Abstract
Recently developed single-cell profiling technologies hold promise to provide new insights including analysis of population heterogeneity and linkage of antigen receptors with gene expression. These technologies produce complex data sets that require knowledge of bioinformatics for appropriate analysis. In this minireview, we discuss several single-cell immune profiling technologies for gene and protein expression, including cytometry by time-of-flight, RNA sequencing, and antigen receptor sequencing, as well as key considerations for analysis that apply to each. Because of the critical importance of data analysis for high parameter single cell analysis, we discuss essential factors in analysis of these data, including quality control, quantification, examples of methods for high dimensional analysis, immune repertoire analysis, and preparation of analysis pipelines. We provide examples of, and suggestions for, application of these innovative methods to transplantation research.
Keywords: basic (laboratory) research/science, flow cytometry, genetics, genomics, immunobiology, informatics, molecular biology, translational research/science
1 |. INTRODUCTION
Cellular and molecular assays have addressed many important questions in transplantation. In particular, protective and pathogenic immunity have been assessed through measurements of immune cell differentiation, antigen specificity, and cellular function. The majority of these analyses have utilized techniques that evaluate bulk cell populations. For example, bulk T cell receptor (TCR) sequencing has demonstrated deletion of alloreactive T cell clones in tolerant transplant recipients.1 Bulk RNA sequencing of kidney allograft biopsies has been used to count single nucleotide variants from donor and recipient and compute a measure of cellular trafficking into the graft.2 These and other bulk analyses have contributed substantially to the state of knowledge in transplantation research.
While bulk techniques have provided substantial insight into fundamental processes, there remain significant areas of transplantation related research that cannot be addressed with these approaches. First, bulk analyses do not address phenotypic heterogeneity, which can be crucial to identifying a cellular subpopulation that contributes to either protection or disease.3 Second, they do not provide a precise definition of individual antigen receptor clones.4 Third, they do not link specific antigen receptors with defined functions. Fourth, in a mixed population of donor and recipient cells, they do not differentiate donor versus recipient gene expression.
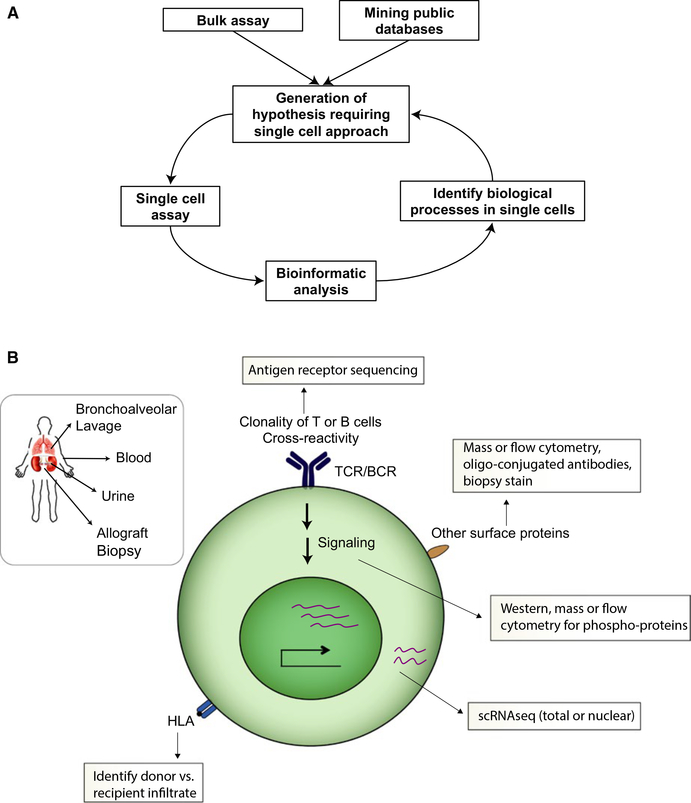
Recent advances in single-cell profiling technologies present novel opportunities to address these limitations, and to study transplant in unprecedented detail (Figure 1A,B). These technologies include cytometry by time-of-flight (CyTOF), RNA sequencing, antigen receptor sequencing, and novel tissue imaging approaches. Each has similarities to preexisting solutions that utilize either a lower parameter number (cytometry, imaging), or bulk assay (RNA and antigen receptor sequencing). Due to the expense and complexity of single-cell assays, it is often advisable to begin with a bulk approach, identify hypotheses that require single-cell approaches, and then proceed with the appropriate single-cell assay (Figure 1A). An analysis workflow for single-cell data will need to be developed in order to take advantage of the richness of data (Figure 1B). Collaborations of bench and bioinformatics scientists are often required, given the complexities inherent to both sides of the process. We present information on available techniques, tools to analyze and interpret the data, and how these methods can move the transplant research field forward. This resource summarizes key considerations and provides references to more comprehensive reviews throughout the text and tables.
FIGURE 1.
Single-cell immune profiling workflow. A, Flow chart of how to use single-cell assays for hypothesis driven research. B, Types of questions that can be analyzed through the single-cell assays. TCR, T cell receptor; BCR, B cell receptor; scRNAseq, single cell RNA sequencing.
2 |. SINGLE-CELL GENE AND PROTEIN EXPRESSION METHODOLOGY
2.1 |. Cytometry—flow and CyTOF
Flow cytometry and fluorescence-activated cell sorting (FACS), which determine multiple but limited parameters per cell, gave birth to single-cell molecular profiling. FACS has been used to analyze a variety of cell phenotypes (Table 1), including cell isolation for a variety of down-stream sequencing applications, including single-cell approaches described below. Cells in suspension are stained with fluorophore-conjugated antibodies and analyzed for emission of fluorescent light. Instruments are available that measure as many as 50 parameters in one sample,5 but due to the limitations of commonly available instrumentation and reagents, and the complexity of controlling for spectral overlap, it is generally more practical to complete experiments using at most 18 to 20 parameters per sample.6
TABLE 1.
Single-cell immune analysis methodology
| Method | Molecules analyzed | Minimum # cells per sample | # parameters | Recommended analysis | Advantages | Disadvantages | Refs. |
|---|---|---|---|---|---|---|---|
| MOLECULAR | |||||||
| Antigen-receptor sequencing | Amplified mRNA from antigen receptor genes | ∼96–1000 (varies) | −80 V regions and 2 C regions. | Clustering | 3,10,16 | ||
| scRNAseq – plate | mRNA, surface protein (index sort only) | 96 or 384 × number of plates | Varies based on primers included. | Dimensionality reduction Clustering |
Incorporates surface protein and mRNA Limited specialized equipment required |
Labor-intensive, lowest cell numbers | 3,4 |
| scRNAseq – emulsion | mRNA, surface protein (CiteSeq only) | ∼1000 | 300–10,000 (or more if sequencing at high depth) | Provides single-cell RNA analysis with relatively little labor | Expensive, surface protein applications limited, requires infrastructure | ||
| NanoString | mRNA, DNA, and/or protein | Varies, at least 6.25 ng RNA input | ∼800 | Dimensionality reduction Clustering |
Can analyze degraded RNA in biopsy specimens Multiple molecule types |
Requires use of specialized NanoString technology | 12 |
| CELLULAR | |||||||
| Flow cytometry (FACS) | Protein, glycosylation epitopes, lipids, phosphorylation | ∼10 000 | 18 (with widely available instruments), 50 (with specialized instrument) | Two-parameter gating Dimensionality reduction Clustering |
Compatible with high cell numbers, sensitive staining Gold standard for isolation of live cells |
Spectral overlap, Practical limit for most applications is 18 parameters | 5,6 |
| Mass cytometry (CyTOF) | At least 3× number for flow cytometry | 40+ | 40+ parameters No spectral overlap |
Poor resolution of positive and negative for some stains Cannot isolate live cells Expense of machine, reagents Lower cell numbers, longer run time than flow cytometry |
7 | ||
| Imaging cytometry | 10 000 | 10 | Gating Image analysis |
Subcellular localization, cellular conjugates | Lower throughput than other cytometric methods, large data files | 22 | |
| TISSUES | |||||||
| Multiplexed ion beam imaging (MIBI) | Proteins | N/A | 100+ | Image analysis (refer to references) | High parameter tissue imaging | Significant infrastructure required to image | 25 |
| Multiplexed fluorescent imaging | N/A | 100+ | Image analysis (refer to references) | High parameter tissue imaging Does not require specialized equipment (except for microscope with appropriate wavelengths) or reagents |
May require specialized programs to analyze data | 23,24 | |
| High parameter fluorescent imaging | N/A | 20+ | Image analysis (refer to references) | 24 |
N/A, not applicable; scRNAseq, single cell RNA sequencing.
CyTOF overcomes this limitation through detection of heavy metal isotopes conjugated to antibodies via time of flight mass spectrometry. The use of heavy metals eliminates spectral overlap, allowing for measurement of 34+ parameters in one sample.7 CyTOF measures the same parameters and cellular functions as flow cytometry, and the higher parameter number increases the depth of analysis, particularly relevant for experiments with limited and valuable patient samples (Table 1). Incorporation of antibodies to total histones and histone modifications allows indepth analysis of epigenetic markers alongside immune phenotyping, in a technique termed Epigenetic landscape profiling using cytometry by Time-Of-Flight (EpiTOF).8 However, the dynamic range of expression of some parameters is greater with fluorescent labels than metal labels, so CyTOF may not be appropriate for all stains. Drawbacks of CyTOF also include lower flow rates and cell numbers compared with flow cytometry, as well as the need for reagents free from heavy metal contaminants (Table 1). Additionally, because CyTOF atomizes cells, this approach cannot isolate viable cells, so FACS remains the gold standard for isolation of purified cells.
2.2 |. RNA sequencing
Single-cell RNAseq (scRNAseq) approaches fall into two broad categories based on the method of isolation: plate-based and microfluidic (Table 1). Several considerations affect data quality regardless of the specific methodology. First, cell viability is critical as dead cells release RNA, decreasing the quality of the RNA and complicating analysis.9 Second, cDNA amplification is required to obtain sufficient sample for sequencing from single cells, which can introduce bias towards amplification of specific sizes of cDNA transcripts, further complicating quantification.4 Many protocols introduce unique molecular identifiers (UMI) during reverse transcription (RT) such that the expression level of a transcript can be represented by the number of distinct UMI for that gene.4
Both plate-based and microfluidic sequencing approaches require cells in suspension. Cell isolation may be followed immediately by library preparation without purification, or by purification through magnetic bead enrichment, FACS sorting, and other methods. Plate-based approaches typically involve a sort of single cells into individual wells of 96- or 384-well plates. Some platforms allow index sorting, a function that records data on fluorescence intensity of each parameter for each sorted cell to incorporate into final analysis.10 In contrast, samples prepared for microfluidic PCR can be isolated in a bulk sort. After cells are put in a single-cell suspension, an emulsion is created to isolate individual cells for RT, amplification, and sequencing.4 The primary advantages of the emulsion approach are significantly higher cell numbers and reduced labor in library preparation.5
These methods are not restricted to cells in suspension but can also be used on tissue specimens including archived biopsy tissues (Table 1). Cells or nuclei can be isolated from fresh or archived biopsy tissue.11 Many scRNAseq protocols can be used on biopsy samples; the NanoString nCounter system is one such approach compatible with formalin-fixed paraffin embedded biopsy specimens and does not require amplification (Table 1).12 An advantage of scRNAseq from tissue is the ability to analyze disease states of both immune cells and adjacent endothelial and epithelial cells in the tissue.13
While microfluidic methods have significant advantages in time and labor savings as well as typically higher throughput, plate-based approaches present solutions not always possible with microfluidic approaches. Specifically, preparation of emulsions requires specialized microfluidic equipment for loading single cells into droplets through microfluidics. Second, until recently, these approaches have traditionally not preserved protein expression data. However, recently developed protocols use oligonucleotide-conjugated surface staining antibodies such that the RT includes DNA barcodes identifying the antibodies.14 The oligonucleotide sequence consists of polyA for RT, a barcode, and sequence for amplification with specific primers.
2.3 |. Barcoding
Many single-cell protocols incorporate the use of barcodes for sample identification and tracking. Barcodes allow for sample pooling, which is advantageous both for consistent sample preparation and for decreased costs. In FACS or CyTOF experiments, barcodes can be created through combinations of different fluorophores and metals conjugated to the same antibody.15 In sequencing experiments, barcodes of 4–8 bp sequences can be incorporated into primers.4
2.4 |. Antigen receptor repertoire analysis
TCR and B cell receptor (BCR) sequences provide a wealth of information on the nature of an immune response, including which V and J subunits are involved, the degree to which there is expansion of antigen-specific clones, and whether the repertoire changes over time.16 BCR sequencing also measures somatic hypermutation. Single-cell approaches measure paired TCR or BCR. Bulk analyses quantitatively measure clonality of the repertoire, but cannot identify paired receptor chains, detect dual productive TCRα rearrangements in one cell, or quantitate somatic hypermutation across both BCR chains. Some single-cell approaches can be coupled to gene expression data to link information on differentiation state and function of cells with a known antigen receptor.3,17
Antigen receptor sequencing utilizes similar approaches to scRNAseq protocols, with some features specific to this application (Table 1). There are two general approaches to antigen receptor sequencing: targeted analysis, and extraction of data from scRNAseq transcriptome analysis.16 In targeted protocols, the antigen receptor gene must be amplified to ensure sequence detection in a high proportion of cells. Linkage of TCR or BCR to gene expression requires either splitting the sample for antigen receptor and transcriptome sequencing separately, or data extraction from transcriptome sequencing. Extracting TCR or BCR from scRNAseq data allows quantification, but requires a more complex computational pipeline. Several groups have produced algorithms for identification of TCR or BCR from scRNAseq datasets.3 Recent studies coupling single-cell antigen receptor sequencing and computational analysis have been used to predict antigen specificity from shared motifs with TCRs of known specificity.3
While TCR clonality as well as V and J regions can be identified from the complementarity determining region (CDR)3 region sequences, somatic hypermutation requires the entire transcript to be sequenced adding additional complexity to analysis of BCR sequence. This can be accomplished through the use of 5’- Rapid Amplification of cDNA Ends (RACE), in which the entire cDNA is amplified.16 In addition, identification of BCR gene segments and rate of somatic hypermutation depends on alignment to a germline database. Thus, there will be some BCRs for which more than one V region may be a statistically valid call.18
2.5 |. Tissue imaging
New imaging approaches are also available for single-cell analysis of biopsy specimens (Table 1). Multiplexed immunohistochemistry (mIHC) involves staining one biopsy specimen with multiple antibodies, with either chemical or fluorescent detection.19,20 CO-Detection by indexing (CODEX) and cyclic immunofluorescence (CycIF) utilize staining with oligonucleotide-conjugated antibodies and an iterative process of sequential primer extension with fluorescently labeled nucleotides in order to image expression of two markers at a time, for as many iterations as necessary.21,22 The result is multiplexed image data with gene expression for many more genes than previously possible. A second approach is multiplexed ion beam imaging (MIBI) which uses metal-conjugated antibodies and secondary ion mass spectroscopy to image tissue sections. Due to the use of metals, MIBI can be used with up to 100 antibodies in one stain.23 Other advances in imaging have increased the number of fluorescent parameters analyzed through improvements in antibody stripping.19
2.6 |. Additional single cell techniques
In addition to the above, other techniques have been adapted to single-cell analysis. These approaches include protein level analysis by western blot,24 cytokine capture with barcoded antibodies,25 metabolite profiling through mass spectrometry,26 and Assay for Transposase-Accessible Chromatin using sequencing.27 Imaging flow cytometry is an innovation in microscopy that links darkfield images of cells with parameters typically identified by FACS, providing a single-cell approach to study cellular signaling and other processes affected by subcellular localization of proteins.28
3 |. ANALYSIS OF SINGLE CELL DATA
Analysis of single-cell data consists of several steps that are common with bulk data analysis: quality control and preprocessing followed by quantification, dimensionality reduction, and visualization. Early errors in data preprocessing and quality checks can introduce subtle errors that propagate throughout the rest of the analysis, making it especially important to create a robust preprocessing and quality control procedure. Single-cell analysis has many of its own unique issues as well. For example, in scRNAseq, one must contend with dropout (when expression values are zero due to technical issues rather than true biology).
3.1 |. Quality control and data processing
Despite best efforts, a systematic shift in measurements, called a batch effect, is virtually unavoidable when data are collected on multiple days and/or machines. Batch effects can lead to erroneous conclusions due to confounding effects. To correct for this, a number of techniques have been developed. For data types with a large number of parameters (eg transcriptomics), the ComBat29 method can be used. ComBat assumes that data come from normal distribution. In cases where data do not meet this assumption, manifold alignment attempts to find a transformation that aligns distributions across batches.30 Normalization and transformation are also integral components of a preprocessing pipeline. Downstream analyses may make assumptions about the distribution and scale of the data, which necessitates transformation. For example, in RNAseq, expression data are bounded at zero. Most methods assume these data are log-transformed before analysis. Therefore, RNAseq values are log transformed after adding a small constant to prevent taking the logarithm of zero.
3.2 |. Quantification
Different single-cell technologies require different quantification approaches. Yet a common key consideration for quantification across all technologies is appropriately controlling for experimental biases. For instance, in case of scRNAseq, samples with UMIs added during RT can be quantified by counting all instances of a gene expressed with the same UMI as one RNA molecule. For scRNAseq without UMI, quantification is intrinsically imprecise, and should use a transformation that reflects the uncertainty of the measurements.
In contrast, quantification of antigen receptor data requires the number of cells with each receptor to identify clonally expanded cells. Accuracy of antigen receptor data depends on the sampling which varies widely. For plate-based approaches with 100–200 cells per sample, measurement of clonal expansion is highly unlikely to be representative for clones that constitute less than 10% of the population due to the high probability of sampling bias. However, even a sample of 50 000 cells from human blood represents 0.0005% of the total lymphocytes in that individual’s blood, which represent ~2% of the lymphocytes in the individual.31 Hence, single-cell data always represent an extremely small sample of the lymphocyte population.
3.3 |. Dimensionality reduction
The high dimensionality of single-cell technologies presents substantial challenges due to increased sparsity, generally referred to as the “curse of dimensionality.” For instance, the spread of distances between points compresses as dimensionality increases which can wreak havoc with distance-based algorithms. This can impede automated subset identification.
A variety of dimensionality reduction techniques have been developed to express the information from these high dimensional samples into substantially smaller number of dimensions (usually 2–3) such that relationships between samples are maintained, but are more interpretable. One of the most commonly used techniques is principal components analysis (PCA) that maps data onto the lower dimensional space such that a large amount of variance in data are explained by a very small number of dimensions, called principal components (PCs), which are readily interpretable as linear combinations of the original dimensions. T-Distributed Stochastic Neighbor Embedding (t-SNE)32 is conceptually similar, but differs from PCA in that the resulting lower dimensional space is not a linear projection and can represent complex nonlinear forms in high dimensional space. Recently described Uniform Manifold Approximation and Projection (UMAP)33,34 is also a nonlinear dimensionality reduction technique, leading to similar results as t-SNE. However, UMAP is significantly faster than t-SNE which allows for investigation of an order of magnitude greater data simultaneously. While these techniques are often used with CyTOF and scRNASeq data, it is important to note that they can be used to analyze almost any type of high dimensional data.
Several additional dimensionality reduction techniques represent high dimensional data as network graphs including SPADE, CITRUS, and SCAFFOLD.35–37 Nodes representing populations in high dimensional space are connected by edges. Due to the nature of the graph structure, rotation of nodes without breaking their connections does not represent a change in the overall structure (one can visualize this by thinking of a mobile for a crib: rotating branches of the mobile does not create a new mobile). Once represented as a network graph, several network analysis techniques can be used to discover a multitude of different properties of the high dimensional dataset. For example, critical nodes can be identified by examining their connections to other nodes and how many paths through the network traverse those nodes.38
3.4 |. Clustering
Unlike bulk methods which compress diverse sub-populations of cells into a single value, single-cell data can be used to examine the natural heterogeneity within a population of cells. Clustering algorithms attempt to find these subsets of cells within or across samples. Typically, this is unsupervised, meaning the researcher allows the algorithm to find clusters in the data without any outside knowledge (such as the disease status, etc.). A myriad of techniques exists for clustering (k-means, hierarchical clustering, DBSCAN39). Some require specification of the number of clusters beforehand whereas others can estimate this from the data. Moreover, as clustering is typically performed in high dimensional space, these clusters might not be apparent from examination of the data in two or three dimensions at a time. In single-cell data, clustering can be used to identify novel populations of cells that are defined by some phenotypic characteristic.
3.5 |. Trajectory analysis
High dimensional single-cell technologies present unique opportunities to investigate developmental or spatial relationships such as stem cell differentiation. These techniques aim to determine a trajectory different cells follow in high dimensional space (Table 2) that assume each cell is a snapshot along various trajectories. Virtually all techniques first build a network using the single-cell data, followed by finding paths from a certain cell type to another along the network. These trajectories allow inferring developmental programs, bifurcation points, and key intermediate stages that might not have been found by looking at the data from a static perspective. We refer to a comprehensive review by Saelens et al who compared 29 of these methods on various datasets.40
TABLE 2.
Single-cell immune analysis computational methodology
| Method | Output | Key hyperparameters | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Dimensionality reduction | Lower-dimensional representation of original data | Visualization of high dimensional data, discovery of subsets of data | Potential information loss | ||
| Principal Component Analysis (PCA) | Original data on new axes where axes are linear combinations of original dimensions | Well-established, easy to interpret, fast, consistent results across applications on the same data | Misses nonlinear patterns in data | 44 | |
| T-distributed Stochastic Neighbor Embedding (t-SNE) | Original data on new axes where axes have no inherent interpretation | Effective number of nearest neighbors (Perplexity) Cycles before algorithm is considered done (Iterations) |
Discovery of nonlinear patterns | Difficult to interpret axes, slow, repeat applications produce different results, requires downsampling | 29 |
| Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) | Original data on new axes where axes have no inherent interpretation | Minimum distance between neighbors in new space Number of neighbors |
Discovery of nonlinear patterns, fast, does not require downsampling | Difficult to interpret, repeat applications produce different results | 30,31 |
| Clustering | Algorithmically-determined groupings of data-points | Distance Metric (how to assign distance between two points) | Unbiased discovery of potentially biologically meaningful groups of data points | ||
| Hierarchical clustering | Data points organized into a tree structure | How distance between clusters is determined (Linkage) | Easily observe multilevel clustering | Determining where to cut tree to produce clusters can be difficult | |
| k-means | k clusters of original data | Number of clusters (k) | Fast, well-established | Need to specify number of clusters beforehand, cannot find clusters that are not simple spheres or ellipses | |
| Density-based spatial clustering of applications with noise (DBSCAN) | Clusters of original data | Min number of points to call a region dense Radius of point’s neighborhood (Epsilon) |
No need to specify number of clusters, can find | Many data points may be classified as “noise” or one large cluster depending on hyperparameters | 45 |
| Repertoire analysis | 38 | ||||
| Diversity | Measure of clonal diversity | Choice of diversity metric (Gini, Entropy, Chao1, Hill, etc.) | Provides a single diversity metric for a sample or population of cells, can be compared across samples and conditions | Can be difficult to interpret intuitively, sensitive to number of samples | |
| Sequence distance | Distance between two TCR or BCR sequences | Choice of distance metric (Levenshtein, etc.) | Distances can be used in downstream applications like clustering or dimensionality reduction for visualization | Distances might not be biologically meaningful | |
| Motif enrichment | Significant sequence motifs | Choice of algorithm (GLIPH, etc.) | Discovery of motifs that may confer specificity | May miss larger motifs depending on hyperparameter choices | |
| Phylogenetics | BCR clonal family trees | Evolutionary model for amino acid mutation | Can infer lineages and branching points during affinity maturation | Can be sensitive to hyperparameters, methods typically optimized for traditional evolutionary models |
TCR, T cell receptor; BCR, B cell receptor; GLIPH, Grouping of Lymphocyte Interactions by Paratope Hotspots.
3.6 |. Single cell repertoire analysis
Advances in sequencing technology also allow examination of the sequences of the complementarity determining region 3 (CDR3) from TCRs and BCRs at the single-cell level. Combining this data with single-cell phenotypic data can be extremely powerful. Due to the inherently discrete nature of sequence data, a slightly different set of techniques must be used (as compared to gene expression data). CDR3 sequences can be compared through a variety of means, the most common of which is Levenshtein distance. This metric represents the number of changes (either amino acids or nucleotides) needed to get from one sequence to another. These distances can then be used to cluster CDR3s or to create CDR3 networks. A common question asked is whether there is an enrichment of a particular motif amongst CDR3s. Algorithms for TCRs such as GLIPH and TCRdist can identify motifs of interest which may confer specificity.3 Identification of groups of cells with similar TCR or BCR is typically followed by analysis of the clonal diversity of each group. For example, 1 might find that all of the activated T cells are oligoclonal. Various metrics such as the Gini coefficient, Hill diversity, and Chao1 have been used to compare clonal diversity under various biological conditions in TCR data. While these methods are generally applicable across TCRs and BCRs, BCRs can also be studied in a phylogenetic context. Due to affinity maturation via somatic hypermutation, a single ancestral B cell clone can diversify, forming a clonal family comprising several clones which recognize a particular antigen. Family trees of BCRs can be constructed using methods from evolutionary biology. We refer readers to a review by Miho et al41 for more details about single-cell repertoire analysis.
3.7 |. Analytic pipelines
A number of analytic pipelines are available that perform the steps described above in logical order with minimal input, and are accessible to those with minimal bioinformatics background. These include CytoBank, FlowJo, SeqGeq, and the 10X Genomics Loupe Browser. For those with bioinformatics experience, published code is available for some analyses in R and Python, including tools available through the online resource GitHub. Resources to learn how to use these tools include R tutorials, help pages, and courses in R and data analysis available through Coursera.
3.8 |. Considerations for interpretation of high dimensional analysis
It is essential to keep in mind the limitations and assumptions that go into each of these algorithms as this is key to interpretation of their results. Misinterpretation can lead to erroneous or misleading conclusions. For example, one must be careful comparing the distances between clusters in t-SNE.42 t-SNE requires the user to specify a value for perplexity, which approximately corresponds to the number of nearest neighbors.32 Different perplexity values will produce different t-SNE plots. Lower perplexity values favor the preservation of fine local structure over global structure whereas higher values preserve global structure. Overall, just as an experimental immunologist would not be wise to use a flow cytometer without understanding antibodies or fluorescence, a researcher should invest the time to understand the inner workings of these algorithms.
4 |. CONSIDERATIONS FOR FEASIBILITY
Many single-cell technologies are labor intensive, expensive, and require specialized technology. Most importantly, they require close collaboration between bench researchers and with those with bioinformatics expertise. It is worth considering all options, such as creating core facilities, collaborating with other institutions, or paying for library preparation and/or sequencing services from a company. Perhaps the biggest factor affecting feasibility of these experiments is the bioinformatic capability to analyze the data (Figure 1A, Table 2). As described above, a variety of tools are available to help researchers with limited bioinformatic experience complete these analyses (Seurat is particularly useful for scRNAseq data).43 Either when collaborating with a bioinformatician or when using premade tools, it is important to understand how the tool works and what fine-tuning will be appropriate for the analysis. For example, dimensionality reductions such as t-SNE and k means clustering can produce slightly different results each time they are completed, and thus should be completed multiple times to account for stochastic effects. Other analyses may need to be customized based on experimental design, for example if a specific transformation or normalization is required, or if adjustments need to be made based on the number of cells. Depending on the specific experiment, however, these tools may or may not be of use. Thus, collaborations with bioinformaticians are strongly recommended whenever possible.
5 |. APPLICATIONS TO TRANSPLANT RESEARCH
The techniques and analyses described have been applied to a variety of biological and clinical questions, but their potential for the study of both protective and pathogenic immunity in the context of transplantation has yet to be fully tapped. Available methods measure immune cell differentiation, antigen specificity, cellular function, and heterogeneity within cell populations. All of these methods can be used to probe cellular function in animal models and in humans. These techniques provide new approaches to investigate crucial and difficult to answer questions in transplantation: identification of key cell populations involved in protective or pathogenic responses, definition of donor and recipient immune infiltrates in the allograft, underlying mechanisms of heterologous immunity, and accurate quantification of T cell or B cell clones in transplant recipients (Figure 1B).
Single-cell protein and/or RNA analysis identifies gene expression across and within cell populations, providing an accurate measure of small subpopulations that might not be detected by bulk analyses. CyTOF has identified a specific subset of T cells associated with operational tolerance in pediatric liver transplant recipients, and specific populations that predict response to desensitization therapy in sensitized kidney transplant candidates.44 The ability to characterize subpopulations is of particular interest for development of biomarkers. Gene and protein expression analyses have been used as biomarkers to predict risk and diagnose rejection as well as additional posttransplant pathology.45–47 Single-cell approaches have the potential to enhance the predictive power of currently available biomarkers and identify new ones especially in situations where differential expression is limited to a subpopulation of cells.
Allograft biopsies can now be analyzed in unprecedented depth with the use of single-cell methods. Kidney biopsies are commonly analyzed by immunofluorescent microscopy for morphology and expression of a limited set of genes. scRNAseq can measure expression of many more genes in single cells in a biopsy, potentially improving diagnostics both before and after transplantation.11 These analyses include both immune infiltrates and cells belonging to the tissue, potentially identifying tissue cells with disease phenotypes.13 scRNAseq datasets can include sequence that differentiates between individuals, such as HLA alleles, and single nucleotide polymorphisms.48 In a sample including both donor and recipient cells, this may provide an opportunity to improve data interpretation through distinguishing the two sources of cells.
In addition to these analyses that have been completed in transplant samples, other exciting experiments are made possible with single-cell methodology. For instance, immune phenotyping and clonal analysis have been used already to define immune responses to viral infection after transplantation.49 Single-cell analyses coupling the two can determine whether virus-specific T cells change expression of key functional genes in the presence of immunosuppression or active infection. As such, single-cell assays will link analysis of phenotypes associated with a specific posttransplant diagnosis to a mechanistic understanding of the underlying processes. This will dramatically increase the potential for development of new transplant therapies based on mechanistic understanding of posttransplant disease.
ACKNOWLEDGMENTS
PK and SS are supported in part by grants from Bill & Melinda Gates Foundation, R01 AI125197–01, 1U19AI109662, U19AI057229, and U19AI090019. LEH is supported by a grant from the American Heart Association/Enduring Hearts, 17POST33660597.
Abbreviations
- 5’-RACE
5’- Rapid Amplification of cDNA Ends
- BCR
B cell receptor
- CDR
complementarity determining region
- CODEX
CO-Detection by indexing
- CyTOF
cytometry by time of flight
- EpiTOF
Epigenetic landscape profiling using cytometry by Time-Of-Flight
- FACS
fluorescence-activated cell sorting
- MIBI
multiplexed ion beam imaging; PCA, principal components analysis
- RT
reverse transcription
- scRNA
seqSingle cell RNAseq
- TCR
T cell receptor
- t-SNE
t-Distributed Stochastic Neighbor Embedding
- UMAP
Uniform Manifold Approximation and Projection
- UMI
unique molecular identifiers
Footnotes
DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Morris H, DeWolf S, Robins R, et al. Tracking donor-reactive T cells: evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med 2015;7(272):272ra210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thareja G, Yang H, Hayat S, et al. Single nucleotide variant counts computed frmo RNA sequencing and cellular traffic into human kidney allografts. Am J Transplant 2018;18(10):2429–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358(6359):58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 2018;50:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cossarizza A, Chang H-D, Radbruch A, Akdis M, Andr I, Annunziato F, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur J Immunol 2017;47:1584–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tung JW, Heydari K, Tirouvanziam R, et al. Modern flow cytometry: a practical approach. Clin Lab Med 2007;27(3):453–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung P, Vallania F, Dvorak M, et al. Single-cell epigenetics - Chromatin modification atlas unveiled by mass cytometry. Clin Immunol 2018;96:40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.10X Genomics. Technical Note – Removal of Dead Cells from Single Cell Suspensions Improves Performance for 10x Genomics® Single Cell Applications. Pleasanton, CA: 10X Genomics; 2017. [Google Scholar]
- 10.Osborne GW. Chapter 21 Recent advances in flow cytometric cell sorting. Methods Cell Biol. 2011;102:533–556. [DOI] [PubMed] [Google Scholar]
- 11.Malone AF, Wu H, Humphreys BD. Bringing renal biopsy interpretation into the molecular age with single-cell RNA sequencing. Semin Nephrol 2018;38(1):31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veldman‐Jones MH, Brant R, Rooney C, et al. Evaluating robustness and sensitivity of the nanostring technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Can Res 2015;75(13):2587–2593. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Malone AF, Donnelly EL, et al. Single‐cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol 2018;29:2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR. Abseq: ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci Rep 2017;7:44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann FJ, Simonds EF, Bendall SC. A universal live cell barcoding-platform for multiplexed human single cell analysis. Sci Rep 2018;8(1):10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol 2017;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han A, Glanville J, Hansmann L, Davis MM. Linking T‐cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol 2014;32(7):684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaari G, Kleinstein SH. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med 2015;7:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. [DOI] [PubMed] [Google Scholar]
- 20.Parra ER. Novel platforms of multiplexed immunofluorescence for study of paraffin tumor tissues. J Cancer Treat Diagn. 2018;2(1):43–53. [Google Scholar]
- 21.Goltsev Y, Samusik N, Kennedy‐Darling J, Vazquez G, Black S, Nolan GP. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174(4):968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J‐ R, Fallahi‐Sichani M, Sorger PK. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 2015;6:8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelo M, Bendall SC, Finck R, et al. Multiplexed ion beam imaging (MIBI) of human breast tumors. Nat Methods. 2014;20(4):436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes AJ, Spelke DP, Xu Z, Kang C‐ C, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11:749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Y, Shi Q, Wei W. Single cell proteomics in biomedicine: high-dimensional data acquisition, visualization, and analysis. Proteomics. 2017;17:3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emara S, Amer S, Ali A, Abouleila Y, Oga A, Masujima T. Single‐cell metabolomics. In: Sussulini A, ed. Metabolomics: from Fundamentals to Clinical Applications. Cham, Switzerland: Springer; 2017:323–343. [DOI] [PubMed] [Google Scholar]
- 27.Buenrostro JD, Wu N, Litzenburger UM, et al. Single‐cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maguire O, O’Loughlin K, Minderman H. Simultaneous assessment of NF-κB/p65 phosphorylation and nuclear localization using imaging flow cytometry. J Immunol Methods. 2015;423:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray data using empirical bayes methods. Biostatistics. 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- 30.Butler A, Satija R. Integrated analysis of single cell transcriptomic data across conditions, technologies, and species. bioRxiv. 2017. 10.1101/164889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trepel F. Number and distribution of lymphocytes in man. A critical analysis. Klin Wschr. 1974;52:511–515. [DOI] [PubMed] [Google Scholar]
- 32.van der Maaten L, Hinton G. Visualizing data using t‐SNE. J Mach Learn Res 2008;9:2579–2605. [Google Scholar]
- 33.McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. J Open Source Softw 2018;3(29):861. [Google Scholar]
- 34.Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single‐cell data using UMAP. Nat Biotechnol 2018;37(1):38. [DOI] [PubMed] [Google Scholar]
- 35.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011;29:886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci 2014;111(26):E2770–E2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spitzer MH, Gherardini PF, Fragiadakis GK, et al. An interactive reference framework for modeling a dynamic immune system. Science. 2015;349(6244):1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal M, Cusick ME, Barabási A‐ L. Interactome networks and human disease. Cell. 2011;144(6):986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu R, Wunsch D II. Survey of clustering algorithms. IEEE Trans Neural Networks. 2005;16(3):645–678. [DOI] [PubMed] [Google Scholar]
- 40.Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods: towards more accurate and robust tools. bioRxiv. 2018. 10.1101/276907. [DOI] [PubMed] [Google Scholar]
- 41.Miho E, Yermanos A, Weber CR, Berger CT, Reddy ST, Greiff V. Computational strategies for dissecting the high-dimensional complexity of adaptive immune repertoires. Front Immunol 2018;9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wattenberg M How to use t‐SNE effectively. Distill 2016. 10.23915/distill.00002. [DOI] [Google Scholar]
- 43.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krams SM, Schaffert S, Lau AH, Martinez OM. Applying mass cytometry to the analysis of lymphoid populations in transplantation. Am J Transplant 2016;17(8):1992–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roedder S, Vitalone M, Khatri P, Sarwal M. Biomarkers in solid organ transplantation: establishing personalized transplantation medicine. Genome Med 2011;3(6):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azad T, Donato M, Heylen L, et al. Inflammatory macrophage‐associated 3-gene signature predicts subclinical allograft injury and graft survival. JCI Insight. 2018;3(2):e95659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med 2013;210(11):2205–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Zhou Q, Wang Y, Ning K. Single‐cell SNP analyses and interpretations based on RNA-Seq data for colon cancer research. Sci Rep 2016;6:34420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suessmuth Y, Mukherjee R, Watkins B, et al. CMV reactivation drives posttransplant T-cell reconstitution and results in defects in the underlying TCRβ repertoire. Blood. 2015;125:3835–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]