Abstract
Background
This study aimed to evaluate the association of circular RNA La‐related RNA‐binding protein 4 (circ‐LARP4) with clinical features and prognosis in osteosarcoma patients, and further explore its effect on chemosensitivity in osteosarcoma cells.
Methods
Seventy‐two osteosarcoma patients with Enneking stage IIA‐IIB who underwent resection were consecutively enrolled, and then, tumor tissues and non‐tumor tissues were obtained. Circ‐LARP4 in tumor tissue/non‐tumor tissue was detected by quantitative polymerase chain reaction. After circ‐LARP4 overexpression and negative control overexpression plasmid transfection, relative cell viability (%) was evaluated by Cell Counting Kit‐8 in MG63 cells treated by different concentrations of cisplatin, methotrexate, and doxorubicin, and IC50 was calculated.
Results
Circ‐LARP4 was downregulated in tumor tissue compared with non‐tumor tissue and had a good value in distinguishing tumor tissue from non‐tumor tissue with an area under curve of 0.829 (95% CI: 0.762‐0.859). Meanwhile, tumor circ‐LARP4 was negatively correlated with the Enneking stage. After resection, circ‐LARP4 high expression patients showed an increased tumor cell necrosis rate to adjuvant chemotherapy compared to circ‐LARP4 low expression patients, and circ‐LARP4 high expression correlated with prolonged disease‐free survival and overall survival. In vitro experiments revealed that circ‐LARP4 overexpression elevated the chemosensitivity of MG63 cells to cisplatin and doxorubicin but not methotrexate, with decreased cisplatin IC50 and doxorubicin IC50 concentrations than negative control. Besides, miR‐424 overexpression attenuated the chemosensitivity in circ‐LARP4 overexpression‐treated MG63 cells.
Conclusion
Circ‐LARP4 high expression correlates with decreased Enneking stage and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging miR‐424 in osteosarcoma.
Keywords: chemosensitivity, circ‐LARP4, Enneking stage, osteosarcoma, survival profile
1. INTRODUCTION
Osteosarcoma, a malignant bone tumor mainly affecting children and younger adults, is a very rare tumor that most frequently attacks the long bones and presents with an extremely miserable survival.1, 2 Usually, the standard treatment for osteosarcoma consists of chemotherapy (including adjuvant chemotherapy and neoadjuvant chemotherapy) and tumor resection. Unfortunately, despite that the survival of patients with localized disease has been predominantly prolonged since the introduction of neoadjuvant chemotherapy, prognosis of patients with metastatic disease remains poor due to the lack of knowledge about the pathogenesis, and also a delay of diagnosis resulting in that most of the patients cannot receive surgery.3 Therefore, management of osteosarcoma still requires more biomarkers that can aid in diagnosis and surveillance of the disease that are crucial to the extension of survival in these patients.
Circular RNAs (circRNAs), a category of endogenous RNAs united by a structure of closed‐loop without a 5ʹ‐3ʹ polarity, are a promising class of non‐coding RNAs capable of regulating transcriptional or posttranscriptional gene expression via functioning as microRNAs (miRNAs) sponges.4, 5, 6, 7 Several individual circRNAs have been revealed to play essential roles in osteosarcoma pathogenesis, such as regulating cancer cell cycle, cell proliferation, and chemoresistance, and serving as prognostic biomarker in osteosarcoma patients.8, 9, 10 CircRNA La‐related RNA‐binding protein 4 (circ‐LARP4) is a novel circRNA derived from the LARP4 gene, a gene encoding La‐related RNA‐binding protein that is capable of regulating cancer cell migration and invasion.11 Furthermore, it is reported that circ‐LARP4 could inhibit cancer cell proliferation and invasion in other cancers apart from osteosarcoma.12 Herein, we speculated that circ‐LARP4 might have the potential to be a biomarker in osteosarcoma patients.
Thus, this study aimed to evaluate the association of circ‐LARP4 expression with clinical features and prognosis in osteosarcoma patients, and further explore its effect on chemosensitivity in osteosarcoma cells.
2. MATERIALS AND METHODS
2.1. Patients
Seventy‐two osteosarcoma patients with Enneking stage IIA ~ IIB who underwent surgery in our hospital were consecutively enrolled in this study. The inclusion criteria were as follows: (a) newly diagnosed as primary osteosarcoma confirmed by open biopsy or core‐needle biopsy, (b) single lesion located in the extremities without distant metastasis, (c) Enneking stage IIA ~ IIB, and (d) voluntary to participate in this study. Patients with following circumstances were excluded: (a) secondary osteosarcoma; (b) primary lesions in extraskeletal organs and tissues; (c) lesions located in axial skeleton; (d) multiple lesions or distant metastasis; (e) history of other malignancies; and (f) pregnant or breastfeeding. The present study was approved by the Ethics Committee of our hospital, and all patients or their guardians signed informed consents before enrollment.
2.2. Collection of samples and baseline data
After completion of the initial diagnosis by X‐ray, computed tomography (CT), whole‐body magnetic resonance imaging (MRI), or positron emission tomography (PET)‐CT, pathological biopsy was performed, including CT‐guided core‐needle biopsy and open biopsy. Then, tumor tissue samples from biopsy were divided into two parts, one was quickly submitted for pathological assessment and staging, and the other was snap‐frozen in the liquid nitrogen and stored at −80°C for further detection. Meanwhile, the corresponding non‐tumor tissue of each enrolled patient was also collected from the open biopsy or definitive surgery, which was resected within at least 5 cm of the tumor margin, and the non‐tumor tissue was snap‐frozen in the liquid nitrogen and stored at −80°C until analysis as well. Patients’ baseline clinical data were recorded after the diagnostic workup was completed, which included age, gender, tumor location, World Health Organization (WHO) classification of sarcoma, pathological fracture status, and Enneking stage. And the surgery type was documented after definitive tumor resection.
2.3. Detection of circ‐LARP4 in clinical samples
The relative expression of circ‐LARP4 in the tumor tissue and non‐tumor tissue was detected by the quantitative polymerase chain reaction (qPCR). The detailed process was presented in “qPCR” subsection.
2.4. Treatment and assessment
After the diagnosis was established, neoadjuvant chemotherapy with MAP regimen (high‐dose methotrexate, cisplatin, and doxorubicin) was administered to all patients for 10 weeks, which consisted of 120 mg/m2 of cisplatin and 75 mg/m2 of doxorubicin (weeks 1 and 6) followed by 12 g/m2 of high‐dose methotrexate (weeks 4, 5, 9, and 10) according to the EURAMOS 1 treatment regimen.13 After the neoadjuvant chemotherapy, patients were re‐assessed by the X‐ray, PET, or bone scan, and then, definitive surgery (limb salvage or amputation) was performed. The histological response to the neoadjuvant chemotherapy was evaluated on the basis of the tumor cell necrosis rate in the resected specimen, and a good response was defined as tumor cell necrosis rate (TCNR) ≥90%; accordingly, a poor response was defined as TCNR <90%.14 Postoperative chemotherapy was based on histological response to neoadjuvant chemotherapy. The original chemotherapy regimen was continued if patients achieved a good response, while intensive regimen or alternative regimen was given to the patients with a poor response.
2.5. Follow‐up
All patients were followed up as clinically indicated or every 2 ~ 3 months for the first 2 years, every 2 ~ 4 months for years 3 ~ 4, and every 6 months for years 5‐10. For the current study, the last follow‐up date was 2019/06/31, and the median follow‐up duration was 22.0 months. Based on the follow‐up data, disease‐free survival (DFS) and overall survival (OS) were calculated for survival analysis. The DFS was defined as time interval from the date of surgery to the date of disease relapse, disease progression, death or last patient contact, whichever came first. The OS was defined as the time interval from the date of surgery to the date of death or last patient contact, whichever came first.
2.6. Cell culture
Human osteosarcoma cell lines MG63 and Saos‐2 were purchased from American Type Culture Collection (ATCC) (Rockefeller) and then cultured in 90% DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) at 37 ℃ under 95% air and 5% CO2 condition.
2.7. Transfection
Circ‐LARP4 overexpression and negative control (NC) overexpression plasmids were constructed by Shanghai QeeJen Bio‐tech Company and then were transferred into MG63 and Saos‐2 cells, which were divided into Circ‐LARP4(+) group and NC(+) group, respectively. Beside, normal MG63 and Saos‐2 cells without transfection served as blank control group.
2.8. Drug sensitivity detection
After 24 hours of transfection, cisplatin (0, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 μmol/L), methotrexate hydrate (0, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 μmol/L), and doxorubicin hydrochloride (0, 0.001, 0.002, 0.004, 0.008, 0.016, and 0.032 μmol/L) were added to treat MG63 cells with three replicate wells, respectively. Also, cisplatin (0, 0.01, 0.02, 0.04, 0.08, 0.16, 0.32, and 0.64 μmol/L), methotrexate hydrate (0, 0.025, 0.05, 0.1, 0.2, 0.4, 0.8, and 1.6 μmol/L), and doxorubicin hydrochloride (0, 0.01, 0.02, 0.04, 0.08, 0.16, and 0.32 μmol/L) were added to treat Saos‐2 cells with three replicate wells. The setting of drug concentration was referred to a previous study.15 Then, after incubation for an additional 48 hours, relative cell viability was measured using Cell Counting Kit‐8 (CCK‐8) (Dojindo, Japan) according to the manufacturer's instructions. Finally, the relative cell viability (%) was calculated by setting corresponding untreated controls (0 μmol/L drug) as 100%, and IC50 of each drug was calculated.
2.9. Interaction of circ‐LARP4 and miR‐424
MiR‐424 was predicted to be a direct target of circ‐LARP4 using Tissue‐Specific CircRNA Database (http://gb.whu.edu.cn/TSCD/) and was reported to bind circ‐LARP4 to exhibit effect on regulating drug resistance in gastric cancer cells.12 Besides, miR‐424 was proposed to decrease sensitivity to chemotherapy drugs such as cisplatin,16 docetaxel,17 doxorubicin, and etoposide.18 Thus, we hypothesized that circ‐LARP4 might regulate drug sensitivity through miR‐424 in osteosarcoma. Thus, at 24 hours after transfection, miR‐424 expression was detected by qPCR.
2.10. Rescue experiments
NC overexpression, miR‐424 overexpression, circ‐LARP4 overexpression, and miR‐424 overexpression plus circ‐LARP4 overexpression plasmids were constructed by Shanghai QeeJen Bio‐tech Company and then were transferred into MG63 and Saos‐2 cells, which were divided into NC(+) group, MiR‐424(+) group, Circ‐LARP4(+) group, and Circ‐LARP4(+)/MiR‐424(+) group, respectively. Then, miR‐424 expression was detected by qPCR at 24 hours after transfection. Besides, after 24 hours of transfection, 1.6 μmol/L cisplatin, 1.6 μmol/L methotrexate hydrate, and 0.008 μmol/L doxorubicin hydrochloride were added to treat MG63 cells with three replicate wells, respectively. And 0.04 μmol/L cisplatin, 0.1 μmol/L methotrexate hydrate, and 0.02 μmol/L doxorubicin hydrochloride were added to treat Saos‐2 cells with three replicate wells, respectively. Then, after incubation for an additional 48 hours, relative cell viability was measured using CCK‐8 (Dojindo, Japan) according to the manufacturer's instructions, and the relative cell viability (%) was calculated by setting NC(+) group as 100%.
2.11. Luciferase reporter assay
Luciferase reporter assay was carried out using Dual‐Luciferase® Reporter (DLR™) Assay System (Promega). Circ‐LARP4 wild‐type (WT) plasmid and circ‐LARP4 mutant (Mut) plasmid were constructed using pGL4 vector (Promega). Circ‐LARP4 WT/Mut plasmid and miR‐424(+)/NC(+) plasmid were co‐transfected into 293T cells using Lipofectamine 2000 (Thermo), which produced four groups: WT + NC(+) cells, WT + MiR‐424(+) cells, Mut + NC(+) cells, and Mut + MiR‐424(+) cells. At 24 hours after transfection, firefly luciferase luminescence was measured according to the Dual‐Luciferase® Reporter Assay System Protocol.
2.12. qPCR
The relative expressions of circ‐LARP4 in the tumor tissue/non‐tumor tissue and miR‐424 in osteosarcoma cells were detected by the qPCR. Firstly, the total RNA was extracted by RNeasy Protect Mini Kit (Qiagen), and then, the RNase R (Epicenter) was used for the digestion of linear RNA for the detection of circ‐LARP4 expression.19 Secondly, the RNA was reversely transcribed by using the iScript™ cDNA Synthesis Kit (with random primer) (Bio‐Rad), and qPCR was performed by QuantiNova SYBR Green PCR Kit (Qiagen). And the reaction condition was as follows: initial denaturation at 95℃ for 5 minutes, and then 40 cycles of denaturation (94℃, 15 seconds), annealing (61℃, 30 seconds), and extending (72 ℃, 30 seconds), and then an extending at 72℃ for 1 minute. Finally, the relative expression of circ‐LARP4 was calculated using GAPDH as internal reference, and the relative expression of miR‐424 was evaluated using U6 as internal reference, with the use of 2−△△Ct formula. In addition, the primers applied in the qPCR were as follows: circ‐LARP4, forward primer (5ʹ‐3ʹ): GAGACCAAGTCATAAGCGTTGTATT, reverse primer (5ʹ‐3ʹ): AAACCAGTTCCTTTAGATGCTACCT; GAPDH, forward primer (5ʹ‐3ʹ): GAGTCCACTGGCGTCTTCAC, reverse primer (5ʹ‐3ʹ): ATCTTGAGGCTGTTGTCATACTTCT; and U6, forward primer (5ʹ‐3ʹ): GCTTCGGCAGCACATATACTAAAAT, reverse primer (5ʹ‐3ʹ): CGCTTCACGAATTTGCGTGTCAT.
2.13. Statistical analysis
Data processing and analysis were performed using SPSS 24.0 (IBM), and graph plotting was conducted using GraphPad Prism 7.02 (GraphPad Software Inc). Data were described as mean and standard deviation (SD), median and interquartile range (IQR) or count (percentage). Comparison was determined by the t test, Wilcoxon signed‐rank test, or chi‐square test. Receiver operating characteristic (ROC) curve was used for assessing the value of circ‐LARP4 expression for differentiating tumor tissue from non‐tumor tissue. Survival curves were estimated using the Kaplan‐Meier method, and the difference of survival was determined by the log‐rank test. IC50 was calculated using Probit regression. P value <.05 was considered statistically significant.
3. RESULTS
3.1. Clinical characteristics
The mean age of osteosarcoma patients was 20.6 ± 13.2 years, in which there were 38 (52.8%) patients who had an age below 18 years (Table 1). And the numbers of male and female patients were 42 (58.3%) and 30 (41.7%), respectively. In addition, the numbers of patients with femur tumor, tibia tumor, and tumor located at other regions were 38 (52.8%), 25 (34.7%), and 9 (12.5%), respectively. The number of patients with a pathological fracture was 14 (19.4%). As for Enneking stage, there were respectively 11 (15.3%) patients with Enneking stage IIA and 61 (84.7%) patients with Enneking stage IIB. Moreover, with respect to the surgery type, the numbers of patients treated by amputation and limb salvage were 21 (29.2%) and 51 (70.8%), respectively. Other information of baseline characteristics is shown in Table 1.
Table 1.
Clinical characteristics of osteosarcoma patients
| Characteristics | Osteosarcoma patients (N = 72) |
|---|---|
| Age (years), mean ± SD | 20.6 ± 13.2 |
| <18 y, No. (%) | 38 (52.8) |
| ≥18 y, No. (%) | 34 (47.2) |
| Gender, No. (%) | |
| Male | 42 (58.3) |
| Female | 30 (41.7) |
| Tumor location, No. (%) | |
| Femur | 38 (52.8) |
| Tibia | 25 (34.7) |
| Others | 9 (12.5) |
| WHO classification of sarcoma, No. (%) | |
| Conventional: chondroblastic | 10 (13.9) |
| Conventional: osteoblastic | 47 (65.3) |
| Conventional: other | 9 (12.5) |
| Telangiectatic | 6 (8.3) |
| Pathological fracture, No. (%) | 14 (19.4) |
| Enneking stage, No. (%) | |
| IIA | 11 (15.3) |
| IIB | 61 (84.7) |
| Surgery type, No. (%) | |
| Amputation | 21 (29.2) |
| Limb salvage | 51 (70.8) |
Abbreviations: SD, standard deviation; WHO, World Health Organization.
3.2. Circ‐LARP4 in tumor tissue and non‐tumor tissue
In osteosarcoma patients, the circ‐LARP4 was upregulated in non‐tumor tissue compared with tumor tissue (P < .001) (Figure 1A), and then, the ROC curve analysis disclosed that circ‐LARP4 had a good value in distinguishing non‐tumor tissue from tumor tissue with an area under curve (AUC) of 0.829 (95% CI: 0.762‐0.859) (Figure 1B).
Figure 1.
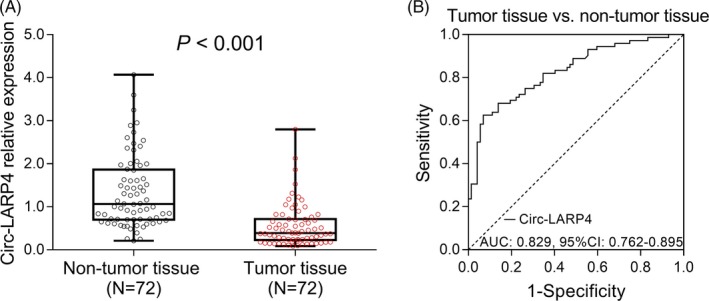
Circ‐LARP4 expression in tumor and non‐tumor tissue from osteosarcoma patients. The expression of circ‐LARP4 in tumor and non‐tumor tissue (A) and the ROC curve analysis of circ‐LARP4 expression for differentiating tumor tissue from non‐tumor tissue (B). Comparison was determined by Wilcoxon signed‐rank test. ROC curve was used for assessing the value of circ‐LARP4 expression for differentiating tumor tissue from non‐tumor tissue. P value <.05 was considered statistically significant. Circ‐LARP4, circular RNA La‐related RNA‐binding protein 4; ROC, receiver operative characteristics; AUC, area under curve
3.3. Correlation of circ‐LARP4 with clinical features
The association of tumor circ‐LARP4 with clinical features was detected, which elucidated that circ‐LARP4 was negatively correlated with the Enneking stage (P = .022), while it was not correlated with age (P = 1.000), gender (P = .633), tumor location (P = .750), WHO classification of sarcoma (P = .755), pathological fracture (P = .234), or surgery type (P = .759) in osteosarcoma patients (Table 2).
Table 2.
Correlation of tumor circ‐LARP4 expression with clinical features
| Clinical features | Circ‐LARP4 low expression (n = 36)* | Circ‐LARP4 high expression (n = 36)* | P value |
|---|---|---|---|
| Age, No. (%) | |||
| <18 y | 19 (52.8) | 19 (52.8) | 1.000 |
| ≥18 y | 17 (47.2) | 17 (47.2) | |
| Gender, No. (%) | |||
| Female | 16 (44.4) | 14 (38.9) | .633 |
| Male | 20 (55.6) | 22 (61.1) | |
| Tumor location, No. (%) | |||
| Femur | 18 (50.0) | 20 (55.5) | .750 |
| Tibia | 14 (38.9) | 11 (30.6) | |
| Others | 4 (11.1) | 5 (13.9) | |
| WHO classification of sarcoma, No. (%) | |||
| Conventional: chondroblastic | 5 (13.9) | 5 (13.9) | .755 |
| Conventional: osteoblastic | 25 (69.5) | 22 (61.1) | |
| Conventional: other | 3 (8.3) | 6 (16.7) | |
| Telangiectatic | 3 (8.3) | 3 (8.3) | |
| Pathological fracture, No. (%) | |||
| No | 27 (75.0) | 31 (86.1) | .234 |
| Yes | 9 (25.0) | 5 (13.9) | |
| Enneking stage, No. (%) | |||
| IIA | 2 (5.6) | 9 (25.0) | .022 |
| IIB | 34 (94.4) | 27 (75.0) | |
| Surgery type, No. (%) | |||
| Amputation | 11 (30.6) | 10 (27.8) | .759 |
| Limb salvage | 25 (69.4) | 26 (72.2) | |
Low expression and high expression were categorized by the median value of circ‐LARP4 relative expression in tumor. Comparison was determined by the chi‐square test. WHO: World Health Organization.
3.4. Correlation of circ‐LARP4 with histological response
After neoadjuvant chemotherapy, the good response rate (TCNR ≥ 90%) was elevated in patients with circ‐LARP4 high expression than that in patients with circ‐LARP4 low expression (55.6% vs 30.6%, P = .032) (Figure 2).
Figure 2.
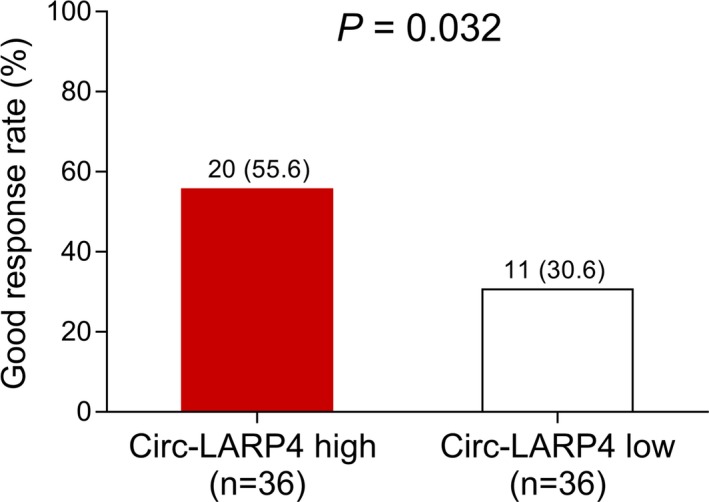
Correlation of circ‐LARP4 with good response rate. The good response rate in patients with circ‐LARP4 high expression and in patients with circ‐LARP4 low expression. Comparison was determined by chi‐square test. P value <.05 was considered statistically significant. Circ‐LARP4, circular RNA La‐related RNA‐binding protein 4
3.5. Association of circ‐LARP4 with survival profiles
With regard to the association of circ‐LARP4 with survival profiles in osteosarcoma patients post‐treatment, the K‐M curve analysis revealed that both DFS (P = .002) (Figure 3A) and OS (P = .015) (Figure 3B) were more prolonged in patients with circ‐LARP4 high expression in tumor tissue than those in patients with circ‐LARP4 low expression.
Figure 3.
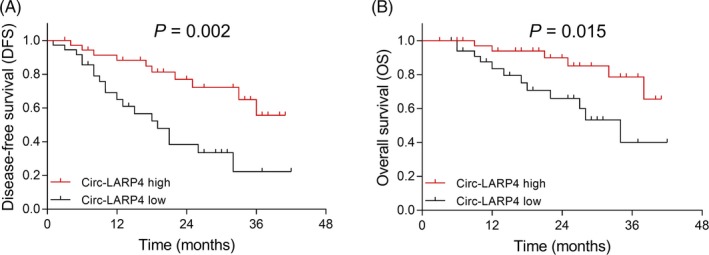
Correlation of circ‐LARP4 expression with DFS and OS. The correlation of circ‐LARP4 expression with DFS and OS. Survival curves were estimated using the Kaplan‐Meier method, and the difference of survival was determined by the log‐rank test. P value <0.05 was considered statistically significant. Circ‐LARP4, circular RNA La‐related RNA‐binding protein 4; DFS, disease‐free survival; OS, overall survival
3.6. Effect of circ‐LARP4 on chemosensitivity in MG63 cells
After transfection, the relative cell viability of MG63 cells was decreased in Circ‐LARP4(+) group compared with NC(+) group when the cells were treated by 0.2 μmol/L (P < .05), 0.4 μmol/L (P < .001), 0.8 μmol/L (P < .01), 1.6 μmol/L (P < .01), and 3.2 μmol/L (P < 0.in 05) but similar by 0.1 μmol/L (P > .05) or 6.4 μmol/L (P > .05) cisplatin (Figure 4A), and the IC50 value of cisplatin in Circ‐LARP4(+) group was nearly half of that in the NC(+) group (1.561 vs 2.888) (Figure 4B). For methotrexate, the relative cell viability was reduced in Circ‐LARP4(+) group than that in NC(+) group when the cells were treated by 0.8 μmol/L (P < .05) methotrexate; however, the values of relative cell viability between the two groups were similar after 0.1 μmol/L (P > .05), 0.2 μmol/L (P > .05), 0.4 μmol/L (P > .05), 1.6 μmol/L (P > .05), 3.2 μmol/L (P > .05), or 6.4 μmol/L (P > .05) methotrexate treatment (Figure 4C), and the IC50 value of methotrexate in the Circ‐LARP4(+) group was similar to that in NC(+) group (2.075 vs. 2.615) (Figure 4D). As to doxorubicin, the relative cell viability was lower in Circ‐LARP(+) group compared with NC(+) group in cells treated by 0.004 μmol/L (P < .05), 0.008 μmol/L (P < .01), and 0.016 μmol/L (P < .05) but similar by 0.001 μmol/L (P > .05), 0.002 μmol/L (P > .05), or 0.032 μmol/L (P > .05) doxorubicin treatment (Figure 4E), and the IC50 value of doxorubicin in the Circ‐LARP4(+) group was decreased compared with NC(+) group (0.007 vs 0.011) (Figure 4F). And these results indicated that circ‐LARP4 elevated the chemosensitivity of osteosarcoma cells to cisplatin and doxorubicin but not methotrexate, which may also explain the prognostic value of circ‐LARP4 in osteosarcoma patients.
Figure 4.
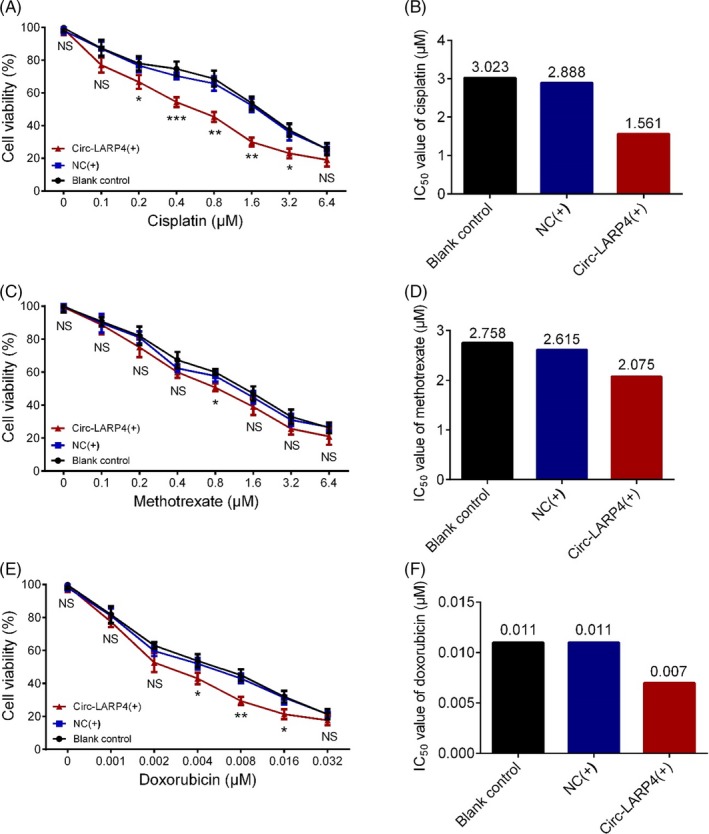
Effect of circ‐LARP4 on chemosensitivity of MG63 cells. Effect of circ‐LARP4 on relative cell viability of MG63 cells under different concentrations of cisplatin treatment (A), the IC50 value of cisplatin in blank control group, NC(+) group and Circ‐LARP4(+) group (B), effect of circ‐LARP4 on relative cell viability under different concentrations of methotrexate treatment (C), the IC50 of methotrexate in blank control, NC(+) group and circ‐LARP4(+) group (D), effect of circ‐LARP4 on relative cell viability under different concentrations of doxorubicin treatment (E), and the IC50 value of doxorubicin in blank control, NC(+) group and circ‐LARP4(+) group (F). Comparison was determined by t test. P value <.05 was considered statistically significant. Circ‐LARP4, circular RNA La‐related RNA‐binding protein 4; NC, negative control
3.7. Effect of circ‐LARP4 and miR‐424 on chemosensitivity in MG63 cells
The miR‐424 expression was downregulated in Circ‐LARP4(+) group compared with NC(+) group after transfections (P < .01) (Figure 5A), which indicated that circ‐LARP4 overexpression could downregulate miR‐424 in osteosarcoma cells. Then in the subsequent rescue experiments, the miR‐424 expression was increased in MiR‐424(+) than that in NC(+) group (P < .001) and was also elevated in Circ‐LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group (P < .001) (Figure 5B). After 1.6 μmol/L cisplatin treatment, the relative cell viability was increased in MiR‐424(+) group compared with NC(+) group (P < .01) and was also higher in Circ‐LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group (P < .01), while was decreased in Circ‐LARP4(+) group compared with NC(+) group (P < .01) (Figure 5C). After the 1.6 μmol/L methotrexate treatment, relative cell viability was upregulated in MiR‐424(+) group compared with NC(+) group (P < .05) and was also increased in Circ‐LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group (P < .05), while showed no difference in Circ‐LARP4(+) group than that in NC(+) group (P > .05) (Figure 5D). And after 0.008 μmol/L doxorubicin treatment, the relative cell viability was elevated in MiR‐424(+) group than that in NC(+) group (P < .05) and was also increased in Circ‐LARP4(+)/MiR‐424(+) group than that in Circ‐LARP4(+) group (P < .05), while was decreased in Circ‐LARP4(+) group compared with NC(+) group (P < .05) (Figure 5E). Additionally, the luciferase reporter assay displayed the binding site of circ‐LARP4 and miR‐424 (Figure S1A,B). These data suggested that circ‐LARP4 might promote chemosensitivity to cisplatin and doxorubicin via sponging miR‐424 in osteosarcoma.
Figure 5.
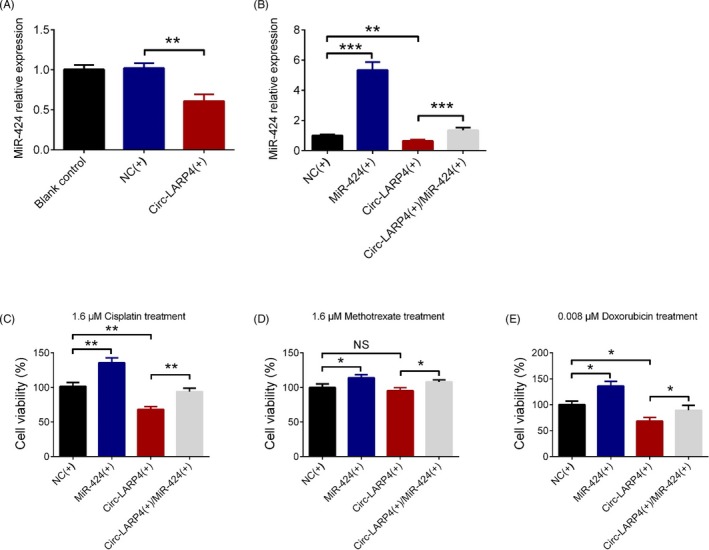
Effect of circ‐LARP4 and miR‐424 on chemosensitivity of MG63 cells. MiR‐424 expression in MG63 cells after circ‐LARP4 overexpression plasmid transfection (A), in further rescue experiments, the miR‐424 expression after transfections (B), the effect of circ‐LARP4 and miR‐424 on relative cell viability in osteosarcoma cells treated by cisplatin (C), methotrexate (D), or doxorubicin (E). Comparison was determined by t test. P value <.05 was considered statistically significant. Circ‐LARP4, circular RNA La‐related RNA‐binding protein 4; miR, microRNA
3.8. Effect of circ‐LARP4 on chemosensitivity in SaoS‐2 cells
In SaoS‐2 cells, the cell viability of cells treated with 0.02 μmol/L (P < .05), 0.04 μmol/L (P < .01), 0.08 μmol/L (P < .05), and 0.016 μmol/L (P < .05) cisplatin was declined in Circ‐LARP(+) group compared with NC(+) group (Figure S2A), and the IC50 value of cisplatin in blank control group, NC(+) group, and Circ‐LARP4(+) group was 0.088, 0.078, and 0.045, respectively (Figure S2B). In addition, the cell viability of cells treated with 0.1 μmol/L (P < .05) and 0.2 μmol/L (P < .05) methotrexate in Circ‐LARP4(+) group was decreased compared with NC(+) group (Figure S2C), and the IC50 value of methotrexate in blank control group, NC(+) group, and Circ‐LARP4(+) group was 0.211, 0.200, and 0.132, respectively (Figure S2D). Besides, cell viability of cells treated with 0.02 μmol/L (P < .05), 0.04 μmol/L (P < .05), 0.08 μmol/L (P < .01), and 0.16 μmol/L (P < .05) doxorubicin in Circ‐LARP4(+) group was decreased than those in NC(+) group (Figure S2E), and the IC50 value of doxorubicin in the three groups was 0.064, 0.054, and 0.028, respectively (Figure S2F). These results indicated that circ‐LARP4 elevated the chemosensitivity to cisplatin, methotrexate, and doxorubicin in Sao‐S2 cells.
3.9. Effect of circ‐LARP4 and miR‐424 on chemosensitivity in SaoS‐2 cells
After transfections, miR‐424 expression was downregulated in Circ‐LARp4(+) group compared with NC(+) group (P < .01) (Figure S3A), while it was upregulated in Circ‐LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group (P < .01) (Figure S3B). Furthermore, cell viability of cells treated with 0.04 μmol/L cisplatin was elevated in MiR‐424(+) group compared with NC(+) group (P < .01) and was also upregulated in Circ‐LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group (P < .01) (Figure S3C). As for methotrexate, the cell viability of cells treated with 0.1 μmol/L methotrexate was increased in MiR‐424(+) group compared with NC(+) group (P < .01) and was higher in Circ‐LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group as well (P < .01) (Figure S3D). Moreover, cell viability in cells treated with 0.02 μmol/L doxorubicin was increased in MiR‐424(+) group compared with NC(+) group (P < .01) and was upregulated in LARP4(+)/MiR‐424(+) group compared with Circ‐LARP4(+) group (P < .05) (Figure S3E). These data suggested that circ‐LARP4 might upregulate chemosensitivity to cisplatin, methotrexate, and doxorubicin in SaoS‐2 cells by sponging miR‐424.
4. DISCUSSION
In this study, we evaluated the expression of circ‐LARP4 in tumor tissue and non‐tumor tissue from osteosarcoma patients, the correlation of circ‐LARP4 expression in tumor tissue with clinical features and survival profiles in osteosarcoma patients, and the effect of circ‐LARP4 on chemosensitivity in osteosarcoma cells, and found that: (a) circ‐LARP4 was upregulated in non‐tumor tissue compared with tumor tissue, and has good value in differentiating tumor tissue from non‐tumor tissue in osteosarcoma patients; (b) circ‐LARP4 high expression correlated with decreased Enneking stage, increased good response rate and also more prolonged DFS as well as OS; and (c) circ‐LARP4 upregulated the chemosensitivity to cisplatin and doxorubicin via sponging miR‐424 in osteosarcoma cells.
CircRNAs are reported in various cancers previously, which present with multiple functions in the regulation of cancer development and progression, and thus, circRNAs are also promising potential assisting biomarkers for cancer management. The functions of circRNAs are mostly found in regulating the cancer cell functions, for instance, circ‐VAPA increases cancer hepatocellular carcinoma cell proliferation by sponging miR‐377‐3p which subsequently disinhibits the expression of PSAP.20 And silence of circ‐ANF609 downregulates cancer cell growth, migration, and invasion through elevating miR‐186‐5p in prostate cancer.21 Besides, there are also reports illuminating that circRNAs participate in cancer pathogenesis via regulating other related processes. For example, it has been reported that circ‐ANKRD12 silencing could induce the changes in molecule and function of the invasive cancer cell phenotypes.22
Circ‐LARP4 is a circRNA derived from the exon 9, exon 10, and intermediate long intron of the LARP4 gene, and this circRNA has been revealed to participate in the regulation of cancer cell functions acting as a miRNA sponge.12 Generally, as one of the transcripts of the LARP4 gene, circ‐LARP4 could directly or indirectly regulate the expression of protein LARP4. As a novel oncogenesis‐related circRNA, the studies of its function in cancer are still very insufficient, not to mention that no study has been done to evaluate the role of circ‐LARP4 in osteosarcoma. A previous study shows that circ‐LAPR4 suppresses cancer cell proliferation and invasion via sponging miR‐424‐5p and mediating the expression of the larger tumor suppressor kinase 1 (LATS1) in gastric cancer.12 Another study elucidates that circ‐LARP4 activates cancer cell senescence through modulating miR‐761/RUNX3/p53/p21 signaling pathway in hepatocellular carcinoma.23 And the LARP4 gene, the location of circ‐LARP4, has also been found to be a tumor suppressor, for example, a recent experiment elucidates that LARP4 gene represses cell motility and migration in ovarian cells.11 Another experiment illuminates that LARP4 inhibits prostate cancer cell migration and invasion.24 As for the prognostic value of circ‐LARP4, a previous cohort study reports that circ‐LARP4 expression is decreased in tumor tissue than that in paired adjacent tissue, and is negatively correlated with Federation of Gynecology and Obstetrics stages and survival in ovarian cancer patients, which is partially in line with our results.25 In this study, we found that circ‐LARP4 was upregulated in non‐tumor tissue compared with tumor tissue, and tumor tissue circ‐LARP4 high expression was associated with decreased Enneking stage, elevated good response rate, and more satisfactory survival profiles in osteosarcoma patients. Here are several possible explanations for these results: (a) First, like its role in other cancers, circ‐LARP4 might serve as a tumor suppressor in osteosarcoma via inhibiting cancer cell proliferation and invasion or inducing cancer cell senescence via regulating multiple tumor‐related signaling pathways/proteins, for instance, regulating the miR‐424‐5p/LATS1 pathway, resulting in that circ‐LARP4 was downregulated in tumor tissue than non‐tumor tissue, which also led to a negative correlation between tumor circ‐LARP4 high expression and Enneking stage in osteosarcoma patients; (b) second, the further functional experiments in our study showed that circ‐LARP4 enhanced chemosensitivity of osteosarcoma cells to cisplatin and doxorubicin, leading to a better histological response, and may explain the result that tumor tissue circ‐LARP4 high expression associated with elevated good response rate; and (c) third, due to that circ‐LARP4 could serve as a tumor suppressor and enhance chemosensitivity in osteosarcoma cells, a tumor tissue high circ‐LARP4 expression might contribute to a better survival in patients due to less severe disease and better response to chemotherapy.11, 12, 24, 25
Furthermore, we speculated that circ‐LARP4 might have influence on the progression and prognosis of osteosarcoma via affecting the chemosensitivity since that chemotherapy is a predominant part in osteosarcoma treatment. Thus, we investigated the effect of circ‐LARP4 on chemosensitivity and its regulatory role of miR‐424 in osteosarcoma cells, and found that circ‐LARP4 elevated the chemosensitivity to cisplatin and doxorubicin in osteosarcoma cells, and further rescue experiment elucidated that circ‐LARP4 might increase chemosensitivity by sponging miR‐424. MiR‐424, belonging to the miR‐424(322)/503 cluster, has been revealed to function as a regulator of development and progression in multiple cancers in prior studies, for instance, miR‐424 enhances the progression of esophageal squamous cell carcinoma via promoting cancer cell proliferation through multilayered regulation.26 And another experiment reveals that miR‐424 and miR‐19a increase epithelial‐to‐mesenchymal transition and migration by targeting transforming growth factor type III receptor in tongue squamous cell carcinoma cells.27 As previously reported in another study, circ‐LARP4 represses cell proliferation and invasion of gastric cancer cells via sponging miR‐424‐5p, which was partially in accordance with our results.12 These results in our study might provide some novel insights about the role of circ‐LARP4 in the progression and prognosis of osteosarcoma.
There were still several limitations in our study: (a) The follow‐up duration in our study was relatively short which should be prolonged in the future study; (b) we did not evaluate circ‐LARP4 in blood sample, which is easier to obtain from patients and more applicable in clinical practice, and thus, the circulating circ‐LARP4 in osteosarcoma patients should be assessed in future studies; and (c) we only included patients in Enneking stage II, and thus, the role of circ‐LARP4 in patients in other stages was not evaluated in this study. However, in order to obtain tumor tissue and adjacent tissue, only patients who had received surgery could be included, who were mostly Enneking stage II patients in clinical practice.
In conclusion, circ‐LARP4 high expression correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging miR‐424 in osteosarcoma.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
Hu Y, Gu J, Shen H, et al. Circular RNA LARP4 correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA‐424 in osteosarcoma. J Clin Lab Anal. 2020;34:e23045 10.1002/jcla.23045
REFERENCES
- 1. Bielack SS, Kempf‐Bielack B, Delling G, et al. Prognostic factors in high‐grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776‐790. [DOI] [PubMed] [Google Scholar]
- 2. Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3‐13. [DOI] [PubMed] [Google Scholar]
- 3. Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23(8):737‐755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzuki H, Tsukahara T. A view of pre‐mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331‐9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cortes‐Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89(4):527‐537. [PMC free article] [PubMed] [Google Scholar]
- 8. Huang L, Chen M, Pan J, Yu W. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR‐1253/FOXF1 pathway. Biochem Biophys Res Comm. 2018;500(2):511‐517. [DOI] [PubMed] [Google Scholar]
- 9. Kun‐Peng Z, Xiao‐Long M, Chun‐Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin H, Jin X, Zhang H, Wang W. Circular RNA hsa‐circ‐0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget. 2017;8(15):25571‐25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egiz M, Usui T, Ishibashi M, et al. La‐Related protein 4 as a suppressor for motility of ovarian cancer cells. Tohoku J Exp Med. 2019;247(1):59‐67. [DOI] [PubMed] [Google Scholar]
- 12. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR‐424‐5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carrle D, Bielack SS. Current strategies of chemotherapy in osteosarcoma. Int Orthop. 2006;30(6):445‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smeland S, Bielack SS, Whelan J, et al. Survival and prognosis with osteosarcoma: outcomes in more than 2000 patients in the EURAMOS‐1 (European and American Osteosarcoma Study) cohort. Eur J Cancer. 2019;109:36‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanas V, Haigl B, Stockhammer V, Sutterluty‐Fall H. MicroRNA‐21 increases proliferation and cisplatin sensitivity of osteosarcoma‐derived cells. PLoS ONE. 2016;11(8):e0161023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu C, Wang H, Chen S, Yang R, Li H, Zhang G. Baicalein inhibits cell growth and increases cisplatin sensitivity of A549 and H460 cells via miR‐424‐3p and targeting PTEN/PI3K/Akt pathway. J Cell Mol Med. 2018;22(4):2478‐2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geretto M, Pulliero A, Rosano C, Zhabayeva D, Bersimbaev R, Izzotti A. Resistance to cancer chemotherapeutic drugs is determined by pivotal microRNA regulators. Am J Cancer Res. 2017;7(6):1350‐1371. [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang D, Shi Z, Li M, Mi J. Hypoxia‐induced miR‐424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014;5:e1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu K, Chen D, Wang Z, et al. Annotation and functional clustering of circRNA expression in rhesus macaque brain during aging. Cell discovery. 2018;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu C, Zhong X, Li J, Xu F. Circular RNA circVAPA promotes cell proliferation in hepatocellular carcinoma. Hum Gene Ther Clin Dev. 2019. 30(4):152‐159. [DOI] [PubMed] [Google Scholar]
- 21. Jin C, Zhao W, Zhang Z, Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up‐regulating microRNA‐186‐5p in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47(1):3350‐3358. [DOI] [PubMed] [Google Scholar]
- 22. Karedath T, Ahmed I, Al Ameri W, et al. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19(1):565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Z, Zuo X, Pu L, et al. circLARP4 induces cellular senescence through regulating miR‐761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110(2):568‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seetharaman S, Flemyng E, Shen J, Conte MR, Ridley AJ. The RNA‐binding protein LARP4 regulates cancer cell migration and invasion. Cytoskeleton. 2016;73(11):680‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zou T, Wang PL, Gao Y, Liang WT. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22(21):7178‐7182. [DOI] [PubMed] [Google Scholar]
- 26. Llobet‐Navas D, Rodriguez‐Barrueco R, Castro V, et al. The miR‐424(322)/503 cluster orchestrates remodeling of the epithelium in the involuting mammary gland. Genes Dev. 2014;28(7):765‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li D, Liu K, Li Z, Wang J, Wang X. miR‐19a and miR‐424 target TGFBR3 to promote epithelial‐to‐mesenchymal transition and migration of tongue squamous cell carcinoma cells. Cell Adh Migr. 2018;12(3):236‐246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


