Abstract
To visualize protein-protein interactions in Candida albicans with the bimolecular fluorescence complementation (BiFC) approach, we created a Tet-on system with the plasmids pWTN1 and pWTN2. Both plasmids bear a hygromycin B-resistant marker (CaHygB) that is compatible with the original Tet-on plasmid pNIM1, which carries a nourseothricin-resistant marker (CaSAT1). By using GFPmut2 and mCherry as reporters, we found that the two complementary Tet-on plasmids act synergistically in C. albicans with doxycycline in a dose-dependent manner and that expression of the fusion proteins, CaCdc11-GFPmut2 and mCherry-CaCdc10, derived from this system, is septum targeted. Furthermore, to allow detection of protein-protein interactions with the reassembly of a split fluorescent protein, we incorporated mCherry into our system. We generated pWTN1-RN and pNIM1-RC, which express the N-terminus (amino acids 1–159) and C-terminus (amino acids 160–237) of mCherry, respectively. To verify BiFC with mCherry, we created the pWTN1-CDC42-RN (or pWTN1-RN-CDC42) and pNIM1-RC-RDI1 plasmids. C. albicans cells containing these plasmids treated with doxycycline co-expressed the N- and C-terminal fragments of mCherry either N-terminally or C-terminally fused with CaCdc42 and CaRdi1, respectively, and the CaCdc42-CaRdi1 interaction reconstituted a functional form of mCherry. The establishment of this Tet-on-based BiFC system in C. albicans should facilitate the exploration of protein-protein interactions under a variety of conditions.
Subject terms: Expression systems, Fungal genetics
Introduction
Protein-protein interactions (PPIs) are a vital mechanism used to mediate a variety of cellular functions in many physiological environments. In the past, several biochemical strategies for the identification and characterization of these PPIs have been developed, such as affinity chromatography, co-immunoprecipitation, cross-linking and the pull-down assay1. In addition to biochemical methods, protein-fragment complementation methods, including the yeast two-hybrid method2, the split-ubiquitin system3 and bimolecular fluorescence complementation (BiFC)4, allow the direct analysis of protein interactions in vivo. BiFC invol ves the reassembly of a fluorescent complex through two non-fluorescent fragments fused to partners that interact5. PPIs in cells can be observed easily using fluorescent microscopy. The detection of PPIs through Förster resonance energy transfer (FRET), like BiFC, is another approach for the visualization of PPIs in living cells that uses two fluorophores, such as cyan and yellow fluorescent proteins (CFP and YFP), linked to the proteins of interest6. In FRET, light energy emitted by the excitation of a CFP fusion protein that is transferred to and excites a YFP fusion protein upon association of interacting fusion proteins7. The benefit of FRET over BiFC is that FRET allows the detection of the dynamic PPIs owing to its reversibility. However, BiFC allows the direct visualization of PPIs to be irreversible in living cells under a fluorescent microscope, eliminating cellular fixation. BiFC has been widely used in many model organisms from yeasts to mammals and plants8–11 to achieve the genome-wide screening for unknown interactions12,13, which cannot be accomplished with FRET.
Candida albicans, an opportunistic fungal pathogen that serves as a commensal on the mucosal surfaces of the gastrointestinal tract, is capable of developing pathogenesis in immunocompromised patients. Morphological transitions to adapt to environmental changes are related to a series of cellular processes that mediate blastopore formation as well as yeast, pseudohyphal or hyphal growth, which is closely linked to the virulence of C. albicans. Due to the incomplete sexual cycles and diploidy of C. albicans in nature, many genetic tools have been developed to study its cell biology14,15. The tetracycline-inducible gene expression system, which is composed of reverse tetracycline transactivator (rtTA) and a tetracycline response element, is a useful tool in C. albicans that allows gene expression to be completely dependent on the presence of tetracycline or its structural analogue doxycycline16,17. Introduction of dominant selectable markers to replace auxotrophic selectable markers is a desirable approach to facilitate the functional study of a gene in C. albicans without interference from nutrient metabolism18–20.
In this study, we improved the Tet-on system with the addition of two dominant selectable markers, CaHygB21 and CaSAT1, for gene manipulation in the wild-type SC5314 strain through integration of the CaADH1 and CaTDH3 loci. Next, we constructed CaCdc11-GFPmut2 and mCherry-CaCdc10 fusion proteins and verified their co-localization to the septin ring structure. Finally, we established an mCherry-based BiFC assay in C. albicans to detect PPIs using CaCdc42 and CaRdi1, which are involved in polarized growth8,22,23. The BiFC system is an effective visual assay to detect PPIs in wild-type C. albicans strains.
Methods
Strains and growth conditions
C. albicans SC531424 was used as the wild-type strain in this study. The wild-type strain and all derivative strains listed in Table 1 were routinely grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) medium adjusted to pH 5.4 supplemented with the appropriate antibiotics and on solid plates containing 2% agar. The antibiotics used were hygromycin B (1 mg/ml, Gold Biotechnology, USA) and nourseothricin (200 µg/ml, Werner Bioagents, Germany). Transformants were selected on YPD agar containing nourseothricin, hygromycin B or both. To induce expression of fluorescent proteins, strains were diluted in YPD containing doxycycline (40 µg/ml, D9891, Sigma) and grown overnight for 3 hours. The concentration of doxycycline was two-fold diluted from 40 to 0.1 µg/ml to test the effect of different doxycycline doses. For cellular morphology-specific growth, cells were grown in YPD overnight and diluted 1:10 in fresh YPD at 30 °C (for yeast cells) or YPD with 10% fetal bovine serum (FBS, HyClone) for growth at 37 °C (for hyphal cells) for 3 hours.
Table 1.
C. albicans strains used in this study.
| Strain | Parent | Genotype | Plasmidsa | Protein expresseda | Reference |
|---|---|---|---|---|---|
| SC5314 | Wild-type strain | (Gillum et al.24) | |||
| WCL101 | SC5314 | ADH1/adh1::PTET-GFPmut2:SAT1 | pNIM1-NC | GFPmut2 | This study |
| WCL102 | SC5314 | TDH3/tdh3::PTET-GFPmut2:HygB | pWTN1 | GFPmut2 | This study |
| WCL103 | SC5314 | TDH3/tdh3::PTET-mCherry:HygB | pWTN2 | mCherry | This study |
| WCL104 | WCL101 | ADH1/adh1::PTET-GFPmut2:SAT1 TDH3/tdh3::PTET-GFPmut2:HygB | pWTN1 & pNIM1-NC | GFPmut2 | This study |
| WCL105 | WCL102 | ADH1/adh1::PTET-GFPmut2:SAT1 TDH3/tdh3::PTET-mCherry:HygB | pWTN2 & pNIM1-NC | mCherry & GFPmut2 | This study |
| WCL106 | SC5314 | TDH3/tdh3::PTET -CDC42-mCherry (1-159):HygB | pWTN1-6H-CDC42-RN | Cdc42-RN | This study |
| WCL107 | SC5314 | ADH1/adh1::PTET -mCherry (160-237) -RDI1:SAT1 | pNIM1-RC-RDI1 | RC-Rdi1 | This study |
| WCL108 | WCL106 | TDH3/tdh3::PTET -CDC42-mCherry (1-159):HygB ADH1/adh1::PTET -mCherry (160-237) -RDI1:SAT1 | pWTN1-6H-CDC42-RN & pNIM1-RC-RDI1 | Cdc42-RN & RC-Rdi1 | This study |
| WCL109 | SC5314 | TDH3/tdh3::PTET -mCherry (1-159):HygB | pWTN1-RN | RN | This study |
| WCL110 | SC5314 | ADH1/adh1::PTET -mCherry (160-237):SAT1 | pNIM1-RC | RC | This study |
| WCL111 | WCL109 | TDH3/tdh3::PTET -mCherry (1-159):HygB ADH1/adh1::PTET -mCherry (160-237):SAT1 | pWTN1-RN & pNIM1-RC | RN & RC | This study |
| WCL112 | WCL109 | TDH3/tdh3::PTET -mCherry (1-159):HygB ADH1/adh1::PTET -mCherry (160-237) -RDI1:SAT1 | pWTN1-RN & pNIM1-RC-RDI1 | RN & RC-Rdi1 | This study |
| WCL113 | WCL106 | TDH3/tdh3::PTET -CDC42-mCheery (1-159):HygB ADH1/adh1::PTET -mCherry (160-237):SAT1 | pWTN1-6H-CDC42-RN & pNIM1-RC | Cdc42-RN & RC | This study |
| WCL114 | SC5314 | TDH3/tdh3::PTET-mCherry-CDC10:HygB | pWTN2-mCherry-CDC10 | mCherry-Cdc10 | This study |
| WCL115 | SC5314 | ADH1/adh1::PTET-CDC11-GFPmut2:SAT1 | pNIM1-CDC11-GFPmut2 | Cdc11-GFPmut2 | This study |
| WCL116 | WCL114 | TDH3/tdh3::PTET-mCherry-CDC10:HygB ADH1/adh1::PTET-CDC11-GFPmut2:SAT1 | pWTN2-mCherry-CDC10 & pNIM1-CDC11-GFPmut2 | mCherry-Cdc10 & Cdc11-GFPmut2 | This study |
| WCL117 | SC5314 | TDH3/tdh3::PTET -mCherry (1-159)-CDC42:HygB | pWTN1-RN-CDC42 | RN-Cdc42 | This study |
| WCL118 | WCL117 | TDH3/tdh3::PTET -mCherry (1-159)-CDC42:HygB ADH1/adh1::PTET -mCherry (160-237) -RDI1:SAT1 | pWTN1-RN-CDC42 & pNIM1-RC-RDI1 | RN-Cdc42 & RC-Rdi1 | This study |
| WCL125 | WCL117 | TDH3/tdh3::PTET -mCherry (1-159)-CDC42:HygB ADH1/adh1::PTET -mCherry (160-237):SAT1 | pWTN1-RN-CDC42 & pNIM1-RC | RN-Cdc42 & RC | This study |
aRN indicated as mCherry (1–159) and RC indicated as mCherry (160–237).
Construction of plasmids
All primers used in this study are listed in Table 2. Details of the construction of plasmids used in this study are described in the Supplemented Materials and Methods.
Table 2.
Synthetic oligonucleotides used in this study.
| Primer | Sequenceb |
|---|---|
| TDH3p SacII F | ATTACCGCGGGTTGCTCCTCGTCGACAA |
| TDH3p XbaI R | CGGGTCTAGACATTGTTAATTAATTTGATTGTAAAG |
| MCS3 F | CTAGATTTAAATATGCATTGACCGCGGATTCAAGAGTCCAAGCT |
| MCS3 R | TGGACTCTTGAATCCGCGGTCAATGCATATTTAAAT |
| MCS4 F | TGCACCAGCTCCGGTACCACTAGTCATGTTTAAACCGG |
| MCS4 R | TCGACCGGTTTAAACATGACTAGTGGTACCGGAGCTGGTGCAGTAC |
| HygB BglII F | GCTTAGATCTCCTGCAGGTATAGTGCTTGCTGTTCGATA |
| HygB NsiI R | CGTTATGCATACGCGTCATTTTATGATGGAATGAATGGG |
| TetO F | AAACTCGAGTTTACCACTCCCTATCAGTGATAGAGAAAAGTGAAAGTCGACATTGAGCT |
| TetO R | CAATGTCGACTTTCACTTTTCTCTATCACTGATAGGGAGTGGTAAACTCGAGTTT |
| miniOP4 SpeI F | GATTACTAGTCATTATTTATATTTGTATGTGTGTAGGAGTT |
| miniOP4 XhoI R | AGGACTCGAGCAAACTTCTTTTTTATTTTTCGCAATAT |
| TDH3t PmeI F2 | AAAAGTTTAAACACCAAACTCGAAGGTGCTC |
| TDH3t KpnI R2 | GATTGGTACCTCTGGTGGAGTAACCGTATT |
| GFPt XbaI F | TTTTCTAGACCGGTCCGACATTTTATGATGGAATGAATGG |
| GFPt SpeI R | TTTACTAGTGTCGACTCGAGATATCCAGCTA |
| mRFP NotI F | ATTTGCGGCCGCTCCGGAGTTTCAAAAGGTGA |
| mRFP AatII R | CATAGACGTCAGGCGCGCCTTTATATAATTCA |
| GS F | ATCAGATCTGGTGGAGGTG |
| GS R | GATCCACCTCCACCAGATCTGAT |
| mRFP N BamHI F | AACTGGATCCATGGTTTCAAAAGGTGAAGAAGATAATATGGC |
| mRFP N NotI R | TCTAGCGGCCGCAGGCCTTTAATCTTCTGGATACATTCTTTCTGATGAAGC |
| mRFP C XhoI F | TCAACTCGAGATGGGTGCTTTAAAAGGTGAAATTAAACAAAGA |
| mRFP C BamHI R | TACTGGATCCTTTATATAATTCATCCATACCACCAGTTGA |
| CaGFP-N-MCS | AATTGTCGACTCGAGATATCCAGCTAGCGGCCGCGTGACCATGAGTAAGGGAGAAGAAC |
| CaGFP-C-MCS | CCGTGATCATTATGCAGGCCTAGATCTTAAGCTGACGTCCGGACCTTTGTATAGTTCATCCATGCC |
| CDC42 EcoRV F | CCTAGATATCCAAACTATAAAATGTGTTGTTGTCGGTG |
| CDC42 BglII R | GGGTAGATCTTAAAATAGTACACTTTTTCGATTTTTTAAT |
| RDI1 SpeI F | GATTACTAGTTCCAATTCTAACGTTGATGATGAT |
| RDI1 StuI R | GCTTAGGCCTCTATTTAGTAATGGAAAAACTCCAAGG |
| RedN KpnI F | CCAAGGTACCACTAGTATGGTTTCAAAAGGTGAAGAAGATAATATGGC |
| RedN EcoRV | CTTTGATATCATCTTCTGGATACATTCTTTCTGATGAAGC |
| CaCDC42 BamHI F | ATAAGGATCCGACGTCATGCAAACTATAAAATGTGTTGTTGTCG |
| CaCDC42 NotI R | ATAAGCGGCCGCAGGCCTCTATAAAATAGTACACTTTTTCGATTTTTTAAT |
| pNIM1 inte F | CATGTCAAAGGATTCAAC |
| pNIM1 inte R | GTATGGTGCCTATCTAAC |
| TDH3 F | TTTGGTTGCGTTAGTCCG |
bRestriction enzyme sites included in primers are highlighted in bold italics.
C. albicans transformation
Each of the strains was made by introducing the appropriate cassette via electroporation essentially as described25 with some previously described modifications20. The plasmid cassettes were excised as KpnI/SacII-digested fragments and extracted with a Gel/PCR DNA Fragments Extraction Kit (Geneaid, Taiwan). One microgram of linear DNA fragments was mixed with 50 μl of competent cells and electroporated in a 0.2 cm cuvette at 1.8 kV via the Gene Pulser Xcell™ system (Bio-Rad, USA). After electroporation, the competent cells were resuspended in 1 ml of ice-cold 1 M sorbitol and transferred to a collection tube. The cells were collected by centrifugation at 4000 r.p.m. and 4 °C and resuspended in 1 ml of YPD medium before incubation at 30 °C for 4 hours. Finally, the cells were plated onto YPD plates supplemented with the appropriate antibiotics and incubated at 30 °C for 1~2 days. The transformants were selected after screening by colony PCR (Supplemental Materials and Methods).
Protein extraction and western blotting
Cells cultured after doxycycline induction were pelleted via centrifugation at 5000 r.p.m. and washed with PBS twice before protein extraction essentially as described previously20. The fluorescent proteins GFPmut2 and mCherry were detected with mouse anti-GFP antibody (11814460001, Roche, Germany) and rabbit anti-mCherry antibody (5993–100, BioVision, USA), respectively, according to the manufacturers’ instructions. β-Actin was used as a loading control and detected with rabbit anti-β-actin antibodies (GTX109639, GeneTex, USA). The PageRuler Prestained Protein Ladder (26616, Thermo Fisher Scientific, USA) was used to indicate the relevant sizes of the protein targets. Signals from the detected proteins were obtained with SuperSignal West Pico Chemiluminescent Substrate (34080, Thermo Fisher Scientific, USA), and images were captured with an ImageQuant LAS 4000 Mini system (GE Healthcare Life Sciences). To quantify the levels of GFPmut2 and mCherry on the blots, images were analysed in triplicate with ImageJ (National Institutes of Health, USA), and then the quantified GFPmut2 levels were normalized to those of β-actin to determine the fold change in expression. Differential expression data were assessed by one-way ANOVA with Bonferroni’s comparison test, and the data were plotted as bar charts by GraphPad Prism 6 (GraphPad Software, USA).
Fluorescence microscopy
In general, C. albicans cells were grown at 30 °C overnight, diluted 1:10 into their respective media containing 40 μg/ml doxycycline, and grown for an additional 3 hours at a temperature specific for the yeast and filamentous forms. For the BiFC assay, the cells were grown in YPD medium with 40 μg/ml doxycycline at 30 °C for 6 hours. To prepare each slide, YPD containing 1% agarose was added to a depression slide, and the top of the agarose was flattened with another slide before hardening. Poly-L-lysine-coated slides were prepared by evenly spreading 10 μl of filter-sterilized 0.1% poly-L-lysine (P8920, Sigma) onto a degreased microscope glass slide with a coverslip, which was then air-dried. A few microlitres of cultured cells were gently added onto the surface of the slide and covered with a coverslip before gently pressing the cover slide against the slide and sealing the slides with clear nail polish. Differential interference contrast (DIC) and fluorescent images were captured by a ZEISS Axio Imager A2 microscope (Zeiss, Germany) with an X-Cite 120Q unit (Excelitas, Massachusetts, United States) with a 120 W mercury vapor short arc lamp. Images were captured with a Retiga EXi (Teledyne QImaging, Canada) and FITC and rhodamine filter sets with excitation/emission wavelengths of 488/509 nm for GFPmut2 and 587/610 nm for mCherry. A Plan Neofluar 100×/1.30 oil objective lens combined with a DIC prism slider was used to capture DIC images. The digital images were analysed by QCapture Pro 7 (Teledyne QImaging, Canada) and processed with ImageJ (National Institutes of Health, USA). To quantify the relative fluorescence intensity of the cells in the presence of doxycycline, randomly selected five fluorescence microscopy images from strains, WCL111, WCL112, WCL113, WCL108, WCL125, and WCL118, were analysed. The relative fluorescence intensity was formulated as the integrated intensity of the fluorescence subtracted the fluorescence of background measured by ImageJ. The fluorescence difference was calculated with one-way ANOVA with Bonferroni’s comparison test, and the data were plotted as bar charts by GraphPad Prism 6 (GraphPad Software, USA).
Nucleotide sequence accession numbers
The sequence of the pWTN2 plasmid has been submitted to GenBank with accession number KP087797.
Results
Construction of an additional tetracycline-regulated gene expression system in C. albicans with the hygromycin B-resistance marker
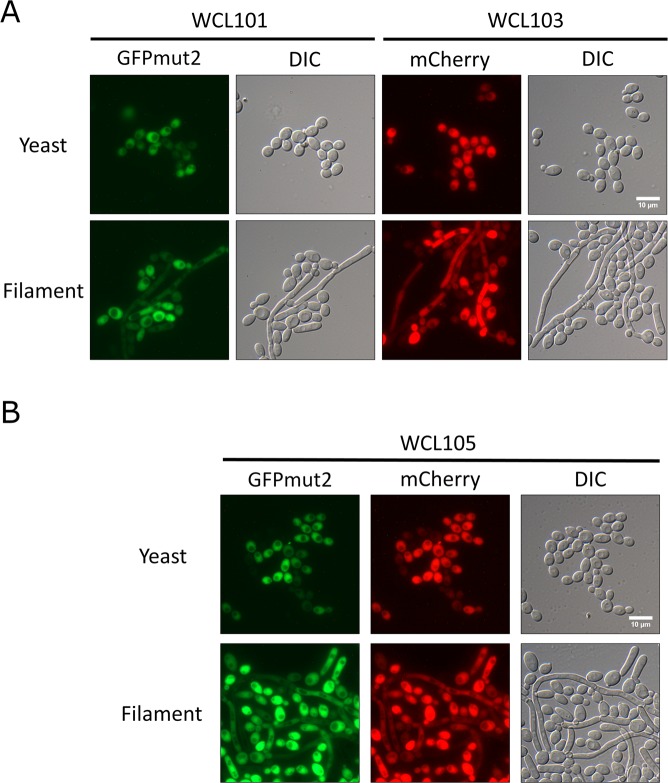
A previous tetracycline-regulated gene expression system using the pNIM1 plasmid, also called the “Tet-on” system, in which the Tet-on cassette was integrated into the CaADH1 locus, was established in C. albicans16. The tetracycline response element (tetO) fused to a minimal CaOP4 promoter and the codon-optimized gene encoding reverse transactivator (rtTA) in the pNIM1 cassette constitute the key components of the Tet-on system. In this system, the nourseothricin-resistant gene CaSAT1 serves as a dominant selection marker, and the homologous region of the CaADH1 locus directs genomic integration of the cassette. That system is a convenient strategy for the expression of a target gene involved in molecular cloning and the subsequent transformation of a restriction enzyme-linearized cassette into wild-type C. albicans. The introduction of Escherichia coli hygromycin B phosphotransferase as an additional dominant selectable marker for hygromycin B resistance has been tested in C. albicans, and the simultaneous presence of hygromycin B and nourseothricin has also been confirmed to be compatible for the selection of C. albicans21. Based on these characteristics, we designed a Tet-on system with two plasmid vectors, pWTN1 and pWTN2, that is capable of gene expression under the control of tetracycline-regulated elements. This system contains a selectable hygromycin B-resistance marker that is flanked with homologous regions of the CaTDH3 locus for its integration (Fig. S1). CaTDH3 encodes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is constitutively expressed in cells and carries out functions associated with the cell wall26; GAPDH is enriched at the stationary phase27, so CaTDH3 was chosen as an alternative locus using its promoter to drive rtTA. To test the function of the new Tet-on system in C. albicans, pWTN1 and pWTN2 were linearized with SacII and KpnI and integrated into the CaTDH3 locus of the wild-type C. albicans SC5314 by transformation, followed by hygromycin B selection; diagnostic colony PCR was then carried out for confirmation (Fig. S2), and the WCL102 and WCL103 strains, respectively, were eventually obtained. The WCL102 strain was capable of expressing GFPmut2, and the WCL103 strain was capable of expressing mCherry, which made the colonies pink in the visual spectrum (Fig. S3). Additionally, the pNIM1-NC plasmid, which was derived from pNIM1, was constructed through the addition of several restriction enzyme sites (Fig. S1). After linearization with KpnI and SacII, the pNIM1-NC plasmid was transformed and integrated at the CaADH1 locus in C. albicans to obtain the WCL101 strain, which was capable of GFPmut2 expression, for use as a control. To assess the compatibility of nourseothricin and hygromycin B in the new system, the pNIM1-NC plasmid was further transformed into the WCL102 and WCL103 strains to obtain the WCL104 and WCL105 strains, respectively, each of which contained either two sets of Tet-on cassettes with two copies of the GFPmut2 coding sequence or two sets of Tet-on cassettes with one copy of the GFPmut2 coding sequence and one copy of the mCherry coding sequence. The doxycycline-inducible expression of GFPmut2 and mCherry in strains WCL101 and WCL103 under yeast and hyphal induction conditions was verified by fluorescence microscopy (Fig. 1A). Additionally, GFPmut2 and mCherry were simultaneously expressed in the WCL105 strain (Fig. 1B). These results indicate that the fluorescent proteins GFPmut2 and mCherry were expressed either individually or concurrently in both the yeast and hyphal forms by the newly constructed Tet-on system.
Figure 1.
Expression of doxycycline-inducible fluorescent proteins in the yeast and filamentous forms of C. albicans. (A) The doxcycline-induced GFPmut2 and mCherry expression in yeast and filamentous forms of C. albicans. C. albicans cells from strains WCL101 and WCL103 were grown to exponential phase and for 3 hours after their transfer to either YPD at 30 °C (yeast) or YPD with 10% FBS at 37 °C (hyphae), both of which contain 40 μg/ml doxycycline. The fluorescent signals from the cells were observed under a fluorescence microscope. The bar indicates 10 μm. (B) Co-expression of GFPmut2 and mCherry induced by doxycycline in yeast and filamentous forms of C. albicans. GFPmut2 and mCherry were co-expressed by doxycycline in C. albicans cells from strain WCL105. The cells were grown in the conditions as in those used in (A). The fluorescent signals from this strain were detected by fluorescent microscopy. The bar indicates 10 μm.
Doxycycline regulates gene expression in a dose- and reverse transactivator-dependent manner
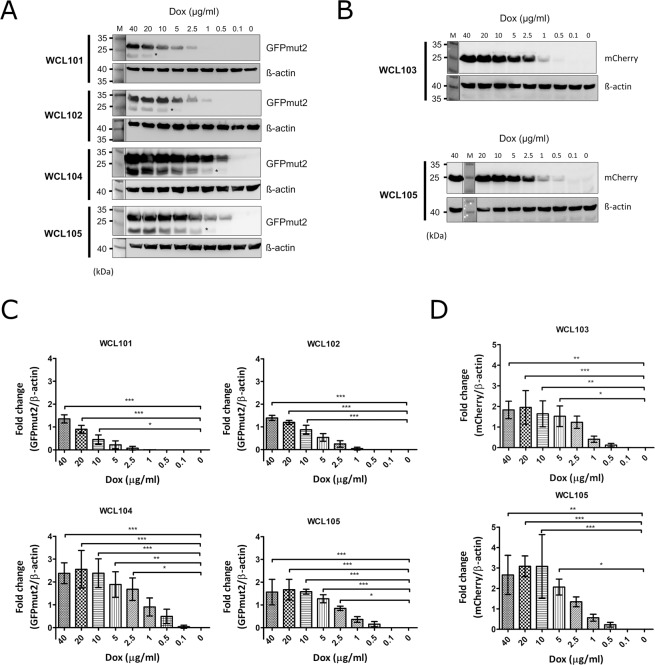
In a previous study, we established an auxotrophic C. albicans strain carrying the Tet-on cassette of the pTET25M plasmid to confirm that the Tet-on system functions in a dose-dependent manner17. As there were two Tet-on systems in C. albicans, the conditions were more complicated than those used for yeast containing one Tet-on system. To determine the strength of gene expression under the two Tet-on systems, the expression of fluorescent proteins was induced in the WCL101, WCL102, WCL104, and WCL105 strains by doxycycline ranging in concentration from 40, 20, 10, 5, 2.5, 1, 0.5, 0.1 to 0 μg/ml and examined by western blotting. Comparison of GFPmut2 expression in the WCL101 and WCL102 strains, in which the individual Tet-on system was integrated at one allele of the CaADH1 or CaTDH3 loci, respectively, revealed that GFPmut2 was expressed in a dose-dependent manner in both strains (Fig. 2A). Additionally, the minimum dose of doxycycline required to induce GFPmut2 expression was different between the WCL101 and WCL102 strains. Through quantitative analysis of western blots performed in triplicate, we observed that the change in GFPmut2 expression was significant in WCL101 cells induced with 10 µg/ml doxycycline and WCL102 cells induced with 5 µg/ml doxycycline (Fig. 2A,C). These results suggested that the CaTDH3 promoter is stronger than the CaADH1 promoter and capable of driving more reverse transactivator (rtTA) expression, leading to the induction of GFPmut2 at a low dose of doxycycline. To better understand the relationship between the level of gene expression and the dose of doxycycline used to induce C. albicans with two Tet-on systems, we compared the amount of GFPmut2 expressed in the WCL101, WCL102, and WCL104 strains. The level of GFPmut2 expression in the WCL104 strain also correlated with the dose of doxycycline and plateaued with induction with 10 to 40 μg/ml doxycycline (Fig. 2A,C). However, the fold change in GFPmut2 expression in the WCL104 strain was as high as 2.5 in the presence of 40 μg/ml doxycycline, which was greater than the approximately 1.5-fold change in GFPmut2 expression in the WCL101 and WCL102 strains (Fig. 2C). The increased GFPmut2 level in the WCL104 strain indicates that the two Tet-on systems had a synergistic effect on target gene expression owing to the two copies of GFPmut2 and the constitutive expression of two rtTA genes. Moreover, the minimal dose of doxycycline required to induce GFPmut2 expression in the WCL104 and WCL105 strains decreased significantly down to 2.5 μg/ml doxycycline (Fig. 2C). A reasonable explanation for this observation is that the increased level of rtTA enriched its binding to doxycycline, leading to the induction of gene expression with a low dose of doxycycline. Although quantitative analysis showed the minimal (optimal) dose of doxycycline required for induction, it appeared that the Tet-on system in the WCL101, WCL102 and WCL105 strains was able to switch on slightly in the presence of doxycycline at 2.5, 1, 0.5, and 0.5 ug/ml, respectively (Fig. 2A). Additionally, to confirm the effects of rtTA on gene expression under two Tet-on systems in an alternative way, we compared the expression of mCherry by using western blotting of the WCL103 and WCL105 strains. As shown in Fig. 2B, mCherry was expressed in the WCL103 and WCL105 strains in a dose-dependent manner, similar to induced GFPmut2 expression in the WCL101 and WCL102 strains. Through quantification assays with statistical analyses (Fig. S4, Tables S1 and S2), the minimal (optimal) dose of doxycycline required to significantly induce mCherry was shown to be 5 μg/ml (Fig. 2D). However, doxycycline at 0.5 μg/ml induced the slight expression of mCherry (Fig. 2B). From these observations, we concluded that the level of rtTA driven by the stronger promoter mainly affects gene expression, differences in the doxycycline dose changed the level of the induced proteins, and the number of target gene copies had a minor effect on the level of the induced gene expression.
Figure 2.
Quantification of the doxycycline-inducible expression level of fluorescent proteins in C. albicans. (A) GFPmut2 was used as an indicator to quantify expression under the Tet-on system in a dose-dependent manner. C. albicans cells from strains WCL101, WCL102, WCL104, and WCL105 were grown in YPD with the indicated concentration of doxycycline for 3 hours, followed by western blotting to analyse the amount of GFPmut2 in the cell lysates. β-Actin was used as an internal control. Asterisks indicate non-specific signals. (B) mCherry was used as another indicator to quantify expression under the Tet-on system in a dose-dependent manner. C. albicans cells from strains WCL103 and WCL105 were grown in YPD with the indicated concentration of doxycycline for 3 hours, and their lysates were subjected to western blotting to analyse the amount of mCherry in the cells. β-Actin was used as an internal control. (C) The change in doxycycline-inducible GFPmut2 expression. The GFPmut2 and β-actin signals on the blots shown in (A) were quantified with ImageJ. The bar charts show the fold change in GFPmut2 expression normalized to the relevant β-actin expression analysed with one-way ANOVA with Bonferroni’s comparison test. (D) The change in doxycycline-induced mCherry expression. The mCherry and β-actin signals on the blots shown in (B). The bar charts were obtained with the same formula used for (C). The numbers of asterisks show the significance of the difference between two values compared by statistical analysis.
Tracing the location of a structural protein in septin via two Tet-on systems
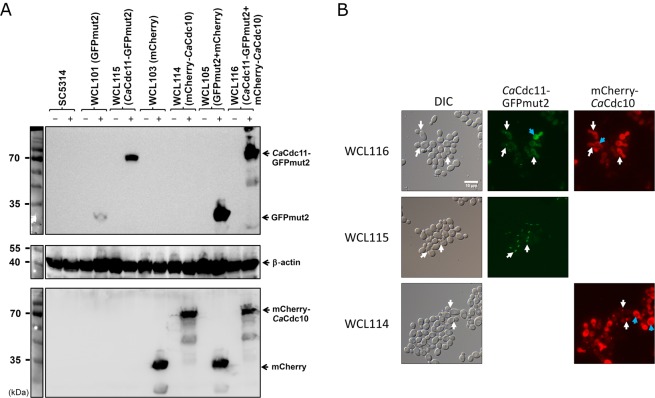
The septin family of proteins in C. albicans, including CaCdc3, CaCdc10, CaCdc11, CaCdc12, CaSep7, CaSpr3, and CaSpr28, are highly conserved cytoskeletal proteins with functions in cytokinesis28. The ring structure formed by septation during budding and filamentous growth is composed of four septin proteins, CaCdc3, CaCdc10, CaCdc11, and CaCdc12, and directs proper bud formation29. To use the fluorescent protein as a signal to label the ring during budding in C. albicans, the septin proteins CaCdc10 and CaCdc11 were incorporated into CaCdc11-GFPmut2 and mCherry-CaCdc10 fusion proteins whose expressions were regulated under the pWTN2 and pNIM1-NC Tet-on systems (Fig. S5), respectively. Cells in the WCL114 and WCL115 strains were capable of expressing the mCherry-CaCdc10 and CaCdc11-GFPmut2 fusion proteins, respectively. These cells were grown in YPD containing 40 µg/ml doxycycline, and the presence of mCherry-CaCdc10 and CaCdc11-GFPmut2 in the WCL114 and WCL115 strains was examined by western blotting (Fig. 3A). The induced expression and location of CaCdc11-GFPmut2 and mCherry-CaCdc10 in the cells after induction were assessed in parallel by the presence of a fluorescent signal at the septin ring by fluorescence microscopy. Focal fluorescence from both the GFPmut2 fusion and mCherry fusion proteins was observed, even though fluorescence was observed in the cytoplasm, particularly in cells from the WCL114 strain, due to the expression of rtTA driven by the strong CaTDH3 promoter (Fig. 3B). To further test the co-localization of CaCdc11-GFPmut2 and mCherry-CaCdc10 at the septin ring in a cell, two Tet-on systems, pWTN2-mCherry-CDC10 and pNIM1-CDC11-GFPmut2, were incorporated and integrated at the CaTDH3 and CaADH1 loci to create the WCL116 strain. The induced expression of both the CaCdc11-GFPmut2 and mCherry-CaCdc10 fusion proteins in the presence of doxycycline was verified by western blotting (Fig. 3A). The colocalisation of GFPmut2 and mCherry at the septin ring detected by fluorescent microscopy (Fig. 3B) suggested that neither GFPmut2 nor mCherry influences the targeting of CaCdc11 and CaCdc10 to the septin ring. However, the high level of GFPmut2 and mCherry expression under the two Tet-on systems resulted in cytoplasmic diffusion of the cells (Fig. 3B, blue arrows). There are two possible explanations for this observation. One is the maintenance of homeostasis of the dynamic proteome through degradation processes, such as the ubiquitin-proteasome pathway30. After the degradation of CaCdc11-GFPmut2 and mCherry-CaCdc10, residual GFPmut2 and mCherry remained in the cytoplasm. As shown in Fig. 3A, some smearing in the samples from the WCL114 and WCL116 strains was observed, suggesting that degradation occurred. The other explanation for the observed cytoplasmic diffusion is that overexpressed CaCdc11-GFPmut2 and mChery-CaCdc10 accumulated in or co-localised at vacuoles (Fig. 3B, blue arrows). Because septins participate in the biogenesis of autophagosomes in yeast31, the overexpression of CaCdc10 and CaCdc11 could have prevented their correct localization. These results helped us to understand that gene overexpression under the Tet-on system not only exerts targeting effects but also brings about side effects due to the high dose of doxycycline (40 μg/ml).
Figure 3.
Expression of the doxycycline-induced CaCdc11-GFPmut2 and mCherry-CaCdc10 fusion proteins localized at the septum of C. albicans. (A) Examination of the expression of CaCdc11-GFPmut2 and mCherry-CaCdc10 in the presence of doxycycline by western blotting. C. albicans cells from strains WCL114, WCL115, and WCL116 were grown in YPD with (+) or without (−) 40 µg/ml doxycycline for 3 hours. The expression of GFPmut2 or/and mCherry in strains WCL101, WCL103, and WCL105 was used as a control. (B) CaCdc11-GFPmut2 and mCherry-CaCdc10 were localized at the septum of strains WCL115 and WCL116, respectively. Co-localisation of mCherry-CaCdc10 and CaCdc11-GFPmut2 was visualized at the septum of the yeast form under a fluorescence microscope. White arrows indicate areas of fluorescent fusion at the septum. Blue arrows indicate the part of fluorescent fusion in vacuoles. The bar indicates 10 μm.
Establishment of a tetracycline-regulated split-mCherry for the BiFC assay in C. albicans
As the CUG codon is noncanonical in C. albicans, the CUG codons in exogenous reporter genes, such as fluorescent proteins and luciferase, must be optimized for their protein expression32,33. With regards to the use of a fluorescent protein, a red fluorescent protein variant, mCherry, was developed for use in the budding yeasts Saccharomyces cerevisiae and C. albicans, both of which surprisingly generate pink colonies (Fig. S3)34. The mCherry protein, a yeast-enhanced monomeric red fluorescent protein (yEmRFP), is an assembly of split amino acid fragments containing amino acids 1–159 (RN, the N-terminal fragment of mCherry) and 160–237 (RC, the C-terminal fragment of mCherry)22, which is similar to the split fragments of EGFP (amino acid sequence 1–157/158–238). Based on its protein complementation, monomeric structure and the formation of pink colonies, we used mCherry as a candidate fluorescence protein for the BiFC assay in C. albicans. To transfer the split fragments of mCherry into the pNIM1-NC and pWTN2 plasmids, intermediate cassettes consisting of the GFPmut2 coding sequence linked to the coding sequence of amino acids 160–237 of mCherry via the DNA encoding a triple GGGGS linker and the coding sequence of amino acids 1–159 of mCherry connected to the GFPmut2 coding sequence through the DNA encoding a triple GGGGS linker were cloned into pNIM1-NC and pWTN2 to generate pNIM1-RC-MCS and pWTN1-6H-MCS-RN, respectively. (see the Materials and Methods).
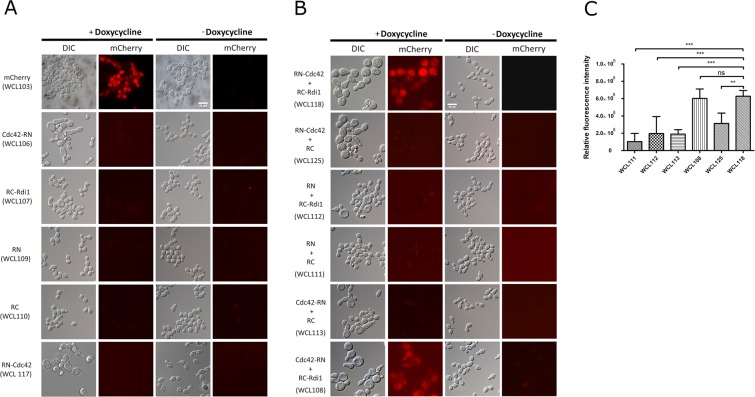
The highly conserved proteins Cdc42 and Rdi1 are known to interact to regulate polarized growth in eukaryotic cells8,23,35,36. These two partners have been applied to the BiFC assay in S. cerevisiae, and a structure of these proteins in complex was resolved via biophysical methods8. By analogy, Cdc42 and Rdi1 were incorporated into the Tet-on-based BiFC system. The coding sequence of CaCDC42 was linked to the sequence encoding the N-terminal amino acids 1–159 of mCherry with a flexible linker containing the triple GGGGS peptide sequence, which was based on the pWTN1-6H-CDC42-RN plasmid (Fig. S6), while the coding sequence of CaRDI1 was connected to a sequence encoding the C-terminal amino acids 160–237 of mCherry, which was based on the pNIM1-RC-RDI1 plasmid (Fig. S6). The C-terminus of Cdc42 interacts with Rdi137. To avoid the interference of interaction between Cdc42 and Rdi1 when the C-terminus of Cdc42 fused with the N-terminus of mCherry derived from the pWTN1-6H-CDC42-RN plasmid was used, we constructed an alternate pWTN1-RN-CDC42 construct (Fig. S6), in which the sequence encoding the N-terminal amino acids 1–159 of mCherry was linked to the coding sequence of CaCDC42 with a flexible linker containing the triple GGGGS peptide sequence. To examine doxycycline-induced protein expression with the Tet-on system by BiFC, strains (Table 1) capable of expressing each single component (WCL106, WCL107, WCL109, WCL110, and WCL117) and two components (WCL108, WCL111, WCL112, WCL113, WCL118, and WCL125) of the Tet-on system were grown in YPD containing 40 µg/ml doxycycline for 3 hours, and protein expression was verified by western blotting (Fig. S7). To detect the red fluorescence and eliminate possible background from the non-fluorescent fragment, strains capable of expressing a single component involved in BiFC (WCL106, WCL107, WCL109, WCL110, and WCL117) were grown in the presence or absence of doxycycline for 6 hours and analysed by fluorescence microscopy (Fig. 4A). As expected, red fluorescence was undetected with and without doxycycline induction in the WCL106, WCL107, WCL109, WCL110 and WCL117 strains, but red fluorescence displayed in the WCL103 capable of expressing mCherry in the presence of doxycycline (Fig. 4A).
Figure 4.
The mCherry-based BiFC system regulated by the Tet-on system in C. albicans. (A) Validation of split mCherry fragments unable to rejoin into a fluorescent complex. C. albicans cells from strains WCL103, WCL106, WCL107, WCL109, WCL110, and WCL117 were grown in YPD with and without 40 μg/ml doxycycline for 6 hours. The cells from strains WCL106, WCL107, and WCL117 increased in size. Red fluorescence in both the presence and absence of doxycycline was undetectable under fluorescent microscopy. The bar indicates 10 μm. (B) Examination of the mCherry-based BiFC system by the interaction of CaCdc42-CaRdi1. C. albicans cells from strains WCL111, WCL112, WCL113, WCL125, WCL108, and WCL118 were grown in YPD with and without 40 μg/ml doxycycline for 6 hours. Red fluorescence in the large cells of strains WCL118 and WCL108 was visualized by fluorescence microscopy. The other strain exhibited a background or undetectable signal. The bar indicates 10 μm. (C) Quantification of relative fluorescence intensity of the strains WCL111, WCL112, WCL113, WCL108, WCL125, and WCL118 with ImageJ. The values of bar charts derived from the integrated density of the original figures (Fig. S8) subtracting the background were analysed with one-way ANOVA with Bonferroni’s comparison test. ns: not statistically significant.
Next, to test the emission of red fluorescence from the BiFC system, strains capable of expressing two components of the BiFC system (WCL108, WCL111,WCL112, WCL113, WCL118, and WCL125) were grown in YPD with or without doxycycline for 6 hours and examined under a fluorescence microscope. Only the cells of strains WCL108 and WCL118 (Cdc42-RN + RC-Rdi1 and RN-Cdc42 + RC-Rdi1, respectively) displayed red fluorescence following growth under doxycycline-induced conditions (Fig. 4B). However, the other strains, WCL125, WCL113, WCL112, and WCL111, exhibited very weak fluorescent signals following growth in the presence of doxycycline (Fig. 4B). The weak fluorescent signal was thought to be background fluorescence because the overexpressed split fragments of mCherry had accumulated in the cells. To validate the formation of fluorescence dependent on the interaction of CaCdc42 and CaRdi1, randomly selected images of 5 fields from the slides on which the strains capable of expressing split-fragments of BiFC under fluorescence microscope were captured (Fig. S8) and quantified the relative fluorescence intensity. By using statistical analysis (Table S3), the relative fluorescence intensity of the strains WCL108 and WCL118 showed a significant difference from the others (Fig. 4C). The unexpected fluorescence occurred in the degraded cells of the strains, likely being the result of the aggregation of the released proteins from the dead cells. Reconstitution of functional mCherry indicated the interaction of the CaCdc42 and CaRdi1 fusion proteins. The reconstituted fluorescence via CaCdc42-CaRdi1 interaction in the WCL118 strain in the presence of doxycycline detected by BiFC was dispersed in the cells. Although Cdc42 and Rdi1 were shown to co-localize in the cytoplasm, at the tips of small buds, and at the mother-bud neck region in the budding yeast38 as an unicellular model, the focal point of the CaCdc42-CaRdi1 interaction in our study was not clear without the use of confocal microscopy. Interestingly, the overexpression of CaCdc42 (WCL113 and WCL125), CaRdi1(WCL112), or both fusion proteins (WCL108 and WCL118) enlarged the cells grown under doxycycline-induced conditions. The phenotypes of these cells resembled those of budding yeast cells overexpressing Cdc42 or Rdi1, in which the cells were increased in size due to the loss of cell polarity39,40. Reconstitution of the split fragments of mCherry in strains WCL118 and WCL108 still occurred in the presence of doxycycline, even if the overexpression of the fusion proteins altered the phenotype. Based on these observations, particularly the results of fluorescence microscopy, we concluded that our newly established BiFC system functions in C. albicans to assay protein-protein interactions.
Discussion
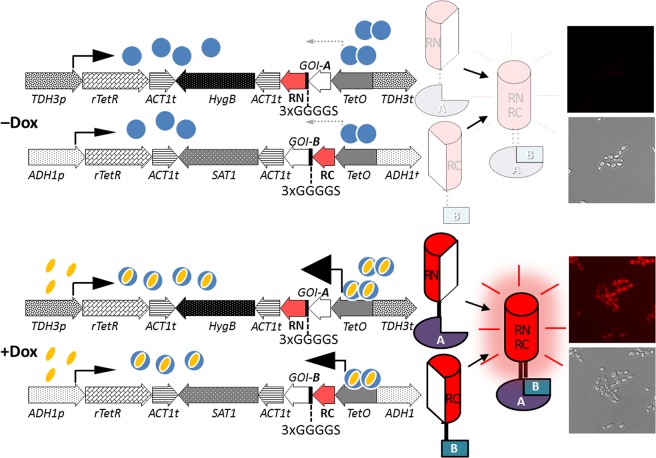
In this study, we took advantage of the dominant selectable marker CaHygB, which comprises the C. albicans codon-optimized coding sequence of hygromycin B phosphotransferase from E. coli driven by the C. albicans TEF2 (CaTEF2) promoter21, and a tetracycline-regulated gene expression system in C. albicans16 to create a new version of the cassette in the form of two plasmids, pWTN1 and pWTN2 (Fig. S1). Each of these plasmids is capable of expressing GFPmut2 or mCherry as a tool for molecular manipulation in C. albicans. Together with the pNIM1-NC plasmid (Fig. S1), which was modified from the pNIM1 plasmid16 carrying the CaSAT1 selection marker for nourseothricin resistance and is capable of expressing GFPmut2, the use of the pWTN2 plasmid (or their derivatives) to generate C. albicans strains confirms that this system, which has the benefit of dominant selection markers, can be used to study the regulation of two proteins simultaneously in cells with diverse morphologies (Fig. 1). Regardless of the identity of the gene regulated by one or two Tet-on systems, GFPmut2 and mCherry were expressed in a dose-dependent manner (Fig. 2). This new system appears to be superior to the current expression system15 for the concurrent dominantly selection and inducible expression of two proteins. A test of the system with the CaCdc10 and CaCdc11 components from the septin family via the production of mCherry-CaCdc10 and CaCdc11-GFPmut2 fusion proteins under tetracycline-regulated promoters showed the co-localization of GFPmut2 and mCherry at the septin ring (Fig. 3). These results indicate the feasibility of introducing the new system to study the function of proteins when localization and imaging are conducted. Furthermore, to extend the new system for the assessment of protein-protein interactions, the BiFC assay was established. A derivative of the DsRed fluorescent protein, mCherry, was divided into two fragments, with one fragment containing amino acids 1–159 and another containing amino acids 160–237, which were generated as two fusion proteins in the Tet-on system22. The well-established interacting proteins CaCdc42 and CaRdi1, which function in polarized growth23, were used to validate the feasibility of BiFC because their homologs in S. cerevisiae have been shown to interact with the BiFC assay8. Indeed, red fluorescence was observed in the WCL108 and WCL118 strains in the presence of doxycycline due to the interaction between CaCdc42 and CaRdi1.
The newly developed Tet-on system functions in a characteristic dose-dependent manner (Fig. 2) and responds immediately to the presence of doxycycline for at most one hour. The regulation of one Tet-on system, with a reverse tetracycline-regulated transactivator (rtTA) and a set of tetracycline response elements, is simple. Because of the constitutive expression of rtTA via two different promoters of CaADH1 and CaTDH3 in these two Tet-on systems, the level of doxycycline-inducible products differs, like the level of GFPmut2 in the WCL101 and WCL102 strains (Fig. 2). The variable strength of the promoters was validated in S. cerevisiae, in which the TDH3 promoter was proven to be stronger than the ADH1 promoter41. This result is consistent with ours, in which the level of GFPmut2 expression was different depending on the promoter (Fig. 2). When rtTA or the dose of doxycycline increases, or the copy number of the target gene is higher, the target protein is expressed at higher levels. Extremely high levels of protein expression result in protein accumulation in the cell, which affects homeostasis42, even though some of the proteins may still retain their normal function in the cell. In the case of mCherry-CaCdc10 and CaCdc11-GFPmut2, mCherry and GFPmut2 were found to colocalise in the cytoplasm and at the septin ring, but their colocalisation was somewhat ambiguous due to the build-up of exceedingly higher amounts of protein in all parts of the cell, which masked focal fluorescence at the septin ring (as in WCL116 in Fig. 3). However, focal fluorescence was easily observed at the septin ring from cells of the WCL114 and WCL115 strains expressing a single fluorescent fusion protein controlled by one Tet-on system, suggesting that induction conditions may need to be optimized in terms of both time and dose to overcome possible functional disturbances of the fusion protein.
In a previous study, S. cerevisiae and C. albicans containing mCherry were developed, and, surprisingly, pink colonies of the derivative strains were observed34. Consequently, we thought that this fluorescent protein could be used in the BiFC assay and applied to detect protein-protein interactions without any fluorescence equipment but with the visible spectrum. However, although the colonies of the WCL108 and WCL118 strains were not pink, red fluorescence was observed. We believe that the structure of the assembled complex is somewhat different from that of the native protein. We used CaCdc42 and CaRdi1 in the BiFC assay in C. albicans as the interruption of CaCdc42 by a cell-penetrating peptide has been shown to inhibit the yeast-to-filament transition43, which can potentially be used as a drug target. Recently, a BiFC system44 based on a set of C. albicans-optimized plasmids containing amino acid biosynthetic genes as selectable markers that uses the C. albicans MET3 promoter to drive expression of split fusion fluorescence proteins was developed. However, this strategy relies on auxotrophic strains for selection, and repression of the repressible MET3 promoter requires methionine and cysteine, which may interfere with or be interfered by the metabolism of the growing cells. Compared with the system mentioned above, our system, in which dominant selection markers and a Tet-on system are used to drive the expression of fusion proteins containing split fragments of fluorescence proteins, is preferential, as shown in a schematic representation (Fig. 5). This newly developed BiFC system can be used not only to assess function but also to conduct genome-wide screening for protein interacting partners in C. albicans. Additionally, once the interaction between two proteins is verified by the BiFC assay, it can be applied further to screen small molecule, peptide, and protein drugs to block their interaction for therapeutic purposes.
Figure 5.
Establishment of tetracycline-induced bimolecular fluorescence complementation to detect protein-protein interaction in Candida albicans. Doxycycline (Dox), a tetracycline analogue, is used to induce gene expression. The sizes of the arrowheads indicate the relative strength of transcriptional activation.
Supplementary information
Acknowledgements
The authors thank Dr. Joachim Morschhäuser (Institut für Molekulare Infektionsbiologie, Universität Würzburg, Würzburg, Germany) for the pNIM1 plasmid, Dr. Brian Wong (Division of Infectious Diseases, Department of Medicine, Oregon Health & Science University, Portland, Oregon, USA) for the pYM70 plasmid and Neta Dean (Department of Biochemistry and Cell Biology, Stony Brook University, Stony Brook, NY, USA) for the CIp-ADH1p-mCherry plasmid. This study was funded by the National Science Council (NSC 101-2629-B-040-001-MY3), which is currently the Ministry of Science and Technology; the Ministry of Science and Technology (MOST 105-2320-B-040-027-MY3 and MOST 108-2320-B-040-009-), Taiwan, R. O. C.; and the National Health Research Institutes of Taiwan, R. O. C. (NHRI-EX100-9808SI).
Author contributions
W.-C.L. designed the study, performed the experiments, analysed the data, and wrote the manuscript. H.S.S. conceived of and designed the study and critiqued the study. J.-C.S. conceived of and designed the study, supervised the project, and wrote the manuscript. All authors have provided their final approval of the manuscript for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59891-7.
References
- 1.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 1995;59:94–123. doi: 10.1128/MMBR.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nat. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 3.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 8.Cole KC, McLaughlin HW, Johnson DI. Use of bimolecular fluorescence complementation to study in vivo interactions between Cdc42p and Rdi1p of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:378–387. doi: 10.1128/EC.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 10.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 11.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant. J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 12.Miller KE, Kim Y, Huh WK, Park HO. Bimolecular Fluorescence Complementation (BiFC) Analysis: Advances and Recent Applications for Genome-Wide Interaction Studies. J. Mol. Biol. 2015;427:2039–2055. doi: 10.1016/j.jmb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonn P, Landegren U. Close Encounters - Probing Proximal Proteins in Live or Fixed Cells. Trends Biochem. Sci. 2017;42:504–515. doi: 10.1016/j.tibs.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Odds FC, Webster CE, Mayuranathan P, Simmons PD. Candida concentrations in the vagina and their association with signs and symptoms of vaginal candidosis. J. Med. Vet. Mycol. 1988;26:277–283. doi: 10.1080/02681218880000391. [DOI] [PubMed] [Google Scholar]
- 15.Xu QR, et al. Molecular genetic techniques for gene manipulation in Candida albicans. Virulence. 2014;5:507–520. doi: 10.4161/viru.28893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai WC, et al. Construction of Candida albicans Tet-on tagging vectors with a Ura-blaster cassette. Yeast. 2011;28:253–263. doi: 10.1002/yea.1833. [DOI] [PubMed] [Google Scholar]
- 18.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staab JF, Sundstrom P. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 2003;11:69–73. doi: 10.1016/S0966-842X(02)00029-X. [DOI] [PubMed] [Google Scholar]
- 20.Lai WC, Sun HF, Lin PH, Ho Lin HL, Shieh JC. A new rapid and efficient system with dominant selection developed to inactivate and conditionally express genes in Candida albicans. Curr. Genet. 2016;62:213–235. doi: 10.1007/s00294-015-0526-6. [DOI] [PubMed] [Google Scholar]
- 21.Basso LR, Jr., et al. Transformation of Candida albicans with a synthetic hygromycin B resistance gene. Yeast. 2010;27:1039–1048. doi: 10.1002/yea.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan JY, et al. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem. Biophys. Res. Commun. 2008;367:47–53. doi: 10.1016/j.bbrc.2007.12.101. [DOI] [PubMed] [Google Scholar]
- 23.Court H, Sudbery P. Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol. Biol. Cell. 2007;18:265–281. doi: 10.1091/mbc.e06-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984;198:179–182. doi: 10.1007/bf00328721. [DOI] [PubMed] [Google Scholar]
- 25.De Backer, M. D. et al. Transformation of Candida albicans by electroporation. Yeast15, 1609–1618, doi:10.1002/(SICI)1097-0061(199911)15:15<1609::AID-YEA485>3.0.CO;2-Y (1999). [DOI] [PubMed]
- 26.Gozalbo D, et al. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 1998;66:2052–2059. doi: 10.1128/IAI.66.5.2052-2059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusch H, et al. A proteomic view of Candida albicans yeast cell metabolism in exponential and stationary growth phases. Int. J. Med. Microbiol. 2008;298:291–318. doi: 10.1016/j.ijmm.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell. 2002;13:2732–2746. doi: 10.1091/mbc.e02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douglas LM, Alvarez FJ, McCreary C, Konopka JB. Septin function in yeast model systems and pathogenic fungi. Eukaryot. Cell. 2005;4:1503–1512. doi: 10.1128/EC.4.9.1503-1512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merlini L, et al. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 2012;8:e1002670. doi: 10.1371/journal.pgen.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barve, G. et al. Septins are involved at the early stages of macroautophagy in S. cerevisiae. J Cell Sci131, 10.1242/jcs.209098 (2018). [DOI] [PMC free article] [PubMed]
- 32.Cormack BP, et al. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology. 1997;143(Pt 2):303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 33.Doyle TC, et al. Expression of firefly luciferase in Candida albicans and its use in the selection of stable transformants. Microb. Pathog. 2006;40:69–81. doi: 10.1016/j.micpath.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Keppler-Ross S, Noffz C, Dean N. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genet. 2008;179:705–710. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chant J. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- 36.Ushinsky SC, et al. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot. Cell. 2002;1:95–104. doi: 10.1128/ec.1.1.95-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richman TJ, et al. Analysis of cell-cycle specific localization of the Rdi1p RhoGDI and the structural determinants required for Cdc42p membrane localization and clustering at sites of polarized growth. Curr. Genet. 2004;45:339–349. doi: 10.1007/s00294-004-0505-9. [DOI] [PubMed] [Google Scholar]
- 39.Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nat. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 40.Richman TJ, Johnson DI. Saccharomyces cerevisiae cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell Biol. 2000;20:8548–8559. doi: 10.1128/mcb.20.22.8548-8559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partow S, Siewers V, Bjorn S, Nielsen J, Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- 42.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Z, Li H, Li Y, Ni F. Inhibition of the pathogenically related morphologic transition in Candida albicans by disrupting Cdc42 binding to its effectors. Chem. Biol. 2007;14:1273–1282. doi: 10.1016/j.chembiol.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Subotic A, et al. A Bimolecular Fluorescence Complementation Tool for Identification of Protein-Protein Interactions in Candida albicans. G3. 2017;7:3509–3520. doi: 10.1534/g3.117.300149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.