Abstract
Small GTPase is a kind of GTP-binding protein commonly found in eukaryotic cells. It plays an important role in cytoskeletal reorganization, cell polarity, cell cycle progression, gene expression and many other significant events in cells, such as the interaction with foreign particles. Therefore, it is of great scientific significance to understand the biological properties of small GTPases as well as the GTPase-nano interplay, since more and more nanomedicine are supposed to be used in biomedical field. However, there is no review in this aspect. This review summarizes the small GTPases in terms of the structure, biological function and its interaction with nanoparticles. We briefly introduced the various nanoparticles such as gold/silver nanoparticles, SWCNT, polymeric micelles and other nano delivery systems that interacted with different GTPases. These current nanoparticles exhibited different pharmacological effect modes and various target design concepts in the small GTPases study. This will help to elucidate the conclusion that the therapeutic strategy targeting small GTPases might be a new research direction. It is believed that the in-depth study on the functional mechanism of GTPases can provide insights for the design and study of nanomedicines.
Keywords: Small GTPase, Nanoparticles, Ras, Rab, Rho
Graphical abstract
1. Introduction
Small GTPase family is a GTP-binding protein family which can be commonly found in eukaryotic cells [1]. It is a kind of GTPases can achieve mutual transformation between GTP and GDP in cytoplasm [2]. These small GTPases act as molecular switches in cells, affecting almost all cellular processes [3]. The most prominent member of small GTPase family is the Ras GTPase, thus the family is also called the Ras superfamily [4], [5].
2. Structure and action mechanism of small GTPases
2.1. Structure of small GTPases
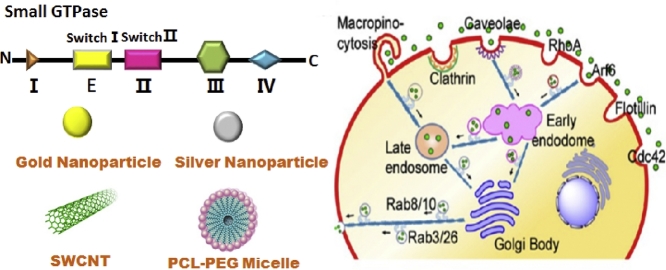
The analysis of the small GTPase protein crystal structure indicates that the GTP binding domain of this type of protein can be subdivided into five relatively conserved motifs G1–G5. G1 motif (I) is a purine nucleotide binding signal; G2 motif (E) is in one of two segments that redirects with GDP or GTP binding function and provides major component of the effector binding surface; G3 motif (II) is related to the binding of nucleotide-related Mg2+; G4 motif (III) brings the hydrogen bond in contact with the guanine ring; G5 motif (Ⅳ) makes indirect associations with the guanine nucleotide [6–8]. Taken together, these elements constitute the conserved ∼20 kDa domain and shared by all Ras superfamily proteins.
2.2. Action mechanism of small GTPases
While participating in physiological activity, the molecular structure of small GTPases present two forms which support mutual transformation, GTP-binding activated state and GDP-binding non-activated state, which can also be called as “ON” state and “OFF” state, respectively [3].
The hydrolysis of GTP to GDP is mediated by GTPase-activated proteins (GAPs), and the exchange of GDP to GTP is mediated by guanine nucleotide exchange factors (GEFs) [9]. GEFs and GAPs coexist in most cells, increasing the diversity of signals that regulate small GTPases activity [8]. Guanine nucleotide dissociation inhibitors (GDIs) are contrary to exchange factors. GDI specifically binds GDP-bound GTPase and inhibits GDP release (Fig. 1B) [10].
Fig. 1.
(A) Schematic diagram of the small GTPase protein sequence. (B) Regulation of small GTPase activity. GTPases cycle between their GTP-bound active and GDP-bound inactive form. They are activated by guanine-nucleotide exchange factors (GEFs), which remove GDP, thus enabling excess cellular GTP to bind. The binding of active GTPase to downstream effector proteins elicits cell responses. GTPase-activating proteins (GAPs), which increase the GTPase activity, are the off-switch. Inactive Rac is sequestered in the cytosol by guanine-nucleotide dissociation inhibitors (GDIs). Reproduced with permission from [78]. Copyright 2018 John Wiley & Sons, Inc.
3. Classification of small GTPase protein
Ras GTP enzyme was initially found to be a mutation of GTPase in various cancers. As time goes by, many GTP enzymes are found to have the similar 3D structure with Ras. At present, the family has over 150 members making it a superfamily. On the FASEB Summer Consultative Conference in 1992, the comprehensive naming principle of Ras-like GTPase was established, dividing the small G protein into five subfamilies of Ras, Rho, Rab, Arf/Sar and Ran according to the sequence, structure and function. The sorting criterion is now widely accepted. Recently, Rad, Rap, Rheb, Rit and Miro are also included into the superfamily [11].
Extensive researches on each family of small GTP enzyme family in term of cancer only focused on how these protein families control the no carcinogenic process (such as immunity and inflammation) at the beginning. The functions of these GTP enzymes are greatly overlapped. Ras takes charge of cell proliferation, Rho is used for cell morphology, Ran is used for nuclear transport, and Rab and Arf are used for vesicle trafficking [8].
3.1. Ras subfamily
Ras superfamily is the first and the most diversified subfamily. It plays a large role in mediating immunity and inflammation [7], [8]. The Ras subfamily protein is almost the generic component in signaling pathway of eukaryotes (including vertebrate, invertebrate and yeast), which plays a key role in the development, proliferation, differentiation and survival of eucaryon.
In addition to the widely expressed major Ras isoforms, the highly conserved H-, K- and N-Ras exhibit diverse biological properties [12], [13]. H-ras, K-ras and N-ras genes are located on chromosomes 11, 12 and 1, respectively. K-ras gene is also known as the p21 gene because it encodes a 21 kDa Ras protein. K-Ras has the largest impact on human cancer.
3.2. Rho subfamily
Rho GTPase family contains well-developed Rac1, RhoA and Cdc42 which modulate the formation of cytoskeleton, cell polarity, cell cycle progression and signal network of gene expression [14], [15], [16]. RhoA, Rac1 and Cdc42 possess the most obvious members [17], [18]. They serve as the molecular switches in separate or correlative signaling pathways [19]. These signaling pathways link the plasma receptors to activate cytoskeleton reorganization and the subsequent biological effect. Cdc42 and Rho can be commonly found in yeast and most mammalian cells, while Rac can only be observed in the latter [20].
Actually, the three kinds of protein play the same significant role in modulating cell polarity, genetic transcription, cell cycle progression, microtubule kinetics, vesicle trafficking, extracellular matrix (ECM) remodeling and various enzymatic activities (using NADPH oxidase activity to generate the reactive oxygen, ROS) [21].
3.3. Rab subfamily
Rab subfamily is the largest subfamily in Small GTPase Protein family [1]. Amid the 93 Small GTPases family members of Arabidopsis, 57 belong to Rab subfamily. Rab subfamily modulates the vesicle trafficking and protein transport of eukaryocyte by regulating complex vesicle trafficking and microtubule system activities.
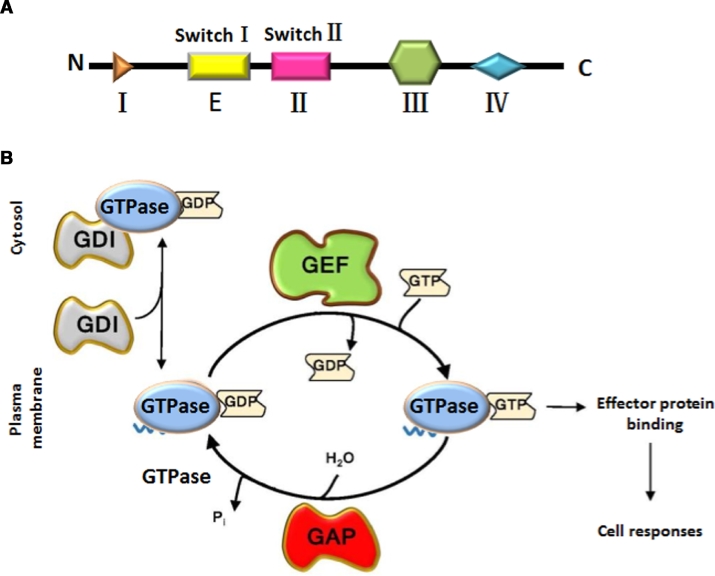
At present, there are 60 kinds of Rab proteins (including isomers) which play an important role in promoting and modulating the berth and integration of trafficking vesicle (Fig. 2). Each organelle has at least one kind of Rab protein and different membranes have different Rab proteins.
Fig. 2.
Overview of Rab GTPases on the endocytic pathway. Rab GTPases function in internalization and transport to degradation, as well as recycling to the plasma membrane and the Golgi. The numbers in the figure indicate other kinds of Rab GTPases, for example, 14 represented of Rab14. Reproduced with permission from [40]. Copyright 2014 Cold Spring Harbor Laboratory Press.
3.4. Arf subfamily
Similar with Rab protein, the ADP-ribosylation factor (Arf) subfamily protein is the major regulatory factor of the biosynthesis of trafficking vesicle in cells [22]. Different from Rab protein which works in single step of membrane transportation, Arf protein can work in various steps. For example, Arf can control (1) formation of coat protein I (COPI)-coated vesicle supporting antiporter between Golgi and ER; (2) formation of clathrin/adaptin 1 (AP1) compound-related vesicle in trans-Golgi network (TGN) and immature vesicle and (3) formation of endosome containing AP3 [6].
3.5. Ran family
Ran is a kind of small GTPases with higher expressive abundance in eukaryocyte [23]. It can modulate the formation of cell spindle apparatus, cell cycle progression, structure and function of karyotheca, material transport of nucleoplasm, cell redox reaction and RNA nuclear export, RNA synthesis and processing, etc. [24].
Ran participates in the modulation of karyoplasm transport of mammal and yeast. Many nucleoporins of mammal and yeast contain Ran binding domain (RBD). RBD, Ran and nuclear import factor form the tripolymer. After nuclear export, when the receptor compound binds with Ran-GTP, the substrate will be released. The remaining receptors and Ran-GTP dimer will return to cytoplasm and re-participate in the transport of nuclear import factors. Ran also regulates the molecule nuclear export. The nuclear export receptor and its substrate bind with Ran-GTP to form the compound. After nuclear export, Ran-GTP will be hydrolyzed into Ran-GDP to separate receptor and substrate [25].
4. Function of small GTPase and its role in nano delivery system
Small GTPase can regulate various vital movements of cells, including cell growth, differentiation, cell movement, lipid vesicle trafficking, etc. [26], [27]. In term of function, Ras subfamily regulates the gene expression; Rho subfamily regulates cytoskeleton reorganization, cell wall synthesis, cell cycle progression, and MAP kinase signal transduction; Rab subfamily and Sar/Arf subfamily regulate the vesicle trafficking and clathrin formation; Ran subfamily regulates karyoplasm transport, microtubule formation, formation of mitotic spindle apparatus and the assembly of karyotheca after cell division. At present, more and more researches suggest the important role of small GTPase in the nanoparticle transport [28], [29], [30], [31].
4.1. Ras
Ras is a GTP-binding protein involved in membrane ruffling, pinocytosis and the formation of stress fibres. The ras oncogenes exhibit transforming properties due to single point mutation in the sequence coding for the active site of the p21 protein [32], [33], [34]. These mutations lead to changes in cellular proliferation and induce tumorigenic properties. In an earlier study, antisense oligonucleotides directed to point mutations (G→U) in codon 12 of Ha-ras mRNA selectively inhibited the proliferation of cells expressing point mutant H-Ras genes [35]. When it was absorbed by PIHCA NPs, the same effect could be achieved when the concentration was 100 times lower than that of the free oligonucleotide. NPs protected oligonucleotides from rapid intracellular degradation, which led to considerably higher intracellular concentrations of intact oligonucleotides. As a result, they markedly inhibited H-Ras dependent tumor growth in nude mice after subcutaneous injection. These experiments showed that using antisense oligonucleotides to inhibit ras oncogenes can block tumor development even though activation of ras oncogenic could be seen in the early stage of tumor progression. This work was very new in 1994 because RNAi emerged in 1990. In another similar study, researchers also silenced the ras gene. They entrapped the antisense siRNA of Ras inside a GSH-responsive nano transporter to be delivered specifically to a tumor cell. Therefore, it knocked down several differentially expressed genes that are regulated by the Ras activation pathway [36]. Efforts to perform in vivo siRNA delivery with a properly designed GSH-responsive nanocarrier might lead to transformed targeted cancer treatments.
In recent studies, Ras is also used to diagnostics cancer. Researchers degraded human genomic DNA was by sonication and inserted the dye fluorescent quenching quantum dots (QDs) after target hybridization in the microfluidic chip. After the K-ras oncogene hybridized with the probe DNA in the channel, the intercalating dye (TOTO-3) could quench the fluorescence of the QD to detect K-ras gene [37]. Wang et al. detected pancreatic cancer patients’ feces of K-ras gene mutations by magnetic nanoprobe showed that nanoparticles were able to detect K-ras mutations in different stages of pancreatic cancer, with comparable sensitivity and specificity to CA19-9 examination for differentiating pancreatic cancer [38]. These methods are both good examples to detect oncogenes and have good clinical application prospect.
There are also studies on other aspects of Ras. Rotblat et al. described a hitherto unknown cellular signaling particle, rasosome [39]. Its size was nanometer and moved rapidly in an ATP-independent random motion in the cytosol and carried multiple copies of palmitoylated Ras proteins. Although it is an endogenous substance, the author still regards it as nanoparticle.
4.2. Rab
Nearly 3/4 of about 70 human Rab-GTPases are involved in endocytic trafficking [40]. Endosomal membrane transport pathways exhibit a significant plasticity due to receptor signaling and Rab-GTPase-regulated scaffolds. The roles of Rab-GTPases are discussed widely, including (1) compartmentalizing the endocytic pathway into early, recycling, late, and lysosomal routes; (2) coordinating individual transport steps from vesicle budding to fusion (3) effector interaction; and (4) signaling cascades [14], [41], [42], [43], [44].
Becker et al. investigated a strategy in a study, using a small GTPase of the Rab family on DNA-Gold nanoparticles in virtue of a site-specifically attached poly (ethylene glycol) linker and thiol place-exchange reaction [45]. Rab proteins act as the central regulators of vesicle trafficking and participate in vesicular budding, targeting, and fusion [42]. Rab6 was originally identified as a key player of microtubule-dependent retrograde traffic, controlling vesicle trafficking from early endosomes to the TGN through the Golgi apparatus, and from the Golgi to the endoplasmatic reticulum [46]. It appears to participate in a complex network of protein–protein interactions.
Another study applied the rapid multicolor 3D live cell confocal fluorescence microscopy, and adopted transient over expression of small GTPases marking various endocytic membranes, indicating the kinetics of nanoparticle trafficking through early endosomes to late endosomes and lysosomes [47]. It is shown that, after the internalization, the 40 nm polystyrene nanoparticles first pass through an early endosome intermediately decorated with Rab5, but they are rapidly transferred to late endosomes and ultimately lysosomes labeled with Rab9 and Rab7, respectively. 100 nm larger nanoparticles also reach acidic Rab9- and Rab7-positive compartments despite a slower rate than the smaller 40 nm nanoparticles. This study revealed that relatively few nanoparticles can access endocytic recycling pathways, considering the lack of significant colocalization with Rab11. It also demonstrated that this quantitative approach could effectively detect rare events in nanoparticle trafficking, specifically nanoparticles in Rab1A-labeled structures, thereby revealing the extensive intracellular interactions between nanoparticles and intracellular environment.
4.3. Rho
4.3.1. Cdc42
Cdc42 signaling can effectively control cell proliferation, cell polarity, survival and invasion [48], [49], [50]. Knockdown of Cdc42 led to a 46% decrease of the internalization of PEGylated silica-coated iron oxide nanoparticles within 24 h incubation in Hela cells, proving the significance of Cdc42 in this endocytosis mechanism [51]. However, knockdown of Dynamin 2, Flotillin-1, Clathrin and PIP5Kα caused no or only minor effects. Endocytosis of iron oxide nanoparticles in HeLa cells was mainly mediated by Caveolin-1 and Cdc42. Cdc42 could mediate the internalization of nanoparticles was referred in two reviews, but there is no actual example [28], [29]. It is shown here for the first time Cdc42 is involved in endocytosis of silica-coated iron oxide nanoparticles in HeLa cells in vitro. We expect that more nanoparticles could be confirmed whether they are related to Cdc42-mediated endocytosis.
4.3.2. Rac1
As the main regulator in actin cytoskeleton reorganization, Rac1 can affect endocytosis and trafficking, cell cycle progression, adhesion and migration [52], [53]. Importantly, Rac1 controls lamellipodia formation and membrane ruffles after being stimulated by extracellular ligands such as epidermal growth factor (EGF), platelet-derived growth factor (PDGF) or insulin [54]. By using pharmacological inhibitors and in virtue of genetic approaches, SWCNT is verified to be endocytosed through Rac1-GTPase mediated macropinocytosis in normal endothelial cells [55].
Berry et al. synthesized magnetic nanoparticles and derivatized with dextran (DD), followed by comparing with similar underivatized plain particles [56]. Despite of the uptake of both the uncoated and the DD-derivatized particles into the cell, the derivatized particles can induce alterations in cell behavior and morphology compared with the plain particles, suggesting that cell response is dependent on the particles coating. The DD nanoparticles led to elevated clathrin localized to the ring-like actin structures due to changes in actin polymerization in response to activation of the small GTPases Rac1 and Cdc42.
Rac1 also participates in transcriptional modulation of gene expression relying on NFκB, JNK y MAPK activation and later induction of AP-1 transcription factors involved in cell proliferation. Diesel et al. developed synthetic silica nanoparticles which amplified an inflammatory response in macrophages [57]. The activation of Rac1 was induced by silica nanoparticles as well as BCG DNA and is regarded as the critical signaling event that induces both cytoskeleton changes as well as activation of inflammatory cell.
Another study identified a key role of Rac1 signaling in the mediation of increased N-Cadherin expression. Using a real-time Rac1-FRET biosensor, high-creep hydrogels increased Rac1 activation under functional support from observed increases in motility and lamellipodial protrusion rates of human mesenchymal stem cells, providing underlying mechanisms for enhanced commitment towards a SMC lineage and the compensatory increase in spread area (isotonic tension) after a creep-induced loss of cytoskeletal tension on viscoelastic substrates [58].
4.3.3. RhoA
It has been proved that Ras homologous A (RhoA) can promote both cell proliferation and cell invasion [59], [60]. Accumulating data indicate that RhoA protein-dependent cell signaling plays a significant role in the malignant process. Over expression of RhoA in cancer suggests a poor prognosis due to the exacerbated tumor cell proliferation and invasion and tumor angiogenesis [61], [62]. Pille et al. applied encapsulated anti-RhoA siRNA in chitosan-coated polyisohexylcyanoacrylate (PIHCA) nanoparticles in xenografted aggressive breast cancers (MDA-MB-231), demonstrating that it can well inhibit cancer aggressivity in vivo, proving its therapeutic promise for treating aggressive breast cancers or cancers of other origins suffering over expressed RhoA [63].
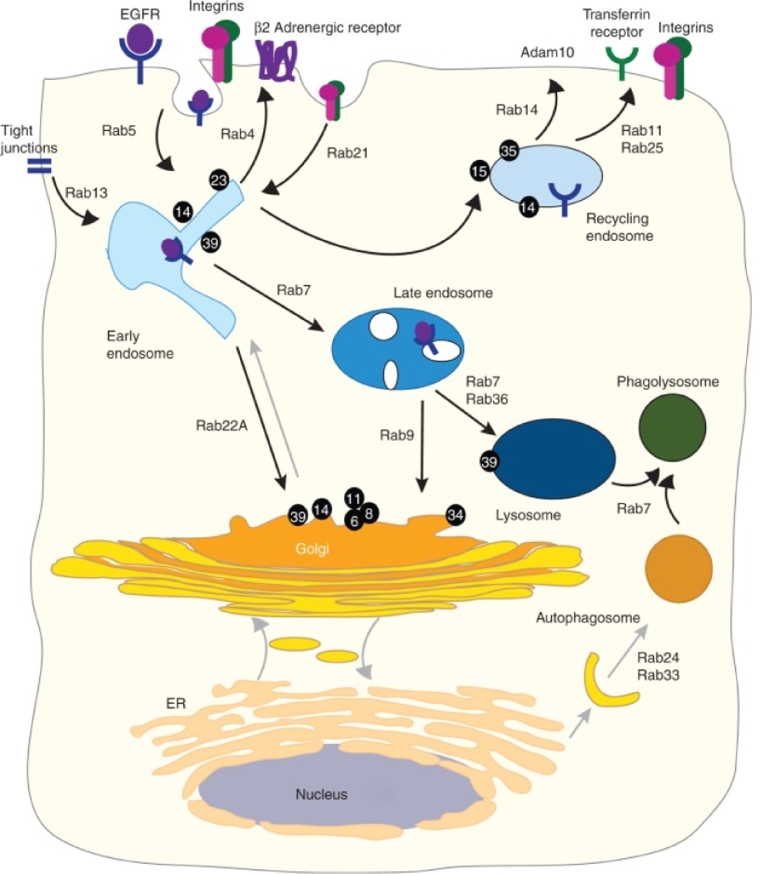
Similarly, because RhoA can obviously inhibit the axonal outgrowth after an injury to the nervous system and the RhoA after activation can counteract regeneration [64], [65]. Microstructured 20 µm thick polymer filaments which act as nerve implants were loaded with chitosan/siRhoA RNA nanoparticles with the purpose to promote nerve regeneration and ensure local delivery of nano therapeutics [66]. Target mRNA was successfully reduced by 65% - 75% and neurite outgrowth was enhanced even in an inhibitory environment. Therefore, the application of nanobio-functionalized implants can work as a novel approach for spinal cord and nerve repair (Fig. 3A,B).
Fig. 3.
(A) Scheme of nanoparticles biofunctionalized nerve implants, are taken up by cells and enable neurite out growth. (B) Neuronal response to nanoparticles by RhoA protein reduction [66]. Copyright (2010), American Chemical Society. (C) Staining of filamentous actin in the AgNP-treated was stronger than that of the control and AgNO3-treated cells [68]. Copyright (2014), Dovepress Ltd. (D) Fibroblasts on RGD-gold squares smaller than 1 mm show altered morphology. Reproduced with permission from [69]. Copyright 2011 Public Library of Science.
Huang et al. designed a mesoporous silica nanoparticle (MSNs), which induces transient but insufficient osteogenic signals in human mesenchymal stem cells (hMSCs) [67]. The uptake of MSNs into hMSCs posed no impact on the cell viability, proliferation and regular osteogenic differentiation of the cells. However, the internalization of MSNs indeed induced actin polymerization and activated the small GTP-binding protein RhoA.
It has also been reported that silver nanoparticles promote osteogenic differentiation of human urine-derived stem cells, induce actin polymerization and increase cytoskeletal tension, and activate RhoA at non-cytotoxic concentrations. However, AgNO3 had no such effect (Fig. 3C) [68].
In another study, in virtue of the nano stencil technique, culture substrates were patterned with gold squares of a width and spacing between 250 nm and 2 mm [69]. The gold was functionalized with RGD peptide as ligand for cellular integrins, and mouse embryo fibroblasts were plated. Small pattern cells showed aberrant fibronectin fibrillogenesis, and the directed migration speed was reduced significantly compared to fibroblasts on 2 mm square patterns. By interfering with RhoA/ROCK signaling eliminated the differences in cellular shape and actin cytoskeleton respectively observed on the large vs. small patterns in the absence of drugs (Fig. 3D).
4.4. Arf6
The role of small GTPase Arf6 in nanoparticle has been seldom studied. In a study of Arf6, the normal rat kidney (NRK) cells and Hela cells co-culture system was employed and the TiO2 NPs transfer was observed [70]. The authors found that the small GTPase Arf6 facilitates the intercellular transfer of smaller NPs and endosomes. However, the transfer vehicle used by TiO2 NPs remains unknown.
4.5. Multiple GTPases
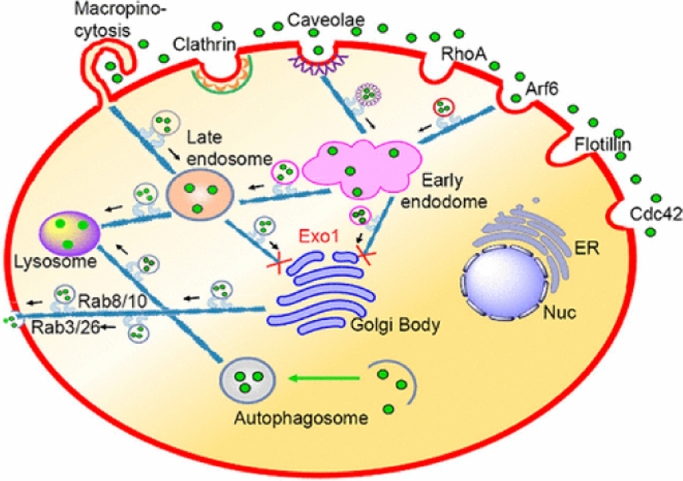
There are also many studies focusing on more than one kind of GTPase. For example, studies found that the aspect ratio (AR) determines the quantity of mesoporous silica nanoparticle uptake. The rods with intermediary AR also induced filopodia, actin polymerization, and activation of small GTP-binding proteins (e.g., Rac1, CDC42) to the greatest, involving the assembly of the actin cytoskeleton and filopodia formation [71]. Ding et al. found the FITC-labeled MSNs-PDA could be colocalized with Caveolae- and Arf6-positive vesicles but could not colocalized with Clathrin-, Flotillin-, Cdc42-, and RhoA-positive vesicles (Fig. 4) [72]. In the EGFP-Rab34 transfected Hela cells, a perfect colocalization was found by Ding et al. between Rab34-marked macropinocytosis and FITC-labeled MSN. This showed that different modifications of the same kind of nanoparticles could also result in different cellular pathways.
Fig. 4.
Schematic representation of the intracellular trafficking network of MSNs. Reproduced with permission from [72]. Copyright 2017 American Chemical Society.
Another research focused on the effect of amphiphilic PCL-PEG nano-micelles on HepG2 cell migration [73]. The nano-micelles with medium PCL and PEG chains increased the expression of Rho GTPases and impeded focal adhesion (FA) components, which accordingly enhanced the motility of HepG2 cells. In contrast, the nano-micelles with large PCL and PEG chains exhibited lower Rho GTPase levels and higher FA components.
It is reported that Zinc Oxide nanoparticles could also induce intercellular adhesion molecule 1 expression through Rac1/Cdc42-MLK3-JNK-c-Jun signaling, which, in comparison, could not be activated in – HUVECs treated by Zinc oxide micro-particles (ZnO-MPs) [74].
5. Conclusion and prospect
Small GTPases are very important regulatory proteins of eukaryotes. They play crucial regulatory function in various movement processes of organism [75], [76], [77]. Different nanoparticles used to investigate the interaction with small GTPases inside cells have been summarized. At present, related researches only cover a few limited types of nanoparticles and GTPases, the action mechanism of the two still requires a systematic interpretation. Small GTPase family has large family members, but their interaction with nanoparticles is still considered very little. We summarized different nanoparticles mentioned in this review to investigate the interaction with small GTPases inside cells and listed them in Table 1 in nanoparticle structure dependent ways. According to this table, we can more clearly see that inorganic nanoparticles are more widely used in the study of small GTPases than organic polymers. But now there are only 21 examples could be found. One of the key issues in this area is how to dig out and identify small GTPase(s) which is/are related to reveal the involved physiological functions and the interaction with nanoparticles. Analysis on structure and function of small GTPases in virtue of methods of biological chemistry, proteomics, molecular biology and genetics, etc. will contribute to a clear understanding about their molecule action mode. It is expected that the exact action mechanisms will be identified in the future, which will be favorable for the designing of better nanomedicines with higher effectiveness for practical application.
Table 1.
Summary of small GTPases related nanoparticles.
| Vehicle | Modification | Mechanism/Aims | GTPases |
|---|---|---|---|
| Polymer nanoparticles | |||
| Micelles [73] | PCL-PEG | The nano-micelles with medium PCL and PEG chains increased the expression of Rho GTPases | Rac1 |
| Cdc42 | |||
| RhoA | |||
| Hydrogels [58] | High-creep | Viscoelastic hydrogels inducted Rac1 activity | Rac1 |
| PIHCA NPs [35] | Anti-Ras siRNA | Inhibition of ras oncogenes by antisense oligonucleotides can block tumor development | Ras |
| PIHCA NPs [63] | Chitosan-coated & anti-RhoA siRNA | Origins suffering over expressed RhoA | RhoA |
| Chitosan NPs [66] | Anti-RhoA siRNA | Blocking of RhoA signal transduction may result in axon regeneration and provide a molecular target for novel therapeutic approaches in the spinal cord | RhoA |
| Inorganic nanoparticles | |||
| Gold nano stencil [69] | RGD peptide | The RhoA/ROCK signal played a role in deformation by the different size of gold nano stencil | RhoA |
| TiO2 NPs [70] | 5/40 nm | Arf6 facilitates the intercellular transfer of smaller 5 nm NPs | Arf6 |
| Mesoporous silica NPs [71] | – | The intermated AR rods activation of small GTP-binding proteins to the greatest | Rac1 |
| CDC42 | |||
| Mesoporous silica NPs [72] | PDA | Arf6 is related to PDA-MSN NPs into the cells | Arf6 |
| Zinc Oxide NPs [74] | – | Induce intercellular adhesion molecule 1 expression through Rac1/Cdc42-MLK3-JNK-c-Jun signaling | Rac1 |
| Cdc42 | |||
| SWCNT [55] | DNA | SWCNT is endocytosed through Rac1 mediated micropinocytosis | Rac1 |
| Quantum Dots [37] | Intercalating dyes | Target hybridization to detect K-ras oncogenes | Ras |
| Gold NPs [45] | Rab6A & DNA | Detailed introduction the immobilization of the Small GTPase Rab6A on DNA–Gold nanoparticles by using a site-specifically attached poly (ethylene glycol) linker and thiol place exchange reaction | Rab |
| Ag NPs [68] | – | AgNPs activated GTP-bound form of RhoA | RhoA |
| Polystyrene NPs [47] | 40/100 nm | Overexpression of small GTPases marking various endocytic membranes, reveal the kinetics of nanoparticle trafficking through early endosomes to late endosomes and lysosomes in living cells | Rab |
| Iron oxide NPs [38] | Probe for K-ras | The magnetic nanoprobe can detect fecal K-ras mutations in different stages of pancreatic cancer | Ras |
| Iron oxide NPs [51] | PEGylated silica-coated | CDC42 is related to the PEGylated SPIONs inside cells | Cdc42 |
| Iron oxide NPs [56] | Derivatized with dextran | The derivatized particles induce alterations in cell behavior and morphology is related to Rac1 | Rac1 |
| Silica NPs [57] | – | Nanoparticles can induce Rac1 related signaling pathways, which amplify an inflammatory response in macrophages | Rac1 |
| Nature naonparticles | |||
| Rasosome [39] | Innate cytosolic nanoparticles | Navigate Ras signaling from different intracellular compartments delivery | Ras |
| Others | |||
| Nano transporter [36] | GSH-responsive | Knock down several differential gene expressions being regulated by Ras-activated pathways like enzyme-linked receptor kinase pathway | Ras |
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
The work was supported by the National Natural Science Foundation of China [81690264], the National Basic Research Program of China [2015CB932100, 2017YFA0205600] and the Innovation Team of the Ministry of Education [BMU20110263].
References
- 1.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313(4):889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 2.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DS, Chen YH. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol. 2012;12(4):458–463. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301(4):1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 5.Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12(2):157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 6.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118(Pt 5):843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 7.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004(250):RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goitre L, Trapani E, Trabalzini L, Retta SF. The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol Biol. 2014;1120:1–18. doi: 10.1007/978-1-62703-791-4_1. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16(13):1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 10.Rak A, Pylypenko O, Durek T. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302(5645):646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 11.Kahn RA, Der CJ, Bokoch GM. The ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J. 1992;6(8):2512–2513. doi: 10.1096/fasebj.6.8.1592203. [DOI] [PubMed] [Google Scholar]
- 12.Haigis KM, Kendall KR, Wang Y. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz MA, Shattil SJ. Signaling networks linking integrins and rho family GTPases. Trends Biochem Sci. 2000;25(8):388–391. doi: 10.1016/s0968-0004(00)01605-4. [DOI] [PubMed] [Google Scholar]
- 16.Rolfe BE, Worth NF, World CJ, Campbell JH, Campbell GR. Rho and vascular disease. Atherosclerosis. 2005;183(1):1–16. doi: 10.1016/j.atherosclerosis.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 18.Bruewer M, Hopkins AM, Hobert ME, Nusrat A, Madara JL. RhoA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am J Physiol Cell Physiol. 2004;287(2):C327–C335. doi: 10.1152/ajpcell.00087.2004. [DOI] [PubMed] [Google Scholar]
- 19.Karnoub AE, Symons M, Campbell SL, Der CJ. Molecular basis for Rho GTPase signaling specificity. Breast Cancer Res Treat. 2004;84(1):61–71. doi: 10.1023/B:BREA.0000018427.84929.5c. [DOI] [PubMed] [Google Scholar]
- 20.Mathur J, Hulskamp M. Signal transduction: Rho-like proteins in plants. Curr Biol. 2002;12(15):R526–R528. doi: 10.1016/s0960-9822(02)01029-1. [DOI] [PubMed] [Google Scholar]
- 21.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 22.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12(6):362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273(36):22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 24.Sazer S, Dasso M. The ran decathlon: multiple roles of Ran. J Cell Sci. 2000;113(Pt 7):1111–1118. doi: 10.1242/jcs.113.7.1111. [DOI] [PubMed] [Google Scholar]
- 25.Kim SH, Arnold D, Lloyd A, Roux SJ. Antisense expression of an Arabidopsis ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell. 2001;13(12):2619–2630. doi: 10.1105/tpc.010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8(23) doi: 10.1186/1478-811X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19(3):625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 28.Lee WH, Loo CY, Traini D, Young PM. Nano- and micro-based inhaled drug delivery systems for targeting alveolar macrophages. Expert Opin Drug Deliv. 2015;12(6):1009–1026. doi: 10.1517/17425247.2015.1039509. [DOI] [PubMed] [Google Scholar]
- 29.Iversen TG, Skotland T, Sandvig K. Endocytosis and intracellular transport of nanoparticles: present knowledge and need for future studies. Nano Today. 2011;6(2):176–185. [Google Scholar]
- 30.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145(3):182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 32.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 33.Miyamoto H, Harada M, Isobe H. Prognostic value of nuclear DNA content and expression of the ras oncogene product in lung cancer. Cancer Res. 1991;51(23 Pt 1):6346–6350. [PubMed] [Google Scholar]
- 34.Mitsudomi T, Steinberg SM, Oie HK. Ras gene mutations in non-small cell lung cancers are associated with shortened survival irrespective of treatment intent. Cancer Res. 1991;51(18):4999–5002. [PubMed] [Google Scholar]
- 35.Schwab G, Chavany C, Duroux I. Antisense oligonucleotides adsorbed to polyalkylcyanoacrylate nanoparticles specifically inhibit mutated Ha-ras-mediated cell proliferation and tumorigenicity in nude mice. Proc Natl Acad Sci USA. 1994;91(22):10460–10464. doi: 10.1073/pnas.91.22.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doss CG, Debottam S, Debajyoti C. Glutathione-responsive nano-transporter-mediated siRNA delivery: silencing the mRNA expression of Ras. Protoplasma. 2013;250(3):787–792. doi: 10.1007/s00709-012-0451-1. [DOI] [PubMed] [Google Scholar]
- 37.Noh HN, Kim JS. Detection of K-ras oncogene from the human genomic DNA using ultrasonication and a quantum dots-based microfluidic chip. J Nanosci Nanotechnol. 2013;13(9):6033–6037. doi: 10.1166/jnn.2013.7654. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Wang J, Chen F, Zhong Z, Qi L. Detection of K-ras gene mutations in feces by magnetic nanoprobe in patients with pancreatic cancer: a preliminary study. Exp Ther Med. 2018;15(1):527–531. doi: 10.3892/etm.2017.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotblat B, Yizhar O, Haklai R, Ashery U, Kloog Y. Ras and its signals diffuse through the cell on randomly moving nanoparticles. Cancer Res. 2006;66(4):1974–1981. doi: 10.1158/0008-5472.CAN-05-3791. [DOI] [PubMed] [Google Scholar]
- 40.Wandinger-Ness A, Zerial M. Rab Proteins and the Compartmentalization of the Endosomal System. Cold Spring Harbor Perspect Biol. 2014;6(11):a–a022616. doi: 10.1101/cshperspect.a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somsel Rodman J, Wandinger-Ness A. Rab GTPases coordinate endocytosis. J Cell Sci. 2000;113(Pt2):183–192. doi: 10.1242/jcs.113.2.183. [DOI] [PubMed] [Google Scholar]
- 42.Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9(4):496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 43.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116(2):153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 44.Schimmoller F, Simon I, Pfeffer SR. Rab GTPases, directors of vesicle docking. J Biol Chem. 1998;273(35):22161–22164. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- 45.Becker CF, Marsac Y, Hazarika P, Moser J, Goody RS, Niemeyer CM. Functional immobilization of the small GTPase Rab6A on DNA-Gold nanoparticles by using a site-specifically attached poly(ethylene glycol) linker and thiol place-exchange reaction. Chembiochem. 2007;8(1):32–36. doi: 10.1002/cbic.200600422. [DOI] [PubMed] [Google Scholar]
- 46.Darchen F, Goud B. Multiple aspects of Rab protein action in the secretory pathway: focus on Rab3 and Rab6. Biochimie. 2000;82(4):375–384. doi: 10.1016/s0300-9084(00)00219-4. [DOI] [PubMed] [Google Scholar]
- 47.Sandin P, Fitzpatrick LW, Simpson JC, Dawson KA. High-speed imaging of Rab family small GTPases reveals rare events in nanoparticle trafficking in living cells. ACS Nano. 2012;6(2):1513–1521. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
- 48.Arias-Romero LE, Chernoff J. Targeting Cdc42 in cancer. Expert Opinion Therapeutic Targets. 2013;17(11):1263–1273. doi: 10.1517/14728222.2013.828037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9(1):86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 50.Etienne-Manneville S. Cdc42–the centre of polarity. J Cell Sci. 2004;117(Pt 8):1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 51.Bohmer N, Jordan A. Caveolin-1 and CDC42 mediated endocytosis of silica-coated iron oxide nanoparticles in HeLa cells. Beilstein J Nanotechnol. 2015;6:167–176. doi: 10.3762/bjnano.6.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardama GA, Alonso DF, Gonzalez N. Relevance of small GTPase Rac1 pathway in drug and radio-resistance mechanisms: opportunities in cancer therapeutics. Crit Rev Oncol Hematol. 2018;124:29–36. doi: 10.1016/j.critrevonc.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther. 2013;12(10):1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29(4):356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharya S, Roxbury D, Gong X, Mukhopadhyay D, Jagota A. DNA conjugated SWCNTs enter endothelial cells via Rac1 mediated macropinocytosis. Nano Lett. 2012;12(4):1826–1830. doi: 10.1021/nl204058u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berry CC, Wells S, Charles S, Aitchison G, Curtis AS. Cell response to dextran-derivatised iron oxide nanoparticles post internalisation. Biomaterials. 2004;25(23):5405–5413. doi: 10.1016/j.biomaterials.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 57.Diesel B, Hoppstadter J, Hachenthal N. Activation of Rac1 GTPase by nanoparticulate structures in human macrophages. Eur J Pharm Biopharm. 2013;84(2):315–324. doi: 10.1016/j.ejpb.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 58.Cameron AR, Frith JE, Gomez GA, Yap AS, Cooper-White JJ. The effect of time-dependent deformation of viscoelastic hydrogels on myogenic induction and Rac1 activity in mesenchymal stem cells. Biomaterials. 2014;35(6):1857–1868. doi: 10.1016/j.biomaterials.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 59.Schmitz AA, Govek EE, Bottner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261(1):1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- 60.Aznar S, Lacal JC. Rho signals to cell growth and apoptosis. Cancer Lett. 2001;165(1):1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- 61.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87(6):635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denoyelle C, Vasse M, Korner M. Cerivastatin, an inhibitor of HMG-CoA reductase, inhibits the signaling pathways involved in the invasiveness and metastatic properties of highly invasive breast cancer cell lines: an in vitro study. Carcinogenesis. 2001;22(8):1139–1148. doi: 10.1093/carcin/22.8.1139. [DOI] [PubMed] [Google Scholar]
- 63.Pille JY, Li H, Blot E. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum Gene Ther. 2006;17(10):1019–1026. doi: 10.1089/hum.2006.17.1019. [DOI] [PubMed] [Google Scholar]
- 64.Shao Z, Browning JL, Lee X. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron. 2005;45(3):353–359. doi: 10.1016/j.neuron.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 65.Mi S, Lee X, Shao Z. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7(3):221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 66.Mittnacht U, Hartmann H, Hein S. Chitosan/siRNA nanoparticles biofunctionalize nerve implants and enable neurite outgrowth. Nano Lett. 2010;10(10):3933–3939. doi: 10.1021/nl1016909. [DOI] [PubMed] [Google Scholar]
- 67.Huang DM, Chung TH, Hung Y. Internalization of mesoporous silica nanoparticles induces transient but not sufficient osteogenic signals in human mesenchymal stem cells. Toxicol Appl Pharmacol. 2008;231(2):208–215. doi: 10.1016/j.taap.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 68.Qin H, Zhu C, An Z. Silver nanoparticles promote osteogenic differentiation of human urine-derived stem cells at noncytotoxic concentrations. Int J Nanomed. 2014;9:2469–2478. doi: 10.2147/IJN.S59753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danen EHJ, Lutz R, Pataky K. Nano-stenciled RGD-gold patterns that inhibit focal contact maturation induce lamellipodia formation in fibroblasts. PLoS One. 2011;6(9):e25459. doi: 10.1371/journal.pone.0025459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schoelermann J, Burtey A, Allouni ZE, Gerdes HH, Cimpan MR. Contact-dependent transfer of TiO(2) nanoparticles between mammalian cells. Nanotoxicology. 2016;10(2):204–215. doi: 10.3109/17435390.2015.1048322. [DOI] [PubMed] [Google Scholar]
- 71.Meng H, Yang S, Li Z. Aspect ratio determines the quantity of mesoporous silica nanoparticle uptake by a small GTPase-dependent macropinocytosis mechanism. ACS Nano. 2011;5(6):4434–4447. doi: 10.1021/nn103344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding L, Zhu X, Wang Y. Intracellular fate of nanoparticles with polydopamine surface engineering and a novel strategy for exocytosis-inhibiting, lysosome impairment-based cancer therapy. Nano Lett. 2017;17(11):6790–6801. doi: 10.1021/acs.nanolett.7b03021. [DOI] [PubMed] [Google Scholar]
- 73.Shen Y, Leng M, Yu H. Effect of amphiphilic PCL-PEG nano-micelles on HepG2 cell migration. Macromol Biosci. 2015;15(3):372–384. doi: 10.1002/mabi.201400376. [DOI] [PubMed] [Google Scholar]
- 74.Li CH, Liao PL, Shyu MK. Zinc oxide nanoparticles-induced intercellular adhesion molecule 1 expression requires Rac1/Cdc42, mixed lineage kinase 3, and c-Jun N-terminal kinase activation in endothelial cells. Toxicol Sci. 2012;126(1):162–172. doi: 10.1093/toxsci/kfr331. [DOI] [PubMed] [Google Scholar]
- 75.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144(6):1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Charest PG, Firtel RA. Big roles for small GTPases in the control of directed cell movement. Biochem J. 2007;401(2):377–390. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 78.Pantarelli C, Welch HCE. Rac-GTPases and Rac-GEFs in neutrophil adhesion, migration and recruitment. Eur J Clin Invest. 2018:e12939. doi: 10.1111/eci.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]