Abstract
Acanthamoeba-infecting Mimiviridae are giant viruses with dsDNA genome up to 1.5 Mb. They build viral factories in the host cytoplasm in which the nuclear-like virus-encoded functions take place. They are themselves the target of infections by 20-kb-dsDNA virophages, replicating in the giant virus factories and can also be found associated with 7-kb-DNA episomes, dubbed transpovirons. Here we isolated a virophage (Zamilon vitis) and two transpovirons respectively associated to B- and C-clade mimiviruses. We found that the virophage could transfer each transpoviron provided the host viruses were devoid of a resident transpoviron (permissive effect). If not, only the resident transpoviron originally isolated from the corresponding virus was replicated and propagated within the virophage progeny (dominance effect). Although B- and C-clade viruses devoid of transpoviron could replicate each transpoviron, they did it with a lower efficiency across clades, suggesting an ongoing process of adaptive co-evolution. We analysed the proteomes of host viruses and virophage particles in search of proteins involved in this adaptation process. This study also highlights a unique example of intricate commensalism in the viral world, where the transpoviron uses the virophage to propagate and where the Zamilon virophage and the transpoviron depend on the giant virus to replicate, without affecting its infectious cycle.
Subject terms: Virology, Cellular microbiology
Introduction
While for decades most of the focus was given to pathogenic viruses and viruses infecting parasites of human, animals and plants for obvious reasons, they are now recognized as major players in the environment and are by far the most abundant entities in all biotopes including oceans, fresh water, soil [1–5] and are even found in association with multicellular organisms’ microbiotes [6–8]. They have also received a lot of attention with the discovery of Mimivirus, the first giant virus with icosahedral capsids visible by light microscopy, enclosing a genome of 1.2 Mb and thousand genes [9, 10]. Many members of the Mimiviridae have then been isolated as well as new families such as the proposed Pandoraviridae [11], Molliviridae [12] and Pithoviridae [13], demonstrating that giant viruses now appear ubiquitous in all kinds of environment. Giant viruses infect unicellular eukaryotes, some regulating the populations of bloom forming algae [14–19]. As of today, the Mimiviridae family appears composed of several distinct subfamilies [10, 15–18, 20–24] one of which, the proposed Megamimivirinae [5, 17, 18], corresponds to the family members specifically infecting Acanthamoeba [20–23]. Members of this subfamilies, collectively refers to as “mimiviruses” throughout this article, can be infected by dsDNA satellite viruses called virophages only able to replicate using the already installed intracytoplasmic viral factory [16, 25–28]. This new type of satellite viruses constitute the Lavidaviridae family [29]. In addition to virophages, mimiviruses can be found associated with a linear plasmid-like 7 kb DNA called a transpoviron [26, 29, 30] making their mobilome uniquely complex among the known large DNA virus families. Sputnik, the first discovered virophage, was found to infect the A-clade Mamavirus [25]. Since it caused the formation of abnormal, less infectious, MamavirusA particles, it was initially proposed that virophages would in general protect host cells undergoing giant viruses’ infections. Such a protective role was quantitatively demonstrated for the protozoan Cafeteria roenbergensis, infected by the CroV [15] virus, in presence of the Mavirus virophage [31, 32]. However, it was later recognized that some virophages replicated without visibly impairing the replication of their associated host virus. This is the case for the mimiviruses of the B or C clades when infected by the Zamilon virophage [28]. On the other hand, members of the A-clade appeared non permissive to Zamilon replication [28]. The resistance to Zamilon infection was linked to a specific locus proposed to encode the viral defence system MIMIVIRE [33–35] but the actual mechanisms governing the virulence of a given virophage vis-à-vis its host virus remain to be elucidated [34, 35]. Transpovirons are commonly detected in Mimivirus genomes and might also be involved in the protection of the mimiviruses against the deleterious effects of virophages infection. In this study we took advantage of a newly isolated virophage (Zamilon vitis) and of its ability to propagate different transpovirons to investigate the specificity of the transpoviron/host virus relationship. We also used B- and C-clade strains of mimiviruses originally devoid of transpoviron to investigate the possible role of the transpoviron in the context of virophage co-infections. Finally, we analyzed the proteome of virophage particles replicated on B- and C-clades host viruses, with and without resident transpoviron, to identify transpoviron proteins that could be involved in the co-evolution process allowing the transpovirons to be replicated by mimiviruses.
Materials and methods
Viruses isolation
The new giant viruses’ strains were isolated from soil recovered from a French vineyard (43°20′25″N-5°24′51″E, MC. vitis), muddy water collected in the Ross river mangrove near Townsville (19°15′42.39″S, 146°49′5.50″E, MB. australiensis) and water collected from a well nearby Bamako (12°31′28.6″N 7°51′26.3″W, MB. maliensis).
Virus production
After being treated with antibiotics the different resuspended samples were added to a culture of Acanthamoeba castellanii (Douglas) Neff (ATCC 30010TM) cells adapted to Fungizone (2.5 μg/ml) and cultures were monitored for cell death as previously described [11–13, 20]. Each virus was then cloned [12] and the viral clones (MB. australiensis, MC. vitis and MB. maliensis) were recovered and amplified prior purification, DNA extraction and cell cycle characterization by electron microscopy (EM). Permissivity to virophage infection on the various giant viruses was analysed using the same protocol to assess the production of virophage after one round of co-infection of each clone with the purified virophage.
Virus purification
All giant viruses were purified on a CsCl gradient as follows. The cell lysate was centrifuged 10 min at 500 × g and the supernatant was centrifuged at 6800 × g for 45 min. The viral pellet was washed once with K36 buffer and the viruses were resuspended in CsCl 1.2 density, loaded on a gradual CsCl gradient of 1.3/1.4/1.5 densities and centrifuged at 100,000 × g for 20 h. The viral disk was then washed three times with K36 buffer. In contrast to Sputnik virophage, Z. vitis and our various giant viruses were still infectious after treatment at 65 °C. We thus could not apply the previously published protocol [26] to separate Z. vitis from the giant viruses. Instead we used several steps of filtration/centrifugation with a final purification on sucrose gradient. The preparation was filtered on 0.2 µm filter and centrifuged at 100,000 × g for 90 min. The pellet was resuspended in 5 ml of 40 mM Tris HCl pH 7.5 and filtered on 0.1 µm filter. The filtrate was then centrifuged at 150,000 × g for 1 h, and the pellet resuspended in 0.2 ml of 40 mM Tris HCl pH 7.5 and loaded on a 70%/60%/50%/40% sucrose gradient and centrifuged at 200,000 × g for 24 h. The band corresponding to the virus was recovered with a syringe and washed once with 40 mM Tris HCl pH 7.5. The purification was controlled by negative staining observation using a FEI Tecnai G2 operating at 200 kV (Fig. S1). Competition experiments were performed using a large excess of virophage particles compared with the giant virus (103 for 1).
Synchronous infections for TEM observations of the infectious cycles
A. castellanii adherent cells in 20 ml culture medium were infected with each giant virus with a MOI of 50 for synchronization. After 1 h of infection at 32 °C, cells were washed three times with 30 ml of PPYG to eliminate the excess of viruses. For each infection time (every hour from 1 to 11 h post infection (pi)), 2.5 ml were recovered and we did include them in resin using the osmium-thiocarbohydrazide-osmium method [36]. Ultrathin sections of 90 nm were observed using a FEI Tecnai G2 operating at 200 kV.
DNA extraction for sequencing
For the MC. vitis clone still associated with the Zamilon vitis virophage we did not try to exhaustively separate the giant virions from Z. vitis virions prior to DNA extraction. For each giant virus clone, genomic DNA was extracted from 1010 virus using the PureLinkTM Genomic DNA mini kit (Invitrogen) according to the manufacturer’s protocol. Finally, for the virophage, after its separation from the giant virus, the DNA was extracted using the same protocol. The purified DNAs were loaded on an agarose gel in search of an extra DNA band suggestive of the presence of an episome that could correspond to a transpoviron. Sequencing of MC. vitis and MoumouvirusB australiensis was performed using Illumina technology on an Illumina MiSeq and the MoumouvirusB australiensis transpoviron (matv) sequence transpoviron DNA was extracted and purified from an agarose gel and sent to Nanopore for sequencing. The MB. maliensis purified DNA was sent to the Novogene company for library preparation and Illumina PE150 sequencing. MoumouvirusB australiensis genome was sequenced using the Pacbio technology.
Genome sequencing, assembly and annotation
Library preparation for Nanopore technology
DNA was quantified using a Qubit 3.0 Fluorimeter (Thermo Fisher Scientific, MA, USA) following the manufacturer’s protocol, and was found to be 12.7 ng·ml−1. 7.5 µl of this DNA was used for library preparation using the RAD002 kit (Oxford Nanopore Technologies, Oxford, UK). Since the input quantity of DNA was lower than recommended for this kit, the active FRM reagent was diluted with three volumes of heat-inactivated FRM, to avoid over-fragmentation of the DNA. The library preparation reaction was set up as follows: the reaction (DNA 7.5 µl, 0.25 × FRM 2.5 µl) was incubated for 1 min at 30 °C followed by 1 min at 75 °C. We added 1 ml of RAD reagent from the RAD002 kit and 0.2 ml of Blunt TA ligase (New England Biolabs, MA, USA) and the reaction was incubated for 5 min at room temperature. The prepared library was then loaded onto a FLO-MIN106 flowcell (version 9.4 nanopores) as per Oxford Nanopore Technologies’ standard protocol.
Library preparation for Illumina technology
Genome sequencing was performed using the instrument Illumina MiSeq. Libraries of genomic DNA were prepared using the Nextera DNA Library Preparation Kit as recommended by the manufacturer (Illumina). The sequencing reaction was performed using the MiSeq Reagent Kit v2 (300-cycles), paired-end reads of 150 nt × 2 (Illumina).
The assembly of MoumouvirusB australiensis genome sequence was performed on one SMRT cell of Pacbio data using the HGAP workflow [37] from the SMRT analysis framework version 2.3.0 with default parameters, resulting in 84,565 corrected reads.
The MinION library was sequenced for 1 h on a MinION Mk1B flowcell (Oxford Nanopore Technologies), generating ~220 Mb of sequence data. Basecalling was performed using the 1D Basecalling for FLO-MIN106 450 bps r1.121 [workflow ID: 1200] workflow (Oxford Nanopore Technologies), and yielded 128,623 reads with a mean length of 1701 bases. Data were filtered to remove reads with a quality score below 8, leaving 76,936 reads, and a mean length of 2369 bases. The reads were assembled using Canu with the default parameters, but with the option stopOnReadQuality = false. The matv resulting from this assembly was further polished using the MB. australiensis Pacbio error corrected reads.
The assembly of the MegavirusC vitis, Zamilon vitis and Megavirus vitis transpoviron (mvtv) genomes was performed using Spades (version 3.9.0) [38] on MiSeq Illumina paired-end reads and Pacbio long reads when available. Spades was used with the following parameters: careful, k = 17, 27, 37, 47, 57, 67, 77, 87, 97, 107, 117, 127, cov-cutoff = off and Pacbio option. For MoumouvirusB maliensis, for which long reads were not available, the assembly was performed using Illumina paired-end reads and Spades (version 3.12.0) with the following parameters: careful, no reads correction, k = 33, 55, 77, 99, 127.
Gene annotation of genomic sequences was done using Augustus [39] trained on already published members of the subfamily gene sets. The tRNAs were searched using tRNAscan-SE [40] with default parameters. Functional annotation of predicted protein-coding genes was done using homology-based sequence searches (BlastP against the NR database [41] and search for conserved domains using the Batch CD-Search tool [42]).
Phylogenetic analyses
The phylogenetic tree of the transpovirons (Fig. 2) was computed using a concatenation of the three conserved genes (orthologous to mvtv_2, mvtv_6 and mvtv_7, see Fig. 2) using the optimal model “LG + G” selected by Prottest [43]. The phylogeny of the giant viruses (including MegavirusC vitis, MoumouvirusB australiensis and MoumouvirusB maliensis, Fig. S2) was performed on a concatenation of the single copy genes shared by all mimiviruses. For that we first predicted the genes using CompareM (https://github.com/dparks1134/CompareM) and clustered them with MCL [44], resulting in 367 conserved genes, and selected among them the 197 present in exactly one copy per virus. Next, the sequences were aligned using Mcoffee [45] and concatenated. Finally, the phylogenetic tree produced from the resulting superalignment was based on the optimal “VT” model selected by Prottest [43]. Average nucleotide identity between the different strains (Fig. S2) was calculated using the OAU tool [46].
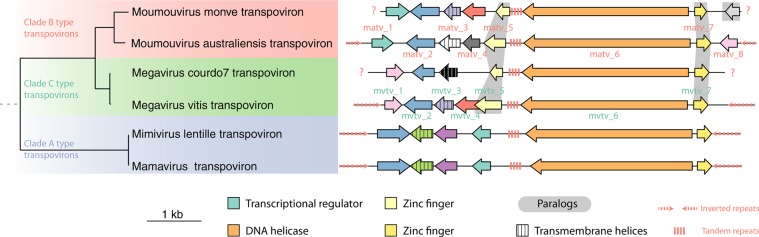
Fig. 2. Phylogeny and genomic organization of transpoviron sequences.
The phylogenetic tree (on the left) was computed from the concatenated sequences of shared orthologous predicted proteins using PhyML [45] with the LG + G model. Bootstrap values (not shown) are all equal to one. The genomic organization (right) shows orthologous genes represented with identical colours and paralogous genes (in a given genome) are highlighted in grey. Gene names are indicated for matv and mvtv.
MS-based proteomic analyses
Characterization of virion proteomes by data dependent acquisition
For proteomic analysis the virions pellets were resuspended in Tris HCl 40 mM pH 7.5, 60 mM DTT, 2% SDS and incubated 10 min at 95 °C. The protein concentration of the lysates were measured using the 660 nm Protein Assay Reagent appended with Ionic Detergent Compatibility Reagent (Thermo Scientific) and 4 µg of proteins were analyzed as previously described [12]. Two replicates were analyzed per sample, except for Z. vitis purified from MB. maliensis and Z. vitis purified from MC. chilensis containing matv. Peptides and proteins were identified and quantified using MaxQuant software (version1.6.2.10) [47]. Spectra were searched against the corresponding MegavirusC chilensis, MegavirusC vitis, MoumouvirusB australiensis, MoumouvirusB maliensis, Zamilon vitis, mvtv, matv and Acanthamoeba castellanii protein sequence databases and the frequently observed contaminants database embedded in MaxQuant. Minimum peptide length was set to 7 aa. Maximum false discovery rates were set to 0.01 at PSM, peptide and protein levels. Intensity-based absolute quantification (iBAQ) [48] values were calculated from MS intensities of unique + razor peptides. Proteins identified in the reverse and potential contaminants databases as well as proteins only identified by site were discarded from the final list of identified proteins. For each analysis, iBAQ values were normalized by the sum of iBAQ values in the analyzed sample [49]. Only proteins identified with a minimum of two unique + razor peptides in one sample were considered.
Dominance effect validation by PCR
Cells were grown in T75 flasks and infected either with MB. australiensis (carrying matv) at MOI 10 and a large excess (103 for 1 giant virus particle) of virophage carrying mvtv, or with MC. vitis (carrying mvtv) at MOI 10 and a large excess of virophage carrying matv. After 40 min of infection, the cells were washed three times and distributed in 12-well plates (1 ml per well). Cells were collected from a well at 45 min, 2 h, 4 h, 6 h, 8 h and 24 h pi. The cells were centrifuged at 1000 × g for 3 min, except for the last point at 24 h that was centrifuged at 16,000 × g for 10 min. Each pellet was resuspended in 100 µl of PPYG medium and cells were frozen in liquid nitrogen to stop the infection and stored at −80 °C. PCR were performed using the Terra PCR Direct Polymerase Mix (Clontech) directly on the cell lysates using the following primers:
qPCR-matv_F TCGCTCATTGATTCACTTTGTAC; qPCR-matv_R AATGTATTATGGGCGAATAATGTT; PCR produced an amplicon of 185 bp.
qPCR-mvtv_F GGCATAAGCAGGTTCGAAAT, qPCR-mvtv_R CATGGCGTGATATTGGTGTG; PCR produced an amplicon of 194 bp.
The PCR experiments were stopped after 20 cycles of amplification and 7 µl of the reaction products were deposited on agarose gel (Fig. S3).
Competition experiments
Cells were grown in T25 flasks (5 ml growth medium) and infected with host viruses carrying one transpoviron at MOI 0.25 and a large excess (103 for 1 giant virus particle) of virophage carrying the complementary transpoviron. After cell lysis, 100 µl of the culture medium containing virophage and host viruses were used to infect another T25 flask containing adherent fresh cells. This process was repeated ten times.
Selective identification of transpoviron in virions capsids
To distinguish the mvtv and matv transpovirons, two sets of primers were designed:
mvtvPFwd: ACCTTCTTGTGCCTTTACTGC, mvtvPRev: CAGGGTTCGGACGGATTACT; PCR produced a 939 bp amplicon.
matvPFwd: TCGCTCATTGATTCACTTTGTAC, matvPRev: CAAAGGGGAGGAAATAATGGAGA; PCR produced a 263 bp amplicon.
PCR were performed using the Terra PCR Direct Polymerase Mix (Clontech) directly on the cell lysates after each round of co-infection. To check the stability of the transpovirons over time, up to ten additional rounds of virus production were performed and the presence of the transpoviron was assessed by PCR.
Total DNA was extracted from purified host viruses and virophages using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific) and serial dilutions of DNA were performed and deposited on a 0.8% agarose gel ran for 45 min at 100 V. For host viruses, we deposited from 1 to 0.25 µg of total DNA and for virophages, from 1 µg to 62.5 ng DNA. DNA bands were revealed using BET staining and images were recorded on a Chemi-smart 2000WL-20M camera (Fischer Bioblock Scientific).
Results
MegavirusC vitis, MoumouvirusB australiensis, MoumouvirusB maliensis and their mobilomes
Three samples recovered from various locations (France, Australia and Mali) produced lytic infections phenotypes when added to cultures of Acanthamoeba castellanii cells. Viral factories were recognizable in the amoeba cells after DAPI staining and we observed the accumulation of spherical particles visible by light microscopy in the culture medium (Fig. 1a). The corresponding virus populations were cloned and amplified as previously described [12]. In all cases, negative staining EM images confirmed the presence of icosahedral virions of ~450 nm in diameter with a stargate structure at one vertex, as was observed previously for all members of the Mimiviridae infecting Acanthamoeba cells [20, 28, 50, 51]. For some clones of the giant virus isolated from the French sample, associated icosahedral virions of ~70 nm diameter were also visible, suggesting the presence of virophages (Fig. 1b–f).
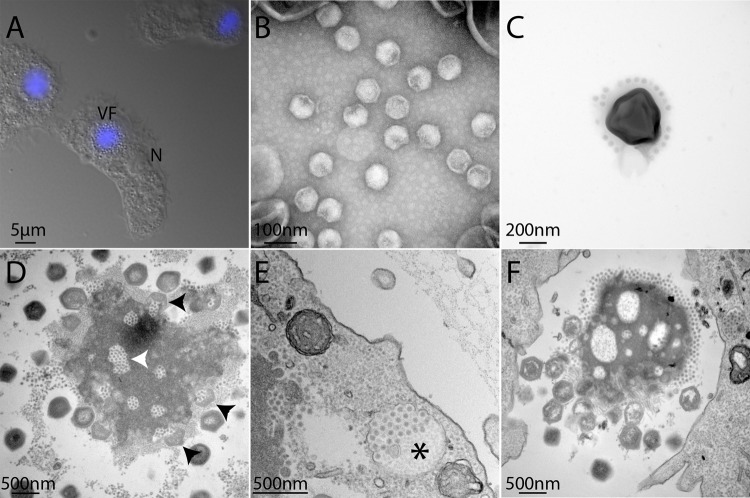
Fig. 1. Microscopy images of MegavirusC vitis and its associated virophage Z. vitis.
a Fluorescence image of DAPI-stained A. castellanii cells infected by MC. vitis and its virophage. Viral particles are visible in the periphery of the viral factory (VF). The cell nucleus (N) remains visible but its fluorescence becomes undetectable due to the intense labelling of the VF DNA. b Transmission electron microscopy (TEM) of Zamilon vitis particles observed by negative staining electron microscopy; c TEM of virophage particles stuck to the giant virus particle (negative staining); d ultrathin section TEM of a MC. vitis viral factory observed in late infection of A. castellanii cells: virophage particles can be seen in holes in the VF (white arrowhead) as well as penetrating a maturing MC. vitis particle (black arrowheads); e neo-synthesized Z. vitis virophage particles gathered in vacuoles (black star) are seen at the periphery of the infected cell suggesting that they are released by exocytosis. f TEM image of an isolated viral factory observed in an ultrathin section of a late infection of A. castellanii cells: virophages accumulate at one pole of the VF as well as in holes in the VF while immature and mature MC. vitis particles are seen at the opposite pole of the VF (Supplementary movie).
Based on its genome sequence, this new isolate was determined to belong to the C clade and named MegavirusC vitis (Fig. S2). Its associated virophage was named Zamilon vitis, in reference to its genomic similarity with the original Zamilon virophage [28]. In addition, the genome sequence assembly process revealed the presence of a 7 kb dsDNA sequence homologous to previously described transpovirons [26]. The MegavirusC vitis associated transpoviron was named mvtv. The Australian isolate was found to be a member of Mimiviridae clade B, and was named MoumouvirusB australiensis (Fig. S2). It came with its own transpoviron that we named matv. Finally, the Bamako (Mali) isolate was determined to be another B-clade member, and named MoumouvirusB maliensis (Fig. S2). This last virus was devoid of transpoviron.
For this study, we included the previously isolated MegavirusC chilensis, as a member of Mimiviridae clade C, devoid of transpoviron (Fig. S2). Underscores A, B and C are used throughout this article to indicate the clade origin of the various mimiviruses.
Comparison of Acanthamoeba cells co-infected by B- or C-clade mimiviruses and Z. vitis
The infectious cycles of MC. vitis (Fig. S4), MC. chilensis, MB. australiensis and MB. maliensis appeared very similar to that of other mimiviruses, with an initial internalization of the virions in vacuoles, followed by the opening of the stargate and the fusion of the internal membrane with that of the vacuole to deliver the nucleoid in the cytoplasm. After 3 h pi, viral factories develop in the cell cytoplasm, delineated by a mesh of fibres excluding all organelles [20, 52]. Later on, neo-synthesized virions are seen budding and maturing at their periphery. Virophages specifically associated to the mimiviruses are thought to penetrate the cells at the same time as their host viruses, either enclosed in the host virus particles, or sticking to their external glycosylated fibrils (Fig. 1c) [25]. The virophages are devoid of a transcription machinery and thus use the transcription apparatus of the host virus to express their genome once released in the cell cytoplasm [25, 53]. During infection of Acanthamoeba cells with MC. vitis or MC. chilensis in the presence of Z. vitis, regions depleted of electron dense material (“holes”) appeared in the viral factory prior to the assembly of any virion. Z. vitis virophage particles then start to accumulate inside these holes (Figs. 1, S5C, D, Supplementary movie) as early as 4 h pi, before the production of host virus particles. Such infectious cycle is reminiscent of the one described for the association between virophage (Sputnik and Zamilon) and mimiviruses [25, 26, 28], with virophages visible at the periphery of the viral factory, some of them seemingly penetrating inside the maturing giant virions (Fig. 1d). The infectious cycle of MoumouvirusB australiensis during co-infection of Acanthamoeba with the Z. vitis virophage was very similar to the one of MC. vitis except that instead of holes, the viral factory appeared to segregate the production of virophages in a separate compartment (Fig. S5E, F). In all cases, during the latest stage, virophages were seen in large vacuoles that appeared to migrate toward the cell membrane to be released through exocytosis (Fig. 1e). However, while Sputnik co-infections lead to aberrant and non-infectious Mimivirus particles [25, 26], Z. vitis, as other Zamilon virophages, does not visibly impede the replication of its host viruses, abnormal particles of which were never observed [28].
The newly isolated mimiviruses’ genomes
The dsDNA genome sequence of MegavirusC vitis was assembled into a single contig of 1,242,360 bp with a G + C content of 25%. It was very close to Megavirus Terra1 [54] and to the C-clade prototype MegavirusC chilensis [20] with whom it shared 99.1 and 96.9% identical nucleotides over the entire genome length, respectively (Fig. S2). MoumouvirusB australiensis genome sequence was assembled in one contig of 1,098,002 bp (25% G + C), and that of MoumouvirusB maliensis in one contig of 999,513 bp (25% G + C). As shown in Fig. S2, MB. australiensis and MB. maliensis belonged to B clade and are closer to each other than to any other moumouviruses, thus initiating a third sub-lineage. Interestingly, the B clade appeared the most divergent among mimiviruses.
Zamilon vitis genome
In addition, we determined the 17,454 bp genome sequence (30% G + C) of Zamilon vitis. It was closely related to that of other virophages infecting the B and C clades, sharing 97.8% identical nucleotides with the prototype Zamilon virophage [28]. The 20 predicted proteins were all conserved in other virophages infecting mimiviruses, sharing 40–80% identical residues with Sputnik [25], their most distant homologue.
The mvtv and matv transpovirons
Finally, we determined the genome sequences of the two new transpovirons. The mvtv DNA sequence was 7417 bp (22% G + C) in length and closely related to the one associated with MegavirusC courdo7 (98% identical nucleotides). The matv DNA sequence was 7584 bp in length (22% G + C) and was related to the one associated with MoumouvirusB monve (89% identical nucleotides). The reconstructed phylogeny of all the known transpoviron genomes clearly showed that they fell into three distinct clusters, mirroring the tripartite clades structure of the host viruses from which they were isolated (Fig. 2).
Transpovirons exhibited terminal inverted repeats (TIRs) of 520 nt for MamavirusA and Lentille virusA transpovirons (lvtv), 380 nt for matv and 510 nt for mvtv. TIRs were missing from the transpovirons associated to MoumouvirusB monve and MegavirusC courdo7, most likely because their published sequences are incomplete [26]. TIRs are well conserved within clades (90% of identical nucleotides between MamavirusA transpoviron and lvtv) but diverged between clades (56% identical nucleotides between lvtv/mvtv and 53% between mvtv/matv). A tandem repeat (TR) of 180–600 nt was present in the centre of all sequenced transpovirons, in an intergenic region 3′ from a conserved helicase (Fig. 2). These TRs were also well conserved within clades (80% identical nucleotides) and divergent between clades (39% identical nucleotides). It is worth mentioning that an evolutionary link between virophages and transpovirons has been proposed [55]. Three predicted proteins were found in all transpovirons (i.e. core genes): a helicase (Mvtv_6/Matv_6), a protein of unknown function (Mvtv_2/Matv_2) and a zinc-finger domain-containing protein (Mvtv_7/Matv_7), (Fig. 2). In addition, all transpovirons encode a small protein with a central transmembrane segment as their only recognizable similarity (Mvtv_3/Matv_3). Seven predicted proteins were shared by at least two transpovirons, including a transcriptional regulator conserved in clades A and B (Matv_1), a protein paralogous to the above core zinc-finger-domain-containing protein and conserved in clades B and C (Mvtv_5/Matv_5), and five other predicted proteins without any functional signature. Finally, four proteins of unknown function were unique and had no detectable homologue in the other transpovirons.
We analyzed the proteome of MC. vitis virions in search of transpoviron proteins specifically associated to this host virus. We identified three transpoviron proteins, Mvtv_3, a putative membrane protein that could be anchored in the giant virus membrane, and Mvtv_2 and Mvtv_4, two putative DNA-binding proteins (Fig. S6, Table S1).
Given that all known virophages infecting mimiviruses have been isolated in the presence of a transpoviron, we also expected the presence of transpoviron-encoded proteins in Z. vitis virions. We thus analyzed the protein composition of the purified Z. vitis particles produced with MC. vitis. The proteomic study confirmed this prediction but suggests a specific interaction between the transpoviron proteins and the virophage in one hand, and the transpoviron proteins and the host virus particles in the other hand. Indeed, in virophage particles, we consistently identified two transpoviron proteins, Mvtv_7, a putative DNA-binding protein and in lesser amount the predicted helicase Mvtv_6 never identified in the MC vitis particles in the absence of virophage particles (Table S1). Four additional proteins were also detected but in much smaller amount, three predicted DNA-binding protein (Mvtv_5, Mvtv_2 and Mvtv_4 in decreasing amounts) and a protein with unknown function, Mvtv_1. These four proteins were also seen in the total proteome of the MC. vitis + Z. vitis virions. In contrast, they were all absent from the proteome of the cloned MC. Vitis particles (Fig. S6). Thus, the transpoviron encodes different subsets of proteins that might be specifically involved in their packaging in two alternative vehicles: the virophage or the host virus particle.
Clade specificity of transpovirons
First, we verified that Z. vitis virophage replication was restricted to host viruses from the B and C clades, as previously described for Zamilon virophages (Table 1) [28]. We also verified by PCR that the MC. vitis clone cleared from virophage and replicated on A. castellanii cells remained associated with its transpoviron mvtv.
Table 1.
Permissivity of the host Megavirinae to Z. vitis virophage and their selectivity for the transpovirons.
| Clade | Giant virus | Permissivity to Z. vitis | Selectivity for transpoviron | |
|---|---|---|---|---|
| Z. vitis + mvtv | Z. vitis + matv | |||
| A | Mimivirus | − | mvtv− | matv− |
| C | MC. chilensis | + | mvtv+ | matv+ |
| MC. chilensis + mvtv | + | mvtv+ | mvtv+ matv+ | |
| MC. chilensis + matv | + | matv+ mvtv+ | matv+ | |
| C | MC. vitis + mvtv | + | mvtv+ | mvtv+ |
| B | MB. australiensis + matv | + | matv+ | matv+ |
| B | MB. maliensis | + | mvtv+ | matv+ |
| MB. maliensis + matv | + | matv+ mvtv+ | matv+ | |
| MB. maliensis + mvtv | + | mvtv+ | mvtv+ matv+ | |
Transpovirons originally associated to a given host virus are underlined. Most abundant transpovirons in host particles are shown in bold
Purified virophage virions carrying the mvtv transpoviron were then used to co-infect A. castellanii with two C-clade megaviruses (MC. vitis/mvtv and MC. chilensis w/o transpoviron) and two B-clade moumouviruses (MB. australiensis/matv and MB. maliensis w/o transpoviron) to assess whether the transpovirons were specific to a given clade of mimiviruses (Fig. 3a).
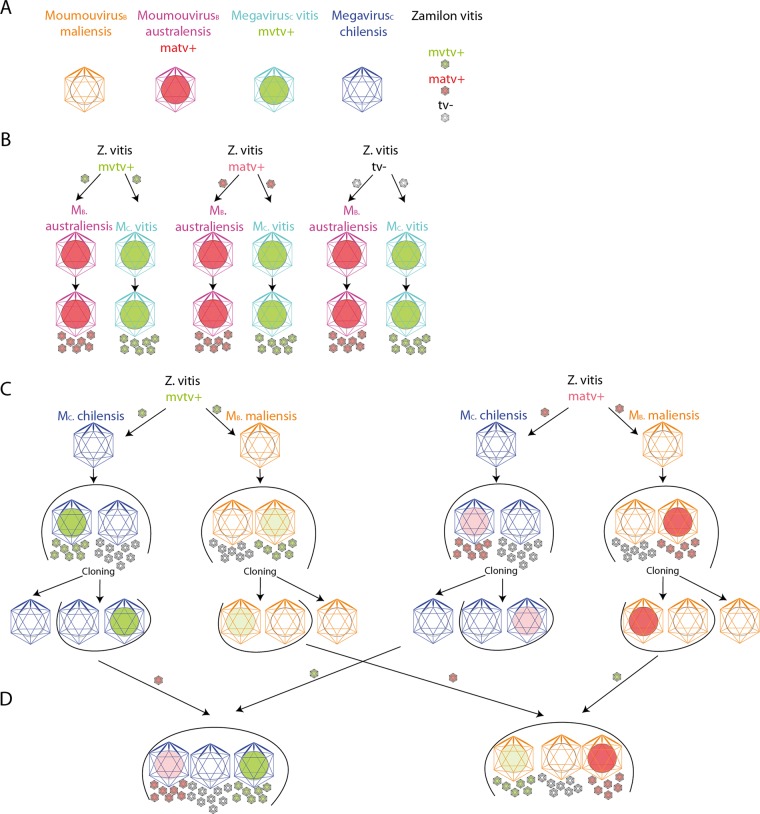
Fig. 3. Dominance effect versus permissive effect.
a Viruses used in this study. MB. maliensis is represented in orange, MB. australiensis in purple, MC. vitis in cyan and MC. chilensis in dark blue. The transpovirons are represented as coloured circles (green for mvtv and pink for matv) inside the giant virus and virophage capsids. b Dominance effect of the resident transpoviron (mvtv in MC. vitis, green circle in cyan capsid; matv in MB. australiensis, pink circle in purple capsid) over the one carried by Z. vitis. Empty Z. vitis (black contour, white capsid) do acquire the resident transpoviron upon replication; c Permissive effect: the two type of transpovirons can be imported and replicated by empty MC. chilensis (dark blue capsid, white circle) and MB. maliensis (orange capsids, white circle), although with different efficiencies. d Combination of the dominance and permissive effect. Colour intensities of the circles (pink for matv, green for mvtv) illustrate the abundance of the transpovirons in the host and virophage particles.
We performed specific PCR for each transpoviron after each round of co-infection and for each additional round of production (up to ten) to assess the presence of matv and mvtv DNA in the cultures after cell lysis. We also performed a proteomic analysis of the resulting virophages to assess the presence of transpoviron proteins (Table S1).
Co-infection of the virophage (carrying mvtv) with MB. australiensis (carrying matv) surprisingly produced a unique population of neo-synthesized virophage particles carrying the matv transpovirons (Table 1, lane 6). The proteomic analysis of the purified virophage capsids also evidenced the replacement of the mvtv proteins by their orthologues in matv, matv_7 and matv_6 and to a lesser extend matv_2. In contrast the orthologues of Mvtv_1 (Matv_8) and Mvtv_5 (Matv_5) were not detected (Table S1). Moreover, PCR performed along the infection cycle of MB. australiensis (carrying matv) and the virophage (carrying mvtv) did not show an increase in mvtv while the matv genome was clearly replicated (Fig. S3A). These results suggested that the host virus strongly favour the replication of its natively associated transpoviron (Table 1, Fig. 3b). If a different one is brought in by the virophage, it is lost and replaced by the one replicated by the host virus, a result we refer to as the “dominance effect”. Consequently, two populations of virophages carrying either mvtv or matv were at our disposal. We confirmed the dominance effect by co-infection of MC. vitis carrying mvtv with virophages carrying matv (Table 1, lane 5). Again, we observed the replacement in the virophage particles of matv (DNA and proteins) by mvtv (DNA and proteins). We also confirmed that the mvtv genome was actively replicated while the amount of matv genome remained stable along the infectious cycle (Fig. S3B). We then used the virophages carrying either mvtv or matv to infect transpoviron-free B-clade (MB. maliensis) and C-clade (MC. chilensis) host viruses. We found that the virophage succeeded in transmitting each transpoviron to each “empty” B- or C-clade host viruses (Table 1, lanes 2 and 7), a result we refer to as the “permissive effect”. However, we observed that matv was preferentially replicated by MB. maliensis and mvtv by MC. chilensis (Table 1). By cloning we showed that the resulting populations of B- and C-clade host viruses were mixtures of transpoviron positive and transpoviron negative particles (Fig. S7). Furthermore, virophage particles produced by transpoviron negatives clones were also devoid of transpoviron (DNA and proteins), indicating that although transpovirons can be carried by virophages, their replication is performed and controlled by the host virus.
We finally took advantage of the permissive effect to produce two populations of clade B (MB. maliensis) and C (MC. chilensis) host viruses, each carrying the matv or mvtv transpoviron. We then challenged them using virophages carrying the other transpoviron. We performed PCR specific for either transpoviron after each round of virophage co-infection (up to ten successive rounds of virophage production) and assessed their presence after cell lysis.
The proteome of the virophages particles was also analysed in order to assess the presence of transpoviron proteins possibly associated to the transpoviron DNA. The results of the various experiments are presented in Table 1 and Table S1 and are interpreted in Fig. 3.
When we infected populations of MC. chilensis carrying matv or mvtv with virophages carrying the complementary transpoviron, the resulting population of MC. chilensis and virophages became positive for the two transpovirons (Table 1, lanes 3 and 4). This apparently violated the strict “dominance effect” observed with MC. vitis and MB. australiensis. However, the persistence of a subpopulation of empty MC. chilensis virion (i.e. devoid of transpoviron) might also explain our results without refuting the dominance rule. In that case, the replication and propagation of the competing virophage-borne transpoviron would be performed by the transpoviron-null (empty) MC. chilensis subpopulation. We investigated this possibility by cloning virions from the mixed mvtv/matv MC. chilensis population and examining their transpoviron content. Strikingly, we never observed MC. chilensis virions simultaneously carrying both types of transpovirons. On the other hand, we observed either mvtv positive (mvtv+) or matv positive (matv+) MC. chilensis clones, as well as others devoid of transpovirons (Fig. 3). This result also suggests that the association between the host virus and the transpovirons are not stable when resulting from a first encounter. In the case of MB. maliensis in the presence of virophage matv, the replacement of the mvtv transpoviron from host particles appeared to be faster, which resulted in the rapid disappearance of mvtv after only six rounds of replication. We also observed that the loss of the transpoviron over host virus replication, in the absence of virophage, was more rapid for MB. maliensis than MC. chilensis suggesting the association between the host virus and the transpoviron was not stable. The cloning step provided virophage-free host virus clones with which to replicate these competition experiments: the matv+ or mvtv+ clones infected by virophages carrying the complementary transpoviron again produced a mixed population of matv+, mvtv+ and transpoviron-null virions (Table 1, lanes 8 and 9). Thus, the persistence of particles devoid of transpoviron allows us to conclude that our results are a combination of the “dominance effect” applied to the subpopulation of transpoviron positive virions and of the “permissive effect” applied to the transpoviron-null subpopulation. In the resulting virophage particles, the only transpoviron proteins consistently identified were Mvtv_7/Matv_7 and Mvtv_6/Matv_6. As expected, they were absent from virophages devoid of transpoviron (Table S1).
To elucidate whether the transpoviron could have a protective role against infection of the host virus by the virophage, we compared the infectious cycles of Acanthamoeba cells infected by MC. chilensis carrying matv, mvtv or without transpoviron. They were strikingly similar both in terms of cycle length and virus production yields. Transpovirons are thus not key, at least in laboratory conditions, in regulating the permissivity of mimiviruses to virophage infection.
Discussion
Mimiviruses are unique in their association with two distinct (often co-existing) dependent entities, virophages and transpovirons, somewhat reminiscent of phages and plasmids afflicting bacteria. As for the virophage, the presence of host virus-like regulatory elements (terminator hairpin and late promoter [56–59]) flanking the transpoviron genes suggest that they also use the host virus transcription machinery rather than that of the cell. The transpoviron might also rely on the host virus DNA replication machinery, in absence of transpoviron-encoded DNA polymerase. Our competition experiments between the mvtv vs. matv transpovirons resulted in the replication of only one transpoviron. Interestingly, the “winner” corresponds to the type originally associated to the host virus (mvtv for MC. vitis and matv for MB. australiensis, Table 1), a phenomenon we called the “dominance effect”. This finding was also confirmed by the immediate replacement of mvtv by matv proteins in virophage particles synthetized with MB. australiensis. However, this result is not simply due to a strict clade-wise specificity. The use of transpoviron-free host virus particles allowed us to demonstrate that MC. chilensis and MB. maliensis can replicate and incorporate each transpoviron, independently (Table 1). Yet, we observed a marked difference in permissivity, with B- and C-clade host viruses favouring their cognate transpoviron types. The central TRs sequences and the TIR flanking the transpovirons replicated by A-clade vs. B- or C-clade host viruses are markedly different. These differences might cause the lack of replication of matv and mvtv in the A-clade Mimivirus. The lesser differences between the B- and C-clade transpovirons might then explain why both of them can still be replicated by MB. maliensis and MC. chilensis. In these host viruses, the competition experiment resulted in the simultaneous replication of both transpovirons. However, the sub-cloning of the mvtv+/matv+ population resulted in mvtv+ only, matv+ only, or transpoviron negative clones. It also appears that neither of the two transpovirons remains stably associated with a host virus for which it was a first encounter, while a preference could emerge once a stable association has been established by co-evolution (i.e. MB. australiensis with matv, MC. vitis with mvtv). Finally, in virophage particles there were fewer copies of matv either produced with MB. australiensis, MB. maliensis or MC. chilensis and fewer copies of mvtv produced with MC. chilensis or MB. maliensis, compared with the number of copies of mvtv produced with MC. vitis. Different transpovirons, even efficiently replicated by the host virus, thus appear to be loaded at different efficiencies in the virophage particles (Fig. S8). A similar result was previously described for the Lentille virus transpoviron that could only be detected in Sputnik2 virophage particles using FISH experiments [26]. A consequence of such suboptimal associations was the production of virophages devoid of transpovirons that could then be used to identify candidate proteins involved in the transpoviron/virophage association. The only difference between virophage particles carrying or not transpovirons was the recurrent presence of two transpoviron-encoded proteins (Mvtv_7/Matv_7 and Mvtv_6/Matv_6) together with the DNA molecule as an episome (Table S1, Fig. S8) suggesting the virophage was a mere vehicle for the transpoviron. These proteins are conserved in all transpovirons, are predicted to be DNA-binding, and were not identified in the proteome of the host virus. Instead, the most abundant transpoviron proteins in MC. vitis virions were two predicted DNA-binding proteins (Mvtv_2 and Mvtv_4) and one predicted membrane protein (Mvtv_3) that could be anchored in the host virus membrane (Table S1). All transpovirons encode a short predicted membrane protein although their primary sequence does appear to be conserved. The dominance effect is reminiscent of plasmids incompatibility or entry exclusion for related plasmids [60–63] or superinfection immunity [64] and superinfection exclusion [65] used by prophages to prevent superinfection by other bacteriophages. For the latter, DNA injection is often inhibited by a membrane-bound protein [66–68], paralleling the eventual role of the transpoviron-encoded membrane protein in giant virus particles carrying a transpoviron.
Since the transpoviron genome is present in both the host virus and the virophage particles, the transpoviron DNA might also adopt a different organization depending on the vehicle (host virus or virophage particles) used for its propagation. The Mvtv_7/Matv_7 and Mvtv_6/Matv_6 proteins could be involved in the packaging or delivery of the transpoviron in and from the virophage particle, while the Mvtv_2/Matv_2 and Mvtv_4 proteins could play a similar role vis-à-vis the host virus and the Mvtv_3 membrane protein may play a role in the dominance effect. Further studies are needed to elucidate the mechanisms at work in packaging and delivery of transpoviron genomes as well as in transpoviron dominance.
The first two types of virophage that have been discovered, Sputnik and Mavirus, respectively infecting Mimivirus [25] and Cafeteria roenbergensis [31] are strongly deleterious to their host viruses, diminishing the production of infectious particles [25, 26, 69] or stopping it altogether [31, 32], effectively protecting the cellular hosts. As parasite of another parasite (the giant virus), these virophages are bona fide hyperparasites [70, 71]. The detection of many additional virophage-related sequences in aquatic environment together with that of Mimivirus-like viruses suggested that they might have a significant ecological role in regulating the population of the giant viruses and of their cellular host (micro-algae or heterotrophic protozoans) [72–74]. However, the hypovirulence (of the host virus) induced by the hyperparasite may ultimately limit its own reproductive success [70]. Thus the evolutionary trajectory of the virophage/host virus/cellular host parasitic cascade may remain antagonistic or end up in a mutualistic or commensal relationship. The uniquely complex tripartite parasitic cascade transpoviron/virophage/host virus analyzed in this work, where none of the actors appears to have a detrimental effect on the others, at least in laboratory conditions, might be at a neutral equilibrium reached as a stable compromise after eons of intricate antagonistic evolution. To the best of our knowledge, the relationship between the transpoviron, the Zamilon virophage and their host giant virus analysed in this work represents the first example of bipartite commensalism in the viral world.
Supplementary information
Acknowledgements
This work was partially supported by the French National Research Agency ANR-14-CE14–0023–01, Institut Français de Bioinformatique (ANR–11–INSB–0013), the Fondation Bettencourt Schueller (OTP51251) and the Provence-Alpes-Côte-d’Azur région (2010 12125). Proteomic experiments were partly supported by the ProFi grant (ANR-10-INBS-08–01). We thank Laurence De Marchi and Daouda Traore for providing the soil samples and Olivier Poirot for helpful discussions. We thank Dr Elsa Garcin for reading and improving the manuscript. We also thank Dr A. Kosta at the IMM imagery platform and F. Richard and A. Aouane for their expert assistance on the IBDM imaging platform and Dr N. Brouilly for data acquisition that allowed producing the Supplementary movie. We also acknowledge the support of the bottom-up platform and informatics group of EDyP.
Data availability
The annotated genomic sequences determined for this work have been deposited in the Genbank/EMBL/DDBJ database under the following accession numbers: M. vitis: MG807319, M. australiensis: MG807320, M. maliensis: MK978772, Z. vitis: MG807318, mvtv: MG807316 and matv, MG807317. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009037 [75].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sandra Jeudy, Lionel Bertaux
Contributor Information
Sandra Jeudy, Email: Sandra.Jeudy@igs.cnrs-mrs.fr.
Jean-Michel Claverie, Email: Jean-Michel.Claverie@univ-amu.fr.
Chantal Abergel, Phone: +33 491 825422, Email: Chantal.Abergel@igs.cnrs-mrs.fr.
Supplementary information
The online version of this article (10.1038/s41396-019-0565-y) contains supplementary material, which is available to authorized users.
References
- 1.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suttle CA. Marine viruses-major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 3.Rowe JM, DeBruyn JM, Poorvin L, LeCleir GR, Johnson ZI, Zinser ER, et al. Viral and bacterial abundance and production in the Western Pacific Ocean and the relation to other oceanic realms. FEMS Microbiol Ecol. 2012;79:359–70. doi: 10.1111/j.1574-6941.2011.01223.x. [DOI] [PubMed] [Google Scholar]
- 4.Wigington CH, Sonderegger D, Brussaard CPD, Buchan A, Finke JF, Fuhrman JA, et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat Microbiol. 2016;1:15024. doi: 10.1038/nmicrobiol.2015.24. [DOI] [PubMed] [Google Scholar]
- 5.Schulz F, Alteio L, Goudeau D, Ryan EM, Yu FB, Malmstrom RR, et al. Hidden diversity of soil giant viruses. Nat Commun. 2018;9:4881. doi: 10.1038/s41467-018-07335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abeles SR, Pride DT. Molecular bases and role of viruses in the human microbiome. J Mol Biol. 2014;426:3892–906. doi: 10.1016/j.jmb.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr JJ. A bacteriophages journey through the human body. Immunol Rev. 2017;279:106–22. doi: 10.1111/imr.12565. [DOI] [PubMed] [Google Scholar]
- 8.Atoni E, Wang Y, Karungu S, Waruhiu C, Zohaib A, Obanda V, et al. Metagenomic virome analysis of culex mosquitoes from Kenya and China. Viruses. 2018;10:30. doi: 10.3390/v10010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scola BL, Audic S, Robert C, Jungang L, Lamballerie X, de, Drancourt M, et al. A giant virus in amoebae. Science. 2003;299:2033.. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 10.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–50. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 11.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–6. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 12.Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci USA. 2015;112:E5327–35. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA. 2014;111:4274–9. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson WH, Schroeder DC, Allen MJ, Holden MTG, Parkhill J, Barrell BG, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–2. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 15.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA. 2010;107:19508–13. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci USA. 2013;110:10800–5. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallot-Lavallée L, Blanc G, Claverie J-M. Comparative genomics of Chrysochromulina ericina virus and other microalga-infecting large DNA viruses highlights their intricate evolutionary relationship with the established Mimiviridae family. J Virol. 2017;91:e00230–17. doi: 10.1128/JVI.00230-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deeg CM, Chow C-ET, Suttle CA. The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. 2017;7: e33014. 10.7554/eLife.33014. [DOI] [PMC free article] [PubMed]
- 19.Sheyn U, Rosenwasser S, Lehahn Y, Barak-Gavish N, Rotkopf R, Bidle KD, et al. Expression profiling of host and virus during a coccolithophore bloom provides insights into the role of viral infection in promoting carbon export. ISME J. 2018;12:704–13. doi: 10.1038/s41396-017-0004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie J-M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci USA. 2011;108:17486–91. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, et al. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol Evol. 2012;4:1324–30. doi: 10.1093/gbe/evs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoosuf N, Pagnier I, Fournous G, Robert C, La Scola B, Raoult D, et al. Complete genome sequence of Courdo11 virus, a member of the family Mimiviridae. Virus Genes. 2014;48:218–23. doi: 10.1007/s11262-013-1016-x. [DOI] [PubMed] [Google Scholar]
- 23.Claverie J-M, Abergel C. Mimiviridae: an expanding family of highly diverse large dsDNA viruses infecting a wide phylogenetic range of aquatic eukaryotes. Viruses. 2018;10:E506. doi: 10.3390/v10090506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, et al. Giant viruses with an expanded complement of translation system components. Science. 2017;356:82–5. doi: 10.1126/science.aal4657. [DOI] [PubMed] [Google Scholar]
- 25.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, et al. The virophage as a unique parasite of the giant Mimivirus. Nature. 2008;455:100–4. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 26.Desnues C, La Scola B, Yutin N, Fournous G, Robert C, Azza S, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci USA. 2012;109:18078–83. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaia M, Pagnier I, Campocasso A, Fournous G, Raoult D, La Scola B. Broad spectrum of mimiviridae virophage allows its isolation using a Mimivirus reporter. PloS One. 2013;8:e61912. doi: 10.1371/journal.pone.0061912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaia M, Benamar S, Boughalmi M, Pagnier I, Croce O, Colson P, et al. Zamilon, a novel virophage with Mimiviridae host specificity. PloS One. 2014;9:e94923. doi: 10.1371/journal.pone.0094923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupovic M, Kuhn JH, Fischer MG. A classification system for virophages and satellite viruses. Arch Virol. 2016;161:233–47. doi: 10.1007/s00705-015-2622-9. [DOI] [PubMed] [Google Scholar]
- 30.Bekliz Meriem, Colson Philippe, La Scola Bernard. The Expanding Family of Virophages. Viruses. 2016;8(11):317. doi: 10.3390/v8110317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–4. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 32.Fischer MG, Hackl T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature. 2016;540:288–91. doi: 10.1038/nature20593. [DOI] [PubMed] [Google Scholar]
- 33.Levasseur A, Bekliz M, Chabrière E, Pontarotti P, La Scola B, Raoult D. MIMIVIRE is a defence system in Mimivirus that confers resistance to virophage. Nature. 2016;531:249–52. doi: 10.1038/nature17146. [DOI] [PubMed] [Google Scholar]
- 34.Claverie J-M, Abergel C. CRISPR-Cas-like system in giant viruses: why MIMIVIRE is not likely to be an adaptive immune system. Virol Sin. 2016;31:193–6. doi: 10.1007/s12250-016-3801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougari S, Abrahao J, Oliveira GP, Bou Khalil JY, La Scola B. Role of the R349 gene and its repeats in the MIMIVIRE defense system. Front Microbiol. 2019;10:1147. doi: 10.3389/fmicb.2019.01147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabre E, Jeudy S, Santini S, Legendre M, Trauchessec M, Couté Y, et al. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat Commun. 2017;8:15087. doi: 10.1038/ncomms15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 38.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–37. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl_2):ii215–25. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 40.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–3. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinforma Oxf Engl. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace IM, O’Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–9. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–6. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 47.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 48.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 49.Shin J-B, Krey JF, Hassan A, Metlagel Z, Tauscher AN, Pagana JM, et al. Molecular architecture of the chick vestibular hair bundle. Nat Neurosci. 2013;16:365–74. doi: 10.1038/nn.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga Mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abrahão J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun. 2018;9:749. doi: 10.1038/s41467-018-03168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrade ACDSP, Rodrigues RAL, Oliveira GP, Andrade KR, Bonjardim CA, La Scola B, et al. Filling knowledge gaps for Mimivirus entry, uncoating, and morphogenesis. J Virol. 2017;91:e01335–17. doi: 10.1128/JVI.01335-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claverie J-M, Abergel C. Mimivirus and its virophage. Annu Rev Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 54.Yoosuf N, Pagnier I, Fournous G, Robert C, Raoult D, La Scola B, et al. Draft genome sequences of Terra1 and Terra2 viruses, new members of the family Mimiviridae isolated from soil. Virology. 2014;452–453:125–32. doi: 10.1016/j.virol.2013.12.032. [DOI] [PubMed] [Google Scholar]
- 55.Krupovic M, Yutin N, Koonin EV. Fusion of a superfamily 1 helicase and an inactivated DNA polymerase is a signature of common evolutionary history of Polintons, polinton-like viruses, Tlr1 transposons and transpovirons. Virus Evol. 2016;2:vew019. doi: 10.1093/ve/vew019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrne D, Grzela R, Lartigue A, Audic S, Chenivesse S, Encinas S, et al. The polyadenylation site of Mimivirus transcripts obeys a stringent “hairpin rule”. Genome Res. 2009;19:1233–42. doi: 10.1101/gr.091561.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Legendre M, Audic S, Poirot O, Hingamp P, Seltzer V, Byrne D, et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–74. doi: 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Legendre M, Santini S, Rico A, Abergel C, Claverie J-M. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol J. 2011;8:99. doi: 10.1186/1743-422X-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Priet S, Lartigue A, Debart F, Claverie J-M, Abergel C. mRNA maturation in giant viruses: variation on a theme. Nucleic Acids Res. 2015;43:3776–88. doi: 10.1093/nar/gkv224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novick RP. Extrachromosomal inheritance in bacteria. Bacteriol Rev. 1969;33:210–63. doi: 10.1128/br.33.2.210-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–95. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcillán-Barcia MP, de la Cruz F. Why is entry exclusion an essential feature of conjugative plasmids? Plasmid. 2008;60:1–18. doi: 10.1016/j.plasmid.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 63.Humbert M, Huguet KT, Coulombe F, Burrus V. Entry exclusion of conjugative plasmids of the IncA, IncC and related untyped incompatibility groups. J Bacteriol. 2019;201:e00731–18. doi: 10.1128/JB.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donnelly-Wu MK, Jacobs WR, Hatfull GF. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–17. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 65.Susskind MM, Wright A, Botstein D. Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium. IV. Genetics and physiology of sieB exclusion. Virology. 1974;62:367–84. doi: 10.1016/0042-6822(74)90399-7. [DOI] [PubMed] [Google Scholar]
- 66.McGrath S, Fitzgerald GF, van Sinderen D. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol Microbiol. 2002;43:509–20. doi: 10.1046/j.1365-2958.2002.02763.x. [DOI] [PubMed] [Google Scholar]
- 67.Mahony J, McGrath S, Fitzgerald GF, van Sinderen D. Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl Environ Microbiol. 2008;74:6206–15. doi: 10.1128/AEM.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun X, Göhler A, Heller KJ, Neve H. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology. 2006;350:146–57. doi: 10.1016/j.virol.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Borges IA, de AssisFL, LKDS Silva, Abrahão J. Rio Negro virophage: sequencing of the near complete genome and transmission electron microscopy of viral factories and particles. Braz J Microbiol Publ Braz Soc Microbiol. 2018;49(Suppl 1):260–1. doi: 10.1016/j.bjm.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parratt SR, Laine A-L. The role of hyperparasitism in microbial pathogen ecology and evolution. ISME J. 2016;10:1815–22. doi: 10.1038/ismej.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duponchel S, Fischer MG. Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses. PLoS Pathog. 2019;15:e1007592. doi: 10.1371/journal.ppat.1007592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108:6163–8. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkins D, Yau S, Williams TJ, Allen MA, Brown MV, DeMaere MZ, et al. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol Rev. 2013;37:303–35. doi: 10.1111/1574-6976.12007. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J, Sun D, Childers A, McDermott TR, Wang Y, Liles MR. Three novel virophage genomes discovered from Yellowstone Lake metagenomes. J Virol. 2015;89:1278–85. doi: 10.1128/JVI.03039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The annotated genomic sequences determined for this work have been deposited in the Genbank/EMBL/DDBJ database under the following accession numbers: M. vitis: MG807319, M. australiensis: MG807320, M. maliensis: MK978772, Z. vitis: MG807318, mvtv: MG807316 and matv, MG807317. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009037 [75].