen Y, Yang Q, Wang P. 2017. Arabidopsis inositol polyphosphate multikinase delays flowering time through mediating transcriptional activation of FLOWERING LOCUS C. Journal of Experimental Botany 68, 5787–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM. 2001. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. The Plant Cell 13, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Development 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Ruelens P, D’hauw M, Maggen T, Dochy N, Torfs S, Kaufmann K, Rohde A, Geuten K. 2017. A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiology 173, 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. 2004. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology 7, 570–574. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. 2003. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harbor Perspectives in Biology 3, a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. 2011. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68, 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D. 2008. The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. The Plant Journal 56, 239–250. [DOI] [PubMed] [Google Scholar]
- Sun T, Goodman HM, Ausubel FM. 1992. Cloning the Arabidopsis GA1 locus by genomic subtraction. The Plant Cell 4, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. [DOI] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Olszewski NE. 2001. Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiology 126, 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Kamphuis LG, Hane JK, Oñate-Sánchez L, Singh KB. 2015. The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS One 10, e0126978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veley KM, Michaels SD. 2008. Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiology 147, 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. The Plant Journal 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Wang C, Tian Q, Hou Z, Mucha M, Aukerman M, Olsen OA. 2007. The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Reports 26, 1357–1366. [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. 2015. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biology 16, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. 1992. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiology 100, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhu D, Lin X, et al. . 2016. RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. The Plant Cell 28, 55–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E. 2017. SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proceedings of the National Academy of Sciences, USA 114, E7018–E7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Zhao H, Xu R, Li QQ. 2008. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. The Plant Journal 54, 899–910. [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. 1999. HOS5—a negative regulator of osmotic stress-induced gene expression in Arabidopsis thaliana. The Plant Journal 19, 569–578. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. 2005. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant & Cell Physiology 46, 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yan Z, Jia J, Yan X, Shi H, Han Y. 2017. Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Molecular Biology 95, 549–565. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. 2003. Living by the calendar: how plants know when to flower. Nature Reviews Molecular Cell Biology 4, 265–275. [DOI] [PubMed] [Google Scholar]
- Yu Y, Liu Z, Wang L, et al. . 2016. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. The Plant Journal 85, 96–106. [DOI] [PubMed] [Google Scholar]
- Yuan S, Zhang ZW, Zheng C, et al. . 2016. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proceedings of the National Academy of Sciences, USA 113, 7661–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, et al. . 2011. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.