Abstract
Alzheimer’s disease (AD) is the most common form of dementia worldwide, characterized by progressive memory impairment, behavioral changes, and, ultimately, loss of consciousness and death. Recently, microRNA (miRNA) dysfunction has been associated with increased production and impaired clearance of amyloid-β (Aβ) peptides, whose accumulation is one of the most well-known pathophysiological markers of this disease. In this study, we identified several miRNAs capable of targeting key proteins of the amyloidogenic pathway. The expression of one of these miRNAs, miR-31, previously found to be decreased in AD patients, was able to simultaneously reduce the levels of APP and Bace1 mRNA in the hippocampus of 17-month-old AD triple-transgenic (3xTg-AD) female mice, leading to a significant improvement of memory deficits and a reduction in anxiety and cognitive inflexibility. In addition, lentiviral-mediated miR-31 expression significantly ameliorated AD neuropathology in this model, drastically reducing Aβ deposition in both the hippocampus and subiculum. Furthermore, the increase of miR-31 levels was enough to reduce the accumulation of glutamate vesicles in the hippocampus to levels found in non-transgenic age-matched animals. Overall, our results suggest that miR-31-mediated modulation of APP and BACE1 can become a therapeutic option in the treatment of AD.
Keywords: Alzheimer’s disease, miR-31, gene therapy, lentiviral vector, APP, BACE1, amyloid-β peptide, cognitive function, memory
Graphical Abstract

Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder that affects millions of individuals worldwide. Accounting for 60%–75% of all elderly dementia cases, AD is currently responsible for a huge socioeconomic burden that makes it one of the major global health concerns of this century. From a pathophysiological point of view, AD is characterized by neuronal dysfunction and death, accompanied by neuroinflammation, leading to progressive cognitive impairment, memory loss, and behavioral alterations.1
Although the exact cellular and molecular mechanisms underlying this disorder remain unclear, the most accepted theory attributes a major role to the aberrant production and accumulation of protein aggregates, including different amyloid-β (Aβ) peptides and tau protein. According to the revised amyloid cascade hypothesis (ACH),2,3 the abnormal accumulation of the Aβ peptide precedes tau pathology,4 resulting from the sequential processing of the amyloid precursor protein (APP) by β-secretase (BACE1) and γ-secretase enzymes.5,6 Overproduction of Aβ or failure to clear Aβ peptides potentiates their aggregation into dimers or oligomers, which cluster together to form fibrils, thus contributing to the formation of insoluble amyloid plaques. Both oligomers and fibrils are thought to be neurotoxic, interfering with synaptic transmission and promoting microglia and astrocyte activation,7 excitotoxicity,8 synaptic loss,7 and, eventually, neuronal death.5 AD treatments currently used in the clinic are based on small molecules and designed to circumvent cognitive impairment by improving or maintaining brain function.9 Nevertheless, in order to efficiently protect cognition and memory and prevent disease onset, it is crucial to develop new strategies for early intervention, aiming to target initial pathological features, such as the abnormal production of Aβ peptides.10
Recent studies have proposed a pivotal role for microRNAs (miRNAs) in aging and neurodegenerative diseases11 by demonstrating that the expression levels of several miRNAs were significantly altered during cellular senescence and neurodegeneration.11,12 These endogenous short regulatory non-coding RNAs (∼18–22 nt) are able to bind target mRNAs, temporarily inhibiting translation or promoting mRNA degradation,11 leading to changes in target protein levels. In support of the putative importance of miRNAs in AD, several studies showed alterations in the miRNA profile of brain, cerebral spinal fluid, and plasma samples of late-onset AD patients, compared to aged-matched controls.13, 14, 15, 16, 17, 18, 19 Additionally, several miRNAs have been predicted to target mRNAs of genes involved in the ACH.11,18,20 Studies demonstrated that the downregulation of specific miRNAs is correlated with overexpression of BACE1 and that miRNA modulation can decrease BACE1 levels, potentially reducing Aβ production.21 These findings strengthen the role of miRNAs in AD pathogenesis and emphasize the relevance of new therapeutic strategies that may arise from the identification of miRNA “signatures” in the AD brain and from the modulation of these specific miRNAs.
In this study, we performed a screening of several miRNAs predicted to target APP and Bace1, with the purpose of identifying miRNAs whose expression could lead to a downregulation of APP and BACE1 proteins and to a consequent decrease in the production of Aβ peptides. To achieve this goal, we used a mouse model of AD (AD triple-transgenic [3xTg-AD] mice) that presents cognitive impairment and the major neuropathological hallmarks of the disease.22 Our results showed that overexpression of miR-31 in the hippocampus significantly reduced plaque load and prevented cognitive decline.
Results
Expression of Selected miRNAs Decreases Both APP and Bace1 mRNA Expression In Vitro
In order to select and validate miRNA candidates capable of binding both APP and Bace1 mRNAs, a computational prediction of miRNA targets was performed employing mathematical algorithms9 available in different online prediction tools: miRANDA-miRSVR,23 TargetScan,24 Diana microT-CDS,25 and miRWalk.26 The top miRNA:mRNA predicted binding sites of each database were organized according to three parameters: (1) binding affinity score, (2) number of databases predicting mRNA:miRNA binding, and (3) experimental data showing miRNA deregulation in AD mouse models and human samples. This analysis resulted in the selection of four miRNAs: miR-17-5p, miR-31-5p, miR-200c-3p, and miR-497-5p (Figure 1A). All four miRNAs were predicted to target the human and mouse 3′ UTR of APP, while two of them, miR-31-5p and miR-497-5p, were also predicted to target the 3′ UTR of mouse Bace1. Interestingly, an additional binding site for miRNA miR-31-5p was also predicted to exist within the CDS sequence of human APP (Figure 1A).
Figure 1.
Expression of miR-31 Decreases APP and Bace1 Expression Levels
(A) Schematic representation of the predicted binding sites of the miRNAs in the 3′ UTR of genes of interest. miR-17-3p, miR-31-5p, miR-200c-3p, and miR-497-3p are predicted to bind the 3′ UTR of human APP mRNA, while miR-31-5p and miR-497-3p are also predicted to bind the 3′ UTR of mouse Bace1 mRNA. Additionally, miR-31-5p has a putative binding site in the CDS of human APP mRNA encoding the APP695 protein isoform. (B–D) Biochemical validation of putative binding sites was performed employing the luciferase assay. (B) miR-17-3p, miR-31-5p, miR-200c-3p, and miR-497-3p reduced luciferase activity upon co-transfection with the human 3′ UTR APP plasmid in HEK293 cells. NMC, miR-17, and miR-200c, n = 4; miR-31 and miR-497, n = 2. (C and D) miR-31-5p was also able to reduce luciferase activity in HT-22 and HEK293 cells, upon co-transfection with the mouse 3′ UTR Bace1 and human CDS APP plasmids, respectively. (C) NMC and miR-31, n = 5; miR-497, n = 2; (D) NMC and miR-31, n = 3. (E) Schematic representation of lentiviral plasmid (pLenti) construction encoding the selected pri-miRNA sequences. (F) Validation of pLenti constructions was performed in HEK293 and HT-22 cells by evaluating the expression of the GFP reporter gene through fluorescence microscopy (original magnification, ×200) and (G–J) quantifying the mRNA levels of human APP and mouse Bace1 by qRT-PCR. (G) mRNA levels of human APP were significantly decreased in HEK293 cells upon transfection with all miRNA-pLenti vectors, while (H) mRNA levels of mouse Bace1 were significantly decreased in HEK293 cells upon transfection with the miR-31 pLenti construct. (G) pNC, pmiR-17, pmiR-31, and pmiR-200c, n = 3; pmiR-497, n = 2; (H) pNC, pmiR-31, and pmiR-497, n = 3. (I and J) The mRNA levels of human APP (I) and mouse Bace1 (J) were also significantly decreased in SH-SY5Y and HT-22 cells, respectively, upon infection with lentiviral particles encoding miR-31. Non-infected cells (Uninf.) and miR-31 lentivirus (LV miR-31), n = 3. Each n represents a temporarily independent experiment performed in duplicate. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001 with respect to the control condition, that is, transfection with (B–D) control mimic (NCM), (G and H) pLenti control vector (pGFP), or (I and J) non-infected cells. (B, C, G, and H) Ordinary one-way ANOVA with Dunnett’s post hoc test. (D, I, and J) Two-tailed unpaired t test.
We next employed the luciferase reporter assay to validate the efficacy of each miRNA:mRNA interaction. Toward this purpose, the sequences encoding human APP 3′ UTR, human APP CDS, and mouse Bace1 3′ UTR were separately cloned in a validation plasmid, upstream of the luciferase gene. The neuronal HT-22 cell line was co-transfected with the different plasmids and with miRNA mimics for the selected miRNAs. A negative control mimic (NCM) was employed as a negative control, and luciferase activity values were normalized with total protein levels. All miRNA mimics were able to significantly decrease luciferase activity (luciferase relative light units [RLU]) following co-transfection with the human pAPP 3′ UTR luciferase construct (Figure 1B; 62% reduction for miR-17-5p mimics [p < 0.001], 34% for miR-31-5p mimics [p = 0.0084], 40% for miR-200c-3p mimics [p < 0.0001], and 71% for miR-497-5p mimics [p < 0.0001]), with respect to NCM, suggesting the existence of at least one functional binding site for each miRNA in the 3′ UTR of human APP.
Regarding Bace1, while miR-31-5p was able to decrease luciferase activity mediated by the Bace1 3′ UTR luciferase construct in approximately 50% (p = 0.0082), validating the presence of a functional miR-31 binding site in Bace1 3′ UTR, miR-497-5p mimic failed to induce a significant reduction in luciferase levels (Figure 1C).
Since an additional miR-31-5p binding site was predicted to exist within the coding region of the human APP gene, the CDS of the APP695 isoform was also cloned into a piS0 vector, upstream of the luciferase reporter gene. Following co-transfection of the pAPP CDS luciferase construct with miR-31-5p mimics (Figure 1D), a mean reduction of 46% (p = 0.0211) was observed in luciferase activity, with respect to NCM. These results indicate that human APP presents an active miR-31-5p binding site within the coding region, which might also contribute to miRNA-dependent mRNA regulation.
We further investigated whether the four selected miRNAs were able to exert a functional effect in vitro, leading to a reduction of APP and Bace1 mRNA expression. Toward this purpose, the primary (pri-) miRNA sequences of each selected miRNA and a negative control (NC) sequence were cloned into a lentiviral backbone plasmid, harboring the GFP reporter gene (LTR-SIN-CMV-GFP-H1-miRNA-LTR plasmids) (Figure 1E). These constructs were then transfected into both human HEK293 and mouse HT-22 cell lines. Transfection efficiency was confirmed by evaluating the number of GFP-positive cells (Figure 1F), while mRNA levels of both human APP and mouse Bace1 were quantified by qRT-PCR in HEK293 (Figure 1G) and HT-22 cells (Figure 1H), respectively. A decrease in APP mRNA levels was observed in HEK293 upon lentiviral-mediated expression of each selected miRNA (Figure 1G; miR-17, mean decrease of 60%, p < 0.0001; miR-31, mean decrease of 55%, p < 0.0001; miR-200c, mean decrease of 38%, p = 0.0047; miR-497, mean decrease of 63%, p < 0.0001). In addition, and as expected, miR-31 expression led to a significant decrease (mean decrease of 42%, p < 0.0001) in Bace1 mRNA levels in HT-22 cells, while miR-497 expression led to a significant but small decrease (mean decrease of 3%, p = 0.0191) in mRNA levels.
Based on our previous observations, and considering that miR-31 expression was able to modulate simultaneously APP and Bace1 mRNA levels, this miRNA was selected for further in vivo studies. Toward this goal, lentiviral particles encoding the precursor sequence of miR-31 (pri-miR-31) were produced in HEK293 cells, and their integrity and infection efficiency were evaluated in vitro, in two neuronal cell lines (mouse HT-22 and human SH-SY5Y), by flow cytometry and qRT-PCR. Infection of SH-SY5Y cells (96.8% infected cells) (Figure S1A) led to a dramatic decrease in human APP mRNA expression, which reached negligible levels (mean decrease of 99.8%, p < 0.0001; Figure 1I), while infection of HT-22 cells (56.6% infected cells) (Figure S1B) also revealed a statistically significant decrease in Bace1 mRNA expression (mean decrease of 73.3%, p < 0.0001; Figure 1J).
Lentiviral-Mediated miR-31 Expression in the Hippocampus and Subiculum Decreases Cognitive Deficits in the 3xTg-AD Mouse Model
We next investigated whether the developed lentiviral particles were able to upregulate miR-31 expression in vivo, contributing to an improvement of cognitive deficits in an animal model of Alzheimer’s disease. Toward this purpose, we employed the 3xTg-AD mouse model,22 which recapitulates, at a late age, the major histopathological features of the disease, including senile plaques, neurofibrillary tangles, and neuroinflammation.27
To evaluate the extent of viral diffusion and validate neuronal infection, a small number (n = 4) of 12-month-old 3xTg-AD animals were stereotaxically injected in the CA1 region of the right hippocampus with 2 μL of lentiviral particles (viral titer of 200 ng/μL) encoding the pri-miR-31 tagged with GFP. These animals received a second injection, in the left hippocampus, of lentiviral particles encoding the negative control miRNA (Figure S1C). This experiment validated the efficacy and diffusion of both viruses, as well as the injection coordinates, since 2 weeks after injection (Figure S1D) widespread GFP expression could be observed throughout the hippocampus and subiculum in both hemispheres.
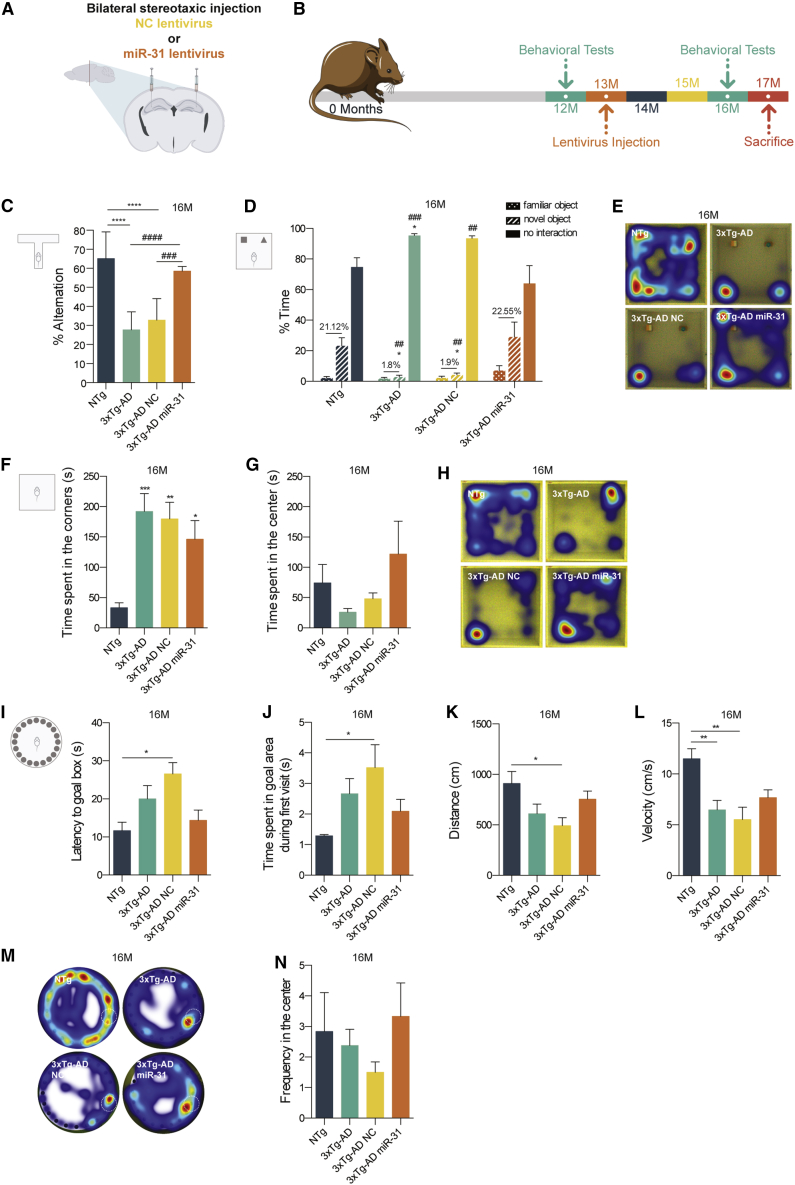
To evaluate the long-term therapeutic potential of miR-31 modulation, a second group of female animals (divided in four temporally independent cohorts) was injected bilaterally with lentiviral particles encoding the pri-miR-31 (3xTg-AD miR-31, n = 9) or the negative control miRNA (3xTg-AD NC, n = 7). These injections were performed at 12 months of age (12M), before the deposition of Aβ plaques, in order to mimic a therapeutic intervention at an early phase of the disease (Figure 2A). Two additional experimental groups were employed in this study: non-transgenic (NTg) animals (n = 8) and untreated 3xTg-AD animals (3xTg-AD, n = 9). The cognitive function of all test animals was analyzed at 12M, immediately before lentiviral injection, and between 16 and 17 months of age (16M) (Figure 2B).
Figure 2.
Decline in Memory and Cognitive Performance Is Arrested in Aged Mice after Bilateral miR-31 Expression in the Hippocampus
(A) Thirteen-month-old 3xTg-AD female mice were stereotaxically injected in the hippocampus of both hemispheres with lentivirus encoding miR-31 or negative control lentivirus (NC). (B) Schematic representation of the experimental time course. Cognitive function was accessed through a battery of behavioral tests, before (12 months) and after (16 months) stereotaxic injection. (C) 16/17-month-old 3xTg-AD miR-31 mice showed a significant increase in the percentage of spontaneous alternation (T-maze test), compared to untreated or 3xTg-AD NC animals. NTg and 3xTg-AD miR-31, n = 7; 3xTg-AD, n = 9; 3xTg-AD NC, n = 6. (D) miR-31 3xTg-AD mice interacted more with the novel object recognition (NOR) with respect to untreated or NC 3xTg-AD animals and (E) revealed an exploratory behavior similar to that of NTg animals (NOR test). (D) NTg, n = 5; 3xTg-AD, 3xTg-AD NC, and 3xTg-AD miR-31, n = 4. (F–H) miR-31 3xTg-AD mice presented (F) a significant decrease in the time spent in the corners of the open field (OF) test (first 5 min) and (G) an increase in the time spent in the center of the OF arena, indicating a less anxious behavior. (F) NTg, n = 8; 3xTg-AD, n = 10; 3xTg-AD NC, n = 7; 3xTg-AD miR-31, n = 9. (G) NTg, n = 7; 3xTg-AD and 3xTg-AD miR-31, n = 8; 3xTg-AD NC, n = 6. (H) NTg and 3xTg-AD miR-31, n = 8; 3xTg-AD, n = 9; 3xTg-AD NC, n = 5. (I–N) miR-31 expression promoted an overall improvement in the long-term memory 3xTg-AD animals. Untreated and 3xTg-AD NC mice, but not 3xTg-AD miR-31 mice, showed (I) a decrease in latency to reach the goal box in the Barnes maze (BM) test, and (J) an increase in the first cumulative duration in the goal area. NTg and 3xTg-AD NC, n = 5; 3xTg-AD, n = 8; 3xTg-AD miR-31, n = 9. (K and L) miR-31 expression also led to an improvement of motor behavior of 3xTg-AD animals, increasing the (K) distance traveled and (L) velocity of miR-31 3xTg-AD mice during BM test. (K) NTg, n = 8; 3xTg-AD, n = 9; 3xTg-AD NC and 3xTg-AD miR-31. n = 7. (L) NTg and 3xTg-AD NC, n = 8; 3xTg-AD, n = 10; 3xTg-AD miR-31, n = 7. (M) miR-31 3xTg-AD animals presented an exploratory strategy similar to that of NTg mice (BM test), (N) performing a higher number of crossings in the arena center. NTg and 3xTg-AD NC, n = 6; 3xTg-AD, n = 8; 3xTg-AD miR-31, n = 9. Data represent mean ± SEM. *p < 0.05 or #p < 0.05, **p < 0.01 or ##p < 0.01, ***p < 0.001 or ###p < 0.001, and ****p < 0.0001 or ####p < 0.0001, with respect to NTg animals (*) or miR-31 3xTg (#). (C, F, G, I–L, and N) One-way ANOVA with Tukey’s post hoc test. (D) Two-way ANOVA with Tukey’s post hoc test. See also Figure S2.
We employed the T-maze spontaneous alternation test to evaluate the left-right discrimination usually presented in healthy animals, which directly translates their reference and working memory capacity. As expected, the spontaneous alternation rate of the 16M NTg animals was around 65%, while the alternation rate of the 3xTg-AD untreated animals dropped to 28%. These results showed a significant difference (p < 0.0001) in the behavioral performance of 3xTg-AD and NC 3xTg-AD mice, suggesting the presence of hippocampal damage in these animals (Figure 2C). Interestingly, the alternation rate of miR-31 3xTg-AD mice was approximately 59%, which corresponds to a significant improvement with respect to both untreated and (p < 0.0001) NC 3xTg-AD animals (p = 0.001) and suggests that lentivirus-mediated miR-31 expression is enough to prevent the dysfunction in spontaneous alternation associated with hippocampal damage.
We next employed the novel object recognition (NOR) test to evaluate short-term memory. This test relies primarily on the innate exploratory behavior of mice in the absence of externally applied rules or reinforcement,28 while also allowing evaluation of overall anxiety. We observed that 16M NTg animals spent more time exploring the novel object (23.2%) than the familiar one (2.08%) (Figure 2D), while 3xTg-AD and NC 3xTg-AD mice only spent 1.8% and 1.9% more time exploring the novel object, respectively (p = 0.0311), and spent significantly less time exploring both objects (p = 0.0275), preferring to remain in the corners of the arena (Figure 2E). These results show that NTg animals, as expected, can recognize and are interested in novelty, while 3xTg-AD and 3xTg-AD NC animals struggle with this task and show clear signs of increased anxiety-like behavior. On the contrary, 3xTg-AD miR-31 animals spent 22.55% more time exploring the novel object than the familiar one (Figure 2D) and only spent 64% of the time not interacting with either object. This result shows a significant difference with respect to untreated (p = 0.0008) and NC (p = 0.0017) groups and illustrates, once again, that 3xTg-AD miR-31-treated animals show a behavior closer to that of NTg mice.
In order to exclude the presence of motor deficits in this animal model, we analyzed mice performance in the open field (OF) test. While 12M 3xTg-AD mice did not present significant differences in distance traveled and in velocity, with respect to NTg mice (Figures S2A and S2C), 16M untreated 3xTg-AD mice showed a tendency to decrease both parameters (Figures S2B and S2D). However, this decrease was not observed in 3xTg-AD miR-31 or NC animals, excluding the presence of relevant motor deficits. Interestingly, while we did not observe differences in the exploratory behavior presented by NTg and 3xTg-AD animals at 12M (Figure S2E), we noticed that, at 16M, untreated 3xTg-AD mice took more time to start exploring the arena than NTg mice, spending significantly more time in the corners during the first 5 min of the trial (Figure S2F). We also analyzed the total time spent in the center of the arena throughout the trial session. Results revealed that, at 16M, both untreated and NC 3xTg-AD mice spent less time in the center than NTg mice, while 3xTg-AD miR-31 mice presented a higher cumulative duration in this area, indicative of an exploratory strategy similar to the NTg group (Figures 2G and 2H). Overall, these results indicate that the anxious behavior presented by 3xTg-AD mice can be ameliorated by miR-31 expression.
Finally, and in order to evaluate the long-term memory and learning capacity of these animals, we employed the Barnes maze (BM) test. We started by monitoring the cognitive and motor abilities of the animals at 12M, immediately before lentiviral injection, in order to ascertain and compare the baseline learning and memory capacity of 3xTg-AD and age-matched NTg animals. In this regard, none of the parameters assessed in the probe day, including distance traveled (Figure S2F), velocity (Figure S2G), and latency to goal box (Figure S2H), showed significant differences between the two experimental groups. In addition, no significant differences were observed in the latency to reach the goal box during the learning trials (Figure S2J), indicating the lack of observable motor or learning deficits at this age. Interestingly, analysis of the heatmaps of the probe trial showed that 12M 3xTg-AD animals already presented a slightly different exploratory strategy, preferring to move in the periphery of the maze, instead of crossing the center of the arena (Figure S2I).
At 16M, all animals were still able to learn the location of the goal box, showing a decrease in the time necessary to reach the target hole across the training trials (Figure S2K). However, during the probe trial, the latency to the goal box of the NTg group was lower than that of untreated and NC 3xTg-AD animals (Figure 2I), but no significant differences were observed with respect to 3xTg-AD miR-31 animals. In this same trial, untreated and NC 3xTg-AD mice spent more time exploring the area where the goal box should be, upon their first visit, with respect to NTg mice (Figure 2J), showing resistance to look for the goal box in a different location, while 3xTg-AD miR-31 animals behaved similarly to NTg mice, quickly exiting the goal box area upon realizing that the exit box was not present. Moreover, while no differences were observed at 12M in the distance traveled and velocity (Figures S2F and S2G), 16M untreated and NC 3xTg-AD mice presented a decrease in distance traveled (Figure 2K) and a significant decrease in velocity (Figure 2L), which are probably linked to the different exploratory strategies adopted by the animals. While NTg and 3xTg-AD miR-31 mice moved indiscriminately in the arena during the probe trial, performing several crossings in the center of the maze (Figure 2M), untreated and NC 3xTg-AD mice explored the maze by preferentially circling the periphery (Figure 2M). This resulted in a higher number of center crossings performed by NTg and 3xTg-AD miR-31 mice with respect to untreated and NC 3xTg-AD mice (Figure 2N).
Taken together, the results from the behavioral tests indicate that miR-31 expression in the hippocampus of aged 3xTg-AD mice ameliorated deficits in short-, work-, and long-term memory and reduced anxiety, strengthening our working hypothesis that miR-31 upregulation in the hippocampus is able to prevent or delay the progression of the cognitive decline associated with AD.
miR-31 Expression Decreases Extracellular and Intracellular Aβ Deposition in 3xTg-AD Animals
In order to determine whether cognitive improvements, triggered by lentiviral-mediated miR-31 expression, correlated with changes in the histopathological hallmarks of AD, we performed immunohistochemistry and stereology analysis in 17M animals. At this age, these animals present both extracellular senile plaques and intracellular neurofibrillary tangles,22 the two most relevant hallmarks of the disease.29
Following immunohistochemical labeling of Aβ peptides in sequential brain sections, employing an antibody specific against human Aβ1–42, we confirmed the presence of extracellular Aβ deposits in untreated 3xTg-AD mice (Figure 3A, left panels, white arrows), which could be found mostly in the subiculum and in the posterior hippocampus. We also observed intraneuronal Aβ1–42 deposits (Figure 3A, left panels, black arrows), which were present in the CA1, CA2, and CA3 regions of the hippocampus and in the V cortical layer. A qualitative assessment of Aβ deposition in 3xTg-AD animals indicated a visible reduction in the number of Aβ plaques in 3xTg-AD miR-31 animals, as well as a decrease in the number of neurons with intracellular Aβ deposits in the subiculum, hippocampus (Figure 3A, right panels), and cortex, with respect to untreated (Figure 3A, left panels) and NC 3xTg-AD animals (Figure 3A, middle panels). These results were further confirmed by confocal microscopy, following labeling of Aβ extracellular deposits with methoxy-XO4. The number and size of bright blue methoxy-XO4-positive deposits in the subiculum were lower in 3xTg-AD miR-31 animals (Figure 3B, right panel), with respect to both untreated (Figure 3B, left panel) and NC 3xTg-AD mice (Figure 3B, middle panel).
Figure 3.
miR-31 Expression Reduces Plaque Load and Intraneuronal Aβ in the Hippocampus and Cortex
(A) Representative microscopy images (original magnification, ×100; scale bar, 200 μm) of Aβ deposition in 50-μm coronal brain slices of 17-month-old (17M) mice from all experimental groups. Aβ immunohistochemistry, using anti-human Aβ 6E10 antibody (see also Figure S3), revealed a reduction of Aβ plaque load (white arrows) and intraneuronal Aβ deposits (black arrows) in the subiculum and CA1 and CA2 hippocampal regions of miR-31 3xTg animals, with respect to 3xTg-AD and 3xTg-AD NC mice. This is quantified in (C)–(E). (B) Representative confocal images (original magnification, ×60; scale bar, 100 μm) of Aβ plaques labeled with methoxy-X04 in the subiculum of 17M 3xTg-AD animals from all experimental groups. miR-31 3xTg animals showed a reduction in the number and size of Aβ deposits, compared to 3xTg untreated or NC 3xTg mice. (C–E) Quantitative analysis of Aβ plaque load and intraneuronal Aβ deposition was performed by stereology. (C) Results revealed a significant reduction of Aβ plaque load/area unit in the subiculum of miR-31 mice, with respect to 3xTg-AD and 3xTg-AD NC animals. 3xTg-AD and 3xTg-AD miR-31, n = 7; 3xTg-AD NC, n = 4. (D) Furthermore, we also observed a reduction in Aβ plaque load/area unit in the hippocampus of miR-31 mice. 3xTg-AD and 3xTg-AD miR-31, n = 9; 3xTg-AD NC, n = 6. (E) Stereological quantification of intraneuronal Aβ deposition/area unit revealed a significant reduction along the different hippocampal subregions, in the subiculum and in the cortex of miR-31 3xTg-AD animals, compared with 3xTg-AD. CA1+CA2: 3xTg-AD and 3xTg-AD miR-31, n = 9; 3xTg-AD NC, n = 5. CA3: 3xTg-AD, n = 9; 3xTg-AD NC, n = 5; 3xTg-AD miR-31, n = 8. Subiculum: 3xTg-AD, n = 9; 3xTg-AD NC, n = 4; 3xTg-AD miR-31, n = 8. Cortex: 3xTg-AD, n = 9; 3xTg-AD NC, n = 5; 3xTg-AD miR-31, n = 8. Data represent mean ± SEM. #p < 0.05, **p < 0.01, ***p < 0.001 with respect to untreated 3xTg-AD animals (*) or NC 3xTg-AD animals (#). (C–E) One-way ANOVA with Tukey’s post hoc test.
In order to have a quantitative assessment of Aβ extracellular deposition, we next performed stereological analysis in the hippocampus and subiculum (Figure S3, first and second lines). Unbiased stereology-based quantification of Aβ plaque load in the subiculum confirmed that lentiviral-mediated miR-31 expression led to a significant decrease in the area occupied by Aβ plaques in this region, with respect to untreated and NC 3xTg-AD mice (p < 0.0001 and p = 0.0259, respectively) (Figure 3C). Moreover, 3xTg-AD miR-31 animals also presented a decrease, albeit not statistically significant, in Aβ load in the hippocampus (Figure 3D).
The quantification of intraneuronal Aβ was performed in the subiculum, hippocampus (CA1+CA2 and CA3 regions), and also in the cortex (Figure S3, first and third lines), and it showed a significant decrease in intraneuronal Aβ labeling in CA1+CA2, CA3, subiculum, and cortex of 3xTg-AD miR-31, with respect to 3xTg-AD animals (p < 0.001, p = 0.014, p = 0.002, and p < 0.0001, respectively) (Figure 3E). Overall, these results indicate that miR-31 expression is able to decrease Aβ deposition, reducing the formation of senile plaques in brain structures directly connected with AD pathophysiology.
Of note, a small but significant unspecific decrease in intraneuronal Aβ was also detected in 3xTg NC animals, both in the cortex and in the CA1+CA2 regions. This might be a direct result of transient changes in neuronal protein production or local immune response triggered by the NC lentivirus, both of which result in the elimination of a small number of infected cells. In order to assess whether lentiviral-mediated miR-31 expression or lentiviral infection, per se, induced changes in microglia and astrocyte activity, we performed immunohistochemistry studies, using Ionized calcium-binding adaptor molecule 1 (IBA-1) and glial fibrillary acidic protein (GFAP) as markers of microglia and astrocytes, respectively, to evaluate changes in the morphology and distribution of these cells. We observed the expected clusters of microglia and astrocytes around Aβ extracellular plaques, in the subiculum and hippocampus, which were more expressive in untreated and NC 3xTg-AD mice (Figures S4A and S4B). We also quantified the expression of key inflammatory cytokines in the hippocampus, such as Tnf-α and lL-6, which are usually released by these cells. However, no differences were observed between all experimental groups (Figures S4C and S4D). Finally, we quantified the mRNA levels of Trem2, a membrane receptor that is expressed by microglia and is found to be downregulated upon microglia activation toward the M1 (classical pro-inflammatory) phenotype. Although the levels of Trem2 mRNA were slightly reduced in the hippocampus of untreated and NC 3xTg-AD animals (Figure S4E), with respect to NTg animals, suggesting the presence of pro-inflammatory microglia cells in this brain region, the differences observed were not statistically significant. Overall, these results indicate that lentiviral injection, per se, does not activate the immune system in the 3xTg-AD model.
miR-31 Expression Reduces the Number and Intensity of VGLUT1 Puncta
To investigate whether Aβ deposition in the hippocampus of 3xTg-AD mice perturbs neurotransmitter release and causes imbalance of excitatory and inhibitory synaptic inputs, we analyzed the number, size, and intensity of VGLUT1 and VGAT puncta in the CA1, CA3, and dentate gyrus regions. The integrated density of VGLUT and VGAT agglomerates was quantified through a range of detection thresholds. In CA1, which is the region of the hippocampus closer to the injection point and presenting the highest number of infected neurons, it was possible to observe a significant increase in the intensity of VGLUT1-positive puncta in untreated and NC 3xTg-AD mice, with respect to NTg, while 3xTg-AD miR-31 animals did not show significant differences (Figures 4A and 4C). We also observed an increase (36%), albeit not statistically significant (p = 0.0584), in the number of VGLUT1 puncta per area unit in 3xTg-AD mice, with respect to NTg animals, but this increase was not observed in NC or miR-31 3xTg-AD animals (Figure 4E). No differences were observed in VGLUT1 puncta size between all experimental groups (Figure 4F). Regarding VGAT, we observed an increase in VGAT puncta intensity in 3xTg-AD NC animals, with respect to NTg mice (Figures 4B and 4D), but no differences were observed in VGAT puncta size and puncta density between all experimental groups (Figures 4G and 4H). No significant differences were observed in VGLUT1 and VGAT puncta intensity, density, and size in CA3 (Figure S5).
Figure 4.
miR-31 Expression Reduces the Number and Intensity of VGLUT1 Puncta in the CA1 Hippocampal Subregion
VGLUT1- and VGAT-positive puncta where labeled by immunohistochemistry in brain slices of 17-months-old (17M) 3xTg-AD and NTg animals in order to quantify the intensity, size, and density of VGLUT1- and VGAT-positive synaptic vesicles in the hippocampus. (A) The intensity of VGLUT1 puncta was significantly decreased in the CA1 subregion of both NTg and miR-31 3xTg-AD mice, with respect to untreated 3xTg-AD animals (threshold of 0–15) and NC 3xTg-AD animals (threshold of 0–9). NTg, n = 3; 3xTg-AD and 3xTg-AD miR-31, n = 4; 3xTg-AD NC, n = 5. (B) No significant differences were observed between the experimental groups regarding the intensity of VGAT puncta. NTg, n = 3; 3xTg-AD, 3xTg-AD NC, and 3xTg-AD miR-31, n = 4. (C and D) Panels show representative confocal images of (C) VGLUT1 and (D) VGAT puncta in the CA1 subregion (original magnification, ×630 plus ×5 digital zoom; scale bars, 4 μm). (E–H) Quantitative analysis of the density and average puncta size of VGLUT1 and VGAT puncta in the CA1 subregion. (E) Results showed a higher density of VGLUT1 puncta in 3xTg-AD mice with respect to NTg. NTg, n = 3; 3xTg-AD and 3xTg-AD miR-31, n = 4; 3xTg-AD NC, n = 5. No significant differences were observed regarding (F) VGLUT1 average puncta size, and VGAT (G) puncta density and (H) average puncta size. (F) NTg, n = 3; 3xTg-AD and 3xTg-AD miR-31, n = 4; 3xTg-AD NC, n = 5. (G and H) NTg, n = 3; 3xTg-AD, 3xTg-AD NC, and 3xTg-AD miR-31, n = 4; Data represent mean ± SEM. *p < 0.05 or #p < 0.05, **p < 0.01 or ##p < 0.01, and ***p < 0.001 or ###p < 0.001 with respect to untreated 3xTg animals (*) or NC 3xTg animals (#). (A and B) two-way ANOVA with Bonferroni’s post hoc test. (E–H) One-way ANOVA with Tukey’s post hoc test. See also Figure S5.
Overall, these results suggest that the increase in Aβ production and extracellular accumulation observed in the 3xTg-AD animal model may lead to an increase in the accumulation of VGLUT1 at synaptic terminals, causing an imbalance in excitatory/inhibitory outputs that can be rescued by miR-31 expression.
miR-31 Expression Promotes a Decrease in APP and BACE1 Expression In Vivo
To explore the mechanisms by which miR-31 expression induces beneficial alterations in 3xTg-AD animals, we investigated whether lentiviral-mediated miR-31 expression was able to decrease the expression of the predicted miR-31 target genes. Toward this purpose, we first evaluated, by qRT-PCR, miR-31 levels in the hippocampus and subiculum of 17M 3xTg-AD mice. As expected, we observed a significant upregulation of this miRNA in miR-31 3xTg-AD mice, with respect to untreated or NC 3xTg-AD animals (Figure 5A). However, it is important to highlight that this increase was moderated, not surpassing the miR-31 physiological levels observed in NTg animals. Interestingly, we also found that miR-31 basal expression was lower in untreated 3xTg-AD animals with respect to NTg mice (Figure 5A), which suggests that miR-31 downregulation may be an intrinsic feature of this model.
Figure 5.
miR-31 Expression Reduces the Levels of Both APP and Bace1 in 3xTg-AD Mice
mRNA and protein extracts were prepared from hippocampal samples of 17-month-old 3xTg-AD and NTg mice. (A) qRT-PCR analysis revealed a significant increase in miR-31 expression in the hippocampus of miR-31 3xTg-AD animals, with respect to untreated and NC 3xTg-AD mice. NTg, n = 3; 3xTg-AD and 3xTg-AD miR-31, n = 7; 3xTg-AD NC, n = 5. (B) The levels of mouse BACE1 mRNA were found to be significantly decreased in miR-31 3xTg-AD animals, with respect to untreated 3xTg-AD mice. NTg, n = 6; 3xTg-AD, n = 7; 3xTg-AD NC, n = 4; 3xTg-AD miR-31, n = 8. (C) A similar decrease in BACE1 protein levels was detected by ELISA in the hippocampus of miR-31 3xTg-AD animals, with respect to untreated 3xTg-AD mice. NTg, n = 3; 3xTg-AD, n = 9; 3xTg-AD NC, n = 4; 3xTg-AD miR-31, n = 8. (D and E) The levels of mouse BACE1 protein were also analyzed by western blot in all experimental groups and were shown to be increased only in untreated 3xTg-AD animals. (D) Quantification revealed a significant decrease in BACE1 protein levels in 3xTg-AD miR-31 animals with respect to untreated 3xTg-AD mice. NTg, n = 6; 3xTg-AD, n = 7; 3xTg-AD NC, n = 5; 3xTg-AD miR-31, n = 7. (F) mRNA levels of human APP were lower in the hippocampus of miR-31 mice, with respect to untreated and NC 3xTg-AD groups. 3xTg-AD, n = 9; 3xTg-AD, NC n = 5; 3xTg-AD miR-31, n = 7. Western blot analysis (G and H) confirmed these results and revealed a significant decrease in human APP protein levels in the hippocampus of miR-31 3xTg-AD animals, compared to untreated and NC 3xTg-AD mice. 3xTg-AD, n = 9; 3xTg-AD NC, n = 4; 3xTg-AD miR-31, n = 7. (I) Mouse APP mRNA expression levels were not significantly different among the 3xTg-AD experimental groups, showing that our strategy is specific for human APP. 3xTg-AD, n = 9; 3xTg-AD NC, n = 5; 3xTg-AD miR-31, n = 8. Data represent mean ± SEM. *p < 0.05 or #p < 0.05 and **p < 0.01 with respect to untreated 3xTg animals (*) or NC 3xTg animals (#). (A–I) One-way ANOVA with Tukey’s post hoc test.
We next evaluated the expression of mouse Bace1 by looking into both mRNA and protein expression in the same hippocampal samples. This analysis revealed that miR-31 3xTg-AD mice presented a significant decrease in both Bace1 mRNA (Figure 5B) and protein levels (Figures 5C–5E), with respect to untreated 3xTg-AD mice. Importantly, lentiviral-mediated miR-31 expression was able to reduce BACE1 expression to values close to those of NTg mice (Figures 5C and 5D).
Furthermore, to understand whether APP knockdown also contributed to the therapeutic effect observed upon miR-31 expression, the mRNA and protein levels of human APP were evaluated in all 3xTg-AD mice by both qRT-PCR and western blot. We found that miR-31 3xTg-AD animals presented a decrease, albeit not statistically significant, in human APP mRNA levels (Figure 5F), as well as a significant 43% and 47% reduction in human APP protein expression (Figures 5G and 5H; Figure S6), with respect to untreated and NC 3xTg-AD animals, respectively. To investigate whether miR-31 was also able to target the mRNA of mouse App, which is ubiquitously present throughout the brain, we designed specific primers sets for this mRNA. Our qRT-PCR results did not show any significant differences in mmu App mRNA levels between the 3xTg-AD groups (Figure 5I).
Finally, in order to understand whether miR-31 presents additional targets in mouse neuronal cells that could contribute to the phenotype observed in miR-31 3xTg-AD mice, we performed PANTHER pathway analysis (functional enrichment analysis) to investigate which other miR-31 predicted targets could be involved in AD pathways. A total of 1,423 mmu-miR-31-5p predicted targets (miRWalk database) were analyzed through this software, revealing seven candidates: Prkce, Cacnb1, Cacnb2, Psen2, Mapk11, Apba2, and Bace2 (Figure S7). However, none of these genes presented higher expression or higher miRNA binding scores, when compared to App or Bace1.
We next looked at other possible relevant targets by cross-referencing information from the miRNA target prediction databases (see Supplemental Materials and Methods) with information regarding the specific transcriptome of the different cellular subsets present in the mouse brain (https://web.stanford.edu/group/barres_lab/brain_rnaseq.html).30 Through this analysis, we obtained a list of 50 miR-31 target genes that displayed high mRNA/miRNA binding scores and, at the same time, were predicted interacting dyads by at least three databases (Figure 6A). From this list, we selected five genes presenting FPKM (fragments per kilobase per million mapped reads) values similar or higher to those of mouse App (200) or Bace1 (20) (Figure 6B). We considered these targets to constitute the most relevant competitors for miR-31 binding in neurons, since they are equally available and present equal or higher binding affinity toward miR-31. Nevertheless, qRT-PCR analysis of these targets in hippocampal samples did not reveal any decrease in the mRNA levels of Atx, Dpy4, and Dusp1 between the four experimental groups (Figure 6B) and, instead, showed a small but significant increase in Dpy5 and Rab3 mRNA levels in 3xTg-AD miR-31 mice, with respect to untreated 3xTg-AD animals. Given the function of these two genes, this increase could be due to a compensatory cellular mechanism stemming from a cellular response to Aβ clearance (Figure 6B).
Figure 6.
miR-31 Does Not Decrease the Expression of Other Predicted Targets
(A) Representative illustration of 50 miR-31 predicted targets, selected by crossing information from miRNA target prediction online databases with information regarding the specific transcriptome of the different cellular subsets of the mouse brain (brain RNA sequencing database: https://web.stanford.edu/group/barres_lab/brain_rnaseq.html). Targets are organized clockwise in a color gradient, according to miR-31 binding score (miRWalk). A group of five genes predicted by three or more databases (miRANDA-miRSVR, TargetScan, Diana microT-CDS, and miRWalk), presenting FPKM values similar to or higher than those of mouse APP (200) or BACE1 (20) and higher predicted binding scores (miRWalk), were selected for further quantification by qRT-PCR. (B) mRNA levels of Atxn7L3b, Dpysl4, Dpysl5, Rab3a, and Dusp1 were quantified in hippocampal RNA extracts of all experimental groups. A significant increase in mouse Dpysl5 and Rab3a mRNA levels was observed in miR-31 3xTg animals with respect to untreated 3xTg mice. Data represent mean ± SEM. ∗p < 0.05 with respect to untreated 3xTg animals. Atxn7L3b: NTg and 3xTg-AD miR-31, n = 7; 3xTg-AD, n = 9; 3xTg-AD NC, n = 5. Dpysl4: NTg and 3xTg-AD miR-31, n = 8; 3xTg-AD, n = 9; 3xTg-AD NC, n = 5. Dpy5sl5: NTg and 3xTg-AD miR-31, n = 8; 3xTg-AD, n = 9; 3xTg-AD NC, n = 4. Rab3a: NTg, n = 7; 3xTg-AD, n = 9; 3xTg-AD NC, n = 4; 3xTg-AD miR-31, n = 8. Dusp1: NTg, n = 7; 3xTg-AD, n = 9; 3xTg-AD NC, n = 5; 3xTg-AD miR-31, n = 8. Western blot analysis of DpysL5 (C and D) and Rab3a (E and F) did not revealed any significant decrease in the levels of both proteins in the hippocampus of miR-31 3xTg-AD animals, compared to untreated and NC 3xTg-AD mice, showing that miR-31 expression is specifically impacting APP and Bace1 levels and not other miR-31 predicted targets. 3xTg-AD, n = 9; 3xTg-AD NC, n = 4; 3xTg-AD miR-31, n = 7. Data represent mean ± SEM. ∗p < 0.05 with respect to untreated 3xTg animals (∗) (B) One-way ANOVA with Dunnett’s post hoc test, (D and F) One-way ANOVA with Tukey's post hoc test.
Overall, these findings show that miR-31 expression leads to a decrease in both human APP and mouse Bace1 expression, suggesting that miR-31-mediated downregulation of these two proteins is likely the mechanism responsible for the observed therapeutic effects.
Discussion
In the present study we provide strong evidence that miR-31 expression alleviates AD neuropathology, in the 3xTg-AD mouse model, by rescuing cognitive function and targeting the amyloidogenic pathway through the downregulation of APP and BACE1 proteins.
Our initial bioinformatics screening allowed the selection of miRNAs with high mRNA:miRNA binding affinity toward APP and Bace1, which had also been previously described to be deregulated in AD patients.18,31, 32, 33 This is the case of miR-31, which has been reported to be decreased in the serum of AD patients,33 illustrating its potential both as a biomarker and a therapeutic target.
In our experiments, miR-31-5p was the only miRNA capable of simultaneously decreasing the mRNA levels of human APP and mouse Bace1 (Figures 1B and 1C). In addition to the hsa APP and mmu Bace1 3′ UTR binding sites, this miRNA was also predicted to have an additional binding site in the CDS of hsa APP (Diana-microT-CDS database)25 (Figure 1A). The regulatory benefits of CDS sites are still mostly unexplored, although a small number of these binding sites have been validated so far in several genes.34, 35, 36, 37 CDS sites are thought to exert their regulatory activity mostly through translation inhibition, instead of through direct mRNA degradation.37 In this work, we present strong evidence that miR-31 is indeed able to bind to the CDS of the human APP gene, leading to a significant decrease in APP mRNA levels (Figure 1D). The existence of this second active binding site in APP helps to explain the strong functional outcomes observed for this miRNA both in vitro and in vivo, in 3xTg-AD mice.
Of note, a gene-silencing technology was recently approved by the US Food and Drug Administration in order to treat a rare condition called hereditary transthyretin amyloidosis38 (ClinicalTrials.gov: NCT01960348), in which mutated forms of the protein transthyretin accumulate in organs and tissues, compromising the nervous system.39 The drug patisiran is the first therapy based on RNA interference (RNAi) approved after a 20-year wait. This accomplishment has been seen as a fresh hope in the field of RNAi-based drugs and the setting stone to many more such drugs in the coming years.40 These recent advances show that the development of efficient tools for gene modulation is no longer the major hindrance in this research field. In fact, in our perspective, the current major limitation is related to ensuring the safety and specificity of the delivery of small RNA-based therapeutics to the target tissues. This is even a bigger concern when considering the specific challenges of targeting the brain, such as the low permeability associated with the blood-brain-barrier, as well as the delicate environment required to maintain the homeostasis of neuronal activity.
The hippocampus, subiculum, and cortex are the brain regions where AD neuropathology is most prominent.41, 42, 43 Damage in these brain regions causes memory loss and cognitive dysfunction, explaining part of the symptomatology presented by AD patients. In the present work, we focused our attention in these regions and we chose to employ only female animals, since even though most research in AD models has been performed in male mice, the prevalence in AD is higher in the females.44,45
We show, using several behavioral tests, that lentiviral-mediated miR-31 expression significantly ameliorates cognitive and memory deficits in 3xTg-AD animals. We observed a particularly striking improvement in the T-maze. It has been described that animals with hippocampal damage may develop side preference in the T-maze showing alternation below 50% in this test (the minimum achievable on a balanced left/right forced schedule).46,47 This was the case of untreated and NC 3xTg-AD animals, which alternated around 30%, illustrating cognitive inflexibility,46 while 3xTg-AD animals expressing miR-31 presented a dramatic improvement, performing similarly to age-matched wild-type animals (Figure 2B). We also observed a reduction in anxiety-like behavior in miR-31 3xTg-AD animals. These animals tended to explore more the NOR, OF, and BM arenas (Figures 2E, 2H, and 2M), with respect to the other 3xTg-AD groups. In addition, these animals also showed an improvement in long-term memory, presenting a shorter latency to reach the escape box in the BM test (Figure 2I). Interestingly, despite having more difficulties remembering the location of the goal box, upon reaching this location, untreated and NC 3xTg-AD animals showed reluctance to leave and further explore the maze, in what can be interpreted as cognitive inflexibility (Figure 2J). Overall, these behavioral results corroborate our hypothesis that 3xTg-AD animals, in addition to presenting the expected cognitive and learning deficits associated with the AD phenotype, are more anxious than NTg animals and present a cognitive inflexibility phenotype. Both of these features are reminiscent of the first symptoms observed in AD patients.48 Moreover, apathy, which was also detected in these animals, remains the most persistent neuropsychiatric symptom throughout the course of the human disease. Nevertheless, this behavior impairment observed in the 3xTg-AD animals was assuaged upon miR-31 lentiviral treatment, allowing these animals to autonomously explore the mazes and novel situations, similarly to NTg animals. We think that additional analysis regarding these behaviors could be suitable to further understand the role and monitor the effects of miR-31 expression in ameliorating anxiety and inflexibility conducts.
miR-31 expression also contributed to ameliorate the neuropathology features observed in this AD model, mediating a decrease in both intracellular Aβ accumulation and extracellular Aβ plaque deposition (Figures 3C–3E). These results directly correlate with the lower mmu BACE1 and hsa APP protein levels detected in miR-31 3xTg-AD animals (Figures 5C and 5F), suggesting that the observed improvement in cognitive function results from the direct targeting of these genes. We hypothesize that the decrease in the expression of these two proteins is able to reduce the production of Aβ peptides through the amyloidogenic pathway, thus delaying Aβ accumulation and protecting the neurons in these key regions from Aβ-mediated synaptic loss and inflammation. Our hypothesis is further supported by the recent work of Hu et al.49 By generating a new BACE1 conditional knockout 5XFAD mouse model, the authors were able to mimic BACE1 inhibition in adults. This approach was able to completely reverse amyloid deposition, leading to a significant improvement of synaptic function and behavior and providing compelling evidence that BACE1 inhibition in the adult brain is able to reverse AD pathology.
Additionally, we observed a robust decrease in amyloid deposition in the subiculum (Figures 3C and 3E). Subiculum is known to be recruited during tasks that involve working and short-term memory50,51. Interestingly, however, it has been demonstrated that the subiculum also displays a role in the regulation of the hypothalamic-pituitary-adrenal (HPA) axis,52, 53, 54 which plays a crucial role in the response to stress and is responsible for corticosterone production. We speculate whether the alleviation of Aβ deposition in this region, which is densely populated by Aβ plaques in this animal model, could improve HPA function, contributing to the amelioration of anxiety-like behaviors, since there is also growing evidence that dysregulation of the HPA axis may be implicated in the onset and progression of AD.55, 56, 57, 58 Moreover, the prefrontal cortex has been shown to be associated with cognitive flexibility. The Aβ accumulation observed in this area in 3xTg-AD mice may explain by itself the behavior inflexibility displayed by these animals. Nevertheless, it has also been suggested that there is a flow input between the prefrontal cortex and the hippocampus, particularly targeting the ventral subiculum, via nucleus accumbens neurons.59 In this context, we consider the hypothesis that the amelioration of Aβ deposition, observed not only in the cortex but also in the subiculum, might influence this neuronal circuit, contributing to the improved cognitive flexibility observed in miR-31 3xTg-AD animals with respect to NC or untreated mice. Finally, we observed an overall accumulation of intraneuronal Aβ in the subiculum, hippocampus regions (CA1+CA2 and CA3), and cortex in the 3xTg-AD animal model, which was significantly suppressed upon lentiviral-mediated miR-31 expression (Figure 3E). These results suggest, once again, that we were able to intervene at an early phase of the disease, successfully delaying AD progression.
miR-31 expression was also shown to protect 3xTg-AD animals from excessive accumulation of glutamate in CA1 pre-synaptic terminals (Figure 4A). Although a decrease in VGLUT1 protein levels has been reported to occur in the brain of late-stage AD patients,60 we observed an increase in the intensity (Figure 4E) and a small increase in the density (Figure 4F) of VGLUT1 labeling in the CA1 region of both untreated and NC 3xTg-AD animals. We think that these somewhat contradictory observations illustrate different stages of the disease. The loss of VGLUT1 reported in human brains may reflect the overall synaptic and neuronal loss that occurs in AD patients, while in AD animal models neuronal death is rarely observed and synaptic loss is usually a late event. In fact, in other AD mouse models, such as APP/PS1 mice, no differences were observed in VGLUT1 total levels between 16M transgenic and wild-type (WT) animals.61 Interestingly, in this same study, the authors reported a significant reduction in glutamate release in APP/PS1 mice at this age. Moreover, a similar result to ours was observed by Timmer et al.62 in the TgSwDI mouse model. These results support our hypothesis that, at the initial stages of the disease, glutamate vesicles become trapped at synaptic terminals, due to a reduction in exocytosis, thus leading to an increase in VGLUT1 labeling in these structures. The reduction in glutamate release may be a direct consequence of Aβ accumulation in the synaptic cleft or, as suggested by Minkeviciene et al.,61 may be due to the presence of PS1 mutations that impact the function of K+ channels known to play a critical role in vesicle exocytosis.
In addition to App and Bace1 genes, our initial bioinformatics analysis provided an extensive list of other possible targets of miR-31. To further explore whether these targets could be a source of off-target effects and also contributed to the phenotype observed in miR-31 3xTg-AD animals, we investigated which target genes were simultaneously expressed in neurons and showed binding affinities toward miR-31 stronger or as strong as App and Bace1. Although, we identified a list of 50 genes with high binding scores (Figure 6A), only five candidate genes presented FPKM values higher than those of Bace1 and close to those of App, therefore constituting direct competitors for miR-31 binding. Nevertheless, none of these genes presented a decrease in mRNA levels following miR-31 expression (Figure 6B), suggesting that they are not the main targets of miR-31 in neuronal cells and do not contribute, at least directly, to the phenotype observed in miR-31 3xTg-AD animals.
We observed a small but unexpected increase in the mRNA levels of two of the tested genes, Dpysl5 and Rab3a, in the brain of 3xTg-AD mice (Figure 6B). Rab3a is a GTP-binding protein implicated in pre-synaptic function and neurotransmitter exocytosis. Interestingly, previous studies reported that Rab3a overexpression in neuroendocrine cells led to inhibition of Ca2+-triggered exocytosis,63 suggesting that Rab3a overexpression may contribute to the observed accumulation of excitatory transporter vesicles at pre-synaptic terminals. With regard to Dpysl5, a member of the collapsin response mediator protein (CRMP) family, its expression has been described to contribute to the generation and survival of newborn neurons in olfactory and hippocampal neurogenic areas.64 Additionally, a study using CRMP5−/− mice revealed an impairment in brain-derived neurotrophic factor (BDNF),65 which is also decreased in AD patients.66 Although the exact signaling pathway of CRMPs in the adult brain still needs to be further elucidated, it is possible that the observed increase in Dpysl5 expression may contribute to the improvement in cognitive function observed in 3xTg-AD animals through the upregulation of BDNF.
We and other authors have recently demonstrated that neuroinflammation is an early and important hallmark of AD, directly contributing to neurodegeneration.67 In 3xTg-AD animals, and as expected, both microglia and astrocytes were found to surround Aβ extracellular plaques in the subiculum and in the hippocampus. These observations correlate with our previous results27 indicating the existence of plaque-driven microglia recruitment and low-grade chronic inflammation in this AD animal model. Despite this, we could not detect an overall increase in microglia and astrocyte number following lentivirus injection (Figures S4A and S4B). Moreover, no differences were observed regarding inflammatory cytokine production or Trem2 expression (Figures S4C–S4E), suggesting that lentiviral-mediated expression, per se, does not trigger an immune response in this AD model, constituting a safe and non-immunogenic therapeutic strategy.
In conclusion, the present study shows that miR-31 expression strongly improves cognitive function and prevents disease progression in the 3xTg-AD mouse model by downregulating APP and Bace1 mRNAs. Thus, a therapeutic strategy based on the modulation of this miRNA could constitute, in the future, a promising approach for AD therapy.
Materials and Methods
Materials
The luciferase reporter plasmids encoding the 3′ UTR of human APP (#SC212695) and mouse BACE1 (#MmiT030538) were purchased from OriGene (USA) and GeneCopoeia (USA), respectively. The pIS0 (#12178), used for insertion of APP CDS, and the pENTR/pSUPER+ (#17338) and pLenti CMV GFP DEST (#19732) vectors, used for lentiviral production, were purchased from Addgene (USA). The pcDNA6.2-GW/miR-neg vector (#K493700), employed for the generation of lentiviral particles encoding the negative control (NC) miRNA, was obtained from Life Technologies (USA). The miRNA mimics and the DharmaFECT Duo transfection reagent were obtained from GE Healthcare Dharmacon (USA). The PCR primers for human APP CDS amplification were obtained from Invitrogen (USA) (Table S1; Supplemental Materials and Methods). The qRT-PCR primers for detection and determination of the selected miRNAs were purchased from Exiqon (Denmark) (Table S1; Supplemental Materials and Methods). The qRT-PCR primers for mRNA quantification were acquired from QIAGEN (Germany) (Table S1; Supplemental Materials and Methods). The following primary antibodies were employed throughout this work: anti-IBA-1 (1:1,000; Wako Pure Chemicals Industries, Japan, #019-19741), anti-GFAP (1:1,000; Chemicon International, USA, #AB5804), anti-Aβ 6E10 (1:2,000; BioLegend, USA), anti-APP (1:2,000; Millipore, USA, #MAB348), anti-VGLUT1 (1:1,000; Millipore, USA, #AB5905), anti-VGAT (1:1,000; Millipore, USA, #AB5052P), and anti-actin (1:10,000; Sigma, USA, #A2066), as well as the following secondary antibodies: Alexa Fluor 568 anti-rabbit immunoglobulin (Ig)G (1:200; Life Technologies, USA), Alexa Fluor 647 anti-guinea pig IgG (1:200; Life Technologies, USA), biotinylated anti-mouse IgG (1:200; Vector Laboratories, Burlingame, CA, USA). Methoxy-X04 (MX04) was obtained from Tocris Bioscience (UK). The ELISA assay kit employed to detect mouse BACE1 protein levels was purchased from Cloud-Clone (China).
Animals
All efforts were conducted to minimize animal suffering and to reduce the number of animals used in this study, in accordance with the guidelines of the local Animal Welfare and Ethics Body (ORBEA), Portuguese National Authority for Animal Health and the European Community Council directive (2010/63/EU) for the care and use of laboratory animals (reference 0421/000/000/2013). Moreover, power analysis was performed to predict the number of animals necessary to detect an effect. 3xTg-AD animals22 were initially provided by the laboratory of Dr. Frank LaFerla (Department of Neurobiology and Behavior, Institute for Brain Aging and Dementia, University of California, Irvine, CA, USA). A colony of these animals was established at the Center for Neuroscience and Cell Biology of the University of Coimbra animal facility. Adult female mice weighing ∼27 g were housed under conventional light and environmental conditions (12-h light/12-h dark cycle, 23°C ± 1°C, 55% ± 5% relative humidity). Food and water were available ad libitum. Age- and sex-matched NTg mice were used as controls. 3xTg-AD animals were found to maintain the same phenotypic and behavioral characteristics initially described by LaFerla and colleagues.22 Details concerning animal sacrifice and brain handling can be found in Supplemental Materials and Methods.
Cell Culture and Transfection
HT-22 (immortalized mouse hippocampal neuronal cell line), HEK293 (immortalized human embryonic kidney 293 cells), and SH-SY5Y (immortalized human neuroblastoma cell line) were cultured at 37°C in a humidified atmosphere containing 5% CO2. All cell lines were routinely tested for mycoplasma throughout this study. HT-22 and HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies, USA), while SH-SY5Y cells were maintained in DMEM plus nutrient mixture F-12 (DMEM/F12) (Gibco). Both media were supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 μg/mL streptomycin, and 1 U/mL penicillin. Co-transfection of the 3′ UTR plasmids and human miRNA mimics was performed employing the DharmaFECT Duo transfection reagent, according to the manufacturer’s instructions. The firefly luciferase reporter assay was performed 48 h after transfection, according to the protocol described by Siebring-van Olst et al. 68 with some variations. Details of the procedures can be found in Supplemental Materials and Methods.
Plasmid Construction and Lentiviral Production
Human APP coding sequence (CDS) was directly amplified from the cDNA of 3xTg-AD mice and cloned into the pIS0 vector (Addgene, USA)69, upstream of the luciferase reporter gene. Colonies with the desired insertion were screened by PCR and further validated by sequencing. Lentiviral plasmids encoding the endogenous precursor sequence of each miRNA (precursor [pre-]miRNAs) and the NC (Invitrogen, USA) were generated by amplifying the precursor sequence and inserting the sequence in the linearized pENTR/pSUPER+ (#575-1, Addgene, USA). The H1-miRNA cassette was then transferred, employing the LR clonase recombination system, into a gateway pLenti CMV GFP destination vector (#736-1, Addgene). Lentiviral particles encoding the precursor sequence of pri-miR-31 were produced in HEK293 cells as previously described.70 Lentiviral particles were resuspended in 1% BSA in sterile PBS. The viral particle content of each batch was determined by quantifying the HIV-1 p24 antigen by ELISA (Retro-Tek kit; Gentaur, France), and batches were kept at −80°C until use.
RNA Extraction and qRT-PCR
Total RNA was extracted from cells/tissue using the miRCURY isolation kit (Exiqon, Denmark), according to the manufacturer’s recommendations. RNA quantification was followed by cDNA conversion and miRNA or mRNA quantification by qRT-PCR performed in the real-time PCR system StepOnePlus (Applied Biosystems, Thermo Fischer Scientific, USA). For miRNA quantification, the miRCURY LNA universal RT miRNA PCR system (Exiqon, Denmark) was employed, in combination with pre-designed LNA primers (Exiqon, Denmark) (Table S1; Supplemental Materials and Methods), and the mouse small nucleolar RNA SNORD 110 was used as a reference. For mRNA quantification, the iQ SYBR Green supermix kit (Bio-Rad, USA) was employed, in combination with pre-designed primers (GeneGlobe, QIAGEN), and the hypoxanthine phosphoribosyltransferase (HPRT) mRNA was used as reference. miRNA and mRNA fold changes were determined by the Pfaffl method. Details of each protocol can be found in Supplemental Materials and Methods.
Stereotaxic Injection into the Hippocampus
Thirteen-month-old mice were anaesthetized with a ketamine/xylazine solution (75 mg/kg ketamine + 10 mg/kg xylazine, intraperitoneally). Lentiviral vectors encoding the miR-31 precursor sequence or a negative control sequence and containing 200 ng of p24 antigen, in a final volume of 2 μL, were stereotaxically injected in both hemispheres of the hippocampus according to the following coordinates: anterior-posterior, −2 mm; medial-lateral, +2 mm and −2 mm; and dorsal-ventral, −2 mm. The viral suspensions were injected at 0.2 mL/min by means of an automatic injector (Stoelting, Wood Dale, IL, USA), and the needle was left in place for an additional 5 min to prevent lentiviral reflux.
Behavior Assays
Animal behavior was assessed before and after lentiviral injection at 12 months and at 16 months of age, respectively. For each test, animals were acclimatized with the test room at least 60 min before the beginning of the first trial. All tests were performed at the same period of the day (during the morning until early afternoon), and all behavioral trials were recorded using a Basler acA1300-60 gc GigE camera (Basler, Germany) and analyzed employing the EthoVision XT v.11.0 software (Noldus, USA). The T-maze test was employed to analyze the working memory, while the NOR was performed to evaluate short-term memory and the BM test was employed to analyze long-term memory. Details of each test can be found in Supplemental Materials and Methods.
Immunohistochemistry and Unbiased Stereology Analysis
Aβ, IBA-1, and GFAP immunolabeling was performed on 50-μm free-floating coronal brain slices by colorimetric immunohistochemistry, employing the Vectastain ABC system and the diaminobenzidine (DAB) substrate (Pierce, Fisher Scientific, USA). For stereological analysis of extracellular and intracellular deposition of Aβ, 12 randomly chosen and equidistant brain slides of each animal, spanning the anteroposterior extent of the hippocampus, were analyzed using Stereo Investigator software and employing the area fraction fractionator tool in a Zeiss Axio Imager Z2 microscope with a motorized XYZ stage. Analysis was performed as previously described,71 with some modifications. For specific details, see Supplemental Materials and Methods.
Immunoblotting Analysis
Total protein extracts were obtained from the hippocampus of 3xTg-AD and wild-type mice from all experimental groups. Western blot analysis of human APP levels was performed as described in Supplemental Materials and Methods. Protein levels were normalized with actin levels and analyzed employing ImageJ software.
Quantitative Analysis of Synaptic Transporters
Labeling of vesicular transporter proteins VGLUT1 and VGAT for puncta analysis by confocal microscopy was performed by fluorescence immunohistochemistry. Three-plane confocal images of areas in the CA1, CA3, and dentate gyrus (0.053 × 0.053 × 0.67 μm3 voxel size) with homogeneous punctuated VGLUT1 and VGAT signals were acquired in the vicinity of GFP-positive neurons in nine slices, spread over the anterior-posterior extent of the hippocampus and separated by 300 μm. Three images were sampled in each area of each slice. Mean integrated density of VGLUT1 and VGAT signals was determined at all possible thresholds and divided by total image area using an in-house macro for FiJi software. Puncta size, number, and intensity were also determined by a previously implemented image analysis protocol.72 Equivalent sections were evaluated for each animal and the analysis parameters were kept constant for all experimental groups. Imaging, measurements, and quantifications were performed blindly to the experimental group. Details of the procedures can be found in Supplemental Materials and Methods.
Statistical Analysis
Statistical analysis was performed with an unpaired Student’s t test and one-way or two-way analysis of variance followed by the adequate post hoc test, for multiple comparisons. Significant outlier (α = 0.05) calculation was performed using Grubb’s test. All data are presented as mean ± SEM. Statistical differences are presented as probability levels of p < 0.05 (∗ or #), p < 0.01 (∗∗ or ##), p < 0.001 (∗∗∗ or ###), and p < 0.0001 (∗∗∗∗ or ####). All tests were two-tailed, and data analysis was performed employing the GraphPad Prism 6 software (GraphPad, USA).
Author Contributions
A.T.B.-V., V.C., J.G., P.C., and A.L.C. performed the experiments. E.F. performed the quantification and statistical analysis of VGLUT1 and VGAT puncta. A.T.B.-V., J.G., L.P.d.A., J.P., and A.L.C. contributed to the design of the experiments. A.T.B.-V., J.G., C.R.d.O., J.P.d.M., J.P., and A.L.C. contributed for the discussion of the results. A.T.B.-V. and A.L.C. wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank Dr. Federico Herrera (Instituto de Tecnologia Química e Biológica António Xavier) for providing the HT-22 cell line, Dr. Ana Luísa Carvalho (Center for Neuroscience and Cell Biology) for supplying some of the antibodies used for VGLUT1 and VGAT quantification, and Dr. Pedro Martins for designing the miR-31 targets images. This work was financed by the European Regional Development Fund (ERDF), through the Centro 2020 Regional Operational Programme under the project CENTRO-01-0145-FEDER-000008: BrainHealth 2020 and through the COMPETE 2020 - Operational Programme for Competitiveness and Internationalisation and Portuguese national funds via FCT - Fundação para a Ciência e a Tecnologia, under the project POCI-01-0145-FEDER-007440 (reference UID/NEU/04539/2013). This work was also supported by the FCT Investigator Programme (IF/00694/2013 to J.P.), the Marie Curie Carrier Integration Grant (PCIG13-GA-2013-618525 to J.P.), HEALTHYAGING 2020 (CENTRO-01-0145-FEDER-000012 to A.T.B.-V.), and Bial Foundation Grant 264/16. A.T.B.-V., J.G., and A.L.C. are recipients of fellowships from the FCT (Grants PTDC/BIM-MEC/0651/2012, SFRH/BPD/120611/2016, and SFRH/BPD/108312/2015). The data, computer scripts and biological materials included in this study are available from the corresponding author upon reasonable request.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.010.
Supplemental Information
References
- 1.Goldman J.S., Hahn S.E., Catania J.W., LaRusse-Eckert S., Butson M.B., Rumbaugh M., Strecker M.N., Roberts J.S., Burke W., Mayeux R., Bird T., American College of Medical Genetics and the National Society of Genetic Counselors Genetic counseling and testing for Alzheimer disease: joint practice guidelines of the American College of Medical Genetics and the National Society of Genetic Counselors. Genet. Med. 2011;13:597–605. doi: 10.1097/GIM.0b013e31821d69b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy J.A., Higgins G.A. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 3.Karran E., Mercken M., De Strooper B. The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 2011;10:698–712. doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- 4.Stancu I.C., Vasconcelos B., Terwel D., Dewachter I. Models of β-amyloid induced Tau-pathology: the long and “folded” road to understand the mechanism. Mol. Neurodegener. 2014;9:51. doi: 10.1186/1750-1326-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim W.S., Kågedal K., Halliday G.M. Alpha-synuclein biology in Lewy body diseases. Alzheimers Res. Ther. 2014;6:73. doi: 10.1186/s13195-014-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohm C., Chen F., Sevalle J., Qamar S., Dodd R., Li Y., Schmitt-Ulms G., Fraser P.E., St George-Hyslop P.H. Current and future implications of basic and translational research on amyloid-β peptide production and removal pathways. Mol. Cell. Neurosci. 2015;66(Pt A):3–11. doi: 10.1016/j.mcn.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zádori D., Veres G., Szalárdy L., Klivényi P., Vécsei L. Alzheimer’s disease: recent concepts on the relation of mitochondrial disturbances, excitotoxicity, neuroinflammation, and kynurenines. J. Alzheimers Dis. 2018;62:523–547. doi: 10.3233/JAD-170929. [DOI] [PubMed] [Google Scholar]
- 9.Peterson S.M., Thompson J.A., Ufkin M.L., Sathyanarayana P., Liaw L., Congdon C.B. Common features of microRNA target prediction tools. Front. Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prince M., Bryce R., Ferri C. 2011. World Alzheimer Report 2011: The benefits of early diagnosis and intervention (Alzheimer’s Disease International)https://www.alz.co.uk/research/WorldAlzheimerReport2011.pdf [Google Scholar]
- 11.Hébert S.S., De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Reddy P.H., Williams J., Smith F., Bhatti J.S., Kumar S., Vijayan M., Kandimalla R., Kuruva C.S., Wang R., Manczak M. MicroRNAs, aging, cellular senescence, and Alzheimer’s disease. Prog. Mol. Biol. Transl. Sci. 2017. 2017:127–171. doi: 10.1016/bs.pmbts.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Hébert S.S., Horré K., Nicolaï L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc. Natl. Acad. Sci. USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X., Muniappan L., Tang G., Ozcan S. Identification of glucose-regulated miRNAs from pancreatic β cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geekiyanage H., Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer’s disease. J. Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunez-Iglesias J., Liu C.C., Morgan T.E., Finch C.E., Zhou X.J. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS ONE. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W.X., Rajeev B.W., Stromberg A.J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P.T. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hébert S.S., Horré K., Nicolaï L., Bergmans B., Papadopoulou A.S., Delacourte A., De Strooper B. MicroRNA regulation of Alzheimer’s amyloid precursor protein expression. Neurobiol. Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Lau P., Bossers K., Janky R., Salta E., Frigerio C.S., Barbash S., Rothman R., Sierksma A.S., Thathiah A., Greenberg D. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idda M.L., Munk R., Abdelmohsen K., Gorospe M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA. 2018;9:e1463. doi: 10.1002/wrna.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boissonneault V., Plante I., Rivest S., Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oddo S., Caccamo A., Shepherd J.D., Murphy M.P., Golde T.E., Kayed R., Metherate R., Mattson M.P., Akbari Y., LaFerla F.M. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Aβ and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 23.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 27.Guedes J.R., Custódia C.M., Silva R.J., de Almeida L.P., Pedroso de Lima M.C., Cardoso A.L. Early miR-155 upregulation contributes to neuroinflammation in Alzheimer’s disease triple transgenic mouse model. Hum. Mol. Genet. 2014;23:6286–6301. doi: 10.1093/hmg/ddu348. [DOI] [PubMed] [Google Scholar]
- 28.Antunes M., Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn. Process. 2012;13:93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez-Bermudez J. Alzheimer’s disease: critical notes on the history of a medical concept. Arch. Med. Res. 2012;43:595–599. doi: 10.1016/j.arcmed.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cogswell J.P., Ward J., Taylor I.A., Waters M., Shi Y., Cannon B., Kelnar K., Kemppainen J., Brown D., Chen C. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 32.Wang W.X., Huang Q., Hu Y., Stromberg A.J., Nelson P.T. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong H., Li J., Huang L., Chen X., Li D., Wang T., Hu C., Xu J., Zhang C., Zen K. Serum microRNA profiles serve as novel biomarkers for the diagnosis of Alzheimer’s disease. Dis. Markers. 2015;2015:625659. doi: 10.1155/2015/625659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr., Jungkamp A.C., Munschauer M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnall-Levin M., Zhao Y., Perrimon N., Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3'UTRs. Proc. Natl. Acad. Sci. USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hausser J., Syed A.P., Bilen B., Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrade C. A peculiar form of peripheral neuropathy; familiar atypical generalized amyloidosis with special involvement of the peripheral nerves. Brain. 1952;75:408–427. doi: 10.1093/brain/75.3.408. [DOI] [PubMed] [Google Scholar]
- 39.Obici L., Kuks J.B., Buades J., Adams D., Suhr O.B., Coelho T., Kyriakides T., European Network for TTR-FAP (ATTReuNET) Recommendations for presymptomatic genetic testing and management of individuals at risk for hereditary transthyretin amyloidosis. Curr. Opin. Neurol. 2016;29(Suppl 1):S27–S35. doi: 10.1097/WCO.0000000000000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ledford H. Gene-silencing technology gets first drug approval after 20-year wait. Nature. 2018;560:291–292. doi: 10.1038/d41586-018-05867-7. [DOI] [PubMed] [Google Scholar]
- 41.Lindberg O., Walterfang M., Looi J.C., Malykhin N., Ostberg P., Zandbelt B., Styner M., Paniagua B., Velakoulis D., Orndahl E., Wahlund L.O. Hippocampal shape analysis in Alzheimer’s disease and frontotemporal lobar degeneration subtypes. J. Alzheimers Dis. 2012;30:355–365. doi: 10.3233/JAD-2012-112210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheff S.W., Price D.A. Synaptic pathology in Alzheimer’s disease: a review of ultrastructural studies. Neurobiol. Aging. 2003;24:1029–1046. doi: 10.1016/j.neurobiolaging.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Scheff S.W., Price D.A. Alzheimer’s disease-related alterations in synaptic density: neocortex and hippocampus. J. Alzheimers Dis. 2006;9(3, Suppl):101–115. doi: 10.3233/jad-2006-9s312. [DOI] [PubMed] [Google Scholar]
- 44.Alzheimer’s A., Alzheimer’s Association 2010 Alzheimer’s disease facts and figures. Alzheimers Dement. 2010;6:158–194. doi: 10.1016/j.jalz.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Prince M., Comas-Herrera A., Knapp M., Guerchet M., Karagiannidou M. World Alzheimer Report 2016: Improving healthcare for people living with dementia. Coverage, quality and costs now and in the future (Alzheimer’s Disease International) 2016. https://www.alz.co.uk/research/WorldAlzheimerReport2016.pdf
- 46.Peñagarikano O., Abrahams B.S., Herman E.I., Winden K.D., Gdalyahu A., Dong H., Sonnenblick L.I., Gruver R., Almajano J., Bragin A. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deacon R.M., Rawlins J.N. T-maze alternation in the rodent. Nat. Protoc. 2006;1:7–12. doi: 10.1038/nprot.2006.2. [DOI] [PubMed] [Google Scholar]
- 48.Tu S., Wong S., Hodges J.R., Irish M., Piguet O., Hornberger M. Lost in spatial translation—a novel tool to objectively assess spatial disorientation in Alzheimer’s disease and frontotemporal dementia. Cortex. 2015;67:83–94. doi: 10.1016/j.cortex.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Hu X., Das B., Hou H., He W., Yan R. BACE1 deletion in the adult mouse reverses preformed amyloid deposition and improves cognitive functions. J. Exp. Med. 2018;215:927–940. doi: 10.1084/jem.20171831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riegert C., Galani R., Heilig S., Lazarus C., Cosquer B., Cassel J.C. Electrolytic lesions of the ventral subiculum weakly alter spatial memory but potentiate amphetamine-induced locomotion. Behav. Brain Res. 2004;152:23–34. doi: 10.1016/j.bbr.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 51.O’Mara S.M., Commins S., Anderson M., Gigg J. The subiculum: a review of form, physiology and function. Prog. Neurobiol. 2001;64:129–155. doi: 10.1016/s0301-0082(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 52.O’Mara S. The subiculum: what it does, what it might do, and what neuroanatomy has yet to tell us. J. Anat. 2005;207:271–282. doi: 10.1111/j.1469-7580.2005.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herman J.P., Cullinan W.E., Morano M.I., Akil H., Watson S.J. Contribution of the ventral subiculum to inhibitory regulation of the hypothalamo-pituitary-adrenocortical axis. J. Neuroendocrinol. 1995;7:475–482. doi: 10.1111/j.1365-2826.1995.tb00784.x. [DOI] [PubMed] [Google Scholar]
- 54.Herman J.P., McKlveen J.M., Ghosal S., Kopp B., Wulsin A., Makinson R., Scheimann J., Myers B. Regulation of the hypothalamic-pituitary-adrenocortical stress response. Compr. Physiol. 2016;6:603–621. doi: 10.1002/cphy.c150015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Csernansky J.G., Dong H., Fagan A.M., Wang L., Xiong C., Holtzman D.M., Morris J.C. Plasma cortisol and progression of dementia in subjects with Alzheimer-type dementia. Am. J. Psychiatry. 2006;163:2164–2169. doi: 10.1176/appi.ajp.163.12.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Souza-Talarico J.N., Caramelli P., Nitrini R., Chaves E.C. Effect of cortisol levels on working memory performance in elderly subjects with Alzheimer’s disease. Arq. Neuropsiquiatr. 2008;66(3B):619–624. doi: 10.1590/s0004-282x2008000500003. [DOI] [PubMed] [Google Scholar]
- 57.Peskind E.R., Wilkinson C.W., Petrie E.C., Schellenberg G.D., Raskind M.A. Increased CSF cortisol in AD is a function of APOE genotype. Neurology. 2001;56:1094–1098. doi: 10.1212/wnl.56.8.1094. [DOI] [PubMed] [Google Scholar]
- 58.Weiner M.F., Vobach S., Olsson K., Svetlik D., Risser R.C. Cortisol secretion and Alzheimer’s disease progression. Biol. Psychiatry. 1997;42:1030–1038. doi: 10.1016/s0006-3223(97)00165-0. [DOI] [PubMed] [Google Scholar]
- 59.Grace A.A. Dopamine regulation of decision making processes. In: Perry E., Collertone D., LeBeau F., Ashton H., editors. New Horizons in the Neuroscience of Consciousness. John Benjamins Publishing Company; 2010. [Google Scholar]
- 60.Kashani A., Lepicard E., Poirel O., Videau C., David J.P., Fallet-Bianco C., Simon A., Delacourte A., Giros B., Epelbaum J. Loss of VGLUT1 and VGLUT2 in the prefrontal cortex is correlated with cognitive decline in Alzheimer disease. Neurobiol. Aging. 2008;29:1619–1630. doi: 10.1016/j.neurobiolaging.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 61.Minkeviciene R., Ihalainen J., Malm T., Matilainen O., Keksa-Goldsteine V., Goldsteins G., Iivonen H., Leguit N., Glennon J., Koistinaho J. Age-related decrease in stimulated glutamate release and vesicular glutamate transporters in APP/PS1 transgenic and wild-type mice. J. Neurochem. 2008;105:584–594. doi: 10.1111/j.1471-4159.2007.05147.x. [DOI] [PubMed] [Google Scholar]
- 62.Timmer N.M., Metaxas A., van der Stelt I., Kluijtmans L.A., van Berckel B.N., Verbeek M.M. Cerebral level of vGlut1 is increased and level of glycine is decreased in TgSwDI mice. J. Alzheimers Dis. 2014;39:89–101. doi: 10.3233/JAD-130437. [DOI] [PubMed] [Google Scholar]
- 63.Schlüter O.M., Khvotchev M., Jahn R., Südhof T.C. Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J. Biol. Chem. 2002;277:40919–40929. doi: 10.1074/jbc.M203704200. [DOI] [PubMed] [Google Scholar]
- 64.Veyrac A., Reibel S., Sacquet J., Mutin M., Camdessanche J.P., Kolattukudy P., Honnorat J., Jourdan F. CRMP5 regulates generation and survival of newborn neurons in olfactory and hippocampal neurogenic areas of the adult mouse brain. PLoS ONE. 2011;6:e23721. doi: 10.1371/journal.pone.0023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamashita N., Mosinger B., Roy A., Miyazaki M., Ugajin K., Nakamura F., Sasaki Y., Yamaguchi K., Kolattukudy P., Goshima Y. CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells. J. Neurosci. 2011;31:1773–1779. doi: 10.1523/JNEUROSCI.5337-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siuda J., Patalong-Ogiewa M., Żmuda W., Targosz-Gajniak M., Niewiadomska E., Matuszek I., Jędrzejowska-Szypułka H., Rudzińska-Bar M. Corrigendum to “Cognitive impairment and BDNF serum levels” [Polish J. Neurol. Neurosurg. 51 (2017) 24–32] Neurol. Neurochir. Pol. 2017;51:537. doi: 10.1016/j.pjnns.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Guedes J.R., Viegas A.T., Pedroso de Lima M.C., Cardoso A.L. Microglia: a double-edged sword for Alzheimer’s disease. In: Giffard E.R., editor. Microglia: Physiology, Regulation and Health Implications. Nova Science Publishers; 2015. [Google Scholar]
- 68.Siebring-van Olst E., Vermeulen C., de Menezes R.X., Howell M., Smit E.F., van Beusechem V.W. Affordable luciferase reporter assay for cell-based high-throughput screening. J. Biomol. Screen. 2013;18:453–461. doi: 10.1177/1087057112465184. [DOI] [PubMed] [Google Scholar]
- 69.Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 70.de Almeida L.P., Zala D., Aebischer P., Déglon N. Neuroprotective effect of a CNTF-expressing lentiviral vector in the quinolinic acid rat model of Huntington’s disease. Neurobiol. Dis. 2001;8:433–446. doi: 10.1006/nbdi.2001.0388. [DOI] [PubMed] [Google Scholar]
- 71.Heggland I., Storkaas I.S., Soligard H.T., Kobro-Flatmoen A., Witter M.P. Stereological estimation of neuron number and plaque load in the hippocampal region of a transgenic rat model of Alzheimer’s disease. Eur. J. Neurosci. 2015;41:1245–1262. doi: 10.1111/ejn.12876. [DOI] [PubMed] [Google Scholar]
- 72.Valero J., Mastrella G., Neiva I., Sánchez S., Malva J.O. Long-term effects of an acute and systemic administration of LPS on adult neurogenesis and spatial memory. Front. Neurosci. 2014;8:83. doi: 10.3389/fnins.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.