Using a toxin/anti-toxin system, comprehensive proteome and metabolome analyses of roots of transgenic Taraxacum koksaghyz plants with or without latex exudation provide insights into latex-specific protein isoforms and metabolic pathways.
Keywords: Articulated laticifers, barnase/barstar, differentially accumulated proteins, latex-non-exuding phenotype, metabolomics, proteomics, Taraxacum koksaghyz
Abstract
Taraxacum koksaghyz has been identified as one of the most promising alternative rubber crops. Its high-quality rubber is produced in the latex of laticifers, a specialized cell type that is organized in a network of elongated tubules throughout the entire plant body. In order to gain insights into the physiological role(s) of latex and hence laticifer biology, we examine the effects of barnase-induced latex RNA degradation on the metabolite and protein compositions in the roots. We established high-quality datasets that enabled precise discrimination between cellular and physiological processes in laticifers and non-laticifer cell types of roots at different vegetative stages. We identified numerous latex-specific proteins, including a perilipin-like protein that has not been studied in plants yet. The barnase-expressing plants revealed a phenotype that did not exude latex, which may provide a valuable genetic basis for future studies of plant–environment interactions concerning latex and also help to clarify the evolution and arbitrary distribution of latex throughout the plant kingdom. The overview of temporal changes in composition and protein abundance provided by our data opens the way for a deeper understanding of the molecular interactions, reactions, and network relationships that underlie the different metabolic pathways in the roots of this potential rubber crop.
Introduction
The Russian dandelion Taraxacum koksaghyz produces considerable amounts of high-quality rubber and other valuable secondary metabolites in the latex of specialized cells called laticifers (van Beilen and Poirier, 2007; Schulze Gronover et al., 2011). The biosynthesis of natural rubber mainly occurs in laticifers of roots, where they form a network of longitudinally elongated, anastomosing tubes (Ulmann, 1951; Hagel et al., 2008; Pickard, 2008; Schulze Gronover et al., 2011). In cross-sections, they appear as symmetrical, concentric circles separated by parenchyma cells, and these circles are gradually formed in the course of plant development and are continuously pushed into the periphery by secondary growth (Ulmann, 1951). Thus, laticifers are present in various differentiated cell stages at the same time within the root. In addition, previous studies have shown that the metabolic activities relating to synthesis of rubber, inulin, and resin in the roots are dependent on their age and on the plant developmental stage (Ulmann, 1951; Kreuzberger et al., 2016).
Natural rubber consists of ~94% of poly(cis-1,4-isoprene) and the remaining 6% is made up of compounds that include associated proteins, carbohydrates, and lipids as well as isoprene-derived compounds such as plant sterols and terpenoids that are most likely to be at least partly responsible for the extraordinary properties of this biopolymer (Schulze Gronover et al., 2011). Commercial natural rubber is mainly produced and harvested from the latex of the rubber tree Hevea brasiliensis, and latex composition, exudation, and its coagulation are subjects of constant research (D’Auzac et al., 1989; Men et al., 2019; Shi et al. 2019). In recent years, molecular analyses have shed more light on the factors that underlie rubber biosynthesis in Taraxacum species. Thus, functional gene studies have confirmed the central role of the mevalonate (MVA) pathway in providing isoprenoid precursors, with 3-hydroxy-3-methyl-glutaryl-coA reductase (HMGR) as the rate-limiting enzyme, (van Deenen et al., 2012), while other proteins such as cis-prenyltransferases (CPTs), the rubber transferase activator CPTL1 (CPT-like protein), the rubber elongation factor (REF), and small rubber particle proteins (SRPPs) have proved to be crucial for rubber biosynthesis and storage (Collins-Silva et al., 2012; Hillebrand et al., 2012; Post et al., 2012; Epping et al., 2015; Laibach et al., 2015). Other studies have identified additional enzymes such as ATP citrate lyase (ACL), acetoacetyl-CoA thiolase (AACT), squalene synthase (SQS), and squalene epoxidase (SQE) to be pivotal for the biosynthesis and structural diversity of isoprene end products in the latex of dandelion species (Pütter et al., 2017; Unland et al. 2018). A potential connection between the reserve carbohydrate inulin and secondary metabolite synthesis in T. koksaghyz has been demonstrated by overexpression of the inulin-degrading enzyme fructan 1-exohydrolase (1-FEH), which results in a significant increase in the rubber content (Stolze et al., 2017). As a result, its easy genetic accessibility and the recent availability of a draft genome sequence (Lin et al., 2018) make T. koksaghyz a powerful model system for further in-depth analysis of rubber biosynthesis and function.
In contrast to rubber and inulin synthesis, little is known about the laticifer system at different root ages and its interplay with the surrounding tissues. In order to address this, we used the bacterial barnase (bn)/barstar (bs) system to generate transgenic T. koksaghyz plants that lacked latex exudation. A modified version of the bn gene was expressed under the control of the REF promoter, which is predominantly active in laticifers (Laibach et al., 2015). To minimize detrimental effects of potential background bn activity in non-target tissues, in particular during plant regeneration in sterile culture, the bn inhibitor bs was co-expressed under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. We then carried out comprehensive metabolomic and proteomic analyses of young, middle-aged, and fully developed roots of the two individual latex-exuding control genotypes, namely the near-isogenic line (NIL) and bs, as well as the non-exuding bn/bs plants. This approach excluded any protein and/or metabolite contamination derived from other non-laticifer cell types of the root and provided for the first time a detailed, in-depth analysis of the systems biology of T. koksaghyz laticifers. In addition, the comprehensive dataset obtained from the analogous NIL provided a comprehensive overview of protein accumulation patterns and metabolite composition in roots of T. koksaghyz at three different vegetation stages.
Materials and methods
Construction of plant transformation vectors
For heterologous expression of barstar (bs) and barnase (bn) in Taraxacum koksaghyz, two vectors were constructed. For bs, the full-length coding sequence (Supplementary Table S1 at JXB online) was amplified via PCR from the pMT1002 vector (Addgene plasmid # 8621; Hartley et al., 1996) using the primer barstar_fwd_XhoI and barstar_rev_XbaI (Supplementary Table S2), and then purified using a PCR clean-up Gel extraction kit (Macherey-Nagel GmbH & Co. KG, Germany). After digestion with XhoI/XbaI, it was then inserted into the binary expression vector pLab12.1, which had also been digested with XhoI/XbaI (Post et al., 2012). For gene expression, the Cauliflower mosaic virus (CaMV) 35S promoter was inserted into the PstI/XhoI sites upstream of the bs coding sequence. For bn, a truncated, codon-optimized version of the bn gene was designed in silico on the basis of the Bacillus amyloliquefaciens coding sequence (403–732 bp; Locus BACBRNA, Accession M14442) and then purchased as a GeneArt® Strings™ DNA Fragment (Supplementary Table S1) from Invitrogen. Sequence modifications were as follows: the signal peptide sequence was removed; five mutations (Q15I, T16R, G65S, T79V, K108R) were introduced to improve bn protein stability, as described previously (Serrano et al., 1993); and a modified sequence (190 bp, deletion of the adenine at eight nucleotides from the 3´-end) of the first catalase intron of Ricinus communis (Mehrotra et al., 2014) was inserted into the Arg59 codon of the bn gene as reported by Hanson et al. (1999). Based on this artificial gene fragment, the bn coding sequence was PCR-amplified using the primer combination barnase_fwd_XhoI and barnase_rev_XbaI, and subsequently cloned into the binary expression vector pLab12.10_pREF (Xing et al., 2014) following the same strategy as described above for the bs gene. In pLab12.10_pREF, gene expression is driven by the latex-specific REF promoter. The REF promoter was replaced by the CaMV 35S promoter for transient agro-infiltration studies.
For bn gene editing, the construction of the CRISPR/Cas9 vector was performed as previously described (Fauser et al., 2014) with slight alterations. A GatewayTM cloning-compatible expression vector was provided by Holger Puchta (Karlsruhe Institute of Technology, Germany). First, the original resistance cassette was replaced by the phosphinothricin gene flanked by the mannopine synthase promoter and terminator, which was derived from the vector pFGCGW (GenBank accession number DQ231581). The protospacer sequence targeting the bn gene was designed using the online CRISPR gRNA design tool ATUM (https://www.atum.bio/eCommerce/cas9/input). Potential ‘off-target’ sequences were validated by blasting the protospacer sequences against the available T. koksaghyz genome (Lin et al., 2018). Preparation of the protospacer was performed by aligning two oligonucleotides with corresponding sequences (Supplementary Table S3). The integrity of all final constructs was validated by sequencing.
Cultivation of plants and processing of root material
Wild-type and transgenic T. koksaghyz plants were cultivated at 18 °C and 20 klux (high pressure sodium lamp, with enhanced yellow and red spectrum) with a 16‐h photoperiod in a controlled greenhouse, as previously described by Unland et al. (2018). Roots were separated by cutting at the root crown, freeze-dried, and pulverized using a ZM 200 Ultra Centrifugal Mill (Retsch, Germany).
Agrobacterium-mediated stable transformation and molecular characterization of transgenic plants
Transgenic T. koksaghyz plants were obtained using accession number MS03 (kindly provided by the Botanical Garden of the University of Muenster, Germany) following established protocols (Stolze et al., 2017). After bs transgenic lines had been established they were transformed with the bn gene. The final selection of transgenic plants harboring both genes was achieved by cultivation on media containing kanamycin and phosphinothricin. Plants transformed with the CRISPR/Cas9 expression vector were selected with phosphinothricin. The identification of transgenic plants was performed by PCR with gene-specific primers (Supplementary Table S2) using a KAPA3G Plant PCR Kit with crude leave extracts (Kapa Biosystems, USA).
Transient gene expression
Agrobacterium-mediated transformation of Nicotiana benthamiana leaves was carried out as previously described (Müller et al., 2010). The bn construct driven by the CaMV 35S promoter and an empty vector control were infiltrated in leaves of 4-week-old N. benthamiana and 12-week-old T. koksaghyz wild-type plants. The N. benthamiana plants were cultivated from seeds (obtained from the Sainsbury Laboratory, University of Cambridge, UK) in the same way as previously described for T. koksaghyz (Unland et al., 2018). The infiltrated plants were incubated for 3–4 d under constant light conditions (13 klux) at 19 °C in a plant growth chamber (CLF Plant Climatics GmbH, Germany).
Metabolite analyses
In all cases, finely pulverized root material was used. 1H-NMR, HPLC, and GC-MS analyses were used for quantification of poly(cis-1,4-isoprene), sugars, squalene/2,3-oxidosqualene, pentacyclic triterpenes, and plant sterols as previously described (Stolze et al., 2017).
For determination of fatty acids, fatty acid methyl ester derivatives were produced by transesterification and subsequently quantified using GC-MS. Samples of 25 mg of freeze-dried, finely ground root tissue were mixed with 975 µl of 3 M HCl in methanol (1:4) and 25 µl pentadecanoic acid (internal standard, stock solution 10 mM in methanol) in HPLC vials and incubated overnight at 60 °C. Then, 800 µl was transferred to a 15-ml falcon tube and thoroughly mixed with 1.2 ml NaCl (5%) and 2 ml hexane. After centrifugation for 5 min at 3700 g, the supernatant was transferred to a new 15-ml falcon tube and the hexane extraction was repeated. The resulting hexane extract (~4 ml) was thoroughly mixed with 1.6 ml NaHCO3 (2%), centrifuged (5 min at 3700 g), evaporated, and finally dissolved in 1 ml hexane. A volume of 1 µl was used for GC-MS analysis. Single triterpenes and fatty acid methyl ester were identified and quantified by GC-MS (as mg g–1 root DW), and corresponding retention indices (RIs) were determined in relation to a C8–C40 alkane calibration standard.
Solvent extraction, spectrometric analysis, and determination of high molecular weight poly(cis-1,4-isoprene) content
Rubber was initially extracted from powdered root samples using the Dionex™ Accelerated Solvent Extractor (ASE)™ 350 system (ThermoFisher Scientific), similar to as described previously by Ramirez-Cadavid et al. (2018). Sample preparation and handling with the ASE system was performed according to the manufacturer’s instructions. All extraction processes were carried out at a constant pressure of 10 MPa and a flush volume of 150%. The samples were initially extracted with actone using an automatic three-extraction cycle program (each cycle lasted 10 min) at 40 °C. This was followed by an extraction with n-hexane at 100 °C using the same three-cyle program as for acetone. For this purpose, powdered root samples were mixed with diatomaceous earth and transferred into 10-ml stainless steel extraction cells that had each been fitted with a cellulosic filter (diameter 27 mm) at the bottom of the cell. The collection vials were weighed before and after the extraction in order to determine the dry weight of the extracts: the n-hexane extracts were dried using a Rocket™ Evaporator (ThermoFisher Scientific) according to the manufacturer’s instructions, and to ensure that the extracts were completely free of solvent, they were subsequently dried for 30 min in a Concentrator Plus (Eppendorf).
Fourier-transform infrared spectroscopy (FTIR) was applied to the dried n-hexane residues to identify the major components using an IRTracer-100 FTIR-spectrophotometer (Shimadzu) equipped with an Attenuated Total Reflection (ATR) module according to the manufacturer’s instructions. The n-hexane extracts were fixed on the measuring surface of the ATR module using a pressure punch. Measurements were made in the range 3600–650 cm–1 with a count of 20 scans.
The amounts and molecular weights of poly(cis-1,4-isoprene) within the n-hexane extracts were determined using a TF2000 thermal field‐flow fractionation system coupled to MALS and ELS detectors (Postnova Analytics, Germany). The extracts were dissolved at 40 °C in toluene to a final concentration of 5 mg ml–1. The samples were then centrifuged for 60 min at 1000 g and the supernatant was diluted 1:3 with toluene. The fractionation was carried out with an injection volume of 100 µl and toluene as the carrier liquid with a flow rate of 0.2 ml min–1. The signals obtained from the ELS detector and the MALS angles 28–156° were used for the calculation with the following parameters: Random coil as calculation type, dn/dc value 0.104 ml g–1, extinction 0.62 ml (g×cm)–1.
Quantitative real-time PCR
For verification of endogenous expression, total RNA was isolated using an innuPREP RNA Mini Kit (Analytik Jena, Germany) and 500 ng samples were subjected to cDNA synthesis using PrimeScript™ RT Master Mix (Clontech). Subsequent quantitative real-time PCR was carried out as previously described (Laibach et al., 2015) using root as well as latex cDNA derived from 12-week-old wild-type plants. For it, three biological replicates were analysed. The housekeeping genes elongation factor 1a (Ef1a) and ribosomal protein L27 (RP) were used for the normalization of gene expression (Pütter et al., 2017). The oligonucleotides used are listed in Supplementary Table S2.
Statistical evaluation of the root composition analysis
Since the data obtained were found not to be normally distributed, a non-parametric and hence more conservative test had to be employed, and therefore we used Mann-Whitney U-tests OriginPro 2016. Since T. koksaghyz exhibits considerable genetic variability (Ulmann, 1951), a significance level of P<0.01 was stringently applied to guarantee the validity of the data and the inferred deductions.
Microscopic examination of root tissue
For light microscopy, the Leica TCS SP5 X confocal unit was used. Fresh root material was cut into pieces (~0.5–1cm), submerged in liquid nitrogen, and fixed with Tissue-Tek O.C.T. Compound (Sakura, Japan) on cork pads. Cross-sections (20 µm) were cut with a Leica CM3050 S cryomicrotome. Laticifers were identified by staining the hydrophobic components in latex with Nile Red (Sigma-Aldrich). Since this stain is highly solvatochromic, meaning that the emitted fluorescent shifts depend on the polarity of the environment (Diaz et al., 2008), lambda scans were performed to record the emission spectrum between 510–590 nm for each genotype. The excitation wavelength was 488 nm (Argon laser). To calculate the emission spectra, four stained images of each genotype were analysed, whilst four independent latex areas were set as regions of interest.
Longitudinal sections of fresh roots were cut with a razor blade and treated with BODIPY 493/503 (ThermoFisher Scientific) to stain and visualize lipophilic substances and hence to identify laticifer cells. As counterstains, we used Fluorescent brightener 28 (FB28) for the cell walls and DAPI (both Sigma-Aldrich) for the nuclear structure. For TEM, fresh roots were cut into pieces, then fixed in osmium-potassium ferrocyanide, and embedded in Epon. For fixation, the root tissues were first incubated overnight in 2.5% glutaraldehyde in 0.1 M Na-cacodylate buffer pH 7.4 (Na-Caco) and then washed three times with with Na-Caco for 20 min each. The tissues were then submerged for 45 min in 1% osmium(VIII) oxide +1.5% potassium ferrocyanide in Na-Caco, and afterward washed three times with Na-Caco for 10 min each. For the final fixation, the tissues were transferred to 1% osmium(VIII)-oxide in Na-Caco for 60 min and then washed five times with Na-Caco for 5 min each. Tissue dehydration was then carried out through a graded ethanol series, after which the samples were processed using the following propylene oxide/Epon series: 20 min in pure propylene oxide; 4 h in 1:2 propylene oxide/Epon mix; 4 h in 1:1 mix; overnight in 2:1 mix; 2× 4 h in pure epon; and overnight in pure Epon. Finally, the tissues were embedded in pure Epon and polymerized for 36 h at 60 °C. Cross-sections and longitudinal sections were imaged using a Philips CM10 TEM.
Protein immunoblotting
To obtain the total root protein contents from the latex-exuding (NIL, bs) and non-exuding (bn/bs) plants, 60 mg samples of finely ground root powder were extracted with 300 µl protein extraction buffer (100 mM sodium phosphate buffer, pH 7, 1 mM EDTA, 0.1% SDS, 0.1% Triton X-100, 20 mM DTT) for 15 min on ice. The extracts were centrifuged twice at 16 000 g for 10 min at 4 °C to remove cell debris. In addition, the total protein content from wild-type latex was extracted as previously described (Post et al., 2012). The protein concentrations were determined using Bradford assays (Bradford, 1976). Subsequently, equal amounts of protein were separated by SDS-PAGE on 12% SDS polyacrylamide gels and transferred to nitrocellulose membranes via western blotting. The transfer of proteins was confirmed by Ponceau S staining (0.1% Ponceau S in 5% acetic acid). For immunodetection, the following primary antibodies were used: a mouse antibody against Arabidopsis AACT2 (dilution 1:1 000; kindly provided by Basil Nikolau of the Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, USA) (Jin et al. 2012); a rabbit antibody against T. brevicorniculatum SRPPs (dilution 1:500) (Hillebrand et al., 2012); and a rabbit antibody against T. koksaghyz fructan:fructan 1-fructosyl transferase (FFT; dilution 1:1 000). The FFT antibody was synthesized by Genosphere Biotechnologies (France) using the following peptide sequence as the epitope: YDAYKDKWTPDNPEFD. Detection was carried out using either horseradish peroxidase-labeled secondary goat anti-mouse (Catalog no. 32 430) or goat anti-rabbit (Catalog no. 32 460) IgG antibodies diluted 1:1500 before use (Sigma-Aldrich).
Protein extraction and peptide desalting
Protein extraction was performed based on an adapted filter-assisted sample preparation (FASP) protocol (Wiśniewski et al., 2009). To obtain the total protein content from the roots, 50-mg samples of finely ground root powder were extracted using 400 µl of protein extraction buffer (4% SDS, 0.1 M DTT, 0.1 M Tris-HCl, pH 7.6) for 5 min at 95 °C. The extracts were centrifuged twice at 18 000 g for 30 min to remove cell debris. Protein extracts (80 µg) were diluted 8-fold with 8 M urea in 0.1 M Tris-HCl, pH 7.6, and the samples were transferred to 0.5-ml 30 kDa MWCO centrifugal filter devices (Vivacon 500 30K, Sartorius). Repeated washes, alkylation with chloroacetamide, and digestion were performed as previously described (Wozny et al., 2018). The digested peptides were desalted and fractionated using SDB StageTips.
LC-MS/MS data acquisition
Dried peptides were dissolved in 2% ACN, 0.1% TFA for analysis. Samples (0.5 µg) were analysed using an EASY-nLC 1200 coupled to a Q Exactive HF mass spectrometer (both ThermoFisher Scientific). Peptides were separated on 20-cm frit-less silica emitters (New Objective, 0.75 µm inner diameter), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9 µm resin (Dr. Maisch HPLC GmbH, Germany). The column was kept at 50 °C in a column oven throughout the run. Peptides were eluted for 115 min using a segmented linear gradient of 5% to 98% solvent B (solvent A: 0% acetonitrile, 0.5% formic acid; solvent B: 80% acetonitrile, 0.5% formic acid) at a flow rate of 300 nl min–1. Mass spectra were acquired in data-dependent acquisition mode with a Top15 method. MS spectra were obtained in an Orbitrap analyser with a mass range of 300–1759 m/z at a resolution of 60 000 FWHM, maximum IT of 55 ms, and a target value of 3×106 ions. Precursors were selected with an isolation window of 1.3 m/z. HCD fragmentation was performed at a normalized collision energy of 25. MS/MS spectra were acquired with a target value of 1×105 ions at a resolution of 15 000 FWHM, maximum IT of 55 ms, and a fixed first mass of m/z 100. Peptides with a charge of +1, ≥6, or with unassigned charge state were excluded from fragmentation for MS2; dynamic exclusion for 30 s prevented repeated selection of precursors.
Proteomic data analysis and annotation
Raw data were processed using the MaxQuant software (Tyanova et al., 2016 a) (v.1.5.2.8; http://www.maxquant.org/). MS/MS spectra were searched using the Andromeda search engine against a dandelion database (Lin et al., 2018) (Genome Warehouse, http://bigd.big.ac.cn/gwh/; accession no. PRJCA000437) with added bs and bn sequences (Supplementary Table S1). Sequences of 248 common contaminant proteins and decoy sequences were automatically added during the search. Trypsin specificity was required and a maximum of two missed cleavages was allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of cysteine residues was set as fixed, and oxidation of methionine and protein N-terminal acetylation were set as variable modifications. Label-free quantification (LFQ) was enabled with a minimum ratio count of 1. Peptide-spectrum matches and proteins were retained if they were below a false discovery rate of 1%. The match between runs was enabled. Downstream data analysis was performed using Perseus v.1.6.0.7 (Tyanova et al., 2016 b). ‘Reverse hits’, ‘contaminants’, and ‘only identified by site’ were removed. The LFQ intensities were log2-transformed. Plotting of the log2 raw and the log2 LFQ intensities confirmed that the automatic normalization procedure of MaxQuant worked reliably (Supplementary Fig. S1), and the log2 LFQ intensities were used for all further analyses. The log2 LFQ intensities were also depicted as histograms to ensure the normal distribution of all samples (Supplementary Fig. S2). Protein intensities that were detected only at one time-point and only in one genotype had to be present in at least three out of four biological replicates in order to be retained in further analysis; otherwise, they were regarded as false positives and removed. The resulting matrices encompassed 8726 protein groups (Supplementary Fig. S3, Supplementary Table S4). For function annotation analysis of the determined protein sequences, homology searches were conducted against the non-redundant NCBI peptide database (NR, http://www.ncbi.nlm.nih.gov) and UniProtKB/Swiss-Prot (http://www.uniprot.org/) (February 2018). The functional annotation tool from DAVID (the database for annotation, visualization, and integrated discovery) was used to identify enriched GO (Gene Ontology) terms (Huang et al., 2009a, 2009b). WEGO (Web Gene Ontology Annotation Plot) was used for visualizing and plotting the GO annotation results (Ye et al., 2006). The PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System of the Gene Ontology Reference Genome Project was used to determine protein classes (Mi et al., 2017). Volcano plots and heatmaps were generated using Perseus v.1.6.0.7 (Tyanova et al., 2016 b). Venn diagrams were plotted using InteractiVenn (http://www.interactivenn.net/).
Proteome data processing for temporal protein abundance in roots and laticifers
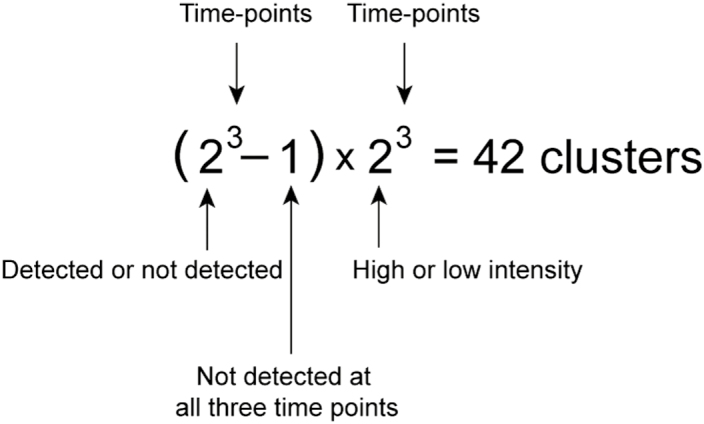
To gain insights into the temporal changes in protein expression of wild-type roots, the initial proteome dataset of the near-isogenic line (NIL) was filtered for proteins that were detected in at least three out of four replicates for at least one time-point. This resulted in the NIL proteome matrix being reduced to 8106 proteins (Supplementary Table S5). In order to generate a meaningful heatmap, the protein intensities declared as NaN (not detected) had to be replaced by a random number. Since the weakest log2 intensity in the whole NIL time-course dataset for 12-week-old plants was 19.14 (protein ID 1662), the 19 was set as the imputation value threshold. Based on the heatmap output, 42 clusters were calculated with the following formula:
The expression profiles of each cluster are depicted in Supplementary Fig. S4. The respective cluster assignments of all 8106 proteins can be found in Supplementary Table S5.
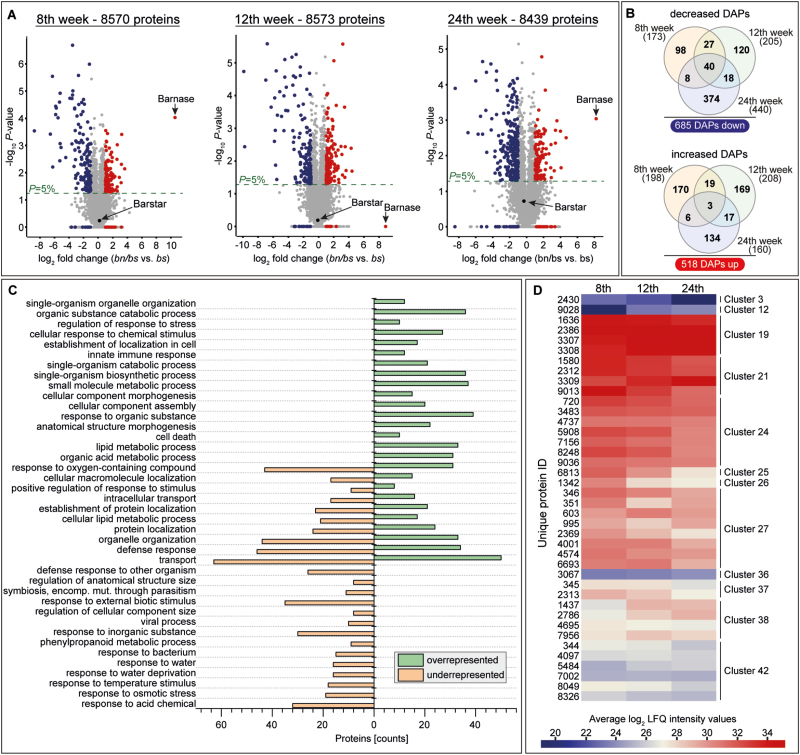
To identify differentially accumulated proteins (DAPs), the proteome data of the bn/bs transgenic plants were compared with the corresponding data of plants harboring bs alone. Prior to this, we checked each time-point for proteins that showed differential abundance between the two control genotypes, NIL and bs. These proteins were removed from further analyses to ensure that they were not mistakenly considered as potential DAPs in the comparison between the bn/bs and the bs transgenic plants. Proteins that showed a log2 ratio of >1 or <–1 and P<0.05 were considered as differentially accumulated. In addition, proteins that were not detectable (invalid log2 ratio) or were almost absent in one of the controls (quantified only for one spectrum out of four replicates, and thus not allowing the generation of a P-value) were also taken into account. However, a prerequisite for this was the detection of these proteins in the other comparative genotype in at least three out of four replicates. By removing these proteins, the following three matrices for further DAP analyses were generated: 8th week (8570 proteins), 12th week (8573 proteins), and 24th week (8439 proteins) (Supplementary Tables S6–S8). The identification of significant DAPs between bn/bs and bs transgenic plants at all three time-points was then conducted with the research criteria detailed above.
Results
Generation of transgenic Taraxacum koksaghyz expressing the bn/bs system
For the generation of barnase (bn) and barstar (bs) transgenic T. koksaghyz plants, we constructed respective expression vectors that included a modified version of the native bn gene. In order to evaluate the enzymatic activity, a CaMV 35S-driven bn gene construct was first expressed in N. benthamiana and T. koksaghyz leaves by agro-infiltration. Necrotic areas were clearly visible at the infiltration points 2–3 d post-infiltration for both species (Supplementary Fig. S5A, B), thereby indicating the functionality of bn.
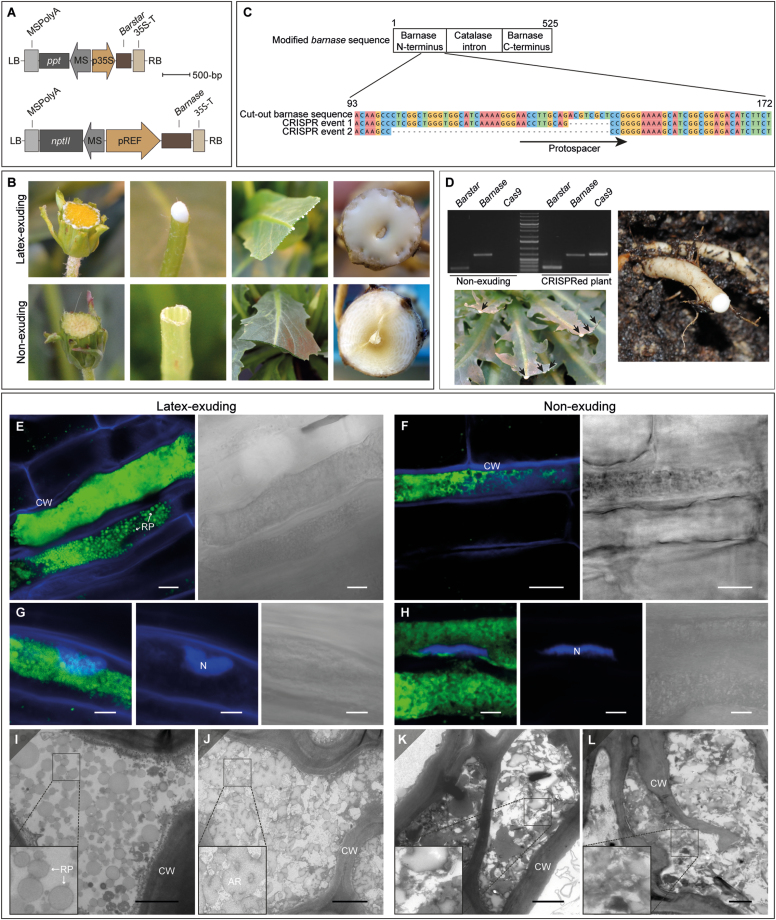
To achieve dominant expression of bn exclusively in laticifers, the gene was placed under the control of the REF promoter, which is almost latex-specific, and then expressed together with the CaMV 35S-controlled bs gene in transgenic T. koksaghyz (Fig. 1A). Seven independent transgenic plants were obtained, of which one did not exude any latex. This non-exuding (bn/bs) plant was crossed with the wild-type to determine whether the non-exuding phenotype co-segregated with the controlled laticifer-specific activity of bn in the resulting T1 generation. According to Mendelian rules, the offspring should have divided into four different genotypes if bn and bs were not linked to the same locus, namely plants without any transgene (near-isogenic lines, NILs), plants with either bs or bn, and plants with both transgenes (bn/bs). As expected, no bn transgenic offspring were identified, and the T1 generation showed the predicted number of latex-exuding (NIL and bs) and non-exuding (bn/bs) plants. In common with the T0 generation, none of the plants showed any abnormal growth (Fig. 1B, Supplementary Fig. S6). To definitively verify that the non-exuding phenotype in the bn/bs plants was only caused by the activity of bn, a loss-of-function mutation in the corresponding transgene was generated using the CRISPR/Cas9 gene editing tool. Two individual plants carrying the loss-of-function versions of the gene were identified (Fig. 1C) and in both of them the latex exudation was reconstituted, demonstrating that this phenotype was indeed mediated by the activity of bn (Fig. 1D).
Fig. 1.
Generation and characterization of transgenic plants of Taraxacum koksaghyz that lack exudation of latex. (A) Schematic representation of T-DNAs carrying either barstar (bs) under the control of the Cauliflower Mosaic Virus 35S promoter (p35S), or the rubber elongation factor promoter (pREF)-controlled and codon-optimized barnase (bn) gene plus integrated catalase intron. Resistance gene cassettes: MS, mannopine synthase promoter; ppt, phosphinothricin resistance gene; nptII, kanamycin resistance gene; MSPolyA, PolyA sequences from mannopine synthase gene. (B) Images of flowers, petioles, leaves, and root crown cuttings. Top: bs-transgenic control plants (latex-exuding). The root had a diameter of 1 cm, which was representative for the control plants (i.e. the near-isogenic line and bs).; Bottom: non-exuding plants expressing bs and bn. The root had a diameter of 1.3 cm. (C) CRISPR/Cas9-induced editing of bn. (D) Verification of bs and bn, and Cas9 expression in offspring derived from crossings between non-exuding and CRISPR/Cas9-gRNA transgenic plants. The images show the reconstituted latex flow in leaves and root of edited plants. (E–H) Confocal laser-scanning microscopy images of exuding bs (E, G) and non-exuding bn/bs (F, H) transgenic plants. Longitudinal sections were stained with either (E, F) BODYPI and Fluorescent brightener 28 (FB28) or (G, H) BODYPI and DAPI. FB28 and DAPI were used to visualize the cell wall and nucleus whilst BODYPI was used to stain lipophilic substances and thus to identify the laticifers. (E, F) Single 2D images; (G, H) are composites made from stacked 2D images (G=8, H=6 images). (I–L) TEM images of cross-sections and longitudinal sections from (I, J) bs controls and (K, L) bn/bs non-exuding transgenic plants. RP, rubber particle; AR, accumulated rubber; CW, cell wall; N, nucleus. Scale bars: (E, F) 10 µm, (G, H) 5 µm, (I–L) 2 µm.
Since the induced non-exuding phenotype was stably inherited, we chose the offspring of the non-exuding × wild-type cross for further in-depth analyses. In addition to the NILs, plants expressing bs only were also analyzed as control to exclude any impact of bs on plant morphology, metabolism and/or physiology.
Microscopic analysis of the laticifer network
We studied the influence of bn activity on laticifer anatomy by use of fluorescence microscopy. Comparison of cross-sections of latex-exuding (NILs and bs) and non-exuding (bn/bs) plants stained with Nile Red indicated the presence of a similar laticifer network with non-polar content in all three genotypes (Supplementary Fig. S7A). However, the emissions of the fluorophore from the sections of the bn/bs transgenic plants were shifted towards longer wavelengths (maximum 564 nm) compared to the controls (maximum 543–549 nm), i.e. from green to more yellowish, indicating an increased polarity of the composition of the stain within the laticifers (Supplementary Fig. S7B). For a higher resolution of the laticifer anatomy, we examined the bs (as a control) and bn/bs plants in more detail by staining longitudinal sections with BODIPY in combination with FB28 (Fig. 1E, F) and DAPI (Fig. 1G, H). In the bs plants the fluorescence was coherent, and distinct fluorescent particles appeared in some regions of the laticifer cytoplasm (Fig. 1E). In contrast, whilst the bn/bs plants also displayed fluorescence, distinct fluorescent particles were not observed (Fig. 1F). DAPI staining identified a large and prolate spherical-shaped nucleus in the laticifers of the bs plants (Fig. 1G), while in the bn/bs plants the nucleus appeared more as an elongated structure (Fig. 1H).
TEM of cross- and longitudinal sections revealed that the bs plants harbored a typical laticifer cell structure with distinct rubber particles and accumulated rubber (Fig. 1I, J). In contrast, the bn/bs plants showed no distinct cytoplasmic structures and, in some parts, a very high electron density (Fig. 1K, L). Nonetheless, these plants did have the characteristic outer laticifer cell wall structure.
Metabolite profiles in roots
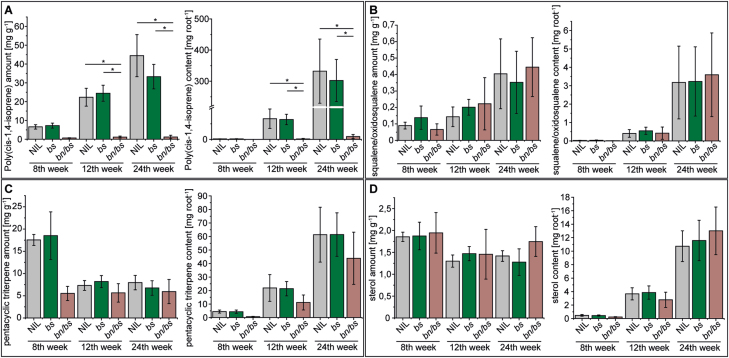
We performed time-series analyses in order to create temporal root-specific metabolic profiles of the latex-exuding and non-exuding plants. The perennial growth of T. koksaghyz is characterized by four vegetative stages (Javorsky, 1944; Ulmann, 1951), which are slow overall growth of the plant (stage 1), increased development of the root system (stage 2), maturity of fruits (stage 3), and mass development of the plant body (stage 4). An age of 8 weeks represents stage 1, 12 weeks represents stages 2 and 3, and 24 weeks represents stage 4, and these time-points were selected for optimal monitoring of the plant development. Whole-roots were subjected to various extraction methods, and metabolite concentrations together with the total content per root were determined. The results showed that throughout the cultivation period considered, the concentration of poly(cis-1,4-isoprene) in the latex-exuding NIL and bs control plants increased continuously from a mean of 6.7 mg g−1 at 8 weeks old up to ~39 mg g−1 at 24 weeks old, with a total mean content of ~320 mg per root system. In contrast, the non-exuding bn/bs plants exhibited a strong reduction in poly(cis-1,4-isoprene) and it remained at approximately the same low level at all three time-points, with a maximum content of 9.2 mg per root system in 24-week-old plants (Fig. 2A, Supplementary Table S9). In addition, a more detailed analysis clearly showed that the poly(cis-1,4-isoprene) formed during growth in the NIL and bs control plants almost exclusively consisted of high molecular-weight cis-1,4-polymer chains (Number average molecular mass (Mn) and mass average molar mass (Mw) ~106 g mol−1). In contrast, the low levels of poly(cis-1,4-isoprene) detected in the bn/bs plants contained only small amounts of high molecular-weight polymers (max. 1.1 mg per root system in 24 week-old bn/bs plants; Supplementary Fig. S8A-C, Supplementary Table S9).
Fig. 2.
Secondary metabolites in roots of transgenic Taraxacum koksaghyz plants that either exude latex (barstar, bs, and the near-isogenic line, NIL) or do not exude latex (barnase, bn/bs). Roots were sampled at 8, 12, and 24 weeks old. Data are means (±SD): for the 8th week five pooled samples of four plants each were sampled, and for the 12th and 24th weeks six individual plants were sampled. Metabolite concentrations and total contents per root were determined for (A) poly(cis-1,4-isoprene), (B) squalene/2-3-oxidosqualene, (C) pentacyclic triterpenes, and (D) sterols. Pentacyclic triterpene concentrations and contents were calculated based on the detection of α- and β-amyrin, lupeol, lup(19,21)-en-3-ol, taraxasterol, and a further two unidentified triterpenes. Sterol concentrations and contents were calculated based on the detection of campesterol, stigmasterol, and sitosterol. Due to non-normal distribution of the data, significance levels among the genotypes at each time-point were assessed using non-parametric Mann–Whitney U-tests (*P<0.01).
We next quantified other MVA pathway-derived metabolites in the roots, namely squalene/2,3-oxidosqualene, triterpenes, and sterols (Fig. 2B–D; Supplementary Tables S10–S12). In all three genotypes, we observed an overall increase in the total amount of these compounds during development due to the increased biomass. The concentration of squalene/2,3-oxidosqualene slightly increased over time, while the concentration of sterols remained more-or-less constant with slight fluctuations (Fig. 2B, D). The highest concentration of triterpenes (18.02 mg g−1) was found in 8-week-old NIL and bs control plants, while bn/bs plants had much lower concentrations (5.53 mg g−1) (Fig. 2C).
To verify that the introduced bn/bs system was not negatively affecting essential physiological functions of the plant, we also performed temporal profiling of fatty acids and sugars. We found that the total amount of fatty acid methyl esters (Supplementary Fig. S9A, Supplementary Table S13) and the storage carbohydrate inulin (Supplementary Fig. S9B) continuously increased throughout the growing season in a similar way for all the genotypes, while the concentrations showed variability but no significant differences among the genotypes. All genotypes at all the time-points also showed a similar degree of polymerization for inulin (Supplementary Fig. S9C).
The highest concentrations of fructose and glucose were found in 8-week-old plants, after which they declined (Supplementary Fig. S9D, E). The concentrations of sucrose in the roots showed high variation among the genotypes across all the time-points. One significant difference was identified for 24-week-old plants, where the concentration of sucrose in the bn/bs plants exceeded that in the bs plants (Supplementary Fig. S9F).
Generation of a proteome dataset for the roots
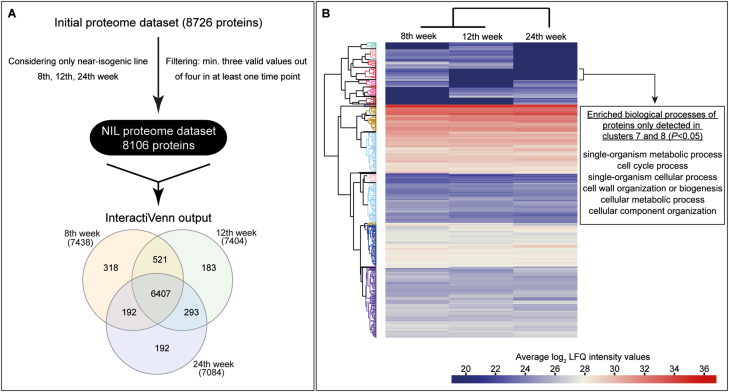
Using the same plant material as for the results presented above, we quantified whole-protein extracts from 8-, 12-, and 24-week-old plants using LC-MS, and the MS/MS spectra obtained were searched against a protein database generated from the previously published T. koksaghyz genome (Lin et al., 2018). After identification and quality-filtering, the proteome matrix encompassed 8726 protein groups containing bs and bn (Supplementary Fig. S3, Supplementary Table S4). Hierarchical cluster analysis showed a relatively closer association between the 8- and 12-week samples compared with those at 24 weeks. Within each time-point, the closest association always appeared between the NIL and bs control plans, whilst the non-exuding bn/bs plants were clearly distant from them. BLAST searches against the NCBI (NR) and UniProtKB/Swiss-Prot databases were conducted, and annotations were found for 8719 proteins (99.94%) and 8626 proteins (98.88%), respectively. Of the NCBI annotations, 99.9% derived from plant proteins, with the largest homology to Helianthus annuus (56.5%). In the UniProtKB/Swiss-Prot annotations, 80.5% were traced back to plant proteins, with the highest homology to Arabidopsis thaliana (75.7%). The most prominent protein identity ranges of the NCBI and UniProtKB/Swiss-Prot annotations were 80–95% and 60–80%, respectively. A GO classification of the proteins was performed, and the following terms were the most abundant: ‘cell’ and ‘cell part’ in cellular components, ‘binding’ and ‘catalytic activity’ in molecular function, and ‘cellular’ and ‘metabolic process’ in biological process.
To first obtain a fundamental picture of the temporal protein abundance of wild-type roots, the proteome data derived from the NIL were analysed in more detail. The initial proteome dataset was filtered for proteins that were detected in at least three out of the four replicates for at least one time-point (see Methods), and in this way the dataset was reduced to 8106 proteins (Fig. 3A, Supplementary Table S5). Of these, 6407 proteins (~79%) were detected at all three time-points. Hierarchical clustering indicated that the proteomes of 8- and 12-week-old NIL plants had more similarities than the proteome of 24-week-old NIL plants, which clustered separately. Based on this heatmap, we calculated that there were 42 clusters (see Methods), where proteins with similar temporal accumulation patterns and accumulation strength were clustered together (Fig. 3B, Supplementary Fig. S4, Supplementary Table S5). This enabled a better overview of protein occurrences that were dependent on the growth stage of the roots. For example, by using these clusters in combination with the GO enrichment analysis, we were able to show that the proteins that were only accumulated at an early stage of root development (8 weeks old) were classified as mainly being involved in biological processes such as metabolic processes and cellular development (Fig. 3B, Supplementary Table S14).
Fig. 3.
Generation of a temporal proteomic map of Taraxacum koksaghyz roots. (A) The filtering procedure used to obtain the proteome dataset of the latex-exuding near-isogenic line (NIL). The Venn diagram illustrates the distribution of the dataset across the three time-points at which sampling was conducted. (B) For hierarchical cluster analysis the mean Log2 Label-Free Quantification (LFQ) intensities per time-point were calculated and compared. Based on all proteins, 42 clusters were calculated to assemble proteins together that possessed a similar expression pattern and strength, as depicted in the tree. The cluster allocations of the whole dataset are given in Supplementary Fig. S4 and Supplementary Table S5. In the example shown, a GO enrichment analysis was conducted for clusters 7 and 8 to identify the corresponding biological processes using the functional annotation tool from DAVID. GO terms with a P<0.05 are depicted (see also Supplementary Table S14).
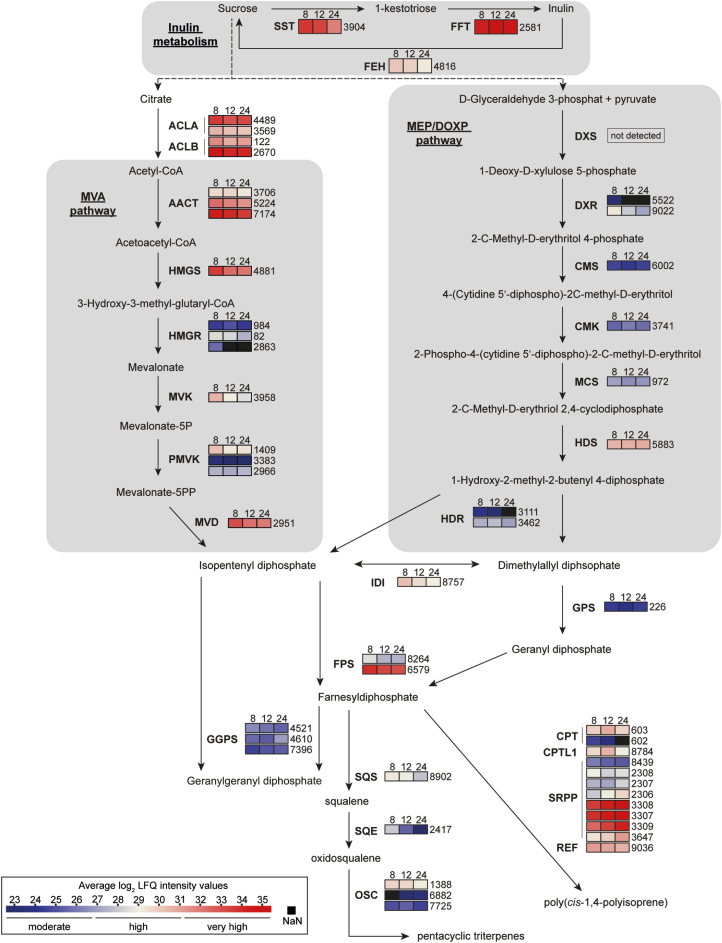
The temporal resolution of the abundance of proteins that participate in inulin metabolism or in biosynthesis of secondary metabolites derived from isopentenyl pyrophosphate [e.g. poly(cis-1,4-isoprene), triterpenes] may help to provide insights into biosynthetic activities during plant growth. We therefore determined the log2 LFQ intensities of the predicted proteins attributed to these pathways, and found that at every time-point, the key enzymes of inulin synthesis 1-SST (protein ID 3904) and FFT (ID 2581) showed a very high abundance that exceeded that of the inulin-degrading enzyme 1-FEH (ID 4816) (Fig. 4). Enzymes such as ACLA (IDs 4489, 3569) and ACLB (IDs 122, 2670) that act upstream of the MVA pathway were present almost constantly at a high to very high level. The amounts of all the MVA-pathway enzymes detected were constant, except for one predicted HMGR (ID 2863), which was found only in 8-week-old plants. The abundance of each enzyme remained the same at all time-points, but the intensity level among them differed from moderate to very high. The first enzyme of the MEP pathway (1-deoxy-D-xylulose-5-phosphate synthase, DXS) was not detected. One DX reductase (DXR; ID 5522) of the MEP pathway was only present in 8-week-old plants and one 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR; ID 3111) homolog was only present in in 8- and 12-week-old plants. The other enzymes belonging to the MEP pathway showed constant moderate-to-high abundances at all the time-points. Downstream enzymes such as isopentenyl diphosphate isomerase (IDI; ID 8757) and farnesyl diphosphate synthases (FPSs; IDs 8264, 6579) had abundances that were high to very high, while the predicted geranyl diphosphate synthase (GPS; ID 226) and geranyl GPSs (GGPSs; IDs 4521, 4610, 7396) had moderate abundances at all time-points. Enzymes related to rubber biosynthesis, such as CPT2 (ID 603), REF (ID 9036), CPTL1 (ID 8784), three predicted SRPP1 homologs (IDs 2307, 2308, 8439), and SRPP2–6 (IDs 2306, 3308, 3307, 3309, 3647) showed log2 LFQ intensities that were high to very high throughout the course of the experiment. CPT3 (ID 602) was only present at a moderate level in 8- and 12-week-old plants. SQS (ID 8902), which synthesizes squalene, was present at a rather high level, but it declined during the course of development. The squalene oxidizing enzyme SQE (ID 2417) showed a high abundance at the 8-week time-point, but it declined drastically during subsequent development. Some oxidosqualene cyclizing (OSC) enzymes were detected: one predicted OSC (ID 1388) showed a constant high abundance, whilst another (ID 7725) was present at a moderate level. A third OSC (ID 6882) was only detected at 12 and 24 weeks, and showed moderate intensity levels.
Fig. 4.
Temporal accumulation profile of key proteins participating in biosynthesis of inulin and IPP-derived compounds in Taraxacum koksaghyz roots of the latex-exuding near-iosgenic line. The colour-coding depicts the protein Log2 Label-Free Quantification (LFQ) intensities detected from the proteome dataset at three time-points representing plants at 8, 12, and 24 weeks old. Only proteins with an annotation identity above 50% are considered (NCBI or UniProtKB/Swiss-Prot). The numbers to the right of the boxes represent the corresponding unique protein IDs of the proteome dataset (Supplementary Table S5). SST, sucrose 1-fructoslytransferase; FFT, fructan:fructan 1-fructosyl transferase; FEH, fructan 1-exohydrolase; ACLA, ATP citrate lyase A; ACLB, ATP citrate lyase B; AACT, acetoacetyl-CoA thiolase; HMGS, 3-hydroxy-3-methylglutaryl-coenzyme A synthase; HMGR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; MVD, mevalonate diphosphate decarboxylase; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; CMS, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5´-diphospho)-2-C-methyl-D-erythritol kinase; MCS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IDI, isopentenyl diphosphate isomerase; FPS, farnesyl diphosphate synthase; GPS, geranyl diphosphate synthase; GGPS, geranylgeranyl diphosphate synthase; SQS, squalene synthase; SQE, squalene epoxidase; OSC, oxidosqualene cyclase; CPT, cis-prenyltransferase; CPTL1, cis-prenyltransferase like; SRPP, small rubber-particle protein; REF, rubber elongation factor.
We then compared the proteome data from transgenic plants expressing either bs alone (latex-exuding) or bn/bs (non-exuding) in order to identify proteins that were either exclusively or preferentially present in laticifers, that play a pivotal role for the cell physiology of lactifers, or that interact with lactifer cells. Prior to making these comparisons, we applied several filtering criteria to exclude potential false positives in order to obtain a meaningful dataset (see Methods). This resulted in matrices for the 8-, 12-, and, 24-week time-points that encompassed 8570, 8573, and 8439 proteins, respectively (Fig. 5A, Supplementary Tables S6–S8). Based on these, 173, 205, and 440 proteins were identified as having lower accumulation at 8, 12, and 24 weeks, respectively, in the bn/bs plants, and 198, 208, 160 proteins were identified as having higher accumulation at 8, 12, and 24 weeks, respectively, in the bn/bs plants. These were designated as differential accumulated proteins (DAPs). In total, the levels of 685 individual proteins were decreased and 518 were increased for the three vegetative stages analysed (Fig. 5B). GO term analyses were performed on these DAPs and a total of 40 annotations were identified corresponding to biological processes that were either over- or under-represented (Fig. 5C). Ten of these annotations were found to be represented by proteins that were both more and less abundant, and these were associated with cell organization, localization, cell lipid metabolism, and stress responses. Annotations that corresponded to less abundant proteins included responses to different biotic and abiotic stimuli, phenylpropanoid metabolism, modulation of cellular components, and structure sizes. Annotations that corresponded to more abundant proteins included different cellular, metabolic, and catabolic processes, and anatomical structure formation and immune responses.
Fig. 5.
Identification of differentially accumulated proteins (DAPs) between roots of transgenic plants of Taraxacum koksaghyz that either exude latex (barstar, bs) or do not exude latex (barnase, bn/bs). Roots were sampled when plants were 8, 12, and 24 weeks old. (A) The protein ratios are displayed as volcano plots: increased DAPs are shown in blue, decreased ones are shown in red. Proteins or protein groups that showed a log2 ratio of >1 or <–1 and *P <0.05 were considered as differentially accumulated. In addition, proteins that were not detectable (invalid log2 ratio) or that were almost absent in one of the genotypes (quantified only for one out of four replicates, thus not allowing for the generation of a P-value) were also taken into account; however, a prerequisite for this was the detection of these proteins in the other comparative genotype in at least three out of four replicates. (B) Venn diagrams illustrating the distribution of the dataset across the three time-points. (C) GO term analysis of the detected DAPs. The biological processes shown met the criteria that at least eight proteins had to be counted and have a calculated P-value <0.05. (D) Heatmap illustrating the temporal protein accumulation patterns in the near-isogenic line (NIL) proteome dataset for the 40 proteins that were detected as differentially less accumulated at all three time-points. The corresponding cluster allocations are shown to the right. See Supplementary Fig. S4 and Supplementary Table S5.
An analysis of all the DAPs that were less abundant identified 22 different protein classes, of which the largest group was hydrolases, consisting of proteases, phosphatases, lipases, galactosidases, esterases, glucosidases, deaminases, deacetylases, and amylases (Supplementary Fig. S10). Of the 685 proteins that were less abundant, 40 were common to all three time points (Fig. 5B). In the NIL proteome dataset, most of these 40 proteins showed a higher temporal abundance and thus appeared in the higher-numbered clusters (Fig. 5D, Supplementary Table S5). At the individual time-points, 98, 120, and 347 proteins were uniquely less abundant at 8, 12, and 24 weeks, respectively, whilst 27, 18, and eight proteins were less abundant between 8–12, 12–24, and 8–24 weeks, respectively (Fig. 5B).
Some proteins that participate in inulin metabolism and terpene-backbone metabolism could be identified as DAPs (Supplementary Fig. S11). We observed a weak decrease of FFT in the 24-week time-point. HMGR1 (ID 82) was significantly decreased at 8 and 12 weeks, while HMGR2 (ID 984) was decreased at 12 and 24 weeks. Proteins related to rubber biosynthesis such as REF, CPT2, and SRPP3–5 were constantly reduced, whereas CPT3 and CPTL1 were only reduced at 8 and 12 weeks. SRPP2 and SQE appeared to be more abundant in the 12th week. Two predicted OSCs were less abundant at some time-points: ID 1388 at the 8th and 12th weeks, and ID 6882 at the 12th week.
In addition to the proteins and protein classes mentioned above, major latex protein (MLP)-like/kirola proteins (IDs 2312, 2313, 4737, 6693), polyphenol oxidases (IDs 1437, 1636), sulfotransferases (IDs 4695, 8049), trypsin inhibitors (IDs 351, 995), late-embryogenesis abundant proteins (IDs 720, 2786), and lectins (IDs 2386, 8246, 8326) were also present among the 40 DAPs that had constantly low levels (Supplementary Tables S6–S8). Other proteins with constantly low levels were annotated to be an aquaporin (ID 2430), a cysteine-rich receptor-like kinase (ID 9028), a 2-oxoglutarate/Fe(II)-dependent oxygenase (ID 6813), a NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (ID 1580), a pectate lyase (ID: 4001), a putative leucine-rich repeat-containing protein DDB (ID 4574), a perilipin (ID 1342), a UDP-glucosyltransferase (ID 4097), and five different cytochrome P450 proteins (CYPs). One of these CYPs was predicted to be a member of the CYP82A3 family (ID 7002). Three of them were predicted to be (S)-N-methylcoclaurine 3’-hydroxylase-like (IDs 344, 345, 346) and it appeared that they were clustered in the T. koksaghyz genome (same contig designation) (Lin et al., 2018). The other one was annotated as costunolide synthase (ID 7156). Several other costunolide synthases together with UDP-glucosyltransferases were detected as being either more or less abundant for at least one time-point or as being unaffected. Apart from the heterologous protein bn, only two other endogenous proteins were always more abundant, and these were annotated as ‘Indole-3-acetic acid-induced protein ARG7’ (ID P32295) and ‘Cytosolic sulfotransferase 13’ (ID Q9ZPQ5) (Supplementary Tables S6–S8). As expected, the heterologous protein bs showed no differential accumulation between the bn/bs and bs plants (Supplementary Table S4).
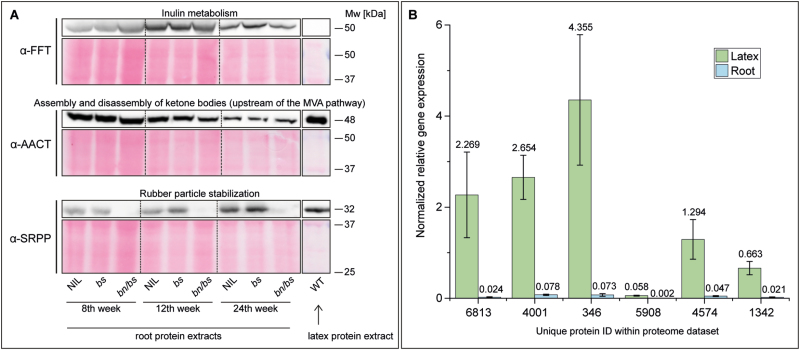
To validate the reliability of the proteome data, we used immunoblotting to check the spatio-temporal abundances of FFT, which participates in inulin metabolism located in the parenchyma cells; of AACT, which provides the precursor for a number of different pathways including that of MVA; and of SRPP3–5, which stabilize the rubber particle shell in latex (Fig. 6A). The FFT protein (~50 kDa) and AACT protein (~48 kDa, three isoforms with IDs 3706, 5224, 7174) were consistently detected in the root material of all the genotypes at all three time-points. The amount of FFT seemed to be highest in the 12th week, while the bands detected for AACT weakened as the plants aged. For SRPP3–5, the protein bands could clearly be detected in the roots of the control plants (NIL, bs) at all three time-points as well as in wild-type latex, and the band intensity of the control plants increased during the course of development. In contrast, this band was absent in the bn/bs plants at all time-points. Hence, the immunoblotting results matched the proteome data obtained by LC-MS/MS analyses.
Fig. 6.
Validation of temporal root protein accumulation and latex specificity in wild-type (WT) Taraxacum koksaghyz and transgenic plants that either exude latex (barstar, bs, and the near-isogenic line, NIL) or do not exude latex (barnase, bn/bs). (A) Western blot analysis. Whole protein extracts from roots of the transgenic plants and extracted latex proteins from the WT were separated using SDS PAGE, transferred to a nitrocellulose membrane, and stained with Ponceau S. Specific primary antibodies were used to target fructan 1-fructosyltransferase (FFT), acetoacetyl-CoA thiolase (AACT), and small rubber particle proteins (SRPPs). Visualization was conducted using a horseradish peroxidase (HRP)-labeled secondary antibody. (B) Relative gene expression levels of six proteins that were detected as being less abundant in bn/bs plants that did not exude latex. Endogenous mRNA levels in latex and in roots were determined in relation to the constitutive genes elongation factor 1a (Ef1a) and ribosomal protein L27 (RP). Data are means (±SD) of three biological replicates; the mean values are shown above the error bars. Protein IDs: 6813, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein; 4001, pectate lyase; 346, (S)-N-methylcoclaurine 3´-hydroxylase isozyme; 5908, alpha/beta hydrolase; 4574, putative leucine-rich repeat-containing protein DDB_G0290503; 1342, perilipin.
Finally, we validated the latex-specificity for six randomly chosen DAPs that were constantly less abundant in the bn/bs plants. The expression of these DAPs was examined by RT-qPCR using mRNA of roots and latex of 12-week-old wild-type plants. The results clearly demonstrated that these genes were specific to the latex (Fig. 6B).
Discussion
Taraxacum koksaghyz represents a promising source of valuable raw materials including natural rubber and other metabolites. However, the large genetic diversity of the species results in great phenotypic variation, meaning that the content of metabolites can differ drastically between individual plants (Ulmann, 1951). Hence, a comprehensive understanding of fundamental developmental and metabolic processes of the plant root and its laticifers is required to successfully establish T. koksaghyz as a suitable crop plant.
To address this issue, we generated transgenic plants expressing the toxin/anti-toxin system barnase (bn)/barstar (bs) that consequently did not exude latex. A well-balanced expression level between bn and bs was crucial for the successful regeneration of transgenic plants that did not exude latex. Although microscopic examination indicated that the tube-like architecture of the laticiferous system was not impaired in the non-exuding plants, neither regular rubber particles nor native nucleus structures could be observed (Fig. 1). Thus, the bn expression driven by the REF promoter had a strong negative impact on latex composition, but not on the development of the actual overall structure of laticifers. This suggests that the REF promoter activity (driving bn) exceeded the CaMV35S promoter activity (driving bs) during the developmental stages when laticifers were already formed. In some species the formation of articulated laticifer initials can already be observed in matured embryos (Evert, 2006; Castelblanque et al., 2016). To date, observations on Taraxacum species have shown the occurrence of laticifer cells within the first 2 weeks, whilst the elongation and anastomosing processes become visible in the following growth stages (Ghaffar et al., 2016). In addition, REF-promoter activity has been shown to be weak in dandelion root tissue at the beginning of plant development, but it then increases over the following weeks (Laibach et al., 2015). We therefore assume that this early laticifer ontology allowed the differentiation of the native tube-like anatomy before the cytotoxic effects of bn affected normal latex production. Compared to the controls, the laticifers of non-exuding plants showed regions with elevated electron density (Fig. 1), which could imply higher levels of protein or membrane aggregations. The large and amorphous electron-permeable structures might represent non-rubber lipids that resulted in the shift in Nile Red fluorescence to longer wavelengths. Nonetheless, the non-exuding plants showed normal growth, which also supports earlier assumptions that latex is not required for plant development (Castelblanque et al., 2016).
Our time-series analyses enabled us to generate temporal root-specific metabolic and proteomic profiles of latex-exuding and non-exuding plants, and their comparison identified several differential accumulated proteins (DAPs) with either relatively high or low abundance (Figs 2, 5, Supplementary Figs S8, S9, Supplementary Tables S6–S13). The protein accumulation patterns that were identified matched the results of immunoblotting (Fig. 6) and hence the reliability of the generated proteome dataset was confirmed. Because laticifers only account for a small proportion of the root (Ulmann, 1951), we assumed that proteins identified as being differentially accumulated (log2 ratio of >1 or <–1, P<0.05) might play essential roles either for laticifer physiology, for the interaction with the surrounding root tissue, or both.
A gradual accumulation of high molecular-weight poly(cis-1,4-isoprene) was observed during the course of development in the latex-exuding near-isogenic line (NIL) and the transgenic bs plants (Fig. 2, Supplementary Fig. S8), as has been described previously for wild-type plants (Ulmann, 1951). This continual biosynthesis was also reflected in the consistent high abundancy of proteins that were annotated to be involved in rubber biosynthesis (Fig. 4). Those proteins, which included HMGRs, CPTs, CPTL1, REF, and SRPP3-5 and are assumed to be preferentially located within the laticifers (Schmidt et al., 2010; Hillebrand et al., 2012; Post et al., 2012; van Deenen et al., 2012; Epping et al., 2015; Laibach et al., 2015), were significantly diminished in bn/bs plants in at least two of the three time-points (Supplementary Fig. S11, Supplementary Tables S6–S8). This was also reflected by the large reduction in poly(cis-1,4-isoprene) content and the lack of high molecular-weight cis-1,4 polymer chains within these plants (Fig. 2, Supplementary Fig. S8).
Molecules such as the sterols and fatty acid methyl esters that fulfil important roles in structural integrity, energy storage, and plant growth also occur as secondary components in latex and are associated with natural rubber (Schulze Gronover et al., 2011). Except for slight variations in concentration that could be explained by the biological variance, these molecules were present in similar amounts in bn/bs and control plants (Fig. 2). In addition, the quantities of mono-, di-, and polysaccharides in the bn/bs plants were not dramatically altered compared to the controls. This indicated that these metabolites were not specifically enriched in the laticifers and also that the bn/bs system did not negatively affect the essential physiological functions of the plant.
Pentacyclic triterpenes showed a reduction in 8-week-old bn/bs plants relative to the controls, but levels reached control values in the following growth stages. It can thus be assumed that the synthesis of triterpenes took place predominantly in laticifers in the first weeks of growth. This was supported by the fact that a partial reduction of OSC proteins, which catalyse a key step in the biosynthesis of isoprene-derived compounds (Phillips et al., 2006), was observed in the 8th and 12th weeks in the bn/bs plants Supplementary Fig. S11, Supplementary Tables S6, S7). Interestingly, an increase of SQE, which plays a crucial role in producing 2,3-oxidosqualene (the substrate of OSC) (Phillips et al., 2006), was evident in the 12-week-old non-exuding plants. Some costunolide synthases were found to be significantly decreased whilst others were increased for at least one-time point in non-exuding plants. These enzymes are involved in the biosynthesis of sesquiterpenoids (KEGG Pathway map00909), which are also a class of isoprene-derived compounds and typically found in the roots of Taraxacum species (Martinez et al., 2015). The increases, which were presumably caused by the lack of analogous pathways in latex, might have promoted the biosynthesis pathway of isoprene-derived compounds in the surrounding root tissues of the bn/bs plants. A similar compensatory reaction has already been reported for this metabolite class (Chadwick et al., 2013). Sesquiterpenoids are typically associated with laticifers, but can also be found in non-laticifer cell types, specifically when produced under situations of biotic stress. Our dataset indicated that the biosynthesis of sesquiterpenoids is not completely restricted to laticifers in T. koksaghyz, since none of the predicted germacrene A synthases and oxidases that provide the precursor molecule for costunolide synthesis were obviously affected.
Our temporal proteome analysis indicated that there were 685 DAP groups that were decreased and 518 that were increased in the non-exuding plants (Fig. 5). Proteins participating in biological processes including cell organization, localization, cell lipid metabolism, and stress responses were present in both groups; thus, some biological processes associated with laticifers and latex were significantly more strongly represented in the surrounding root tissue of the non-exuding plants, as has already been noted above for some enzymes of the isoprene pathway. In addition, cell wall-modifying enzymes, for example belonging to the phenylpropanoid pathway, were less abundant in these plants, suggesting that they function in altering the laticifer cell wall structure.
By conducting an analysis of protein classes, we were able to show that typical latex proteins, in particular proteases, phosphatases, lipases, and oxidoreductases such as polyphenol oxidases (Konno, 2011) were decreased in the non-exuding plants for at least one of the time-points that we sampled. Of the 685 less-abundant proteins, 40 were identified at decreased levels at all the time-points (Fig. 5, Supplementary Tables S6–S8). These included the proteins of rubber biosynthesis mentioned above and also other well-known latex-associated proteins such as MLP-like proteins, protease inhibitors, and lectins (Konno, 2011). Interestingly, a glyceraldehyde-3-phosphate dehydrogenase isoform was also consistently less abundant in the non-exuding plants, supporting the notion that latex has high biosynthetic activity. We also identified other proteins that were less abundant at all the time-points that have not previously been investigated in the latex of T. koksaghyz. These included cytochrome P450 enzymes (CYPs), of which one belonged to the costunolide synthases (discussed above). Another candidate belonged to the CYP82 family, a subfamily of the clan CYP71 (Nelson et al., 2004), which includes triterpenoid-decorating enzymes (Miettinen et al., 2017). Three predicted proteins were characterized as (S)-N-methylcoclaurine 3´-hydroxylase-like, which are usually involved in alkaloid metabolism (Pauli and Kutchan, 1998). Given that alkaloids have not previously been reported as typical metabolites in the roots of T. koksaghyz, identification of the function of these CYP P450 enzymes might reveal novel metabolites and/or derivatives. Interestingly, the protein levels of several other CYP P450 enzymes were altered for at least one time-point, suggesting that this enzyme class has important functions for the root and laticifer systems since they are considered to be the most versatile oxidizing enzymes. The abundance of different sulfotransferases and UDP-glucosyltransferases were also altered. Both these enzyme classes have been shown to perform scaffold modifications of different secondary metabolites (Sawai and Saito, 2011; Hirschmann et al., 2014). Two other consistently decreased proteins were identified that may be of particular interest for further studies, the first of which was a 2-oxoglutarate/iron-dependent oxygenase. These proteins are considered to be more functionally diverse than CYPs and are probably involved in numerous metabolic pathways as well as in regulating epigenetic regulation (Farrow and Facchini, 2014). The second was a protein with a REF domain that showed a low sequence homology to the perilipin protein from Mus musculus. Interestingly, perilipins are located on the surface of lipid droplets (LDs) in mammalian cells, and seem to be responsible for forming nascent LDs and stabilizing mature LDs (Itabe et al., 2017). Proteins containing a REF domain are also often associated with LD structures in plants and fulfill similar functions, such as those in T. koksaghyz attached to rubber particles (Laibach et al., 2015). So far, perilipins and related proteins have not been described as typical proteins in plants and in latex. Hence, we have identified a new plant-derived perilipin-like protein that might be another interesting factor associated with LD-like structures, such as rubber particles.
The proteins discussed above represent only a few examples from our reliable dataset, which can be mined for future research and from which we have already been able to validate the latex-specificity of six randomly chosen proteins at the transcriptional level (Fig. 6). We detected numerous other proteins at lower levels in the non-exuding plants at only one or two of the three vegetative stages investigated (Fig. 5, Supplementary Tables S6–S8). Thus, we were able to identify other latex-associated proteins, the occurrence of which was temporally dependent on the growth phase of the plant. This provides insights into latex-associated metabolic and developmental processes that change during plant growth, and hence provides a basis for further studies on the molecular framework of laticifer development and function. In addition, our comprehensive root proteome, with the calculated 42 protein clusters, gives a more precise view of protein occurrences that depend on the age and vegetative phase of the root and thus it can be used in future to gain a better understanding of fundamental metabolic and developmental processes of the plant root. Finally, subjecting the non-exuding plants to biotic and abiotic stresses might help to uncover the ecological functions of latex, but this would need expanded statistical analysis in order to allow for the extensive genetic diversity that is associated with T. koksaghyz plants.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Validation of the proteome dataset normalization procedure using MaxQuant.
Fig. S2. Histograms of the log2 LFQ intensities of all 36 proteome samples.
Fig. S3. Workflow used to generate the initial proteome dataset.
Fig. S4. Temporal protein accumulation patterns of the near-isogenic line depicted as 42 clusters.
Fig. S5. Examination of the cytotoxic activity of the generated barnase.
Fig. S6. Plant morphology over time of the different genotypes.
Fig. S7. CLSM analysis of cross-sections from the three genotypes.
Fig. S8. Content of high molecular-weight poly(cis-1,4-isoprene) in the roots.
Fig. S9. Primary metabolites in the roots.
Fig. S10. Protein class annotation of less-abundant proteins in the bn/bs plants.
Fig. S11. Differentially accumulated proteins within inulin and terpene-backbone metabolism.
Table S1. Sequences of genes used in this study.
Table S2. Oligonucleotides used in this study.
Table S3. CRISPR protospacer sequences.
Table S4. Matrix of the initial proteome dataset.
Table S5. Proteome matrix of the near-isogenic line.
Table S6. Proteome matrix for the identification of differentially accumulated proteins between bn/bs versus bs plants at 8 weeks old.
Table S7. Proteome matrix for the identification of differentially accumulated proteins between bn/bs versus bs plants at 12 weeks old.
Table S8. Proteome matrix for the identification of differentially accumulated proteins between bn/bs versus bs plants at 24 weeks old.
Table S9. Quantification of poly(cis-1,4-isoprene) by nuclear magnetic resonance spectroscopy and thermal field-flow fractionation.
Table S10. Quantification of squalene and 2,3-oxidosqualene by GC-MS.
Table S11. Quantification of triterpenes by GC-MS.
Table S12. Quantification of sterols by GC-MS.
Table S13. Quantification of fatty acid methy ester by GC-MS.
Table S14. GO enrichment analysis of clusters 7 and 8 performed using the functional annotation tool from DAVID.
Data deposition
The mass spectrometry proteomics data are available at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2016) with the dataset identifier PXD011773.
Acknowledgements
We give grateful thanks for the technical assistance of Daniela Ahlert, Sascha Ahrens, Paulina Pieloch (University of Muenster, Institute of Plant Biology and Biotechnology), Gianina Ramos (Fraunhofer Institute for Molecular Biology and Applied Ecology, Muenster), and Ursula Malkus (University Hospital and University of Muenster, Institute of Medical Physics and Biophysics). We would also like to thank Dr Celeste Riley Brennecka (University of Muenster, Institute for Planetology) for critical reading of the manuscript.
Glossary
Abbreviations
- AACT
acetoacetyl-CoA thiolase
- ACL
ATP citrate lyase
- bs
barstar
- bn
barnase
- CaMV
Cauliflower mosaic virus
- CPT
cis-prenyltransferase
- CPTL1
CPT-like protein 1
- CYP
cytochrome P450
- DAP
differentially accumulated protein
- FEH
fructan 1-exohydrolase
- FFT
fructan:fructan 1-fructosyl transferase
- GO
gene ontology
- HMGR
3-hydroxy-3-methyl-glutaryl-coenzyme A reductase
- LD
lipid droplet
- MEP
2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate
- MVA
mevalonate
- NIL
near-isogenic line
- OSC
oxidosqualene cyclase
- REF
rubber elongation factor
- SRPP
small rubber particle protein
- SQE
squalene epoxidase
- SQS
squalene synthase
Funding
This study was financially supported by the Deutsche Forschungsgemeinschaft (INST 211/744-1 FUGG) for I.F.
References
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Castelblanque L, Balaguer B, Martí C, Rodríguez JJ, Orozco M, Vera P. 2016. Novel insights into the organization of laticifer cells: a cell comprising a unified whole system. Plant Physiology 172, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick M, Trewin H, Gawthrop F, Wagstaff C. 2013. Sesquiterpenoids lactones: benefits to plants and people. International Journal of Molecular Sciences 14, 12780–12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Silva J, Nural AT, Skaggs A, et al. . 2012. Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, result in qualitative and quantitative changes in rubber metabolism. Phytochemistry 79, 46–56. [DOI] [PubMed] [Google Scholar]
- D’Auzac J, Jacob JL, Chrestin H. 1989. Physiology of rubber tree latex. The laticiferous cell and latex–a model of cytoplasm. Boca Raton, FL: CRC Press [Google Scholar]
- Diaz G, Melis M, Batetta B, Angius F, Falchi AM. 2008. Hydrophobic characterization of intracellular lipids in situ by Nile Red red/yellow emission ratio. Micron 39, 819–824. [DOI] [PubMed] [Google Scholar]
- Epping J, van Deenen N, Niephaus E, Stolze A, Fricke J, Huber C, Eisenreich W, Twyman RM, Prüfer D, Schulze Gronover C. 2015. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nature Plants 1, 15048. [Google Scholar]
- Evert RF. 2006. Internal secretory structures. In: Esau’s plant anatomy, vol. 3 Evert RF, ed. Hoboken, NJ: John Wiley & Sons, Inc, 473–501. [Google Scholar]
- Farrow SC, Facchini PJ. 2014. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Frontiers in Plant Science 5, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H. 2014. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. The Plant Journal 79, 348–359. [DOI] [PubMed] [Google Scholar]
- Ghaffar MAA, Meulia T, Cornish K. 2016. Laticifer and rubber particle ontogeny in Taraxacum kok-saghyz (rubber dandelion) roots. Microscopy and Microanalysis 22, 1034–1035.27681364 [Google Scholar]
- Hagel JM, Yeung EC, Facchini PJ. 2008. Got milk? The secret life of laticifers. Trends in Plant Science 13, 631–639. [DOI] [PubMed] [Google Scholar]
- Hanson B, Engler D, Moy Y, Newman B, Ralston E, Gutterson N. 1999. A simple method to enrich an Agrobacterium-transformed population for plants containing only T-DNA sequences. The Plant Journal 19, 727–734. [DOI] [PubMed] [Google Scholar]
- Hartley RW, Both V, Hebert EJ, Homerova D. 1996. Barstar inhibits extracellular ribonucleases of Streptomyces and allows their production from recombinant genes. Protein & Peptide Letters 3, 225–231 [Google Scholar]
- Hillebrand A, Post JJ, Wurbs D, Wahler D, Lenders M, Krzyzanek V, Prüfer D, Gronover CS. 2012. Down-regulation of small rubber particle protein expression affects integrity of rubber particles and rubber content in Taraxacum brevicorniculatum. PLoS ONE 7, e41874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmann F, Krause F, Papenbrock J. 2014. The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Frontiers in Plant Science 5, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Itabe H, Yamaguchi T, Nimura S, Sasabe N. 2017. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids in Health and Disease 16, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javorsky L. 1944. Die neue Kautschukpflanze Kok-Saghyz und ihr Anbau in Soviet-Russland. Der Tropenpflanzer 1, 1–48. [Google Scholar]
- Jin H, Song Z, Nikolau BJ. 2012. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. The Plant Journal 70, 1015–1032. [DOI] [PubMed] [Google Scholar]
- Konno K. 2011. Plant latex and other exudates as plant defense systems: roles of various defense chemicals and proteins contained therein. Phytochemistry 72, 1510–1530. [DOI] [PubMed] [Google Scholar]
- Kreuzberger M, Hahn T, Zibek S, Schiemann J, Thiele K. 2016. Seasonal pattern of biomass and rubber and inulin of wild Russian dandelion (Taraxacum koksaghyz L. Rodin) under experimental field conditions. European Journal of Agronomy 80, 66–77. [Google Scholar]
- Laibach N, Hillebrand A, Twyman RM, Prüfer D, Schulze Gronover C. 2015. Identification of a Taraxacum brevicorniculatum rubber elongation factor protein that is localized on rubber particles and promotes rubber biosynthesis. The Plant Journal 82, 609–620. [DOI] [PubMed] [Google Scholar]
- Lin T, Xu X, Ruan J. et al. . 2018. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. National Science Review 5, 78–87. [Google Scholar]
- Martinez M, Poirrier P, Chamy R, Prüfer D, Schulze-Gronover C, Jorquera L, Ruiz G. 2015. Taraxacum officinale and related species—An ethnopharmacological review and its potential as a commercial medicinal plant. Journal of Ethnopharmacology 169, 244–262. [DOI] [PubMed] [Google Scholar]
- Mehrotra AK, Bhullar S, Burma PK. 2014. Development of intron-containing barnase gene (barnase-int) encoding a toxic protein to facilitate its cloning in bacterial cells. Journal of Plant Biochemistry and Biotechnology 23, 435–439. [Google Scholar]
- Men X, Wang F, Chen GO, Zhang HB, Xian M. 2019. Biosynthesis of natural rubber: current state and perspectives. International Journal of Molecular Sciences 20, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K, Pollier J, Buyst D, et al. . 2017. The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nature Communications 8, 14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Noll GA, Ernst AM, Rüping B, Groscurth S, Twyman RM, Kawchuk LM, Prüfer D. 2010. Recombinant artificial forisomes provide ample quantities of smart biomaterials for use in technical devices. Applied Microbiology and Biotechnology 88, 689–698. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. 2004. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiology 135, 756–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli HH, Kutchan TM. 1998. Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3´-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. The Plant Journal 13, 793–801. [DOI] [PubMed] [Google Scholar]
- Phillips DR, Rasbery JM, Bartel B, Matsuda SP. 2006. Biosynthetic diversity in plant triterpene cyclization. Current Opinion in Plant Biology 9, 305–314. [DOI] [PubMed] [Google Scholar]
- Pickard WF. 2008. Laticifers and secretory ducts: two other tube systems in plants. New Phytologist 177, 877–888. [DOI] [PubMed] [Google Scholar]
- Post J, van Deenen N, Fricke J, et al. . 2012. Laticifer-specific cis-prenyltransferase silencing affects the rubber, triterpene, and inulin content of Taraxacum brevicorniculatum. Plant Physiology 158, 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütter KM, van Deenen N, Unland K, Prüfer D, Schulze Gronover C. 2017. Isoprenoid biosynthesis in dandelion latex is enhanced by the overexpression of three key enzymes involved in the mevalonate pathway. BMC Plant Biology 17, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Cadavid DA, Valles-Ramireza S, Cornish K, Michel FC Jr. 2018. Simultaneous quantification of rubber, inulin, and resins in Taraxacum kok-saghyz (TK) roots by sequential solvent extraction. Industrial Crops and Products 122, 647–656. [Google Scholar]
- Sawai S, Saito K. 2011. Triterpenoid biosynthesis and engineering in plants. Frontiers in Plant Science 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Lenders M, Hillebrand A, van Deenen N, Munt O, Reichelt R, Eisenreich W, Fischer R, Prüfer D, Gronover CS. 2010. Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochemistry 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze Gronover C, Wahler D, Prüfer D. 2011. Natural rubber biosynthesis and physic-chemical studies on plant derived latex. Biotechnology of Biopolymers doi: 10.5772/17144. [DOI] [Google Scholar]
- Serrano L, Day AG, Fersht AR. 1993. Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. Journal of Molecular Biology 233, 305–312. [DOI] [PubMed] [Google Scholar]
- Shi M, Li Y, Deng S, Wang D, Chen Y, Yang S, Wu J, Tian WM. 2019. The formation and accumulation of protein-networks by physical interactions in the rapid occlusion of laticifer cells in rubber tree undergoing successive mechanical wounding. BMC Plant Biology 19, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolze A, Wanke A, van Deenen N, Geyer R, Prüfer D, Schulze Gronover C. 2017. Development of rubber-enriched dandelion varieties by metabolic engineering of the inulin pathway. Plant Biotechnology Journal 15, 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. 2016a The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols 11, 2301–2319. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016b The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Ulmann M. 1951. Wertvolle Kautschukpflanzen des gemässigten Klimas. Akademie-Verlag [Google Scholar]
- Unland K, Pütter KM, Vorwerk K, van Deenen N, Twyman RM, Prüfer D, Schulze Gronover C. 2018. Functional characterization of squalene synthase and squalene epoxidase in Taraxacum koksaghyz. Plant Direct 2, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Poirier Y. 2007. Establishment of new crops for the production of natural rubber. Trends in Biotechnology 25, 522–529. [DOI] [PubMed] [Google Scholar]
- van Deenen N, Bachmann AL, Schmidt T, Schaller H, Sand J, Prüfer D, Schulze Gronover C. 2012. Molecular cloning of mevalonate pathway genes from Taraxacum brevicorniculatum and functional characterisation of the key enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Molecular Biology Reports 39, 4337–4349. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, et al. . 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44, D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nature Methods 6, 359–362. [DOI] [PubMed] [Google Scholar]
- Wozny D, Kramer K, Finkemeier I, Acosta IF, Koornneef M. 2018. Genes for seed longevity in barley identified by genomic analysis on near isogenic lines. Plant, Cell & Environment 41, 1895–1911. [DOI] [PubMed] [Google Scholar]
- Xing S, van Deenen N, Magliano P, Frahm L, Forestier E, Nawrath C, Schaller H, Gronover CS, Prüfer D, Poirier Y. 2014. ATP citrate lyase activity is post-translationally regulated by sink strength and impacts the wax, cutin and rubber biosynthetic pathways. The Plant Journal 79, 270–284. [DOI] [PubMed] [Google Scholar]
- Ye J, McGinnis S, Madden TL. 2006. BLAST: improvements for better sequence analysis. Nucleic Acids Research 34, 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.