Abstract
Symbiont-mediated nutritional mutualisms can contribute to the host fitness of insects, especially for those that feed exclusively on nutritionally unbalanced diets. Here, we elucidate the importance of B group vitamins in the association of endosymbiotic bacteria Wolbachia with two plant-sap feeding insects, the small brown planthopper, Laodelphax striatellus (Fallén), and the brown planthopper, Nilaparvata lugens (Stål). Infected planthoppers of both species laid more eggs than uninfected planthoppers, while the experimental transfer of Wolbachia into uninfected lines of one planthopper species rescued this fecundity deficit. The genomic analysis showed that Wolbachia strains from the two planthopper species encoded complete biosynthesis operons for biotin and riboflavin, while a metabolic analysis revealed that Wolbachia-infected planthoppers of both species had higher titers of biotin and riboflavin. Furthermore, experimental supplementation of food with a mixture of biotin and riboflavin recovered the fecundity deficit of Wolbachia-uninfected planthoppers. In addition, comparative genomic analysis suggested that the riboflavin synthesis genes are conserved among Wolbachia supergroups. Biotin operons are rare in Wolbachia, and those described share a recent ancestor that may have been horizontally transferred from Cardinium bacteria. Our research demonstrates a type of mutualism that involves a facultative interaction between Wolbachia and plant-sap feeding insects involving vitamin Bs.
Subject terms: Agricultural genetics, Bacterial genetics
Introduction
The intracellular bacteria Wolbachia infect a wide variety of invertebrates [1] and contain many supergroups [2–4], which are remarkably different in their biology and host distribution. Most insect Wolbachia strains, belonging to supergroup A or B [2], are facultative symbionts, with a few exceptions in bed bugs and parasitoid wasps [5, 6]. As parasites, Wolbachia have been known to manipulate host reproduction in order to facilitate their maternal transmission through inducing male-killing, feminization, parthenogenesis, and cytoplasmic incompatibility (CI) [7]. Recently, extensive evidence also shows that Wolbachia can benefit a number of insects as mutualists (reviewed in [8]). For example, Wolbachia can protect arthropod hosts against a variety of pathogens and abiotic stresses [9–11]. Some Wolbachia are also essential for successful egg development, such as in bed bugs, parasitic wasps and collembolan species [5, 6, 12, 13], and some Wolbachia can enhance the fecundity of female host insects [5, 14–16]. For instance, in the parasitoid wasp, Asobara tabida, Wolbachia strain wAtab3 (supergroup A) is required for oogenesis and is obligatory for female reproduction [5]. Wolbachia-infected (WI) females lay significantly more eggs than Wolbachia-uninfected (WU) individuals in Aedes albopictus mosquitoes, Drosophila melanogaster, and Psocoptera booklice [14–16]. However, the molecular mechanisms underlying Wolbachia-mediated fecundity enhancement are not fully elucidated.
For insects that feed exclusively on certain types of diet (such as blood, plant-sap, and grain), insects cannot obtain complete nourishment due to some nutrients (like essential amino acids and most vitamins) being missing from food sources. As a result, those insects rely on symbiotically-mediated supplementation from microbial partners [17]. The vitamin Bs are water-soluble organic micronutrients that function as coenzymes in insects and other animals. The significance of vitamin B supplementation by symbionts has been well established, particularly in blood-feeding insects [18]. For instance, the tsetse fly, Glossina morsitans, relies on their obligatory symbiont, Wigglesworthia, to synthesize pyridoxamine (B6) and folate (B9) [19, 20]. The obligatory symbiont, Francisella, contributes to the fitness of African soft tick, Ornithodoros moubata, by synthesizing biotin (B7), riboflavin (B2), folate, and the cofactors coenzyme A and flavin adenine dinucleotide [21]. The obligatory symbiont, Wolbachia (supergroup F), synthesizes biotin and riboflavin to increase the fitness of the bed bug, Cimex lectularius [6]. In contrast to essential amino acids, vitamin Bs were thought to be unimportant nutrition factors for plant-sap feeders until recent evidence from metagenomics studies in aphids and experimental studies in red cotton bugs showed that vitamin Bs contributed to host survival and development [17, 22].
The small brown planthopper, Laodelphax striatellus (Fallén) (Delphacidae), and brown planthopper, Nilaparvata lugens (Stål) (Delphacidae), are two of the most destructive insect pests on rice that feed exclusively on plant-sap. Large populations of planthoppers cause a noncontagious plant disease, named “hopperburn”, which causes the damaged leaves to become brown and dry, affecting plant yield [23]. It is estimated that, since the 1980s, the occurrence area of N. lugens covered about 50% of the area of cultivated rice, and caused 0.3 billion dollars economic loss per year [24]. Previous studies have showed that planthoppers harbor primary yeast-like symbiont (YLS) that may provide essential amino acids to host insects [25]. However, YLS genomes do not have complete vitamin B synthesis genes. In N. lugens, genomic analysis showed a symbiont, Candidatus Arsenophonus nilaparvatae (Arsenophonus hereafter) encodes genes needed for vitamin B biosynthesis [26]. Individuals infected with Wolbachia (53%) usually do not harbor Arsenophonus [27]; L. striatellus lacking Arsenophonus are more than 90% infected with CI-inducing Wolbachia [28–31].
Previously we reported that Wolbachia enhance host fecundity in planthoppers [29, 32]. In this study, we confirmed the positive effects of Wolbachia on female fecundity of L. striatellus and N. lugens, and demonstrated that Wolbachia-mediated provision of biotin and riboflavin is critical for both rice planthoppers by using a combination of experiments and genomic analyses.
Materials and methods
Insect rearing
The L. striatellus and N. lugens planthoppers were collected in rice farms at Nanjing and Sanya, China, respectively. Because of the almost 100% Wolbachia infection rate in L. striatellus [31], we first obtained WU planthoppers using tetracycline treatment (1 mg/ml) [30] and then microinjected Wolbachia from wild-type individuals back to WU planthopper embryos/nymphs (Fig. S1, Supplementary Materials and Methods). After confirming that the new-established L. striatellus line was reinfected with Wolbachia and behaved similarly to naturally-infected lines through their effects on host planthopper fecundity (Figs. S1 and S2), we used this reinfected line for all further comparisons to assess Wolbachia effects.
The infection rate of Wolbachia in N. lugens was around 50% [27]. WI and WU N. lugens were then isolated from natural populations and their nuclear genetic background was homogenized by backcrossing for at least seven generations. All planthoppers were maintained on rice seedlings in a climate-controlled room (25 °C, 60% RH, and a photoperiod of 8D: 16L [dark: light]). Before experiments, Wolbachia infection status in planthoppers was checked by PCR amplification of Wolbachia surface protein (wsp) gene with primers (81F/691R) [33]. Diagnostic PCR with specific primers showed the infection rate of Aresnophonus in N. lugens populations (either WI or WU insects) was 0%. Planthopper lines were further maintained for over 20 generations.
Fecundity assay
A group of 50 new nymphs were reared under different temperature conditions on rice seedlings. When the nymphs developed into adults, one virgin female (1-day-old) was placed in an oviposition chamber at room temperature (25 °C) to mate with two virgin males for 3 days, and the males were then removed after mating. Female fecundity was recorded daily in the next 2 weeks, with 30 replicates performed for each treatment. The temperature conditions for assays were 20, 25, and 27 °C for L. striatellus and 20, 25, and 30 °C for N. lugens.
Wolbachia density quantification
The abdomens or ovaries of planthopper nymphs were dissected in PBS. The genomic DNA was extracted with a Wizard Genomic DNA Purification Kit (Promega, Fitchburg, WI, USA). The relative Wolbachia density was measured as previously described [29]. In brief, gene copies of Wolbachia surface protein (wsp) were quantified with qPCR with SYBR Premix Ex Taq (Takara, Kusatsu, Shiga, Japan) using ABI 7300 Real-Time PCR system and normalized relative to single-copy genes of the planthopper (beta-actin for L. striatellus and 40S ribosomal protein S11 for N. lugens) (Table S1). Three technical replicates were done for each gene per sample.
Vitamin titer analysis
Planthoppers developing into 3rd instar nymphs were collected for vitamin analysis. Sample preparation was based on the method described previously [34] and outlined in “Supplementary Materials and Methods”. In brief, the sample was quantitatively analyzed using the LC (Agilent, Santa Clara, CA, USA)-MS (Thermo Scientific, Waltham, MA, USA) system. Biotin and riboflavin were measured for quantification under the electrospray ionization-positive mode using D-biotin (for biotin) and 13C4,15N2-riboflavin (for riboflavin) as an internal standard. All vitamin analyses were undertaken in triplicate.
Wolbachia gene expression analysis
Expression of Wolbachia biotin and riboflavin synthesis genes was quantified with reverse transcription quantitative PCR (RT-qPCR). Total RNA was extracted from pools of 3rd instar nymphs per sample using Trizol (Thermo Scientific, Waltham, MA, USA). RNA was reverse-transcribed into cDNA using HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech, Nanjing, Jiangsu, China) following the manufacturer’s instructions. After that, cDNA amplification was quantitatively assessed with SYBR Green dye using ChamQTM SYBR® qPCR Master Mix (Vazyme Biotech, Nanjing, Jiangsu, China) in a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The qPCR cycling conditions involved an initial step of 95 °C for 3 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The specificity of the amplified products was determined by the presence of a single peak in the melting curve. Each cDNA sample was quantified in duplicate for each gene. The planthopper 60S ribosomal protein L14 (rpl14) gene was chosen as the reference control. The 2−∆∆CT method was used to quantify relative gene expression [35]. The efficiency of primers was checked by qPCR by diluting cDNA as a gradient. All primer sequences used in this study are shown in Table S1.
Nutrition supplement experiment
After hatching, planthopper nymphs were supplemented with artificial diets containing different doses of biotin and riboflavin (Table S2) in a rearing chamber under different temperatures. The emerged adults were then transferred to oviposition chambers under room temperature to measure female fecundity [36]. One end of the oviposition chamber was covered with a normal artificial diet sachet and the other side was filled with oviposition medium (5% sucrose and 0.004 M salicylic acid, pH 6.5). The rearing chamber, oviposition chamber, and artificial diets were developed by Fu et al. [37].
Statistical analysis
The number of deposited eggs, Wolbachia densities, and vitamin titers were analyzed with a Shapiro–Wilk normality test followed by parametric or nonparametric analysis depending on whether data were normally distributed. For parametric analysis, Student’s t test was used for the comparison of two treatments. One-way or three-way analysis of variances (ANOVA) with the Tukey post hoc tests were used for multiple treatment comparisons. For nonparametric analysis, we ran Wilcoxon rank-sum tests for comparisons of two treatments or Kruskal–Wallis tests with pairwise comparisons involving Bonferroni corrections where multiple treatments were compared. Multiple comparisons were completed using the “agricolae” package [38]. All statistical analyses were performed in R 3.5 [39].
Genome sequencing and vitamin gene annotation
Approximately 2000 adult planthoppers were collected for DNA extraction. The details of the extraction procedure can be found in the “Supplementary Materials and Methods”. A combination of 454 pyrosequencing, Illumina sequencing, and targeted Sanger sequencing was performed to obtain the wLug and wStriCN genomes in BGI Group (BGI, Shenzhen, Guangdong, China). The draft genome was generated by assembling Illumina reads into contigs and scaffolds with SOAPdenovo version 1.05 [40] and then by closing gaps with 454 reads and Sanger sequencing results of PCR amplicons. The assembled draft genomes were annotated using Prokka version 1.14 [41] with default parameters. Vitamin synthesis genes were annotated based on KO (KEGG Ortholog database) [42] and eggNOG database [43]. More details of gene prediction and annotation are provided in the “Supplementary Materials and Methods”.
Molecular phylogenetic analysis
To assess the phylogenetic relationships of wLug and wStriCN, we inferred their phylogeny using five multilocus sequence typing (MLST) genes (gatB, coxA, hcpA, fbpA, and ftsZ) and protein-coding sequences from all available genomes of Wolbachia A, B and F supergroups from NCBI (Table S6). The protein sequences were grouped with OrthoFinder version 2.2.6 [44], concatenated with FASconCAT-G version 1.04 [45], and trimmed with Gblocks version 0.91b [46] before alignment. Multiple sequence alignment was performed using MAFFT version 7.402 [47]. The maximum likelihood phylogenetic tree was constructed with IQ-TREE version 1.6.5 [48] using the best-fitting nucleotide substitution model (option “-m AUTO”). Node support was calculated with 1000 ultrafast bootstraps. The tree was visualized with FigTree software version 1.4.3 (https://github.com/rambaut/figtree/releases).
Evolution of biotin and riboflavin synthesis genes
The biotin and riboflavin synthesis pathway genes were identified with ortholog analysis by OrthoFinder version 2.2.6 [44] and a manual check of gene annotations from eggNOG database [43]. The biotin and riboflavin operon structures were visualized with the R package “genoPlotR” [49]. The distance matrix of concatenated biotin operon sequences was calculated with MEGA7 [50]. The comparison between the phylogenic trees was made with Dendroscope version 3.5.9 [51].
Results
Wolbachia stably enhance female fecundity in L. striatellus and N. lugens
We first compared the biological performance of the L. striatellus strain that was reinfected with Wolbachia after antibiotic curing with the naturally-infected L. striatellus strain and determined whether the new-established planthopper strain performed similarly to the naturally-infected planthoppers. We found that the newly-established WI planthoppers had a similar preoviposition period and deposited similar numbers of eggs compared with the natural WI planthoppers (100.1% of natural WI, p = 0.997, Fig. S1C). When compared with the tetracycline cured strain (WU), the artificially infected planthoppers (labeled as WI in Fig. 1a) had significantly higher fecundity at room temperature (146% of WU, p < 0.001, 25 °C, see also Fig. S1C). Therefore, Wolbachia appears to have a direct effect on fecundity.
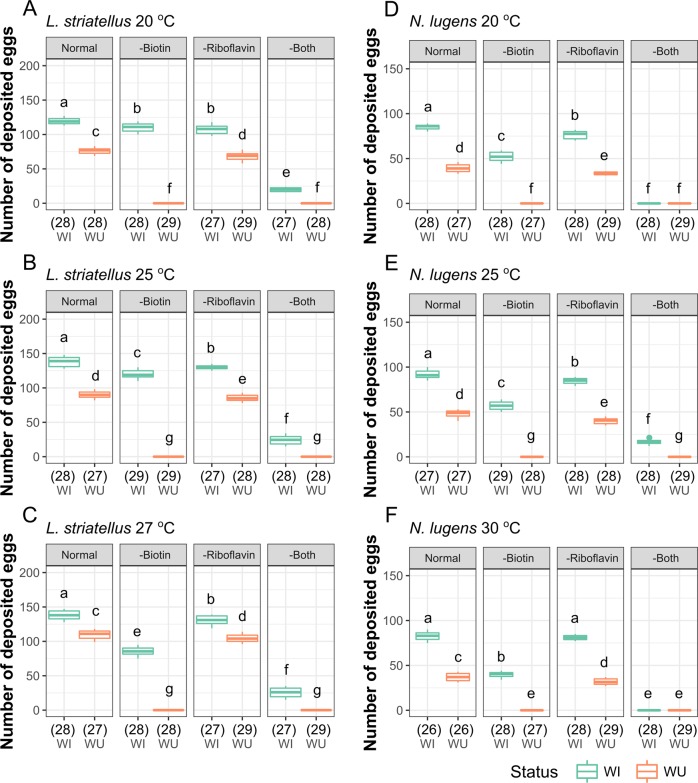
Fig. 1.
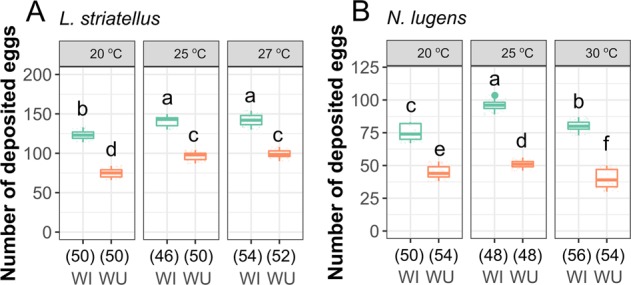
Biological effect of Wolbachia on fecundity of L. striatellus and N. lugens at different temperatures. Number of deposited eggs for L. striatellus (a) and N. lugens (b). WI and WU represent infected and uninfected lines, respectively. The numbers of replicates are listed in parentheses under the treatments. Different letters indicate significant statistical differences at p < 0.05
Since Wolbachia density is sensitive to temperature [52, 53], we reared planthoppers under different temperatures, and then measured the correlation between female fecundity and Wolbachia density in hosts. The density of Wolbachia in L. striatellus was relatively smaller at lower temperatures (20 °C for nymph and 18 °C for adult) and higher at 25 °C (Figs. S3 and S4). The fecundity of WI planthoppers was also significantly reduced at lower temperature compared with that of planthoppers reared at room temperature (Fig. 1a), which indicates that fecundity changes are determined by Wolbachia density as well. Overall, regardless of the rearing temperatures, more eggs were deposited by WI females than WU females (Fig. 1a and Table S3). Similar to the pattern in L. striatellus, WI N. lugens laid more eggs than uninfected planthoppers at all temperatures (Fig. 1b). The proportion of eggs that hatched was similar between WI and WU planthoppers (p = 0.9123 for L. striatellus and p = 0.8609 for N. lugens, Fig. S5), suggesting the laid eggs were of similar quality. These results suggest that Wolbachia is beneficial for reproduction in both species independent of temperature conditions.
Genome sequencing of wLug and wStriCN
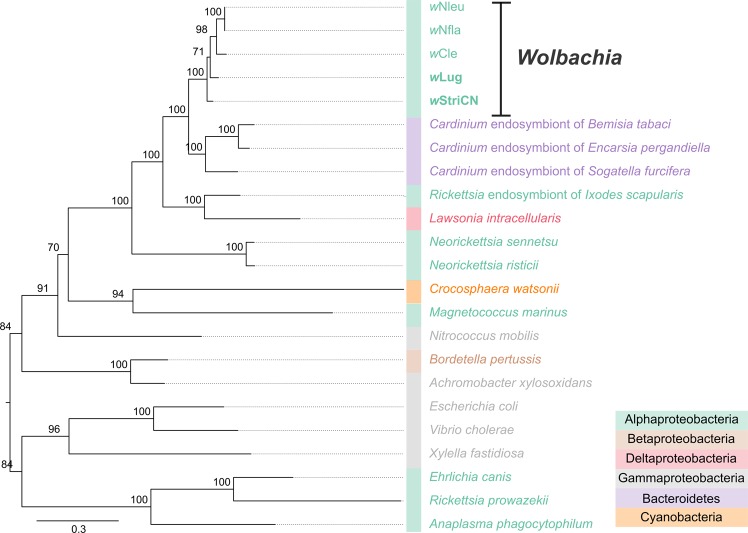
Given that the two Wolbachia have similar effects on their respective host species, we hypothesized that they might share similar genomic features which provide benefits for egg production. We combined Illumina, 454 and targeted Sanger sequencing data and obtained draft genomes of wLug (Wolbachia from N. lugens) (Fig. S6A) and wStriCN (Wolbachia from L. striatellus China strain) (Fig. S6B and Table S4). The assembled genomes of wLug and wStriCN contain two scaffolds and are 1.54 and 1.78 Mb, respectively (Table S4). The wLug and wStriCN genomes have 1516 and 1747 coding DNA sequences, covering 84.51 and 85.34% of their genomes (Tables S5 and S6). Clusters of Orthologous Groups analyses showed similar gene function classifications between wLug and wStriCN (Table S7). Phylogenetic analysis of genome-wide single-copy genes of insect-associated Wolbachia genomes and MLST genes allocated both wLug and wStriCN to supergroup B (Figs. S7 and S8). Both analyses confirmed that wLug and wStriCN cluster in different parts of the B group phylogeny.
Identification of vitamin B synthetic pathways from wLug and wStriCN genomes
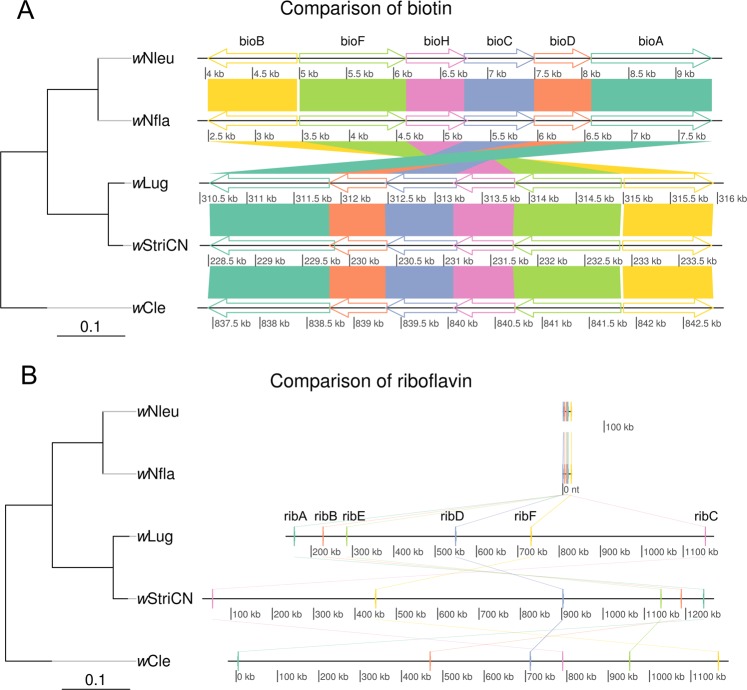
Previous studies showed that Wolbachia was essential for reproduction of the bloodsucking insect host via provisioning of B vitamins [34, 54]. Therefore, we focused on vitamin B biosynthetic pathways. By searching Wolbachia encoded protein sequences against EggNOG and KEGG databases, we found the biosynthetic pathways for folate, pyridoxine, and thiamine in both wLug and wStriCN genomes were incomplete (Table S8). Neither of the wLug and wStriCN genomes had biosynthetic pathways for nicotinate and pantothenate (Table S8). However, we found complete biosynthetic pathways for biotin and riboflavin nutrition synthesis in both wLug and wStriCN (Table S8). The biotin synthesis pathway includes genes bioC, bioH, bioF, bioA, bioD, and bioB. These genes locate closely together on the same position of the wLug and wStriCN genomes (Fig. 2a). Based on the 30 analyzed Wolbachia genomes in this study, apart from wLug and wStriCN, the complete biotin synthesis pathway has only so far been identified in Wolbachia of the bed bug C. lectularius (wCle) [34], cuckoo bees Nomada flava (wNfla) and N. leucophthalma (wNleu) [55] (Table S9). Furthermore, we found wLug and wStriCN encode compact operons of the riboflavin synthesis genes, which are ribA, ribD, ribB, ribE, ribC, and ribF (Table S9). Moreover, being different from wNfla and wNleu, riboflavin synthesis genes in wLug and wStriCN are scattered on Wolbachia genome scaffolds (Fig. 2). From the genomic information obtained, we hypothesized that Wolbachia might provide biotin and riboflavin to N. lugens and L. striatellus.
Fig. 2.
Comparison of the organization of biotin (a) and riboflavin (b) synthesis genes in Wolbachia. The phylogeny of the only five Wolbachia strains that have complete biotin operons is shown. Bootstrap values of the Maximum Likelihood (ML) phylogenetic tree are 100% for all branches. Locations of vitamin B synthesis genes on the scaffolds are shown
The biotin and riboflavin titers are higher in WI L. striatellus and N. lugens individuals
To validate the hypothesis that Wolbachia provide most biotin and riboflavin in planthoppers, we quantified gene expression of Wolbachia biotin and riboflavin biosynthesis genes and measured biotin and riboflavin titers in WI and WU planthoppers. RT-qPCR analysis results showed the Wolbachia biotin and riboflavin biosynthesis genes were expressed much higher in WI than in WU planthoppers (Figs. S9 and S10). In addition, LC–MS results showed that the WI insects exhibited significantly higher titers of biotin and riboflavin in both species, indicating that both wLug and wStriCN are capable of provisioning biotin and riboflavin (Fig. 3), which is concordant with the genomic data (Table S9). We note that biotin and riboflavin were not completely absent in WU planthoppers, suggesting that planthoppers may have access to other sources of vitamins.
Fig. 3.
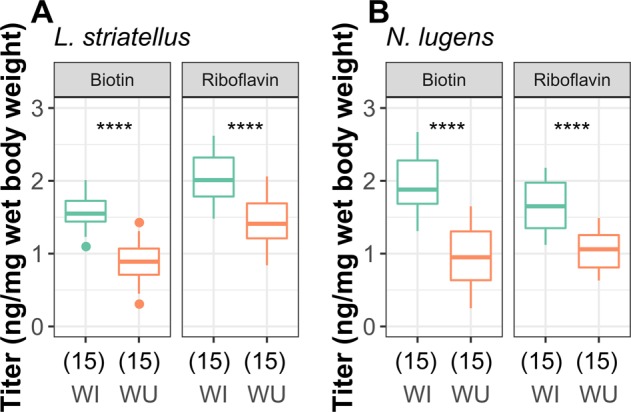
Quantification of biotin and riboflavin in Wolbachia-infected (WI) and -uninfected (WU) L. striatellus and N. lugens. Vitamin quantities were compared with Wilcoxon rank-sum tests. The numbers of replicates are given in parentheses under the treatments. ****p < 0.0001
Biotin and riboflavin rescue the fecundity deficiency in WU L. striatellus and N. lugens
Hypothesizing that riboflavin and biotin from Wolbachia may be important for fecundity in L. striatellus and N. lugens, we undertook a three-factor experiment at three temperatures. We fed WI and WU planthopper nymphs on planthopper artificial diets containing different doses of biotin and riboflavin (Table S3) and measured the number of eggs laid after emergence. In concordance with results on rice plants, WI L. striatellus, and N. lugens exhibited higher fecundity than WU planthoppers on the normal artificial diet across all three temperatures tested for each species (Fig. 4). Three-way ANOVAs at each temperature showed that Wolbachia infection status, as well as biotin and riboflavin concentration, all significantly affected the fecundity of planthoppers. However there were significant interaction effects (p < 0.001, Table 1), reflecting the fact that combinations of the supplemented vitamins influenced the relative difference in fecundity between WI and WU.Note: * p < 0.05, ** p < 0.01, *** p < 0.001.
Fig. 4.
Number of deposited eggs for L. striatellus and N. lugens reared on artificial diets supplemented with less biotin, riboflavin, and less biotin and riboflavin under different temperatures. a–c L. striatellus at 20, 25, and 27 °C; d–f N. lugens at 20, 25, and 30 °C. WI and WU represent infected and uninfected lines, respectively. Normal is the artificial diet that has a normal concentration (1×) of biotin and riboflavin. The other treatments (labeled with “−”) lack the vitamins. The detailed recipes are shown in Table S2. The numbers of replicates are given in parentheses under the treatments. Different letters indicate significant statistical differences at p < 0.05
Table 1.
Three-way ANOVAs on factors influencing planthopper fecundity at each tested temperature
| L. striatellus | 20 °C | 25 °C | 27 °C | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Effects | df | Mean Sq | F value | df | Mean Sq | F value | df | Mean Sq | F value |
| Status | 1 | 68152 | 2931.92*** | 1 | 46278 | 1739.10*** | 1 | 34189 | 1109.683*** |
| Biotin | 1 | 36526 | 1571.39*** | 1 | 44741 | 1681.35*** | 1 | 19904 | 646.02*** |
| Riboflavin | 1 | 278 | 11.97*** | 1 | 25 | 0.92 | 1 | 1904 | 61.81*** |
| Status:Biotin | 1 | 7960 | 342.44*** | 1 | 10788 | 405.39*** | 1 | 1611 | 52.27*** |
| Status:Riboflavin | 1 | 107 | 4.59* | 1 | 169 | 6.37* | 1 | 168 | 5.44* |
| Biotin:Riboflavin | 1 | 21 | 0.92 | 1 | 1328 | 49.91*** | 1 | 223 | 7.25** |
| Status:Biotin:Riboflavin | 1 | 277 | 11.93*** | 1 | 714 | 26.85*** | 1 | 11 | 0.36 |
| Residuals | 215 | 23 | 218 | 27 | 214 | 31 | |||
| N. lugens | 20 °C | 25 °C | 30 °C | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Effects | df | Mean Sq | F value | df | Mean Sq | F value | df | Mean Sq | F value |
| Status | 1 | 67554 | 3825.34*** | 1 | 50496 | 629.84*** | 1 | 47235 | 3127.04*** |
| Biotin | 1 | 79693 | 4512.76*** | 1 | 98210 | 1224.97*** | 1 | 61265 | 4055.86*** |
| Riboflavin | 1 | 70 | 3.99* | 1 | 1043 | 13.01*** | 1 | 1192 | 78.94*** |
| Status:Biotin | 1 | 11158 | 631.82*** | 1 | 6398 | 79.80*** | 1 | 6393 | 423.22*** |
| Status:Riboflavin | 1 | 8606 | 487.34*** | 1 | 9378 | 116.97*** | 1 | 9023 | 597.33*** |
| Biotin:Riboflavin | 1 | 20449 | 1157.97*** | 1 | 18130 | 226.14*** | 1 | 13767 | 911.40*** |
| Status:Biotin:Riboflavin | 1 | 7478 | 423.44*** | 1 | 6880 | 85.82*** | 1 | 9643 | 638.42*** |
| Residuals | 214 | 18 | 214 | 80 | 209 | 15 | |||
Note: *p < 0.05, **p < 0.01, ***p < 0.001.
When biotin and riboflavin were excluded from the diet, we observed a significant decrease in the fecundity of L. striatellus and N. lugens (Fig. 4 and Table S3). Compared with WI planthoppers, WU planthoppers were particularly sensitive to the reduction of biotin; for both species, the lack of biotin resulted in no eggs being laid regardless of temperature (Fig. 4 and Table S3). The absence of riboflavin also led to a reduction in the fecundity of WU planthoppers across all treatments. In addition, the reduction in fecundity in the absence of biotin was minor in WI planthoppers (87% of normal for L. striatellus and 62% of normal for N. lugens at 25 °C) compared with the complete loss of fecundity (zero deposited eggs) for the WU planthoppers (Fig. 4 and Table S3).
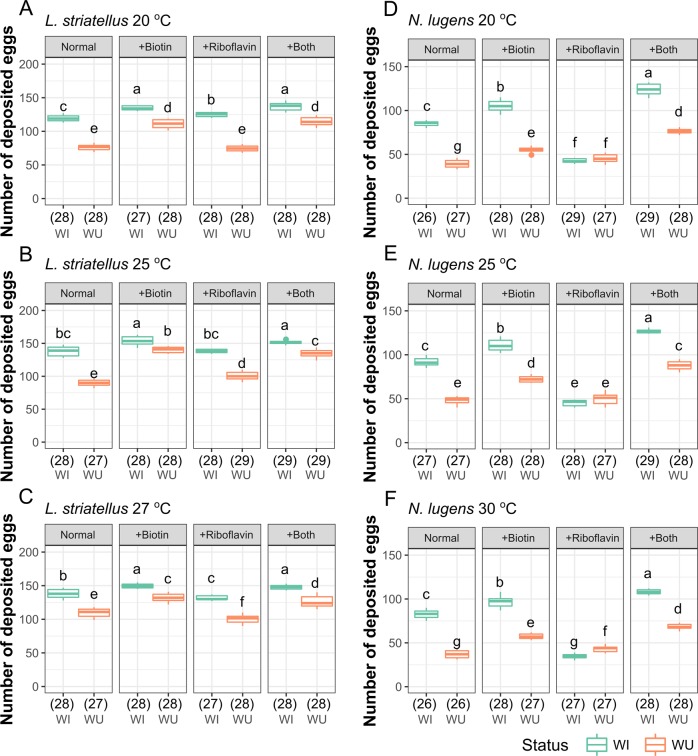
We also analyzed whether supplementing vitamins can rescue the fecundity defect of WU planthoppers. For both L. striatellus and N. lugens, when WU females were reared on diets with more biotin, they laid significantly more eggs (156% of normal for WU L. striatellus and 151% of normal for WU N. lugens at 25 °C, Fig. 5 and Table S3). When reared on diets supplemented with both biotin and riboflavin, the fecundity of WU planthoppers was mostly recovered to that of WI planthoppers (97.6% of normal WI L. striatellus and 95.5% of normal WI N. lugens at 25 °C, Fig. 5b). For WI L. striatellus, the benefit of supplementation by biotin was significant under all three temperature conditions, but not always for riboflavin (Fig. 5a–c). In WI N. lugens, supplementation by biotin consistently increased fecundity (125% of normal at 25 °C), but supplementation by riboflavin decreased fecundity (49.0% of normal at 25 °C, Fig. 5d–f and Table S3). Taken together, the above results suggest that Wolbachia-provided biotin (and perhaps riboflavin as well) are important for female fecundity in L. striatellus and N. lugens.
Fig. 5.
Number of deposited eggs for L. striatellus and N. lugens reared on artificial diets supplemented with more biotin, riboflavin and more biotin and riboflavin under different temperatures. a–c L. striatellus at 20, 25, and 27 °C; d–f N. lugens at 20, 25, and 30 °C. WI and WU represent infected and uninfected lines respectively. Normal is the artificial diet that has a normal concentration (1×) of biotin and riboflavin. The other treatments (labeled with “+”) contain 3× fold of the normal concentration of corresponding vitamins. The detailed recipes are shown in Table S2. The numbers of replicates are listed in parentheses under the treatments. Different letters indicate significant statistical differences at p < 0.05
Evolutionary origin of biotin and riboflavin operons in wLug and wStriCN
Apart from wLug and wStriCN, complete biotin synthesis pathways exist only in wCle [34], wNfla, and wNleu [55] (Table S8), while riboflavin synthesis genes exist in most other insect-associated Wolbachia [54]. To understand the evolution of biotin and riboflavin synthesis genes in Wolbachia, we analyzed available complete and draft genomes of insect-associated Wolbachia. Phylogenetic analysis showed that both wLug and wStriCN belong to supergroup B, while wNfla and wNleu belong to supergroup A and wCle belongs to supergroup F (Fig. S7). The existence of similar biotin synthesis genes in distinct Wolbachia supergroups may reflect either horizontal transfer of biotin synthesis genes into the respective supergroups, or a large-scale gene loss in various distinct Wolbachia supergroups of an ancient biotin gene complex [55].
When we investigated the phylogenetic history of biotin synthesis genes across diverse bacteria, the incongruence between phylogenies of biotin operons and bacterial taxonomy supports the frequent horizontal transfer of biotin synthesis genes (Fig. 6). The distinct monophyletic clade of Wolbachia biotin genes implies a relatively recent acquisition. To identify putative donors of biotin operon genes, protein sequences of Wolbachia biotin synthesis genes were blasted against the NCBI “refseq_protein” database. The top blast hits were from the intracellular symbiont Cardinium (Table S10), which is consistent with the phylogenetic tree of biotin operon genes (Fig. 6).
Fig. 6.
Phylogenetic analysis of biotin synthesis genes of Wolbachia. The ML tree was calculated with a concatenated protein sequence (BioA, BioD, BioC, BioH, BioF, and BioB) of bacteria from six different phyla using an LG + I + G4 substitution model. Taxonomic affiliations are coded by color. The Wolbachia sequence obtained in this study is shown in bold. Bootstrap values are indicated at nodes (only values > 50% are shown). The GenBank accession numbers of proteins used in the analysis are listed in Table S9
In contrast, the riboflavin synthesis genes were conserved across most sequenced Wolbachia genomes [54] (Table S9). The phylogeny of riboflavin operons mirrored the Wolbachia genomic phylogeny to some extent (Fig. 7), suggesting that the riboflavin synthesis genes were present in a common ancestor of Wolbachia. Meanwhile, the incongruent phylogenies found within supergroups implies that vertical transmission is not strictly followed among closely-related Wolbachia strains [54].
Fig. 7.
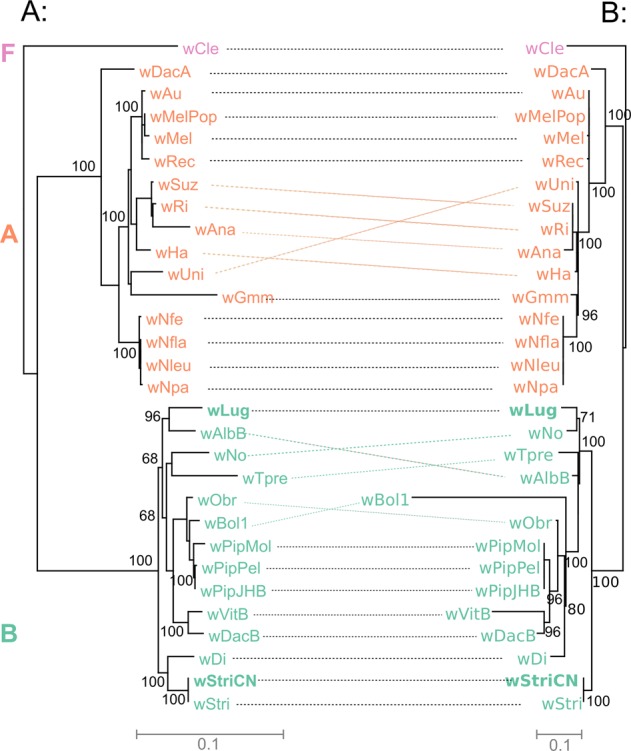
Phylogenetic comparisons between Wolbachia core genes and riboflavin synthesis genes. a Phylogeny of Wolbachia supergroups A and B. The ML tree is based on concatenated protein sequences of 268 single-copy genes. b Phylogeny of riboflavin synthesis genes. The ML tree is based on concatenated protein sequences of ribA, ribB, ribD, ribC, ribE, and ribF. The trees were rerooted with the outgroup wCle. Numbers on nodes correspond to bootstrap support values (>50%). The scale bar represents the average number of substitutions per site. Full names and accession numbers of Wolbachia strains are given in Table S9
Discussion
In this study, we investigated the mechanism of Wolbachia-mediated benefit in plant-sap feeding planthoppers. Our experimental and genomic evidence demonstrated that B supergroup Wolbachia enhance planthopper fitness, likely by synthesizing essential nutrients (biotin and riboflavin). We confirmed that both wLug and wStriCN increase female fecundity in the Hemipteran planthoppers N. lugens and L. striatellus [29, 32], which was evident across a range of temperatures at which planthoppers were reared. Our data also showed that Wolbachia density in planthopper nymphs was reduced under nonoptimal temperature conditions, with likely flow-on effects on the extent to which Wolbachia enhanced host reproduction.
Our genomic analysis, metabolic analysis and nutrient supplementing experiments indicate that the Wolbachia-mediated benefit in plant-sap feeding planthoppers involves provision of biotin and riboflavin. Biotin and riboflavin are vitamin B family members that function as coenzymes and are generally not synthesized by insects. Many insects, especially those who feed exclusively on a limited type of diet, derive their vitamin B requirements from their microbial symbionts [18]. For instance, the blood-feeding tsetse fly G. morsitans and African soft tick O. moubata rely on the symbionts Wigglesworthia and Francisella to synthesize vitamin Bs [19–21]. However, the importance of vitamin Bs for plant-sap feeders may be underestimated. Recently, a metagenomics study revealed that the primary symbiont of aphids, Buchnera, has lost the ability to synthesize riboflavin, while other coresident symbionts in aphids have intact routes for the biosynthesis of this compound [22]. Experimental studies showed that gut symbionts of red cotton bugs, Dysdercus fasciatus, contribute toward host fitness through the supplementation of vitamin Bs [17]. In combination with our results, it is becoming clear that vitamin Bs are important in plant-sap feeders.
Comparative genomic analysis of diverse insect-associated Wolbachia strains showed the synthesis pathway for riboflavin is highly conserved [54]. In contrast, the biotin synthesis pathway so far has only been detected in five Wolbachia strains, i.e., wCle, wNleu, wNfla, wLug, and wStriCN (Table S6). wNleu and wNfla were reported to encode complete biotin synthesis operons via genomic analysis [55], but no biological experiments have yet been completed. The biological roles of Wolbachia-provided riboflavin and biotin have been experimentally demonstrated for the wCle infection in C. lectularius, a blood-feeding bed bug [6, 34, 54]. However, wCle belongs to supergroup F, phylogenetically distant from most other insect-associated Wolbachia (Fig. S7). Moreover, wCle is an obligate bacteriocyte-associated nutritional mutualist for the bed bug [6], whose localization, infection dynamics, and biological roles are more comparable to those of obligate mutualistic bacteriocyte symbionts like Wigglesworthia in tsetse flies and Buchnera in aphids. In our case, both wLug and wStriCN are facultative symbionts and belong to supergroup B (Fig. S7). Given that supergroup B Wolbachia are prevalent in many arthropods living on limited types of diets [1], our findings raise the possibility of Wolbachia-provided nutritional mutualism in other insects.
Comparative genomic analysis indicate that biotin operons are found in five Wolbachia genomes scattered across three supergroups, supporting the notion that the biotin operons were possibly laterally transferred from another organism. The Wolbachia biotin synthesis genes formed a well-supported monophyletic group for the biotin operon phylogeny, suggesting the biotin operon might originate from the same ancestor [55]. BLAST results show that the most closely-related protein sequences of the biotin operon are from Cardinium, an intracellular endosymbiont [56]. Cardinium was reported to coinfect with Wolbachia in some planthoppers [31]. A recent genome analysis revealed that Cardinium from Sogatella furcifera, another rice planthopper, also harbors complete synthesis genes of biotin [57]. Therefore, we speculate that horizontal transfers of biotin synthesis genes might have occurred several times (in species coinfected by Cardinium and Wolbachia).
In contrast, riboflavin synthesis genes are likely to have been present in the ancestor of Wolbachia, as they are conserved in most Wolbachia genomes and evolved mostly convergent with Wolbachia phylogeny [54]. Riboflavin provisioning appears to be critical for Wolbachia and therefore maintained throughout diverse Wolbachia strains. Recent studies show Wolbachia infection levels decreased in mosquito cells lacking riboflavin [58], suggesting the importance of riboflavin for Wolbachia. Our data also showed that adding riboflavin increased the fecundity of WU planthoppers. However, though the mechanism is still unknown, this treatment resulted in adverse effects on WI N. lugens, suggesting a particular titer of this vitamin is required in N. lugens. Biotin supplementation was more critical for recovering host fitness in WU planthoppers than riboflavin supplementation (Fig. 4) and this also appears to be the situation in bed bugs [34, 54], pointing to the value of acquiring biotin synthesis operons horizontally for Wolbachia in these insects.
Rice planthoppers harbor primary mutualistic YLS. YLS synthesize all essential amino acids and ergosterol for planthoppers and are important for host fitness [25, 59, 60]. Previous studies have reported that high temperature can dramatically suppress the population of YLS and result in negative fitness consequences for host planthoppers [61]. In the current study, regardless of rearing temperatures, WI planthoppers consistently laid more eggs than the WU individuals, suggesting that the Wolbachia-mediated fecundity advantage is independent of temperature (and perhaps any effects of YLS). It is worth noting that genome analysis showed both YLS and N. lugens genomes had gene deficiencies in several vitamin B biosynthesis pathways [25]. Based on our results, we speculate that Wolbachia-provided vitamin Bs compensate with YLS to benefit rice planthopper fitness. On the other hand, field investigations showed that N. lugens individuals that lack Wolbachia usually harbor Arsenophonus [27, 29]. Although no experiments have yet been undertaken, genomic analysis showed Arsenophonus encodes all genes needed for vitamin B biosynthesis [26], which may have similar nutritional mutualistic roles as Wolbachia.
Overall, our results demonstrate that two members of supergroup B Wolbachia can increase the fecundity of plant-sap sucking planthoppers. In addition, our results point to the beneficial effect of Wolbachia-synthesized biotin and riboflavin for plant-sap feeders. Removal of these bacteria from planthoppers might provide a new target for pest control. This is the first report of supergroup B Wolbachia acting as nutritional mutualists in plant-sap sucking insects, highlighting the importance of beneficial phenotypes across the Wolbachia phylogeny.
Supplementary information
Acknowledgements
We thank Professor John Werren of the University of Rochester, USA for his comments on the paper, and Dr Jing-Tao Sun, Tong-Pu Li, and Pei-Wen Mo of Nanjing Agricultural University, China for their help on this project. This work was supported by the National Natural Science Foundation of China (Nos. 31672035 and 31871976) to X-YH and the startup grant from Nanjing Agricultural University to X-LB (No. 804015).
Author contributions
JFJ, XLB, and XYH designed the research; JFJ, XLB, YG, DSZ, KJZ, HJH, JTG, XZ, ZX, AH, and XYH performed the research; JFJ, XLB, DSZ, JJH, ZX, AH, and XYH wrote and edited the paper. All authors read and approved the paper.
Data availability
The draft genome sequences data are freely available in NCBI GenBank under accession numbers MUIY00000000 (wLug) and MUIX00000000 (wStriCN).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jia-Fei Ju, Xiao-Li Bing
Supplementary information
The online version of this article (10.1038/s41396-019-0559-9) contains supplementary material, which is available to authorized users.
References
- 1.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo N, Paraskevopoulos C, Bourtzis K, O’Neill S, Werren J, Bordenstein S, et al. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 2007;57:654. doi: 10.1099/ijs.0.64515-0. [DOI] [PubMed] [Google Scholar]
- 3.Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae) Appl Environ Microbiol. 2009;75:1036–43. doi: 10.1128/AEM.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bing XL, Xia WQ, Gui JD, Yan GH, Wang XW, Liu SS. Diversity and evolution of the Wolbachia endosymbionts of Bemisia (Hemiptera: Aleyrodidae) whiteflies. Ecol Evol. 2014;4:2714–37. doi: 10.1002/ece3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Boulétreau M. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci USA. 2001;98:6247–52. doi: 10.1073/pnas.101304298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosokawa T, Koga R, Kikuchi Y, Meng X-Y, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci USA. 2010;107:769–74. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Neill SL, Hoffmann AA, Werren JH (eds). Influential passengers, inherited microorganisms and arthropod reproduction. (Oxford University Press, New York, 1997)
- 8.Zug R, Hammerstein P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev. 2015;90:89–111. doi: 10.1111/brv.12098. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira L, Ferreira, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iturbe-Ormaetxe I, Walker T, O’ Neill SL. Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 2011;12:508–18. doi: 10.1038/embor.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brownlie JC, Cass BN, Riegler M, Witsenburg JJ, Iturbe-Ormaetxe I, McGraw EA, et al. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 2009;5:e1000368. doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer N, Charif D, Henri H, Bataille M, Prevost G, Kraaijeveld K, et al. A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity. 2009;103:248–56. doi: 10.1038/hdy.2009.63. [DOI] [PubMed] [Google Scholar]
- 13.MJTN Timmermans. Ellers J. Wolbachia endosymbiont is essential for egg hatching in a parthenogenetic arthropod. Evol Ecol. 2008;23:931. [Google Scholar]
- 14.Dobson SL, Rattanadechakul W, Marsland EJ. Fitness advantage and cytoplasmic incompatibility in Wolbachia single- and superinfected Aedes albopictus. Heredity. 2004;93:135. doi: 10.1038/sj.hdy.6800458. [DOI] [PubMed] [Google Scholar]
- 15.Dong P, Wang J-J, Zhao Z-M. Infection by Wolbachia bacteria and its influence on the reproduction of the stored-product psocid, Liposcelis tricolor. J Insect Sci. 2006;6:24. doi: 10.1673/2006_06_24.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry AJ, Palmer MR, Rand DM. Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity. 2004;93:379. doi: 10.1038/sj.hdy.6800514. [DOI] [PubMed] [Google Scholar]
- 17.Salem H, Bauer E, Strauss AS, Vogel H, Marz M, Kaltenpoth M. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc R Soc B. 2014;281:20141838. doi: 10.1098/rspb.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas AE. The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr Opin Insect Sci. 2017;23:65–9. doi: 10.1016/j.cois.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Michalkova V, Benoit JB, Weiss BL, Attardo GM, Aksoy S. Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl Environ Microbiol. 2014;80:5844–53. doi: 10.1128/AEM.01150-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder AK, Rio RVM. “Wigglesworthia morsitans” folate (vitamin B9) biosynthesis contributes to tsetse host fitness. Appl Environ Microbiol. 2015;81:5375–86. doi: 10.1128/AEM.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duron O, Morel O, Noël V, Buysse M, Binetruy F, Lancelot R, et al. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr Biol. 2018;28:1896–902. doi: 10.1016/j.cub.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 22.Meseguer AS, Manzano-Marín A, Coeur d’Acier A, Clamens A-L, Godefroid M, Jousselin E. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol Ecol. 2017;26:2363–78. doi: 10.1111/mec.13910. [DOI] [PubMed] [Google Scholar]
- 23.Backus EA, Serrano MS, Ranger CM. Mechanisms of hopperburn: an overview of insect taxonomy, behavior, and physiology. Annu Rev Entomol. 2005;50:125–51. doi: 10.1146/annurev.ento.49.061802.123310. [DOI] [PubMed] [Google Scholar]
- 24.Min S, Lee SW, Choi B-R, Lee SH, Kwon DH. Insecticide resistance monitoring and correlation analysis to select appropriate insecticides against Nilaparvata lugens (Stål), a migratory pest in Korea. J Asia Pac Entomol. 2014;17:711–16. [Google Scholar]
- 25.Xue J, Zhou X, Zhang C-X, Yu L-L, Fan H-W, Wang Z, et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014;15:521. doi: 10.1186/s13059-014-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan H-W, Lu J-B, Ye Y-X, Yu X-P, Zhang C-X. Characteristics of the draft genome of “Candidatus Arsenophonus nilaparvatae”, a facultative endosymbiont of Nilaparvata lugens. Insect Sci. 2016;23:478–86. doi: 10.1111/1744-7917.12318. [DOI] [PubMed] [Google Scholar]
- 27.Qu L-Y, Lou Y-H, Fan H-W, Ye Y-X, Huang H-J, Hu M-Q, et al. Two endosymbiotic bacteria, Wolbachia and Arsenophonus, in the brown planthopper Nilaparvata lugens. Symbiosis. 2013;61:47–53. [Google Scholar]
- 28.Kittayapong P, Jamnongluk W, Thipaksorn A, Milne JR, Sindhusake C. Wolbachia infection complexity among insects in the tropical rice-field community. Mol Ecol. 2003;12:1049–60. doi: 10.1046/j.1365-294x.2003.01793.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Zhang K-J, Hong X-Y. Population dynamics of noncytoplasmic incompatibility-inducing Wolbachia in Nilaparvata lugens and its effects on host adult life span and female fitness. Environ Entomol. 2010;39:1801–09. doi: 10.1603/EN10051. [DOI] [PubMed] [Google Scholar]
- 30.Noda H, Miyoshi T, Zhang Q, Watanabe K, Deng K, Hoshizaki S. Wolbachia infection shared among planthoppers (Homoptera: Delphacidae) and their endoparasite (Strepsiptera: Elenchidae): a probable case of interspecies transmission. Mol Ecol. 2001;10:2101–06. doi: 10.1046/j.0962-1083.2001.01334.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang K-J, Han X, Hong X-Y. Various infection status and molecular evidence for horizontal transmission and recombination of Wolbachia and Cardinium among rice planthoppers and related species. Insect Sci. 2013;20:329–44. doi: 10.1111/j.1744-7917.2012.01537.x. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y, Hoffmann AA, Xu X-Q, Zhang X, Huang H-J, Ju J-F, et al. Wolbachia-induced apoptosis associated with increased fecundity in Laodelphax striatellus (Hemiptera: Delphacidae) Insect Mol Biol. 2018;27:796–807. doi: 10.1111/imb.12518. [DOI] [PubMed] [Google Scholar]
- 33.Braig HR, Zhou W, Dobson SL, O’Neill SL. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol. 1998;180:2373–78. doi: 10.1128/jb.180.9.2373-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikoh N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc Natl Acad Sci USA. 2014;111:10257–62. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Koyama K. Artificial rearing and nutritional physiology of the planthoppers and leafhoppers (Hemiptera, Delphacidae and Deltocephalidae) on a holidic diet. JARQ. 1988;20:20–7. [Google Scholar]
- 37.Fu Q, Zhang Z, Hu C, Lai F, Sun Z. A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stal) Appl Entomol Zool. 2001;36:111–16. [Google Scholar]
- 38.De Mendiburu F, Simon R. Agricolae—ten years of an open source statistical tool for experiments in breeding, agriculture and biology. PeerJ Preprints. 2015;3:e1404v1. [Google Scholar]
- 39.R Team C. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018.
- 40.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2009;20:265–72. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [Google Scholar]
- 42.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–93.. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kück P, Longo GC. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 2014;11:81. doi: 10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 47.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guy L, Roat Kultima J, Andersson SGE. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–5. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–7. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 52.Wiwatanaratanabutr I, Kittayapong P. Effects of crowding and temperature on Wolbachia infection density among life cycle stages of Aedes albopictus. J Invertebr Pathol. 2009;102:220–4. doi: 10.1016/j.jip.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Ross PA, Wiwatanaratanabutr I, Axford JK, White VL, Endersby-Harshman NM, Hoffmann AA. Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PLoS Pathog. 2017;13:e1006006. doi: 10.1371/journal.ppat.1006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moriyama M, Nikoh N, Hosokawa T, Fukatsu T. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. mBio. 2015;6:e01732–15. doi: 10.1128/mBio.01732-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerth M, Bleidorn C. Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat Microbiol. 2016;2:16241. doi: 10.1038/nmicrobiol.2016.241. [DOI] [PubMed] [Google Scholar]
- 56.Penz T, Schmitz-Esser S, Kelly SE, Cass BN, Müller A, Woyke T, et al. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 2012;8:e1003012. doi: 10.1371/journal.pgen.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Z, Fu Y, Guo D, Wu Y, Ajayi OE, Wu Q. Bacterial endosymbiont Cardinium cSfur genome sequence provides insights for understanding the symbiotic relationship in Sogatella furcifera host. BMC Genom. 2018;19:68. doi: 10.1186/s12864-018-5078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fallon AM, Baldridge GD, Carroll EM, Kurtz CM. Depletion of host cell riboflavin reduces Wolbachia levels in cultured mosquito cells. Vitr Cell Dev Biol Anim. 2014;50:707–13. doi: 10.1007/s11626-014-9758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wetzel JM, Ohnishi M, Fujita T, Nakanishi K, Naya Y, Noda H, et al. Diversity in steroidogenesis of symbiotic microorganisms from planthoppers. J Chem Ecol. 1992;18:2083–94. doi: 10.1007/BF00981929. [DOI] [PubMed] [Google Scholar]
- 60.Hongoh Y, Ishikawa H. Uric acid as a nitrogen resource for the brown planthopper, Nilaparvata lugens: studies with synthetic diets and aposymbiotic insects. Zool Sci. 1997;14:581–6. [Google Scholar]
- 61.Chen C-C, Cheng L-L, Hou RF. Studies on the intracellular yeast-like symbiote in the brown planthopper, Nilaparvata lugens Stål. Z Angew Entomol. 1981;92:440–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The draft genome sequences data are freely available in NCBI GenBank under accession numbers MUIY00000000 (wLug) and MUIX00000000 (wStriCN).