Abstract
Patients with acute myeloid leukaemia who relapse following therapy have few treatment options and face poor outcomes. Immune checkpoint inhibition, for example, by antibody-mediated programmed death-1 (PD-1) blockade, is a potent therapeutic modality that improves treatment outcomes in acute myeloid leukaemia. Here, we show that systemically delivered blood platelets decorated with anti-PD-1 antibodies (aPD-1) and conjugated to haematopoietic stem cells (HSCs) suppress the growth and recurrence of leukaemia in mice. Following intravenous injection into mice bearing leukaemia cells, the HSC–platelet–aPD-1 conjugate migrated to the bone marrow and locally released aPD-1, significantly enhancing anti-leukaemia immune responses, and increasing the number of active T cells, production of cytokines and chemokines, and survival time of the mice. This cellular conjugate also promoted resistance to re-challenge with leukaemia cells. Taking advantage of the homing capability of HSCs and in situ activation of platelets for the enhanced delivery of a checkpoint inhibitor, this cellular combination-mediated drug delivery strategy can significantly augment the therapeutic efficacy of checkpoint blockade.
The traditional treatment for acute myeloid leukaemia (AML)—a clonal malignancy comprising an increase in myeloblasts in the bone marrow1–3—includes anthracycline and cytarabine-based chemotherapy regimens4,5. However, the efficacy of traditional chemotherapy for AML is far from satisfactory, as most patients who achieve complete remission will ultimately relapse due to the incomplete elimination of leukaemia cells6–9. The prognosis of patients with relapsed leukaemia is dismal10–12. Although relapsed leukaemia could be potentially cured by haematopoietic stem cell (HSC) transplantation, the cost of such transplantation is often associated with high mortality induced by infections or graftversus-host disease13,14. The emerging technologies of engineering T cells provide a new approach to treat AML15. T cells from patients themselves could be removed from the circulation and genetically modified to express an artificial T-cell receptor (designated as a chimeric antigen receptor) in vitro that is designed to specifically recognize the tumour-associated antigens16–18. Chimeric antigen receptor-modified T cells enable the redirection of T-cell specificity and achieve impressive treatment outcomes against blood cancers in the clinic19,20,21. However, alleviation of the side effects, such as cytokine storm and B-cell aplasia, remains clinically challenging15,20. The development of new treatment approaches that can effectively eliminate leukaemia cells and avoid side effects is therefore highly desirable to enhance the therapeutic efficacy and prognosis of patients with AML.
Programmed death-1 (PD-1) is an immune inhibitory co-receptor expressed on a variety of immune cells such as T cells, B cells and natural killer cells22. When bound by its ligands, PD-L1 and PD-L2, PD-1 functions by inhibiting an activated T-cell response23,24. Tumour cells upregulate PD-L1 in response to inflammation, thereby suppressing an anti-tumour immune response25. Blockade of PD-1 using monoclonal anti-PD-1 antibodies (aPD-1) inhibits tumour-mediated immune suppression and has been demonstrated to improve outcomes in a variety of cancers26. Preclinical studies suggest that blocking the PD-1 pathway may improve outcomes in AML27–29. Thus, the use of aPD-1 represents a promising strategy in the therapeutic armamentarium for AML. Here, we describe a HSC–platelet cellular combination delivery system that can facilitate transport of aPD-1 to the bone marrow and subsequent release of aPD-1 by in situ platelet activation (Fig. 1a). The construction of HSC–platelet assembly is mediated by conjugation of platelets with the HSC plasma membrane through a click reaction (Supplementary Fig. 1). The immune checkpoint inhibitor aPD-1 is covalently decorated on the surface of platelets. Furthermore, the release of aPD-1 can be promoted through the potential generation of platelet-derived microparticles (PMPs) after activation of platelets30, which further enhances the binding of aPD-1 to T cells. After intravenous injection, we have demonstrated that HSC–platelet–aPD-1 assembly (designated as S–P–aPD-1) could effectively accumulate in the bone marrow, where the residual leukaemia cells locate after traditional treatment31. Using C1498 and WEHI-3 leukaemia-bearing mice as AML models, we found that S–P–aPD-1 could significantly inhibit leukaemia growth by inducing a potent immune response through the activation of T cells and generation of multiple cytokines and chemokines. Furthermore, such an immune response is durable as it can induce resistance to re-challenging leukaemia cells.
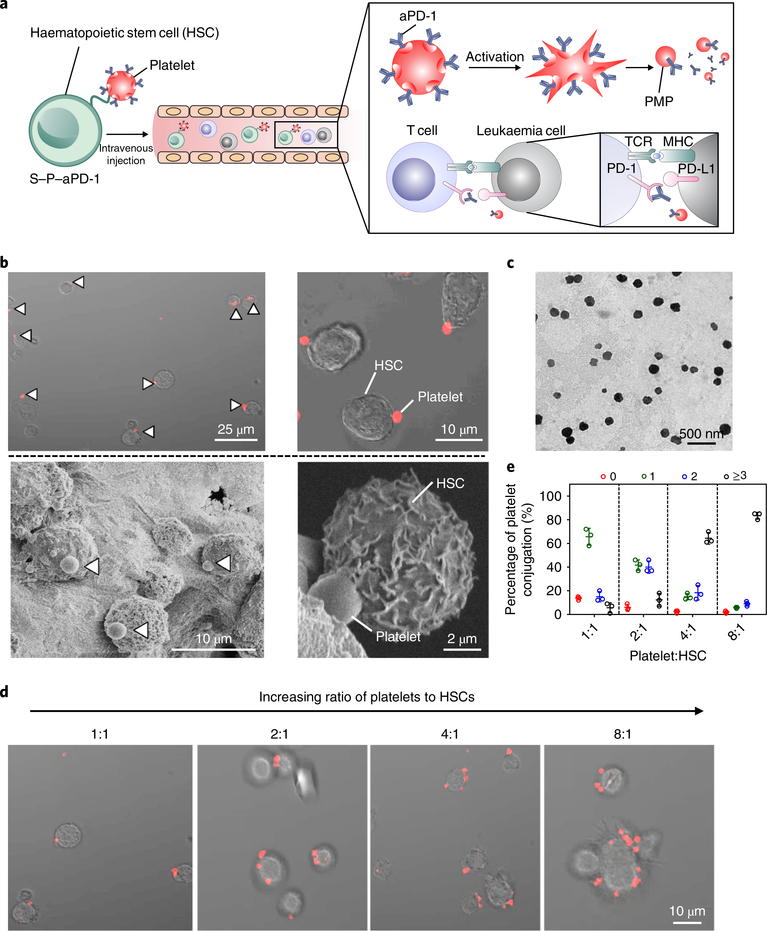
Fig. 1 |. Characterization of the S–P–aPD-1 cellular combination delivery system.
a, Schematic of HSC–platelet assembly-assisted aPD-1 delivery. After intravenous delivery, the S–P–aPD-1 could home to the bone marrow and the platelets could be locally activated and release aPD-1 to bind T cells for an enhanced immune response. MHC, major histocompatibility complex; TCR, T-cell receptor. b, Confocal microscopy (top) and SEM characterization (bottom) of S–P–aPD-1 conjugates. The platelets were labelled with rhodamine B for confocal observation. White arrows indicate the presence of platelets. c, Transmission electron microscopy (TEM) characterization of PMPs from S–P–aPD-1 after activation by 0.5 U ml−1 thrombin. d, Fluorescence imaging of S–P–aPD-1 with different ratios of platelets to HSCs. e, Quantitative analysis of conjugated platelets on HSCs (the number of bound platelets per cell is indicated as 0, 1, 2, 3.). The quantification was based on counting HSCs (numbers = 200) under the confocal microscope. The experiments were repeated three times. Data are presented as means ± s.d.
Engineering platelets decorated with aPD-1
Platelets were collected from the whole mouse blood and treated with prostaglandin E1 (PGE1) to inhibit platelet activation32. aPD-1 was conjugated to the platelets using sulfosuccinimidyl-4-(N-maleimidomethyl)-cyclohexane-1-carboxylate (Sulfo-SMCC) as a linker using a covalent conjugation method (Supplementary Fig. 2). Both flow cytometry analysis and confocal images confirmed the successful decoration of aPD-1 on the platelets (Supplementary Figs. 3 and 4). Furthermore, we quantified the conjugation amount of aPD-1 on the platelets using an enzyme-linked immunosorbent assay (ELISA), which showed that the maximum conjugation amount of aPD-1 that could be achieved was ~0.3 pg per platelet (Supplementary Fig. 5a). Such coupling showed negligible cytotoxicity and did not induce the lysis of platelets, which was confirmed by the integrity of platelets after 24 h (Supplementary Fig. 5b). In addition, we characterized the functionality of platelets via collagen binding and platelet aggregation. As shown in Supplementary Fig. 6, P–aPD-1 effectively bound to collagen, with insignificant difference compared with native platelets. Both platelets and P–aPD-1 aggregated after activation, indicating that aPD-1 conjugation did not alter platelet functionality. Furthermore, we validated that after activation, a remarkable amount of aPD-1 was released potentially due to the generation of PMPs, which was significantly higher than that of the non-activated platelets, indicating the well-preserved biofunctionality of platelets after decoration of aPD-1 (Supplementary Fig. 5c)33.
Integration of HSCs with platelets
HSCs were isolated from the femur and tibia of C57BL/6J mice (Supplementary Fig. 7) and cultured in 40 μM N-azidoacetylgalactosamine-tetraacylated (Ac4GalNAz) containing medium for 72 h. Ac4GalNAz has been identified to label many cells through N-acetylgalactosamine metabolism and incorporate into mucin-type O-linked glycoproteins34,35. The cell viability study confirmed insignificant cytotoxicity of Ac4GalNAz at the studied concentration (Supplementary Fig. 8). The presence of azide groups on the surface of HSCs was determined by the addition of an alkynyl-based probe, fluorescein (FAM) alkyne35. The flow cytometry analysis showed increased fluorescence signal on Ac4GalNAz-treated HSCs when reacted with FAM alkyne through a click reaction, which was significantly higher than that of the HSC control (Supplementary Fig. 9). Furthermore, the Ac4GalNAz-treated HSCs displayed obvious fluorescence under confocal observation, while negligible fluorescence signal was found on the HSC control (Supplementary Fig. 10).
Next, we conjugated the platelets with dibenzocyclooctyne-PEG4-N-hydroxysuccinimidyl ester (DBCO-PEG4-NHS ester)36, which reacts with the amine groups on the platelet surface. The decoration of DBCO-PEG4-NHS ester was determined by the covalent attachment of an azide-based fluorescence probe, azide-fluor 488. The flow cytometry analysis showed brighter fluorescence signals in DBCO-PEG4-NHS ester-treated platelets than in non-treated platelets and platelets physically mixed with azide probe (Supplementary Fig.11).
To decorate the HSCs with platelets through the click reaction, DBCO-PEG4-NHS ester-treated platelets were subjected to the azide-incorporated HSCs and incubated for 45 min. To avoid the aggregation of platelet–HSC assembly, excessive azide-PEG was added to block the free DBCO groups on the platelets37. As observed from the confocal and scanning electron microscopy (SEM) imaging, a direct conjugation at the cell–cell interface was verified and the cellular decoration was tuned at a 1:1 ratio (platelets:HSCs) (Fig. 1b). Additionally, both HSC and platelet morphologies were well maintained after conjugation. The platelet function was preserved, as evidenced by preservation of several key proteins and generation of PMPs after activation (Supplementary Fig. 12 and Fig. 1c). To further investigate the effect of numbers of conjugated platelets on the surface of HSCs, we increased the reaction ratio of platelets to HSCs and quantified the numbers of platelets bound to the HSCs. At a ratio of 1:1 (platelets:HSCs), the majority of assemblies were one platelet on one HSC (about 66%). By increasing platelet numbers, the conjugated platelets on the surface of HSCs were increased correspondingly. At a ratio of 8:1, over 80% of the HSCs were bound with more than three platelets (Fig. 1d,e). However, the HSC viability decreased along with the increased numbers of conjugated platelets (Supplementary Fig. 13). We therefore selected a reaction ratio of 1:1 (platelets:HSCs) for the following studies.
In vivo treatment efficacy of S–P–aPD-1
Retention of residual leukaemia cells in the bone marrow is one of the main reasons for AML relapse38. Thus, selective accumulation of anti-leukaemia drugs in the bone marrow is crucial to enhance the therapeutic index39,40. To investigate the bone marrow homing capability of S–P–aPD-1, we first evaluated the in vivo pharmacokinetics of S–P–aPD-1. As shown in Fig. 2a, S–P–aPD-1 displayed a significantly longer half-life than free aPD-1, which could be ascribed to the longer persistence in the circulation of HSCs with platelets. We further demonstrated that aPD-1 and Cy5.5-aPD-1 showed an insignificant difference in clearance in vivo (Supplementary Fig. 14). We then tested the bone marrow homing capability of S–P–aPD-1. aPD-1 was labelled with Cy5.5 and conjugated to HSC, platelets and HSC–platelet, respectively. Free aPD-1, HSC–aPD-1 (designated as S–aPD-1), platelet–aPD-1 (designated as P–aPD-1), HSC + platelet–aPD-1 mixture (designated as S + P–aPD-1) and S–P–aPD-1 were intravenously injected into C57BL/6J mice at the equivalent aPD-1 dose and the leg bones were taken out for imaging after 6 h. Both S–aPD-1 and S–P–aPD-1 showed higher fluorescence signals in the bone marrow than P–aPD-1, S + P–aPD-1 mixture and free aPD-1 groups, which suggested superior bone marrow accumulation capability of HSCs (Fig. 2b). The quantitative results showed over 25-fold greater fluorescence signal from bone tissue in S–aPD-1- and S–P–aPD-1-treated mice than the other groups (Fig. 2c). In addition to the bone marrow, the S–P–aPD-1 conjugates could also accumulate in the liver, spleen and lungs (Supplementary Fig. 15). Moreover, the simple blend of HSC and P–aPD-1 did not enhance the accumulation of aPD-1 in the bone marrow. The bone marrow accumulation of HSC and HSC–platelet assembly was further confirmed by fluorescence imaging (Supplementary Fig. 16a–c), and demonstrated the preservation integrity of the membranes of both HSC and platelets localized within the bone marrow. Furthermore, the fluorescence signals of PMPs were found in the bone marrow treated with HSC–platelets, whereas insignificant PMPs were observed in the platelet and PMPs groups (Supplementary Figs. 16d and 17). The existence of PMPs suggested the in situ potential generation of PMPs in the bone marrow after treatment with HSC–platelets, which might be triggered by the leukaemia microenvironment in the bone marrow41–44.
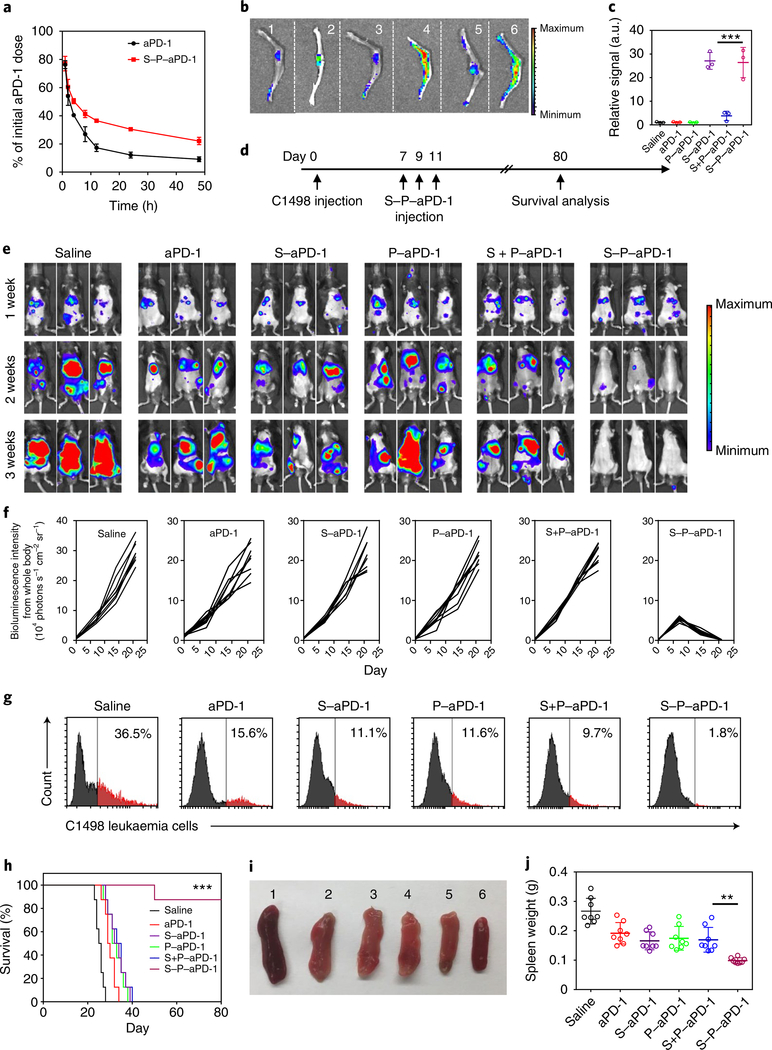
Fig. 2 |. In vivo treatment efficacy of S–P–aPD-1.
a, In vivo pharmacokinetics of free aPD-1 and S–P–aPD-1 at the aPD-1 dose of 1 mg per kg (n = 3). Data are presented as means ± s.d. b, Fluorescence images of bone tissues from mice treated with saline (1), Cy5.5-labelled free aPD-1 (2), P–aPD-1 (3), S–aPD-1 (4), S + P–aPD-1 (5) and S–P–aPD-1 (6). The experiments were repeated three times. c, Region-of-interest analysis of fluorescent intensities from bone tissues. Data are presented as means ± s.d. (n = 3). ***P < 0.0001, one-way ANOVA, followed by Tukey’s HSD post hoc test. d, Schematic of building a C1498 leukaemia model and treatment plan. e, Bioluminescence images of mice treated with saline, free aPD-1, S–aPD-1, P–aPD-1, S + P–aPD-1 and S–P–aPD-1 (HSCs/platelets: 5 × 107 cells in 100 μl PBS; aPD-1: 0.5 mg per kg). The experiments were repeated three times. f, Region-of-interest analysis of bioluminescence intensities from whole mouse bodies. g, Flow cytometry analysis of the number of C1498 cells in peripheral blood. The experiments were repeated three times. h, Survival curves for treated and control mice (n = 8). Statistical significance was calculated by log-rank test (***P < 0.0001). i, Morphologies of spleens from mice receiving different treatments (saline (1); free aPD-1 (2); S–aPD-1 (3); P–aPD-1 (4); S + P–aPD-1 (5); and S–P–aPD-1 (6)). The experiments were repeated three times. j, Weights of spleens after the various treatments. Data are presented as means ± s.d. (n = 8). **P = 0.0037, one-way ANOVA, followed by Tukey’s HSD post hoc test.
To investigate the treatment efficacy of S–P–aPD-1 against AML, the C1498 cell line was intravenously injected into C57BL/6J mice45. The surface expression of PD-L1 on C1498 cells after administration was confirmed by flow cytometry (Supplementary Fig. 18). The C1498 leukaemia-bearing mice were then treated with three doses of saline, HSCs, platelets, S–P–Rat-immunoglobulin G (IgG), free aPD-1, S–aPD-1, P–aPD-1, S + P–aPD-1 or S–P–aPD-1 every other day after one week at the aPD-1 dose of 0.5 mg per kg (Fig. 2d). In addition, another group of mice were treated with daily administration of aPD-1 for 6 d (aPD-1 dose: 0.25 mg per kg). The development of leukaemia was monitored by the bioluminescence of C1498 cells in vivo. As shown in Fig. 2e,f, the mice treated with S–P–aPD-1 displayed decreased bioluminescence signals after two weeks, and the bioluminescence signals could barely be detected after three weeks. Furthermore, seven of the eight mice in this group displayed a strong immune response with insignificant detectable leukaemia cell signals. In contrast, the mice treated with aPD-1 and daily administration of aPD-1 did not show significant response. The modest treatment efficacy of the P–aPD-1 groups could be attributed to the lack of bone marrow homing capability of the platelets. Furthermore, the lack of immune responses in the S–aPD-1 group could be ascribed to the inefficient activation of T cells due to the steric hindrance of cell–cell interaction46, internalization of aPD-1 by HSCs (Supplementary Fig. 19) and/or difficulty of aPD-1 release. In contrast, S–P–aPD-1 effectively accumulated in the bone marrow and released aPD-1 to unleash leukaemia-specific T cells. The survival rate was about 87.5% after 80 d in mice receiving S–P–aPD-1. However, no mice survived beyond 40 d for all other treatments with aPD-1, or beyond 30 d for the saline, HSC and platelet control groups (Fig. 2h and Supplementary Fig. 20c). We also analysed the presence of C1498 cells in the peripheral blood using flow cytometry. As shown in Fig. 2g and Supplementary Figs. 20b and 21, mice receiving the S–P–aPD-1 treatment displayed lower numbers of C1498 cells compared with the other treatment groups with aPD-1, as well as the saline, HSC and platelets control groups. To further demonstrate that the enhanced immune response observed in mice treated with S–P–aPD-1 was not mainly due to the long circulation of aPD-1 and not secondary to an immune reaction against the rat-derived aPD-1 antibody, S–P–Rat-IgG and daily aPD-1 treatments were also tested. As shown in Supplementary Fig. 20a–c, the S–P–Rat-IgG group and treatment groups receiving a daily administration of aPD-1 clearly did not have inhibited leukaemia growth.
Furthermore, the spleens of the mice receiving different treatments were removed and imaged (Fig. 2i and Supplementary Fig. 20d). The spleen from S–P–aPD-1-treated mice displayed a normal morphology, whereas the spleens from other treatment groups were enlarged47,48. The quantitative results showed that the spleens from the mice treated with S–P–aPD-1 were ½ ~ 1/3 the weight of other spleens (Fig. 2j and Supplementary Fig. 20e). We then used haematoxylin and eosin staining to investigate the development of leukaemia in the main organs. As shown in Supplementary Fig. 22, leukaemia cells were found in the bone marrow, liver, spleen and lung tissues in mice treated with saline. In contrast, the mice treated with S–P–aPD-1 displayed negligible numbers of leukaemia cells in the main organs.
T-cell-mediated immune response
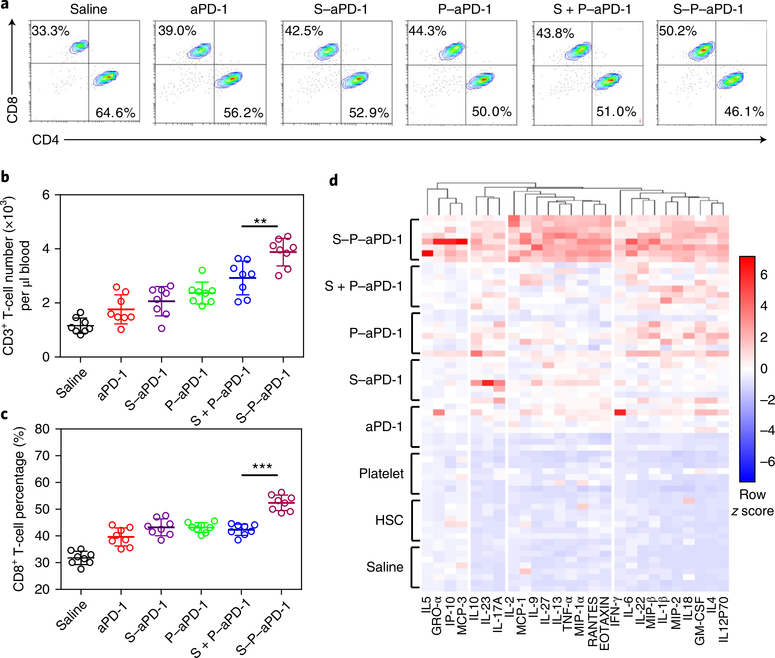
To understand the cellular mechanisms underlying the observed therapeutic effects of S–P–aPD-1, upon treatment, the T cells in the peripheral blood were collected and analysed by flow cytometry. An approximately fourfold increase in the total number of CD3+ T cells was observed in mice receiving the S–P–aPD-1 treatment compared with the saline, HSC and platelet control groups (Fig. 3b and Supplementary Fig. 23). CD3+ T cells in the S–P–aPD-1 treatment group showed a 1.9 ~ 2.4-fold increase compared with the other treatment groups with aPD-1 (Fig. 3b). Moreover, mice receiving S–P–aPD-1 treatment displayed a 1.5-fold increase in CD8+ T cells compared with the saline, HSC and platelet control groups, and an approximately 1.3-fold increase compared with the other treatment groups with aPD-1 (Fig. 3a,c and Supplementary Fig. 23). Furthermore, IFNγ+ CD8+ T cells in the S–P–aPD-1 treatment group increased by 1.9 ~ 2.6-fold compared with the other treatment groups with aPD-1, and 6.4 ~ 17.8-fold compared with the saline, HSC and platelet control groups (Supplementary Fig. 24). Increases in CD3+, CD8+ and IFNγ+ CD8+ T cells were suggestive of boosted T-cell immune responses in the S–P–aPD-1-treated mice. We further analysed T-cell subsets in the bone marrow. PD-1-expressing T cells were detected in the bone marrow of leukaemia-bearing mice by flow cytometry (Supplementary Fig. 25). Mice receiving the S–P–aPD-1 treatment displayed 1.8 ~1.9-fold increases in CD8+ T cells compared with the saline, HSC and platelet control groups, and approximately 1.3-fold increases compared with the other treatment groups with aPD-1 (Supplementary Fig. 26). Moreover, the CD8+ T cells in the bone marrow of mice after treatment with S–P–aPD-1 were about 44% Granzyme B positive (Supplementary Fig. 27), which was significantly higher than the other treatment groups, suggesting that effector T-cell number was increased after S–P–aPD-1 treatment. Besides this, the number of effector T cells exhibiting early activation status in the S–P–aPD-1 treatment group was higher than in the other treatment groups, as evidenced by higher percentages of CD8+CD44+CD69+ (Supplementary Fig. 28) and CD8+CD44+CD25+ (Supplementary Fig. 29) T cells. IFNγ+ CD8+ T cells in the S–P–aPD-1 treatment group showed 2.6 ~ 2.9-fold increases compared with the other aPD-1 treatment groups, and 5.3 ~ 21.2-fold increases compared with the saline, HSC and platelet control groups (Supplementary Fig. 30). In contrast, CD8+ and IFNγ+ CD8+ T-cell subsets in the bone marrow of non-leukaemia-bearing mice did not show significant increases after treatment with S–P–aPD-1 (Supplementary Fig. 31). Luminex-based quantification of cytokines and chemokines49 revealed four clusters of co-regulated proteins, with pro-inflammatory factors increased in the peripheral blood after treatment with aPD-1 (Fig. 3d). Furthermore, the majority of cytokines and chemokines were upregulated in the S–P–aPD-1 group compared with the other treatment groups with aPD-1. The increased serum cytokine levels might reflect the alteration of other immune cell subsets, including monocytes and myeloid cells, as AML is characterized by prevention of maturation of monocytes and myeloid cells3,50. The development of leukaemia could potentially cause abnormalities of monocytes and other myeloid cells, including granulocytes in AML patients, as shown by previous studies51,52.
Fig. 3 |. Analysis of t cells, cytokines and chemokines.
a, Flow cytometry analysis of CD8+ T cells (gated on CD3+ T cells) in peripheral blood. The experiments were repeated three times. b, Quantitative analysis of the number of CD3+ T cells. Data are presented as means ± s.d. (n = 8). ** P = 0.0048, one-way ANOVA, followed by Tukey’s HSD post hoc test. c, Quantitative analysis of the percentage of CD8+ T cells. Data are presented as means ± s.d. (n = 8). ***P < 0.0001, one-way ANOVA, followed by Tukey’s HSD post hoc test. d, Luminex-based quantification of cytokines and chemokines (n = 8).
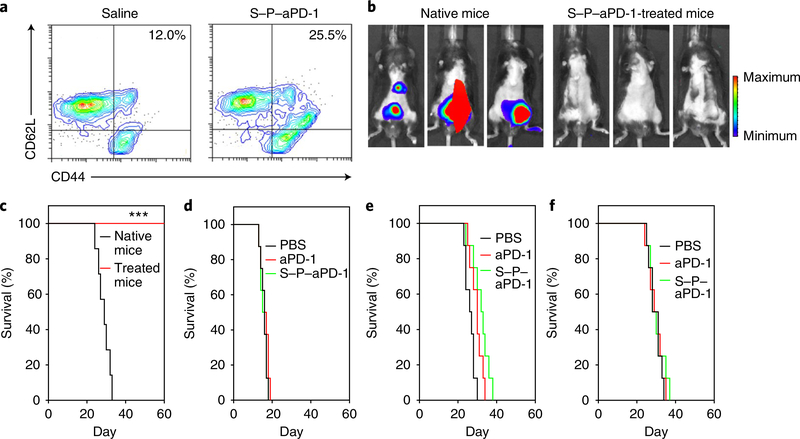
S–P–aPD-1 treatment caused a shift of native CD8+ T cells to the active phenotype and to the central memory CD8+ T-cell phenotype in vivo. The CD44hiCD62Lhi central memory T-cell subset showed a 2.1-fold increase in the S–P–aPD-1-treated mice compared with the saline control (Fig. 4a). Similarly, the CD44hiCD122hi memory T-cell subset also showed an increase (Supplementary Fig. 32). The functionality of the memory subsets in S–P–aPD-1-treated mice was shown in re-challenged experiments. Mice showing control of leukaemia growth after treatment with S–P–aPD-1 were re-challenged with 1 × 106 C1498 cells at day 80. Leukaemia grew rapidly in native mice, while S–P–aPD-1-treated mice remained leukaemia free at day 60 (Fig. 4b,c). To further confirm the key role of the T-cell-mediated immune response in S–P–aPD-1 treatment, T-cell-knockout mice (rag−/−) were injected with C1498 cells and then administered with S–P–aPD-1. As shown in Fig. 4d, control mice, as well as mice treated with free aPD-1 and S–P–aPD-1 showed equal leukaemia growth. In addition, CD8 T-cell depletion experiments in mice bearing C1498 leukaemia showed that the therapeutic effects of S–P–aPD-1 were abrogated in the absence of CD8+ T cells (Supplementary Fig. 33 and Fig. 4e). We also investigated the treatment efficacy of S–P–aPD-1 in PD-1 knockout mice (PD−/−). A significantly diminished anti-leukaemia effect was found in PD−/− mice (Fig. 4f), which was associated with partial exhaustion of CD8+ T cells, as evidenced by the increase of TIM-3+ and LAG-3+ T cells (Supplementary Fig. 34).
Fig. 4 |. S–P–aPD-1 induced a durable immune response.
a, Flow cytometry analysis of CD44hiCD62Lhi T cells (gated on CD8+ T cells) in saline- and S–P–aPD-1-treated mice (n = 3). b, Bioluminescence images of native and treated mice re-challenged with 1 × 106 C1498 cells at 3 weeks. c, Survival curves for native and treated mice after re-challenge with C1498 cells (n = 7). ***P < 0.001, log-rank test. d–f, Survival curves for rag−/− mice (d), CD8-depleted mice (e) and PD−/− mice (f) treated with PBS, free aPD-1 and S–P–aPD-1 at an aPD-1 dose of 0.5 mg per kg (n = 8).
To assess the effectiveness of S–P–aPD-1 in treating another type of leukaemia, we used the WEHI-3 myelomonocytic leukaemia cell line in BALB/cJ mice. As illustrated in Supplementary Fig. 35, in this leukaemia model, mice receiving S–P–aPD-1 therapy showed better leukaemia control, and 62.5% of the treated mice were alive at day 50. In contrast, mice in the control groups succumbed by day 40 and displayed larger spleens (Supplementary Fig. 35).
In summary, our study describes a ‘cell-combination’ strategy for drug delivery in which one cell type drives homing to the leukaemia site and the other delivers the drug. This combined approach allows the achievement of effective immune responses and inhibition of leukaemia growth. We obtained these effects by taking advantage of HSCs’ homing capability and in situ activation of platelets. S–P–aPD-1 promotes the delivery of aPD-1 in the bone marrow where the leukaemia is localized, and effectively unleashes leukaemia-specific T cells that control leukaemia growth and recurrence. S–P–aPD-1 seems to induce a potent immune response while mitigating toxicities. Furthermore, such a cell-assembly-mediated drug-delivery approach could be adopted to incorporate other bioparticulates to treat other diseases for which spatiotemporal drug delivery is essential53–55. However, whether the platelet activation of the S–P–aPD-1 could cause potential side effects in patients due to the release of pro-inflammatory molecules or systemic T-cell activation during the circulation of S–P–aPD-1 in the blood stream remains to be studied.
Methods
Cell lines and cells
The murine leukaemia cell line—C1498 tagged with luciferase and DsRed fluorescence—was kindly provided by B. Blazar (University of Minnesota). The WEHI-3 cells were purchased from the University of North Carolina tissue culture facility. The C1498 and WEHI-3 cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco; Invitrogen) supplemented with 10% foetal bovine serum (Invitrogen), 100 U ml−1 penicillin (Invitrogen) and 100 U ml−1 streptomycin (Invitrogen). The HSCs and progenitor cells (designated as HSCs in all the studies) were isolated from the femur and tibia of C57BL/6J mice. Bone marrow was first pre-enriched with a lineage depletion kit (Miltenyi). The resulting cells were subsequently incubated and selected with anti-Sca-1 microbeads (Miltenyi) to obtain lin-Sca-1+ HSCs with over 90% purity. Cells were cultured in Serum-Free Expansion Medium (SFEM; STEMCELL Technologies) with the addition of human interleukin-6 (50 ng ml−1; Thermo Fisher Scientific), human Flt3 ligand (100 ng ml−1; Thermo Fisher Scientific), murine stem cell factors (50 ng ml−1; Thermo Fisher Scientific) and low-density lipoprotein (40 μg ml−1; Thermo Fisher Scientific). Cells were cultured in an incubator (Thermo Fisher Scientific) at 37 °C under an atmosphere of 5% CO2 and 90% relative humidity. The cells were sub-cultivated approximately every 2–3 d at 80% confluence at a split ratio of 1:3.
Antibody
The aPD-1 was obtained from BioLegend (catalogue number 114114, clone: RMP1–14). The antibodies used for immunostaining were specific for CD3 (BioLegend; catalogue number 100236, clone: 17A2), CD4 (BD Biosciences; catalogue number 553046, clone: RM4–5), CD8a (BioLegend; catalogue number 100708, clone: 53–6.7), IFN-γ (BioLegend; catalogue number 505806, clone: XMG1.2), CD122 (BioLegend; catalogue number 123207, clone: TM-β1), CD41 (BioLegend; catalogue number 133904, clone: MWReg30), CD9 (BioLegend; catalogue number 124807, clone: MZ3), CD61 (BioLegend; catalogue number 104307, clone: 2C9.G2 (HMβ3–1)), CD62P (BioLegend; catalogue number 148305, clone: RMP-1), CD36 (BioLegend; catalogue number 102605, clone: HM36), CD154 (BioLegend; catalogue number 106505, clone: MR1), CD62L (BioLegend; catalogue number 104405, clone: MEL-14), CD44 (BioLegend; catalogue number 103024, clone: IM7), CD34 (BioLegend; catalogue number 128609, clone: HM34), CD38 (BD Biosciences; catalogue number 558813, clone: 90/CD38), CD117 (BioLegend; catalogue number 105812, clone: 2B8), CD366 (BD Biosciences; catalogue number 566346, clone: 5D12/TIM-3), CD223 (BioLegend; catalogue number 125207, clone: C9B7W), CD274 (BioLegend; catalogue number 124311, clone: 10F.9G2), Granzyme B (BioLegend; catalogue number 372204, clone: QA16A02), CD25 (BioLegend; catalogue number 101904, clone: 3C7) and CD69 (BioLegend; catalogue number 104508, clone: H1.2F3). The cells after staining were subjected to fluorescence-activated cell-sorting analysis following the manufacturers’ instructions. Multicolour flow cytometry was used with appropriate compensation. All antibodies were used following the manufacturers’ instructions. The fluorochromes conjugated on the antibody were exactly matched to the same fluorochrome channel. After staining, cells were analysed on a fluorescence-activated cell-sorting Calibur instrument (CytoFLEX; BD Biosciences), using the FlowJo or CytExpert software packages. CD8+ T-cell depletion antibody (clone: YTS169.4) was purchased from Bio X Cell. The rat IgG was purchased from Invitrogen. Recombined mPD-1 was obtained from R&D Systems. The secondary antibodies used for immunostaining were goat anti-rat IgG (H+L) (Thermo Fisher Scientific; catalogue number A18866).
Mice
C57BL/6J mice, T-cell knockout mice (B6.129S7-Rag1tm-Mom/J), PD-1 knockout mice (B6.Cg-Pdcd1tm1.1Shr/J) and Blab/c mice were purchased from the Jackson laboratory. Six- to eight-week-old male animals were used throughout all the experiments. All the animal studies strictly followed the animal protocol approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill and North Carolina State University. The number of mice used for each experiment was determined by G*Power analysis software.
Preparation of aPD-1-conjugated platelets
Murine platelets were isolated from whole mouse blood. Briefly, whole blood was collected from the C57BL/6J mice (non-terminal collection from the orbital sinus) with dipotassium ethylenediaminetetraacetic acid-treated tubes. The platelet-rich plasma (PRP) was then collected by centrifuging the whole blood at 100g for 20 min at room temperature. Thereafter, PGE1 was added to PRP at a final concentration of 1 μM, and PRP was centrifuged for another 20 min at 100g to further get rid of red blood cells. To isolate the platelets, the PRP was centrifuged at 800g for 20 min. Next, the pellet was collected and resuspended in phosphate buffered saline (PBS) containing 1 μM PGE1. Some 20–25 ml of blood was collected from the mice and ~10 × 108 platelets were isolated each time. Platelets were resuspended in 1 ml PBS with 1 μM PGE1 at the concentration of 1 × 108 platelets per 100 μl PBS. For the in vitro activation of platelets, the platelet solution was centrifuged at 800g for 20 min and suspended in PBS buffer for the following activation experiments. The number of platelets was counted using a haemocytometer under a microscope.
To decorate aPD-1 on the surface of platelets through an SMCC linker (Sulfo-SMCC; Pierce), the free thiol groups on the surface of the platelets after incubation with Traut’s reagent were first examined using flow cytometry (1 × 104 events were collected for analysis)33,56. Briefly, 1 × 106 platelets were mixed with 0.1 mg ml−1 maleimeide fluorescence probe (Mal-FITC; Sigma–Aldrich) in PBS and centrifuged at 800g for 10 min after 15 min reaction. The platelets were then washed by PBS, centrifuged at 800g for 20 min and subjected to flow cytometry. Red blood cells that have been reported to have minimal free thiol groups were used as the control group here. Next, aPD-1 was reacted with Sulfo-SMCC at a molar ratio of 1:1.2 for 2 h at 4 °C. The mixture was then centrifuged in an ultrafiltration tube (molecular weight cut-off = 3 kDa) to discard the excess SMCC linker. Thereafter, SMCC-aPD-1 (0.4 pg per platelet) was added to platelets and maintained at room temperature for 1 h to obtain P–aPD-1. The excess antibodies were removed by centrifugation at 800g for 20 min. The pellet was washed and stored in PBS containing 1 μM PGE1 at room temperature before use in the experiments. To further evaluate the aPD-1 conjugation efficiency, various volumes of aPD-1 were reacted with platelets, as described above, and the resulting P–aPD-1 were centrifuged at 800g, washed with PBS and lysed using ultrasonication in 0.1% Triton buffer. The amount of aPD-1 conjugated to the platelets was measured via ELISA (Rat IgG Total ELISA Kit; eBioscience). To evaluate the effects of aPD-1 on the platelets, the stability of platelets was investigated by counting the numbers of platelets using a microscope at 24 h post-reaction. To study the aPD-1 release, 0.5 U ml−1 thrombin was added to 1 × 108 P–aPD-1 suspension (500 μl) to activate the platelets at 37 °C without stirring. At pre-arranged time intervals, 50 μl samples were collected and centrifuged at 800g for 20 min and the supernatant was detected by ELISA following the method described above. The non-activated P–aPD-1 was used as a control.
To further investigate the functionality of platelets after conjugation of aPD-1, the collagen-binding and platelet aggregation studies were performed. Murine collagen type I/III (Bio-Rad) was reconstituted to a concentration of 2.0 mg ml−1, added to a confocal dish and incubated overnight at 4 °C. The plate was further blocked with 2% bovine serum albumin for 2 h and washed with PBS for the collagen-binding study. The blank plate was only blocked but without the addition of collagen. 1 × 107 P–aPD-1 or platelets stained with wheat germ agglutinin Alexa Fluor 594 were added to plate with or without collagen pre-coating. The plate was then washed with PBS after 1 min of incubation and subjected to confocal imaging. For the aggregation studies, P–aPD-1 or platelets labelled with wheat germ agglutinin Alexa Fluor 594 were incubated in complete medium with 0.5 U ml−1 thrombin, then subjected to confocal laser scanning microscopy (LSM 710; Zeiss).
To visualize the decoration of aPD-1 on the surface of platelets, aPD-1 was conjugated with fluorescein isothiocyanate (FITC) and platelets were stained with rhodamine B. The P–aPD-1 was then observed via confocal laser scanning microscopy. To further characterize the P–aPD-1, the P–aPD-1 was stained with phycoerythrin (PE)-stained rat anti-IgG antibody and subjected to flow cytometry (1 × 104 events were collected for analysis). The unstained platelet and simple mixture of platelet and isotype control antibody (anti-human CD8 antibody; BioLegend, clone: SK1) were used as controls.
Preparation of S–P–aPD-1 assembly
HSCs were isolated from the femur and tibia of C57BL/6J mice and cultured in 40 μM Ac4GalNAz (Thermo Fisher Scientific) containing medium for 72 h. The HSCs were stained with APC-anti-c-kit antibody, FITC-anti-CD34 antibody and PE-anti-CD38 antibody and subjected to flow cytometry for analysis (1 × 104 events were collected for analysis). To detect the presence of azide groups on the surface of HSCs, 1 × 106 HSCs were incubated with PBS containing 50 μM Copper(ii)-TBTA complex, 2 mM sodium ascorbate and 25 μM FAM alkyne in the dark. After 15 min, the resulting HSCs were washed with PBS three times and subjected to flow cytometry analysis (1 × 104 events were collected for analysis) and confocal observation. To functionalize the platelets with triple bonds for the click reaction, 1 × 106 platelets were treated with 20 μM DBCO-PEG4-NHS ester for 30 min at room temperature and then decorated with aPD-1, as described above. To examine the presence of triple bonds on the platelets, the resulting platelets were reacted with 20 μM azide-FITC probe for 15 min in the dark and then subjected to flow cytometry analysis (5 × 103 events were collected for analysis).
For conjugation of platelets to HSCs, 1 × 107 DBCO-functionalized platelets stained with rhodamine B were added to 1 × 107 Ac4GalNAz-treated HSCs and incubated for 45 min at 37 °C. Thereafter, excess azide-PEG (50 μM) was added to the HSC/platelet mixture and incubated for an additional 15 min to quench the additional DBCO on the surface of the platelets. After centrifugation at 400g for 5 min, the resulting platelet–HSC assembly was observed via confocal microscopy.
To investigate the conjugated number of platelets on the surface of HSCs, the reacted amount of platelets stained with rhodamine B was increased from 1:1 (platelets:HSCs) to 8:1. After addition of excess azide-PEG, the resulting S–P–aPD-1 was washed with PBS three times and subjected to confocal microscopy. The percentage of conjugated platelets was quantified on the HSCs by counting 200 S–P–aPD-1 assemblies under confocal observation at different reaction ratios.
For SEM characterization, S–P–aPD-1 was first fixed with 3.5% glutaraldehyde for 4 h, washed with PBS three times and then dehydrated with ethanol in a graded series (30, 50, 70, 85 and 90% each time for 15 min and 100% twice for 30 min), then treated with tert-butanol. After drying under vacuum, S–P–aPD-1 was coated with gold/palladium and examined by SEM (Verios 460L).
To test the bioactivity of platelets after conjugation with HSCs, S–P–aPD-1 was treated with 0.5 U ml−1 thrombin for 30 min at 37 °C without stirring, then centrifuged at 400g for 5 min. The supernatant was collected and stained with 2% uranyl acetate, then observed with a transmission electron microscope (JEOL 2000FX; Hitachi). The functionality of platelets on S–P–aPD-1 was also detected by examination of key protein expression on the platelets. Briefly, S–P–aPD-1 was stained with various rat anti-mouse antibodies (CD61, CD41, CD9, CD36, CD62P and CD154; BioLegend) and analysed by flow cytometry (1 × 104 events were collected for analysis). The platelets were also stained with anti-human CD8 antibody for isotype control. CD62P and CD154 detection was performed after the addition of thrombin for platelet activation.
To test the viability of the HSCs after incubation of Ac4GalNAz, they were seeded in a 96-well plate at a density of 1 × 104 cells well−1 and incubated with 10, 20, 40 or 80 μM Ac4GalNAz for 72 h. Then, we added 20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg ml−1). After 4 h incubation, the plate was centrifuged at 500g and the medium was replaced with 150 μl of dimethyl sulfoxide. The absorbance was measured at a wavelength of 570 nm using a microplate reader. To measure the cell viability of HSCs conjugated with platelets, the HSCs were incubated with different amounts of platelets and cultured for another 48 h. Then, the cell viability was investigated by MTT assay as described above.
For conjugation of aPD-1 on the surface of HSCs, 1 × 106 HSCs were reacted with SMCC-aPD-1 stained with rhodamine B at 4 °C for 1 h. After centrifugation at 400g for 5 min, S–aPD-1 was incubated at 37 °C for 2 and 6 h. After staining with endo-lysosomal tracker green, Hoechst 33258 and trypan blue, S–aPD-1 was subjected to confocal microscopy for observation.
In vivo pharmacokinetics and biodistribution
All animals were treated in accordance with the Guide for the Care and Use of Laboratory Animals, approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill and North Carolina State University. Six mice were randomly divided into two groups and intravenously injected with Cy5.5-aPD-1 (aPD-1: 1 mg per kg), S–P–aPD-1-Cy5.5 (aPD-1: 1 mg per kg; HSCs/platelets: 1 × 108; in 200 μl PBS for each mouse). At predetermined time points (1, 2, 4, 8, 12, 24 and 48 h), a 10 μl blood sample was collected from the tail, diluted in 100 μl water and subjected to sonication. The aPD-1 was measured by fluorescence of Cy5.5. The concentration of aPD-1 in 0.5 min was set as 100%. Furthermore, to investigate the in vivo clearance of free aPD-1 and Cy5.5-aPD-1, six mice were randomly divided into two groups and intravenously injected with aPD-1 and Cy5.5-aPD-1 at an aPD-1 dose of 1 mg per kg. At predetermined time points (0.083, 0.5, 1, 3, 6, 12, 24 and 48 h), a 20 μl blood sample was collected and centrifuged for further ELISA assay. For ELISA assay, the 96-well plate was pre-incubated with 2 μg ml−1 mPD-1 protein overnight and blocked with 2% bovine serum albumin for 2 h. Then, the blood sample was diluted and added to the platelets for incubation for 1 h. For the following procedures, we followed the manufacturer’s procedure relating to the rat IgG ELISA detection kit. For the in vivo biodistribution study, the mice were intravenously injected with PBS, Cy5.5-labelled free aPD-1, S–aPD-1, P–aPD-1, a mixture of HSCs and aPD-1, and S–P–aPD-1 at a Cy5.5 dose of 30 nmol per kg. After 6 h, the bones and other major organs were taken out and fluorescence images were recorded using an IVIS Spectrum system (PerkinElmer). The fluorescence intensities of regions of interest were analysed by Living Image Software. For bone marrow accumulation of HSC–platelet assemblies, the leukaemia-bearing mice were intravenously injected with HSC–platelet assemblies (HSCs were stained with FITC, while platelets were stained with rhodamine B; HSC–platelets: 5 × 107). At 12 h post-injection, the mice were euthanized. The bones were collected and bone tissues were fixed in the 10% formalin. After 48 h, the bones were decalcified in ethylenediaminetetraacetic acid solution for 2 d, followed by incubation in 30% sucrose. Thereafter, the bone tissues were frozen in O.C.T. medium for sectioning. After staining with Hoechst, the bone slides were subjected to confocal microscopy for observation. To observe the potential generation of PMP in the bone marrow, the rhodamine B-labelled platelets, PMPs and HSC–platelets were injected intravenously into the leukaemia-bearing mice. After 24 h, the bone tissues were collected, sectioned as described above and subjected to confocal microscopy observation. Quantification of the fluorescence signal was performed using ImageJ software.
In vivo leukaemia treatment
To build a leukaemia model, 1 × 106 luciferase-tagged C1498 cells were intravenously injected into mice. The expression of PD-L1 on C1498 cells was analysed by flow cytometry after staining with anti-PD-L1 antibody (5 × 104 events were collected for analysis). After 7 d, the 64 mice were randomly divided into 8 groups and intravenously administered with saline, HSCs, platelets, free aPD-1, S–aPD-1, P–aPD-1, S + P–aPD-1 or S–P–aPD-1 (5 × 107 cells) at aPD-1 dose of 0.5 mg per kg via the tail vein. Non-leukaemia-bearing mice were injected with S–P–aPD-1 (5 × 107 cells) at aPD-1 dose of 0.5 mg per kg, also via the tail vein. We further added an S–P–rat-IgG group and treatment groups receiving daily administration of aPD-1 at an IgG dose of 0.5 mg per kg and an aPD-1 dose of 0.25 mg per kg. The treatment was repeated every other day three times. The growth of leukaemia was monitored by detection of bioluminescence signals from C1498 cells. We used D-Luciferin (Xenogen) as a substrate for luciferase and each mouse was injected intraperitoneally with D-Luciferin at a dose of 150 mg per kg in 100 μl PBS. Bioluminescence images were collected after 5 min injection of D-Luciferin with an IVIS Spectrum Imaging System (PerkinElmer) and the acquisition time of the bioluminescence signal was 5 min. The bioluminescence signals were recorded at 1, 2 and 3 weeks after injection of the C1498 cells. Living Image Software version 4.3.1 (PerkinElmer) was used to quantitate the bioluminescence signal. To correct the background bioluminescence, we subtracted signals acquired from leukaemia-free mice (injected with D-Luciferin). After 3 weeks, a 100 μl blood sample was collected and lysed with red blood cell lysis buffer. The remaining cells were subjected to flow cytometry for investigation of the numbers of C1498 cells in the peripheral blood (2 × 104 events were collected for the flow cytometry assay). The survival time of each mouse was recorded and the spleen was weighed and imaged. The following euthanasia criteria were used in leukaemia-bearing mice: (1) diminished activity and hunched, with a rough hair coat (lack of grooming); (2) moribund animals; (3) muscle atrophy or emaciation; (4) abnormal distension; (5) incontinence or prolonged diarrhoea (longer than 48 h); and (6) neurological disorders (seizures, circling). The main tissues (heart, liver, spleen, lung, liver and bone) were taken out for the haematoxylin and eosin staining. The slides were observed using an optical microscope (DM5500B; Leica).
To investigate the cellular mechanism underlying the treatment efficacy of S–P–aPD-1, T-cell knockout mice (rag−/−), CD8+ T-cell depleted mice (CD8−/−) and PD-1 knockout mice (PD−/−) were injected with PBS, free aPD-1 and S–P–aPD-1 at an aPD-1 concentration of 0.5 mg per kg. The survival time of each mouse was recorded. To deplete CD8+ T cells, C57BL/6J mice were injected with 500 μg of anti-CD8 monoclonal antibodies (clone YTS169.4; Bio X Cell) intraperitoneally every 72 h, beginning 2 d before S–P–aPD-1 treatment, for the duration of the treatment. Depletion was confirmed by flow cytometry analysis of T cells isolated from the peripheral blood (CytoFLEX; Beckman Coulter) (5 × 104 events were collected for the flow cytometry assay). All flow cytometry data were analysed using CytExpert and Flowjo software.
For the leukaemia re-challenge study, C57BL/6J mice were injected with 1 × 106 C1498 cells 80 d after the first injection of C1498 cells. Bioluminescence signals were then monitored weekly and the survival time was recorded.
To build another leukaemia model, 1 × 106 WEHI-3 cells were intravenously injected into mice. After 7 d, the 64 mice were randomly divided into 8 groups and intravenously administered with saline, HSCs, platelets, free aPD-1, S–aPD-1, P–aPD-1, S + P–aPD-1 or S–P–aPD-1 (5 × 107 cells) at an aPD-1 concentration of 0.5 mg per kg via the tail vein. The survival time of each mouse was recorded and the spleen was weighted.
Cytokine and chemokine detection and T-cell analysis
The plasma levels of multiple cytokines and chemokines were measured using Luminex-based detection. Peripheral blood was harvested at day 12 and centrifuged at 500g for 10 min. The supernatant was aliquoted and stored at −80 °C until analysis. Samples were diluted with Luminex assay buffer following manufacturer’s instructions. Cytokine values were z-score normalized per sample before clustering. Samples were divided into four groups arbitrarily, based on their cytokine profile by k-means clustering, using the pheatmap function. To investigate the PD-1 expression in T cells in the bone marrow, the bone marrow was collected and stained with anti-CD8 and anti-PD-1 antibodies (5 × 104 events were collected for analysis). The anti-human CD8 antibody was used as an isotype control antibody. For T-cell analysis, peripheral blood and bone marrow were harvested at day 12. The blood sample was lysed first with red blood cell lysis buffer and then stained with anti-CD3, anti-CD8, anti-CD4, anti-Granzyme B, anti-CD44, anti-CD25, anti-CD69 and anti-IFNγ antibodies for 30 min. The Granzyme B staining was performed according to BioLegend’s intracellular staining protocol. To count CD3+ T-cell numbers, 150 μl blood was lysed first using red blood cell lysis buffer and subjected to flow cytometry analysis after staining with APC-anti-CD3 antibody with the addition of 50 μl counting beads. We collected 1 × 105 events for CD8 and CD4 analysis, and 5 × 104 events for IFNγ analysis. For memory T-cell analysis, the splenocytes were taken out of the saline- and S–P–aPD-1-treated mice after 30 d and stained with anti-CD8, anti-CD44, anti-CD62L and anti-CD122 antibody. Thereafter, the stained cells were subjected to flow cytometry for analysis (5 × 104 events were collected for analysis). To detect T-cell exhaustion in PD−/− mice, the lymphocytes of lymph nodes from normal mice and PD−/− mice were collected, stained with anti-CD8, anti-TIM-3 and anti-LAG-3, and subjected to flow cytometry analysis (5 × 104 events were collected for analysis). To analyse T cells from the spleen and lymph nodes, tissues were first mechanically disrupted and the cells were filtered through a 40 μm strainer for further analysis.
Statistics
All results are presented as means ± s.d. Statistical analysis was evaluated using GraphPad Prism (7.0). The log-rank test was performed for statistical analysis of survival time, and one-way analysis of variance (ANOVA) followed by Tukey’s honest significant difference (HSD) post hoc test for multiple comparisons was performed for other statistical analyses. The differences between experimental groups and control groups were considered statistically significant at P < 0.05. *P < 0.05; **P < 0.01; and ***P < 0.001.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information. Source data for the figures are available in Figshare at https://doi.org/10.6084/m9.figshare.7033481.
Supplementary Material
Acknowledgements
This work was supported by grants from the start-up packages of UNC/NC State and UCLA, the Sloan Research Fellowship of the Alfred P. Sloan Foundation, the National Key R&D Program of China (2017YFA0205600), the National Natural Science Foundation of China (51728301, 81690263) and the China Scholarship Council (CSC). We acknowledge B. Blazar at the University of Minnesota for providing the C1498-Luc cell line and M. Liu at New York University for assistance in cytokine analysis. Z.G. acknowledges support from W. Gu and P. Zhang.
Footnotes
Competing interests
Patents describing the cell-combination drug-delivery system documented in this article have been filed with the US Patent Office. Q.H. and Z.G. are inventors of the following provisional patent application: US 62/653,843. J.F.Z. has received honoraria from Agios, Celgene and Tolero, consultancy from Celgene and Asystbio Laboratories, and research funding from Merck, Takeda and Tolero. The other authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-018-0310-2.
References
- 1.Huntly BJ & Gilliland DG Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat. Rev. Cancer 5, 311–321 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Dick JE Acute myeloid leukemia stem cells. Ann. NY Acad. Sci 1044, 1–5 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Estey E & Döhner H Acute myeloid leukaemia. Lancet 368, 1894–1907 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Döhner H et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European Leukemia Net. Blood 115, 453–474 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Lowenberg B, Downing JR & Burnett A Acute myeloid leukemia. N. Engl. J. Med. 1999, 1051–1062 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Advani R et al. Treatment of refractory and relapsed acute myelogenous leukemia with combination chemotherapy plus the multidrug resistance modulator PSC 833 (Valspodar). Blood 93, 787–795 (1999). [PubMed] [Google Scholar]
- 7.Fernandez HF et al. Anthracycline dose intensification in acute myeloid leukemia. N. Engl. J. Med 361, 1249–1259 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leith CP et al. Frequency and clinical significance of the expression of the multidrug resistance proteins MDR1/P-glycoprotein, MRP1, and LRP in acute myeloid leukemia. A Southwest Oncology Group Study. Blood 94, 1086–1099 (1999). [PubMed] [Google Scholar]
- 9.Gottesman MM, Fojo T & Bates SE Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Leith CP et al. Acute myeloid leukemia in the elderly: assessment of multidrug resistance (MDR1) and cytogenetics distinguishes biologic subgroups with remarkably distinct responses to standard chemotherapy. A Southwest Oncology Group study. Blood 89, 3323–3329 (1997). [PubMed] [Google Scholar]
- 11.Ofran Y & Rowe JM Treatment for relapsed acute myeloid leukemia: what is new? Curr. Opin. Hematol 19, 89–94 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ding L et al. Clonal evolution in relapsed acute myeloid leukemia revealed by whole genome sequencing. Nature 481, 506–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagasse E et al. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat. Med 6, 1229–1234 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Wilson A & Trumpp A Bone-marrow haematopoietic-stem-cell niches. Nat. Rev. Immunol 6, 93–106 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Maude SL et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellebrecht CT et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 353, 179–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C-Y, Roybal KT, Puchner EM, Onuffer J & Lim WA Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brentjens RJ et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med 5, 177ra138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingwell K CAR T therapies drive into new terrain. Nat. Rev. Drug Discov 16, 301–304 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Jackson HJ, Rafiq S & Brentjens RJ Driving CAR T-cells forward. Nat. Rev. Clin. Oncol 13, 370–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kershaw MH, Westwood JA & Darcy PK Gene-engineered T cells for cancer therapy. Nat. Rev. Cancer 13, 525–541 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Topalian SL, Drake CG & Pardoll DM Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida Y, Agata Y, Shibahara K & Honjo T Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 11, 3887–3895 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keir ME et al. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med 203, 883–895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Q et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood 116, 2484–2493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClanahan F et al. PD-L1 checkpoint blockade prevents immune dysfunction and leukemia development in a mouse model of chronic lymphocytic leukemia. Blood 126, 203–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Gajewski TF & Kline J PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood 114, 1545–1552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamath S, Blann A & Lip G Platelet activation: assessment and quantification. Eur. Heart J. 22, 1561–1571 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Giralt SA & Champlin RE Leukemia relapse after allogeneic bone marrow transplantation: a review. Blood 84, 3603–3612 (1994). [PubMed] [Google Scholar]
- 32.Hu C-MJ et al. Nanoparticle biointerfacing by platelet membrane cloaking. Nature 526, 118–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C et al. In situ activation of platelets with checkpoint inhibitors for post-surgical cancer immunotherapy. Nat. Biomed. Eng. 1, 0011 (2017). [Google Scholar]
- 34.Hang HC, Yu C, Kato DL & Bertozzi CR A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl Acad. Sci. USA 100, 14846–14851 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi P et al. Spatiotemporal control of cell–cell reversible interactions using molecular engineering. Nat. Commun. 7, 13088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao M et al. Clickable protein nanocapsules for targeted delivery of recombinant p53 protein. J. Am. Chem. Soc 136, 15319–15325 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Eeftens JM, van der Torre J, Burnham DR & Dekker C Copper-free click chemistry for attachment of biomolecules in magnetic tweezers. BMC Biophys. 8, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leopold LH & Willemze R The treatment of acute myeloid leukemia in first relapse: a comprehensive review of the literature. Leuk. Lymphoma 43, 1715–1727 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Swami A et al. Engineered nanomedicine for myeloma and bone microenvironment targeting. Proc. Natl Acad. Sci. USA 111, 10287–10292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Q et al. Engineered nanoplatelets for enhanced treatment of multiple myeloma and thrombus. Adv. Mater. 28, 9573–9580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruggeri ZM, Orje JN, Habermann R, Federici AB & Reininger AJ Activation-independent platelet adhesion and aggregation under elevated shear stress. Blood 108, 1903–1910 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazaki Y et al. High shear stress can initiate both platelet aggregation and shedding of procoagulant containing microparticles. Blood 88, 3456–3464 (1996). [PubMed] [Google Scholar]
- 43.Yan M & Jurasz P The role of platelets in the tumor microenvironment: from solid tumors to leukemia. Biochim. Biophys. Acta 1863, 392–400 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Velez J et al. Platelets promote mitochondrial uncoupling and resistance to apoptosis in leukemia cells: a novel paradigm for the bone marrow microenvironment. Cancer Microenviron. 7, 79–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q et al. Depletion of endogenous tumor-associated regulatory T cells improves the efficacy of adoptive cytotoxic T-cell immunotherapy in murine acute myeloid leukemia. Blood 114, 3793–3802 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hlavacek WS, Posner RG & Perelson AS Steric effects on multivalent ligand–receptor binding: exclusion of ligand sites by bound cell surface receptors. Biophys. J 76, 3031–3043 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costinean S et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl Acad. Sci. USA 103, 7024–7029 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandolfi A et al. PAK1 is a therapeutic target in acute myeloid leukemia and myelodysplastic syndrome. Blood 126, 1118–1127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moynihan KD et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat. Med 22, 1402–1410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Döhner H, Weisdorf DJ & Bloomfield CD Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Ayar SP, Ravula S & Polski JM Granulocyte, monocyte and blast immunophenotype abnormalities in acute myeloid leukemia with myelodysplasia-related changes. Ann. Clin. Lab. Sci. 44, 3–9 (2014). [PubMed] [Google Scholar]
- 52.Stetler-Stevenson M et al. Diagnostic utility of flow cytometric immunophenotyping in myelodysplastic syndrome. Blood 98, 979–987 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Lu Y, Aimetti AA, Langer R & Gu Z Bioresponsive materials. Nat. Rev. Mater 1, 16075 (2016). [Google Scholar]
- 54.Wang Q et al. Non-genetic engineering of cells for drug delivery and cell-based therapy. Adv. Drug Deliv. Rev 91, 125–140 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Chen Z, Hu Q & Gu Z Leveraging engineering of cells for drug delivery. Acc. Chem. Res 51, 668–677 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Cheng H et al. Stem cell membrane engineering for cell rolling using peptide conjugation and tuningof cell-selectin interaction kinetics. Biomaterials 33, 5004–5012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and its Supplementary Information. Source data for the figures are available in Figshare at https://doi.org/10.6084/m9.figshare.7033481.