Abstract
While genome erosion is extensively studied in intracellular symbionts, the metabolic implications of reductive evolution in microbes subsisting extracellularly remain poorly understood. Stammera capleta—an extracellular symbiont in leaf beetles—possesses an extremely reduced genome (0.27 Mb), enabling the study of drastic reductive evolution in the absence of intracellularity. Here, we outline the genomic and transcriptomic profiles of Stammera and its host to elucidate host-symbiont metabolic interactions. Given the symbiont’s substantial demands for nutrients and membrane components, the host’s symbiotic organ shows repurposing of internal resources by upregulating nutrient transporters and cuticle-processing genes targeting epithelial chitin. Facilitated by this supplementation and its localization, Stammera exhibits a highly streamlined gene expression profile and a fermentation pathway for energy conversion, sharply contrasting the respiratory metabolism retained by most intracellular symbionts. Our results provide insights into a tightly regulated and metabolically integrated extracellular symbiosis, expanding our understanding of the minimal metabolism required to sustain life outside of a host cell.
Subject terms: Molecular evolution, Microbial ecology
The study of reductive genome evolution in bacterial endosymbionts has yielded valuable insights into the minimal gene set necessary to sustain life [1, 2]. Many insects harbor such microbial symbionts with reduced genomes, which complement the metabolism of their host through the biosynthesis of essential amino acids and vitamins [3]. Vertical transmission of these beneficial microbes often inflicts severe population bottlenecks, inducing rapid genomic changes due to genetic drift [4]. As such, neutral or slightly deleterious mutations can become fixed, leading to the AT-biased genomic composition and a degenerated codon usage [3], as well as severely reduced biosynthetic capabilities. Consequently, intracellular symbionts require extensive nutritional support from the host, blurring the line between organelles and autonomous organisms [1].
While the metabolic implications of reductive genome evolution are well-characterized in intracellular symbionts, our understanding of the corresponding selective pressures in extracellular bacteria is limited. Similar to their intracellular counterparts, some extracellular symbionts are restricted to a defined environment within symbiotic organs, relaxing selection to maintain many biosynthetic pathways [5]. Concordantly, recent studies have described a few extracellular insect symbionts with reduced genomes [5, 6], which represent interesting cases to study the general patterns governing reductive genome evolution. However, the extent of this loss and the corresponding metabolic consequences are generally thought to be less extreme than in intracellular symbionts, as evident by their disparate genome sizes [7, 8].
Recently, we reported on an extracellular symbiont with a drastically reduced genome that encodes for the smallest gene set of any organism subsisting outside of a host cell [2]. This gammaproteobacterial microbe, ‘Candidatus Stammera capleta’ (henceforth Stammera), is vertically transmitted and located in specialized foregut-associated symbiotic organs of the tortoise leaf beetle (Coleoptera: Chrysomelidae: Cassida rubiginosa), an obligate folivore [2]. Through the production and secretion of two pectinases, Stammera aids its host in the digestion of plant cell wall polysaccharides. Thus, in contrast to most obligate nutritional symbioses, the function for the host is reduced to two isolated enzymatic tasks, instead of pathways with multiple enzymes (e.g., amino acid biosynthesis) [3] (Table S1). In the present study, we investigated the metabolic potential of Stammera in the context of the host’s as well as symbiont’s transcriptomic profiles to better understand how a microbe can afford such a limited metabolism outside the nutritional comforts of a host cell.
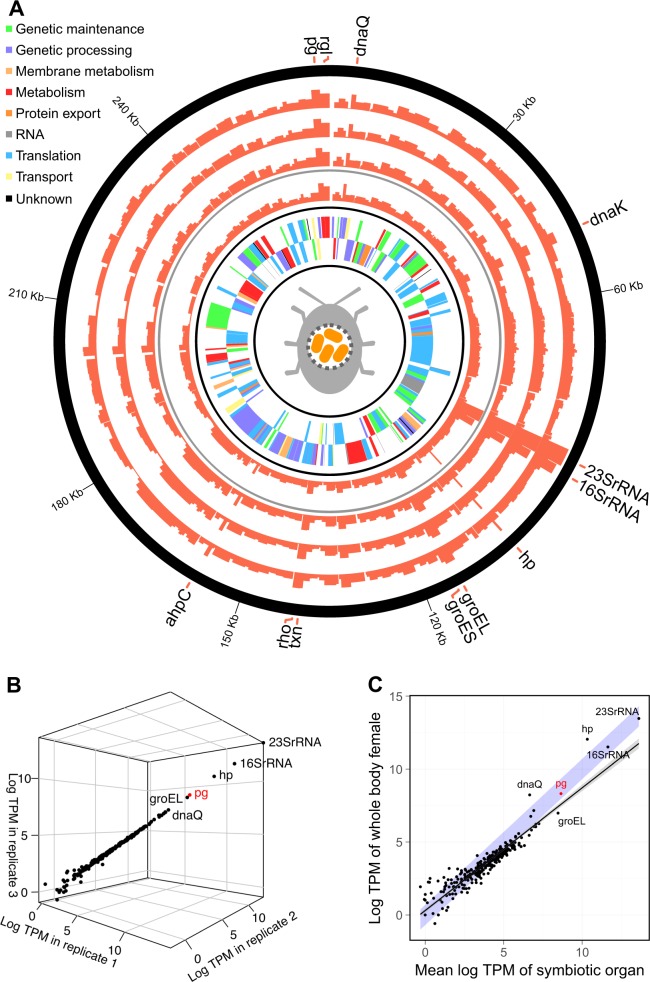
The highly consistent gene expression profile of Stammera (mean pairwise Pearson correlation of three replicates: R2 = 0.98; p < 0.001) demonstrates a restricted control at the transcriptional level over the reduced genome (Fig. 1). Among the most highly expressed transcripts are chaperone-encoding genes (groEL, groES, dnaK) (Fig. 1; Table S2). Reflecting their capacity to ameliorate protein dysfunction, their preferential expression may indicate the need for protein maintenance and refolding. This is consistent with other symbionts possessing reduced genomes [9, 10], underlining the general challenges in maintaining stability of cellular functions in AT-enriched chromosomes as well as accumulated mutations [11]. Strikingly, the expression of polygalacturonase (pg), a pectinase, exceeded the expression of all other protein-coding genes (Fig. 1), reflecting the mutualistic role of Stammera towards its host. Rhamnogalacturonan lyase (rgl), a supplementary pectinase, was also preferentially expressed (Fig. 1A). The elevated expression patterns in Stammera are consistent with the transcriptional profiles reported for primary endosymbionts, where genes coding for chaperones and host-beneficial factors (e.g,. essential amino acids) are among the most highly expressed [12].
Fig. 1.
Expression profile in transcripts per million (TPM) of the annotated Stammera capleta genome. The inner two layers in (a) represent the identified genes in the forward and reverse direction, the next layer represents the Stammera gene expression profile in the whole body sample of a female Csassida rubiginosa (excluding the gut-associated symbiotic organ, but including the symbiont transmission organs associated with the reproductive tract), and the outer three layers represent the three replicates of Stammera gene expression profiles in the gut-associated symbiotic organs of two male and one female C. rubiginosa, respectively. b Correlation of the relative expression of each Stammera gene for the three replicates of symbiotic organs (mean pairwise Pearson correlation of gene expression profiles between replicate RNAseq data sets: R2 = 0.98; p < 0.001). c Correlation of the mean relative Stammera gene expression in the gut-associated symbiotic organ replicates and the whole body female sample (Pearson correlation, R2 = 0.9; p < 0.001). The line represents a linear regression and the blue area indicates a perfect correlation between both variables. Genes encoding for a hypothetical protein and the polygalacturonase are abbreviated as hp and pg, respectively between both variables. Genes are abbreviated as: hypothetical protein (hp), polygalacturonase (pg), rhmanogalacturonan lyase (rgl), alkyl hydroperoxide reductase C (ahpC), transcription termination factor Rho (rho), and thioredoxine (txn).
In many insect symbioses, the host is able to control the symbiont population by leveraging its immune system. Among the differentially expressed transcripts in the symbiotic organ were immune signaling genes that were generally downregulated compared with whole body samples (Fig. S1). Interestingly, an isoform of a peptidoglycan recognition protein, as well as a galectin and cathepsin B were strongly upregulated in the symbiotic organ (Fig. S1). In weevils, antimicrobial peptides are known to regulate symbiont populations to avoid overgrowth [13]. Galectins, in particular, are discussed as relevant cell surface-binding proteins that could potentially control microbial colonization [14]. Immunoregulation thus represents an interesting avenue to understand how symbiont populations fluctuate in response to the metabolic demands of their hosts.
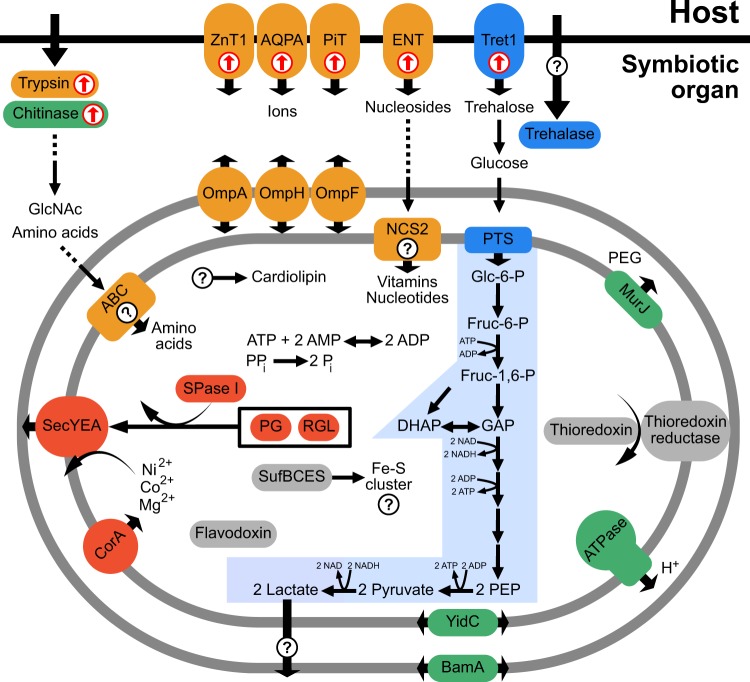
Stammera’s degraded genome raises the question of how the functional deficiencies of this extracellular bacterium are compensated. Several host genes for managing internal insect resources, such as chitin remodeling and transport of nutrients, are upregulated in the symbiotic organ (Figs. S1 and S2). Given its connection to the foregut [2] the symbiotic organ is likely surrounded by chitin. The turn-over of foregut-lining chitin is a common process in insects and releases several nutrients such as N-acetylglucosamines [15] that serve as precursors for microbial membrane biosynthesis [16], which is eroded in Stammera (Fig. 2, Table S1). In addition, the host cells of the symbiotic organ exhibit an upregulated expression of trypsin (Fig. 2). Trypsin-like proteases degrade insect cuticular proteins [17], which could enrich Stammera’s environment with amino acids (Fig. 2), since Stammera cannot synthesize these nutrients (Fig. 2 and Table S1). The upregulation of nucleoside and ion transporters in the symbiotic organ (Fig. 2) may reflect additional supplementation of essential compounds to Stammera, as previously described in nutritional partnerships involving sap-feeding Hemiptera and their bacterial endosymbionts [18, 19]. The most highly upregulated transport protein in the symbiotic organ is a transporter for trehalose (Fig. 2), one of the most abundant sugars in insects [20], potentially supporting the energy metabolism of Stammera (Fig. 2). Taken together, we predict that digestive enzymes and transporters of the symbiotic organ could enrich Stammera with essential nutrients by remodeling the extracellular host environment.
Fig. 2.
Stammera capleta reconstructed metabolism and nutritional supplementation by the host, as inferred from genomic and transcriptomic data, respectively. Carbohydrate metabolism is marked in blue, production and export of pectinase enzymes in red, cofactor metabolism in orange, membrane metabolism in green, and individual metabolic enzymes in gray. Cellular processes of which the functions are unknown, or no corresponding gene was found are highlighted with a question mark. Host genes that were found to be significantly upregulated in the symbiotic organ are highlighted with red arrows
Although often highly reduced, some intracellular symbionts appear to rely on retained genes in the TCA cycle for the production of amino acids and vitamins [1, 5], tying respiratory energy generation with the host-beneficial factors that underlie the host-symbiont partnership. For Stammera, the sole functional contribution to the host lies in the production of pectinases, relaxing selection for the symbiont to maintain vitamin and amino acid biosynthetic pathways and, correspondingly, the TCA cycle alongside the enzymes of the respiratory complex. Instead, the reduction equivalents (e.g., NADH) generated during glycolysis, can be oxidized through a lactate dehydrogenase (LDH). This metabolism requires 18 times more glucose than aerobic respiration to generate the same amount of ATP [21], resulting in a high carbohydrate demand, which could be met by the host’s carbohydrate-rich diet and the upregulation of a trehalose transporter (Fig. 2). Interestingly, a fermentative LDH is missing in related symbionts, but is present in free-living relatives (Table S3), indicating that a conditional fermentative and respiratory metabolism was restricted to sole respiration in most endosymbionts (Fig. 3). Lactate fermentation has been discussed in the context of parasitic eukaryotes [22], but is unknown in insect symbionts with reduced genomes, likely because lactate accumulation would result in toxicity within host cells [23]. In the case of Stammera, however, the extracellular localization supports the erosion to a sole fermentative metabolism by minimizing acid stress. Thus, Stammera’s central metabolism is reduced considerably beyond that of some intracellular symbionts, maintaining glycolysis and an LDH. Further functional studies are needed to validate the hypothesis this study poses on the evolution of Stammera’s metabolism (Fig. 3).
Fig. 3.
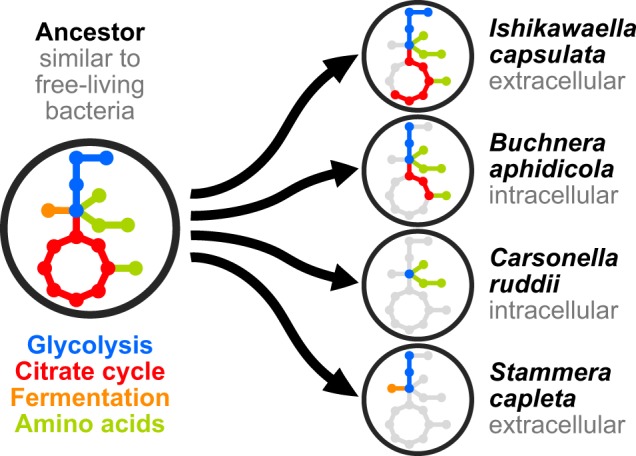
Proposed reductive evolutionary scenarios leading to variable degrees of metabolic pathway loss in representative Gammaproteobacteria symbionts. The putative relatives are similar to free-living microbes, such as Escherichia coli, that possess the complete pathways for glycolysis, aerobic respiration, as well as mixed acid fermentation
In conclusion, despite sharing many metabolic features with intracellular symbionts possessing drastically reduced genomes, Stammera relies on a fermentative metabolism for autonomous energy conversion, permitted by its extracellular location, host supplementation, and pectinolytic function.
Supplementary information
Acknowledgements
We would like to thank Gottfried Unden for valuable discussion on Stammera’s metabolism. The authors gratefully acknowledge the computing time granted on the supercomputer Mogon at Johannes Gutenberg University Mainz (hpc.uni-mainz.de). Funding from the Alexander von Humboldt Foundation (to HS), Wissenschaftskolleg zu Berlin (to HS), Smithsonian Institution (to HS) and European Research Council (CoG 819585 "SYMBeetle" to MK) is gratefully acknowledged.
Author contributions
EB, MK, and HS conceived the study. EB performed the genome and transcriptome annotation. EB, MK, and HS analyzed data. EB wrote the paper. All authors edited and commented on the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Martin Kaltenpoth, Hassan Salem
Contributor Information
Martin Kaltenpoth, Email: mkaltenpoth@uni-mainz.de.
Hassan Salem, Email: hassan.s.salem@gmail.com.
Supplementary information
The online version of this article (10.1038/s41396-019-0562-1) contains supplementary material, which is available to authorized users.
References
- 1.Tamames J, Gil R, Latorre A, Peretó J, Silva FJ, Moya A. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evolut Biol. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salem H, Bauer E, Kirsch R, Berasategui A, Cripps M, Weiss B, et al. Drastic genome reduction in an herbivore’s pectinolytic symbiont. Cell. 2017;171:1520–31.e13. doi: 10.1016/j.cell.2017.10.029. [DOI] [PubMed] [Google Scholar]
- 3.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 4.Kuo C-H, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19:1450–54. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol Evol. 2011;3:702–14. doi: 10.1093/gbe/evr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenyon LJ, Meulia T, Sabree ZL. Habitat visualization and genomic analysis of “Candidatus Pantoea carbekii,” the primary symbiont of the brown marmorated stink bug. Genome Biol Evol. 2015;7:620–35. doi: 10.1093/gbe/evv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 8.Salem Hassan, Florez Laura, Gerardo Nicole, Kaltenpoth Martin. An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc Biol Sci. 2015;282:20142957. doi: 10.1098/rspb.2014.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haines LR, Haddow JD, Aksoy S, Gooding RH, Pearson TW. The major protein in the midgut of teneral Glossina morsitans morsitans is a molecular chaperone from the endosymbiotic bacterium Wigglesworthia glossinidia. Insect Biochem Mol Biol. 2002;32:1429–38. doi: 10.1016/S0965-1748(02)00063-2. [DOI] [PubMed] [Google Scholar]
- 10.Hara E, Fukatsu T, Kakeda K, Kengaku M, Ohtaka C, Ishikawa H. The predominant protein in an aphid endosymbiont is homologous to an E. coli heat shock protein. Symbiosis. 1990;8:271–83. [Google Scholar]
- 11.Wernegreen JJ, Moran NA. Evidence for genetic drift in endosymbionts (Buchnera): Analyses of protein-coding genes. Mol Biol Evol. 1990;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]
- 12.Bennett GM, Chong RA. Genome-wide transcriptional dynamics in the companion bacterial symbionts of the glassy-winged sharpshooter (Cicadellidae Homalodisca vitripennis) reveal differential gene expression in bacteria occupying multiple host organs. G3: Genes, Genomes, Genet. 2017;7:3073–82. doi: 10.1534/g3.117.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–5. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 14.Baum LG, Garner OB, Schaefer K, Lee B. Microbe–host interactions are positively and negatively regulated by galectin–glycan interactions. Front Immunol. 2014;5:284. doi: 10.3389/fimmu.2014.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukamizo T, Kramer KJ. Mechanism of chitin hydrolysis by the binary chitinase system in insect moulting fluid. Insect Biochem. 1985;15:141–45. doi: 10.1016/0020-1790(85)90001-0. [DOI] [Google Scholar]
- 16.Skarzynski T, Mistry A, Wonacott A, Hutchinson SE, Kelly VA, Duncan K. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure. 1996;4:1465–74. doi: 10.1016/S0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 17.Samuels RI, Charnley AK, Reynolds SE. A cuticle-degrading proteinase from the moulting fluid of the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 1993;23:607–14. doi: 10.1016/0965-1748(93)90034-P. [DOI] [PubMed] [Google Scholar]
- 18.Poliakov, A, Russell, CW, Ponnala, L, Hoops, HJ, Sun, Q, Douglas, AE, et al. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol Cell Proteom. 2011; mcp.M110: 007039. [DOI] [PMC free article] [PubMed]
- 19.Ramsey JS, MacDonald SJ, Jander G, Nakabachi A, Thomas GH, Douglas AE. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010;19:241–8. doi: 10.1111/j.1365-2583.2009.00945.x. [DOI] [PubMed] [Google Scholar]
- 20.Thompson SN. Trehalose - the insect ‘blood’sugar. Adv Insect Physiol. 2003;31:85. [Google Scholar]
- 21.Deken RHde. The Crabtree effect: a regulatory system in yeast. Microbiology. 1966;44:149–56. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- 22.Xia N, Yang J, Ye S, Zhang L, Zhou Y, Zhao J, et al. Functional analysis of toxoplasma lactate dehydrogenases suggests critical roles of lactate fermentation for parasite growth in vivo. Cell Microbiol. 2018;20:e12794. doi: 10.1111/cmi.12794. [DOI] [PubMed] [Google Scholar]
- 23.Legrand C, Bour JM, Jacob C, Capiaumont J, Martial A, Marc A, et al. Lactate dehydrogenase (LDH) activity of the number of dead cells in the medium of cultured eukaryotic cells as marker. J Biotechnol. 1992;25:231–43. doi: 10.1016/0168-1656(92)90158-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.