Abstract
Objective
This study aimed to investigate the predictive value of long noncoding RNA nuclear enriched abundant transcript 1 (lncRNA NEAT1) for acute ischemic stroke (AIS) risk and to explore the correlation of lncRNA NEAT1 with disease severity, inflammation, recurrence and target microRNAs in patients with AIS.
Methods
210 patients with AIS and 210 controls were enrolled, and their peripheral blood samples were collected within 24 hours after admission and collected on the enrollment, respectively. lncRNA NEAT1 expression was detected by quantitative polymerase chain reaction (qPCR). For patients with AIS, disease severity was evaluated by National Institute of Health Stroke Scale (NIHSS) score; plasma concentrations of inflammatory factors and lncRNA NEAT1 target microRNAs were measured by enzyme‐linked immune sorbent assay and qPCR, respectively; stroke recurrence and death were recorded; and recurrence‐free survival (RFS) was calculated.
Results
lncRNA NEAT1 expression was elevated in patients with AIS compared with controls, and it had a good predictive value for AIS risk (area under the curve [AUC]: 0.804 [95% confidence interval [CI]: 0.763‐0.845]). In patients with AIS, lncRNA NEAT1 expression positively correlated with NIHSS score and inflammatory factor levels including C‐reactive protein (CRP), tumor necrosis factor (TNF)‐α, interleukin (IL)‐6, IL‐8, and IL‐22, while it negatively correlated with anti‐inflammatory cytokine IL‐10 level. Besides, lncRNA NEAT1 predicted increased recurrence/death risk (AUC: 0.641 [95% CI: 0.541‐0.741]), and its high expression correlated with worse RFS. Additionally, lncRNA NEAT1 expression negatively correlated with microRNA‐124 and microRNA‐125a expressions.
Conclusion
LncRNA NEAT1 may serve as a novel biomarker for assisting AIS management and prognosis.
Keywords: acute ischemic stroke, inflammation, long noncoding RNA, nuclear enriched abundant transcript 1, recurrence‐free survival
1. INTRODUCTION
Acute ischemic stroke (AIS) is a common and serious disease that occurs when there is a sudden loss in cerebral blood flow due to thromboembolic occlusion in the cerebral or cervical artery.1, 2 Acute ischemic stroke results in irreversible infarction of brain tissue as well as functional impairment, and patients with AIS requires early recognition and quick medical care to achieve the best outcomes.3 Currently, the common managements for AIS are mainly focusing on restoring reperfusion into the ischemic brain area, such as intravenous alteplase and endovascular thrombectomy, whereas the narrow treatment time window is still the major limitation for the application of therapies, and prognosis of patients with AIS remains poor.4, 5, 6 A growing evidence suggests that early identification of AIS risk and prediction of treatment outcomes in patients with AIS may improve the prognosis of these patients.
Long noncoding RNA (lncRNA) is a group of noncoding transcripts with more than 200 nucleotides in length, and lncRNAs are known as crucial regulators of plentiful biological processes.7, 8 Long noncoding RNA nuclear enriched abundant transcript 1 (NEAT1), which is an lncRNA that is essential for the formation of paraspeckles and interacts with many intracellular regulatory factors, has been widely investigated in the cancer field.9, 10, 11 Recently, some studies have revealed that lncRNA NEAT1 is overexpressed in myocardial infarction patients and tuberculosis patients.12, 13 Furthermore, the biological roles of lncRNA NEAT1 in cardiovascular and cerebrovascular diseases have been disclosed in some experiments, such as the promotion of lncRNA NEAT1 on inflammatory response in myocardial ischemia‐reperfusion injury mice models and the inhibition of lncRNA NEAT1 knockdown on atherosclerosis‐correlated events in vitro.14, 15 For stroke, previous study displays that lncRNA NEAT1 facilitates oxygen‐glucose deprivation/reoxygenation (OGD/R) injury and neuroinflammation damage of microglial cells, meanwhile, lncRNA NEAT1 enhances intima thickening or vascular occlusion through modulating the phenotype conversion of vascular smooth muscle cells, together with excessive apoptosis of epithelial cells, indicating that lncRNA NEAT1 is a therapeutic target for ischemic stroke.16, 17 Based on these indications, we hypothesized that lncRNA NEAT1 might play a critical role in AIS development, progression, and prognosis, however, the related evidence was limited.
Therefore, this study was conducted to investigate the predictive value of lncRNA NEAT1 for AIS risk and to explore the correlation of lncRNA NEAT1 expression with disease severity, inflammation, recurrence and potential target microRNAs in patients with AIS.
2. METHODS
2.1. Participants
From January 2013 to December 2015, 210 patients with AIS and 210 controls were consecutively enrolled in the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. The inclusion criteria for patients with AIS were as follows: (a) newly diagnosed as AIS according to patient's medical history, laboratory and neurological examination, computed tomography (CT) scan, magnetic resonance imaging (MRI), and/or diffusion weighted imaging; (b) admission to the hospital within 24 hours following the onset of symptoms; (c) age above 18 years old. The exclusion criteria for patients with AIS were as follows: (a) cerebral hemorrhage; (b) severe infections, inflammatory or autoimmune diseases; (c) history of hematological malignancies or solid tumors; (d) died within 24 hours after admission; (e) received immunosuppressive therapy within 6 months; and (f) pregnant or lactating woman. Controls were screened from the high‐stroke‐risk population, which was defined as subjects with at least three of following risk factors: hypertension, atrial fibrillation or valvulopathy, tobacco use, hyperlipidemia, diabetes mellitus, lack of physical exercise, overweight or obesity, and family history of stroke, while controls were excluded if they had history of stroke, hematological malignancies or solid tumors, severe infections, inflammatory or autoimmune diseases, or were pregnant or lactating woman. This study was approved by the Ethics Committee of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology and was conducted according to the Declaration of Helsinki. All participants or their guardians signed the informed consents before enrollment.
2.2. Data and sample collection
All the participants' clinical data were collected after enrollment, which including age, gender, body mass index (BMI), smoke, hypertension, diabetes mellitus, hyperlipidemia, hyperuricemia, and chronic kidney disease (CKD). Besides, for the patients with AIS, C‐reactive protein (CRP) and National Institute of Health Stroke Scale (NIHSS) score that were used to assess the severity of AIS were also recorded at baseline. Peripheral blood samples from patients with AIS were collected within 24 hours after admission, and peripheral blood samples of controls were collected on the enrollment. All blood samples were collected into anticoagulant tube and centrifuged at 1600 g for 10 minutes at 4℃. Then the supernatants were separated and further centrifuged at 12 000 g for 10 minutes at 4°C. Subsequently, the plasma was obtained and stored at −80°C until determination.
2.3. LncRNA NEAT1 detection
The relative expression of lncRNA NEAT1 in plasma of patients with AIS and controls were detected by real‐time quantitative polymerase chain reaction (qPCR). The detail process was displayed in “qPCR” subsection.
2.4. Inflammatory cytokines measurement
For patients with AIS, the levels of inflammatory cytokines in plasma including tumor necrosis factor‐α (TNF‐α), interleukin‐1β (IL‐1β), IL‐6, IL‐8, IL‐10, IL‐17, and IL‐22 were measured by enzyme‐linked immune sorbent assay (ELISA) using commercial human ELISA Kits (Abcam) following the manufacturer's protocol.
2.5. MiRNAs detection
MiR‐124, miR‐125a, and miR‐146b are reported to be direct target genes of lncRNA NEAT1,18, 19, 20 which are also well‐known as inflammation‐related miRNAs. Hence, we hypothesized that lncRNA NEAT1 might function via miR‐124, miR‐125a, and miR‐146b in AIS as well, and we detected the relative expression of plasma miR‐124, miR‐125a and miR‐146b in patients with AIS. The detail process was displayed in “qPCR” subsection.
2.6. qPCR
Total RNA was extracted from plasma samples using TRIzol Reagent (Invitrogen), and then transcription to cDNA was conducted using PrimeScript RT reagent Kit (Takara). The qPCR process was performed with the application of QuantiNova SYBR Green PCR Kit (Qiagen). GAPDH was applied as the internal reference for lncRNA NEAT1, and U6 was applied as the internal reference for miRNAs. Sequences of primers used in qPCR were as follows: LncRNA NEAT1, forward primer: TGTCCCTCGGCTATGTCAGA, reverse primer: GAGGGGACGTGTTTCCTGAG; miRNA‐124, forward primer: ACACTCCAGCTGGGCGTGTTCACAGCGGACCT, reverse primer: TGTCGTGGAGTCGGCAATTC; miRNA‐125a, forward primer: ACACTCCAGCTGGGTCCCTGAGACCCTTTAAC, reverse primer: TGTCGTGGAGTCGGCAATTC; miRNA‐146b, forward primer: ACACTCCAGCTGGGTGAGAACTGAATTCCA, reverse primer: TGTCGTGGAGTCGGCAATTC; GAPDH, forward primer: TGACCACAGTCCATGCCATCAC reverse primer: GCCTGCTTCACCACCTTCTTGA; U6, forward primer: CTCGCTTCGGCAGCACATATACTA, reverse primer: ACGAATTTGCGTGTCATCCTTGC.
2.7. Follow‐up
All patients with AIS received appropriate treatments decided by physicians. And all patients were followed up to 36 months or stroke recurrence or death, and the median follow‐up duration was 36.0 months ranging from 0.0 to 36.0 months. Stroke recurrence and death were recorded during follow‐up, and the patients with AIS were further divided into recurrence/death group and non‐recurrence group. Recurrence‐free survival (RFS) was defined as the duration from admission to the death or stroke recurrence. For the patients who lost follow‐up, calculation of RFS was censored on the date they were last known to be alive.
2.8. Statistical analysis
All the statistical analyses were performed with the use of SPSS 24.0 statistical software (IBM), and figures were made using SPSS 24.0 statistical software (IBM) or GraphPad Prism 7.00 software (GraphPad Software). Continuous variables were checked for normality by using Kolmogorov‐Smirnov test. The normal distributed variables were presented as mean ± standard deviation (SD), and the skewed distributed variables were expressed as median and interquartile range (IQR). Categorical variables were presented as count (percentage). Comparisons between groups were determined by Student's t test, Wilcoxon rank sum test or chi‐square test. Correlations between continuous variables were analyzed by Spearman's rank correlation test. Correlations between categorical variables were analyzed by chi‐square test. The discrimination performance of continuous variables was evaluated using receiver operating characteristic (ROC) curves and the area under the curve (AUC) with 95% confidence interval (CI). Recurrence‐free survival was displayed by Kaplan‐Meier (K‐M) curve and the difference of RFS between groups was determined by log‐rank test. All tests were two‐tailed, and P value < 0.05 was considered significant.
3. RESULTS
3.1. Study flow
Both patients with AIS and controls were enrolled in our study. For the AIS patients' recruitment, a total of 322 acute stroke patients were screened, while 122 of them were excluded (Figure 1). Then, 210 newly onset patients with AIS were recruited, and the plasma samples were collected within 24 hours after admission for further detection. All patients with AIS were followed up to 36 months or until they suffered stroke recurrence or death, whereas, 26 patients were censored on the data of last known to be alive. Finally, 210 patients with AIS were included in the final analysis. For the controls' recruitment, 251 controls with vascular risk factors were screened, while 41 controls were excluded. Subsequently, 210 controls were recruited, and their plasma samples were collected on the enrollment, followed by the detection of lncRNA NEAT1.
Figure 1.
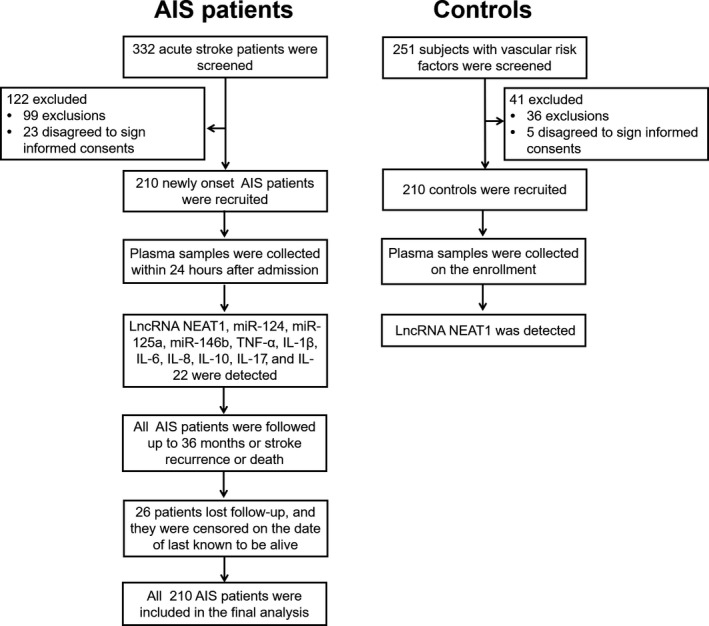
Study flow
3.2. Comparison of characteristics between patients with AIS and controls
210 patients with AIS with mean age of 62.2 ± 13.0 years and 210 controls with mean age of 60.5 ± 10.4 years were enrolled in this study (Table 1). There were 159 (75.7%) males and 51 (24.3%) females in patients with AIS, and 165 (78.6%) males and 45 (21.4%) females in controls, meanwhile, the mean BMI of patients with AIS and controls was 24.8 ± 2.9 kg/m2 and 24.4 ± 2.6 kg/m2, respectively. No difference of age (P = .138), gender (P = .486) or BMI (P = .234) was found between patients with AIS and controls. Besides, no difference of smoke (P = .922), hypertension (P = .105), diabetes mellitus (P = .234), hyperlipidemia (P = .558), hyperuricemia (P = .532), or CKD (P = .157) was observed between the two groups, and the detailed information of these characteristics was displayed in Table 1. For patients with AIS, the mean NIHSS score was 8.3 ± 3.6, moreover, median values of biochemical indexes including CRP, TNF‐α, IL‐1β, IL‐6, IL‐8, IL‐10, IL‐17, and IL‐22 were 35.6 (27.5‐49.7) (mg/L), 78.7 (56.2‐124.5) (pg/mL), 6.7 (4.6‐9.2) (pg/mL), 61.4 (49.9‐80.7) (pg/mL), 67.6 (50.2‐97.2) (pg/mL), 15.8 (11.2‐23.3) (pg/mL), 129.4 (82.7‐170.8) (pg/mL), and 75.8 (54.6‐108.8) (pg/mL), respectively.
Table 1.
Clinical characteristics of AIS patients and controls
| Items | AIS patients (N = 210) | Controls (N = 210) | P value |
|---|---|---|---|
| Age (y), mean ± SD | 62.2 ± 13.0 | 60.5 ± 10.4 | .138 |
| Gender, No. (%) | .486 | ||
| Male | 159 (75.7) | 165 (78.6) | |
| Female | 51 (24.3) | 45 (21.4) | |
| BMI (kg/m2), mean ± SD | 24.8 ± 2.9 | 24.4 ± 2.6 | .234 |
| Smoke, No. (%) | .922 | ||
| No | 117 (55.7) | 118 (56.2) | |
| Yes | 93 (44.3) | 92 (43.8) | |
| Hypertension, No. (%) | .105 | ||
| No | 33 (15.7) | 46 (21.9) | |
| Yes | 177 (84.3) | 164 (78.1) | |
| Diabetes mellitus, No. (%) | .234 | ||
| No | 160 (76.2) | 170 (81.0) | |
| Yes | 50 (23.8) | 40 (19.0) | |
| Hyperlipidemia, No. (%) | .558 | ||
| No | 107 (51.0) | 113 (53.8) | |
| Yes | 103 (49.0) | 97 (46.2) | |
| Hyperuricemia, No. (%) | .532 | ||
| No | 139 (66.2) | 145 (69.0) | |
| Yes | 71 (33.8) | 65 (31.0) | |
| CKD, No. (%) | .157 | ||
| No | 176 (83.8) | 186 (88.6) | |
| Yes | 34 (16.2) | 24 (11.4) | |
| NIHSS score, mean ± SD | 8.3 ± 3.6 | – | – |
| Biochemical indexes, median (IQR) | |||
| CRP (mg/L) | 35.6 (27.5‐49.7) | – | – |
| TNF‐α (pg/mL) | 78.7 (56.2‐124.5) | – | – |
| IL‐1β (pg/mL) | 6.7 (4.6‐9.2) | – | – |
| IL‐6 (pg/mL) | 61.4 (49.9‐80.7) | – | – |
| IL‐8 (pg/mL) | 67.6 (50.2‐97.2) | – | – |
| IL‐10 (pg/mL) | 15.8 (11.2‐23.3) | – | – |
| IL‐17 (pg/mL) | 129.4 (82.7‐170.8) | – | – |
| IL‐22 (pg/mL) | 75.8 (54.6‐108.8) | – | – |
Comparison was determined by Student's t test or chi‐square test.
Abbreviations: AIS, acute ischemic stroke; BMI, body mass index; CKD, chronic kidney disease; CRP, C‐reactive protein; IL, interleukin; IQR, interquartile range; NIHSS, National Institute of Health Stroke Scale; SD, standard deviation; TNF‐α, tumor necrosis factor‐α.
3.3. Comparison of lncRNA NEAT1 expression between patients with AIS and controls
LncRNA NEAT1 expression was elevated in patients with AIS (2.107 [1.269‐3.046]) compared with Controls (0.987 [0.409‐1.495]) (P < .001) (Figure 2A). And, ROC curve displayed that lncRNA NEAT1 had a good predictive value for AIS risk (AUC = 0.804 [95%CI: 0.763‐0.845]) (Figure 2B), with the best cutoff point value of 1.471, where the AUC reached the maximum value, and the sensitivity as well as specificity at best cutoff point was 64.3% and 82.9%, respectively.
Figure 2.
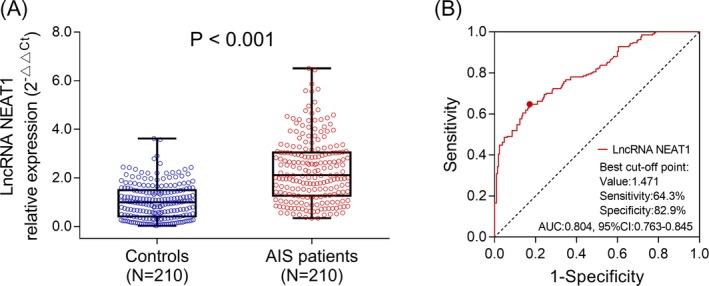
LncRNA NEAT1 expression in AIS patients and controls. LncRNA NEAT1 expression detected by qPCR in AIS patients and controls (A). Predictive value of lncRNA NEAT1 for AIS risk evaluated by ROC curve (B). Comparison between two groups was determined by Wilcoxon rank sum test. LncRNA NEAT1, long noncoding RNA nuclear enriched abundant transcript 1; AIS, acute ischemic stroke; qPCR, quantitative polymerase chain reaction; ROC curve, receiver operating characteristic curve. P < .05 was considered significant
3.4. Correlation of lncRNA NEAT1 expression with clinical characteristics in patients with AIS
Among the continuous variables in patients with AIS, lncRNA NEAT1 expression was positively correlated with NIHSS score (P < .001, r = .503), CRP (P < .001, r = .385), TNF‐α (P = .001, r = .228), IL‐6 (P < .001, r = .275), IL‐8 (P = .018, r = .163), and IL‐22 (P = .002, r = .218), while it was negatively correlated with IL‐10 (P < .001, r = −.256) (Table 2). No correlation of lncRNA NEAT1 expression with other continuous variables including age (P = .611, r = .035), BMI (P = .583, r = −.038), IL‐1β (P = .431, r = .055), and IL‐17 (P = .390, r = .060) was observed. For categorical variables, lncRNA NEAT1 high expression was numerically associated with absence of smoke in patients with AIS (P = .071), but without significant difference (Table 3). And, no correlation of lncRNA NEAT1 expression with other categorical variables including gender (P = .148), hypertension (P = .850), diabetes mellitus (P = .195), hyperlipidemia (P = .490), hyperuricemia (P = .307), or CKD (P = .261) was found in patients with AIS.
Table 2.
Correlation of lncRNA NEAT1 with continuous variables in AIS patients
| Items | LncRNA NEAT1 relative expression | |
|---|---|---|
| P value | Correlation coefficient (r) | |
| Age | .611 | .035 |
| BMI | .583 | −.038 |
| NIHSS score | <.001 | .503 |
| CRP | <.001 | .385 |
| TNF‐α | .001 | .228 |
| IL‐1β | .431 | .055 |
| IL‐6 | <.001 | .275 |
| IL‐8 | .018 | .163 |
| IL‐10 | <.001 | −.256 |
| IL‐17 | .390 | .060 |
| IL‐22 | .002 | .218 |
Correlation was determined by Spearman's rank correlation test.
Abbreviations: AIS, acute ischemic stroke; BMI, body mass index; CRP, C‐reactive protein; IL, interleukin; lncRNA, long non‐coding RNA; NIHSS, National Institute of Health stroke scale; SD, standard deviation; TNF‐α, tumor necrosis factor‐α.
Table 3.
Correlation of lncRNA NEAT1 with categorical variables in AIS patients
| Items | LncRNA NEAT1 expressiona | P value | |
|---|---|---|---|
| High | Low | ||
| Gender, No. (%) | .148 | ||
| Male | 75 (47.2) | 84 (52.8) | |
| Female | 30 (58.8) | 21 (41.2) | |
| Smoke, No. (%) | .071 | ||
| No | 65 (55.6) | 52 (44.4) | |
| Yes | 40 (43.0) | 53 (57.0) | |
| Hypertension, No. (%) | .850 | ||
| No | 16 (48.5) | 17 (51.5) | |
| Yes | 89 (50.3) | 88 (49.7) | |
| Diabetes mellitus, No. (%) | .195 | ||
| No | 84 (52.5) | 76 (47.5) | |
| Yes | 21 (42.0) | 29 (58.0) | |
| Hyperlipidemia, No. (%) | .490 | ||
| No | 56 (52.3) | 51 (47.7) | |
| Yes | 49 (47.6) | 54 (52.4) | |
| Hyperuricemia, No. (%) | .307 | ||
| No | 73 (52.5) | 66 (47.5) | |
| Yes | 32 (45.1) | 39 (54.9) | |
| CKD, No. (%) | .261 | ||
| No | 85 (48.3) | 91 (51.7) | |
| Yes | 20 (58.8) | 14 (41.2) | |
Correlation was determined by chi‐square test.
Abbreviations: AIS, acute ischemic stroke; CKD, chronic kidney disease; lncRNA, long non‐coding RNA.
High or low expression was classified by the median value of LncRNA NEAT1 in AIS patients.
3.5. Comparison of LncRNA NEAT1 expression between recurrence/death patients and non‐recurrence survivors
LncRNA NEAT1 expression was increased in recurrence/death patients compared with non‐recurrence survivors (P = .003) (Figure 3A). Besides, ROC curve showed that lncRNA NEAT1 could predict death/recurrence risk with the AUC of 0.641 (95% CI: 0.541‐0.741), meanwhile, CRP as well as NIHSS score also predicted death/recurrence risk in patients with AIS with the AUC of 0.709 (95%CI: 0.621‐0.797) as well as 0.653 (95%CI: 0.552‐0.754), respectively (Figure 3B), suggesting that lncRNA NEAT1 presented with equal predictive value for death/recurrence risk compared with CRP and NIHSS score.
Figure 3.
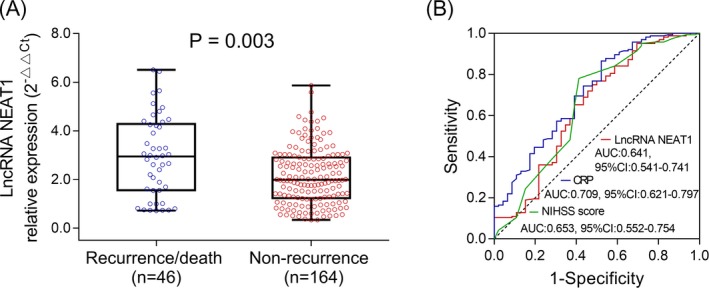
LncRNA NEAT1 expression in recurrence/death patients and non‐recurrence survivors. LncRNA NEAT1 expression detected by qPCR in recurrence/death patients and non‐recurrence survivors (A). Predictive value of lncRNA NEAT1 for recurrence/death evaluated by ROC curves (B). Comparison between two groups was determined by Wilcoxon rank sum test. LncRNA NEAT1, long noncoding RNA nuclear enriched abundant transcript 1; qPCR, quantitative polymerase chain reaction; ROC curves, receiver operating characteristic curves. P < .05 was considered significant
3.6. Comparison of RFS between lncRNA NEAT1 high expression and lncRNA NEAT1 low expression patients
K‐M curves showed that RFS in lncRNA NEAT1 high expression patients was shorter compared with lncRNA NEAT1 low expression patients (P = .019; Figure 4).
Figure 4.
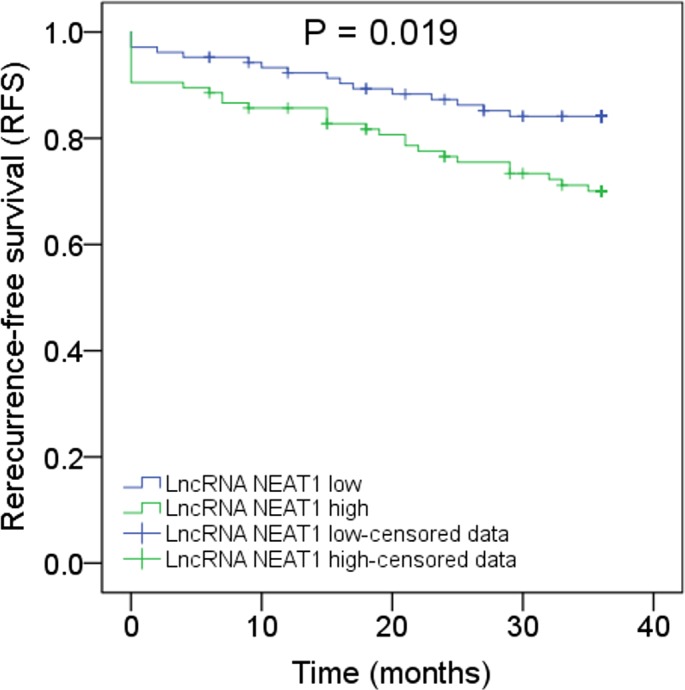
Correlation of lncRNA NEAT1 expression with RFS. Analysis of RFS in lncRNA NEAT1 high expression patients and lncRNA NEAT1 low expression patients. K‐M curves were used to display the RFS. Comparison between two groups was determined by log‐rank test. LncRNA NEAT1, long noncoding RNA nuclear enriched abundant transcript 1; RFS, recurrence‐free survival K‐M curves, Kaplan‐Meier curves. P < .05 was considered significant
3.7. Correlation of lncRNA NEAT1 expression with miR‐124, miR‐125a, and miR‐146b expressions in patients with AIS
LncRNA NEAT1 expression was negatively correlated with miR‐124 (P < .001, r = −.483) and miR‐125a (P < .001, r = −.508) expressions in patients with AIS, while no correlation of lncRNA NEAT1 expression with miR‐146b (P = .175, r = −.094) expression was found (Figure 5).
Figure 5.
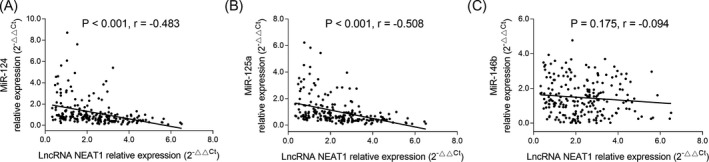
Association of lncRNA NEAT1 expression with several targeted genes. Correlation of lncRNA NEAT1 expression with miR‐124 expression in AIS patients (A). Correlation of lncRNA NEAT1 expression with miR‐125a expression in AIS patients (B). Correlation of lncRNA NEAT1 expression with miR‐146b expression in AIS patients (C). Correlation of lncRNA NEAT1 expression with target genes was assessed by Spearman's rank correlation test. LncRNA NEAT1, long noncoding RNA nuclear enriched abundant transcript 1; miR, microRNA; AIS, acute ischemic stroke. P < .05 was considered significant
4. DISCUSSION
LncRNA NEAT1, as an abundant and ubiquitously expressed lncRNA, has been reported to be a pivotal factor in the pathology of various diseases, including cardiovascular and cerebrovascular diseases.14, 15, 16 For example, a study shows that lncRNA NEAT1 is overexpressed in myocardial IR injury mice models and lncRNA NEAT1 knockdown represses levels of pro‐inflammatory cytokines (TNF‐α, IL‐1β, and IL‐6), decreases troponin level, reduces cardiocytes apoptosis, and inhibits release of lactate LDH in vitro.14 Besides, a study displays that lncRNA NEAT1 knockdown inhibits levels of IL‐6, IL‐1β, cyclooxygenase‐2 (COX‐2), and TNF‐α via regulating miR‐342‐3p in human macrophages THP‐1 cells.15 For stroke, a study discloses that lncRNA NEAT1 upregulation enhances OGD/R injury‐induced inflammatory response in cerebral microglial cells through Wnt/β‐catenin signaling pathway.16 These studies reveal that lncRNA NEAT1 may promote the disease occurrence and enhance inflammation response via targeted genes or downstream signaling pathways in cardiovascular and cerebrovascular diseases.
In clinical practice, only a few studies explore the role of lncRNA NEAT1 in disease risk or disease condition of cardiovascular and cerebrovascular diseases or other inflammatory disease.13, 21 For example, lncRNA NEAT1 has been found to be overexpressed in post‐myocardial infarction patients and patients with tuberculosis, indicating that lncRNA NEAT1 may act as a biomarker for increased risk of these diseases.12, 13 For the disease condition or inflammation, a study displays that lncRNA NEAT1 expression is associated with left ventricular ejection fraction, low‐density lipoprotein (LDL) cholesterol, and high‐density lipoprotein (HDL) cholesterol in post‐myocardial infarction patients.13 And, another study shows that lncRNA NEAT1 expression is positively associated with Acute Physiology and Chronic Health Evaluation (APACHE) II score, TNF‐α, IL‐1β, IL‐6, and IL‐8, but is negatively correlated with IL‐10 in sepsis patients.21 These data suggest that lncRNA NEAT1 may positively correlate with disease severity and inflammation level in cardiovascular and cerebrovascular diseases as well as inflammatory disease. Moreover, a previous study has disclosed that lncRNA NEAT1 promotes the OGD/R injury and neuroinflammation damage in the cell model of stroke.16 Taken together, we speculated that lncRNA NEAT1 might also be involved in the disease risk, progression, and inflammation of AIS, whereas related exploration is seldomly reported. In our study, we enrolled 210 patients with AIS and 210 controls to investigate the correlation of the lncRNA NEAT1 expression with AIS risk, and we found that lncRNA NEAT1 was overexpressed in patients with AIS, meanwhile, it could predict AIS risk. Furthermore, we also explored the association of lncRNA NEAT1 expression with disease condition as well as inflammation level in patients with AIS, and we observed that lncRNA NEAT1 high expression was associated with increased NIHSS score, raised CRP level, elevated pro‐inflammatory cytokines levels (including TNF‐α, IL‐6, IL‐8, and IL‐22), and reduced anti‐inflammatory cytokine level (IL‐10) in patients with AIS. The possible reasons for these results might be as follows: (a) lncRNA NEAT1 might enhance the dysfunction of cerebral microglial cells through Wnt/β‐catenin signaling pathway, thus, it contributed to impairing the normal cerebrovascular physiology that led to advanced disease severity, and consequently, elevated NIHSS score was found in lncRNA NEAT1 high expression patients with AIS16; (b) lncRNA NEAT1 might induce the loss of motor neurons and lead to the devastating neurodegenerative disorder, thus aggravated the abnormal cognitive deficits and functional activities of patients with AIS and increased the NIHSS score22; (c) lncRNA NEAT1 promoted intima thickening or vascular occlusion by modulating the phenotype conversion of vascular smooth muscle cells, together with excessive apoptosis of epithelial cells, which might enhance the atherosclerotic lesion and aggravate the severity of AIS, thus lncRNA NEAT1 correlated with increased NIHSS score17; (d) lncRNA NEAT1 might enhance the reactive oxygen species (ROS) level to disturb the normal initiation of inflammatory cascades, which further induced excess production of inflammatory cytokines, and thus, it led to increased levels of inflammatory factors (including CRP, TNF‐α, IL‐6, IL‐8, and IL‐22) as well as reduced anti‐inflammatory cytokine level in patients with AIS.23
As to the predictive value of lncRNA NEAT1 for prognosis, previous studies mainly focus on the prognostic value of lncRNA NEAT1 in different types of cancers, little is known about the role of lncRNA NEAT1 in prognosis of cardiovascular and cerebrovascular diseases or inflammatory disease, especially AIS.24, 25 In our study, we explored the role of lncRNA NEAT1 in disease recurrence as well as survival profile of patients with AIS. We observed that lncRNA NEAT1 could predict raised recurrence/death risk, and its predictive value for recurrence/death risk was equal with CRP and NIHSS score, furthermore, lncRNA NEAT1 expression was negatively associated with RFS in patients with AIS. The following reasons might explain these results: (a) lncRNA NEAT1 might accelerate the disease progression and promote the inflammatory response through regulating some target genes or pathways, therefore, its overexpression caused worse disease severity and further reduced RFS in patients with AIS14, 15; (b) lncRNA NEAT1 might facilitate the resistance to therapy of patients with AIS and consequently decreased the RFS, while detailed mechanism remained largely unclear.
MicroRNAs are evolutionarily conserved noncoding RNA oligonucleotides and are involved in the pathology of numerous diseases.26, 27 According to previous studies, miR‐124, miR‐125a, and miR‐146b, which are frequently investigated miRNAs, are well known as inflammation‐related miRNAs, moreover, they have been identified as direct target genes of lncRNA NEAT1.18, 19, 20 Thus, we hypothesized that lncRNA NEAT1 might function via regulating miR‐124, miR‐125a, and miR‐146b in AIS as well. To verify this hypothesis, we assessed the correlation of lncRNA NEAT1 expression with the expression of plasma miR‐124, miR‐125a, and miR‐146b in patients with AIS. And, we observed that lncRNA NEAT1 expression negatively correlated with miR‐124 and miR‐125a expressions, while no correlation of lncRNA NEAT1 expression with miR‐146b expression was found in patients with AIS. These data suggested that lncRNA NEAT1 might affect the disease occurrence, disease severity, inflammation level, and prognosis of AIS through negatively regulating miR‐124 and miR‐125a.
Some limitations existed in our study: (a) the follow‐up duration (median 36.0 months) was relatively short, and long‐term effect of lncRNA NEAT1 on prognosis was not investigated; (b) this was a single‐center study, which might lack wide representativeness; (c) although we observed the negative correlation of lncRNA NEAT1 with miR‐124 and miR‐125a, the detailed molecular mechanisms of lncRNA NEAT1 in AIS had not been explored. Further multi‐center study with longer follow‐up duration is needed to validate our results, and the molecular mechanisms need to be further explored.
In conclusion, lncRNA NEAT1 may serve as a novel biomarker that predicts higher AIS risk and contributes to the surveillance of disease severity, inflammation level, and prognosis in AIS.
ACKNOWLEDGMENTS
This study was supported by Hubei Municipal Commission of Health and Family Planning (WJ7F018).
Li P, Duan S, Fu A. Long noncoding RNA NEAT1 correlates with higher disease risk, worse disease condition, decreased miR‐124 and miR‐125a and predicts poor recurrence‐free survival of acute ischemic stroke. J Clin Lab Anal. 2020;34:e23056 10.1002/jcla.23056
Ping Li and Shuyuan Duan contributed equally to this work.
REFERENCES
- 1. Urdaneta AE, Bhalla P. Cutting edge acute ischemic stroke management. Emerg Med Clin North Am. 2019;37(3):365‐379. [DOI] [PubMed] [Google Scholar]
- 2. Heit JJ, Zaharchuk G, Wintermark M. Advanced neuroimaging of acute ischemic stroke: penumbra and collateral assessment. Neuroimaging Clin N Am. 2018;28(4):585‐597. [DOI] [PubMed] [Google Scholar]
- 3. Boustia F, Crespy A, Janot K, Herbreteau D. Management of patients with acute ischemic stroke regarding endovascular treatment. Presse Med. 2019;48(6):664‐671. [DOI] [PubMed] [Google Scholar]
- 4. Nepal G, Kharel G, Ahamad ST, Basnet B. Tenecteplase versus Alteplase for the Management of acute ischemic stroke in a low‐income country‐Nepal: cost, efficacy, and safety. Cureus. 2018;10(2):e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papanagiotou P, Ntaios G. Endovascular Thrombectomy in Acute Ischemic Stroke. Circ Cardiovasc Interv. 2018;11(1):e005362. [DOI] [PubMed] [Google Scholar]
- 6. Agostoni E, Carolei A, Micieli G, Provinciali L, Toni D, Vidale S. The organisation of the acute ischemic stroke management: key notes of the Italian Neurological Society and of the Italian Stroke Organization. Neurol Sci. 2018;39(3):415‐422. [DOI] [PubMed] [Google Scholar]
- 7. Kopp F. Molecular functions and biological roles of long non‐coding RNAs in human physiology and disease. J Gene Med. 2019;21(8):e3104. [DOI] [PubMed] [Google Scholar]
- 8. Alishahi M, Ghaedrahmati F, Kolagar TA, et al. Long non‐coding RNAs and cell death following ischemic stroke. Metab Brain Dis. 2019;34(5):1243‐1251. [DOI] [PubMed] [Google Scholar]
- 9. Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641‐27650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parasramka M, Yan IK, Wang X, et al. BAP1 dependent expression of long non‐coding RNA NEAT‐1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer. 2017;16(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up‐regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20(16):3373‐3377. [PubMed] [Google Scholar]
- 12. Huang S, Huang Z, Luo Q, Qing C. The expression of lncRNA NEAT1 in human tuberculosis and its antituberculosis effect. Biomed Res Int. 2018;2018:9529072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gast M, Rauch B, Haghikia A, et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovasc Res. 2019: 115(13):1886-1906.cvz085. [DOI] [PubMed] [Google Scholar]
- 14. Du XJ, Wei J, Tian D, et al. NEAT1 promotes myocardial ischemia‐reperfusion injury via activating the MAPK signaling pathway. J Cell Physiol. 2019;234(10):18773‐18780. [DOI] [PubMed] [Google Scholar]
- 15. Wang L, Xia JW, Ke ZP, Zhang BH. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR‐342‐3p in human macrophages THP‐1 cells. J Cell Physiol. 2019;234(4):5319‐5326. [DOI] [PubMed] [Google Scholar]
- 16. Han D, Zhou Y. YY1‐induced upregulation of lncRNA NEAT1 contributes to OGD/R injury‐induced inflammatory response in cerebral microglial cells via Wnt/beta‐catenin signaling pathway. Vitro Cell Dev Biol Anim. 2019;55(7):501‐511. [DOI] [PubMed] [Google Scholar]
- 17. Zhou H, Wang B, Yang YX, et al. Long noncoding RNAs in pathological cardiac remodeling: a review of the update literature. Biomed Res Int. 2019;2019:7159592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao MY, Wang GQ, Wang NN, Yu QY, Liu RL, Shi WQ. The long‐non‐coding RNA NEAT1 is a novel target for Alzheimer's disease progression via miR‐124/BACE1 axis. Neurol Res. 2019;41(6):489‐497. [DOI] [PubMed] [Google Scholar]
- 19. Huang X, Zhong R, He X, et al. Investigations on the mechanism of progesterone in inhibiting endometrial cancer cell cycle and viability via regulation of long noncoding RNA NEAT1/microRNA‐146b‐5p mediated Wnt/beta‐catenin signaling. IUBMB Life. 2019;71(2):223‐234. [DOI] [PubMed] [Google Scholar]
- 20. Yan H, Liang H, Liu L, Chen D, Zhang Q. Long noncoding RNA NEAT1 sponges miR125a5p to suppress cardiomyocyte apoptosis via BCL2L12. Mol Med Rep. 2019;19(5):4468‐4474. [DOI] [PubMed] [Google Scholar]
- 21. Huang Q, Huang C, Luo Y, He F, Zhang R. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am J Emerg Med. 2018;36(9):1659‐1663. [DOI] [PubMed] [Google Scholar]
- 22. Nishimoto Y, Nakagawa S, Hirose T, et al. The long non‐coding RNA nuclear‐enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain. 2013;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dua K, Malyla V, Singhvi G, et al. Increasing complexity and interactions of oxidative stress in chronic respiratory diseases: An emerging need for novel drug delivery systems. Chem Biol Interact. 2019;299:168‐178. [DOI] [PubMed] [Google Scholar]
- 24. Idogawa M, Nakase H, Sasaki Y, Tokino T. Prognostic effect of long noncoding RNA NEAT1 expression depends on p53 mutation status in cancer. J Oncol. 2019;2019:4368068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghafouri‐Fard S, Taheri M. Nuclear Enriched Abundant Transcript 1 (NEAT1): A long non‐coding RNA with diverse functions in tumorigenesis. Biomed Pharmacother. 2019;111:51‐59. [DOI] [PubMed] [Google Scholar]
- 26. Momen‐Heravi F, Bala S. miRNA regulation of innate immunity. J Leukoc Biol. 2018. 10.1002/JLB.3MIR1117-459R. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27. Verjans R, van Bilsen M, Schroen B. MiRNA deregulation in cardiac aging and associated disorders. Int Rev Cell Mol Biol. 2017;334:207‐263. [DOI] [PubMed] [Google Scholar]


