Abstract
BACKGROUND: KRAS gene mutations are well known as a key driver of advanced non–small cell lung cancer (NSCLC). The impact of KRAS-mutant subtypes on the survival benefit from salvage chemotherapy is controversial. Here, we present a real-world study in patients across China with advanced NSCLC with KRAS mutations using a website-based patient self-report system. METHODS: We identified a total of 75 patients diagnosed with KRAS-mutant (determined by molecular sequencing) advanced NSCLC between 2014/5/9 and 2019/5/30. KRAS mutation subtypes were divided into G12C and non-G12C groups for statistical analysis. The clinicopathological characteristics and treatment survival benefit in all patients with a KRAS mutation were evaluated. Programmed death-ligand 1 (PD-L1) expression data were collected from 30 patients in the same cohort. RESULTS: In this study, 23 patients with stage IIIB NSCLC and 52 patients with stage IV NSCLC were enrolled with 58 men and 17 women; the median age was 60 years (39–84). All patients received regular chemotherapy/radiotherapy/targeted therapy/immune therapy as per the disease condition. Four main KRAS mutation subtypes were detected: G12C (33%), G12V (19%), G12A (12%), and G12D (12%). Three predominant KRAS comutations were detected: TP53-KRAS (31%), EGFR-KRAS (11%), and STK11-KRAS (8%). Compared with the KRAS non-G12C mutation subtype, patients with the KRAS G12C mutation had potentially longer progression-free survival (PFS) after first-line chemotherapy (4.7 vs. 2.5 months, p < 0.05). Pemetrexed-based chemotherapy appeared to be superior to taxanes- and gemcitabine-based chemotherapies in all patients (PFS: 5.0 vs. 1.5 and 2.3 months, respectively, p > 0.05). Cox regression analysis showed that the KRAS G12C mutation and pemetrexed-based first-line chemotherapy were positive influencers for PFS after first-line (hazard ratios = 0.31 and 0.55, respectively, P < 0.05), but not second-line chemotherapies. CONCLUSION: The KRAS G12C mutation could be a predictive biomarker for better survival benefit from first-line chemotherapy in patients with advanced NSCLC and KRAS mutations. The first-line chemotherapy regimen could possibly influence the outcome in patients with KRAS mutations. Larger and prospective clinical trials are warranted to confirm our conclusions.
Introduction
Advanced non–small cell lung cancer (NSCLC) is the most common and life-threatening cancer around the world regardless of gender [1]. Cytotoxic chemotherapy has long been the only choice for these patients [2]. Fortunately, tyrosine kinase inhibitors (TKIs) that target epithelial growth factor receptor (EGFR) mutations have led to a new era of targeted therapy that is more effective and has lower toxicity than chemotherapy in treating advanced NSCLC [3].
KRAS mutations are another definite oncogenic driver, similar to EGFR, in advanced NSCLC. Point mutations at positions 12, 13, or 61 in the KRAS gene lead to an amino acid replacement and cause constitutive activation of the RAS signaling pathway. This could further interact with multiple effectors including the mitogen-activated protein kinase, phosphoinositide 3-kinase, and signal transducer and activator of transcription cascades [4]. KRAS mutations occur in about 20–30% in Western and 10–15% in Eastern populations with lung adenocarcinomas [5]. Exclusive occurrence with an EGFR mutation is also a feature of KRAS mutations in patients with advanced NSCLC [6,7].
Unfortunately, KRAS mutations have not been targeted successfully similar to EGFR mutations [8]. This is partly due to the high diversity in KRAS-mutant subtypes that leads to different biological outcomes and responses to treatment [5]. KRAS-G12C is the most common mutant subtype in all KRAS mutations and is associated with a poor outcome in early-stage NSCLC [9]. Gene expression profiles in lung cancer cell lines and primary tumors revealed that the KRAS-G12C mutant had an epithelial-to-mesenchymal transition and a KRAS-independent phenotype [9]. In smokers with KRAS mutations, a higher frequency of KRAS-G12C was observed in women than in men harboring the same mutation [6]. Co-occurrence with TP53 or STK11 mutations is very common in KRAS mutations [7,10,11], although none were targetable until now. Chemotherapy remains the primary real-world treatment for NSCLC despite increasing evidence to support KRAS direct and nondirect targeted therapies, including targeting KRAS downstream pathways [12,13] and antiangiogenesis therapies [14].
The role of KRAS mutations as a prognostic or predictive factor for systemic treatment in NSCLC remains uncertain [15]. The association of specific KRAS-mutant subtypes with survival benefit from chemotherapy in patients with advanced NSCLC is worth studying. Thus, we present a website-based patient self-report study from 75 patients with stage III-IV NSCLC and KRAS mutations across China to address these issues.
Methods
Patients
A total of 75 patients with NSCLC, confirmed by pathological diagnosis, from across China from May 2014 to May 2019, were reviewed retrospectively using a website-based patient self-report system. Histology subtyping was determined in accordance with the 2004 World Health Organization classification. Tumor staging was based on the 7th edition of the Lung Cancer Staging system from the American Joint Committee on Cancer. Age, smoking status, Eastern Cooperative Oncology Group performance status, histology, disease stage, brain or bone metastasis, and molecular information were documented at first diagnosis. All clinical information, including diagnosis, treatment, and clinical outcome, was collected through the system and confirmed by local professional oncologists. Patients were followed up from the date of diagnosis until the date of death from all causes, or until the last approachable follow-up. Tumor response was evaluated in accordance with the Response Evaluation Criteria in Solid Tumors (RECIST, version 1.1). The treatment response was evaluated during the first month of initial therapy and every two months thereafter. This study was approved by the Chinese Academy of Sciences University Cancer Hospital (Zhejiang Cancer Hospital) Ethics Committee, and a written informed consent was obtained from each patient to use the clinical data for research before the medical intervention started.
Molecular detection
Targeted region capture–combined next-generation sequencing was performed for the 75 patients with NSCLC. Genomic DNA sequencing libraries were prepared using the protocols recommended by the TruSeq DNA Library Preparation Kit (Illumina, San Diego, CA, USA). For samples close to the minimum input requirement, additional precapture polymerase chain reaction cycles were performed to generate sufficient product for hybridization. The libraries were hybridized to custom designed probes (Integrated DNA Technology, Coralville, IA, USA) including all exons of 170 genes and selected introns of anaplastic lymphoma kinase, RET, and ROS1 for the detection of genomic rearrangements. DNA sequencing was performed on a HiSeq3000 sequencing system (Illumina) with 2 × 75 bp paired-end reads. The reads were aligned to the human genome–build GRCh37 using a Burrows-Wheeler aligner (BWA). Somatic single-nucleotide variant and indel calls were generated using MuTect and GATK, respectively. Somatic copy number alterations were identified using CONTRA. Genomic rearrangements were identified using software developed in-house to analyze chimeric read pairs.
Statistical analyses
Kaplan-Meier curves and the two-sided log-rank test were used for univariate survival analyses. The Cox proportional hazards model was used to complete the univariate and multivariate survival analyses with the hazard ratio (HR) and corresponding 95% confidence interval. Progression-free survival (PFS) was defined as the time from the date of initial treatment to the date of systemic progression or death, or censored at the date of the last follow-up, whichever came first to trigger the event. Significance between groups was defined as p values < 0.05. Statistical analyses were performed using the R software/environment.
Results
Patient characteristics and treatments
A total of 23 Chinese patients with stage IIIB and 52 Chinese patients with stage IV NSCLC were enrolled. There were 58 men and 17 women. The median age was 60 years (range: 39–84). Of the 75 patients, four main subtypes of KRAS mutations were detected including G12C (33%), G12V (19%), G12A (12%), and G12D (12%). Three predominant KRAS comutations were detected including TP53-KRAS (31%), EGFR-KRAS (11%), and STK11-KRAS (8%). All patients received regular chemotherapy/radiotherapy/targeted therapy/immune therapy based on the disease condition. Baseline and clinicopathological information of all patients is summarized in Table 1. Detailed treatments of all patients are listed in Table 2.
Table 1.
Demographic Characteristics of Patients with KRAS Mutation (N = 75)
| Factors | Number, n (%) |
|---|---|
| Gender | |
| Female | 17 (22.7%) |
| Male | 58 (77.3%) |
| Age, years | |
| <60 | 37 (49.3%) |
| ≥60 | 38 (50.7%) |
| Smoking history | |
| Yes | 59 (78.7%) |
| No | 16 (21.3%) |
| Performance score | |
| 0–2 | 61 (81.3%) |
| 3–4 | 14 (18.7%) |
| Histology | |
| Adenocarcinoma | 68 (90.7%) |
| *Nonadenocarcinoma | 7 (9.3%) |
| PD-L1 expression | |
| Yes | 21 (28%) |
| No | 9 (12%) |
| Unknown | 45 (60%) |
| TMB<10 mutations/Mb | |
| Yes | 9 (12%) |
| No | 5 (6.7%) |
| Unknown | 61 (81.3%) |
| Brain metastasis | |
| Yes | 18 (24%) |
| No | 56 (74.7%) |
| Unknown | 1 (1.3%) |
| Bone metastasis | |
| Yes | 33 (44%) |
| No | 42 (56%) |
| Pleural effusion | |
| Yes | 6 (8%) |
| No | 69 (92%) |
| Immune therapy | |
| Yes | 31 (41.3%) |
| No | 40 (53.3%) |
| Unknown | 4 (5.4%) |
| MEK inhibitor | |
| Yes | 9 (12%) |
| No | 66 (88%) |
| TKI | |
| Yes | 14 (18.7%) |
| No | 57 (76%) |
| Unknown | 4 (5.3%) |
| Angiogenesis inhibitors | |
| Yes | 18 (24%) |
| No | 57 (76%) |
| Brain radiation | |
| Yes | 12 (16%) |
| No | 51 (68%) |
| Unknown | 12 (16%) |
| 1st-line chemotherapy | |
| Taxanes-based | 7 (9.3%) |
| Pemetrexed-based | 51 (68%) |
| Gemcitabine-based | 3 (4%) |
| Other | 1 (1.3%) |
| No | 13 (17.4%) |
| 2nd-line chemotherapy | |
| Yes | 32 (42.7%) |
| No | 43 (57.3%) |
TMB, tumor mutation burden.
*Nonadenocarcinoma included two cases with adenosquamous carcinoma, two squamous carcinoma, three non–small cell lung cancer with unknown histologic subtype.
Table 2.
More Details of Chemotherapy in All 75 Patients with KRAS Mutations
| Case | KRAS mutation | KRAS comutation | 1st-line chemotherapy | 2nd-line chemotherapy |
|---|---|---|---|---|
| 1 | Q61H | EGFR-KRAS | Pemetrexed + carboplatin | Nab-paclitaxel + cisplatin |
| 2 | G12C | TP53-KRAS | Pemetrexed | No |
| 3 | G13D | EGFR-KRAS | Pemetrexed + carboplatin + bevacizumab | No |
| 4 | Q61L | TP53-KRAS | Pemetrexed + cisplatin + bevacizumab | Nab-paclitaxel + bevacizumab |
| 5 | G12C | Others | Pemetrexed + cisplatin + bevacizumab | Pemetrexed + cisplatin + bevacizumab |
| 6 | G12C | TP53-KRAS | Pemetrexed | No |
| 7 | G12D | STK11-KRAS | Pemetrexed + bevacizumab | No |
| 8 | G12S | TP53-KRAS | Pemetrexed + lobaplatin | No |
| 9 | G12V | Others | Pemetrexed + carboplatin | No |
| 10 | G12V | Others | Pemetrexed + cisplatin | Pemetrexed + bevacizumab |
| 11 | G12V | Others | Pemetrexed + carboplatin | Liposome paclitaxel |
| 12 | G12C | TP53-KRAS | Paclitaxel + carboplatin + bevacizumab | Bevacizumab |
| 13 | G12S | TP53-KRAS | Pemetrexed + carboplatin | Bevacizumab |
| 14 | Unknown | EGFR-KRAS | No | No |
| 15 | G12V | Others | Pemetrexed + cisplatin + bevacizumab | Docetaxel + lopaplatin |
| 16 | G12C | Others | Pemetrexed + carboplatin + bevacizumab | Docetaxel |
| 17 | G12C | Others | Pemetrexed + carboplatin + bevacizumab | No |
| 18 | G12C | TP53-KRAS | No | No |
| 19 | G12C | TP53-KRAS | Pemetrexed + carboplatin + bevacizumab | No |
| 20 | G12V | others | Pemetrexed + cisplatin | Pemetrexed + bevacizumab |
| 21 | G12C | others | Pemetrexed + carboplatin + bevacizumab | No |
| 22 | G12C | others | Pemetrexed + nedaplatin | No |
| 23 | G12A | TP53-KRAS | Pemetrexed + cisplatin | Pemetrexed + carboplatin + bevacizumab |
| 24 | G12C | TP53-KRAS | Gemcitabine + cisplatin | Docetaxel |
| 25 | G12V | TP53-KRAS | Pemetrexed | No |
| 26 | G12C | others | Pemetrexed + cisplatin + bevacizumab | No |
| 27 | Q61H | others | Pemetrexed + cisplatin + bevacizumab | No |
| 28 | G13D | others | Pemetrexed + cisplatin | Docetaxel + bevacizumab |
| 29 | G12D | STK11-KRAS | Pemetrexed + nedaplatin | No |
| 30 | G12V | others | Pemetrexed + carboplatin | No |
| 31 | G12A | others | Pemetrexed + cisplatin | Paclitaxel + nedaplatin |
| 32 | G12A | others | Pemetrexed + cisplatin | Paclitaxel + nedaplatin |
| 33 | unknown | others | Cisplatin/carboplatin | Other |
| 34 | G12C | TP53-KRAS | Pemetrexed + carboplatin | No |
| 35 | G12D | others | Pemetrexed + cisplatin + bevacizumab | Pemetrexed + cisplatin + bevacizumab |
| 36 | G12C | TP53-KRAS | Nab-paclitaxel | No |
| 37 | G12C | EGFR-KRAS | Pemetrexed + nedaplatin | No |
| 38 | G12C | others | Pemetrexed + carboplatin | No |
| 39 | unknown | others | Nab-paclitaxel + nedaplatin | No |
| 40 | G12A | EGFR-KRAS | No | No |
| 41 | G12D | others | Pemetrexed + carboplatin | Pemetrexed + nedaplatin |
| 42 | G12V | STK11-KRAS | No | No |
| 43 | G12S | others | No | No |
| 44 | G12D | TP53-KRAS | Pemetrexed + carboplatin | No |
| 45 | G12C | others | Nab-paclitaxel + cisplatin | No |
| 46 | G12C | TP53-KRAS | Pemetrexed + cisplatin | No |
| 47 | G12C | others | Pemetrexed | Docetaxel + gemcitabine |
| 48 | G12V | STK11-KRAS | Pemetrexed | No |
| 49 | G12C | TP53-KRAS | Liposome paclitaxel + nedaplatin | Gemcitabine + cisplatin |
| 50 | unknown | others | Pemetrexed | Cisplatin |
| 51 | G12A | STK11-KRAS | Gemcitabine + carboplatin | Pemetrexed + carboplatin |
| 52 | G12C | others | Pemetrexed + nedaplatin | Docetaxel + bevacizumab |
| 53 | G12V | TP53-KRAS | Pemetrexed + carboplatin | No |
| 54 | unknown | EGFR-KRAS | No | Nab-paclitaxel + carboplatin |
| 55 | G12D | others | No | No |
| 56 | unknown | TP53-KRAS | Paclitaxel + carboplatin | No |
| 57 | unknown | others | Gemcitabine + cisplatin | Nab-paclitaxel + lopaplatin |
| 58 | G12D | STK11-KRAS | Paclitaxel + carboplatin | No |
| 59 | G12A | others | No | No |
| 60 | G12V | TP53-KRAS | Pemetrexed + cisplatin | Paclitaxel + carboplatin |
| 61 | unknown | TP53-KRAS | Pemetrexed + carboplatin | Vinorelbine |
| 62 | G12A | EGFR-KRAS | No | No |
| 63 | G12C | others | Pemetrexed | Docetaxel |
| 64 | unknown | others | Pemetrexed + carboplatin | Docetaxel + bevacizumab |
| 65 | unknown | EGFR-KRAS | Pemetrexed + nedaplatin | No |
| 66 | G12C | others | Pemetrexed + nedaplatin | Docetaxel + lopaplatin |
| 67 | G12C | TP53-KRAS | Pemetrexed + carboplatin + bevacizumab | No |
| 68 | G12D | others | No | No |
| 69 | G12A | others | No | No |
| 70 | G12A | others | No | No |
| 71 | G12V | TP53-KRAS | Pemetrexed + nedaplatin | Nab-paclitaxel + nedaplatin |
| 72 | G13S | others | Pemetrexed + cisplatin | No |
| 73 | G12D | TP53-KRAS | Pemetrexed + nedaplatin + bevacizumab | No |
| 74 | G12C | others | Pemetrexed + carboplatin + bevacizumab | Nab-paclitaxel + carboplatin + bevacizumab |
| 75 | G12V | others | No | No |
Molecular detection
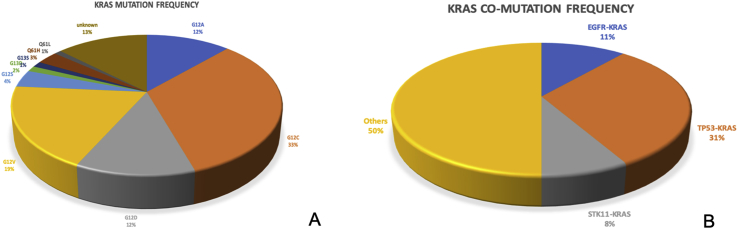
Four main KRAS mutation subtypes were detected including G12C (33%), G12V (19%), G12A (12%), and G12D (12%). Other identified KRAS mutations included G12S, G12R, G13C, G13D, and G13S. Three patients had complex KRAS mutation subtypes: G12D + G13V, G12C + G12D + G13V, and G12C + G13V + G13S + G12V. Three predominant KRAS comutations were detected including TP53-KRAS (31%), EGFR-KRAS (11%), and STK11-KRAS (8%). The distribution of molecular mutations is shown in Figure 1. Detailed KRAS mutation information of all patients is listed in Table 2.
Figure 1.
(A) KRAS mutation subtypes and (B) comutation KRAS subtypes identified in 75 patients.
Survival analysis
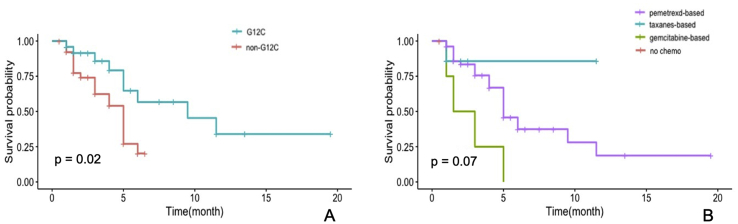
The median PFS times after first-, and first- and second-line chemotherapy were 3 and 4 months, respectively. In the univariate analysis, patients with the KRAS G12C mutation had potentially longer PFS after the first-line chemotherapy than those with other KRAS mutations (4.7 vs. 2.5 months, p < 0.05); pemetrexed-based chemotherapy tended to be superior to taxanes- and gemcitabine-based chemotherapies in all patients (PFS: 5.0 vs. 1.5 and 2.3 months, respectively, p > 0.07) (Figure 2, Table 3).
Figure 2.
(A) Progression-free survival curves after first-line chemotherapy in patients with or without the KRAS G12C mutation (4.7 vs. 2.5 months, p < 0.05). (B) Progression-free survival curves in patients who accepted pemetrexed-, or taxane-, or gemcitabine-based chemotherapy (5.0 vs. 1.5 and 2.3 months, respectively, p < 0.05).
Table 3.
Univariate Analysis for Progression-Free Survival in Patients with KRAS Mutation and Advanced NSCLC
| Variables | PFSa |
PFSb |
PFSc |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| KRAS G12C (with vs. without) | 0.357 | 0.15–0.85 | 0.02 | 0.65 | 0.11–3.83 | 0.63 | 0.46 | 0.2–1.05 | 0.06 |
| Gender (female vs. male) | 1.63 | 0.73–3.69 | 0.23 | 2.09 | 0.38–11.52 | 0.4 | 1.04 | 0.44–2.47 | 0.92 |
| Age (≥60 vs.<60 years) | 0.6 | 0.28–1.27 | 0.18 | 0.4 | 0.07–2.26 | 0.3 | 0.52 | 0.25–1.1 | 0.09 |
| Smoke, (never vs. current/former) | 1.202 | 0.45–3.17 | 0.71 | – | – | – | 2.06 | 0.71–5.95 | 0.18 |
| Histology (adenocarcinoma vs. nonadenocarcinoma) | 1.61 | 0.37–6.92 | 0.52 | 1.85 | 0.21–16.38 | 0.56 | 1.86 | 0.43–8.12 | 0.41 |
| ECOG PS (3–4 vs. 0–2) | 1.44 | 0.58–3.54 | 0.43 | 2.11 | 0.38–11.62 | 0.39 | 1.72 | 0.7–4.23 | 0.24 |
| PD-L1 expression (yes vs. no) | 0.47 | 0.1–2.11 | 0.32 | – | – | – | 0.24 | 0.04–1.41 | 1.11 |
| Brain metastasis (yes vs. no) | 1.26 | 0.54–2.93 | 0.59 | – | – | – | 1.0 | 0.43–2.35 | 1.00 |
| Bone metastasis (yes vs. no) | 1.58 | 0.76–3.31 | 0.22 | 1.16 | 0.23–5.78 | 0.85 | 1.49 | 0.72–3.1 | 0.29 |
| d1st-line chemo | 0.59 | 0.33–1.04 | 0.07 | 0.65 | 0.28–1.52 | 0.32 | 1.07 | 0.73–1.58 | 0.72 |
| e2nd-line chemo | 28.86 | 3.92–212.4 | – | – | – | - | 23.92 | 3.25–176.14 | – |
1st-line chemotherapy.
2nd-line chemotherapy.
Data on 1st-line and 2nd-line chemotherapy and the multivariate analysis were not available. PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; PD-L1, Programmed death-ligand 1.
1st-line chemo: pemetrexed-based vs. taxanes-based vs. gemcitabine-based vs. no-chemo, no-chemo as reference.
2nd-line chemo: chemo vs. no-chemo, no-chemo as reference.
Adjusting for age and smoking status, the KRAS G12C mutation and chemotherapy regimens remained positive impactors on PFS of the first-line chemotherapy, but not PFS of first- and second-line chemotherapies (Table 4).
Table 4.
Multivariate Survival Analysis for Progression-Free Survival in Patients with KRAS Mutation
| Variables | PFSa |
PFSb |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| KRAS G12C (with vs. without) | 0.31 | 0.13–0.77 | 0.01 | 0.46 | 0.19–1.07 | 0.07 |
| Age (≥60 vs.<60) | 0.63 | 0.91–1 | 0.15 | 0.49 | 0.23–1.06 | 0.07 |
| Smoke, (never vs. current/former) | 1.202 | 0.46–3.62 | 0.62 | 2.58 | 0.84–7.87 | 0.10 |
| c1st-line chemo | 0.55 | 0.3–1 | 0.04 | 1.10 | 0.74–1.63 | 0.64 |
1st-line chemotherapy.
Data on 1st-line and 2nd-line chemotherapy and the multivariate analysis were not available. PFS, progression-free survival; HR, hazard ratio; CI, confidence interval.
1st-line chemo included pemetrexed-, taxanes- and gemcitabine-based chemotherapy, nonpemetrexed chemotherapy as reference.
Discussion
Patients with KRAS mutations have distinct treatment responses and survival benefits compared with those without such mutations in advanced NSCLC [16]. Furthermore, different KRAS mutation subtypes may also have specific prognostic value in treatment outcomes. Here, we presented a retrospective study to address the characteristics of KRAS mutation subtypes and efficacy of first-line chemotherapy in advanced NSCLC from patients across China. KRAS G12C was the most common subtype detected, and patients with KRAS G12C mutation tend to have longer PFS time after first-line chemotherapy than patients with other KRAS mutation. Both KRAS G12C mutation and pemetrexed-based chemotherapy were positive impactors for survival benefit from first-line therapy in patients with KRAS mutation.
Although previous studies failed to find a positive connection between KRAS mutations and sensitivity or resistance to chemotherapy in patients with advanced NSCLC [17,18], the inconsistent chemotherapy regimens in different studies may have affected this conclusion. In our study, the survival benefit from first-line chemotherapy in patients with the KRAS G12C mutation appeared to be better than non-G12C KRAS patients. This result was consistent with a previous report that patients with a KRAS codon 12 mutation seemed to have a better outcome than those with a codon 13 mutation [19]. Moreover, patients with the KRAS G12C mutation had shorter PFS and overall survival than non-G12C KRAS patients when both carried a KRAS mutation but without an EGFR mutation and under TKI treatment [20].
Although patients with the KRAS-G12C mutation do not benefit from TKI treatment, they may respond to chemotherapy because of the aggressive phenotype. About 68% of patients in our study accepted pemetrexed-based treatment as first-line chemotherapy. We found an almost significant difference between PFS in patients with and without the KRAS-G12C mutation after first-line chemotherapy. Recently, one study from Korea concluded that the KRAS-G12C mutation was a marker for poor prognosis of pemetrexed treatment in NSCLC [21]. Of note, the KRAS-G12C mutation was only compared with KRAS wild-type, but not other KRAS mutations, in that study. We analyzed the prognostic value of KRAS-G12C after two types of chemotherapy, and the same trend was observed in PFS benefit, although not statistically significant. Thus, we suggest that identifying the KRAS mutation subtype should be done before commencing chemotherapy in patients with a KRAS mutation. The predictive value of KRAS-mutant subtypes in the chemosensitivity of pemetrexed needs to be confirmed by launching a prospective clinical trial in the future.
To date, molecular inhibitors targeting KRAS mutations directly or indirectly have shown promising efficacy in patients with NSCLC with such mutations [22,23]. The latest released results from a phase I study presented at the 2019 ASCO Annual Meeting indicated that the KRAS-G12C inhibitor, AMG 510, achieved a 50% response rate in patients with advanced NSCLC and the KRAS-G12C mutation [24]. However, the combination of a molecular inhibitor with chemotherapy failed to achieve a positive result [25].
Immunotherapy is another remarkable strategy for immune-responsive lung cancer. Tao et al. [26] reported that the programmed death-ligand 1 (PD-L1) expression status appeared to be independent of the KRAS mutation subtype and that concurrent PD-L1 expression and G12C mutation was associated with a particularly poor prognosis. Although we found PD-L1–positive expression was doubled in patients with KRAS mutation, a lack of data on its expression and treatment outcomes prohibited any prognostic impact conclusion of PD-L1–positive expression in patients with KRAS mutation from this study.
Our study has several limitations that should be acknowledged. First, a bias in patient selection may be inevitable in a retrospective clinical study. However, more actionable information for clinical practice can be attained from a real-world study than that from registered clinical trials. Second, the pemetrexed-based chemotherapy used in our study had higher efficacy than taxane-based chemotherapy, which is considered the most effective treatment in patients with KRAS mutation and NSCLC [27]. Furthermore, pemetrexed may be the most suitable maintenance therapy after completion of first-line chemotherapy [28]. The duration and efficacy of different first-line chemotherapies are worth exploring in patients with KRAS mutations.
In summary, this study identified that the KRAS G12C mutation appeared to be a positive biomarker to predict the survival benefit from first-line chemotherapy, compared with patients without the KRAS G12C mutation. A regimen of first-line chemotherapy should be considered in patients with advanced NSCLC and KRAS mutations. Larger and prospective clinical trials are warranted to confirm our conclusions.
Conflict of Interest
None has any conflict of interest.
Acknowledgments
This study was supported in part by grants from the Zhejiang Public Welfare Technology Research Program (GJ20H160001), Science and Technology Planning project of Zhejiang Province (LGF19H160002), Medical Scientific Research Foundation of Zhejiang Province of China (2019RC027), Zhejiang Traditional Chinese Medicine Science Fund Project (2020ZB037), and Xisike-Hanson Cancer Research Foundation (Y-HS2019-20).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.12.004.
Contributor Information
Min Wang, Email: wangmin@mail.zjxu.edu.cn.
Chun-wei Xu, Email: xuchunweibbb@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bunn P.A., Kelly K. New chemotherapeutic agents prolong survival and improve quality of life in non-small cell lung cancer: a review of the literature and future directions. Clin Cancer Res. 1998;4(5):1087–1100. [PubMed] [Google Scholar]
- 3.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Bos J.L. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 5.Scheffler M., Ihle M.A., Hein R., Merkelbach-Bruse S., Scheel A.H., Siemanowski J. K-ras mutation subtypes in NSCLC and associated co-occurring mutations in other oncogenic pathways. J Thorac Oncol. 2019;14(4):606–616. doi: 10.1016/j.jtho.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Dogan S., Shen R., Ang D.C., Johnson M.L., D'Angelo S.P., Paik P.K. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbons D.L., Byers L.A., Kurie J.M. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12(1):3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janku F., Stewart D.J., Kurzrock R. Targeted therapy in non-small-cell lung cancer-is it becoming a reality? Nat Rev Clin Oncol. 2010;7(7):401. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 9.Nadal E., Chen G., Prensner J.R., Shiratsuchi H., Sam C., Zhao L. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2014;9(10):1513–1522. doi: 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 10.Pao W., Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 11.Ding L., Getz G., Wheeler D.A., Mardis E.R., McLellan M.D., Cibulskis K. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camidge D.R., Ou S.-H.I., Shapiro G., Shapiro Geoffrey I., Otterson G.A., Villaruz L.C. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC) J Clin Oncol. 2014;32(Supl) abstr8001. [Google Scholar]
- 13.Takezawa K., Pirazzoli V., Arcila M.E., Nebhan C.A., Song X., de Stanchina E. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2(10):922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly R.J., Rajan A., Force J., Lopez-Chavez A., Keen C., Cao L. Evaluation of KRAS mutations, angiogenic biomarkers, and DCE-MRI in patients with advanced non–small-cell lung cancer receiving sorafenib. Clin Cancer Res. 2011;17(5):1190–1199. doi: 10.1158/1078-0432.CCR-10-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J.M., Hwang D.W., Ahn J.S., Ahn M.J., Park K. Prognostic and predictive value of KRAS mutations in advanced non-small cell lung cancer. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Román M., Baraibar I., López I., Nadal E., Rolfo C., Vicent S. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer. 2018;17(1):33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camps C., Jantus-Lewintre E., Cabrera A., Blasco A., Sanmartín E., Gallach S. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer. 2011;72(3):365–369. doi: 10.1016/j.lungcan.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Kalikaki A., Koutsopoulos A., Hatzidaki D., Trypaki M., Kontopodis E., Stathopoulos E. Clinical outcome of patients with non-small cell lung cancer receiving front-line chemotherapy according to EGFR and K-RAS mutation status. Lung Cancer. 2010;69(1):110–115. doi: 10.1016/j.lungcan.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Metro G., Chiari R., Duranti S., Siggillino A., Fischer M.J., Giannarelli D. Impact of specific mutant KRAS on clinical outcome of EGFR-TKI-treated advanced non-small cell lung cancer patients with an EGFR wild type genotype. Lung Cancer. 2012;78(1):81–86. doi: 10.1016/j.lungcan.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Fiala O., Pesek M., Finek J., Benesova L., Belsanova B., Minarik M. The dominant role of G12C over other KRAS mutation types in the negative prediction of efficacy of epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Cancer Genet. 2013;206(1-2):26–31. doi: 10.1016/j.cancergen.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Park S., Kim J.-Y., Lee S.-H., Suh B., Keam B., Kim T.M. KRAS G12C mutation as a poor prognostic marker of pemetrexed treatment in non-small cell lung cancer. Korean J Intern Med. 2017;32(3):514. doi: 10.3904/kjim.2015.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenschein G.R., Jr., Smit E.F., Planchard D., Kim D.W., Cadranel J., De Pas T. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC) Ann Oncol. 2015;26(5):894–901. doi: 10.1093/annonc/mdv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jänne P., Smith I., McWalter G., Mann H., Dougherty B., Walker J. Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non-small-cell lung cancer. Br J Canc. 2015;113(2):199. doi: 10.1038/bjc.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fakih M., O'Neil B., Price T.J., Falchook G.S., Desai J., Kuo J. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol. 2019;37(suppl) abstr 3003. [Google Scholar]
- 25.Jänne P.A., van den Heuvel M.M., Barlesi F., Cobo M., Mazieres J., Crinò L. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non–small cell lung cancer: the SELECT-1 randomized clinical TrialSelumetinib + Docetaxel vs docetaxel alone in advanced non–small cell lung CancerSelumetinib + Docetaxel vs docetaxel alone in advanced non–small cell lung cancer. J Am Med Assoc. 2017;317(18):1844–1853. doi: 10.1001/jama.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao L., Sun J., Mekhail T., Meng L., Fang C., Du Y. The prognostic value of KRAS mutation subtypes and PD-L1 expression in patients with lung adenocarcinoma. J Clin Oncol. 2019;37(suppl) doi: 10.1016/j.cllc.2020.07.004. abstr e20022. [DOI] [PubMed] [Google Scholar]
- 27.Mellema W.W., Masen-Poos L., Smit E.F., Hendriks L.E., Aerts J.G., Termeer A. Comparison of clinical outcome after first-line platinum-based chemotherapy in different types of KRAS mutated advanced non-small-cell lung cancer. Lung Cancer. 2015;90(2):249–254. doi: 10.1016/j.lungcan.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Pennell N.A. Selection of chemotherapy for patients with advanced non-small cell lung cancer. Clevel Clin J Med. 2012;79(Electronic Suppl 1):eS46–eS50. doi: 10.3949/ccjm.78.s2.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.