Abstract
Objectives:
Hypertensive heart disease (HHD) and hypertrophic cardiomyopathy (HCM) are associated with increased left ventricular wall thickness (LVWT). Contemporary guidelines define HCM as LVWT ≥15 mm unexplained by other disease, which complicates the diagnosis in cases of co-occurrences.
Background:
Conventional global native T1 mapping involves calculation of mean T1 values, as a surrogate for fibrosis. However, there may be differences in its spatial localization, such as diffuse and more focal fibrosis in HHD and HCM, respectively. As radiomic texture analysis (TA) can quantify spatial distributions of pixel intensity levels, we hypothesize that TA would allow differentiation between HHD and HCM.
Methods:
We identified 232 subjects (53 HHD, 108 HCM, 71 controls) for TA who consecutively underwent free-breathing multi-slice native T1 mapping. Four sets of texture descriptors were applied to capture spatially dependent and independent pixel statistics. Six texture features were sequentially selected with best discriminatory capacity between HHD and HCM and tested using Support Vector Machine (SVM) classifier. Each disease group was randomly split 4:1 (Feature Selection:Test Validation), where the pattern’s reproducibility was analyzed in the test validation dataset.
Results:
The selected texture features provided the maximum diagnostic accuracy of 86.2% (c-statistic 0.820, CI 0.769–0.903) using SVM. For the test validation dataset the pattern’s accuracy remained high at 80.0% (c-statistic 0.89, CI 0.77–1.0). Global native T1 with an accuracy of 64% separated only modestly between HHD and HCM patients (c-statistic 0.549, CI 0.452–0.640).
Conclusions:
Radiomics analysis of native T1 images discriminates between HHD and HCM patients and provides incremental value over global native T1 mapping.
Keywords: Left ventricular hypertrophy, hypertrophic cardiomyopathy, hypertensive heart disease, radiomics, cardiovascular magnetic resonance, T1 mapping
Introduction
The differential diagnosis between hypertensive heart disease (HHD) (1) and hypertrophic cardiomyopathy (HCM) (2,3) occurs frequently in clinical practice with a representative 19–23% co-occurrence in gene-positive HCM patients (4,5). Particular the presence of hypertension and left ventricular wall thickness (LVWT) ≥15 mm, the major diagnostic criterion of HCM, is a diagnostic challenge and requires elaborate cardiac imaging (6). Considering that both disease have late gadolinium enhancement (7), reduced left ventricular (LV) strain (8), diastolic dysfunction (9), asymmetric (10) as well as concentric LV hypertrophy (11) in common, the diagnosis based on cardiac imaging alone remains often uncertain.
The histological correlates of compensated LV hypertrophy, the predominant form in HHD, are hyperplasia of normally arranged cardiomyocytes (12) and a structured interstitial fibrosis supporting the increased biomechanical load (13). HCM on the other hand is characterized by myocyte disarray (11,14), arranged frequently around central cores of collagen (15), and a heterogeneous distribution of collagen rich areas (14). The distribution of fibrosis in HCM is therefore of more focal nature than the rather diffuse fibrosis in HHD.
Several parametric mapping techniques in cardiovascular magnetic resonance (CMR) imaging are capable of quantifying myocardial fibrosis (16). Of these, native T1 is best established in HHD (9,17) and HCM (9,18), including a strong correlation with histological findings (19). Hinojar et al. also demonstrated that native T1 discriminates between HCM and HHD (9). Furthermore, CMR and histology findings have similar regional distributions: on histology fibrosis is often more severe in the ventricular septum (11), a finding identical to increased native T1 values (9,18).
Traditional visual inspection of CMR images and averaging intensity levels in large regions of interest, performed in native T1 imaging, often ignores subtle changes such as fibrosis patterns. Radiomics, motivated by the concept that biomedical imaging contains information reflecting on disease specific processes and accessible by quantitative image analyses (20), has the potential to extract such information. CMR examples are the detection of ventricular myocardial scar (21) or the differentiation between HCM patients and controls (22), both based on qualitative non-contrast T1-weighted image texture analyses (TA). Another CMR image approach for TA is the use of semi-quantitative late gadolinium enhancement (23) or quantitative CMR images (24), which better reflect on physical tissue properties leading to improved disease substrate registration. TA itself consists of a wide range of mathematical computation techniques quantifying spatial distributions of pixel intensities levels (20–25), which in combination with a variety of robust predictive models can provide reliable diagnostic tools (20). CMR examples are the differentiation between HCM and control subjects with gray-level non-uniformity, a run-length matrix feature, using a Boruta machine learning algorithm (22), and identification of high and low arrhythmic risk patients using local binary patterns and Support Vector Machine (SVM) classifiers (25). Independent from the methodology chosen, CMR based TA has proven potential for several clinical applications (20–25). We hypothesized that a radiomic approach using TA on quantitative CMR images would allow the detection of HCM and HHD specific patterns which reflect on myocardial fibrosis and differentiate between both diseases characterized by an increased LVWT phenotype.
Methods
Study populations
We included 232 consecutive subjects, who were referred for CMR imaging between July 2014 and March 2018. Informed consent was obtained from all participants and the imaging protocol was approved by the Institutional Review Board. Inclusion criteria for the three patient groups (controls, HHD and HCM patients) were based on established diagnostic criteria and CMR measurements (2,3,26–28).
HCM (n=108) was diagnosed in one of two ways: normal LV cavity size with wall thickness ≥15 mm, or a wall thickness above the normal range with high clinical suspicion (i.e. electrocardiogram abnormalities, apical variant phenotype, HCM family history + LVWT ≥13 mm), both not explained by loading conditions (2,3). HCM patients with previous septal ablation or myectomy were excluded.
HHD (n=53) was defined as increased LVWT (≥12 mm) (28) associated with the diagnosis of arterial hypertension (26) in the absence of severe chronic kidney disease, LV cavity dilatation, and cardiac disease that could result in a similar magnitude of hypertrophy (i.e. moderate-to-severe valvular heart disease, inherited/acquired cardiomyopathies).
The control group (n=71, Supplemental Methods) demonstrated normal cardiac dimensions/volumes, normal cardiac function, and absence of late gadolinium enhancement. With the exception of premature ventricular beats and mild valvular disease, none of the control subjects had a history of cardiac disease including cardiac surgery or interventions.
Exclusion criteria for all subjects were the established diagnosis of amyloidosis, iron-deposition or Anderson-Fabry disease, evidence of inflammatory processes in the myo- or pericardium, history of ST-segment elevation myocardial infarction, and athletic activity with sufficient duration, intensity and frequency to explain abnormal LVWT.
Cardiovascular Magnetic Resonance
CMR was performed using a 1.5T scanner (Achieva, Philips Healthcare, Best, The Netherlands) equipped with a 32-element cardiac coil. Native T1 mapping was performed using the slice-interleaved T1 mapping (STONE) sequence which enables acquisition of 5 slices in the short-axis plane during a 90 seconds free breathing scan (29). Five short-axis slices of native T1 mapping images were analyzed using custom software (MedIACare, Boston, Massachusetts, US). Motion correction was performed using the adaptive registration of varying contrast-weighted images for an improved tissue characterization approach (30). The Supplemental Methods contains native T1 mapping parameters, and clinical CMR details.
A single reader (U.N.) extracted the LV myocardium from T1 maps by manually delineating the LV epicardial and endocardial borders. Three month later two readers (U.N. & H.E.) repeated segmentation of the test validation dataset for intra- and interobserver reproducibility assessment. Both readers were blinded to clinical information and, to omit artifacts from the registration process, excluded motion-corrupted regions. Myocardial regions of each slice were reshaped to rectangles of standard size (32×192 pixels). The lower left corner of the rectangle corresponds to the inferior right ventricular-LV insertion point providing comparable orientation and position of all T1 segments. For each study participant, 5 rectangular T1 maps at different slice levels are stacked to provide a single map with whole heart coverage (Figure 1). Using a PC with Intel Xeon 2.60 GHz CPU and 32 GB RAM, manual epi- and endocardial border definition as well as computation of TA features (0.5 seconds) required in total ~3 minutes per patient.
Figure 1: Hypertrophic Cardiomyopathy (HCM) vs Hypertensive Heart Disease (HHD) comparison of stacked myocardial T1 maps.
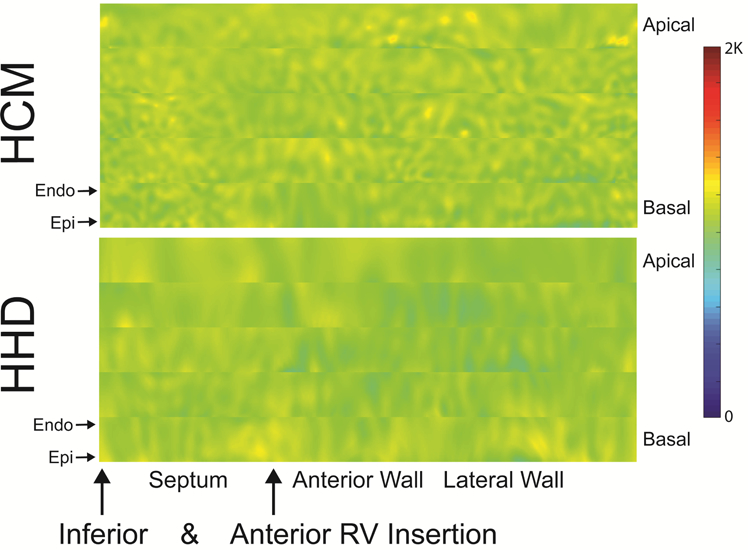
Myocardial T1 maps of rectangular shape for 5 slices of the heart from base to apex, stacked on each other (epicardial on top of endocardial region). HCM is characterized by a patchy pattern predominantly in the septal region, whilst HHD presents with a rather smooth homogeneous profile in most parts of the myocardium. Endo, endocardial orientation; Epi, epicardial orientation; RV, right ventricular.
Feature Extraction and Selection
Image TA represents a statistical assessment technique of pixel signal intensity distributions and relationships between neighboring pixels. As such, it not only allows quantification of image structure but also provides means for pattern recognition, which can be used to enhance diagnostic accuracy in medical imaging. For our purpose, we extracted texture features from each stack of normalized rectangular myocardial T1 maps using four sets of distinct texture descriptors (Table 2, Supplemental Methods). To develop and validate a tissue feature pattern for the differentiation between HHD and HCM, both groups were randomly divide ~4:1 (Feature Selection vs Test Validation dataset) (Figure S1). In order to reduce the dimensionality of extracted features in the feature selection dataset, we used a sequential forward feature selection strategy (Supplemental Methods). To replicate our initial result in an additional cohort, a step supporting generalizability, we tested the selected features within the remaining 20% of participants. This was accomplished using the images of the test validation dataset for feature quantification and SVM classifier trained on the feature selection dataset. Additionally, a subset of patients (equal LVWT subgroup) was matched for potential cofounding factors to examine the test’s diagnostic value in patients with similar morphology. Therefore HHD patients with LVWT ≥15 mm (n=10) and HCM patients with LVWT <15 mm (n=13) were matched exactly by gender, presence of LV hypertrophy and maximal LVWT, whilst grouping patients with similar age, LV mass index, global and septal T1 (Table S1). To improve comprehension of disease specific tissue characteristic identified by the selected features, controls subjects were compared with HHD and HCM patients of the feature selection dataset. The feature extraction and selection processes were performed with Matlab (The MathWorks Inc., Natick, Massachusetts, US).
Table 2.
Overview of all computed texture categories with corresponding features
| Texture Category | Texture Feature (no) |
|---|---|
| Histogram | Mean, variance, skewness, kurtosis, 5th to 10th central moments (n=10) |
| Run-length matrix (computed for four angles [vertical, horizontal, 45°, and 135°]) (38) | Short & long run emphasis, gray-level non-uniformity, run-length non-uniformity, run percentage, low & high gray-level run emphasis, short run low & high gray-level emphasis, long run low & high level emphasis (n=44) |
| Co-occurrence matrix (computed for four directions [vertical, horizontal, 45°, and 135°] with 10 displacements [from 1 pixel to 10 pixels], where all directions were averaged) (39) | Correlation, energy, contrast, homogeneity of T1 mapping values, entropy, sum of squares (n=60) |
| Local binary patterns (computed based on binarisation using center pixel value as threshold) (33) | Histogram representation of these patterns in a 3x3 pixel neighborhood was used to create rotationally-invariant Fourier descriptors (n=38) |
Statistics
Data were analyzed using SPSS (version 18.0; International Business Machines Corp., Armonk, New York, USA). Normality of data distribution was determined using the Kolmogorov-Smirnov test and visual inspection of Q-Q plots. The Chi-squared test, two-sample t-test or the Mann-Whitney U-test was conducted as appropriate. Areas under the receiver operating characteristic curves were compared using the DeLong method (31). To provide comparability between our radiomics and global native T1 analyses, test accuracy, sensitivity, specificity, and c-statistics were calculated using probabilities derived from linear SVM classifier. To represent the selected features, the Hx-index was calculated based on a linear regression model (Hx=β0+β1x1+ … +β6x6) using all selected features as independent and the diagnosis HHD or HCM as binary dependent variables. The intraclass correlation coefficient for a 2-way mixed-effects model with absolute agreement was calculated to assess the intra- and interobserver reproducibility of selected features. Based on intraclass correlations coefficients, agreement was defined as fair (0.4–0.59), good (0.6–0.74), and excellent (≥0.75) (32).
Results
Subject characteristics are presented in Table 1. The HCM cohort consisted of patients with asymmetric septal hypertrophy (n=61, 57%), concentric hypertrophy (n=27, 25%), and apical variant (n=20, 19%) (6).
Table 1.
Patient Characteristics, Global Morphological, and Functional Measures based on Cardiovascular Magnetic Resonance.
| Total (n=232) | Controls (n=71) | HHD (n=53) | HCM (n=108) | |
|---|---|---|---|---|
| Age, years | 55±14 | 53±14 | 60±10 | 55±14* |
| Sex, male n (%) | 166 (72) | 37 (51) | 44 (81) | 85 (75) |
| Body surface area, m2 | 2.0±0.2 | 1.9±0.2 ǁ† | 2.1±0.2 | 2.0±0.2* |
| Systolic Blood Pressure, mmHg | 130±17 | 125±16 §† | 140±16 | 128±16 † |
| Diastolic Blood Pressure, mmHg | 78±13 | 75±10 §* | 85±12 | 77±14 † |
| Heart Rate, bpm | 67±10 | 69±11 | 67±11 | 67±9 |
| New York Heart Association, stage | ||||
| Stage II, n (%) | 22 (9) | 0 (0) | 9 (17) | 13 (12) |
| Stage III, n (%) | 3 (1) | 0 (0) | 0 (0) | 3 (3) |
| Caucasian, n (%) | 163 (70) | 60 (82) | 36 (67) | 67 (62) |
| Hypertension, n (%) | 134 (58) | 26 (36)† | 54 (100) | 54 (50)† |
| Antihypertensive treatment, n (%) | 136 (59) | 21 (29)†§ | 47 (89) | 68 (63)† |
| ACEI/ARB, n (%) | 76 (32) | 14 (20)† | 31 (58) | 31 (29)† |
| Beta-Blocker, n (%) | 75 (32) | 7 (10)†§ | 28 (53) | 40 (37)‡ |
| Calcium Channel Blocker, n (%) | 56 (24) | 10 (14)† | 25 (47) | 21 (19)† |
| Diuretics, n (%) | 40 (17) | 8 (11)* | 17 (32) | 15 (14)* |
| Dyslipidemia, n (%) | 132 (57) | 32 (44)‡ | 38 (72) | 62 (57)‡ |
| Diabetes mellitus, n (%) | 31 (13) | 3 (4)*# | 13 (24) | 15 (14)* |
| Serum Creatinine, mg/dl | 1.0±0.3 | 0.8±0.2 † | 1.1±0.3 | 1.0±0.2 ‡ |
| Estimated Glomerular Filtration | 81±22 | 92±23* | 73±19 | 80±20 ‡ |
| Rate (Modification of Diet in Renal Disease equation), % | ||||
| Transthoracic Echocardiography (n=163) | ||||
| Diastolic dysfunction, grade | ||||
| Grade I (inverted E/A ratio), n (%) | 54 (23) | 8 (11) | 20 (38) | 26 (24) |
| Grade II (pseudonormalization), n (%) | 23 (10) | 0 (0) | 3 (6) | 20 (19)† |
| E/E’ | 10±4 | 7±2 §† | 10±3 | 11±4 |
| Deceleration time (ms) | 208±52 | 208±53 | 211±47 | 206±55 |
| LV outflow tract-gradient, n (%) | 37 (16) | 0 (0) | 6 (11) | 31 (29) |
| Systolic Anterior Motion, n (%) | 34 (15) | 0 (0) | 3 (6) | 31 (29)* |
| Cardiovascular Magnetic Resonance | ||||
| Left Atrial Diameter, mm | 38±8 | 33±7 §† | 40±7 | 40±8 |
| LV end-diastolic volume, mL | 147±34 | 140±30 ‡ | 155±41 | 146±32 |
| LV ejection fraction % | 64±7 | 62±5 # | 65±6 | 66±7 |
| LV mass index, mg/m2 | 64±25 | 45±11 §† | 64±17 | 92±51* |
| Maximal LVWT, mm | 14±5 | 8±2 †§ | 13±2 | 18±4 † |
| LVWT ≥ 15 mm, n (%) | 107 (46) | 0 (0) | 10 (19) | 97 (85) † |
| Left ventricular hypertrophy, n (%) | 73 (31) | 0 (0) | 10 (19) | 63 (55)* |
| Late Gadolinium Enhancement, n (%) | 59 (25) | 0 (0) | 3 (6) | 56 (52)† |
| T1 mapping | ||||
| Global native T1 (ms) | 1078±33 | 1069±29 § | 1066±25 | 1091±35 † |
| Septal native T1 (ms) | 1094±39 | 1078±33 § | 1089±32 | 1106±42* |
ACEI, angiotensin-converting enzyme inhibitor; ARB, angiotensin 2 receptor blocker; HCM, hypertrophic cardiomyopathy; HHD, hypertensive heart disease; LV, left ventricular; LVWT, left ventricular wall thickness
P<0.001 when compared with HHD subgroup
P<0.01 when compared with HHD subgroup
P<0.05 when compared with HHD subgroup
P<0.001 when compared with HCM subgroup
P<0.01 when compared with HCM subgroup
P<0.05 when compared with HCM subgroup
T1 mapping
Global native T1 was significantly higher in HCM compared to HHD patients (P<0.001, Table 1). Exams of HHD and HCM patients with LV hypertrophy had increased global native T1 (HHD, 1086±31 vs. 1060±20 ms, P=0.002; HCM, 1104±39 vs. 1080±26 ms, P=0.001). Septal native T1 was higher than global native T1 (P<0.001 for all). In HCM but not in HHD patients, global native T1 and LV mass index (R=0.30, P=0.001) or maximal LVWT (R=0.30, P=0.002) were mildly correlated. This finding persisted for septal native T1 (LV mass index: R=0.28, P=0.003; maximal LVWT: R=0.25, P=0.10).
Texture Feature selection
During the feature selection process six of the 152 original features were selected for their, in combination, superior differentiation capacity between HHD and HCM. The resulting features were: 2 run-length matrix features (run-length non-uniformity [135°] [RLN(135°)], short run high gray-level emphasis [135°] [SRHGE(135°)]) and 4 local binary pattern (LBP) histogram indices (15, 20, 25, 28) (33). These features contributed to the TA pattern in the following order (highest to lowest): LBP(28), RLN(135°), LBP(15), LBP(25), SRHGE(135°), LBP(20). The distribution of these six features in each study cohort is described in Table 3. With the exception of LBP(20) (P≥0.05 for all), selected LBPs (ρ: 0.731 to 0.788, P<0.001 for all) and run length matrix features (ρ=0.904, P<0.001) were well correlated within their category, but only moderately between categories (ρ: −0.212 to −0.328, when P<0.05) (Figure 2). The variability of individual texture feature correlations within and between categories for all 152 features is illustrated in Figure S2.
Table 3.
Differences of selected texture features between controls, HHD, and HCM.
| Controls (n=71) | HHD (n=44) | HCM (n=87) | |
|---|---|---|---|
| LBP(15), 10−2 | 2.9±0.6 | 2.9±0.3 | 2.6±0.4*† |
| LBP(20), 10−2 | 1.3 [0.7;2.3] | 1.1 [0.7;1.9] | 1.1 [0.3;2.2] |
| LBP(25), 10−2 | 3.4±0.4 | 3.2±0.3 | 3.0±0.4* |
| LBP(28), 10−2 | 1.1±0.3 | 1.0±0.3 | 0.7±0.3*† |
| RLN(135°), 104 | 1.8±0.3 | 1.8±0.3 | 2.1±0.5*† |
| SRHGE(135°), 104 | 2.5 [2.0;3.0] | 2.4 [2.1;2.7] | 2.8 [2.4;3.3] |
LBP, local binary pattern; RLN, run-length non-uniformity; SRHGE, short run high gray-level emphasis.
P<0.00033 when compared with controls
P<0.00033 when compared with HHD
P-values were Bonferroni corrected (0.05/152) to account for type I error rates.
Figure 2. Correlogram to illustrate the relationship among the selected texture features.
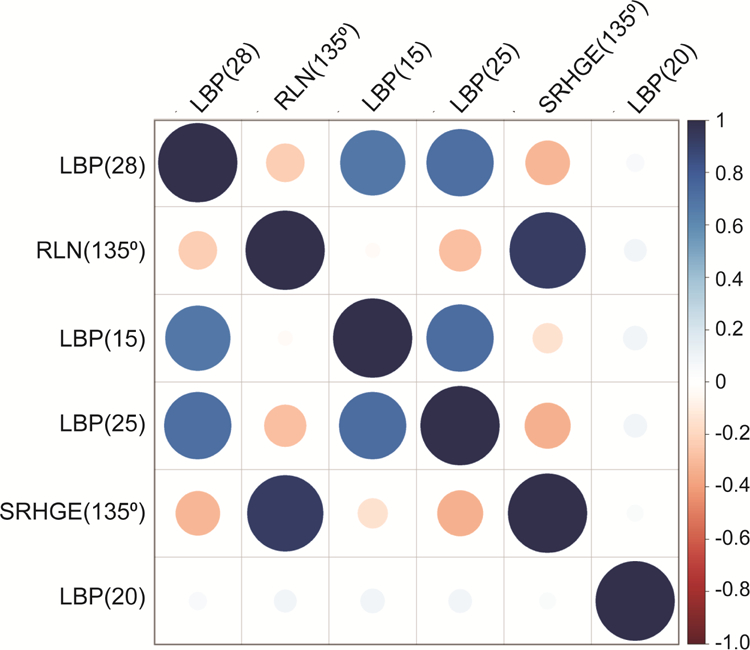
Smaller/lighter circles indicate lower correlation compared to larger/darker circles. Red circles represent negative correlations whilst blue circles represent positive correlations.
Test qualities of selected texture features
The six selected texture features provided in combination a diagnostic accuracy of 86.2% (c=0.82, CI 0.77–0.90) in the feature selection dataset and of 80.0% (c=0.89, CI 0.77–1.0) in the test validation dataset (Figure S1). The combination of the two most contributing feature LBP(28) and RLN(135°) provided a diagnostic accuracy of 80.0% (c=0.77, CI 0.71–0.86) for the feature selection dataset. The accuracy in the equal LVWT subgroup was 73.9% (c=0.70, CI 0.59–0.88). Test sensitivities and specificities are summarized in Table 4. Overall accuracies of global native T1 and septal T1 for differentiation between HCM and HHD were only 64.0% (c=0.55, CI 0.45–0.64) and 66.4% (c=0.45, CI 0.35–0.56), respectively. The discriminatory power of the selected features over global native T1 is illustrated in Figure 3, where the Hx-index (HCM vs. HHD, 0.58±1.68 vs. −2.00±1.75, P<0.001), and global native T1 (HCM vs. HHD, 1091±36 vs. 1067±26 ms, P<0.001) are plotted for HHD and HCM patients in the feature selection dataset.
Table 4.
Diagnostic capacity of the selected tissue features for HCM detection
| Patient Groups | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC (95% CI) |
|---|---|---|---|---|
| Feature selection (n=131) | 86.2 | 90.8 | 77.2 | 0.82 (0.77–0.90) |
| Test Validation (n=30) | 80.0 | 94.0 | 61.5 | 0.89 (0.77–1.00) |
| Equal LVWT subgroup (n=46) | 73.9 | 73.9 | 73.9 | 0.70 (0.59–0.88) |
Figure 3. Global myocardial native T1 values and the Hx-index measured over 5 slices in HHD (green) and HCM (blue) patients of the feature selection dataset.
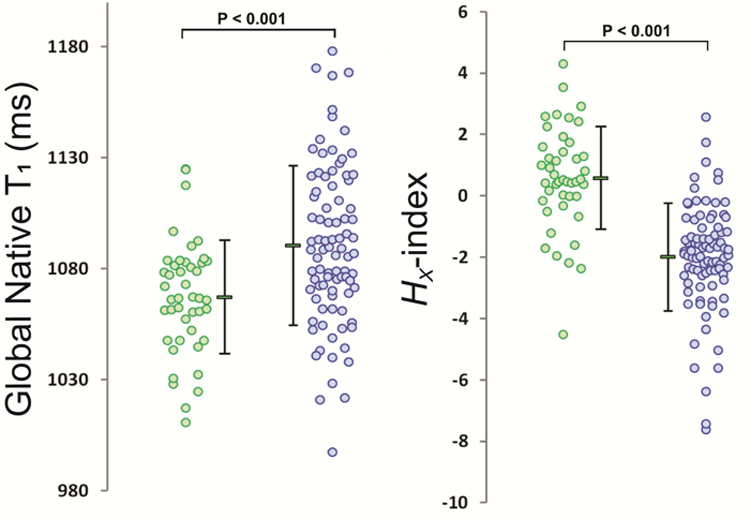
The Hx-index was calculated by a linear combination of the six selected texture analysis features using regression analyses. Depicted dots represent individual patients and the corresponding mean and standard deviation is represented adjacently. The Hx-index showed improved discrimination (i.e. smaller overlap) between HHD and HCM in comparison to global native T1.
For overall comparison of HCM with control subjects, the test accuracy of the selected features was 81.0% (c=0.85, CI 0.79–0.91), whereas their capacity to differentiate between HHD and control subjects was only minimal (accuracy 66.1%; c=0.67, CI 0.58–0.78).
The intraobserver reproducibility was good to excellent for all selected features (Table 5). The interobserver reproducibility was good to excellent for five features, but only fair for LBP(20) (Table 5). With the exception of LBP(20), Bland-Altman analyses showed narrow limits of agreement for the selected features on intra- and interobserver level (Figure S3).
Table 5.
Intraclass correlation coefficients for the intra- and interobserver reproducibility of the selected texture features (highest to lowest).
| LBP(28) | RLN(135°) | LBP(15) | LBP(25) | SRHGE(135°) | LBP(20) | |
|---|---|---|---|---|---|---|
| Intraobserver | 0.93 | 0.98 | 0.87 | 0.88 | 0.99 | 0.64 |
| Interobserver | 0.86 | 0.96 | 0.71 | 0.88 | 0.96 | 0.53 |
LBP, local binary pattern; RLN, run-length non-uniformity; SRHGE, short run high gray-level emphasis.
Discussion
Our proof-of-concept study demonstrates that TA is feasible for native T1 images and represents the first study applying TA/radiomics on T1 mapping to a common clinical diagnostic challenge, the phenotype differentiation between HHD and HCM. The selected image pre-processing and feature extraction strategy provides robust tissue feature quantification as illustrated by the good to excellent intra- and interobserver reproducibility and the narrow limits of agreements by Bland-Altman analyses for the majority of selected features. One feature, LBP(20), was more sensitive to myocardial segmentation. However the five more robust feature had in combination an accuracy of 85.5% for differentiation between HHD and HCM, which emphasizes the minimal contribution of LBP(20) to the test’s discriminatory power.
Whilst global native T1 is able to differentiate between HHD and HCM (9), the overlap of measured values is extensive (Figure 3) leading to an accuracy of 64% only. Compared to results reported by Hinojar et al. (9) test accuracies were lower in our cohort. This observation is probably contributed to three differences: multiple instead of single slice coverage, use of the STONE instead of the MOLLI T1 mapping sequences (19), phenotype severity and heterogeneity (see Supplemental Material). The selected radiomics approach improves the differentiation capacity of quantitative CMR images significantly, as documented by the large difference between selected feature’s and native T1’s area under the curve (P<0.001). Since global native T1 belongs to the histogram features (Table 2), our observation suggests that the selected features provide incremental diagnostic information beyond native T1 values.
A major challenge of TA is the large quantity of available texture features. To reduce the effect of overfitting, we selected a variety of well-established texture features (20–23,25,34). The use of quantitative mapping data in opposite to qualitative CMR improves standardization as well as reflection on physical tissue properties (35). We identified six features which in combination accomplished the highest accuracy for differentiation between HHD and HCM. The accuracy of the selected features decreased in the test validation dataset, a frequent observation in cardiac ‘omics’ research (36). Also, the ability of the selected features to differentiate between healthy controls and HCM but only to small extent between healthy controls and HHD patients implies the detection of HCM specific myocardial changes (Table 3).
The two most contributing TA features were LBP(28) and RLN(135°). The former represents a rotation invariant image descriptor computed from discrete Fourier transforms of LBP histograms, which maintains spatial orientation information (33). RLN(135°) measures the diversity of pixel intensities on a line with a 135° angle towards the horizontal. Higher values therefore reflect on increased pixel level inhomogeneity in the myocardium of HCM patients. The combination of LBP(28) and RLN(135°) or all six selected texture features leads to an increment in test accuracy, suggesting that each feature contains different information despite intra- and interclass correlations. Radiomic analysis is prompted by the concept that biomedical images contain information reflecting on disease specific changes accessible with quantitative image analyses (20). Our results proved the concepts validity for the differentiation between HHD and HCM. Our findings probably reflect on disease specific differences in fibrosis patterns, particular as native T1 is a surrogate marker of fibrosis (19) and as the pathohistological substrate of HCM is different from HHD (11–14). As shown in the equal LVWT subgroup, these findings appeared independent of a number of co-founding factors, including global/septal native T1. As CMR is standard of care for clinical assessment of HCM (2,3), supplemental radiomics analyses could be easily implemented in routine practice. Multidimensional hyperplanes defined by SVM during a feature selection process could be used to diagnose individual patients. Alternatively, cut-off values for a one-dimensional index comparable to our Hx-index (Table S4) could be used. However, these cut-offs should be validated in an independent cohort, as proof of overall robustness requires external validation.
Limitation
Our study has several limitations. Our definition of HHD relied on presence of increased wall thickness (28) instead of LV hypertrophy (1). This approach was chosen to adjust for the diagnostic criteria of HCM, LVWT ≥15 mm rather than LV hypertrophy (2). Second the influence of sarcomeric and non-sarcomeric HCM (37), different feature selection methods (21), T1 sequences (19), field strength, and image pre-processing (20–23,25) on TA features needs to be investigated. Third, we included rotation invariant LBPs and direction-dependent features to capture two dimensional spatial orientation of texture features, however integration of three-dimensional features might provide additional diagnostic information in future studies. Finally, we performed our radiomics TA in a small single center cohort using one 1.5T CMR scanner with internal validation only, whilst larger multicenter, multivendor, multifield studies might provide better generalizability of results including overall proof of robustness in the context of external validation.
Conclusion
TA is feasible for native myocardial T1 imaging and helps to discriminate between HHD and HCM populations. Additionally, the identified six tissue feature pattern provides incremental value over global native T1 mapping. For verification multicenter multivendor studies in larger cohorts are warranted before implementing radiomics analyses of quantitative native T1 imaging in clinical practice.
Supplementary Material
Perspectives.
Competences in Medical Knowledge:
Radiomic analysis of quantitative cardiovascular magnetic resonance images enables discrimination between hypertensive heart disease and hypertrophic cardiomyopathy.
Texture-based features derived from native T1 mapping improve the disease-specific classification accuracy of averaged T1 values.
Translational Outlook:
Further verification multicenter multivendor studies in larger cohorts are warranted before implementing radiomics analyses of quantitative native T1 imaging in clinical practice.
Acknowledgments
The authors thank Beth Goddu, RTR(MR), Patrick Pierce, BSRT(MR) and Sophie Berg, RNBSN for their help with CMR scanning and patient management.
Funding: The project described was supported in part by National Institutes of Health 1R01HL129185-01, 1R01HL129157, and 1R21HL127650 (Bethesda, MD, USA); American Heart Association (AHA) 15EIA22710040 (Waltham, MA, USA).
Abbreviations
- CMR
Cardiovascular magnetic resonance
- HCM
Hypertrophic cardiomyopathy
- HHD
Hypertensive heart disease
- LBP
Local binary pattern
- LV
Left ventricle/ventricular
- LVWT
Left ventricular wall thickness
- RLN
Run-length non-uniformity
- SRHGE
Short run high gray level emphasis
- SVM
Support vector machine
- TA
Texture analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Relationship to industry: Nothing to disclose
References
- 1.Lip GYH, Felmeden DC, Li-Saw-Hee FL, Beevers DG Hypertensive heart disease: A complex syndrome or a hypertensive “cardiomyopathy”? Eur Heart J 2000;21(20):1653–65. Doi: 10.1053/euhj.2000.2339. [DOI] [PubMed] [Google Scholar]
- 2.Elliott PM, Uk C, Anastasakis A, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy. Eur Heart J 2014;35(39):2733–79. Doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 3.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: Executive summary: A report of the American College of cardiology foundation/American heart association task force on practice guidelines. Circulation 2011;124(24):2761–96. Doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 4.Gruner C, Ivanov J, Care M, et al. Toronto hypertrophic cardiomyopathy genotype score for prediction of a positive genotype in hypertrophic cardiomyopathy. Circ Cardiovasc Genet 2013;6(1):19–26. Doi: 10.1161/CIRCGENETICS.112.963363. [DOI] [PubMed] [Google Scholar]
- 5.Bos JM, Will ML, Gersh BJ, Kruisselbrink TM, Ommen SR, Ackerman MJ Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc 2014;89(6):727–37. Doi: 10.1016/j.mayocp.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afonso LC, Bernal J, Bax JJ, Abraham TP Echocardiography in hypertrophic cardiomyopathy. The role of conventional and emerging technologies. JACC Cardiovasc Imaging 2008;1(6):787–800. Doi: 10.1016/j.jcmg.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Rudolph A, Abdel-Aty H, Bohl S, et al. Noninvasive detection of fibrosis applying contrast-enhanced cardiac magnetic resonance in different forms of left ventricular hypertrophy. Relation to remodeling. J Am Coll Cardiol 2009;53(3):284–91. Doi: 10.1016/j.jacc.2008.08.064. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues JCL, Rohan S, Ghosh Dastidar A, et al. Hypertensive heart disease versus hypertrophic cardiomyopathy: multi-parametric cardiovascular magnetic resonance discriminators when end-diastolic wall thickness ≥ 15 mm. Eur Radiol 2017;27(3):1125–35. Doi: 10.1007/s00330-016-4468-2. [DOI] [PubMed] [Google Scholar]
- 9.Hinojar R, Varma N, Child N, et al. T1 mapping in discrimination of hypertrophic phenotypes: Hypertensive heart disease and hypertrophic cardiomyopathy: Findings from the International T1 Multicenter Cardiovascular Magnetic Resonance Study. Circ Cardiovasc Imaging 2015;8(12):e003285 Doi: 10.1161/CIRCIMAGING.115.003285. [DOI] [PubMed] [Google Scholar]
- 10.Canepa M, Pozios I, Vianello PF, et al. Distinguishing ventricular septal bulge versus hypertrophic cardiomyopathy in the elderly. Heart 2016;102(14):1087–94. Doi: 10.1136/heartjnl-2015-308764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes SE The pathology of hypertrophic cardiomyopathy. Histopathology 2004;44:412–27. Doi: 10.1111/j.1365-2559.2004.01835.x. [DOI] [PubMed] [Google Scholar]
- 12.Coelho-Filho OR, Shah RV, Mitchell R, et al. Quantification of cardiomyocyte hypertrophy by cardiac magnetic resonance: Implications for early cardiac remodeling. Circulation 2013;128(11):1225–33. Doi: 10.1161/CIRCULATIONAHA.112.000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi MA Pathologic fibrosis and connective tissue matrix in left ventricular hypertrophy due to chronic arterial hypertension in humans. J Hypertens 1998;16(7):1031–41. Doi: 10.1097/00004872-199816070-00018. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M, Fujiwara H, Onodera T, Wu DJ, Hamashima Y, Kawai C Quantitative analysis of myocardial fibrosis in normals, hypertensive hearts, and hypertrophic cardiomyopathy. Heart 1986;55(6):575–81. Doi: 10.1136/hrt.55.6.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies MJ, McKenna WJ Hypertrophic cardiomyopathy - pathology and pathogenesis. Histopathology 1995;26(6):493–500. Doi: 10.1111/j.1365-2559.1995.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 16.Messroghli DR, Moon JC, Ferreira VM, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2 and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imagin. J Cardiovasc Magn Reson 2017;19(1):1–24. Doi: 10.1186/s12968-017-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues JCL, Amadu AM, Dastidar AG, et al. Comprehensive characterisation of hypertensive heart disease left ventricular phenotypes. Heart 2016;102(20):1671–9. Doi: 10.1136/heartjnl-2016-309576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato S, Nakamori S, Bellm S, et al. Myocardial Native T1 Time in Patients With Hypertrophic Cardiomyopathy. Am J Cardiol 2016;118(7):1057–62. Doi: 10.1016/j.amjcard.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Child N, Suna G, Dabir D, et al. Comparison of MOLLI, shMOLLLI, and SASHA in discrimination between health and disease and relationship with histologically derived collagen volume fraction. Eur Hear J - Cardiovasc Imaging 2017;(June):1–9. Doi: 10.1093/ehjci/jex309. [DOI] [PubMed] [Google Scholar]
- 20.Gillies RJ, Kinahan PE, Hricak H Radiomics: Images are more than pictures, They are data. Radiology 2016;278(2):563–77. Doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baessler B, Mannil M, Oebel S, Maintz D, Alkadhi H, Manka R Subacute and chronic left ventricular myocardial scar: Accuracy of texture analysis on nonenhanced cine MR images. Radiology 2018;286(1):103–12. Doi: 10.1148/radiol.2017170213. [DOI] [PubMed] [Google Scholar]
- 22.Baeßler B, Mannil M, Maintz D, Alkadhi H, Manka R Texture analysis and machine learning of non-contrast T1-weighted MR images in patients with hypertrophic cardiomyopathy—Preliminary results. Eur J Radiol 2018;102(November 2017):61–7. Doi: 10.1016/j.ejrad.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Larroza A, Materka A, López-Lereu MP, Monmeneu JV, Bodí V, Moratal D Differentiation between acute and chronic myocardial infarction by means of texture analysis of late gadolinium enhancement and cine cardiac magnetic resonance imaging. Eur J Radiol 2017;92(April):78–83. Doi: 10.1016/j.ejrad.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Baessler B, Luecke C, Lurz J, et al. Cardiac MRI Texture Analysis of T1 and T2 Maps in Patients with Infarctlike Acute Myocarditis. Radiology 2018:180411 Doi: 10.1148/radiol.2018180411. [DOI] [PubMed] [Google Scholar]
- 25.Kotu LP, Engan K, Borhani R, et al. Cardiac magnetic resonance image-based classification of the risk of arrhythmias in post-myocardial infarction patients. Artif Intell Med 2015;64(3):205–15. Doi: 10.1016/j.artmed.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Chobanian AV, Bakris GL, Black HR, et al. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension 2003;42(6):1206–52. Doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 27.Olivotto I, Maron MS, Autore C, et al. Assessment and significance of left ventricular mass by cardiovascular magnetic resonance in hypertrophic cardiomyopathy. J Am Coll Cardiol 2008;52(7):559–66. Doi: 10.1016/j.jacc.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 28.Salton CJ, Chuang ML, O’Donnell CJ, et al. Gender differences and normal left ventricular anatomy in an adult population free of hypertension. A cardiovascular magnetic resonance study of the Framingham Heart Study Offspring cohort. J Am Coll Cardiol 2002;39(6):1055–60. Doi: S0735109702017126 [pii]. [DOI] [PubMed] [Google Scholar]
- 29.Weingärtner S, Roujol S, Akçakaya M, Basha TA, Nezafat R Free-breathing multislice native myocardial T1 mapping using the slice-interleaved T1 (STONE) sequence. Magn Reson Med 2015;74(1):115–24. Doi: 10.1002/mrm.25387. [DOI] [PubMed] [Google Scholar]
- 30.Roujol S, Foppa M, Weingärtner S, Manning WJ, Nezafat R Adaptive registration of varying contrast-weighted images for improved tissue characterization (ARCTIC): Application to T1 mapping. Magn Reson Med 2015;73(4):1469–82. Doi: 10.1002/mrm.25270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delong ER, DeLong DM, Clarke-Pearson DL Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988;44(3):837–45. [PubMed] [Google Scholar]
- 32.Khan JN, Singh A, Nazir SA, Kanagala P, Gershlick AH, McCann GP Comparison of cardiovascular magnetic resonance feature tracking and tagging for the assessment of left ventricular systolic strain in acute myocardial infarction. Eur J Radiol 2015;84(5):840–8. Doi: 10.1016/j.ejrad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Ahonen T, Matas J, He C, Pietikäinen M Rotation invariant image description with local binary pattern histogram fourier features. Lect Notes Comput Sci (Including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 2009:61–70. Doi: 10.1007/978-3-642-02230-2_7. [DOI] [Google Scholar]
- 34.Kolossváry M, Kellermayer M, Merkely B, Maurovich-Horvat P Cardiac computed tomography radiomics. J Thorac Imaging 2018;33(1):26–34. Doi: 10.1097/RTI.0000000000000268. [DOI] [PubMed] [Google Scholar]
- 35.Kumar V, Gu Y, Basu S, et al. Radiomics: The process and the challenges. Magn Reson Imaging 2012;30(9):1234–48. Doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 2016;315(23):2532–41. Doi: 10.1001/jama.2016.5951. [DOI] [PubMed] [Google Scholar]
- 37.Tower-Rader A, Mohananey D, To A, Lever HM, Popovic ZB, Desai MY Prognostic Value of Global Longitudinal Strain in Hypertrophic Cardiomyopathy. JACC Cardiovasc Imaging 2018. Doi: 10.1016/j.jcmg.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 38.Tang X Texture information in run-length matrices. IEEE Trans Image Process 1998;7(11):1602–9. Doi: 10.1109/83.725367. [DOI] [PubMed] [Google Scholar]
- 39.Haralick R, Shanmugan K, Dinstein I Textural features for image classification. IEEE Trans Syst Man Cybern 1973:610–21. Doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


