Abstract
BACKGROUND
Early screening for colorectal cancer (CRC) is important in clinical practice. However, the currently methods are inadequate because of high cost and low diagnostic value.
AIM
To develop a new examination method based on the serum biomarker panel for the early detection of CRC.
METHODS
Three hundred and fifty cases of CRC, 300 cases of colorectal polyps and 360 cases of normal controls. Combined with the results of area under curve (AUC) and correlation analysis, the binary Logistic regression analysis of the remaining indexes which is in accordance with the requirements was carried out, and discriminant analysis, classification tree and artificial neural network analysis were used to analyze the remaining indexes at the same time.
RESULTS
By comparison of these methods, we obtained the ability to distinguish CRC from healthy control group, malignant disease group and benign disease group. Artificial neural network had the best diagnostic value when compared with binary logistic regression, discriminant analysis, and classification tree. The AUC of CRC and the control group was 0.992 (0.987, 0.997), sensitivity and specificity were 98.9% and 95.6%. The AUC of the malignant disease group and benign group was 0.996 (0.992, 0.999), sensitivity and specificity were 97.4% and 96.7%.
CONCLUSION
Artificial neural network diagnosis method can improve the sensitivity and specificity of the diagnosis of CRC, and a novel assistant diagnostic method was built for the early detection of CRC.
Keywords: Colorectal cancer, Serum, Index, Biomarker, Artificial neural network
Core tip: We aimed to combine the serum index together by several multiparameter method, such as, the binary logistic regression, discriminant analysis, classification tree and artificial neural network analysis. Finally, a multiparameter diagnostic model based on artificial neural network which showed better diagnostic value was built for the early detection of colorectal cancer.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer[1] and the fourth major cause of cancer-related deaths worldwide. CRC has a high incidence and high mortality rate and is a public health burden in most industrialized countries. In recent years, the incidence of CRC in Asia is rising rapidly[2]. In eastern Asia, the incidence of countries, such as China, Japan, South Korea and Singapore, has increased two to four times in recent decades. Among Asian ethnic groups, the incidence of CRC in China is significantly higher than that of other ethnic groups. According to the 2003 China Cancer Database, CRC is one of the three fastest growing morbidity cancers[1].
CRC is a common malignant tumor in the gastrointestinal tract. Early symptoms are not obvious. As cancer increases, it shows changes in bowel habits, blood in the stool, diarrhea, alternating diarrhea or constipation, local abdominal pain and other symptoms. In the advanced stage, CRC shows anemia and body weight loss or other systemic symptoms. A typical CRC is developed from a focal change in benign, precancerous polyps. These polyps are local growths or the accumulation of abnormal cells in the intestinal mucosa that protrude into the intestinal lumen[3]. With time, the dividing cells in these polyps may accumulate enough genetic changes to gain the ability to invade the intestinal wall, which is a hallmark of CRC that may eventually become more susceptible to change and spread to regional lymph nodes, eventually spread to distant transfer sites[4]. This multistep development process accumulates over time and allows early precancerous polyps to be screened and tested before the average risk of CRC is cancerous, which may lead to a dramatic decline in the incidence of CRC[5].
Clinical screening for CRC involves (1) Colonoscopy. Many studies have confirmed that colonoscopy, as a useful screening tool, can reduce the incidence of CRC by 76% and mortality by 65%[6]; (2) Sigmoidoscopy. Compared with colonoscopy, sigmoidoscopy has the advantages of low cost, short preparation time and no need for sedation[7]; (3) Computed tomography colon imaging; (4) FOBT and fecal immunochemical tests. These tests have advantages of low cost, noninvasiveness and good tolerance. FOBT and fecal immunochemical tests are widely used on a global scale, but are also susceptible to food, drugs and other factors, and the stool collection is inconvenient, resulting in a high false positive rate[8]; and (5) Screening for biomarkers: (a) Carcinoembryonic antigen (CEA); (b) Circulating tumor cell; (c) Circulating tumor DNA/RNA; and (d) Abnormal DNA methylation[9].
Early screening for CRC plays an important role in combating and controlling the growth of CRC morbidity and mortality worldwide[10]. However, the currently available screening methods are severely inadequate because of their high cost and cumbersome preparation procedures that ultimately result in a low participation rate. People are often reluctant to use colonoscopy[11]. Therefore, developing an unconventional method of testing based on blood biomarkers as the first test procedure may be the ideal method. This method will make it possible to identify high-incidence individuals among the general public. Colonoscopy will become the second type of examination and continue to screen high-incidence populations. This strategy will encourage participation rates and will help achieve the goal of early screening for CRC and will reduce the globally expected increase in CRC incidence[12]. Blood-based screening experiments attract the public because of their noninvasive and low patient harm features. This screening is easy to perform and can be repeated in shorter time intervals, which in turn leads to higher participation rates[13].
In this study, we conducted a retrospective analysis. Through t test, ROC curve analysis, binary logistic regression analysis, and simultaneous use of discriminant analysis, classification tree and artificial neural network analysis of multiple methods combined detection were used to determine the tumor marker diagnostic value for detection of CRC.
MATERIALS AND METHODS
Patients selection
The serum samples of the patients involved in this study were from blood samples of patients and confirmed by imaging and pathology. According to the blood collection record, all serum biochemical and immunological indexes of the CRC disease group and the healthy control group were counted from the medical records of each subject for the subsequent statistical analysis. As shown in Table 1, these analyses included 300 cases of colorectal benign polyps and 350 cases of malignant colorectal cancer (166 cases in early stage and 136 in late stage, 48 cases unconfirmed). All patients had clear imaging and pathological diagnosis and did not receive radiotherapy, chemotherapy or other immunotherapy before surgery. A total of 360 patients in the healthy control group received physical examination, were examined by tumor markers and imaging examinations, had no diseases related to the study, and the tumor markers and imaging examinations were all qualified. The serum index in the hospital was collected and used to analyze.
Table 1.
The clinical characteristics of patients with colorectal cancer, n (%)
| Malignant (n = 350) | Benign (n = 300) | Controls (n = 360) | |
| Age, yr | |||
| < 40 | 10 (2.86) | 16 (5.33) | 63 (17.5) |
| 40-60 | 149 (42.57) | 143 (47.67) | 258 (71.67) |
| ≥ 60 | 191 (54.57) | 141 (47) | 39 (10.83) |
| Gender | |||
| Male | 217 (62) | 200 (66.67) | 212 (58.89) |
| Female | 133 (38) | 100 (33.33) | 148 (41.11) |
| T | |||
| T1-2 | 67 (19.14) | ||
| T3-4 | 235 (67.14) | ||
| Lymph node | |||
| Yes | 165 (47.14) | ||
| No | 119 (34) | ||
| Metastasis | |||
| Yes | 30 (8.57) | ||
| No | 320 (91.43) | ||
| 0 | 2 (0.58) | ||
| TNM Stage | |||
| I | 56 (16) | ||
| II | 108 (30.86) | ||
| III | 106 (30.29) | ||
| IV | 30 (8.57) | ||
| Non | 48 (13.7) | ||
Statistical analysis
Statistical analysis was performed on comparing colorectal cancer and healthy controls, malignant disease group and benign disease group. Data from various indexes of colorectal cancer and healthy control groups as well as malignant disease groups and benign disease groups were compared by t test. The diagnostic value was evaluated by the area under the curve of the ROC curve, and the cutoff value was determined by the Youden index. The combination of indexes is analyzed by statistical methods, such as binary logistic regression analysis, discriminant analysis, classification tree and artificial neural network. All data were statistically analyzed by SPSS (version 20.0, SPSS Inc. Chicago, IL) software. All statistical tests were bilateral, and P < 0.01 was considered statistically significant.
RESULTS
Significant analysis and ROC curve analysis of CRC and healthy control group, malignant disease group and benign disease group
There were significant differences in 32 indexes between CRC and healthy controls. There were significant differences in 37 indexes between the malignant disease group and the benign disease group. The ROC curves were performed on 36 indexes with significant differences in colorectal cancer and healthy controls and 42 indexes with significant differences between the malignant disease group and the benign disease group. Among these indexes, 32 indexes of colorectal cancer and healthy control group had P values < 0.01. There were 37 indexes in the malignant disease group and the benign disease group with P values < 0.01.
Among these results, the largest area under the curve in CRC and healthy control group were for RDV and CEA, with area under the curve values of 0.781 and 0.846, respectively. When the RDV cutoff value was 12.625, the sensitivity and specificity were 61.7%, 82.2%; when the CEA cutoff value was 1.915, the sensitivity and specificity were 81.4%, 71.7%, respectively. The largest area under the curve in the malignant disease group and the benign disease group were for CRP and H-FABP, with area under the curve values of 0.798 and 0.762, respectively. When the CRP cutoff value was 0.145, the sensitivity and specificity were 62.0%, 89.7%; when the H-FABP cutoff value was 1.965, the sensitivity and specificity were 73.7%, 88.3%, respectively.
Binary logistic analysis of CRC and healthy control group, malignant disease group and benign disease group
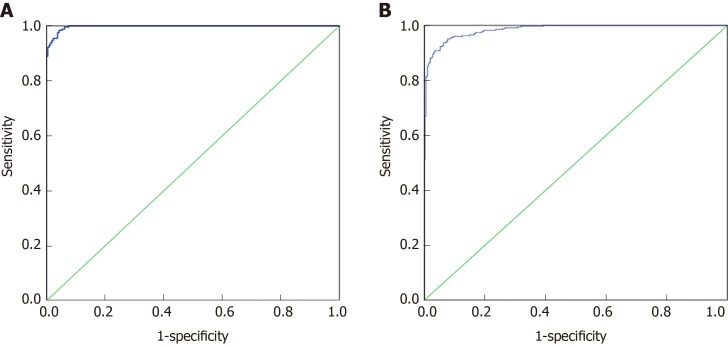
Seventy percent of the data from 32 indexes in CRC and healthy controls and 37 indexes in the malignant disease group and benign disease group were used for the establishment of a binary logistic regression analysis model. As shown in the Figure 1A, the area under the curve for the CRC and the healthy control group was 0.989 (0.982, 0.995). When the cutoff value was 0.479, its sensitivity and specificity were 90.2% and 90.1%, respectively. As shown in the Figure 1B, the area under the curve of the malignant disease group and the benign disease group was 0.929 (0.901, 0.958), and when the cutoff value was 0.329, its sensitivity and specificity were 98.4% and 95.7%, respectively. Binary logistic regression analysis was more effective in distinguishing colorectal cancer from healthy controls than in malignant and benign disease groups.
Figure 1.
Binary Logistic analysis of colorectal cancer and healthy control group, malignant disease group and benign disease group. A: ROC curve of the binary logistic regression analysis model of the colorectal cancer and healthy control group, respectively; B: ROC curve of the binary logistic regression analysis model of the colorectal cancer and healthy control group.
Discriminant analysis of CRC and healthy control group, malignant disease group and benign disease group
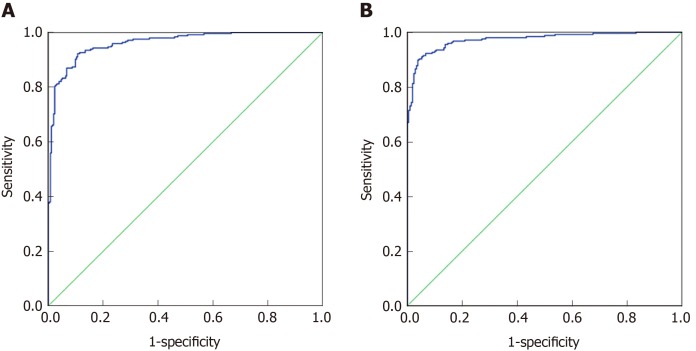
Seventy percent of the data from 32 indexes in CRC and healthy controls and 37 indexes in malignant disease groups and benign disease groups were used to establish a discriminant analysis model. As shown in Figure 2A, the area under the curve for CRC and healthy controls was 0.961 (0.946, 0.977), and when the cutoff value was 0.33, its sensitivity and specificity were 92.2% and 89.3%, respectively. As shown in Figure 2B, the area under the curve for the malignant disease group and the benign disease group was 0.973 (0.960, 0.986), and when the cutoff value was 0.467, its sensitivity and specificity were 91% and 94.8%, respectively. Discriminant analysis differentiated between the malignant disease group and the benign disease group better than the CRC and healthy control group.
Figure 2.
Discriminant analysis results of colorectal cancer and healthy control group, malignant disease group and benign disease group. A: ROC curve of the discriminant analysis model of the colorectal cancer and healthy control group, respectively; B: ROC curve of the discriminant analysis model of the colorectal cancer and healthy control group.
Classification tree analysis of CRC and healthy control group, malignant disease group and benign disease group
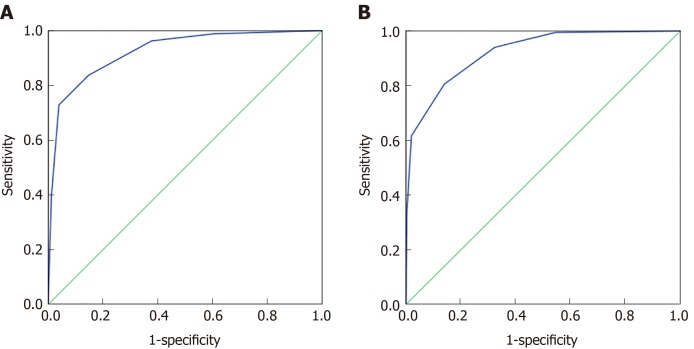
The 70% data for 32 indexes in the CRC and healthy control groups and the 37 indexes in the malignant disease group and the benign disease group were established by the classification tree model, and 30% of the data were used for model validation. As shown in Figure 3A, the prediction accuracy rate of the healthy control group was 85.5%, the prediction accuracy rate of the CRC group was 77.1%, and the overall prediction accuracy rate was 81.3%. The area under the curve of CRC and the healthy control group was 0.924 (0.905, 0.944), and when the cutoff value was 0.4324, its sensitivity and specificity were 83.7% and 85.3%, respectively. As shown in Figure 3B, the predictive accuracy rate was 82.8% in the benign disease group, 75.2% in the malignant disease group, and 78.8% in the overall prediction rate. The area under the curve of the malignant disease group and benign disease group was 0.922 (0.903, 0.941), and when the cutoff value was 0.564, its sensitivity and specificity were 80.6% and 86%, respectively. The classification tree analysis distinguished the CRC from the healthy control group basically in the same way as the malignant disease group and the benign disease group.
Figure 3.
Classification tree analysis results of colorectal cancer and healthy control group, malignant disease group and benign disease group. A: ROC curve of the classification tree analysis model of the colorectal cancer and healthy control group, respectively; B: ROC curve of the classification tree analysis model of the colorectal cancer and healthy control group.
Artificial neural network analysis of CRC and healthy control group, malignant disease group and benign disease group
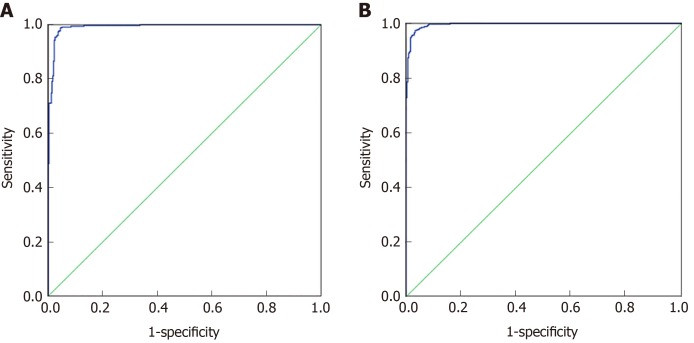
Seventy percent of the data from 27 indicators in CRC and healthy controls and 30 indicators in the malignant disease group and benign disease group were used for artificial neural network model establishment, and 30% of the data were used for model validation. As shown in Figure 4A, the prediction accuracy rate of the healthy control group was 88.6%, the prediction accuracy rate of the CRC group was 84.8%, and the overall prediction accuracy rate was 87.8%. The area under the curve of CRC and the healthy control group was 0.992 (0.987, 0.997), and when the cutoff value was 0.065, its sensitivity and specificity were 98.9% and 95.6%, respectively. As shown in Figure 4B, the predictive accuracy rate was 90.0% in the benign disease group, 88.9% in the malignant disease group, and 89.4% in the overall prediction. The area under the curve of the malignant disease group and benign disease group was 0.996 (0.992, 0.999), and when the cutoff value was 0.443, its sensitivity and specificity were 97.4% and 96.7%, respectively. The effect of artificial neural network analysis on CRC and the healthy control group was basically the same as that of the malignant disease group and the benign disease group, but the prediction accuracy rate was higher than that of the classification tree method.
Figure 4.
Artificial neural network analysis results of colorectal cancer and healthy control group, malignant disease group and benign disease group. A: ROC curve of the artificial neural network analysis model of the colorectal cancer and healthy control group, respectively; B: ROC curve of the artificial neural network analysis model of the colorectal cancer and healthy control group.
Through comparison of these four methods, we obtained the ability to distinguish colorectal cancer from healthy control group, malignant disease group and benign disease group: Artificial neural network > binary logistic regression > discriminant analysis > classification tree.
DISCUSSION
Colorectal cancer is the second most common cancer disease in women and the third most common cancer in men. The number of new cases worldwide was estimated as 1.2 million in 2008, and the deaths were approximately 600000[14].
Tumor markers can be present in cells, tissues, blood, and feces and can thus be qualitatively or quantitatively detected by related techniques[15]. With the development of molecular diagnosis, lots of novel detection method have been developed[16-20]. Tumor markers can be an important tool for cancer detection and patient prognosis. Gene mutations are the main factor in the development of CRC, and many discoveries have been made in recent years. APC, VEGF, Septin9 and other DNA in feces, blood and other biological fluids can be used as the primary detection and prognostic indicator[21-25]. In addition to genetic alterations such as mutations, microsatellite instability[26,27] and hypermethylation of tumor suppressor genes in promoter regions have also been extensively studied[28,29]. MicroRNAs and their putative target gene dysregulation may affect the development of colorectal cancer[30]. Protein markers, such as IMP3[31,32] and COX-2[33], have attracted much attention in CRC screening, and their concentrations may be related to CRC[34]. There are very few tumor markers that simultaneously satisfy high sensitivity and specificity, mainly because tumor markers are difficult to distinguish between benign diseases and malignant diseases when the levels are elevated[35].
A large number of studies have found that the clinical significance of detecting the increase[15] in the level of a single tumor marker is very limited. Therefore, people have improved the diagnostic value of tumor markers by two methods: continuous detection and joint detection. Continuous testing is used in the detection of malignant tumors, but it is mainly used for the detection of therapeutic effects and early diagnosis of prognosis. Joint detection is a very promising as an early detection method for malignant tumors by detecting multiple indicators to improve the sensitivity and specificity of tumor marker diagnosis, such as binary logistic regression analysis, discriminant analysis, classification tree analysis, and artificial neural network, which have improved the shortcomings of tumor markers that are difficult to simultaneously meet the sensitivity and specificity. Several indicators are combined, and statistical methods are used to improve the diagnostic value of tumor markers.
In this study, the diagnostic value of the combined diagnostic analysis for distinguishing between healthy controls and disease groups was superior to that of single-index tests; the diagnostic value of combined diagnostic analysis for distinguishing between benign disease groups and malignant disease groups was significantly better than the single-index test. Through comparison of these four methods, we obtained the ability to distinguish colorectal cancer from healthy control group, malignant disease group and benign disease group: Artificial neural network > binary logistic regression > discriminant analysis > classification tree.
However, there are still some limitations in our study. First, the sample size in our study was relatively small, and it may affect the results of our study. Second, although we have built a multiparameter diagnostic model, and it has better diagnostic value when compared with the conventional biomarker, but the diagnostic model has not been validated. Third, the diagnostic value should be performed on multi-center and larger sample size to validate its diagnostic value.
In conclusion, through multiparameter joint diagnostic analysis, we found that the combined diagnosis method can improve the sensitivity and specificity of the diagnosis, but some joint diagnosis methods may not be improved. Therefore, the optimal strategy is determined by comparing various joint diagnosis methods, followed by verification of the sample and confirmation of its value for use.
ARTICLE HIGHLIGHTS
Research background
Early screening for colorectal cancer (CRC) is important in clinical practice. However, the currently methods are inadequate because of high cost and low diagnostic value.
Research motivation
Blood-based screening method attract the public because of their noninvasive, and multiparameter method was demonstrated to increase the diagnostic value.
Research objectives
We aimed to conduct a retrospective analysis. By multiparameter methods combined detection were used to determine the tumor marker diagnostic value for detection of CRC.
Research methods
350 CRC, 300 colorectal polyps and 360 normal controls were enrolled. Combined with the results of area under curve, the binary Logistic regression analysis, and discriminant analysis, classification tree and artificial neural network were used to analyze the diagnostic value.
Research results
For distinguishing CRC from healthy control group, malignant disease group and benign disease group. Artificial neural network had the best diagnostic value when compared with the other methods. The area under the curve of CRC and the control group was 0.992 (0.987, 0.997), sensitivity and specificity were 98.9% and 95.6%. The area under the curve of the malignant disease group and benign group was 0.996 (0.992, 0.999), sensitivity and specificity were 97.4% and 96.7%.
Research conclusions
Artificial neural network diagnosis method can provide a novel assistant diagnostic method was built for the early detection of CRC.
Research perspectives
Although we have built a multiparameter diagnostic model, the sample size was relatively small, and the diagnostic model has not been validated. Multi-center and larger sample size to validate its diagnostic value.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Peking University Cancer Hospital and Institute review board.
Informed consent statement: All study participants or their legal guardian provided written informed consent prior to study enrollment.
Conflict-of-interest statement: The authors have no conflicts of interest to declare.
STROBE statement: The authors have read the STROBE Statement – checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Manuscript source: Unsolicited manuscript
Peer-review started: December 18, 2019
First decision: January 13, 2020
Article in press: February 8, 2020
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Snowdon VK, Takamatsu S, Voigt M S-Editor: Zhang L L-Editor: A E-Editor: Ma YJ
Contributor Information
Wen-Yue Song, School of Life Science and Biopharmaceutics, Shenyang Pharmaceutical University, Shenyang 110016, Liaoning Province, China; Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Interventional Therapy Department, Peking University Cancer Hospital and Institute, Beijing 100142, China.
Xin Zhang, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Interventional Therapy Department, Peking University Cancer Hospital and Institute, Beijing 100142, China.
Qi Zhang, School of Life Science and Biopharmaceutics, Shenyang Pharmaceutical University, Shenyang 110016, Liaoning Province, China; Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Interventional Therapy Department, Peking University Cancer Hospital and Institute, Beijing 100142, China.
Peng-Jun Zhang, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Interventional Therapy Department, Peking University Cancer Hospital and Institute, Beijing 100142, China.
Rong Zhang, School of Life Science and Biopharmaceutics, Shenyang Pharmaceutical University, Shenyang 110016, Liaoning Province, China. zhangrong7110@163.com.
Data sharing statement
No additional data are available.
References
- 1.Pourhoseingholi MA. Increased burden of colorectal cancer in Asia. World J Gastrointest Oncol. 2012;4:68–70. doi: 10.4251/wjgo.v4.i4.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung J. Colorectal cancer screening: It’s time for action in Asia. Cancer Detect Prev. 2007;31:1–2. doi: 10.1016/j.cdp.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Bluegrass Coalition for Colorectal Screening. KMA Cancer Committee, Colorectal cancer facts. J Ky Med Assoc. 2005;103:118. [Google Scholar]
- 4.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–976. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stracci F, Zorzi M, Grazzini G. Colorectal cancer screening: tests, strategies, and perspectives. Front Public Health. 2014;2:210. doi: 10.3389/fpubh.2014.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moody L, He H, Pan YX, Chen H. Methods and novel technology for microRNA quantification in colorectal cancer screening. Clin Epigenetics. 2017;9:119. doi: 10.1186/s13148-017-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ. 2014;348:g2467. doi: 10.1136/bmj.g2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabeneck L, Rumble RB, Thompson F, Mills M, Oleschuk C, Whibley A, Messersmith H, Lewis N. Fecal immunochemical tests compared with guaiac fecal occult blood tests for population-based colorectal cancer screening. Can J Gastroenterol. 2012;26:131–147. doi: 10.1155/2012/486328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hundt S, Haug U, Brenner H. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2007;16:1935–1953. doi: 10.1158/1055-9965.EPI-06-0994. [DOI] [PubMed] [Google Scholar]
- 10.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 11.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S, Johnson D, Johnson CD, Levin TR, Pickhardt PJ, Rex DK, Smith RA, Thorson A, Winawer SJ American Cancer Society Colorectal Cancer Advisory Group; US Multi-Society Task Force; American College of Radiology Colon Cancer Committee. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570–1595. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Kahi CJ, Anderson JC, Rex DK. Screening and surveillance for colorectal cancer: state of the art. Gastrointest Endosc. 2013;77:335–350. doi: 10.1016/j.gie.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Ganepola GA, Nizin J, Rutledge JR, Chang DH. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J Gastrointest Oncol. 2014;6:83–97. doi: 10.4251/wjgo.v6.i4.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 15.Ratto C, Sofo L, Ippoliti M, Merico M, Doglietto GB, Crucitti F. Prognostic factors in colorectal cancer. Diseases of the Colon and Rectum. 1998;41:1033–1049. doi: 10.1007/BF02237397. [DOI] [PubMed] [Google Scholar]
- 16.Gao W, Long L, Tian X, Xu F, Liu J, Singh PK, Botella JR, Song C. Genome Editing in Cotton with the CRISPR/Cas9 System. Front Plant Sci. 2017;8:1364. doi: 10.3389/fpls.2017.01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J, Li K, Jin L, Xu R, Miao K, Yang F, Qi C, Zhang L, Botella JR, Wang R, Miao Y. A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants. Plant Methods. 2018;14:40. doi: 10.1186/s13007-018-0305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei KJ, Lin YM, An GY. miR156 modulates rhizosphere acidification in response to phosphate limitation in Arabidopsis. J Plant Res. 2016;129:275–284. doi: 10.1007/s10265-015-0778-8. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q, Qiao J, Zhang S, He S, Shi Y, Yuan Y, Zhang X, Cai Y. Changes in DNA methylation assessed by genomic bisulfite sequencing suggest a role for DNA methylation in cotton fruiting branch development. PeerJ. 2018;6:e4945. doi: 10.7717/peerj.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Zhang Y, Liu J, Wang L, Liu P, Yin Z, Guo S, Ma J, Lu Z, Wang T, She Y, Miao Y, Ma L, Chen S, Li Y, Dai S. Proteomic discovery of H2O2 response in roots and functional characterization of PutGLP gene from alkaligrass. Planta. 2018;248:1079–1099. doi: 10.1007/s00425-018-2940-8. [DOI] [PubMed] [Google Scholar]
- 21.Chen TH, Chang SW, Huang CC, Wang KL, Yeh KT, Liu CN, Lee H, Lin CC, Cheng YW. The prognostic significance of APC gene mutation and miR-21 expression in advanced-stage colorectal cancer. Colorectal Dis. 2013;15:1367–1374. doi: 10.1111/codi.12318. [DOI] [PubMed] [Google Scholar]
- 22.Iacopetta B, Grieu F, Li W, Ruszkiewicz A, Caruso M, Moore J, Watanabe G, Kawakami K. APC gene methylation is inversely correlated with features of the CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2006;119:2272–2278. doi: 10.1002/ijc.22237. [DOI] [PubMed] [Google Scholar]
- 23.Falchook GS, Kurzrock R. VEGF and dual-EGFR inhibition in colorectal cancer. Cell Cycle. 2015;14:1129–1130. doi: 10.1080/15384101.2015.1022071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulendran M, Stebbing JF, Marks CG, Rockall TA. Predictive and prognostic factors in colorectal cancer: a personalized approach. Cancers (Basel) 2011;3:1622–1638. doi: 10.3390/cancers3021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan S, Liu Z, Yu S, Bao Y. Diagnostic Value of Methylated Septin9 for Colorectal Cancer Screening: A Meta-Analysis. Med Sci Monit. 2016;22:3409–3418. doi: 10.12659/MSM.900590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 27.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 28.Samowitz WS. The CpG island methylator phenotype in colorectal cancer. J Mol Diagn. 2007;9:281–283. doi: 10.2353/jmoldx.2007.070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 30.Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, Fujita S, Yamamoto S, Baba H, Matsumura Y. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila) 2010;3:1435–1442. doi: 10.1158/1940-6207.CAPR-10-0036. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Yan D, Tang H, Zhou C, Fan J, Li S, Wang X, Xia J, Huang F, Qiu G, Peng Z. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16:3499–3506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 32.Lin L, Zhang J, Wang Y, Ju W, Ma Y, Li L, Chen L. Insulin-like growth factor-II mRNA-binding protein 3 predicts a poor prognosis for colorectal adenocarcinoma. Oncol Lett. 2013;6:740–744. doi: 10.3892/ol.2013.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernández-Marcelo T, Sánchez-Pernaute A, Pascua I, De Juan C, Head J, Torres-García AJ, Iniesta P. Clinical Relevance of Telomere Status and Telomerase Activity in Colorectal Cancer. PLoS One. 2016;11:e0149626. doi: 10.1371/journal.pone.0149626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangadhar T, Schilsky RL. Molecular markers to individualize adjuvant therapy for colon cancer. Nat Rev Clin Oncol. 2010;7:318–325. doi: 10.1038/nrclinonc.2010.62. [DOI] [PubMed] [Google Scholar]
- 35.Symonds EL, Young GP. Blood Tests for Colorectal Cancer Screening in the Standard Risk Population. Current Colorectal Cancer Reports. 2015;11:397–407. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.