Abstract
Phthalates are associated with multiple, adverse reproductive outcomes including increased risk of uterine leiomyoma (fibroids). Phthalates can interact with epigenetic modifications including microRNAs (miRNAs), which help regulate processes crucial to fibroid pathogenesis. However, no prior study has examined the influence of phthalates on miRNA expression in fibroid tumors. We conducted a preliminary, cross-sectional study to examine the associations between phthalate exposures and miRNA expression levels in fibroid tumors and to explore potential effect modification by race/ethnicity. We quantified expression levels of 754 miRNAs in fibroid tumor samples and analyzed spot urine samples for phthalate metabolites collected from 45 pre-menopausal women undergoing surgery for fibroid treatment at an academic hospital. Associations between miRNA levels in fibroids and phthalate biomarkers were evaluated using linear regression adjusting for age, race/ethnicity, and body mass index (BMI). Statistical tests were adjusted for multiple comparisons. We also performed in silico Ingenuity Pathway Analysis to identify the biological pathways that are regulated by phthalate-associated miRNAs. Mono-hydroxybutyl phthalate and mono(2-ethyl-5-hydroxyhexyl) phthalate were positively associated with miR-10a-5p (β = 0.76, 95% CI = [0.40, 1.11]) and miR-577 (β = 1.06, 95% CI = [0.53, 1.59]), respectively. A total of 8 phthalate-miRNA associations varied by race/ethnicity (qinteraction < 0.10). Pathway analysis revealed that mRNA gene targets of phthalate-associated miRNAs were significantly associated with multiple fibroid-related processes including angiogenesis, apoptosis, and proliferation of connective tissues. Collectively, these data suggest that exposures to some phthalates are associated with miRNA in fibroids, and that associations may vary by race/ethnicity. Validation of these findings may provide insight into mechanisms underlying associations between phthalates and fibroids and contribute to novel hypotheses regarding racial/ethnic disparities in fibroids.
Keywords: Endocrine-disrupting chemicals, epigenetics, gynecology, health disparities, non-coding RNAs, women’s health
Introduction
Uterine leiomyoma (fibroids)—smooth muscle tumors that develop during reproductive years in most women—are a devastating disorder for millions of women who experience symptoms (eg, heavy bleeding and pelvic pain) and/or reproductive dysfunction (eg, infertility).1,2 Fibroids are more common and more severe in U.S. Black women.3,4 Surgery remains the mainstay of fibroid treatment.5
Fibroid biology remains poorly understood, which has hampered progress in identifying risk factors, prognosis, and non-invasive treatments. While the exact mechanisms of fibroid pathobiology are unclear, several cellular activities have been implicated in fibroid formation and growth. Abnormal myometrial and fibroid stem cells exhibit an exaggerated response to estrogen and progesterone, stimulating processes such as cell proliferation, inhibition of apoptosis, and extracellular matrix (ECM) formation.6 As with other types of tumors, these processes may contribute to cellular transformation, fibroid tumor initiation and growth.6 For example, large quantities of ECM (secreted by fibroblasts) separate the smooth muscle cells, contributing to tumor expansion, solid-state signaling, and the characteristic “stiffness” of fibroid tissue.6,7 Growth factors (eg, fibroblast growth factor 2 [FGF2] and vascular endothelial growth factor [VEGF]) embedded within the ECM may also regulate fibroid formation.7,8 Additional studies suggest that fibroids exhibit both pro-angiogenic and anti-angiogenic properties, since fibroids have reduced blood flow and altered vasculature compared with normal myometrium.9,10
MicroRNAs (miRNAs) have the potential to accelerate prevention and treatment efforts since these epigenetic alterations help regulate mechanisms important to fibroid development. MicroRNAs are small (<22 nucleotides), non-coding RNA molecules that regulate post-transcriptional gene expression through gene silencing.11 Approximately 50 to 100 miRNAs are differently expressed in fibroids compared with myometrium in patient-matched, surgical specimens.12-15 Predicted gene targets of these miRNAs modulate processes that may play a significant role in fibroid pathobiology including cell proliferation, apoptosis, and ECM turnover16 In vitro studies have validated the functional role of several miRNAs that are dysregulated in vivo.17-19
MicroRNA regulation may also be involved in biological pathways linking environmental chemicals to fibroid pathogenesis. Our research group recently reported that higher exposure to certain phthalates, such as di(2-ethylhexyl) phthalate (DEHP), is associated with greater uterine volume, a measure of fibroid severity, among women with symptomatic fibroids.20 Phthalates are a class of multi-functional chemicals commonly used in consumer products such as personal care products, home furnishings, and food contact materials.21 Phthalates can migrate out of products and enter the human body through ingestion, inhalation, or dermal application. Once inside the body, phthalates are rapidly metabolized and excreted into urine.22 Urinary concentrations of phthalate metabolites are common biomarkers of exposure22 and are detected in nearly all reproductive-aged women.21,23 Certain phthalates are reproductive toxicants in animal models24,25 and are also associated with adverse reproductive outcomes in humans.26-28 While the relationship between phthalates and miRNA expression in fibroids is currently unknown, phthalate exposures are associated with miRNA expression in extracellular vesicles within follicular fluid29 and the placenta.30
The objective of our preliminary study is to examine the association between phthalate exposures and miRNA expression in fibroid tumors among women undergoing surgical treatment for fibroids. Our secondary objective is to examine whether associations between phthalate exposure and miRNA expression vary by race/ethnicity as prior studies suggest racial/ethnic differences in both phthalate exposures31,32 and fibroid miRNA expression.13,33
Methods
Study population
In 2014-2017, we recruited women into the Fibroids, Observational Research on Genes and the Environment (FORGE) study who were seeking care for symptomatic fibroids at the George Washington University (GW) Medical Faculty Associates and intending to undergo surgical management at GW Hospital, an urban academic hospital that serves the Washington, DC metropolitan area. Eligible women were non-pregnant, pre-menopausal, English-speaking, and ⩾18 years of age. The sample for our current study is limited to women with phthalate metabolite and miRNA data (N = 45). The GW Institutional Review Board approved the study. All participants provided written informed consent prior to enrollment.
Clinical and demographic data
We abstracted data from patients’ medical records on race/ethnicity, age, body mass index (BMI), last menstrual period, and insurance type.
Phthalate exposure assessment
Sample collection and analytical methods have been previously described20 and are briefly summarized below. We collected spot urine samples from participants in sterile polypropylene cups during a pre-operative appointment. For 1 participant, urine was collected after surgery. Each urine sample was analyzed for specific gravity (SG) using a handheld refractometer (Atago Co., Ltd) and then stored in polypropylene cryovials at −80°C. The Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) quantified 14 phthalate metabolites by online solid-phase extraction high-performance liquid chromatography isotope dilution tandem mass spectrometry.34 This analysis includes the 13 metabolites detected in >50% of the population. The names of the phthalate metabolites, their parent compounds, and detection frequencies are presented in Table 1. Common sources of parent phthalate compounds are provided in Supplemental Table S1. Biomarker concentrations were adjusted for urine dilution using the following formula: (phthalate biomarker concentration) × ([1.017 − 1] / [SG − 1]), where 1.017 is the median specific gravity in this study sample and SG is the specific gravity of the individual’s urine sample.35 Biomarker concentrations below the limit of detection (LOD) were replaced with the LOD divided by the square root of 2 prior to SG adjustment.36 In addition to examining individual metabolites, we also calculated 2 summary measures using previously described methods: the molar sum of DEHP metabolites (ΣDEHP)21 and a potency-weighted sum of anti-androgenic phthalate metabolites (ΣAA phthalates).31
Table 1.
Descriptive statistics of specific gravity–adjusted phthalate biomarker concentrations in urine (N = 45).
| Parent compound: phthalate metabolite | % >LOD | GM (GSD) (ng/mL) |
||
|---|---|---|---|---|
| Overall | Black (N = 28) |
White/Latina (N = 17) |
||
| Diethyl phthalate (DEP) | ||||
| Monoethyl phthalate (MEP) | 100 | 108.15 (6.86) | 205.53 (5.10)a | 37.56 (7.04)a |
| Di-n-butyl phthalate (DnBP) | ||||
| Mono-n-butyl phthalate (MnBP) | 96 | 8.65 (2.66) | 9.07 (3.04) | 8.00 (2.09) |
| Mono-hydroxybutyl phthalate (MHBP) | 73 | 1.06 (1.94) | 1.04 (2.00) | 1.08 (1.89) |
| Diisobutyl phthalate (DiBP) | ||||
| Monoisobutyl phthalate (MiBP) | 96 | 7.17 (2.26) | 8.22 (2.30) | 5.72 (2.12) |
| Mono-hydroxyisobutyl phthalate (MHiBP) | 89 | 2.50 (2.18) | 2.45 (2.24) | 2.59 (2.12) |
| Butylbenzyl phthalate (BBzP) | ||||
| Monobenzyl phthalate (MBzP) | 93 | 2.87 (3.13) | 3.42 (3.72) | 2.16 (2.07) |
| Di-n-octyl phthalate (DnOP) | ||||
| Mono(3-carboxypropyl) phthalate (MCPP) | 76 | 1.49 (3.35) | 1.29 (3.17) | 1.88 (3.64) |
| Diisononyl phthalate (DiNP) | ||||
| Monocarboxyoctyl phthalate (MCOP) | 98 | 13.13 (4.78) | 12.16 (4.87) | 14.89 (4.83) |
| Diisodecyl phthalate (DiDP) | ||||
| Monocarboxynonyl phthalate (MCNP) | 100 | 1.96 (2.83) | 2.16 (3.12) | 1.66 (2.37) |
| Di(2-ethylhexyl) phthalate (DEHP) | ||||
| Mono(2-ethylhexyl) phthalate (MEHP) | 62 | 1.78 (2.30) | 1.77 (2.15) | 1.80 (2.61) |
| Mono(2-ethyl-5-hydroxyhexyl) phthalate (MEHHP) | 98 | 6.39 (2.49) | 6.63 (2.58) | 6.01 (2.39) |
| Mono(2-ethyl-5-oxohexyl) phthalate (MEOHP) | 98 | 4.11 (2.65) | 4.30 (2.88) | 3.82 (2.32) |
| Mono(2-ethyl-5-carboxypentyl) phthalate (MECPP) | 100 | 10.46 (2.31) | 10.81 (2.45) | 9.91 (2.14) |
| ∑DEHP | – | 23.39 (2.29) | 24.38 (2.37) | 21.84 (2.22) |
| ∑Anti-androgenic (AA) phthalates | – | 45.88 (2.40) | 50.55 (2.46) | 39.11 (2.32) |
Abbreviations: GM, geometric mean; GSD, geometric standard deviation; LOD, limit of detection.
P < .05 from t-test by race/ethnicity.
Fibroid and myometrium tissue collection
Fibroid tissue was collected during hysterectomy or myomectomy procedures. For patients with multiple fibroids, we sampled the largest fibroid. Fibroid tissue samples were cut approximately 1 cm in thickness, and large fibroids were sampled 1 to 2 cm away from the peripheral zone. Samples were collected within 1 hour of surgery and snap frozen in liquid nitrogen until analysis. Pathology reports were reviewed to confirm that tumor samples were non-cancerous and actually represented leiomyomatous tissues. Among the subset of participants who underwent hysterectomy (N = 19), myometrium was also collected.
Expression analysis of miRNAs
The Laboratory of Precision Environmental Health at the Columbia University Mailman School of Public Health performed sample processing and RNA isolation. Snap-frozen tissue samples were incubated in RNAlater-ICE Frozen Tissue Transition Solution (Thermo Fisher) according to the manufacturer’s protocol. Tissues were homogenized using a TissueRuptor (Qiagen). Then, total RNA was isolated using miRCURY RNA Isolation Tissue Kit (Exiqon). RNA was quantified on a NanoPhotometer Pearl (Implen), and quality was verified with a Nano Assay run on a Bioanalyzer 2100 (Agilent Technologies).
All RNA samples were processed with RNA Clean & Concentrator Kits (Zymo Research). An aliquot of 100 ng of concentrated RNA from each sample was screened for 754 miRNAs and 4 internal controls (ath-miR159a, RNU48, RNU44, and U6) using TaqMan OpenArray (Thermo Fisher). QuantStudio 12K Flex is a fixed-content panel containing validated human TaqMan assays derived from Sanger miRbase release v.14. All 754 assays have been functionally validated with miRNA artificial templates. Samples were first reverse transcribed to cDNA using Megaplex RT Primers Pools A and B. The cDNA products were pre-amplified with Megaplex PreAmp Primers, Human Pools A and B, according to the manufacturer’s protocol. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed on the QuantStudio 12K Flex Real-Time PCR System (Life Technologies). Relative quantification cycle values (Cq) were defined as the amplification cycle at which the fluorescence levels for each of the miRNAs exceed the background fluorescence threshold.
Statistical analysis
We calculated descriptive statistics for demographic and environmental variables. SG-adjusted phthalate biomarkers were natural log-transformed prior to statistical analysis. Comparisons of phthalate biomarkers by race/ethnicity were performed using t-tests.
To extract miRNA data, we used Thermo Fisher Cloud Relative Quantification software. We performed quality control on raw qPCR data according to the following criteria, suggested by the manufacturer: (a) amplification scores ⩾ 1.1 and (b) Cq confidence ⩾ 0.8. Data that did not meet these criteria were set to missing. Cq values <40 were considered unexpressed. Further analyses were limited to 388 miRNAs detected in ⩾80% of fibroid samples. We used the global mean method to normalize miRNA expression data using the following formula37: ΔCq_miRNAi = Cq_miRNAi – Cq_miRNAi_global_mean. Non-detected values were coded as 40 in the calculation. Normalization was conducted separately for miRNAs in fibroid and myometrium.
We tested differences in miRNA expression between participant-matched samples of fibroid and myometrium using paired t-tests. This analysis was limited to 377 miRNAs that were detected in ⩾80% of both fibroid and myometrium samples. We calculated the fold change in fibroid compared with myometrium miRNA expression using the 2−ΔΔCt formula.38 We further compared differences in expression of 388 miRNAs in fibroids by race/ethnicity using t-tests.
Next, we evaluated associations between phthalate biomarkers and miRNA expression in fibroids with multivariable linear regression models in which we modeled phthalate exposures as log-transformed continuous variables. All models were adjusted a priori for age (continuous), BMI (continuous), and race/ethnicity (Black vs White/Latina). White and Latina women were collapsed into 1 category since there was only 1 Latina participant.
We conducted 2 additional sets of analyses on the 285 phthalate biomarkers and miRNA combinations with significant associations (P < .05) in adjusted models (described above). First, we tested these associations using miRNA expression in myometrium adjusting for age, race/ethnicity, and BMI. Next, we evaluated for potential effect modification of phthalate-miRNA associations by race/ethnicity. We conducted linear regression models of fibroid miRNA expression and phthalate biomarkers adjusted for age and BMI with an interaction term for phthalate biomarker and race/ethnicity as well as the main effects of phthalate biomarker and race/ethnicity.
To control the false discovery rate (FDR) in multiple testing, we applied the Benjamini-Hochberg procedure to the number of miRNAs tested within each of the individual phthalate metabolites.39 We did not apply FDR adjustment to the number of phthalate biomarkers because phthalate metabolites show moderate-to-high correlation with each other20 and the conventional Benjamini-Hochberg approach assumes that all hypotheses are independent.39 Data analysis was performed in R version 3.3.3 (R Foundation for Statistical Computing, Vienna, Austria). For multiple comparison adjustment, we chose a less conservative threshold of q-value < 0.10 due to our modest sample size. MicroRNA names reflect mature sequence names from miRbase release 22.
MicroRNA targets and biological network analysis
We used the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems) to identify miRNA targets and explore downstream biological networks. MicroRNAs significantly associated with phthalate biomarkers (FDR q < 0.10) were included. We used the miRNA Target Filter tool to link messengerRNA (mRNA) targets to each miRNA from TarBase, Target Scan, miRecords, and Ingenuity Expert Findings. We filtered the pathway and network analyses using the experimentally observed confidence level. Because IPA does not identify pathways specific to fibroids, we searched for pathways known a priori to be associated with fibroid pathogenesis.7,16 The statistical significance of biological disease and function pathways was calculated using the Fisher exact test with an alpha of 0.05.
Results
Most women were Black (62%), overweight or obese (76%), privately insured (64%), and undergoing a myomectomy (58%). Compared with White/Latina women, Black women were more likely to be obese, publicly insured, and having a hysterectomy. Fibroid characteristics did not differ by race/ethnicity (Table 2). Phthalate exposures were ubiquitous; 9 phthalate metabolites were detected in >90% of participants. Mono-ethyl phthalate (MEP) concentrations were significantly higher in Black women (Table 1).
Table 2.
Demographic and clinical characteristics of FORGE participants by race/ethnicity.
| Characteristics | Black/African American (n = 28) |
White or Latinaa (n = 17) |
Total (N = 45) |
|---|---|---|---|
| N (column percentage) or median (IQR) | |||
| Age (years) | 39 (33-46) | 40 (36-43) | 39 (36-46) |
| BMI (kg/m2) | 30.4 (26.6-40.7) | 24.8 (21.7-27.1) | 28.5 (25.1-35.5) |
| Private insurance | 17 (61%) | 12 (71%) | 29 (64%) |
| Surgery type | |||
| Myomectomy | 12 (43%) | 14 (82%) | 26 (58%) |
| Hysterectomy | 16 (57%) | 3 (18%) | 19 (42%) |
| No. of fibroids | 4 (2-6) | 3 (1-7) | 4 (2-6) |
| Size of largest fibroid | 7.3 (5.9-11.2) | 8.3 (3.5-11.0) | 7.7 (5.8-11.0) |
| Uterine volume | 664 (239-1228) | 606 (327-799) | 646 (243-1082) |
Abbreviations: FORGE, Fibroids, Observational Research on Genes and the Environment study; BMI, body mass index (kg/m2); IQR, interquartile range.
One participant self-identified as Latina.
In total, 35 miRNAs were under-expressed and 39 miRNAs were over-expressed in fibroid compared with myometrium (FDR q < 0.10) (Table 3). Expressions of the following miRNAs were ⩾3 fold greater in myometrium: miR-10a-5p, miR-10a-3p, miR-140-3p, miR-144-5p, miR-150-5p, miR-205-5p, miR-27a-5p, miR-29b-2-5p, miR-29c-5p, miR-451a, and miR-95-3p. Conversely, expressions of these miRNAs were ⩾3 fold greater in fibroid: miR-135a-5p, miR-135b-5p, miR-137-3p, miR-302b-3p, miR-335-3p, miR-34a-5p, miR-34a-3p, miR-34b-5p, miR-34c-5p, miR-483-5p, miR-488-3p, miR-488-5p, miR-508-3p, miR-577, miR-592, miR-651-5p, miR-885-5p, and miR-9-3p.
Table 3.
MiRNAs showing differential expression in fibroid versus myometrium (N = 19).
| Under-expressed |
Over-expressed |
||
|---|---|---|---|
| Target | Fold change | Target | Fold change |
| let-7b-5p | −1.9 | miR-1201 | 2.0 |
| miR-10a-5p | −3.7 | miR-130b-3p | 1.5 |
| miR-10a-3p | −3.4 | miR-135a-5p | 13.1 |
| miR-1247-5p | −2.8 | miR-135b-5p | 17.2 |
| miR-126-3p | −2.5 | miR-137-3p | 4.0 |
| miR-126-5p | −2.0 | miR-181a-2-3p | 2.1 |
| miR-139-3p | −1.7 | miR-184 | 2.3 |
| miR-139-5p | −2.9 | miR-301b-3p | 1.7 |
| miR-140-5p | −2.1 | miR-302b-3p | 4.7 |
| miR-140-3p | −3.0 | miR-323-3p | 2.0 |
| miR-144-5p | −3.4 | miR-335-5p | 2.6 |
| miR-149-5p | −1.9 | miR-335-3p | 3.9 |
| miR-150-5p | −3.4 | miR-337-3p | 1.8 |
| miR-185-5p | −1.5 | miR-34a-5p | 3.8 |
| miR-193a-5p | −2.8 | miR-34a-3p | 3.7 |
| miR-203a-3p | −2.2 | miR-34b-3p | 1.9 |
| miR-205-5p | −3.3 | miR-34b-5p | 3.0 |
| miR-22-3p | −1.7 | miR-34c-5p | 3.8 |
| miR-27a-5p | −3.0 | miR-369-5p | 1.9 |
| miR-29a-3p | −2.5 | miR-378a-5p | 1.8 |
| miR-29b-3p | −2.7 | miR-380-5p | 1.7 |
| miR-29b-2-5p | −3.3 | miR-381-3p | 2.0 |
| miR-29c-3p | −2.1 | miR-409-5p | 2.1 |
| miR-29c-5p | −3.2 | miR-411-3p | 1.6 |
| miR-30a-3p | −1.8 | miR-455-5p | 2.2 |
| miR-30e-3p | −1.7 | miR-483-5p | 3.5 |
| miR-324-3p | −1.6 | miR-488-3p | 3.2 |
| miR-338-3p | −2.4 | miR-488-5p | 3.6 |
| miR-451a | −3.7 | miR-499a-5p | 1.7 |
| miR-452-5p | −1.8 | miR-503-5p | 2.0 |
| miR-511-5p | −2.2 | miR-508-3p | 7.3 |
| miR-616-5p | −1.4 | miR-548c-5p | 1.7 |
| miR-628-3p | −2.6 | miR-551b-3p | 2.9 |
| miR-652-3p | −1.8 | miR-577 | 5.5 |
| miR-95-3p | −3.2 | miR-592 | 8.8 |
| miR-651-5p | 6.9 | ||
| miR-885-5p | 3.8 | ||
| miR-889-3p | 2.0 | ||
| miR-9-3p | 6.0 | ||
Abbreviation: miRNAs, microRNAs.
All differences are FDR q < 0.10 after multiple comparison adjustment. The negative inverse fold change values are shown for targets under-expressed in fibroid tissue (−1/fold change).
In adjusted models, there were 285 significant associations between phthalate biomarkers and miRNAs (P < .05) (Supplemental Table S2), and 34 of these associations were significant at P < .005 (Table 4). After adjusting for multiple testing, we found 2 miRNAs associated with phthalate biomarkers. A 1-unit increase in the natural-log concentrations of mono-hydroxybutyl phthalate (MHBP) was associated with a 0.76 (95% CI = [0.40, 1.11]) ΔCq increase in miR-10a-5p expression (FDR q = 0.05), where a higher ΔCq indicates lower relative expression. The association of mono(2-ethyl-5-hydroxyhexyl phthalate (MEHHP) and miR-577 (β= 1.06, 95% CI = [0.53, 1.59]) was marginally significant (FDR q = 0.10). Phthalate biomarkers associated with miRNA targets in fibroids were not associated with these targets in myometrium (no association had FDR q < 0.10; data not shown).
Table 4.
Association between phthalate biomarkers and miRNAs in fibroid tissue.a
| Phthalate | miRNA name | Estimateb | 95% CI | P-value | FDR q-valuec |
|---|---|---|---|---|---|
| MEP | miR-429 | −0.60 | [–0.93, –0.27] | .0006 | 0.24 |
| miR-200b-3p | −0.46 | [–0.77, –0.16] | .004 | 0.53 | |
| miR-548c-5p | −0.29 | [–0.47, –0.10] | .004 | 0.53 | |
| MnBP | miR-338-5p | 1.55 | [0.53, 2.58] | .004 | 0.73 |
| MHBP | miR-10a-5p | 0.76 | [0.40, 1.11] | .0001 | 0.05 |
| miR-30a-3p | 0.65 | [0.25, 1.06] | .002 | 0.40 | |
| miR-142-5p | 0.70 | [0.24, 1.15] | .004 | 0.40 | |
| miR-142-3p | 0.75 | [0.25, 1.25] | .004 | 0.40 | |
| MHiBP | miR-10a-5p | 0.58 | [0.23, 0.93] | .002 | 0.61 |
| MCPP | miR-154-5p | 1.13 | [0.53, 1.72] | .0004 | 0.17 |
| miR-7-5p | 0.54 | [0.19, 0.90] | .004 | 0.54 | |
| MCOP | miR-21-3p | −0.29 | [–0.47, –0.12] | .002 | 0.35 |
| miR-582-3p | −0.28 | [–0.45, –0.11] | .002 | 0.35 | |
| miR-449b-5p | 0.37 | [0.13, 0.62] | .004 | 0.46 | |
| MCNP | miR-590-3p | 0.70 | [0.27, 1.14] | .002 | 0.87 |
| MEHP | miR-635 | 0.70 | [0.27, 1.14] | .002 | 0.67 |
| MEHHP | miR-577 | 1.06 | [0.53, 1.59] | .0003 | 0.10 |
| miR-1179 | 0.88 | [0.36, 1.40] | .002 | 0.27 | |
| miR-7-2-3p | 0.61 | [0.25, 0.98] | .002 | 0.23 | |
| miR-192-3p | 0.48 | [0.18, 0.78] | .003 | 0.28 | |
| miR-7-5p | 0.65 | [0.22, 1.09] | .004 | 0.32 | |
| MEOHP | miR-1179 | 0.85 | [0.37, 1.33] | .0009 | 0.27 |
| miR-577 | 0.88 | [0.36, 1.40] | .002 | 0.27 | |
| miR-572 | −0.50 | [–0.81, –0.19] | .002 | 0.27 | |
| miR-26b-3p | 0.47 | [0.16, 0.77] | .004 | 0.29 | |
| miR-7-5p | 0.62 | [0.22, 1.03] | .004 | 0.29 | |
| MECPP | miR-577 | 1.01 | [0.41, 1.62] | .002 | 0.61 |
| miR-93-5p | 0.39 | [0.13, 0.65] | .004 | 0.62 | |
| ΣDEHP | miR-577 | 1.13 | [0.53, 1.72] | .0004 | 0.17 |
| miR-1179 | 0.92 | [0.33, 1.51] | .003 | 0.42 | |
| ΣAA phthalates | miR-7-5p | 0.80 | [0.31, 1.29] | .002 | 0.35 |
| miR-7-2-3p | 0.68 | [0.26, 1.10] | .003 | 0.35 | |
| miR-577 | 0.97 | [0.36, 1.59] | .003 | 0.35 | |
| miR-572 | −0.56 | [–0.93, –0.20] | .004 | 0.35 |
Abbreviations: AA, anti-androgenic; DEHP, di(2-ethylhexyl) phthalate; MCNP, monocarboxynonyl phthalate, MCOP, monocarboxyoctyl phthalate; MCPP, mono(3-carboxypropyl) phthalate; MECPP, mono(2-ethyl-5-carboxypentyl) phthalate; MEHP, mono(2-ethylhexyl) phthalate; MEHHP, mono(2-ethyl-5-hydroxyhexyl) phthalate; MEOHP, mono(2-ethyl-5-oxohexyl) phthalate; MEP, mono-ethyl phthalate; MHBP, mono-hydroxybutyl phthalate; miRNAs, microRNAs; MnBP, mono-n-butyl phthalate.
Association at P < .005 are shown. The full list of phthalate-miRNA associations significant at P < .05 are shown in Supplemental Table S2. All associations are adjusted for age, BMI, and race/ethnicity. Lower ΔCq indicates higher relative miRNA expression.
For every 1-unit increase in the natural-log phthalate metabolite concentration, there is an effect size increase/decrease in the miRNA ΔCq.
FDR q-values are adjusted for multiple testing for the 388 miRNAs but not for the number of phthalate biomarkers.
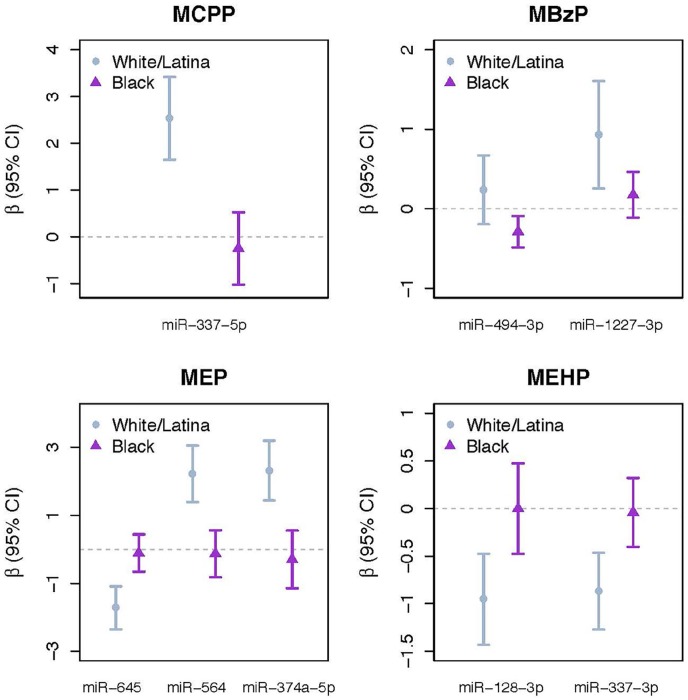
After multiple testing adjustment, 8 phthalate-miRNA associations varied significantly between White/Latina and Black women (interaction term FDR q < 0.10) (Figure 1). Among Black women, there was an association between monobenzyl phthalate (MBzP) and miR-494-3p. Among white/Latina women, there were associations between: mono(3-carboxypropyl phthalate) (MCPP) and miR-337-5p; MBzP and miR-1227-3p; MEP and miR-645; MEP and miR-564; MEP and miR-374-5p; mono(2-ethylhexyl) phthalate (MEHP) and miR-128-3p; and MEHP and miR-337-3p. There were no significant differences in miRNA expression by race/ethnicity after multiple testing adjustment (data not shown).
Figure 1.
Association between phthalate biomarkers and miRNAs in fibroid tissue with significant effect measure modification by race/ethnicity (FDR-adjusted q value < 0.10 for race/ethnicity × phthalate interaction term). Results are from models with an interaction term for phthalate biomarker and race/ethnicity, adjusted for age and BMI as well as including the main effects of phthalate biomarker (natural log-transformed, specific gravity-adjusted) and race/ethnicity. FDR q-values are adjusted for multiple testing for number of miRNAs but not for the number of phthalate biomarkers. Lower ΔCq signifies higher miRNA expression. BMI indicates body mass index; FDR, false discovery rate; miRNAs, microRNAs.
A total of 10 miRNAs that were significantly associated with phthalate biomarkers either in main analysis (miR-10a-5p, miR-577) or in certain racial/ethnic groups (miR-494-3p, miR-337-5p, miR-1227-3p, miR-645, miR-564, miR-374a-5p, miR-128-3p, miR-337-3p) were selected for downstream target prediction and functional enrichment analysis. We identified 923 mRNA targets that were experimentally observed or highly predicted targets of the 10 miRNAs. Inclusion of only experimentally observed mRNA targets yielded 26 mRNA targets for 3 miRNAs (miR-10a-5p, miR-128-3p, miR-494-3p). These mRNA targets were significantly associated with multiple fibroid-related processes including angiogenesis, apoptosis, differentiation of muscle cells, proliferation of connective tissues, and cell viability (Table 5, Supplemental Figure S1). They were also associated with relevant disease pathways such as tumorigenesis of the reproductive tract (P = 2.52 × 10−9), smooth muscle tumors (P = 4.70 × 10−5), and differentiation of muscle cells (P = 5.00 × 10−15).
Table 5.
miRNA-mRNA gene targets and significant biological disease and function pathways related to fibroid pathogenesis.
| Biological pathways | miRNA name | mRNA gene target | P-value |
|---|---|---|---|
| Functions | |||
| Cell viability | miR-128-3p, miR-494-3p, miR-10a-5p | BMI1, E2 F3, HMOX1, KIT, KLF4, NF1, NTRK3, PTEN, SNAP25, TGFBR1, TXNIP, WEE1 | 5.38E-06 |
| Cell survival | miR-128-3p, miR-494-3p, miR-10a-5p | ADORA2B, BMI1, E2 F3, HMOX1, KIT, KLF4, NF1, NTRK3, PTEN, SNAP25, TGFBR1, TXNIP, WEE1 | 1.46E-06 |
| Cell cycle progression | miR-128-3p, miR-494-3p, miR-10a-5p | ADORA2B, BMI1, DBI, E2 F3, FGF16, HMOX1, KIT, KLF4, KMT2A, NF1, NTRK3, PTEN, TGFBR1, WEE1 | 2.60E-10 |
| Apoptosis | miR-128-3p, miR-494-3p, miR-10a-5p | ADORA2B, AGO1, BMI1, E2 F3, HMOX1, HOXA1, KIT, KLF4, KMT2A, LDLR, NF1, NTRK3, PTEN, TGFBR1, TXNIP, VSNL1, WEE1 | 1.65E-06 |
| Angiogenesis | miR-128-3p, miR-494-3p, miR-10a-5p | ADORA2B, AGO1, HMOX1, HOXA1, HOXD10, KIT, KLF4, KMT2A, NF1, PTEN, TGFBR1 | 2.88E-07 |
| Cell transformation | miR-128-3p, miR-494-3p, miR-10a-5p | BMI1, KIT, KMT2A, NTRK3, PTEN, USF2 | 2.61E-05 |
| Proliferation of connective tissue cells | miR-128-3p, miR-494-3p, miR-10a-5p | BMI1, HMOX1, KLF4, KMT2A, NF1, NTRK3, PTEN, TXNIP | 5.35E-06 |
| Development of connective tissue cells | miR-128-3p, miR-494-3p, miR-10a-5p | KIT, KLF4, LDLR, TGFBR1, TXNIP | 4.06E-05 |
| Proliferation of fibroblast cell lines | miR-128-3p, miR-494-3p | BMI1, E2 F3, HMOX1, KIT, KMT2A, NTRK3, PTEN, TXNIP | 1.65E-07 |
| Proliferation of stem cells | miR-128-3p, miR-494-3p, miR-10a-5p | AGO1, BMI1, E2 F3, HOXA1, KIT, NF1, PTEN, TGFBR1 | 3.90E-10 |
| Disease pathways | |||
| Tumorigenesis of reproductive tract | miR-128-3p, miR-494-3p, miR-10a-5p | AFF1, DCX, E2 F3, FGF16, HOXA1, HOXD10, KIT, KLF4, KMT2A, LDLR, NF1, NTRK3, PTEN, RELN, SCN3A, TGFBR1, TXNIP, WEE1 | 2.52E-09 |
| Benign tumors | miR-128-3p, miR-494-3p, miR-10a-5p | AFF1, DCX, HMOX1, KIT, KLF4, NF1, PTEN, RELN, WEE1 | 1.90E-06 |
| Differentiation of muscle cells | miR-128-3p, miR-494-3p | LDLR, HMOX1 | 5.00E-15 |
| Smooth muscle tumor | miR-128-3p, miR-494-3p, miR-10a-5p | DCX, KIT, NF1, PTEN, WEE1 | 4.70E-05 |
| Proliferation of muscle cells | miR-128-3p, miR-494-3p, miR-10a-5p | E2 F3, FGF16, HMOX1, KIT, KLF4, NF1, PTEN | 2.38E-06 |
Abbreviations: ADORA2B, adenosine A2b receptor; AFF1, AF4/FMR2 family member 1; AGO1, argonaute RISC component 1; BMI1, BMI1 proto-oncogene, polycomb ring finger; DBI, diazepam binding inhibitor, acyl-CoA binding protein; DCX, doublecortin; E2 F3, E2 F transcription factor 3; FGF16, fibroblast growth factor 16; HOXA1, homeobox A1; HOXD10, homeobox D10; HMOX1, heme oxygenase 1; KIT, KIT proto-oncogene receptor tyrosine kinase; KMT2A, lysine methyltransferase 2A; KLF4, kruppel like factor 4; LDLR, low density lipoprotein receptor; NF1, neurofibromin 1; NTRK3, neurotrophic receptor tyrosine kinase 3; PTEN, phosphatase and tensin homolog; RELN, reelin; SCN3A, sodium voltage-gated channel alpha subunit 3; SNAP25, synaptosome associated protein 25; TGFBR1, transforming growth factor β receptor 1; TXNIP, thioredoxin interacting protein; USF2, upstream transcription factor 2, c-fos interaction; VSNL1, visinin like 1; WEE1, WEE1 G2 checkpoint kinase.
Discussion
In this cross-sectional study of pre-menopausal women seeking surgery for their symptomatic fibroids, we found that biomarkers of certain phthalates were associated with miRNA expression in fibroid tumors, but not in the myometrium. Furthermore, some of the associations between phthalate biomarkers and miRNAs in fibroids varied by race/ethnicity. These associations persisted even after adjustment for multiple comparisons.
Regulation of miRNAs varied between fibroid and myometrium. There were no associations between phthalate biomarkers and miRNAs in myometrium, and we found significant differences between patient-matched samples of fibroid and myometrium in expression of 74 miRNAs. Consistent with other studies, we observed that miR-10a, miR-150, miR-29b, miR-29c, and miR-451 are under-expressed and miR-34a is over-expressed in fibroids.12,13,15,33,40,41 (These names may not reflect current miRBase v22 names.) These reproducible associations have emerged across studies despite the use of different technologies for miRNA quantification. While the functional significance of aberrant miRNA expression is an area of ongoing research, several studies have demonstrated that downregulation of the miR-29 family contributes to ECM formation in fibroid cells.17,18
MiR-10a-5p expression was associated with concentrations of MHBP, an oxidative metabolite of DnBP (di-n-butyl phthalate), which is found in some personal care products (eg, nail polish) and certain medications (see Supplemental Table S1). Urinary concentrations of DnBP metabolites in this study population were generally similar to those measured from a national sample of U.S. women.42 DnBP is among the most potent anti-androgenic phthalates.25 While DnBP metabolites were not associated with fibroid severity in our prior study, we did observe a positive association between the weighted sum of anti-androgenic phthalates, which includes MHBP and other DnBP metabolites, and greater uterine volume.20
Our secondary analysis suggests that the influence of phthalate exposures on miRNA expression may vary by race/ethnicity. These results are not solely explained by biological differences between groups since we did not observe significant differences in miRNA expression by race/ethnicity. Our results are in contrast to 2 prior studies that found different miRNA profiles in the fibroids of Black and White women.13,33 These discrepancies may partly be explained by methodological differences as only one of the prior studies adjusted for multiple comparisons.13 Similarly, these findings are not entirely explained by racial/ethnic differences in phthalate exposures since we only observed meaningful differences by race/ethnicity for 1 phthalate metabolite. Our findings may indicate that the effect of phthalates on miRNA regulation may vary by the growth status of the fibroid. Peddada et al43 found that growth rates for fibroids were much higher for older (>35 years) Black women than for older (>35 years) White women, and most of our study population was >35 years of age. Alternatively, these findings may indicate that the epigenome is sensitive to interactions between chemical and non-chemical stressors. Race/ethnicity in the United States is predominately a social construct that influences a wide range of physical and social environmental exposures (eg, diet, socio-economic status, and psychosocial stress) due to residential segregation, racism, and cultural factors.32,44-46 Prior studies suggest that social adversity early in life can alter the biological response to environmental pollutants later in life.47,48 Future studies aimed at understanding racial/ethnic disparities in fibroid outcomes should consider the joint contribution of social and environmental exposures and their influence on the epigenome.
Our findings, in combination with prior experimental studies, begin to elucidate the mechanisms underlying phthalate toxicity in fibroids. We found that multiple miRNAs associated with phthalate exposures can interact with biological pathways important to fibroid pathogenesis. Moreover, several of the mRNA gene targets identified in our analysis (eg, PTEN, ADORA2B, KIT, E2 F3) are associated with proteins that were over-expressed in phthalate-treated fibroid cells.49-52 Kim and colleagues53,54 reported that exposing human fibroid cells to DEHP enhanced cell proliferation, blocked apoptosis, and induced protein expression of HIF-1α, COX-2, PCNA, and BLC2. It is plausible that the toxicity demonstrated in vitro is partly mediated through miRNA regulation. For example, PTEN is a validated target of miR-494-3p and a known tumor suppressor.55,56 Knockout of the PTEN gene can increase expression of COX-2,50 which regulates proliferation of smooth muscle cells in fibroids.57
While multiple prior studies have compared miRNA expression between fibroid and myometrium, to our knowledge, ours is the first to examine the associations between environmental chemical exposures and miRNA expression in human fibroid tissue. Our study is also novel in that we considered interactions between race/ethnicity and environmental chemical exposure. Another important strength is that we quantified the presence of a large number of miRNAs and corrected for multiple comparisons. Our study sample was diverse with respect to race/ethnicity, socio-economic status, and choice of surgical intervention, which may help account for the vast heterogeneity of this disorder.
As this is a preliminary study, there are important limitations. Our sample size was modest, which may lead to unstable or spurious results. Because all the study participants were undergoing surgery for symptomatic fibroids, these results may not be generalizable in women with asymptomatic fibroids. Our findings may be limited by residual confounding from other dietary or environmental factors. We only measured 1 tumor sample per participant so we could not examine within-individual variability in fibroid miRNA expression. We relied on a spot urine sample to estimate phthalate exposures, which may result in exposure misclassification because phthalates have a short half-life in the body.58 In addition, our study was cross-sectional so we cannot infer causality. Future studies should prospectively assess the contribution of environmental exposures to changes in miRNA expression and development of clinically relevant outcomes through large epidemiologic studies. Finally, because most in silico programs do not have biological pathways mapped specifically for fibroids, previous studies have relied on cancer biology models to predict the biological impact of miRNAs on fibroid pathogenesis.16 Our analysis was limited to the examination of several biological pathways that we defined a priori based on this literature. While it is plausible that cellular mechanisms relevant to malignant tumor growth also apply to growth of benign tumors such as fibroids, further research is needed to validate the predicted pathways for fibroid-associated miRNA-gene targets.
In conclusion, our study suggests that regulation of miRNAs in fibroids may be influenced by exogenous factors, such as environmental chemicals. Additional multidisciplinary translational research, such as ours, that integrates population, clinical, and basic sciences has the potential to advance understanding of the biological mechanisms underlying fibroid etiology. This knowledge may help stimulate new hypotheses about drivers of racial/ethnic disparities as well as inform prevention and treatment modalities for this complex disorder with a large public health impact.
Supplemental Material
Supplemental material, Supplemental_Materials_2020.01.27 for Phthalate Exposures and MicroRNA Expression in Uterine Fibroids: The FORGE Study by Ami R Zota, Ruth J Geller, Brianna N VanNoy, Cherie Q Marfori, Sana Tabbara, Lisa Y Hu, Andrea A Baccarelli and Gaby N Moawad in Epigenetics Insights
Acknowledgments
The authors acknowledge Dr James Robinson, Dr Jessica Opoku-Anane, and Dr Maria (Vicky) Vargas for their assistance with study design and recruitment of study participants. They thank Dr Charles Macri, who helped to initiate collaborations between the Medical Faculty Associates and Milken School of Public Health at the George Washington University. They acknowledge Susanna Mitro, Tyiesha Johnson, Angela Stoehr, Darah Wright, and Brenda Trejo who contributed to participant recruitment, data collection, and database management. They thank Dr Raja Mazumder, Dr Anelia Horvath, and staff at the McCormick Genomic and Proteomic Center for providing access to the Ingenuity Pathway Analysis software. They acknowledge Kasey Brennan for contributing to miRNA analysis. They also acknowledge Dr Antonia Calafat, Xiaoyun Ye, Manori Silva, and Tao Jia who contributed to the quantification of the phthalate metabolites.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Development (Grant No. R21HD096248), National Center for Advancing Translational Sciences (Grant Nos UL1TR001876 and KL2TR001877), The George Washington University Milken School of Public Health, and The George Washington University Office of the Vice President for Research (Cross-disciplinary Research Fund). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Dr Moawad is a speaker for Intuitive Surgical.
Author Contributions: ARZ conceived and designed the study, interpreted the data, and wrote the manuscript. RJG and BVN performed data analysis. AAB and LYH conducted miRNA analysis. CQM, AAB, ST, and GNM assisted in study design and interpretation of the results. All authors critically reviewed and approved the final manuscript.
ORCID iDs: Brianna N VanNoy  https://orcid.org/0000-0002-0059-6268
https://orcid.org/0000-0002-0059-6268
Cherie Q Marfori  https://orcid.org/0000-0002-2158-723X
https://orcid.org/0000-0002-2158-723X
Supplemental material: Supplemental material for this article is available online.
References
- 1. Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol. 2003;188:100-107. [DOI] [PubMed] [Google Scholar]
- 2. Walker CL, Stewart EA. Uterine fibroids: the elephant in the room. Science. 2005;308:1589-1592. [DOI] [PubMed] [Google Scholar]
- 3. Jacoby VL, Fujimoto VY, Giudice LC, Kuppermann M, Washington AE. Racial and ethnic disparities in benign gynecologic conditions and associated surgeries. Am J Obstet Gynecol. 2010;202:514-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kjerulff K, Langenberg P, Seidman J, Stolley P, Guzinski G. Uterine leiomyomas. Racial differences in severity, symptoms and age at diagnosis. J Reprod Med. 1996;41:483-490. [PubMed] [Google Scholar]
- 5. Stewart EA. Uterine fibroids. N Engl J Med. 2015;372:1646-1655. [DOI] [PubMed] [Google Scholar]
- 6. Bulun SE. Uterine fibroids. N Engl J Med. 2013;369:1344-1355. [DOI] [PubMed] [Google Scholar]
- 7. Stewart EA, Laughlin-Tommaso SK, Catherino WH, Lalitkumar S, Gupta D, Vollenhoven B. Uterine fibroids. Nat Rev Dis Primers. 2016;2:16043. [DOI] [PubMed] [Google Scholar]
- 8. Stewart EA, Nowak RA. Leiomyoma-related bleeding: a classic hypothesis updated for the molecular era. Hum Reprod Update. 1996;2:295-306. [DOI] [PubMed] [Google Scholar]
- 9. Fleischer R, Weston GC, Vollenhoven BJ, Rogers PAW. Pathophysiology of fibroid disease: angiogenesis and regulation of smooth muscle proliferation. Best Pract Res Clin Obstet Gynaecol. 2008;22:603-614. [DOI] [PubMed] [Google Scholar]
- 10. Luo X, Chegini N. The expression and potential regulatory function of microRNAs in the pathogenesis of leiomyoma. Semin Reprod Med. 2008;26:500-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [DOI] [PubMed] [Google Scholar]
- 12. Marsh EE, Lin Z, Yin P, Milad M, Chakravarti D, Bulun SE. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil Steril. 2008;89:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang TS, Zhang XM, Obijuru L, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336-347. [DOI] [PubMed] [Google Scholar]
- 14. Chuang TD, Panda H, Luo X, Chegini N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr Relat Cancer. 2012;19:541-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2008;12:227-240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Karmon AE, Cardozo ER, Rueda BR, Styer AK. MicroRNAs in the development and pathobiology of uterine leiomyomata: does evidence support future strategies for clinical intervention? Hum Reprod Update. 2014;20:670-687. [DOI] [PubMed] [Google Scholar]
- 17. Marsh EE, Steinberg ML, Parker JB, Wu J, Chakravarti D, Bulun SE. Decreased expression of microRNA-29 family in leiomyoma contributes to increased major fibrillar collagen production. Fertil Steril. 2016;106:766-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qiang W, Liu Z, Serna VA, et al. Down-regulation of miR-29b is essential for pathogenesis of uterine leiomyoma. Endocrinology. 2014;155:663-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng Y, Laser J, Shi G, et al. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663-673. [DOI] [PubMed] [Google Scholar]
- 20. Zota AR, Geller RJ, Calafat AM, Marfori CQ, Baccarelli AA, Moawad GN. Phthalates exposure and uterine fibroid burden among women undergoing surgical treatment for fibroids: a preliminary study. Fertil Steril. 2019;111:112-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zota AR, Calafat AM, Woodruff TJ. Temporal trends in phthalate exposures: findings from the National Health and Nutrition Examination Survey, 2001-2010. Environ Health Perspect. 2014;122:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wittassek M, Angerer J. Phthalates: metabolism and exposure. Int J Androl. 2008;31:131-138. [DOI] [PubMed] [Google Scholar]
- 23. Woodruff TJ, Zota AR, Schwartz JM. Environmental chemicals in pregnant women in the United States: NHANES 2003-2004. Environ Health Perspect. 2011;119:878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lovekamp-Swan T, Davis BJ. Mechanisms of phthalate ester toxicity in the female reproductive system. Environ Health Perspect. 2003;111:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Howdeshell KL, Rider CV, Wilson VS, Gray LE., Jr. Mechanisms of action of phthalate esters, individually and in combination, to induce abnormal reproductive development in male laboratory rats. Environ Res. 2008;108:168-176. [DOI] [PubMed] [Google Scholar]
- 26. Fu Z, Zhao F, Chen K, et al. Association between urinary phthalate metabolites and risk of breast cancer and uterine leiomyoma. Reprod Toxicol. 2017;74:134-142. [DOI] [PubMed] [Google Scholar]
- 27. Hauser R, Gaskins AJ, Souter I, et al. Urinary phthalate metabolite concentrations and reproductive outcomes among women undergoing in vitro fertilization: results from the EARTH study. Environ Health Perspect. 2016;124:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Messerlian C, Souter I, Gaskins AJ, et al. Urinary phthalate metabolites and ovarian reserve among women seeking infertility care. Hum Reprod. 2016;31:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez RM, Hauser R, Liang L, et al. Urinary concentrations of phenols and phthalate metabolites reflect extracellular vesicle microRNA expression in follicular fluid. Environ Int. 2019;123:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LaRocca J, Binder AM, McElrath TF, Michels KB. First-trimester urine concentrations of phthalate metabolites and phenols and placenta miRNA expression in a cohort of US women. Environ Health Perspect. 2016;124:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varshavsky JR, Zota AR, Woodruff TJ. A novel method for calculating potency-weighted cumulative phthalates exposure with implications for identifying racial/ethnic disparities among U.S. reproductive-aged women in NHANES 2001-2012. Environ Sci Technol. 2016;50:10616-10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zota AR, Shamasunder B. The environmental injustice of beauty: framing chemical exposures from beauty products as a health disparities concern. Am J Obstet Gynecol. 2017;217:418.e1-418.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Z, Guo H, Wu J, et al. Differential expression of miRNAs in uterine leiomyoma and adjacent myometrium of different races. Am J Clin Exp Obstet Gynecol. 2015;2:45-56. [Google Scholar]
- 34. Silva MJ, Samandar E, Preau JL, Jr, Reidy JA, Needham LL, Calafat AM. Quantification of 22 phthalate metabolites in human urine. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860:106-112. [DOI] [PubMed] [Google Scholar]
- 35. Boeniger MF, Lowry LK, Rosenberg J. Interpretation of urine results used to assess chemical exposure with emphasis on creatinine adjustments: a review. Am Ind Hyg Assoc J. 1993;54:615-627. [DOI] [PubMed] [Google Scholar]
- 36. Hornung RW, Reed LD. Estimation of average concentration in the presence of nondetectable values. Appl Occup Environ Hyg. 1990;5:46-51. [Google Scholar]
- 37. Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402-408. [DOI] [PubMed] [Google Scholar]
- 39. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B (Methodological). 1995;57:289-300. [Google Scholar]
- 40. Danielson LS, Menendez S, Attolini CS, et al. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am J Pathol. 2010;177:908-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Georgieva B, Milev I, Minkov I, Dimitrova I, Bradford AP, Baev V. Characterization of the uterine leiomyoma microRNAome by deep sequencing. Genomics. 2012;99:275-281. [DOI] [PubMed] [Google Scholar]
- 42. Centers for Disease Control and Prevention (CDC), National Center for Health Statistics (NCHS). National Health and Nutrition Examination Survey Data, 2013-2014. Hyattsville, MD: U.S. Department of Health and Human Services; 2016. [Google Scholar]
- 43. Peddada SD, Laughlin SK, Miner K, et al. Growth of uterine leiomyomata among premenopausal black and white women. Proc Natl Acad Sci USA. 2008;105:19887-19892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zota AR, Phillips CA, Mitro SD. Recent fast food consumption and bisphenol A and phthalates exposures among the U.S. population in NHANES, 2003-2010. Environ Health Perspect. 2016;124:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morello-Frosch R, Zuk M, Jerrett M, Shamasunder B, Kyle AD. Understanding the cumulative impacts of inequalities in environmental health: implications for policy. Health Aff (Millwood). 2011;30:879-887. [DOI] [PubMed] [Google Scholar]
- 46. Rehkopf DH, Needham BL. The impact of race and ethnicity in the social epigenomic regulation of disease. Nutritional Epigenomics. 2019;14:51-65. [Google Scholar]
- 47. Brunst KJ, Sanchez-Guerra M, Chiu Y-HM, et al. Prenatal particulate matter exposure and mitochondrial dysfunction at the maternal-fetal interface: effect modification by maternal lifetime trauma and child sex. Environ Int. 2018;112:49-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hazlehurst M, Nurius P, Hajat A. Individual and neighborhood stressors, air pollution and cardiovascular disease. Int J Environ Res Public Health. 2018;15:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ning C, Wen J, Zhang Y, et al. Excess adenosine A2B receptor signaling contributes to priapism through HIF-1α mediated reduction of PDE5 gene expression. FASEB J. 2014;28:2725-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Daikoku T, Hirota Y, Tranguch S, et al. Conditional loss of uterine PTEN unfailingly and rapidly induces endometrial cancer in mice. Cancer Res. 2008;68: 5619-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leone G, DeGregori J, Yan Z, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiang X, Zhou J, Yuen NK, et al. Imatinib targeting of KIT-mutant oncoprotein in melanoma. Clin Cancer Res. 2008;14:7726-7732. [DOI] [PubMed] [Google Scholar]
- 53. Kim JH. Analysis of the in vitro effects of di-(2-ethylhexyl) phthalate exposure on human uterine leiomyoma cells. Exp Ther Med. 2018;15:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim JH, Kim SH, Oh YS, et al. In vitro effects of phthalate esters in human myometrial and leiomyoma cells and increased urinary level of phthalate metabolite in women with uterine leiomyoma. Fertil Steril. 2017;107:1061-1069. [DOI] [PubMed] [Google Scholar]
- 55. Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283. [DOI] [PubMed] [Google Scholar]
- 56. Liu K, Liu S, Zhang W, et al. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 2015;34:1003-1010. [DOI] [PubMed] [Google Scholar]
- 57. Ke X, Dou F, Cheng Z, et al. High expression of cyclooxygenase-2 in uterine fibroids and its correlation with cell proliferation. Eur J Obstet Gynecol Reprod Biol. 2013;168:199-203. [DOI] [PubMed] [Google Scholar]
- 58. Perrier F, Giorgis-Allemand L, Slama R, Philippat C. Within-subject pooling of biological samples to reduce exposure misclassification in biomarker-based studies. Epidemiology. 2016;27:378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Materials_2020.01.27 for Phthalate Exposures and MicroRNA Expression in Uterine Fibroids: The FORGE Study by Ami R Zota, Ruth J Geller, Brianna N VanNoy, Cherie Q Marfori, Sana Tabbara, Lisa Y Hu, Andrea A Baccarelli and Gaby N Moawad in Epigenetics Insights