Abstract
Urban sprawl increasingly affects the ecology of natural populations, including host–microbiota interactions, with observed differences in the gut microbiota between urban and rural hosts. While different mechanisms could explain this pattern, dietary uptake constitutes a likely candidate. To assess the contribution of diet in explaining urban–rural variation in gut microbiota, we performed an aviary experiment in which urban and rural house sparrows were fed with mimics of urban or rural diets. Before the experiment, rural sparrows hosted more diverse gut communities, with a higher relative abundance of Enterococcaceae and Staphylococcaceae and lower abundance of genes involved in xenobiotic degradation and lipid metabolism than their urban counterparts. The experimental diets significantly altered gut microbiota α- and β-diversity and taxonomic composition, with the strongest shifts occurring in individuals exposed to contrasting diets. Overall, diet-induced shifts resembled initial differences between free-ranging urban and rural hosts. Furthermore, rural diet had a positive impact on urban host body mass but only in hosts with the highest initial gut diversity. Overall, our results indicate that diet constitutes an important factor contributing to differences in gut microbiota along the urbanization gradient and provide new insights on possible fitness consequences of a reduced gut diversity in urban settings.
Keywords: microbiome, Passer domesticus, 16S metabarcoding, cloaca, bacterial communities, plasticity
1. Introduction
Human activities are increasingly recognized as dominant drivers of contemporary environmental change, and the magnitude, variety and longevity of these effects have given rise to the launch of a new epoch: the Anthropocene [1]. While consequences of anthropogenic change at population and species levels are well documented, putative effects on interspecific interactions are still less well understood [2,3]. One type of interaction that received increasing attention within the context of anthropogenic change is that between animal hosts and their associated gut microbiota, driven by the general expectation that gut microbiomes are at least partly shaped by the host's environment. In support of this, gut microbiomes were earlier shown to vary with habitat fragmentation and degradation [4,5], pollution of air, water and soil [6,7], and climate change [8]. Urbanization is increasingly recognized as a key component of human-induced change, as it combines several such facets of global change [9]. In order to better understand whether and how urbanization drives variation in the gut microbiome, the study of wild animal populations present along urbanization gradients could provide relevant insights on host–microbiome interactions in urban settings and consequences for host fitness (see [10,11]). Along these lines, recent studies on bird populations have shown consistent differences in community diversity and structure, as well as taxonomic and functional composition of gut microbiota in relation to urbanization [12,13].
Different non-exclusive mechanisms may explain this pattern linking gut microbiota characteristics to urbanization levels. First, digestive tracts may host representative samples of microbial communities present in the surrounding environment [14], resulting in local variation in gut bacterial communities [15,16]. As bacterial communities in water and soil have been shown to vary with urbanization [17,18], hosts living along such gradients would hence be exposed to different microbial pools likely to colonize their guts. In birds, passive uptake of microbiota present in the environment may occur through feather preening for instance, with correlations between feather and gut microbiota having been shown in two birds species [19]. Second, gut microbiomes are known to covary with genetic, immune and morphological traits of hosts [20–22], which by the process of ‘host filtering’ may select for specific communities [23]. Because such traits have been shown to be impacted by urbanization (e.g. in birds [24–27]), urban-induced variations in the gut microbiota may reflect shifts in host traits along the urban gradient.
A third major mechanism is host diet which depends both on host environment and host-specific traits, such as nutritional status, food preference or behaviour. Diet comprises an active intake of food, which is likely to be associated with specific bacteria, and thus plays a predominant role in shaping the gut microbiota [28]. A suite of studies have shown gut microbial differences between hosts specialized on different food resources (e.g. [29] in birds). Diet-related variation in gut taxonomic composition has been shown at the intra-specific level as well [30]. Dietary shifts in urban populations have been widely observed in mammals [31] and birds [32]. These might result from an over-abundance of non-native plants, human food waste or food from artificial feeders [33–35] or reflect changes in availability of natural foods as secondary effects of urbanization on trophic dynamics [36], or even a change in food requirements and/or feeding strategies [26]. As a result, diet-mediated shifts in gut microbiota can be expected to occur along urban gradients.
Here, we integrate correlative and experimental approaches to assess the contribution of the diet in urban-rural variation in gut microbiota composition of house sparrows (Passer domesticus), an avian commensal of human settlements, with a particular focus on gut community plasticity in response to contrasting diets. In sparrows, diet is known to vary according to urbanization [37]. Recently, Teyssier et al. [13] showed that gut microbiota in Flemish urban sparrows showed lower diversity, a different taxonomic composition and lower levels of seasonal variation, than rural birds. Building on these findings, we here report on an aviary experiment in which sparrows from three urban and three rural populations in southern France were fed with mimics of either urban or rural diets. Gut microbiota were sampled upon capture and after six weeks of diet treatment. This approach allowed us to describe gut microbiota characteristics from free-ranging sparrow populations in relation to urbanization, to quantify effects of subsequent diet treatments on microbial diversity, taxonomic and inferred functional potential, and to assess how experimentally induced dietary shifts in gut microbiota match natural variation recorded among free-ranging populations. At the individual level, it allowed us to test whether effects of diet treatment on host condition vary in relation to their microbiome characteristics, and to what extent these relationships differ between urban and rural birds. Because gut microbiota diversity is pivotal for host fitness (for instance, lower diversity reduces host capacity to assimilate nutrients [38] and to resist pathogen invasion [39] and probably plays an essential role in the host's capacity to respond to environmental change [40]), we specifically examined the impact of initial gut diversity on the sparrows' response to the experimental diets.
2. Methods
(a). Study area and sampling
Between 18 September and 5 October 2014, a total of 114 house sparrows were trapped with mist-nets in sites where house sparrows naturally occur (no artificial feeding sites were installed) in three urban and rural areas in south-western France. Urbanization ratio (UR) of the different areas were characterised using the percentage of build-up area within a 100 m radius around capture sites, corresponding to the average home range of house sparrows [41], using 2.2. MIPY Geo (MIPYGeo Grand Public) and CORINE Land Cover maps. Urbanization ratio (UR) was 100% in the three urban plots (Toulouse: 43°36′17″ N, 1°26′50″ E; Tarbes: 43°13′57″ N, 0°4′41″ E; Pau: 43°18′ N, 0°22′42″ W) and only a few per cent in the three rural plots (Caraman: 43°31′42″ N, 1°44′11″ E; UR = 3.77%; Montégut: 43°25′57″ N, 0°58′11″ E; UR = 5.03%; Cologne: 43°42′57″ N, 0°55′39″ E; UR = 3.95%). Upon capture, each individual was ringed, aged, sexed, weighed and measured (tarsus), and their cloacal microbiota were sampled (see below). After sampling, all birds were transferred to experimental aviaries located at the Station d'Ecologie Théorique et Expérimentale (Moulis, France).
(b). Microbiota sampling
Gut bacterial communities were sampled by gently inserting a sterile pipette tip into the cloaca of each bird, injecting 200 µl of sterile phosphate-buffered saline, and then drawing it out again. Samples were immediately placed in sterile vials, kept in a coolbox in the field and later stored at −20°C. Prior to sampling, the exterior of the cloaca was cleaned with alcohol to avoid contamination from external bacteria. Control samples were collected by pipetting 200 µl of the saline solution into a sterile vial to check for possible contamination of the pipette tips and the saline solution during sampling and preparation. While each part of the digestive tract harbours specific bacterial communities, microbial shifts incurred in the higher intestine of birds are believed to lead to concurrent shifts in cloacal communities (e.g. [42]), making cloacal sampling a reliable non-invasive technique to study inter-individual variability in gut communities (e.g. on various bird species [43–45]).
(c). Diet experiment
The diet experiment lasted six weeks in which birds were kept in an outdoor aviary composed of identical cages of 4 m × 1 m × 3 m equipped with roosting boxes and bamboo plants for perching. Birds from the same capture site were kept in small groups (average 4.75 ± 0.74, range: 3–6), with age and sex distributions as constant as possible across groups. Birds were then randomly assigned to one of the two experimental diets: an ‘urban diet' treatment composed of 30% maize, 25% bread, 25% cake and 20% potato chips, and a ‘rural diet' treatment composed of 49% maize, 24% wheat, 24% sunflower seed and 3% dried mealworm (see electronic supplementary material, figure S1 for details of the experimental design). These diets were selected according to the literature review ([37] and references therein). A detailed description of the nutritional composition of each diet is given in Salleh et al. [46]. When both diets were simultaneously provided to free-ranging individuals in a standardized design, urban individuals preferred the ‘urban diet' and vice versa (L.d.N. 2015, unpublished data), suggesting that our experimental diets realistically mimicked the natural ones. Birds were fed ad libitum. Gut microbiota of all birds was resampled at the end of the experiment. Animal welfare, maintenance and experimental procedures followed French regulations and guidelines (DREAL permit no. 31-2014-09). Sparrows were caught under capture and ringing permit no. 15038 (French National Natural History Museum).
(d). PCR amplification and high-throughput sequencing
Bacterial DNA was extracted using the Qiagen DNeasy Blood & Tissue Kit and the standard protocol designed for purification of total DNA from Gram-positive bacteria (Qiagen, Venlo, Netherlands). The V5-V6 region of the bacteria 16S rRNA gene was amplified by PCR. The library construction (PCR-free Biooscientific library preparation kit) and the sequencing (Illumina MiSeq 250 bp paired-end v3 chemistry) were performed at the Genopole of Toulouse (France). Further details regarding DNA amplification and sequencing are described in the electronic supplementary material.
(e). Bioinformatic analysis
Illumina sequencing data were processed and filtered using the OBITools package [47]. OTUs clustering (SWARM algorithm with 97% similarity threshold) and taxonomic assignation (SILVA 132-16S gene data bank) were performed using FROGS, a Galaxy pipeline [48]. After removal of contaminants and singletons, our dataset comprised 186 OTUs with an average of 5637.7 ± 107.12 (s.e.) reads per samples. Full details on data processing and filtering are provided in electronic supplementary material.
Inferred functional potential of bacterial communities were analysed using PICRUSt ([49] further details in electronic supplementary material).
(f). Statistical analyses
Microbiota α-diversity was measured using OTU richness, Chao1 index (accounting for undetected rare OTUs) and Shannon diversity index. Variations in α-diversity, body condition and microbiota inferred functional potential were analysed with generalized linear mixed effect models. Full details on models construction, parameterization and final model selection are provided in electronic supplementary material.
Variations in microbiota β-diversity were analysed using permutational multivariate analysis of variance (PERMANOVA; Adonis function) on dissimilarity matrices based on Jaccard, Bray–Curtis and Unifrac distances. Analyses were performed with R using the VEGAN package [50] and details of the parameterization of the adonis models found in electronic supplementary material, methods. Inter-group dissimilarities were analysed with linear models including all pairwise Jaccard distances between the different diet-origin combinations. As Jaccard and Bray–Curtis distances yielded similar results, thus indicating that relative abundances do not contribute much to β-diversity in our dataset, only results using Jaccard distances are shown.
Differences in taxonomic composition were analysed using linear discriminant analysis on effect size (LEfSe, [51]) using a non-parametric Kruskal–Wallis test to detect abundance differences and linear discriminant analysis (LDA) to estimate the effect size of each differentially abundant features using the Galaxy pipeline.
3. Results
(a). Variation in microbiota according to urbanization (before the experiment)
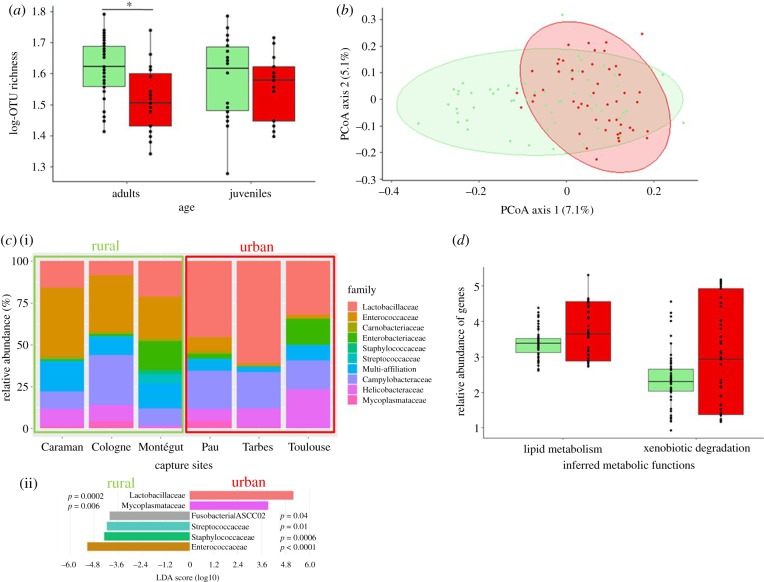
Adult sparrows from urban populations showed less diverse gut microbiota communities compared to rural ones, whereas juveniles did not differ in α-diversity (GLMM: urbanization × age: OTU richness, T1,32 = 7.41, p = 0.01; Chao1, T1.32 = 7.41, p = 0.01, figure 1a; electronic supplementary material, figure S5). With the Shannon index, these differences were not significant (GLMM: urbanization: T1,4 = 4.49, p = 0.1) indicating that species evenness is not an important factor explaining urban-rural variation. Inter-individual microbial similarity was significantly explained by urbanization ratio and capture site (PERMANOVA: urbanization F = 3.23, R2 = 0.03, p = 0.001, figure 1b; capture site, F = 2.24, R2 = 0.1, p = 0.001; electronic supplementary material figure S6; see also electronic supplementary material, table S2). Urbanization also shaped the taxonomic composition of gut microbiota, with rural hosts showing significantly higher abundances of two Firmicutes families in particular, Enterococcaceae and Staphylococcaceae, and urban hosts showing higher abundances of Lactobacillaceae (see figure 1c for stats and electronic supplementary material, figures S7 and S8 for a full overview of taxonomic differences). Inferred gene content associated with two metabolic functions, xenobiotic degradation (F1,102 = 5.94, p = 0.02) and lipid metabolism (F1,102 = 5.8, p = 0.02), were significantly over-represented in the gut microbiome of urban sparrows when compared with their rural counterparts (figure 1d), while no differences related to urbanization were found in the 10 other KEGG2 functional features (see electronic supplementary material, figure S9).
Figure 1.
(a) Bacterial species richness (log of OTU richness) of free-ranging house sparrows according to urbanization and bird age. (b) PCoA ordination based on a presence–absence similarity matrix of the gut microbiota of free-ranging house sparrows according to the urbanization ratio of the capture site. Numbers in parenthesis refer to the variance explained by the ordination axis. Coloured circles refer to 95% CI ellipses. (c)(i) Gut taxonomic composition (family level) of free-ranging house sparrows according to urbanization and capture site. (ii) Linear discriminant analysis (LDA) effect sizes representing the six families that most significantly differ in abundance between rural and urban populations at capture (see electronic supplementary material, figure S4 for full results). (d) Relative abundance of genes involved in two inferred metabolic functions according to urbanization. For all figures, median represented by the black line, 25 and 75% quartiles by the lower and upper box and 90% confidence interval by the whiskers. Red colour represents birds of urban origin and green rural origin. (Online version in colour.)
(b). Variation in microbiota induced by the diet experiment
Our experimental treatment involved six weeks of captivity which induced strong shifts in α- and β-diversity, taxonomic composition and inferred functional potential of the microbiota of all individuals (i.e. irrespective of their origin or diet). Moreover, inter-host similarity significantly increased over the course of the experiment. Captivity explained most of the variation observed in the gut microbiota during the experiment (detailed results can be found in electronic supplementary material, results). In the following section, we examine to which extent the experimental diet influenced the remaining variance (not explained by captivity per se), by focusing specifically on differences associated with the diet treatment.
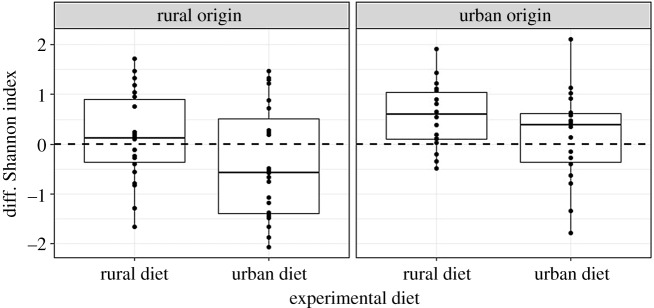
Diet treatment caused significant changes in gut α-diversity, with the urban diet inducing a decrease in diversity over the time of the experiment (mean diff. Shannon = −0.14 ± 0.15), and the rural diet inducing an increase (mean diff. Shannon = 0.33 ± 0.15; table 1). However, the strength of these diet-induced changes was also significantly influenced by the birds' origin (table 1). Overall, diet effects were strongest when the experimental diet did not match the origin, with the strongest decrease observed in birds of rural origin fed on an urban diet (diff. Shannon: −0.39 ± 0.19) and the strongest increase in urban birds fed on a rural diet (diff. Shannon + 0.58 ± 0.22; figure 2). Diet treatments also resulted in significant shifts in gut microbiota composition (PERMANOVA: F = 2.05, R2 = 0.01, p = 0.001), although the influence of origin was still maintained (F = 2.2, R2 = 0.01, p = 0.002). Post-experimental microbiota composition was most dissimilar to pre-experimental microbiota when sparrows were fed on a contrasting diet (PERMANOVA: origin × diet: F = 1.5, R2 = 0.01, p = 0.03; electronic supplementary material, figure S10). Corollarily, the microbiota of urban birds fed on a rural diet shifted more towards the pre-experimental microbiota of rural birds (intergroup similarity: 31.04 ± 0.16%) than did the microbiota of urban birds fed on an urban diet (intergroup similarity 28.3 ± 0.17%, F1,2182 = 143.2, p < 0.0001).
Table 1.
Summary of statistical models explaining the change in the Shannon index (diff. Shannon) between pre- and post-experimental treatment.
| factor | estimate | t-value | p-value |
|---|---|---|---|
| diet | −0.64 | T1,15 = −2.45 | 0.03 |
| origin | 0.18 | T1,15 = 0.62 | 0.03 |
| diet × origin | 0.33 | T1,15 = 0.69 | 0.42 |
Figure 2.
Gut microbiota changes of Shannon diversity index (diff. Shannon) between pre- and post-experimental treatment according to the experimental diet and bird origin.
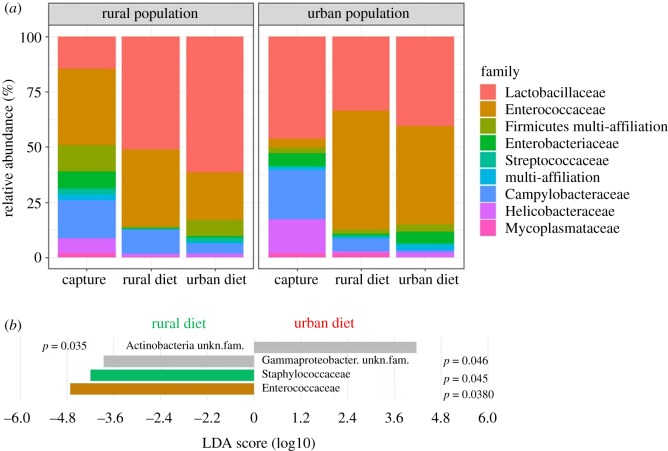
The diet treatment also caused significant shifts in taxonomic composition with birds fed on a rural diet showing significantly higher abundances of Enterococcaceae, and Staphylococcaceae in particular (figure 3). Similarly to the patterns in α- and β-diversity, the strongest taxonomic shifts occurred when sparrows were fed on a contrasting diet with respect to their origin (figure 3; electronic supplementary material, figure S11).
Figure 3.
(a) Gut taxonomic composition (family level) of house sparrows according to their origin and diet treatment. (b) Linear discriminant analysis (LDA) effect sizes representing families that significantly differ in abundance after exposure to rural or urban experimental diets. (Online version in colour.)
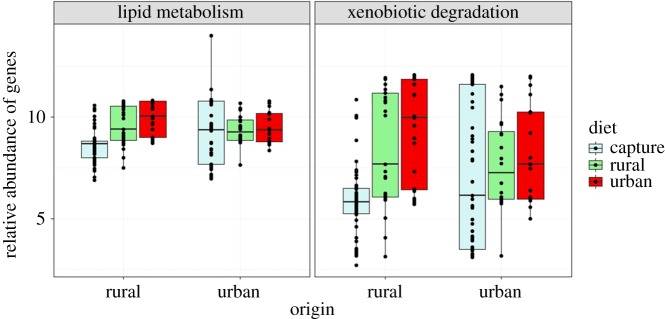
In terms of metabolic inferred functional potential, the most significant shifts were induced by the captivity regardless of the diet (see electronic supplementary material, figure S4 for details). When focusing on both functions that significantly differed between rural and urban free-living sparrows (i.e. lipid metabolism and xenobiotic degradation), we observed that diet treatments did not induce any significant change in urban birds (lipid: F1,34 = 0.52, p = 0.47; xenobiotics: F1,34 = 1.2, p = 0.3), but significantly increased these metabolic features in rural birds (lipid: F1,43 = 50.9, p < 0.0001; xenobiotics: F1,43 = 51.1, p < 0.0001; figure 4), with such shifts being most pronounced when fed on an urban diet.
Figure 4.
Relative abundance of genes involved in two inferred metabolic functions according to bird origin and the diet treatment. (Online version in colour.)
(c). Impact of microbiota variation on host condition
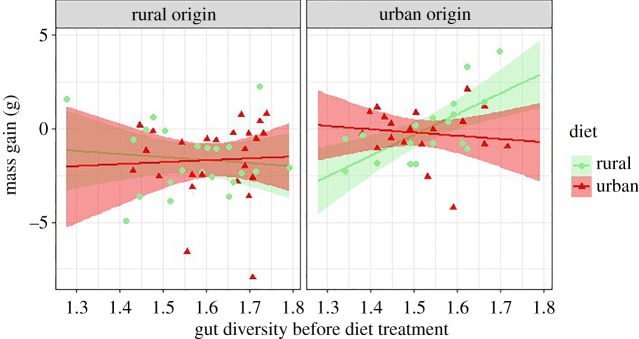
There was no significant correlation between individual host body condition and microbiota α-diversity (OTU richness, Chao1 and Shannon) neither before nor after the experimental treatment (p > 0.05 for all). However, changes in body mass during the experiment covaried with the diet treatment in interaction with bird origin and initial gut diversity (GLMM: gut diversity × diet × origin, OTU richness: F1,60 = 4.14, p = 0.05; Chao1: F1,60 = 4.14, p = 0.05; non-significant trend with Shannon: F1,60 = 3.18, p = 0.08) but not with the initial β-diversity (GLMM: PCo 1: F1,63 = 3.38, p = 0.07; PCo 2: F1,63 = 1.91, p = 0.17). Urban individuals exposed to a rural diet gained most mass, in particular when characterized by high α-diversity in gut microbiota before the diet treatment (figure 5).
Figure 5.
Mass gain (in grams) during the experiment in relation to the initial gut microbiota diversity (log OTU richness) and bird origin. Coloured areas around the regression line refer to the 95% CI. (Online version in colour.)
4. Discussion
(a). Effect of urbanization on the gut microbiota of free-ranging sparrows
Comparison of the gut microbiota of sparrows sampled before the experiment shows a clear contrast in gut community features according to urbanization. First, urbanization of the sites of capture affected α-diversity, with urban adult birds hosting less diverse communities than rural birds. Second, it influenced gut community β-diversity in that community OTU composition was most explained by the level of urbanization of the site of capture. Third, it was associated with significant taxonomic shifts, with urban gut microbiomes being characterised by lower levels of Enterococcaceae, Staphylococcaceae and higher levels of Lactobacillaceae. Last, sequences involved in lipid metabolism and xenobiotic degradation were overrepresented in urban gut communities. These results largely corroborate the findings of recent study on house sparrows in Flanders (Belgium), where urban individuals showed a similar decrease in gut diversity, lower levels of Staphylococceae, higher levels of Lactobacillaceae and increased levels of xenobiotic degradation functions [13]. These similarities are all the more striking as both studies were carried out in substantially different geographical areas and contexts, suggesting that the patterns observed are not due to specific regional contexts, but to a consistent urbanization effect in this species. A recent study on Californian white-crowned sparrows (Zonotrichia leucophrys) showed higher gut diversity in urban populations [12]. New World cities comprise much greener and less built-up habitats when compared with European ones [52] suggesting that ‘urban environments' in different geographical regions may not constitute true replicates. This is supported by the strong positive correlation between green cover and gut diversity found by Phillips et al. [12] which actually corroborates the findings of Teyssier et al. [13] and this study rather than contradicts them. While addressing effects of human presence rather than of urbanization per se, another recent study on two species of Darwin's finch reported a lower gut diversity in areas where humans were present, for one of both species [53]. These similarities irrespective of host taxa, geographical region and ecological context, especially with regard to reduced α-diversity, beg the question as to the mechanisms that explain this trend, with dietary changes comprising the most likely candidate.
(b). Effect of the diet experiment on the gut microbiota
One of the main patterns we observed is that the experiment in itself induced large shifts in the gut microbiota in all birds, regardless of diet type and origin. In particular, it induced a general increase in bacterial richness, explained most of the variation in β-diversity, and substantially modified taxonomic composition and inferred functional potential. Furthermore, we observed a significant and general increase in microbial similarity between birds sharing the same cage over the course of the experiment. The housing in aviaries for over six weeks, involving drastic changes in the birds' environment (and associated environmental bacteria), but also inevitable modifications in behaviour, social interactions, as well as various physiological features all probably play a part in this general captivity effect, which has been well described in birds (e.g. [54,55]). Although it does not allow us to distinguish between the various aforementioned mechanisms, this captivity effect highlights the primordial importance of non-dietary factors in explaining microbiome variation at the intraspecific scale.
Despite this strong and pervasive effect of captivity, which homogenized the gut communities and could have potentially masked all other types of signal, our experimental urban and rural diets induced specific and significant shifts in the α-diversity, the taxonomic and functional features as well as the overall composition of the microbiota.
Overall, the rural diet caused an increase in microbiota diversity whereas the urban diet led to reductions, with the strongest decrease impacting rural birds fed on an urban diet. These diet-induced diversity changes were likely related to differences in nutrient composition between experimental diets. Chemical analyses showed our urban diet to contain lower energy and protein content and, in particular, only half as much fibre than the rural diet [46]. Dietary fibres contain abundant complex carbohydrates (also known as MACs—microbiota-accessible carbohydrates) which serve as the primary source of carbon and energy for the distal gut microbiota and as a result play a crucial role in shaping gut communities [56]. Numerous studies in mammalian models, including humans, have shown that low-MAC diets (low fibre, high fat and processed food) lead to a drastic and sometimes long-term reduction in gut microbiota diversity ([56] and references therein). Such diets select for a small number of bacterial taxa that are favoured by low-MAC diets and outcompete the numerous taxa required to metabolize the complex polysaccharides contained in fibres. Since our urban diet contained both less fibre and more processed food, the same process is likely to be at play in our experiment.
The taxonomic shifts induced by our experimental diets were characterized by the increase, under the rural diet, in the relative abundance of two families of Firmicutes (Enterococcaceae and Staphylococcaceae), a phyla usually positively associated with the metabolism of dietary plant polysaccharids [57]. Quite remarkably, these two families happen to be the taxa which most significantly characterised the free-ranging rural birds prior to the experiment. Experiments in poultry showing an increase in Enterococcaceae [58] and Staphylococcaceae [59] with a wheat-dominated diet support the possibility that wheat content may be the main driver of such increases in our experiment (25% wheat in our rural diet versus 0% in the urban diet).
While the experiment itself (aviary conditions and/or artificial diet) was the main factor inducing shifts in most inferred metabolic function features, we observed an increase in inferred lipid metabolism and xenobiotic degradation potential in rural birds and this was more pronounced when they were fed on an urban diet. This suggests that the microbiota not only shifted taxonomically but also in functional potential, likely in response to the more fatty urban diet and possibly also due to the presence of pollutants in the urban diet [60]. This increase was not observed in urban birds, likely because these features were already high before the start of the experiment. Overall, post-experimental gut communities of sparrows fed on urban and rural diets resemble those of wild-caught urban and rural sparrow prior to experimental treatment, respectively. This finding provides evidence that the variation in gut microbiota we observe along urban gradients is indeed, to a certain extent, mediated by diet.
(c). Gut microbiota plasticity in response to diet change and effects on host body condition
Gut bacterial communities have been shown to be extremely plastic entities that undergo rapid changes in structure, diversity and relative abundances of different taxa in response to environmental changes [61]. In particular, there is evidence that the gut microbiota responds rapidly to short-term diet changes [62]. Moreover, it has been hypothesized that this microbiota plasticity could facilitate host acclimation in the face of rapid environmental variation [40]. In our data, a certain fraction of the microbiota remains stable despite the diet experiment: after six weeks of the experiment, bird origin still explains a significant part of the variation in α-diversity, taxonomic composition (unchanged levels of Lactobacillaceae in urban birds) and β-diversity (post-experiment origin effect in PERMANOVA), indicating that a part of the original urban versus rural enterotype is maintained, possibly due to long-term dietary effects [28], or genetic or early-life effects [63] for instance. Yet, another substantial fraction of the microbiota responded strongly to the diet change as illustrated by the fact that in all microbiota metrics (α-diversity, β-diversity, taxonomic and functional composition), changes were strongest when the experimental diet deviated most from the pre-experimental one. Interestingly, our results showed that rural sparrows fed on an urban diet underwent more important microbiota changes than urban ones fed on a rural diet. This pattern could simply be due to the possibility that, despite our efforts to mimic the natural diets as faithfully as possible, the experimental rural diet was less different to the natural urban diet than the contrary. An alternative explanation is that the rural gut microbiota is more plastic than the urban gut microbiota. Such higher plasticity would make sense in the light of the diversity differences between urban and rural microbiotas: the probability of hosting taxa adapted to the novel diet is higher when there is a larger pool of selectable taxa (and functions). Furthermore, the seasonal variation in diet in rural environments, which translated into seasonal variation in gut microbiota in rural but not in urban sparrows [13], could increase the probability of regularly acquiring new taxa, thus facilitating such plasticity in rural birds. In support of this consideration, it has been shown in mice that the negative effects of a low-fibre diet on the gut microbiota is largely reversible within one generation, but that a long-term and multigenerational low-fibre diet leads to an irreversible loss of diversity which cannot be restored by the reintroduction of a high-fibre diet [56], indicating a loss in plasticity. Likewise, it has been suggested in humans that the stable homogeneity of the industrialised diet (as opposed to the seasonal variation in hunter–gatherers) could have contributed to the extinction of seasonally volatile taxa within the gut [64].
When we investigated the microbiota-mediated effect of diet change on host condition, we found that urban birds gained weight on a rural diet when their initial gut diversity was high, whereas the rural diet induced a weight loss when the initial gut microbiota was low. In other words, urban birds only benefited from the positive impact of a rural diet (more fibre, energy and protein) when their gut microbiota was high at the start of the experiment, suggesting that the reduced taxonomic and functional diversity in urban birds (found in this study and [13]) prevented them from properly metabolizing the complex polysaccharides present in the rural diet. Long-term exposure to an urban diet could thus not only lead to a loss of diversity, but also a loss of plasticity of the gut microbiota, thereby reducing the capacity of urban hosts to benefit from novel (and potentially beneficial) diets. Further studies specifically testing this hypothesis, through experimental reductions in gut diversity for instance, as were recently examined in house sparrow nestlings [65] would be useful to validate this supposition.
In conclusion, we here provide experimental evidence that differential food sources available to sparrows in cities and rural areas likely contribute to the observed variation in diversity, taxonomic and functional composition of their gut microbiota. Our data further suggest that the loss of diversity and shifts in composition and inferred function induced by urban diets may negatively impact urban hosts through a reduction in gut microbiota plasticity, thus hampering the host's capacity to cope with change. A broader perspective brought by this study is that by providing urban dwellers access to processed, low-fibre food, the industrialization and Westernization of human diets could possibly induce collateral negative effects on other species found in urban environments.
Supplementary Material
Acknowledgements
We are grateful to H. Matheve and P. Vantieghem for field assistance and A. Chaine for providing access to the aviaries and logistical help. We also thank S. Manzi, A. Iribar-Pelozuelo, N. Parthuisot for laboratory assistance and L. Zinger, S. Leclaire and S. Jacob for their help with bioinformatics and statistical analyses. We are grateful to the genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing computing and storage resources.
Ethics
Animal welfare, maintenance, and experimental procedures followed French regulations and guidelines (DREAL permit no. 31-2014-09). Sparrows were caught under capture and ringing permit no. 15038 (French National Natural History Museum).
Data accessibility
All sequences and associated metadata have been deposited on the NCBI SRA: PRJNA596683. The nucleotide sequences have been made available through NCBI https://www.ncbi.nlm.nih.gov/.
Authors' contributions
J.W., L.L., L.d.N. and A.T. conceived and designed the study. A.T., N.S.H. and J.W. carried out the fieldwork and the aviary experiment. A.T. performed the laboratory work and bioinformatic analyses. A.T. and J.W. performed the statistical analyses. A.T., J.W. and L.L. led the writing and E.M. contributed substantially to the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This research was funded by the Interuniversity Attraction Poles Programme Phase VII (P07/4) initiated by the Belgian Science Policy Office and the French Laboratory of Excellence project ‘Tulip’ (ANR-10-LABX-41 and ANR-11-IDEX-0002-02).
References
- 1.Lewis SL, Maslin MA. 2015. Defining the anthropocene. Nature 519, 171–180. ( 10.1038/nature14258) [DOI] [PubMed] [Google Scholar]
- 2.Alberti M. 2015. Eco-evolutionary dynamics in an urbanizing planet. Trends Ecol. Evol. 30, 114–126. ( 10.1016/j.tree.2014.11.007) [DOI] [PubMed] [Google Scholar]
- 3.Calegaro-Marques C, Amato SB. 2014. Urbanization breaks up host-parasite interactions: a case study on parasite community ecology of Rufous-Bellied Thrushes (Turdus rufiventris) along a rural-urban gradient. PLoS ONE 9, e103144 ( 10.1371/journal.pone.0103144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amato KR, et al. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353. ( 10.1038/ismej.2013.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barelli C, et al. 2015. Habitat fragmentation is associated to gut microbiota diversity of an endangered primate: implications for conservation. Sci. Rep. 5, 14862 ( 10.1038/srep14862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alderete TL, Jones RB, Chen Z, Kim JS, Habre R, Lurmann F, Gilliland FD, Goran MI. 2018. Exposure to traffic-related air pollution and the composition of the gut microbiota in overweight and obese adolescents. Environ. Res. 161, 472–478. ( 10.1016/j.envres.2017.11.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Y, Wu S, Zeng Z, Fu Z. 2017. Effects of environmental pollutants on gut microbiota. Environ. Pollut. 222, 1–9. ( 10.1016/j.envpol.2016.11.045) [DOI] [PubMed] [Google Scholar]
- 8.Bestion E, Jacob S, Zinger L, Gesu LD, Richard M, White J, Cote J. 2017. Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat. Ecol. Evol. 1, 0161 ( 10.1038/s41559-017-0161) [DOI] [PubMed] [Google Scholar]
- 9.Merckx T, et al. 2018. Body-size shifts in aquatic and terrestrial urban communities. Nature 558, 113–116. ( 10.1038/s41586-018-0140-0) [DOI] [PubMed] [Google Scholar]
- 10.Amato KR. 2013. Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microbiome Sci. Med. 1, 10–29. ( 10.2478/micsm-2013-0002) [DOI] [Google Scholar]
- 11.Hird SM. 2017. Evolutionary biology needs wild microbiomes. Front. Microbiol. 8, 725 ( 10.3389/fmicb.2017.00725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips JN, Berlow M, Derryberry EP. 2018. The effects of landscape urbanization on the gut microbiome: an exploration into the gut of urban and rural white-crowned sparrows. Front. Ecol. Evol. 6, 148 ( 10.3389/fevo.2018.00148) [DOI] [Google Scholar]
- 13.Teyssier A, Rouffaer LO, Saleh Hudin N, Strubbe D, Matthysen E, Lens L, White J. 2018. Inside the guts of the city: urban-induced alterations of the gut microbiota in a wild passerine. Sci. Total Environ. 612(Supp. C), 1276–1286. ( 10.1016/j.scitotenv.2017.09.035) [DOI] [PubMed] [Google Scholar]
- 14.Sullam KE, Essinger SD, Lozupone CA, O'connor MP, Rosen GL, Knight R, Kilham S, Russell J. 2012. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363–3378. ( 10.1111/j.1365-294X.2012.05552.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bletz MC, et al. 2016. Amphibian gut microbiota shifts differentially in community structure but converges on habitat-specific predicted functions. Nat. Commun. 7, 13699 ( 10.1038/ncomms13699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klomp JE, Murphy MT, Smith SB, McKay JE, Ferrera I, Reysenbach AL. 2008. Cloacal microbial communities of female spotted towhees Pipilo maculatus: microgeographic variation and individual sources of variability. J. Avian Biol. 39, 530–538. ( 10.1111/j.0908-8857.2008.04333.x) [DOI] [Google Scholar]
- 17.Medeiros JD, Cantão ME, Cesar DE, Nicolás MF, Diniz CG, Silva VL, Vasconcelos ATR, Coelho C. 2016. Comparative metagenome of a stream impacted by the urbanization phenomenon. Braz. J. Microbiol. 47, 835–845. ( 10.1016/j.bjm.2016.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Marshall CW, Cheng M, Xu H, Li H, Yang X, Zheng T. 2017. Changes in land use driven by urbanization impact nitrogen cycling and the microbial community composition in soils. Sci. Rep. 7, 44049 ( 10.1038/srep44049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Veelen HPJ, Falcao Salles J, Tieleman BI. 2017. Multi-level comparisons of cloacal, skin, feather and nest-associated microbiota suggest considerable influence of horizontal acquisition on the microbiota assembly of sympatric woodlarks and skylarks. Microbiome 5, 156 ( 10.1186/s40168-017-0371-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonder MJ, et al. 2016. The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407 ( 10.1038/ng.3663) [DOI] [PubMed] [Google Scholar]
- 21.Hernández-Gómez O, Briggler JT, Williams RN. 2018. Influence of immunogenetics, sex and body condition on the cutaneous microbial communities of two giant salamanders. Mol. Ecol. 27, 1915–1929. ( 10.1111/mec.14500) [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. 2014. Host adaptive immunity alters gut microbiota. ISME J. 9, 770 ( 10.1038/ismej.2014.165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. 2018. Is host filtering the main driver of phylosymbiosis across the tree of life? Bik H, editor. mSystems 3, e00097-18 ( 10.1128/mSystems.00097-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailly J, et al. 2016. Negative impact of urban habitat on immunity in the great tit Parus major. Oecologia 182, 1053–1062. ( 10.1007/s00442-016-3730-2) [DOI] [PubMed] [Google Scholar]
- 25.Chávez-Zichinelli CA, MacGregor-Fors I, Rohana PT, Valdéz R, Romano MC, Schondube JE. 2010. Stress responses of the house sparrow (Passer domesticus) to different urban land uses. Landsc. Urban Plan. 98, 183–189. ( 10.1016/j.landurbplan.2010.08.001) [DOI] [Google Scholar]
- 26.Liker A, Papp Z, Bókony V, Lendvai ÁZ. 2008. Lean birds in the city: body size and condition of house sparrows along the urbanization gradient. J. Anim. Ecol. 77, 789–795. ( 10.1111/j.1365-2656.2008.01402.x) [DOI] [PubMed] [Google Scholar]
- 27.Perrier C, Lozano del Campo A, Szulkin M, Demeyrier V, Gregoire A, Charmantier A. 2018. Great tits and the city: distribution of genomic diversity and gene–environment associations along an urbanization gradient. Evol. Appl. 11, 593–613. ( 10.1111/eva.12580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu GD, et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. ( 10.1126/science.1208344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodawatta KH, Sam K, Jønsson KA, Poulsen M. 2018. Comparative analyses of the digestive tract microbiota of New Guinean passerine birds. Front. Microbiol. 9, 1830 ( 10.3389/fmicb.2018.01830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Knight R, Caporaso JG, Svanbäck R. 2014. Individuals' diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 17, 979–987. ( 10.1111/ele.12301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panek M, Budny M. 2017. Variation in the feeding pattern of red foxes in relation to changes in anthropogenic resource availability in a rural habitat of western Poland. Mamm. Biol. 82, 1–7. ( 10.1016/j.mambio.2016.09.002) [DOI] [Google Scholar]
- 32.Murray MH, Kidd AD, Curry SE, Hepinstall-Cymerman J, Yabsley MJ, Adams HC, Ellison T, Welch CN, Hernandez SM. 2018. From wetland specialist to hand-fed generalist: shifts in diet and condition with provisioning for a recently urbanized wading bird. Phil. Trans. R. Soc. B 373, 20170100 ( 10.1098/rstb.2017.0100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imhoff ML, Tucker CJ, Lawrence WT, Stutzer DC. 2000. The use of multisource satellite and geospatial data to study the effect of urbanization on primary productivity in the United States. IEEE Trans. Geosci. Remote Sens. 38, 2549–2556. ( 10.1109/36.843042) [DOI] [Google Scholar]
- 34.Murray MH, Hill J, Whyte P, St Clair CC. 2016. Urban compost attracts coyotes, contains toxins, and may promote disease in urban-adapted wildlife. Ecohealth 13, 285–292. ( 10.1007/s10393-016-1105-0) [DOI] [PubMed] [Google Scholar]
- 35.Plummer KE, Siriwardena GM, Conway GJ, Risely K, Toms MP. 2015. Is supplementary feeding in gardens a driver of evolutionary change in a migratory bird species? Glob. Change Biol. 21, 4353–4363. ( 10.1111/gcb.13070) [DOI] [PubMed] [Google Scholar]
- 36.El-Sabaawi R. 2018. Trophic structure in a rapidly urbanizing planet. Funct. Ecol. 32, 1718–1728. ( 10.1111/1365-2435.13114) [DOI] [Google Scholar]
- 37.Anderson TR. 2006. Biology of the ubiquitous house sparrow: from genes to populations. Oxford, UK: Oxford University Press. [Google Scholar]
- 38.Le Chatelier E, et al. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. ( 10.1038/nature12506) [DOI] [PubMed] [Google Scholar]
- 39.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. ( 10.1038/nri3535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP. 2016. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. ( 10.1016/j.tree.2016.06.008) [DOI] [PubMed] [Google Scholar]
- 41.Vangestel C, Braeckman BP, Matheve H, Lens L. 2010. Constraints on home range behaviour affect nutritional condition in urban house sparrows (Passer domesticus). Biol. J. Linn. Soc. 101, 41–50. ( 10.1111/j.1095-8312.2010.01493.x) [DOI] [Google Scholar]
- 42.Newbold LK, Oliver AE, Cuthbertson L, Walkington SE, Gweon HS, Heard MS, Van Der Gast CJ. 2015. Rearing and foraging affects bumblebee (Bombus terrestris) gut microbiota. Environ. Microbiol. Rep. 7, 634–641. ( 10.1111/1758-2229.12299) [DOI] [PubMed] [Google Scholar]
- 43.Lucas FS, Heeb P. 2005. Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caeruleus nestlings. J. Avian Biol. 36, 510–516. ( 10.1111/j.0908-8857.2005.03479.x) [DOI] [Google Scholar]
- 44.Ruiz-Rodriguez M, Lucas FS, Heeb P, Soler JJ. 2009. Differences in intestinal microbiota between avian brood parasites and their hosts. Biol. J. Linn. Soc. 96, 406–414. ( 10.1111/j.1095-8312.2008.01127.x) [DOI] [Google Scholar]
- 45.van Dongen WF, et al. 2013. Age-related differences in the cloacal microbiota of a wild bird species. BMC Ecol. 13, 11 ( 10.1186/1472-6785-13-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salleh HN, et al. 2016. Predictable food supplies induce plastic shifts in avian scaled body mass. Behav. Ecol. 27, 1833–1840. [Google Scholar]
- 47.Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E. 2016. Obitools: a unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. ( 10.1111/1755-0998.12428) [DOI] [PubMed] [Google Scholar]
- 48.Escudié F, et al. 2018. FROGS: find, rapidly, OTUs with galaxy solution. Bioinformatics 34, 1287–1294. ( 10.1093/bioinformatics/btx791) [DOI] [PubMed] [Google Scholar]
- 49.Langille MGI, et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. ( 10.1038/nbt.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oksanen J, et al. 2007. The vegan package. Community Ecol. Package 10, 631–637. [Google Scholar]
- 51.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 ( 10.1186/gb-2011-12-6-r60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bagan H, Yamagata Y. 2014. Land-cover change analysis in 50 global cities by using a combination of Landsat data and analysis of grid cells. Environ. Res. Lett. 9, 064015 ( 10.1088/1748-9326/9/6/064015) [DOI] [Google Scholar]
- 53.Knutie SA, Chaves JA, Gotanda KM. 2019. Human activity can influence the gut microbiota of Darwin's finches in the Galapagos Islands. Mol. Ecol. 28, 2441–2450. [DOI] [PubMed] [Google Scholar]
- 54.Waite DW, Taylor MW. 2014. Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front. Microbiol. 5, 223 ( 10.3389/fmicb.2014.00223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, Zheng S, Sharshov K, Cao J, Sun H, Yang F, Wang X, Li L. 2016. Distinctive gut microbial community structure in both the wild and farmed Swan goose (Anser cygnoides). J. Basic Microbiol. 56, 1299–1307. ( 10.1002/jobm.201600155) [DOI] [PubMed] [Google Scholar]
- 56.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. ( 10.1038/nature16504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73, 1073–1078. ( 10.1128/AEM.02340-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hübener K, Vahjen W, Simon O. 2002. Bacterial responses to different dietary cereal types and xylanase supplementation in the intestine of broiler chicken. Arch. Für Tierernaehrung. 56, 167–187. ( 10.1080/00039420214191) [DOI] [PubMed] [Google Scholar]
- 59.van der Hoeven-Hangoor E, van der Vossen JMBM, Schuren FHJ, Verstegen MWA, Oliveira JE, Montijn RC, Hendriks WH. 2013. Ileal microbiota composition of broilers fed various commercial diet compositions. Poult. Sci. 92, 2713–2723. ( 10.3382/ps.2013-03017) [DOI] [PubMed] [Google Scholar]
- 60.Defois C, Ratel J, Denis S, Batut B, Beugnot R, Peyretaillade E, Engel E, Peyret P. 2017. Environmental pollutant Benzo[a]Pyrene impacts the volatile metabolome and transcriptome of the human gut microbiota. Front. Microbiol. 8, 1562 ( 10.3389/fmicb.2017.01562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Candela M, Biagi E, Maccaferri S, Turroni S, Brigidi P. 2012. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol. 20, 385–391. ( 10.1016/j.tim.2012.05.003) [DOI] [PubMed] [Google Scholar]
- 62.David LA, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. ( 10.1038/nature12820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sbihi H, Boutin RC, Cutler C, Suen M, Finlay BB, Turvey SE. 2019. Thinking bigger: how early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy 74, 2103–2115. ( 10.1111/all.13812) [DOI] [PubMed] [Google Scholar]
- 64.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390. ( 10.1038/s41579-019-0191-8) [DOI] [PubMed] [Google Scholar]
- 65.Kohl KD, Brun A, Bordenstein SR, Caviedes-Vidal E, Karasov WH. 2018. Gut microbes limit growth in house sparrow nestlings (Passer domesticus) but not through limitations in digestive capacity. Integr. Zool. 13, 139–151. ( 10.1111/1749-4877.12289) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences and associated metadata have been deposited on the NCBI SRA: PRJNA596683. The nucleotide sequences have been made available through NCBI https://www.ncbi.nlm.nih.gov/.