Abstract
Proteomics has been used to investigate cross-talk between the intestinal microbiome and host biological processes. In this study, an in ovo technique and a proteomics approach was used to address how early bacterial colonization in the gastrointestinal tract (GIT) could modulate inflammatory and immune responses in young broilers. Embryos at 18 embryogenic days were inoculated with saline (S), 102 CFU of Citrobacter freundii (CF), Citrobacter species (C2), or lactic acid bacteria mixture (L) into the amnion. At 10 days posthatch, ileum samples from 12 birds per treatment were selected for tandem mass spectrometry analysis. Our further findings indicated that treatment-specific influences on early GIT microbiota resulted in different immune responses in mature broilers. Predicted functional analyses revealed activation of inflammation pathways in broilers treated in ovo with L and CF. Exposure to L enhanced functional annotation related to activation, trafficking of immune cells, and skeletal growth based-network, while CF inhibited biological functions associated with immune cell migration and inflammatory response. These results highlighted that proper immune function was dependent on specific GIT microbiota profiles, in which early-life exposure to L-based probiotic may have modulated the immune functions, whereas neonatal colonization of Enterobacteriaceae strains may have led to immune dysregulation associated with chronic inflammation.
Keywords: segmented filamentous bacteria, probiotic in ovo, immunity, inflammation, Enterobacteriaceae, pioneer colonizers, Ingenuity Pathway Analysis
Introduction
Pioneer colonization of intestinal microbiota has a major effect on driving the maturation and composition course of microbial communities over time (Juricova et al., 2013; Rodrigues et al., 2019; Wilson et al., 2019). Later, the cross-talk between microbiota composition and immune cells has been highly associated with the establishment of immune competence (Crhanova et al., 2011; Chung et al., 2012; Schokker et al., 2017; Duan, 2018). Germ-free mouse models have been essential to reveal a strong influence of intestinal microbial communities on the proper immune function. The lack of intestinal microbiota in these mice caused extensive deficits in the development of the gut-associated lymphoid tissues, abnormal production of immune cells, and other immunological deficiencies (Round and Mazmanian, 2009). In this context, a recent study with broilers has shown that the use of antibiotics during early life perturbed microbiota colonization, subsequently triggering an alteration in systemic immune programming (Schokker et al., 2017). Nevertheless, the specific microbial populations involved in immune-modulatory functions are beginning to be deciphered with the advancement of metagenomic analyses.
It has been reported by Kogut (2019) that the avian neonatal phase is an important window of opportunity to manipulate the intestinal microbiome toward beneficial bacterial growth. In fact, our previous studies showed that early exposure of embryos to lactic acid bacteria or Enterobacteriaceae strains resulted in different microbiome profiles at day of hatch and 10 days of age, suggesting that neonatal exposure to beneficial bacteria may be critical for influencing gastrointestinal tract (GIT) populations throughout the maturation of the poultry microbiota (Wilson et al., 2019). However, whether this pioneer intestinal microbiome modulation can affect the host immunological functions remain unclear.
In this study, the influence of early intestinal bacterial colonization on the inflammatory and immune response of young broilers was investigated. For this purpose, two non-pathogenic Enterobacteriaceae isolates and a lactic acid bacteria-based probiotics were introduced in ovo, and mass spectrometry-based proteome analysis was performed on ileum tissue. To test our hypothesis, we focused on intestinal inflammatory and immune-related proteins, screened the biological functions predicted by Ingenuity Pathway Analysis (IPA), and linked inflammation biomarkers to intestinal microbial signatures established in the ileal microbiome of 10-day-old broiler chickens.
Materials and Methods
Experimental Design
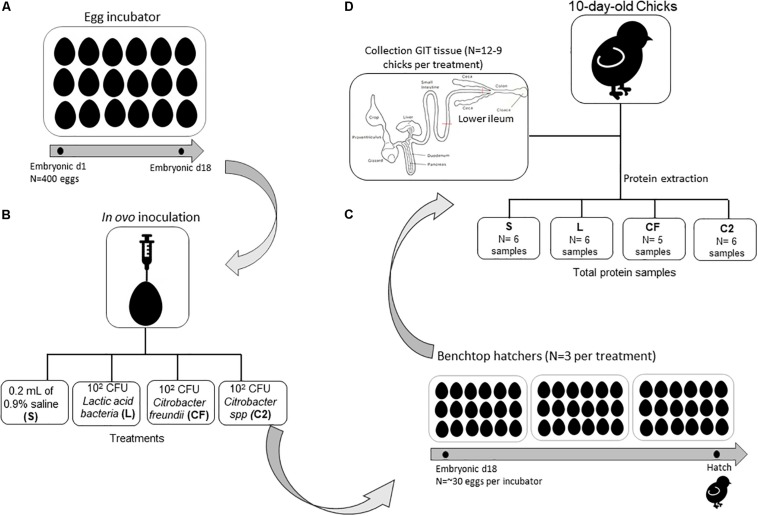
A total of 400 eggs from commercial Ross 708 broiler breeder flocks were obtained from a local hatchery. Per standard operating procedures, the eggs were sanitized before storage and incubation. All eggs were incubated under standard conditions at the Ohio Agricultural Research and Development Center’s poultry research farm. Once eggs were confirmed fertile, at embryonic day 18, the air-cell end of each egg was treated with iodine (povidone–iodine 10% topical solution, Drug Mart, Medina, OH, United States) before a small hole was punched into the shell with an inoculation needle. In ovo inoculations contained one of the following: 0.2 ml of 0.9% sterile saline (S), which served as the control group, or approximately 102 cells of Citrobacter freundii (CF), Citrobacter spp. (C2), or lactic acid bacteria mixture (L) administered into the amnion (Figure 1). After inoculation, up to 30 eggs were allocated by treatments into three separate benchtop hatchers (Hova-Bator model 1602N, Savannah, GA, United States) for a total of 12 hatchers. All hatchers were disinfected with 10% bleach before use. Strains CF and C2 were selected from our previous study as non-pathogenic bacteria from the gut of healthy birds (Bielke et al., 2003), and the homology of strains was confirmed by next-generation sequencing. The L culture was composed of a mixed inoculum of Lactobacillus salivarius and Pediococcus sp. Bacterial inoculations were prepared as described by Wilson et al. (2019). Preliminary experimental observations concluded that the inclusion of isolates at ∼102 CFU did not affect hatchability compared to the S control treatment (data not published). All experimental procedures were approved by the Ohio State University’s Institutional Animal Care and Use Committee.
FIGURE 1.
Schematic overview of experimental design and gastrointestinal (GIT) tissue collection for proteome analyses. (A) Four hundred eggs were incubated under standard conditions in one single-stage egg incubator until embryonic day 18. (B) In ovo inoculations contained one of the following: 0.2 ml of 0.9% sterile saline (S), which served as the control group, or ∼102 cells of Citrobacter freundii (CF), Citrobacter spp. (C2), or a lactic acid bacteria mixture (L) was administered into the amnion. (C) After inoculation, up to 30 eggs were allocated by treatments into three separate benchtop hatchers per treatment. (D) Ten days posthatch, chicks were selected for ileum sample collection (pooled samples of two birds per treatment) to perform proteome analyses.
Sample Collection
Immediately posthatch, chicks were comingled on a treatment basis, and 128 chicks were placed into treatment-separated brooder battery cages with ad libitum access to a standard corn–soy diet and water (Nutrient Requirements of Poultry, 1994). At 10 days posthatch, 12 chicks per treatment were randomly selected for ileal proteome analysis, however, only nine birds were sampled from CF. Chicks were euthanized via cervical dislocation, and the region proximal to the ileocecal junction and distal to Meckel’s diverticulum, designated as lower ileum, was aseptically collected post mortem (Figure 1). Ileum tissue was placed into 1.5-ml tubes, flash frozen in liquid nitrogen at the time of collection, and stored at −80°C until further use.
Once thawed, 0.1 g of ileal tissue from each sample was individually placed in 5 ml of buffer (8 M urea/2 M thiourea, 2 mM dithiothreitol, 50 mM Tris, 5% sodium dodecyl sulfate). The extraction protocol was a modified version previously described by Iqbal et al. (2004) and Kong et al. (2016). In brief, samples were homogenized for 5 s (PRO250 Homogenizer, Pro Scientific, Oxford, CT, United States), then 500 μl of homogenate was added to 2-ml tubes containing 0.1 g stainless steel beads (SSB14B Next Advance, Averill Park, NY, United States). Samples were homogenized for a total of 3 min in 30-second intervals (MiniBeadbeater-16, Model 607, BioSpec Products, Bartlesville, OK, United States) and centrifuged at 4°C at 14,000 rpm (21,952 × g) for 20 min. The supernatant was collected, aliquoted, and placed into −80°C until further use.
To ensure proper extraction, concentration of total protein was quantified with the Bradford assay (Bradford reagent, VWR, Suwanee, GA, United States) and a standard bovine serum albumin curve (VWR, Suwanee, GA, United States) on a Synergy HTX multimode plate reader (BioTek U.S., Winooski, VT, United States). Samples were mixed to create pooled samples of two birds per treatment (n = 6 samples for L and C2; n = 5 samples CF; Figure 1) and sent to the Ohio State University Proteomics Core lab for in-solution digestion and mass spectrometry.
Proteomics Analyses
Samples were precipitated with trichloroacetic acid and then resuspended in 50 mM ammonium bicarbonate. A total of 5 ml of dithiothreitol (5 μg/μl in 50 mM ammonium bicarbonate) was added, and the samples were incubated at 56°C for 15 min. After incubation, 5 μl of iodoacetamide (15 mg/ml in 50 mM ammonium bicarbonate) was added, and the samples were kept in the dark at room temperature for 30 min. Sequencing grade-modified trypsin (Promega; Madison, WI, United States) prepared in 50 mM ammonium bicarbonate was added to each sample at an estimated 1:20/1:100 enzyme/substrate ratio and incubated at 37°C overnight. The reaction was quenched the following day by adding acetic acid for acidification. Once samples were quenched, the peptide concentration was measured by Nanodrop (Thermo Scientific Nanodrop 2000; Waltham, MA, United States).
Capillary-liquid chromatography-nanospray tandem mass spectrometry (capillary-LC/MS/MS) of global protein identification was performed on a Thermo Fisher Fusion mass spectrometer (Thermo Scientific, Waltham, MA, United States). Samples were separated on a Thermo Nano C18 column (UltiMateTM 3000 HPLC system, Thermo Scientific; Waltham, MA, United States). The MS/MS data sequences were scanned and based on the preview mode data-dependent TopSpeedTM method with collision-induced dissociation and electron-transfer dissociation as fragmentation methods. The raw data were searched on Sequest via Proteome Discoverer (Proteome DiscovererTM software, Thermo Scientific, Waltham, MA, United States). The data were searched against the most recent Uniprot Gallus gallus database for the identification of proteins. Only proteins with <0.05 false discovery rate were reported. Proteins with a Mascot score of 50 or higher with a minimum of two unique peptides from one protein having a −b or −y ion sequence tag of five residues or better were accepted. Any modifications or low-score peptide/protein identifications were manually checked for validation.
Biological Interpretation
Label-free quantitation was performed using the spectral count approach, in which the relative protein quantitation is measured by comparing the number of MS/MS spectra identified from the same protein in each of the multiple LC/MS/MS datasets. Comparisons between in ovo bacterial treatments and S control group were performed in Scaffold (Scaffold 4.8.4, Proteome Software, Portland, OR, United States). Student’s t test (p < 0.05) was performed to identify significance across the fold-change values. Differentially expressed proteins (DEPs) at the level of p ≤ 0.10 were uploaded into IPA system1 to retrieve further inflammatory and immune information in terms of gene ontology, upstream regulators, and causal networks. The IPA functionalities for differentially expressed genes in chicken are based primarily on mammalian biological mechanisms (Kong et al., 2011). The statistical measure Z score was displayed to make predictions about potential activation (Z score ≥ 2.00) or inhibition (Z score < −2.00) of regulators using the information of the protein regulation direction. Qualified predictions were also made for high (Z score ≥ | 1.90| or more) medium (Z score = | 1.70–1.90|) or low (Z score = | 1.50–1.70|), respectively (Kong et al., 2016). Casual networks were performed to show mechanistic hypotheses to explain the expression changes observed in the datasets based on cause–effect relationships reported in the literature (Krämer et al., 2014). The p value of the overlap, which measures any significant statistical overlap between the samples in the dataset and the genes that are regulated by the corresponding transcriptional regulator, was also determined and recorded. Fisher’s exact test calculated the value at a significance of p < 0.05.
Correlation Analyses
To further identify the specific bacterial groups that primarily accounted for the differences observed in the expression of inflammation-related proteins in the ileum of broilers, the microbiome data of same samples were added into the analysis, from BioProject ID PRJNA552855, previously published by Rodrigues et al. (2019). The most dominant genus-level operational taxonomic units detected in the four treatments were identified (Supplementary Table S1). Based on the inflammatory annotation generated by the IPA system, the DEPs involved in the inflammation signaling also were determined. Given the considerable number of variables, only overexpressed proteins or those with at least one upregulated DEP within each treatment dataset were selected. Owing to the unlikelihood of perfectly linear relationships and the presence of significant variation between relative bacterial abundance and protein log2-fold change, we applied Spearman’s correlation coefficient (R) using RStudio software.
Results
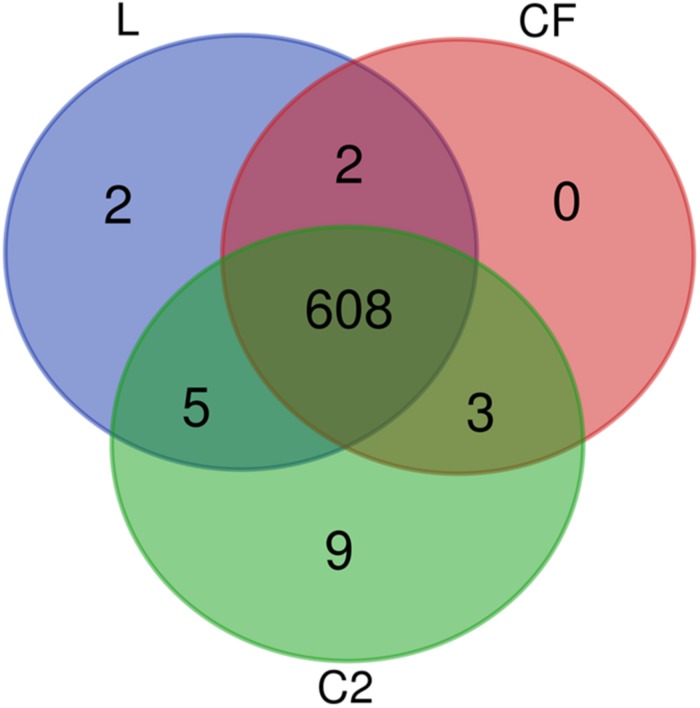
From the proteomic datasets, we identified 617 proteins in L, 613 proteins in CF, and 625 proteins in C2 (Supplementary Tables S2–S4, respectively). Accordingly, 608 proteins were common across all three treatment conditions (Figure 2).
FIGURE 2.
Venn diagram depicting unique and shared proteins identified in the ileum of 10-day-old broilers treated in ovo with lactic acid bacteria mixture (L), Citrobacter freundii (CF), or Citrobacter spp. (C2).
Subsequently, DEPs significantly lower or equal to 0.1 were identified. A total of 61 DEPs were displayed in L (Table 1), 44 DEPs in CF (Table 2), and 63 in C2 (Table 3). Furthermore, we only evaluated the biological interactions associated with inflammatory and immune response signaling.
TABLE 1.
Differentially expressed proteins (DEPs) from ileal samples of broilers treated with lactic acid bacteria mixture (L) in ovo.
| UniProt ID | Proteins | Protein name | Fold change | p Value |
| Q10751 | ACE | Angiotensin I converting enzyme | –1.000 | 0.003 |
| P02612 | MYL9 | Myosin light chain 9 | 2.400 | 0.004 |
| Q5ZIV5 | PDCD10 | Programmed cell death 10 | 6.800 | 0.005 |
| P20678 | Cyp2c23 | Cytochrome P450, family 2, subfamily c, polypeptide 23 | –10.000 | 0.007 |
| Q9PW72 | PDLIM4 | PDZ and LIM domain 4 | –2.000 | 0.013 |
| P24797 | ATP1A2 | ATPase Na + /K + transporting subunit alpha 2 | –1.600 | 0.014 |
| P50594 | MAGOH | Mago homolog, exon junction complex subunit | 4.400 | 0.015 |
| Q90597 | MX2 | MX dynamin like GTPase 2 | –1.600 | 0.016 |
| P08940 | LECT2 | Leukocyte cell derived chemotaxin 2 | 3.500 | 0.017 |
| P67883 | RPL30 | Ribosomal protein L30 | –1.660 | 0.019 |
| Q5ZMU6 | PPP4R2 | Protein phosphatase 4 regulatory subunit 2 | ∞ | 0.021 |
| P31696-10 | AGRN | Agrin | –5.000 | 0.024 |
| P02701 | AVD | Avidin | 6.700 | 0.027 |
| E1C2P3 | HSPA14 | Heat shock protein family A (Hsp70) member 14 | 5.000 | 0.029 |
| P05180 | Cyp2c23 | Cytochrome P450, family 2, subfamily c, polypeptide 23 | –3.300 | 0.031 |
| P47990 | XDH | Xanthine dehydrogenase | –2.500 | 0.031 |
| Q5ZIW2 | CNOT10 | CCR4-NOT transcription complex subunit 10 | –3.300 | 0.032 |
| P05122-5 | CKB | Creatine kinase B | 3.300 | 0.033 |
| P02789 | TF | Transferrin | –1.250 | 0.035 |
| O57535 | NME2 | NME/NM23 nucleoside diphosphate kinase 2 | –1.420 | 0.037 |
| Q6IVA4 | UBA5 | Ubiquitin like modifier activating enzyme 5 | 3.500 | 0.038 |
| Q5ZKG5 | ACP1 | Acid phosphatase 1 | –2.000 | 0.041 |
| Q07598 | SCP2 | Sterol carrier protein 2 | –2.000 | 0.042 |
| Q5ZKP6 | ADA | Adenosine deaminase | –2.000 | 0.043 |
| P09540 | MYL4 | Myosin light chain 4 | 1.800 | 0.045 |
| Q5ZLN5 | TARDBP | TAR DNA binding protein | –2.000 | 0.046 |
| P08636 | RPS17 | Ribosomal protein S17 | 1.300 | 0.046 |
| P61160 | ACTR2 | ARP2 actin related protein 2 homolog | –1.250 | 0.051 |
| Q5ZJL9 | SAMHD1 | SAM and HD domain deoxynucleoside triphosphate triphosphohydrolase 1 | 2.600 | 0.052 |
| Q5ZK88 | PSPC1 | Paraspeckle component 1 | –1.660 | 0.053 |
| Q90629 | PRSS2 | Serine protease 2 | –2.500 | 0.054 |
| Q02391 | GLG1 | Golgi glycoprotein 1 | –1.600 | 0.055 |
| P14105 | MYH9 | Myosin heavy chain 9 | –1.250 | 0.060 |
| P84173 | PHB | Prohibitin | –1.420 | 0.063 |
| P26990 | ARF6 | ADP ribosylation factor 6 | 2.400 | 0.063 |
| P47807 | MYO1A | Myosin IA | –1.600 | 0.066 |
| Q5ZLN1 | PGAM1 | Phosphoglycerate mutase 1 | –1.420 | 0.068 |
| Q5ZHX7 | CYB5R2 | Cytochrome b5 reductase 2 | –2.500 | 0.072 |
| Q5ZLR5 | UQCRFS1 | Ubiquinol-cytochrome c reductase, Rieske iron–sulfur polypeptide 1 | 1.400 | 0.074 |
| P17790 | BSG | Basigin (Ok blood group) | 2.100 | 0.075 |
| Q9YH06 | HMGB1 | High mobility group box 1 | 1.700 | 0.078 |
| Q5ZLT0 | XPO7 | exportin 7 | –2.000 | 0.084 |
| Q5ZKD7 | MOV10 | Mov10 RISC complex RNA helicase | –1.420 | 0.084 |
| P27003 | S100A10 | S100 calcium binding protein A10 | ∞ | 0.084 |
| P10184 | SPINK5 | Serine peptidase inhibitor, Kazal type 5 | –2.000 | 0.085 |
| P05083 | ASL | Argininosuccinate lyase | 2.200 | 0.085 |
| P23228 | HMGCS1 | 3-Hydroxy-3-methylglutaryl-CoA synthase 1 | –2.500 | 0.086 |
| Q5ZM33 | HP1BP3 | Heterochromatin protein 1 binding protein 3 | 1.500 | 0.086 |
| Q5ZK62 | ACAP2 | ArfGAP with coiled coil, ankyrin repeat PH domains 2 | –1.000 | 0.093 |
| P16924 | P4HA1 | Prolyl 4-hydroxylase subunit alpha 1 | –1.420 | 0.095 |
| Q5ZKV8 | KIF2A | Kinesin family member 2A | 3.500 | 0.095 |
| O42265 | PSMA1 | Proteasome subunit alpha 1 | –1.660 | 0.097 |
| P68139 | ACTA1 | Actin, alpha 1, skeletal muscle | 3.200 | 0.098 |
| Q90615 | ITGA1 | Integrin subunit alpha 1 | 1.800 | 0.099 |
| Q5ZHP5 | CHMP4B | Charged multivesicular body protein 4B | –1.420 | 0.100 |
| P70079 | CKMT1 | Creatine kinase U-type, mitochondrial | –1.250 | 0.100 |
| P09652 | TUBB3 | Tubulin beta 3 class III | 2.500 | 0.100 |
| P24802 | PLOD1 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 3.000 | 0.100 |
| Q5ZKC1 | EIF2A | Eukaryotic translation initiation factor 2A | ∞ | 0.100 |
TABLE 2.
Summary of differentially expressed proteins (DEPs) identified in the ileum of broilers exposed to Citrobacter freundii (CF) in ovo.
| UniProt ID | Proteins | Protein name | Fold change | p Value |
| Q6JHU8 | P3H1 | Prolyl 3-hydroxylase 1 | 3.200 | 0.003 |
| Q5ZKC9 | YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 1.500 | 0.003 |
| Q5ZI89 | TRAPPC11 | Trafficking protein particle complex 11 | –1.000 | 0.007 |
| Q01841 | TGM2 | Transglutaminase 2 | –1.420 | 0.010 |
| P00940 | TPI1 | Triosephosphate isomerase 1 | 1.500 | 0.015 |
| Q10751 | ACE | Angiotensin I converting enzyme | –10.000 | 0.016 |
| P55080 | MFAP1 | Microfibril-associated protein 1 | 7.500 | 0.018 |
| Q5ZIV5 | PDCD10 | Programmed cell death protein 10 | 5.800 | 0.021 |
| Q9PW72 | PDLIM4 | PDZ and LIM domain 4 | –5.000 | 0.021 |
| Q5ZM35 | TWF2 | Twinfilin actin binding protein 2 | 7.100 | 0.022 |
| Q90615 | ITGA1 | Integrin subunit alpha 1 | 1.600 | 0.026 |
| Q02391 | GLG1 | Golgi glycoprotein 1 | –4.000 | 0.028 |
| Q5ZLN0 | LRRC40 | Leucine rich repeat containing 40 | 2.200 | 0.031 |
| Q5ZJ08 | YARS | Tyrosyl-tRNA synthetase | –4.000 | 0.031 |
| Q02020 | FGB | Fibrinogen beta chain | –1.600 | 0.032 |
| P26446 | PARP1 | Poly(ADP-ribose) polymerase 1 | –3.300 | 0.036 |
| Q9W6H0 | OGN | Osteoglycin | 1.600 | 0.037 |
| Q5ZKU5 | RAB14 | RAB14, member RAS oncogene family | –4.000 | 0.038 |
| P51890 | LUM | Lumican | –2.500 | 0.042 |
| Q5ZHN4 | RP2 | RP2, ARL3 GTPase activating protein | –3.300 | 0.051 |
| P00548 | PKM | Pyruvate kinase M1/2 | –1.250 | 0.052 |
| P0CB50 | PRDX1 | Peroxiredoxin 1 | –1.600 | 0.052 |
| P19966 | TAGLN | Transgelin | 2.100 | 0.057 |
| P87362 | BLMH | Bleomycin hydrolase | –2.500 | 0.059 |
| Q08392 | Gsta1 | Glutathione S-transferase alpha 1 | 1.700 | 0.059 |
| P13914 | NAT1 | N-acetyltransferase 1 | 2.600 | 0.059 |
| Q5ZIY5 | PPP2R2D | Protein phosphatase 2 regulatory subunit B delta | 2.000 | 0.062 |
| P05083 | ASL | Argininosuccinate lyase | 2.200 | 0.064 |
| P23228 | HMGCS1 | 3-Hydroxy-3-methylglutaryl-CoA synthase 1 | –3.300 | 0.064 |
| Q90YH9 | TES | Testin LIM domain protein | –2.500 | 0.067 |
| Q5F381 | EPCAM | Epithelial cell adhesion molecule | –1.420 | 0.069 |
| P56517 | HDAC1 | Histone deacetylase 1 | –4.000 | 0.070 |
| A0A1N8W591 | HDAC1 | Histone deacetylase | 1.200 | 0.075 |
| Q90611 | MMP2 | Matrix metallopeptidase 2 | –1.000 | 0.076 |
| Q5F418 | PSMD1 | Proteasome 26S subunit, non-ATPase 1 | –4.000 | 0.076 |
| Q5ZIL2 | VPS29 | VPS29, retromer complex component | –1.000 | 0.076 |
| Q5F464 | LPP | LIM domain containing preferred translocation partner in lipoma | 1.300 | 0.079 |
| Q5ZJ27 | HOOK1 | Hook microtubule tethering protein 1 | –1.000 | 0.090 |
| P02701 | AVD | Avidin | 6.200 | 0.099 |
| Q05744 | CTSD | Cathepsin D | 1.800 | 0.100 |
| P12902 | HMG-14A | Non-histone chromosomal protein HMG-14A | –3.300 | 0.100 |
| Q98TF8 | RPL22 | Ribosomal protein L22 | 2.000 | 0.100 |
TABLE 3.
Differentially expressed proteins (DEPs) in ileum of 10-day-old broilers exposed to Citrobacter spp. (C2) in ovo.
| UniProt ID | Proteins | Protein name | Fold change | p Value |
| P05122 | CKB | Creatine kinase B | –1.250 | 0.002 |
| P47836 | RPS4X | Ribosomal protein S4 X-linked | –1.600 | 0.002 |
| Q2IAL7 | CATHL2 | Cathelicidin-2 | 4.500 | 0.003 |
| P05122-5 | CPK-B | Creatine kinase B Isoform 4 | –10.000 | 0.006 |
| P09652 | TUBB3 | Tubulin beta 3 class III | 2.900 | 0.006 |
| Q9YGQ1 | EEF1B2 | Eukaryotic translation elongation factor 1 beta 2 | 1.800 | 0.008 |
| Q5ZMU6 | PPP4R2 | Protein phosphatase 4 regulatory subunit 2 | ∞ | 0.016 |
| P24802 | PLOD1 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 3.400 | 0.020 |
| Q5ZHP5 | CHMP4B | Charged multivesicular body protein 4B | –1.600 | 0.021 |
| Q5ZIW2 | CNOT10 | CCR4-NOT transcription complex subunit 10 | –5.000 | 0.028 |
| Q5ZKA4 | EIF3J | Eukaryotic translation initiation factor 3 subunit J | 1.600 | 0.032 |
| Q90615 | ITGA1 | Integrin subunit alpha 1 | 1.600 | 0.032 |
| Q8UVD9 | KHSRP | Far upstream element-binding protein 2 | –1.420 | 0.034 |
| P05083 | ASL | Argininosuccinate lyase | 2.300 | 0.035 |
| Q5ZHN4 | RP2 | RP2, ARL3 GTPase activating protein | –3.300 | 0.037 |
| Q5ZKC1 | EIF2A | Eukaryotic translation initiation factor 2A | ∞ | 0.039 |
| P41125 | RPL13 | Ribosomal protein L13 | 1.700 | 0.041 |
| Q5ZLR5 | UQCRFS1 | Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 1.600 | 0.044 |
| P27731 | TTR | Transthyretin | 1.800 | 0.049 |
| Q5ZLN1 | PGAM1 | Phosphoglycerate mutase 1 | –1.420 | 0.050 |
| P23228 | HMGCS1 | 3-Hydroxy-3-methylglutaryl-CoA synthase 1 | –3.300 | 0.051 |
| P08106 | HSPA2 | Heat shock protein family A (Hsp70) member 2 | –1.420 | 0.051 |
| Q5ZK33 | LETM1 | Leucine zipper and EF-hand containing transmembrane protein 1 | 2.400 | 0.051 |
| P49712 | ATP6V1B2 | ATPase H + transporting V1 subunit B2 | 1.600 | 0.052 |
| Q5ZJU3 | ASNS | Asparagine synthetase (glutamine-hydrolyzing) | 1.700 | 0.053 |
| P24797 | ATP1A2 | ATPase Na + /K + transporting subunit alpha 2 | –1.420 | 0.053 |
| Q90629 | PRSS2 | Serine protease 2 | –2.500 | 0.053 |
| O73885 | HSPA8 | Heat shock protein family A (Hsp70) member 8 | –1.420 | 0.054 |
| P62846 | RPS15 | Ribosomal protein S15 | 2.000 | 0.060 |
| P26990 | ARF6 | ADP ribosylation factor 6 | 2.600 | 0.061 |
| Q90593 | HSPA5 | Heat shock protein family A (Hsp70) member 5 | –1.250 | 0.061 |
| P30622 | CLIP1 | CAP-Gly domain containing linker protein 1 | ∞ | 0.062 |
| P09540 | MYL4 | Myosin light chain 4 | 1.400 | 0.063 |
| O75083 | WDR1 | WD repeat domain 1 | 3.000 | 0.063 |
| P10184 | SPINK5 | Serine peptidase inhibitor, Kazal type 5 | 1.500 | 0.065 |
| O59725 | mic60 | MICOS complex subunit Mic60 | 8.000 | 0.066 |
| Q5ZL77 | RIC8A | RIC8 guanine nucleotide exchange factor A | ∞ | 0.066 |
| P02675 | FGB | Fibrinogen beta chain | 1.800 | 0.068 |
| Q90835 | EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 1.400 | 0.069 |
| Q5ZLD7 | VPS53 | VPS53, GARP complex subunit | ∞ | 0.071 |
| P12276 | FASN | Fatty acid synthase | 1.600 | 0.072 |
| E1C2P3 | HSPA14 | Heat shock protein family A (Hsp70) member 14 | 3.200 | 0.072 |
| Q5ZJJ2 | RPA1 | Replication protein A1 | –2.500 | 0.073 |
| P02314 | HMGN2 | Non-histone chromosomal protein HMG-17 | –2.500 | 0.075 |
| Q5ZLN0 | LRRC40 | Leucine rich repeat containing 40 | 2.000 | 0.076 |
| O93256 | KRT19 | keratin 19 | 9.700 | 0.077 |
| Q9YHT2 | SDHB | Succinate dehydrogenase complex iron sulfur subunit B | 1.900 | 0.078 |
| P51890 | LUM | Lumican | –4.000 | 0.079 |
| Q5ZLL5 | COQ5 | Coenzyme Q5, methyltransferase | 1.800 | 0.084 |
| P04354 | CALB1 | Calbindin 1 | –1.600 | 0.086 |
| Q5ZI72 | HNRNPDL | Heterogeneous nuclear ribonucleoprotein D like | 1.800 | 0.086 |
| P02001 | HBAD | Hemoglobin subunit alpha-D | 1.800 | 0.088 |
| P08251 | ATP1B1 | ATPase Na+/K+transporting subunit beta 1 | –2.500 | 0.089 |
| P09102 | P4HB | Prolyl 4-hydroxylase subunit beta | –1.250 | 0.090 |
| P12957 | Cald1 | Caldesmon 1 | –1.250 | 0.094 |
| P68139 | ACTA1 | Actin, alpha 1, skeletal muscle | 3.200 | 0.096 |
| P02612 | MYL9 | Myosin light chain 9 | 1.700 | 0.096 |
| P26583 | HMGB2 | High mobility group box 2 | –2.500 | 0.097 |
| P08940 | LECT2 | Leukocyte cell derived chemotaxin 2 | 5.100 | 0.097 |
| Q5ZKQ5 | CASC4 | Cancer susceptibility 4 | –1.600 | 0.100 |
| O13268 | PSMA7 | Proteasome subunit alpha 7 | –1.420 | 0.100 |
| Q5ZHX1 | RAP1B | RAP1B, member of RAS oncogene family | 1.400 | 0.100 |
| P27003 | S100A10 | S100 calcium binding protein A10 | ∞ | 0.100 |
Biological Functions
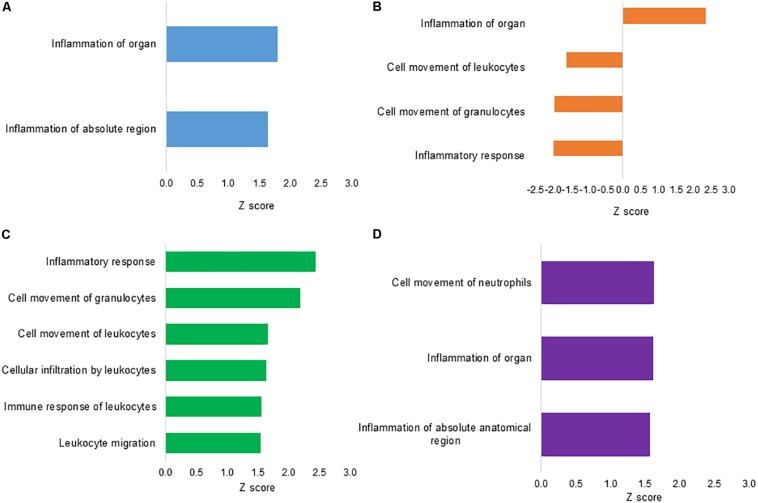
To assess the functional annotation associated with DEPs, the disease processes and cellular functions were predicted by the IPA approach. Figure 3 summarizes the major predicted effects on functional annotation coordinated to the inflammatory and immune responses. In L treatment, inflammation of organ (Z score = 1.80) and inflammation of absolute region (Z score = 1.65) were predicted to be activated in relation to S control group (Figure 3A). In CF, a predicted high activation Z score value was assigned only to inflammation of organ (Z score = 2.32). Biological functions associated with inflammatory response (Z score = −1.96), cell movement of granulocytes (Z score = −1.95) and leukocytes (Z score = −1.60) were predicted to be inhibited when compared to S control group (Figure 3B). There was no qualified prediction of downstream functional effects in C2.
FIGURE 3.
Downstream effects analysis based on differentially expressed proteins from ileal samples of broilers early exposed to lactic acid bacteria mixture (L), Citrobacter freundii (CF), or Citrobacter spp. (C2). Biological functions involved with inflammation and immune response processes of (A) L vs. S, following (B) CF vs. S, (C) L vs. CF, and (D) L vs. C2. The vertical axis shows the predicted biological function, while the horizontal axis shows the Z score calculated for the particular effect.
To better understand the differential regulation of inflammatory and immune signaling response across treatments, the L protein expression metadata generated by IPA were further compared to CF and C2 profiles (IPA comparison analyses). As illustrated in Figure 3C, when L was compared to CF, the biological functional analysis predicted the activation of leukocyte migration (Z score = 1.54), immune response of leukocytes (Z score = 1.56), cellular infiltration by leukocytes (Z score = 1.64), cell movement of leukocytes (Z score = 1.67), cell movement of granulocytes (Z score = 2.19), and inflammatory response (Z score = 2.44). When examining L versus C2, there were predicted activation of inflammation of absolute anatomical region (Z score = 1.57), inflammation of organ (Z score = 1.62), and cell movement of neutrophils (Z score = 1.63; Figure 3D).
Inflammatory-Related Proteins
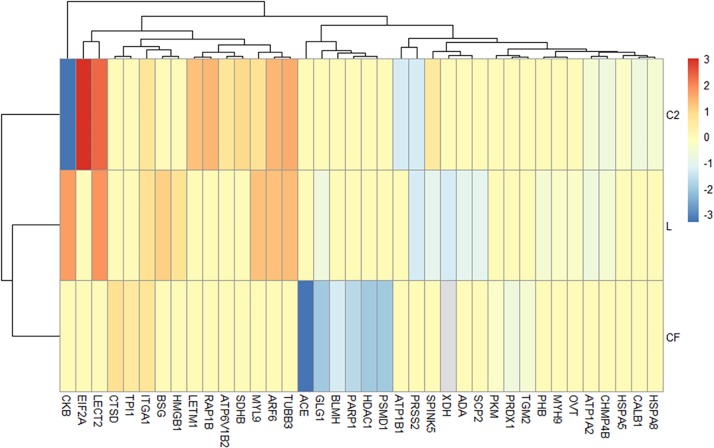
Based on the IPA diseases and function annotation, the proteomic signatures associated with inflammatory downstream effects were identified (Figure 4 and Supplementary Table S5). Venn diagrams show the ileal DEPs related to inflammatory molecular functions (Supplementary Figure S1) in each in ovo treatment. The exposure to L in ovo downregulated the expression of eleven proteins (PRSS2, XDH, ADA, SCP2, SPINK5, ATP1A2, GLG1, PHB, MYH9, OVT, and ACE) and overexpressed eight (HMGB1, ITGA1, BSG, ARF6, MYL9, TUBB3, CKB, and LECT2). Similarly, there were nine downregulated (ACE, GLG1, HDAC1, PSMD1, PARP1, BLMH, PRDX1, TGM2, and PKM) and three upregulated DEPs (TPI1, ITGA1, and CTSD) found in CF versus S control. In the C2 treatment, the expression of eight proteins (CKB, ATP1B1, PRSS2, CALB1, CHMP4B, ATP1A2, HSPA8, and HSPA5) related to the inflammatory mechanism was downregulated. Likewise, seven DEPs (RAP1B, SPINK5, ATP6V1B2, SDHB, LECT2, TUBB3, MYL9, LETM1, ARF6, ITGA1, and EIF2A) were found to be upregulated in the ileum of C2-treated chicks. The Venn diagram showed that nine proteins (ATP1A2, TUBB3, LECT2, MYL9, PRSS2, CKB, ARF6, SPINK5, and CHMP4B) were present throughout L and C2 treatments, and only ITGA1 was shared across L, CF, and C2 (Supplementary Figure S1). It is also important to note that some of the above proteins were implicated in more than one process.
FIGURE 4.
Differentially expressed proteins (DEPs) profiling identified by Ingenuity Pathway Analysis associated with the inflammatory annotation. (A) Heatmap plot represents the log2-fold changes of proteins expressed in ileal samples of broilers treated with lactic acid bacteria mixture (L), Citrobacter freundii (CF), or Citrobacter spp. (C2) in relation to control treatment. Orange shades represent underexpression, while blue shades indicate overexpression of the particular protein. Light gray cells indicate no expression. Hierarchical clustering in the rows is based on the protein composition similarity between treatments, while that in the columns is based on protein abundance.
Causal Network
Network analysis, derived from Ingenuity Knowledge Base, was drawn to determine the likely relevant causal relationships for changes in DEPs within the in ovo datasets. The top-enriched network in L treatment contained proteins associated with nucleic acid metabolism, skeletal and muscular development, and small molecule biochemistry (score, 44; 19 molecules, Supplementary Figure S2A). The DEPs identified in this network included ACP1, ACTA1, ACTR2, ADA, AGRN, ATP1A2, BSG, CKB, KIF2A, MX2, MYH9, MYL4, MYL9, P4HA1, PGAM1, RPS17, TARDBP, UBA5, and XDH.
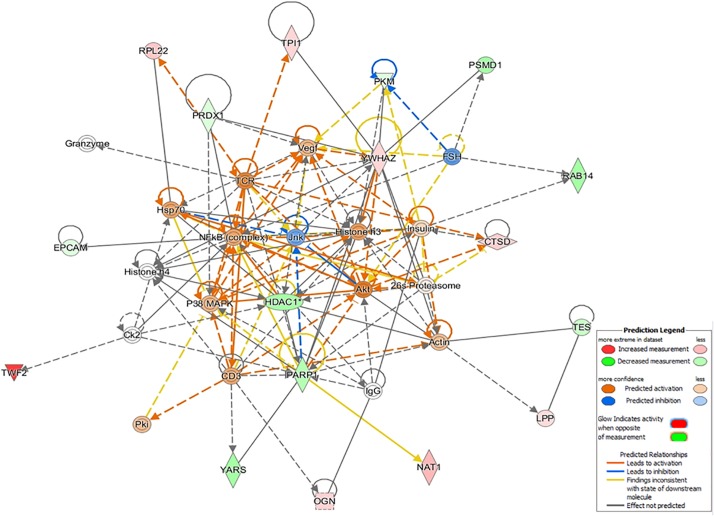
Interestingly, only in the CF dataset, the top-enriched network was composed of several proteins related to immunological and inflammatory functions, as well as the hub regulators Hsp70, nuclear factor-κB (complex), Vegf, and CD3. This network complex was characterized as cancer, organismal injury, abnormalities, and respiratory disease (42 score, 17 molecules, Figure 5), and the determined DEPs were CTSD, EPCAM, HDAC1, LPP, NAT1, OGN, PARP1, PKM, PRDX1, PSMD1, RAB14, RPL22, TES, TPI1, TWF2, YARS, and YWHAZ.
FIGURE 5.
The top-enriched IPA causal network in Citrobacter freundii (CF) relates to cancer, organismal injury, abnormalities, and respiratory disease. The networks are built graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The red nodes represent the upregulation of proteins in the dataset, while green represents downregulation. The color intensity represents the relative magnitude of change in protein expression.
In C2, the most enriched network was linked to cellular function and maintenance, endocrine system disorders, and small molecule biochemistry (score 47, 20 molecules, Supplementary Figure S2B). This network consists of 20 proteins in our proteomic data set (ATP1A2, ATP1B1, ATP6V1B2, Cald1, CLIP1, HMGCS1, HSPA5, ITGA1, KRT19, LRRC40, LUM, MYL4, P4HB, PGAM1, PRSS2, RAP1B, RPS15, RPS4X, S100A10, and SDHB).
Correlation Analysis
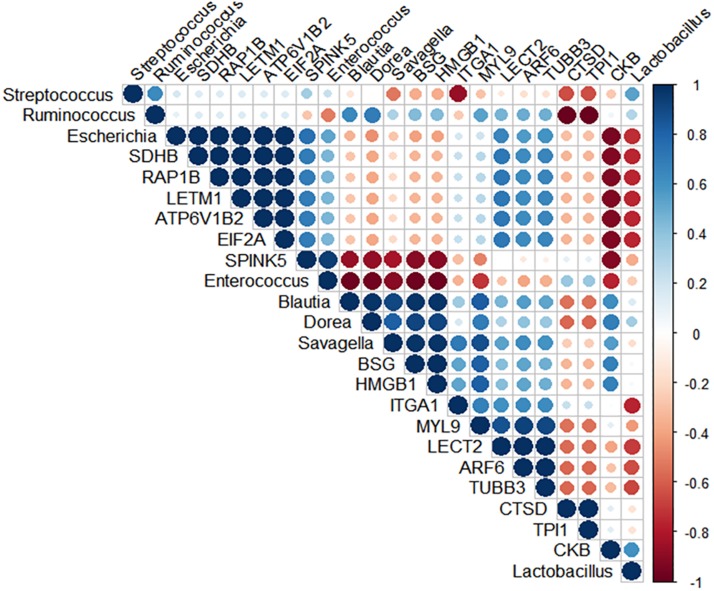
Spearman’s rank correlation was performed to search for potential positive relationships between the microbial composition and the intestinal inflammatory related-proteins (Figure 6 and Supplementary Table S6). Correlation analysis of the broilers’ intestinal proteome and bacterial abundance revealed a positive relationship between Candidatus Savagella, also known as segmented filamentous bacteria (SFB), and myosin light chain 9 (MYL9; R = 0.95, p = 0.051) and Integrin alpha-1 (ITGA1; R = 0.95, p = 0.051). In addition, Enterococcus was positively associated with serine peptidase inhibitor Kazal type 5 (SPINK5; R = 0.95, p = 0.051).
FIGURE 6.
Spearman’s rank correlation matrix of the dominant microbial populations and inflammation-related proteins in the ileum of 10-day-old broilers. Strong correlations are indicated by large circles. The colors of the scale bar denote the nature of the correlation, indicating a perfect positive correlation (dark blue) and –1 indicating perfect negative correlation (dark red). Candidatus Savagella population was positively correlated with MYL9 (R = 0.95, p = 0.051) and ITGA1 (R = 0.95, p = 0.051), as well as Enterococcus abundance had a positive association with SPINK5 (R = 0.95, p = 0.051).
Discussion
Given the magnitude of understanding the cross-talk between intestinal microbiota and host physiology, the impact of the early intestinal colonization on the microbiota in young broilers has previously been addressed by our lab (Rodrigues et al., 2019; Wilson et al., 2019). To comprehensively complement these previous studies, we used a proteomic approach to examine the ileal protein composition in response to the early exposure to L-based probiotics or Enterobacteriaceae strains. The findings presented here indicate that the intestinal pioneer colonization may modulate the immunological functions of young broilers.
Predicted function analyses showed enhanced annotation related to inflammation signaling in broilers in ovo treated with L (Z score = 1.80; Figure 3A) and CF (Z score = 2.32; Figure 3B). The prediction of inflammatory signaling found in L treatment was supported by the upregulation of a marker of inflammation, such as high mobility group box 1 (HMGB1; Yang and Tracey, 2010). Furthermore, treatment-specific biological functions found in L were associated with inflammatory response (Figure 3C). In general, the inflammatory response may be beneficial for the host as an acute and transient mechanism to mediate clearance of inciting agents in the GIT. Alternatively, the failure to control inflammation could inflict chronic and severe tissue damage (Xiao, 2017; Ptaschinski and Lukacs, 2018).
Currently, the predominant knowledge of intestinal inflammation in poultry is based on dysbiosis and mucosal barrier leakage studies (Kuttappan et al., 2015; Bielke et al., 2017; Ducatelle et al., 2018). These physiological conditions are primarily caused by the proliferation and colonization of pathogens in the intestine. However, the recent multiomics pipelines have identified commensal bacteria not only influencing metabolic and immune function but also triggering a state of tolerance with an inflammation-like response (Buffie and Pamer, 2013; Kogut et al., 2018). It has been proposed that SFB, a Clostridiaceae member, have a dominant effect on the mucosal immune system via stimulation of T cells, such as Th17, and increased proinflammatory cytokines and immunoglobulin A (IgA) production. Owing to the fact that colonization of SFB stimulates IgA release, it has been postulated as one of the mechanisms by which SFB might control pathogen overgrowth (Chen et al., 2018). On the other hand, the excessive immune reactions driven by SFB may also be accompanied by a physiological inflammation status (Ivanov et al., 2009; Ivanov and Littman, 2010; Chung et al., 2012; Buffie and Pamer, 2013). Our microbiome results previously published by Wilson et al. (2019) and Rodrigues et al. (2019) showed the succession of microbial communities colonization through the maturation of microbiota in chicks treated with L in ovo, with no evidence of potential pathogens overgrowth. Nonetheless, the previous findings showed a reduction in Enterococcus abundance and a particular prominent population of SFB in lower ileum of 10-day-old broilers in L treatment (Supplementary Table S1; Rodrigues et al., 2019). In this context, further action was taken to identify a potential microbial signature, which could be highly associated with the observed ileal inflammatory signaling. Spearman’s coefficient analyses revealed a strong positive correlation between SFB population and ITGA1 and MYL9. The protein MYL9 plays a crucial role in the complex system that regulates the contraction of smooth muscle. Recently, Hayashizaki et al. (2016) reported that MYL9 and MYL12 are functional ligands for CD69, suggesting that CD69-MYL9/12 interaction is also involved in recruiting activated cells to inflamed tissues. Indeed, high expression of MYL9/12 was related to sepsis-induced acute kidney injury, inflamed mouse airways, and patients with eosinophilic chronic rhinosinusitis (Wu et al., 2015; Hayashizaki et al., 2016). Similarly, Zhang et al. (2018) identified ITGA1 as an inflammatory-associated gene. Notably, we speculate that the high SFB population may be driving the predicted ileal mucosa proinflammatory status found in broilers early exposed to L. Nevertheless, the mechanism underlying SFB-mediated inflammation is yet unknown and must be further explored. Other groups have hypothesized that the interaction of commensal microbiota and young broilers during the development of the immune system could result in a transient inflammation without tissue damage. To the best of our knowledge, the link between SFB colonization and intestinal inflammation in broilers has not been proposed before. Future research is warranted to validate the SFB-associated proteomic signature generated from mass spectrometry. Such studies have the potential to develop markers of colonization and physiological effects mediated by SFB in human medicine and livestock.
Further support of early-life microbiota in programming immunological functions was shown by the biological annotation and causal network analyses. Treatment with L promoted a differential regulation of systemic immune processes in molecular profiles compared to CF and C2. The exposure of embryos to L enhanced the immune response function annotation associated with activation and trafficking of immune cells (Figures 3C,D), and the top-enriched network was related to skeletal growth (Supplementary Figure S2A). Citrobacter treatments, particularly CF, promoted inhibition of functions linked to immune cell migration and inflammatory response (Figures 3C,D, 5). Besides, based on the up- and downexpression pattern of proteins and the cause–effect relationships existing in Ingenuity Knowledge Base, a network related to injury and abnormalities in the organ was predicted in CF. This network is concerned primarily with the canonical nuclear factor-κB pathway, which is activated during the onset of inflammation (Lawrence, 2009). Our recent research has shown that the neonatal Enterobacteriaceae colonization mediated intestinal proteomic changes accompanied by inflammation in newly hatched chicks (Wilson et al., 2020). Chronic inflammation develops when the immune system is unable to clear a persisting insult, which generates a harmful environment and results in tissue impair (Meirow and Baniyash, 2017). Under all conditions, long-term inflammation can suppress immunity by decreasing immune cell numbers and function and/or increasing active immunosuppressive mechanisms (Dhabhar, 2009). Based on the cumulative findings, it is thought that the introduction of CF in ovo produced specific host–microbe interactions, which may have led to dysregulated immunity and immunosuppression by inducing long-term inflammation in 10-day-old broilers.
The maturation of the immune system starts in the first week of life in broilers (Crhanova et al., 2011), and although the immune maturation process can be driven by genetics and environmental conditions, the intestinal microbial composition has been identified to play a significant role in modulating immune responses (Crhanova et al., 2011; Chung et al., 2012; Arsenault et al., 2014; Gollwitzer and Marsland, 2015). Given these pieces of evidence, it is believed that manipulating the intestinal bacteria colonization with early-life exposure to L-based probiotics may modulate the development and maturation of immune functions of broilers. Madej and Bednarczyk (2016) and Pender et al. (2017) have reported that the modification of early-life intestinal microbiota through Lactobacillus-based probiotics in ovo stimulated an immunomodulatory effect in broiler chickens. In addition, the supplementation of host-tailored probiotics has shown to enhance colonization of ileal SFB, which may have contributed to confer immunostimulatory benefits for turkeys (Ward et al., 2019). Our analyses here show that the high colonization of SFB (Rodrigues et al., 2019) may be potentially associated with the enhancement of immune response in L-treated broilers at 10 days of age. These results increased the evidence that host-specific microbiota may drive the intestinal immune maturation (Ivanov et al., 2009; Crhanova et al., 2011; Chung et al., 2012; Buffie and Pamer, 2013; Hedblom et al., 2018).
In summary, the proteomics and bioinformatics analyses presented here suggest that despite shared predicted inflammation pathways by 10 days of age, triggers and inflammatory response were treatment specific in L and CF birds. Based on the microbiome profile previously published by Rodrigues et al. (2019), it is speculated that the high population of SFB in the ileum may be associated with the inflammation-like response found in L treatment, while the early intestinal colonization by Enterobacteriaceae may have caused a low-grade chronic inflammation in the intestine of CF birds. Supporting this, it was highlighted that proper immune function was dependent on specific intestinal microbiota. For instance, exposure of L-based probiotics may have shaped the development of immune functions, whereas the complexity of intestinal microbiota caused by early colonization of Enterobacteriaceae strains may have dysregulated the immunological response in 10-day-old broilers.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015504.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
Author Contributions
DR, MT, KW, AD, KC, and WB carried out the project. DR performed analyses, interpreted the results, and wrote the manuscript in consultation with LB and WB. All authors contributed to experimental design, discussed the results, and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Proteomics Lab and Dr. Wilbur Ouma for their bioinformatics assistance. The authors would also like to thank the OARDC Poultry Research Farm for providing eggs and participating in the animal husbandry portion of the study. Lastly, the authors would like to acknowledge and thank the OARDC Research Enhancement Committee Grants Program (SEEDS) and Arkansas Biosciences Institute for providing the funding of this research.
Funding. This research was supported by the OARDC Research Enhancement Competitive Grants Program (SEEDS) grant no. 2016035, and Arkansas Biosciences Institute (ABI: Little Rock, AR).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00020/full#supplementary-material
References
- Arsenault R. J., Trost B., Kogut M. H. (2014). A comparison of the chicken and turkey proteomes and phosphoproteomes in the development of poultry-specific immuno-metabolism kinome peptide arrays. Front. Vet. Sci. 1:22. 10.3389/fvets.2014.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielke L. R., Elwood A. L., Donoghue D. J., Donoghue A. M., Newberry L. A., Neighbor N. K., et al. (2003). Approach for selection of individual enteric bacteria for competitive exclusion in turkey poults. Poult. Sci. 82 1378–1382. 10.1093/ps/82.9.1378 [DOI] [PubMed] [Google Scholar]
- Bielke L. R., Hargis B. M., Latorre J. D. (2017). “Impact of enteric health and mucosal permeability on skeletal health and lameness in poultry,” in Understanding the Gut-Bone Signaling Axis: Mechanisms and Therapeutic Implications Advances in Experimental Medicine and Biology, eds McCabe L. R., Parameswaran N. (Cham: Springer; ), 185–197. 10.1007/978-3-319-66653-2_9 [DOI] [PubMed] [Google Scholar]
- Buffie C. G., Pamer E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13 790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Chen H., Shu X., Yin Y., Li J., Qin J., et al. (2018). Presence of segmented filamentous bacteria in human children and its potential role in the modulation of human gut immunity. Front. Microbiol. 9:1403. 10.3389/fmicb.2018.01403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Pamp S. J., Hill J. A., Surana N. K., Edelman S. M., Troy E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149 1578–1593. 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crhanova M., Hradecka H., Faldynova M., Matulova M., Havlickova H., Sisak F., et al. (2011). Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar enteritidis infection. Infect. Immun. 79 2755–2763. 10.1128/IAI.01375-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhabhar F. S. (2009). Enhancing versus suppressive effects of stress on immune function: implications for immunoprotection and immunopathology. Neuroimmunomodulation 16 300–317. 10.1159/000216188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M. (2018). Microbiota and immune cell crosstalk: dialogues driving health and disease. Clin. Transl. Immunol. 7:e1020. 10.1002/cti2.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle R., Goossens E., De Meyer F., Eeckhaut V., Antonissen G., Haesebrouck F., et al. (2018). Biomarkers for monitoring intestinal health in poultry: present status and future perspectives. Vet. Res. 49:43. 10.1186/s13567-018-0538-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollwitzer E. S., Marsland B. J. (2015). Impact of early-life exposures on immune maturation and susceptibility to disease. Trends Immunol. 36 684–696. 10.1016/j.it.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Hayashizaki K., Kimura M. Y., Tokoyoda K., Hosokawa H., Shinoda K., Hirahara K., et al. (2016). Myosin light chains 9 and 12 are functional ligands for CD69 that regulate airway inflammation. Sci. Immunol. 1:eaaf9154. 10.1126/sciimmunol.aaf9154 [DOI] [PubMed] [Google Scholar]
- Hedblom G. A., Reiland H. A., Sylte M. J., Johnson T. J., Baumler D. J. (2018). Segmented filamentous bacteria – metabolism meets immunity. Front. Microbiol. 9:1991. 10.3389/fmicb.2018.01991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M., Pumford N. R., Tang Z. X., Lassiter K., Wing T., Cooper M., et al. (2004). Low feed efficient broilers within a single genetic line exhibit higher oxidative stress and protein expression in breast muscle with lower mitochondrial complex activity. Poult. Sci. 83 474–484. 10.1093/ps/83.3.474 [DOI] [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139 485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Littman D. R. (2010). Segmented filamentous bacteria take the stage. Mucosal Immunol. 3 209–212. 10.1038/mi.2010.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juricova H., Videnska P., Lukac M., Faldynova M., Babak V., Havlickova H., et al. (2013). Influence of Salmonella enterica serovar enteritidis infection on the development of the cecum microbiota in newly hatched chicks. Appl. Env. Microbiol. 79 745–747. 10.1128/AEM.02628-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut M. H. (2019). The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Technol. 250 32–40. 10.1016/j.anifeedsci.2018.10.008 [DOI] [Google Scholar]
- Kogut M. H., Genovese K. J., Swaggerty C. L., He H., Broom L. (2018). Inflammatory phenotypes in the intestine of poultry: not all inflammation is created equal. Poult. Sci. 97 2339–2346. 10.3382/ps/pey087 [DOI] [PubMed] [Google Scholar]
- Kong B., Song J., Lee J. Y., Hargis B. M., Wing T., Lassiter K., et al. (2011). Gene expression in breast muscle associated feed efficiency in a single male broiler line using a chicken 44k microarray. I. Top differentially expressed genes. Poult. Sci. 90 2535–2547. 10.3382/ps.2011-01435 [DOI] [PubMed] [Google Scholar]
- Kong B.-W., Lassiter K., Piekarski-Welsher A., Dridi S., Reverter-Gomez A., Hudson N. J., et al. (2016). Proteomics of breast muscle tissue associated with the phenotypic expression of feed efficiency within a pedigree male broiler line: I. highlight on mitochondria. PLoS One 11:e0155679. 10.1371/journal.pone.0155679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Tugendreich S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30 523–530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttappan V. A., Vicuña E. A., Latorre J. D., Wolfenden A. D., Téllez G. I., Hargis B. M., et al. (2015). Evaluation of gastrointestinal leakage in multiple enteric inflammation models in chickens. Front. Vet. Sci. 2:66. 10.3389/fvets.2015.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. (2009). The Nuclear Factor NF-κB Pathway in Inflammation. Cold Spring Harb. Perspect. Biol. 1:a001651. 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej J. P., Bednarczyk M. (2016). Effect of in ovo-delivered prebiotics and synbiotics on the morphology and specific immune cell composition in the gut-associated lymphoid tissue. Poult. Sci. 95 19–29. 10.3382/ps/pev291 [DOI] [PubMed] [Google Scholar]
- Meirow Y., Baniyash M. (2017). Immune biomarkers for chronic inflammation related complications in non-cancerous and cancerous diseases. Cancer Immunol. Immunother. 66 1089–1101. 10.1007/s00262-017-2035-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutrient Requirements of Poultry, (1994). Nutrient Requirements of Poultry. Washington, DC: The National Academies Press. [Google Scholar]
- Pender C. M., Kim S., Potter T. D., Ritzi M. M., Young M., Dalloul R. A. (2017). In ovo supplementation of probiotics and its effects on performance and immune-related gene expression in broiler chicks. Poult. Sci. 96 1052–1062. 10.3382/ps/pew381 [DOI] [PubMed] [Google Scholar]
- Ptaschinski C., Lukacs N. W. (2018). “Chapter 2 - Acute and Chronic Inflammation Induces Disease Pathogenesis,” in Molecular Pathology (Second Edition), eds Coleman W. B., Tsongalis G. J. (Cambridge, MA: Academic Press; ), 25–43. 10.1016/b978-0-12-802761-5.00002-x [DOI] [Google Scholar]
- Rodrigues D. R., Wilson K., Brings W., Duff A., Chasser K., Bielke L. R. (2019). Intestinal pioneer colonizers as drivers of ileal microbial composition and diversity of broiler chickens. Front. Microbiol. 10:2858 10.3389/fmicb.2019.02858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2009). The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9 313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schokker D., Jansman A. J. M., Veninga G., de Bruin N., Vastenhouw S. A., de Bree F. M., et al. (2017). Perturbation of microbiota in one-day old broiler chickens with antibiotic for 24 hours negatively affects intestinal immune development. BMC Genomics 18:241. 10.1186/s12864-017-3625-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. L., Weber B. P., Mendoza K. M., Danzeisen J. L., Llop K., Lang K., et al. (2019). Antibiotics and host-tailored probiotics similarly modulate effects on the developing avian microbiome, mycobiome, and host gene expression. mBio 10:e2171-19. 10.1128/mBio.02171-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. M., Rodrigues D. R., Briggs W. N., Duff A. F., Chasser K. M., Bielke L. R. (2019). Evaluation of the impact of in ovo administered bacteria on microbiome of chicks through 10 days of age. Poult. Sci. 98 5949–5960. 10.3382/ps/pez388 [DOI] [PubMed] [Google Scholar]
- Wilson K. M., Rodrigues D. R., Briggs W. N., Duff A. F., Chasser K. M., Bottje W. G., et al. (2020). Impact of in ovo administered pioneer colonizers on intestinal proteome in day of hatch chicks. Poult. Sci. (in press). 10.1016/j.psj.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Dong X.-J., Li Y.-Y., Zhao Y., Xu Q.-L., Su L. (2015). Identification of phosphorylated MYL12B as a potential plasma biomarker for septic acute kidney injury using a quantitative proteomic approach. 8. Int. J. Clin. Exp. Pathol. 8 14409–14416. [PMC free article] [PubMed] [Google Scholar]
- Xiao T. S. (2017). Innate immunity and inflammation. Cell. Mol. Immunol. 14 1–3. 10.1038/cmi.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Tracey K. J. (2010). Targeting HMGB1 in inflammation. Biochim. Biophys. Acta BBA Gene Regul. Mech. 1799 149–156. 10.1016/j.bbagrm.2009.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang N., Xu A. (2018). Screening of genes associated with inflammatory responses in the endolymphatic sac reveals underlying mechanisms for autoimmune inner ear diseases. Exp. Ther. Med. 16 2460–2470. 10.3892/etm.2018.6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015504.