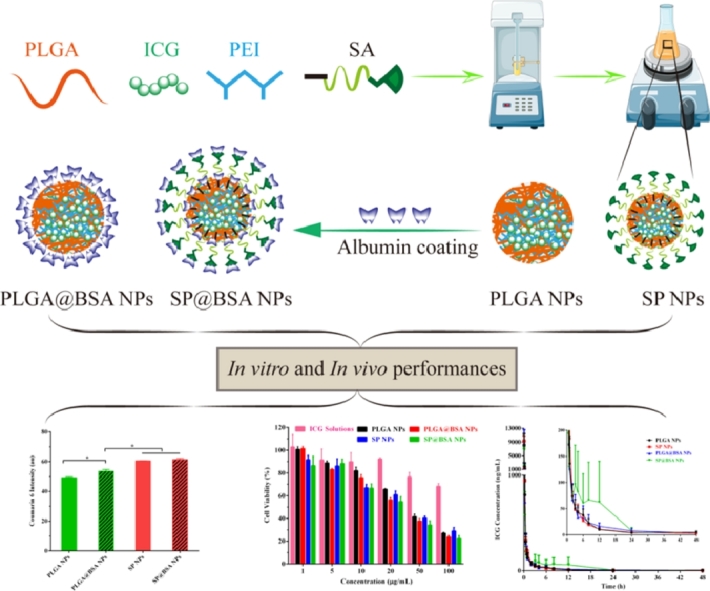
Abstract
Preformed albumin corona of albumin-nonselective nanoparticles (NPs) is widely exploited to inhibit the unavoidable protein adsorption upon intravenous administration. However, very few studies have concerned the preformed albumin corona of albumin-selective NPs. Herein, we report a novel type of albumin-selective NPs by decorating 6-maleimidocaproyl polyethylene glycol stearate (SA) onto PLGA NPs (SP NPs) surface, taking albumin-nonselective PLGA NPs as control. PLGA NPs and SP NPs were prepared by emulsion-solvent evaporation method and the resultant NPs were in spherical shape with an average diameter around 180 nm. The corresponding albumin-coating PLGA NPs (PLGA@BSA NPs) and albumin-coating SP NPs (SP@BSA NPs) were formulated by incubating SP NPs or PLGA NPs with bovine serum albumin solution, respectively. The impact of albumin corona on particle characteristics, stability, photothermal effect, cytotoxicity, cell uptake, spheroid penetration and pharmacokinetics was investigated. In line with previous findings of preformed albumin coating, PLGA@BSA NPs exhibited higher stability, cytotoxicity, cell internalization and spheroid penetration performances in vitro, and longer blood circulation time in vivo than those of albumin-nonselective PLGA NPs, but albumin-selective SP NPs is capable of achieving a comparable in vitro and in vivo performances with both SP@BSA NPs and PLGA@BSA NPs. Our results demonstrate that SA decorated albumin-selective NPs pave a versatile avenue for optimizing nanoparticulate delivery without preformed albumin corona.
Keywords: Preformed albumin corona, Albumin-nonselective PLGA NPs, Albumin-selective SP NPs, In vitro and in vivo performances
Graphic abstract
1. Introduction
The rapid development of nanomaterials in biomedical and biotechnological applications offers nanoparticles a great potential in disease diagnosis and therapy [1], [2]. On account of the high surface free energy, non-specific plasma-protein opsonization occurs immediately upon NPs injection into biological fluids, forming the so-called “protein corona”. The subsequently formed corona changes the original features of NPs including particle size, zeta potential and surface composition, resulting in a “biological identity” that is recognized by cells in vivo, whereas the “pristine identity” was extensively masked [3], [4], [5]. Actually, the following biological corona could also change particle-cell responses, such as NPs cellular uptake and in vivo distribution, and even trigger the accompanying rapid clearance and nanotoxicity [1], [6], [7], [8]. To address these challenges, in vitro preforming albumin corona on NPs is widely employed to inhibit opsonin proteins adsorption and prolong the blood circulation capability. And this strategy has been observed on a diverse range of albumin-nonselective NPs, including Poly-3-hydroxybutyrate-co-3-hydroxyhexanoate (PHBHHx) NPs [9], quantum dots [10], [11], superparamagnetic iron oxide NPs [1], [6], [12], [13], nanocrystals [14], [15], graphene oxide [16], [17], Au NPs [18], manganese dioxide [15], [19] and so forth. As expected, this in vitro preformed albumin corona indeed confers albumin-nonselective NPs decreased plasma proteins adsorption and complement activation, enhanced circulation stability, and improved tumor accumulation in vivo. However, the complex and expensive formulation development process also made in vitro coating strategy less attractive.
These issues motivates researchers to engineer novel albumin-selective NPs. Albumin, the most abundant protein in plasma (55%), has been tremendously explored for its native circulation capacity, low-immunogenicity and good stability [20], [21]. And indeed, successful albumin-based nanomedicines, such as covalent-binding Doxorubicin-EMCH (DOXO-EMCH), have been applied in clinical trials. Precise one maleimide group was selectively reacted with one cysteine-34 residue of endogenous circulating albumin within 2–5 min [21], [22].
Indocyanine green (ICG) is chosen as model drug due to its approval by the U.S. Food and Drug Administration (FDA) and effective photon dynamic/thermal conversion capacity [23], [24], [25], [26]. Despite the above advantages, nevertheless, its clinical application is extremely limited owing to its short-circulation, unstable properties such as aggregation and degradation in aqueous solution.
Given the rapid conjugation of maleimide group with the cysteine-34 residue of albumin and the dilemma of ICG application, we herein report a strategy to decorate maleimide groups on PLGA NPs surfaces to fabricate albumin-selective NPs, and then to unravel the impact of preformed albumin corona on in vitro and in vivo performances of the engineered albumin-selective NPs, taking albumin-nonselective PLGA NPs as control. In this study, a novel bioconjugate 6-maleimidocaproyl polyethylene glycol stearate (SA) was synthesized, and then SA was decorated on PLGA NPs by emulsion-solvent evaporation method. The resultant NPs possessed a higher ICG loading efficiency due to electrostatic interaction between ICG and polyethyleneimine (PEI). Besides the above NPs characterization, the impact of albumin corona on stability, photothermal conversion capability, cytotoxicity, cell uptake, spheroid penetration and in vivo pharmacokinetics of albumin-selective SP NPs, was also evaluated.
2. Materials and methods
2.1. Materials
Poly(lactic-co-glycolic acid) (PLGA) was purchased from Jinan Daigang Biomaterial Co., Ltd. Indocyanine Green (ICG) was obtained from Bomei Pharmaceutical New Technology Development Co., Ltd. Polyethyleneimine (PEI, Mw = 10 kDa) was bought from Aladdin (Shanghai, China). Polyethylene Glycol Monostearate was purchased from Tokyo chemical industry Co., Ltd. 3-(4, 5-dimthyl-2-thiazolyl)−2, 5-dipphenyl-2H-terazolium bromide (MTT) and trypsin-EDTA were purchased from Sigma-Aldrich (USA). Coumarin-6 was purchased from Sigma (St. Louis, MO, USA). All other solvents and reagents utilized in this study were analytical or HPLC grade.
2.2. Synthesis of 6-maleimidocaproyl polyethylene glycol stearate (SA)
Synthesis of 6-maleimidocaproic acid
to Istvan Toth et al. report [27]. Briefly, 6-aminocaproic acid (30 mmol) and maleic anhydride (30 mmol) were refluxed in AcOH (about 70 ml) for 6 h. Then, 30 mmol Ac2O was added dropwise and refluxed for a further 2 h. After reaction, the objective products were purified using silica column chromatography after drying in vacuum. The chemical structure of 6-maleimidocaproic acid was confirmed by MS, NMR with CDCl3 as solvent and IR.
Synthesis of 6-maleimidocaproyl chloride
To synthesize 6-maleimidocaproyl chloride, 6-maleimidocaproic acid was dissolved in oxalyl chloride and refluxed with stirring at 70 °C for 2 h, and the extra oxalyl chloride was removed using rotary evaporator. The chemical structure of 6-maleimidocaproyl chloride was confirmed by MS, NMR with CDCl3 as solvent and IR
-
Synthesis of SA
6-maleimidocaproyl chloride (20 mmol) was dissolved in 40 ml dehydrated dichloromethane and then 20 ml dehydrated dichloromethane containing polyethylene glycol stearate (8 mmol) was added dropwise into 6-maleimidocaproyl chloride solution with stirring at 40 °C for 24 h. After reaction, the mixture was separated over SiO2. The chemical structure of SA was confirmed by NMR with CDCl3 as solvent and IR.
2.3. Preparation of SP and SP@BSA NPs
SP NPs were prepared by emulsion-solvent evaporation method. 9 mg SA, 30 mg PLGA, 1.5 mg ICG and 1.5 mg PEI were dissolved in 0.6 ml mixed solution of dehydrated dichloromethane and methanol (2:1, v/v) and then 10 ml deionized water was added into it. Next, the mixture was ultrasound using probe-type sonifier (JY92-2D, Scientz, China) for 5 min in ice bath; the organic solvents were evaporated with magnetic stirring in darkness for 6 h. The excess methanol was removed under vacuum at 40 °C in dark. PLGA NPs were prepared in the same way as described above without SA. Coumarin-6 labeled PLGA (C6-labeled PLGA NPs) or SP NPs (C6-labeled SP NPs) were prepared similarly except for changing DTX to coumarin-6. Finally, the organic solvent-free NPs were stored at 4 °C.
The preformed albumin corona SP NPs (SP@BSA NPs) or preformed albumin corona PLGA NPs (PLGA@BSA NPs) were constructed according to Peng et al. [1], [9]. Briefly, 1 ml SP NPs (3 mg/ml) was incubated with bovine serum albumin (BSA) solution (20 mg/ml in saline) at 37 °C for 2 h, and then SP@BSA NPs and PLGA@BSA NPs were isolated by centrifugation (13 000 rpm) at 4 °C for 15 min, respectively.
2.4. Characterizations of SP and PLGA NPs
2.4.1. Size distribution and zeta potential
The particle size, size distribution and zeta potential of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs were measured by dynamic light scattering (DLS) method with zetasizer instrument (Nano ZS, Malvern Co., UK), and the measurements were repeated in triplicate.
2.4.2. Morphology of SP and PLGA NPs
The morphology of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs were explored by transmission electron microscope (TEM) (H-600, Hitachi, Japan). The NPs were negatively stained with 0.2% phosphotungstic acid aqueous solutions before observation under TEM.
2.4.3. Infrared spectra (IR) of SP@BSA and PLGA@BSA NPs
The albumin corona of SP@BSA NPs and PLGA@BSA NPs were monitored by infrared spectrometer (Bruker IFS55, Germany). Infrared spectra of SP@BSA NPs and PLGA@BSA NPs (freeze without protectant) were recorded at the frequency ranging from 400 cm−1 to 4000 cm−1 to determine the frequency changes.
2.5. Encapsulation efficiency of SP NPs and PLGA NPs
The ICG loading efficiency of SP/PLGA NPs was evaluated as the percentage of entrapped ICG with respect to the total ICG added. In this study, the encapsulation efficiency was determined by measuring the amount of centrifugal supernatant after NPs isolation [25].
Where ICGTotal is the amount of ICG added in the preparation; ICGSupernatan t is the amount of ICG in the centrifugal supernatant after NPs isolation.
2.6. In vitro ICG release profiles
The in vitro ICG release performance was estimated against pH 7.4 PBS by the dialysis method. Briefly, 2 ml ICG, SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs were sealed in a dialysis bag (MWCO = 14 kDa, Spectrum Laboratories, USA) and incubated in 30 ml of release medium at 37 °C under orbital shaking (100 rpm), respectively. At designated intervals, 2 ml samples were withdrawn and replaced with equivalent fresh medium. The maximum ICG absorbance was evaluated using Thermo scientific varioskan flash. And the corresponding ICG leakage profiles were fitted by zero-order, first-order, Higuchi, Weibull and Ritger-Peppas model, respectively.
2.7. Circular dichroism spectroscopy
Circular dichroism (CD) was applied to assess the conformational changes of BSA on the surfaces of SP@BSA NPs and PLGA@BSA NPs [28]. Both the measurements were probed using CD spectropolarimeter (Bio-Logic MOS 450, France, Grenoble) in a 1 cm path length quartz cell from 190 to 400 nm at 37 °C [29].
2.8. Measurements of temperature
SP NPs, PLGA NPs, SP@BSA NPs, PLGA@BSA NPs and free ICG solution (100 µg/ml of ICG) were exposed to an 808 nm laser product (Changchun, China) at 1.5 W/cm2 for 3 min. During irradiation, the temperature was measured every 30 s.
2.9. Colloids and UV stability of NPs
The colloidal stability of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs was evaluated by monitoring particle size and zeta potential of the NPs [30].
The absorption spectra stability of ICG, SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs at different times were detected on an UV–vis spectrometer (Shimadzu, Japan) [25].
2.10. Gel electrophoresis and confocal laser scanning microscope
The harvest of soft/hard corona from PLGA NPs and SP NPs was carried out by the centrifugation method [31]. Briefly, SP@BSA NPs and PLGA@BSA NPs were washed by repeated centrifugation (16 000 g for 10 min) and resuspension in water with supernatants collected after each wash, respectively. After five times wash, the sample was repeatly centrifugated and resuspended in buffer containing 6% sodium dodecyl sulfate (SDS) solution to remove the hard adsorption from the NPs surface twice as above. Supernatant was diluted by 50% in Laemmli buffer (solarbio, china), boiled for 5 min, and then loaded onto 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The supernatant from the first wash was diluted 10 times with additional amount of water to avoid overloading the gel. Proteins were stained with Coomassie brilliant blue for 20 min. All the samples were run in triplicate.
To testify that albumin-selective SP NPs could covalently conjugate albumin, we first incubated C6-labeled SP NPs with rhodamine-labeled BSA at 37 °C for 2 h. Then the NPs were centrifuged as above to remove soft/hard corona. The covalently linked BSA corona was observed using confocal laser scanning microscope (CLSM, FluoViewFV1000, Olympus, Japan) [31], [32].
2.11. Cytotoxicity evaluation
The in vitro cytotoxicity of free ICG solution, SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs was determined by MTT assay on 4T1 cell lines. Briefly, cells were incubated in 200 µL culture medium in 96-well plates at a density of 5000 cells/well for 24 h. Then, the cells were exposed to serial dilutions of ICG, SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs, irradiated with 808 nm NIR at 1.5 W/cm2 for 3 min and further incubated for 48 h. The cells without any treatment were used as control. At the end of the incubation, 20 µl of MTT (5 mg/ml) was added, and the plates were incubated for additional 4 h at 37 °C. The medium was then discarded, and the formed formazan crystals were dissolved in DMSO (200 µl). The absorbance of samples was determined at the wavelength of 570 nm by microplate reader. Each concentration samples was tested in triplicate.
2.12. Cellular uptake and uptake mechanisms
Cellular uptake was evaluated in 4T1 cell. Cells were seeded in 12-well plates containing 1 ml of media at a density of 8 × 104 cells/well for 24 h. The medium was replaced with fresh medium containing SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs (ICG concentration of 30 µg/ml) at 37 °C for 2 h. The cells were washed thrice with ice-cold PBS, and analyzed using a flow cytometer (FACSCalibur, BD Biosciences).
Endocytotic pathways of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs were studied using 4T1 cells. 4T1 cells were seeded in 12-well plates containing 1 ml of media at a density of 8 × 104 cells/well for 24 h. Next, the cells were pre-incubated with serum-free medium consisting of different inhibitors, sodium azide (6 µg/ml), chlorpromazine (5 µg/ml), quercetin (6 µg/ml), indomethacin (3 µg/ml), colchicine (8 µg/ml) and ammonium chloride (10 mM) for 1 h. Thereafter, the medium was replaced by SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs with serum medium containing the corresponding inhibitors for another 2 h at 37 °C, respectively. Additionally, cells were also treated with the NPs at 4 °C for 2 h. Finally, the cells were evaluated for quantitative analysis by flow cytometry.
2.13. 4T1 spheroid penetration
The penetration capability of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs was assessed in three-dimensional 4T1 spheroids by CLSM. In short, 1 × 104/well cells were seeded into 96-well round-bottom plates and centrifuged at 1500 g for 15 min. Then, the cells were cultured at 37 °C for 5 d to form compact spheroids (diameter approximately 500 µm). Next, SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs at an equivalent ICG concentration of 30 µg/ml were added and incubated at 37 °C for 4 h, respectively. The tumor spheroid was rinsed with ice-cold PBS three times before observation under CLSM.
2.14. Pharmacokinetic profiles
The Sprague-Dawley rats (200–250 g) were randomly divided into four groups for pharmacokinetics studies according to the Guide for Care and Use of Laboratory Animals of Shenyang Pharmaceutical University. SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs were intravenously (I.V.) injected at an equivalent ICG dose of 4 mg/kg, respectively. At predetermined time points, 0.3 ml blood samples were collected by centrifugation at 13 000 rpm for 10 min, and frozen at −80 °C. The concentration of ICG in the blood samples was determined by a validated ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) method on an ACQUITY UPLC™ system (Waters Co., Ltd., Milford, MA, USA). The related pharmacokinetic parameters were achieved using DAS 2.0 software.
2.15. Statistical analysis
All results were expressed as mean or mean ± SD (standard deviation). One-way ANOVA or t test was applied to probe the significance in the experiments. * P < 0.05, ** P < 0.01 and *** P < 0.001.
3. Results and discussions
3.1. Synthesis and characterization of SA
6-maleimidocaproic acid was synthesized by 6-aminocaproic acid and maleic anhydride according to Istvan Toth et al. report [27]. Then, the product was used as a reactant to formulate highly active 6-maleimidocaproyl chloride. The novel SA conjugate was synthesized by esterificating polyethylene glycol stearate (n = 45) and 6-maleimidocaproyl chloride. The concrete synthesis scheme was illustrated in Fig. S1. Furthermore, the corresponding chemical structures of 6-maleimidocaproic acid, 6-maleimidocaproyl chloride and SA were confirmed by mass spectrum, NMR spectrum in CDCl3 or infrared spectra, respectively (Fig. S2-6 and Supporting information).
3.2. Preparation and characterization of NPs
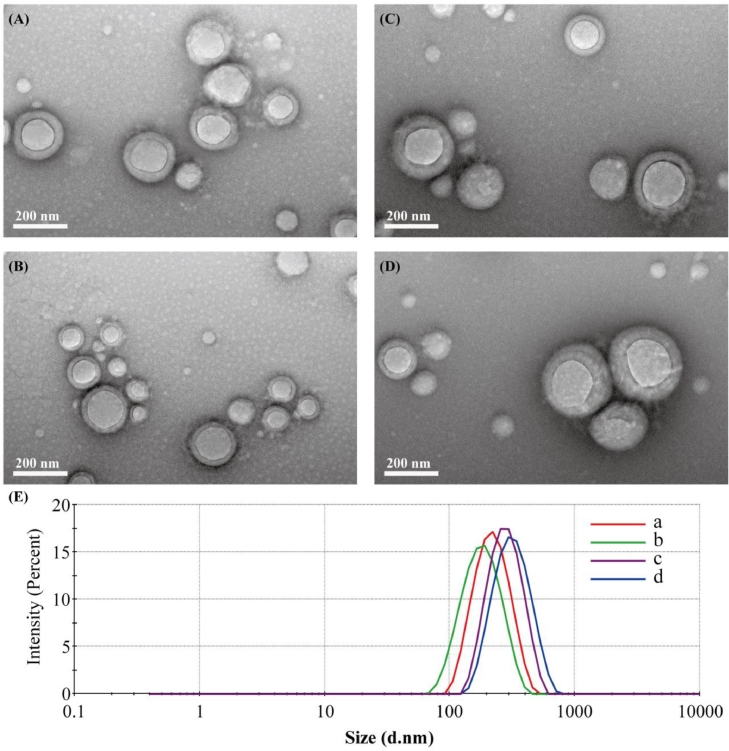
The application of ICG in clinical practices is greatly limited by its high solubility, poor stability such as aggregation and degradation in aqueous solution, and the challenge to efficiently load ICG into nanoparticle core is supposed to be difficult [23], [33], [34], [35]. In this work, Our formulated PLGA NPs and SP NPs exhibited an enhanced ICG encapsulation efficiency 79% and 85% via electrostatic interaction between ICG and PEI, respectively, which were higher than that (60%) reported by Shao and co-workers [36]. As revealed in Fig. 1 and Table 1, the designed NPs both exhibited spherical morphology with a diameter of around 180 nm, implying that the introduction of SA into this formulation had no significant difference on particle size. Moreover, the zeta potential of SP NPs and PLGA NPs was around 8 mV, indicating that ICG/PEI complexes were almost entrapped in nanoparticle hydrophobic core. Compared with the spectrum of free ICG, a significant red-shift of ICG absorption peak (ranging from 770 to 805 nm) emerged in the spectra of both SP NPs and PLGA NPs. These results evidenced that ICG were loaded into the NPs and there were obvious ICG intermolecular aggregation in SP NPs and PLGA NPs due to the substantially electrostatic interaction between ICG and PEI. Meanwhile, the existence of external soft/hard BSA corona did not influence the state of entrapped ICG.
Fig. 1.
Transmission electron microscopy (TEM) images of (A) PLGA NPs; (B) SP NPs (C) PLGA@BSA NPs and (D) SP@BSA NPs and (E) particle size distribution of NPs (a) PLGA NPs; (b) SP NPs (c) PLGA@BSA NPs and (d) SP@BSA NPs.
Table 1.
Physicochemical characterization of PLGA NPs, PLGA@BSA NPs, SP NPs and SP@BSA NPs. (mean ± SD, n = 3).
| Preparations | Size (nm) | Zeta (mV) | PDI | EE (%) |
|---|---|---|---|---|
| PLGA NPs | 186.5 ± 14.1 | 7.82 ± 1.09 | 0.27 ± 0.06 | 78.87 ± 3.11 |
| PLGA@BSA NPs | 222.1 ± 7.0 | −5.05 ± 0.30 | 0.34 ± 0.05 | – |
| SP NPs | 173.4 ± 2.7 | 8.74 ± 0.22 | 0.13 ± 0.03 | 84.79 ± 2.48 |
| SP@BSA NPs | 271.8 ± 6.9 | −8.06 ± 0.36 | 0.16 ± 0.04 | – |
Substantive studies have demostrated that incubation of NPs with proteins results in immidiate protein adsoprtion on NPs, and the chaperoned corona confers NPs a new physichemical characteristic and even a different fate in vivo [3], [13], [31], [37]. In coincidence with these performances, an increase in particle size and a down-converted surface charge were observed for PLGA@BSA NPs and SP@BSA NPs after forming albumin corona on the surface. Interstingly, the albumin coating PLGA@BSA NPs size (222 nm) is smaller than SP@BSA NPs (270 nm), which might be ascribed to the presence of PEG shell and maleimide-thiol bind between albumin and maleimide at the top of PEG chain [21]. The zeta potential of SP@BSA NPs and PLGA@BSA NPs were converted to −8 mV and −5 mV after preformed albumin coating, respectively. Furthermore, the IR spectrum of SP@BSA NPs displayed a broad absorption peak from 2900 to 3550 cm-1 due to the exsitence of hydroxyl/amino groups of BSA, and exhibited a blue-shift of carbonyl (from 1740 to 1660 cm-1) owing to the strength vibration of BSA amide bonds in comparsion with the spectrum of SP NPs (Fig. S5). Additionally, the IR changes of PLGA@BSA NPs were analyzed in the supporting information (Fig. S6) as well. All the results indicated the formation of albumin corona on SP NPs and PLGA NPs.
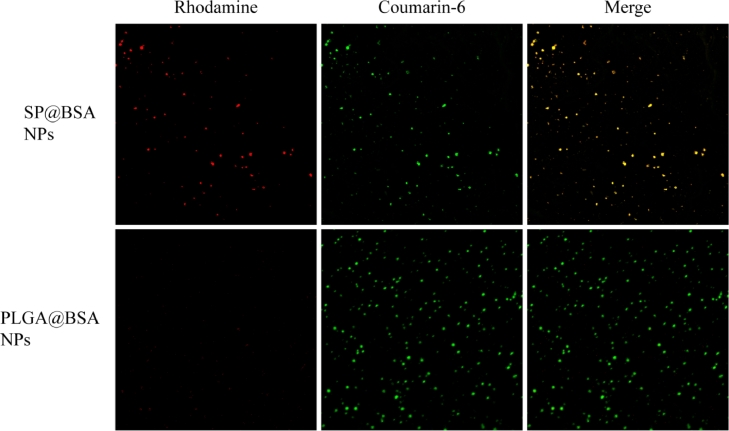
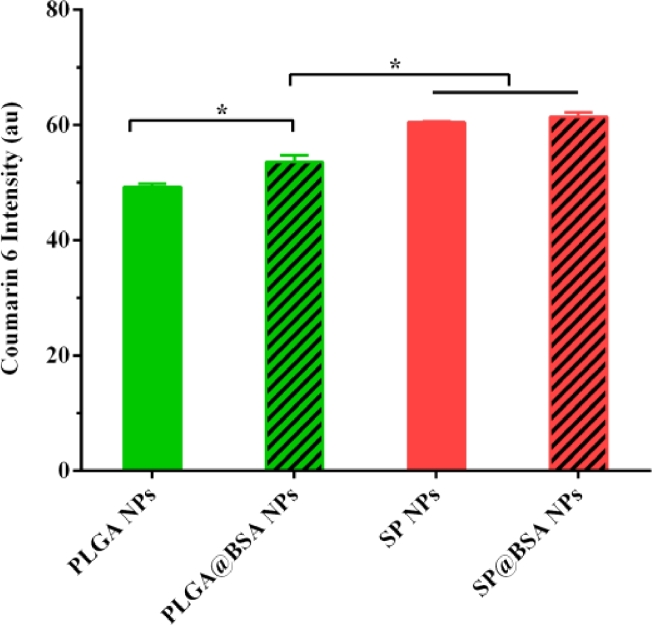
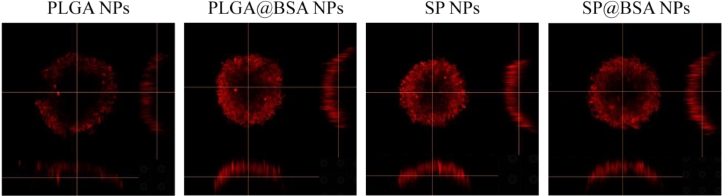
We also collected the soft and hard corona by repeated centrifugation and resuspension in water and 6% SDS solution, respectively. The BSA in the supernatant was detected using SDS-PAGE with a Coomassie protein stain. It should be obviously noticed that BSA was non-selectively adsorped on SP NPs and PLGA NPs to form soft and hard albumin corona. Different from the only non-specific adsorption on PLGA@BSA NPs, there was BSA residue on SP@BSA NPs due to the covalent maleimide-thiol linker between BSA and maleimide at the top of PEG chain even after the removal of soft and hard corona (Fig. 2) [13]. To further testify that albumin-selective SP NPs could covalently conjugate albumin, we employed CLSM to observe SP@BSA NPs and PLGA@BSA NPs after the removal of the soft/hard corona. As shown in Fig. 3, the rhodamine-labeled BSA red fluorescent is very weaker while the green fluorescent of C6-labeled PLGA NPs is very stronger (Fig. 3), which meant that there was none or negligible BSA on soft/hard corona-removed PLGA@BSA NPs. In contrast, the rhodamine-labeled BSA and soft/hard corona-removed C6-labeled SP NPs were overlapped and remained as a single NP or NPs complexes. This results visually verify that albumin-selective SP NPs could covalently conjugate to BSA [32]. Meanwhile, CD spectroscopy was also utilized to measure the structure of BSA corona on SP@BSA NPs and PLGA@BSA NPs. Compared with the spectral absorption profiles of BSA, negligible structure secondary change of SP@BSA NPs and PLGA@BSA NPs (Fig. S7) was detected, indicating the structure and activity of BSA on NPs were little influenced, which might avoid the recognition by macrophage cells in vivo.
Fig. 2.
The characterization of covalent-albumin corona on SP@BSA NPs by SDS-PAGE, taking PLGA@BSA NPs as control. After each centrifugation and resuspension, the supernatant (S) was loaded onto the SDS-PAGE. S1 was diluted 10 times with additional amount of water to avoid overloading the gel. SDS was used to remove the hard adsorption BSA from the NP surface. Meanwhile, the residual NPs were also loaded onto gel to identify the covalent- conjugated BSA.
Fig. 3.
The CLSM colocalization image of rhodamine-labeled BSA and C6-labeled SP NPs images of PLGA NPs and SP NPs after removal soft/hard corona, taking C6-labeled PLGA NPs as control.
3.4. Release tests in vitro
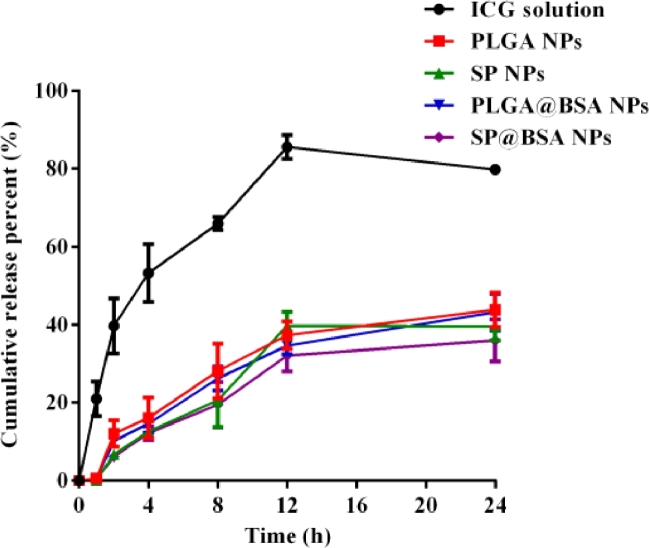
The release leakage profiles of ICG solution, SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs against pH 7.4 phosphate buffer were presented in Fig. 4. Compared with the rapid release of free ICG solution (over 80%) across the dialysis over 24 h, the ICG release rates of the four NPs formulations were significantly slower due to PEI/ICG electrostatic interaction within the hydrophobic core of NPs, the shelled barrier of PLGA vehicles and the further protection of PEG or albumin corona of NPs [23]. Meanwhile, the release profiles were best fitted to first order model (Table S1), suggesting that the release mechanism of PLGA NPs was fickian diffusion, whereas those of SP NPs, SP@BSA NPs and PLGA@BSA NPs might involve the combination of fickian diffusion and matrix erosion according to Ritger-Peppas model.
Fig. 4.
In vitro release Profiles of ICG, PLGA NPs, SP NPs, PLGA@BSA NPs and SP@BSA NPs against pH 7.4 PBS.
3.5. Photothermal effect and stability
The photothermal convention efficiency of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs were investigated by monitoring the temperature variation over time under 808 nm laser at an energy density of 1.5 W/cm2 for 3 min. The temperature of the ICG solution, PLGA NPs, SP NPs, PLGA@BSA NPs and SP@BSA NPs approximately increased to 45.3, 46.5, 43.1, 45.5 and 44.5 °C, separately. In contrast, the control PBS only sustained at 27.7 °C after irradiation for 30 s (Fig. S8). Therefore, all the formulations could be effective in ablating tumor cells due to the excellent photothermal conversion capability [38].
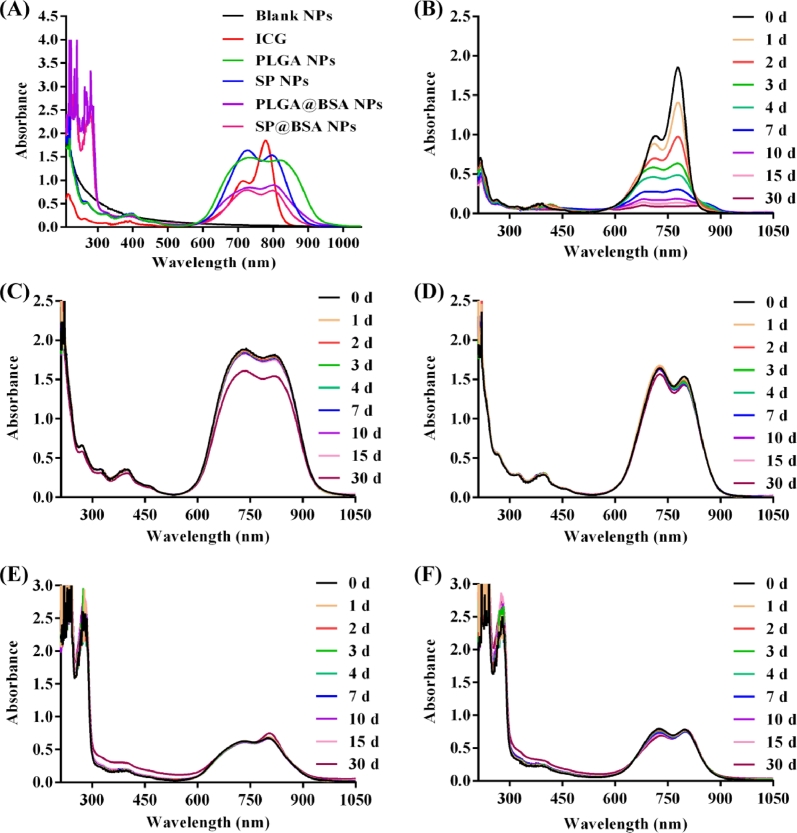
The photostability of SP NPs, PLGA NPs, SP@BSA NPs and PLGA@BSA NPs was probed by recording the absorption spectra with ICG in aqueous phase as control. As shown in Fig. 5, the absorbance capacity of free ICG was lowered by 84% after 7 d due to its degradation and self-aggregation, while that of ICG in SP NPs, SP@BSA NPs and PLGA@BSA NPs was only truncated less than 5% even after 30 d storage, indicating that PEI/ICG complex within the hydrophobic core could efficiently salvage ICG from degradation and self-aggregation [23], [25]. Meawhile, SP NPs and SP@BSA NPs sustained constant or slightly variable size and zeta poteintial over 30 d (Fig. S9). Notwithstanding, the absorption capacity of PLGA NPs reduced by 25% after 30 d and its swollen size and broadened size distribution were also observed after 4 d in comparison with PLGA@BSA NPs, which might result from the lack of protection albumin corona and protection of PEG hydration layer around the particles. All of SP NPs, SP@BSA NPs and PLGA@BSA NPs demonstrated excellent photostability and colloidal stability due to the combination of electrostatic interaction, the shelled barrier of PLGA vehicles and the further protection of PEG or albumin corona of NPs.
Fig. 5.
UV–vis absorption spectra of NPs (A), and UV–vis absorption stability of ICG (B), PLGA NPs (C), SP NPs (D), PLGA@BSA NPs (E) and SP@BSA NPs (F).
3.6. Cell uptake and spheroid penetration
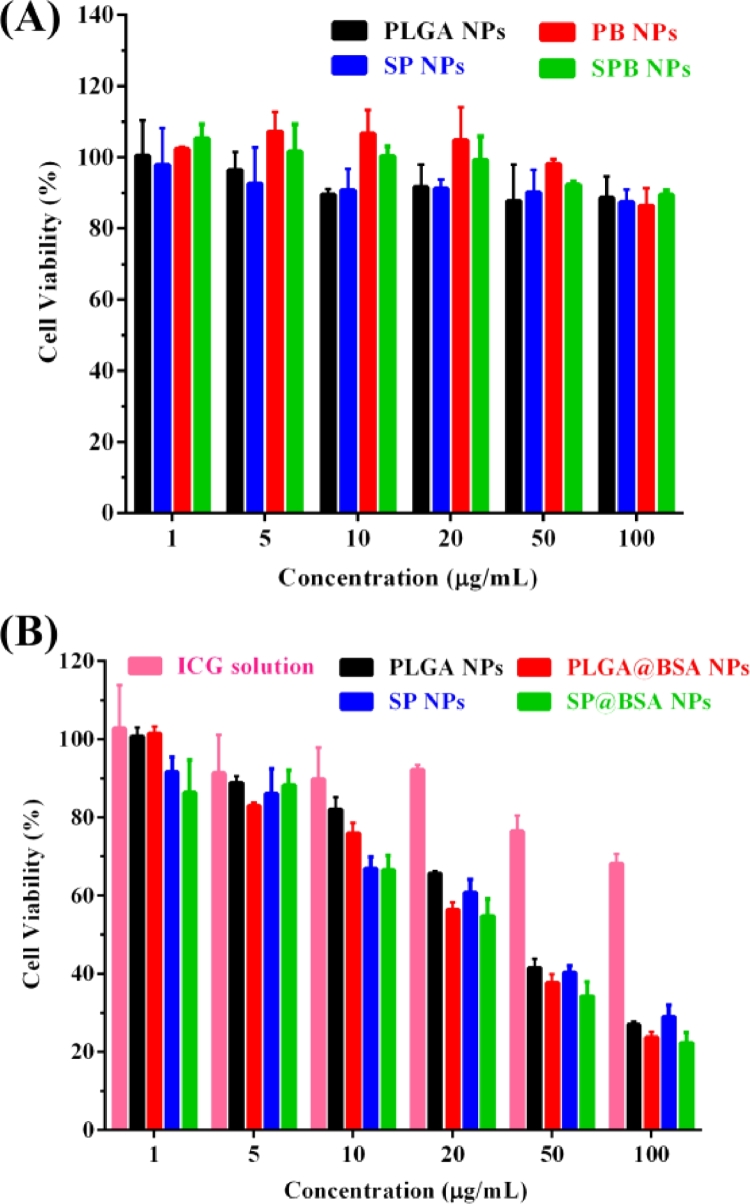
The cytotoxicity of blank NPs formulations (without ICG) on 4T1 cell was measured under irradition (808 nm, 1.5 W/cm2, 3 min) to assess carrier compatibility. As illustrated in Fig. 6A, four NPs vehicles exhibited little cytotoxicity at all dosages tested. Secreted protein acidic and rich in cysteine (SPARC), a high affinity albumin-binding protein, is reported to facilitate albumin or albumin-bound NP accumulation in the tumor cells, and its expression level is higher on tumor cell line [20], [21], [39], [40]. In our study, the cytotoxicity of PLGA NPs was significantly reduced in comparision with PLGA@BSA NPs (Fig. 6B and Table S2), which might be due to SPARC-mediated internalization process. Whereas, the cytotoxicity of SP NPs and SP@BSA NPs was nearly identified, indicating that SP NPs could covalently link to the cysteine-34 residue of serum albumin and form albumin corona on NPs surfaces, which was consistence with our designed maleimide-based prodrugs such as maleimide-5-Fluorouracil (EMC-5-FU) and the above results of western bloting [41]. We then quantificationally evaluated the cellular uptake of NPs to 4T1 cells. In accordance with the above in vitro cytotoxicity results, Fig. 7 revealed that SP NPs, SP@BSA NPs and PLGA@BSA NPs had a similar endocytosis efficiency, which was higher than that of albumin-nonselective PLGA NPs. Moreover, the uptake mechanism of NPs into 4T1 cells was also investigated, where sodium azide and 4 °C were exploited to evaluate the influence of energy on the cellular uptake. Chlorpromazine, colchicine, indomethacine, quercetin and ammonium chloride were utilized to block the clathrin, macropinocytosis, caveolin, clathrin/caveolin-independent and lysosome-mediated internalization pathways, respectively. As shown in Fig. S10, the cellular endocytosis of NPs was associated with multiple endocytotic pathways, including clathrin-mediated, caveolae-mediated, and caveolae/clathrin-independent endocytosis and macropinocytosis [42]. Then, spheroid penetration of NPs was also performed. The results suggested SP NPs, SP@BSA NPs and PLGA@BSA NPs had a similar penetration efficiency, whose fluorescence intensity were stronger, deeper than albumin-nonselective PLGA NPs in vitro 4T1 tumor spheroids (Fig. 8) [43], [44].
Fig. 6.
Cytotoxicity of blank vehicles (A) and NPs (B) on 4T1 under an 808 nm NIR laser at 1.5 W/cm2 for 3 min and further incubation for 48 h.
Fig. 7.
Cellular internalization of the NPs on 4T1 cells for 2 h.
Fig. 8.
Nanoparticle penetration on 4T1 cell spheroids post-incubation with PLGA NPs, SP NPs, SP@BSA NPs and PLGA@BSA NPs for 4 h.
3.7. Pharmacokinetics
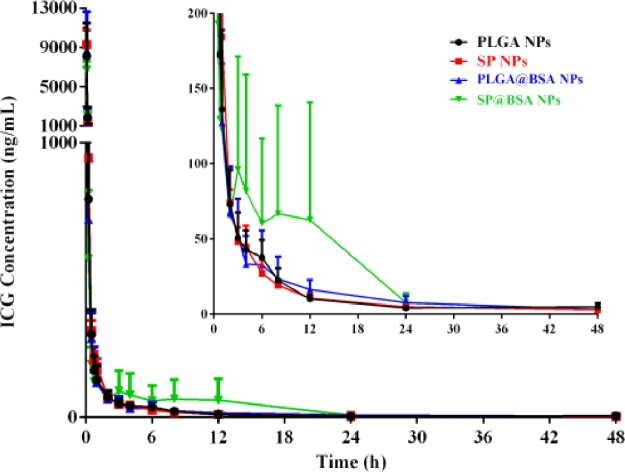
Four groups of rats were intravenously injected with PLGA NPs, SP NPs, SP@BSA NPs and PLGA@BSA NPs formulations at the dose equivalent to 4 mg/kg ICG. The plasma concentrations of ICG were measured by the validated UPLC-MS/MS method. As shown in Table 2 and Fig. 9, the AUC was slightly changed in the investigation. However, the circulation capability of NPs was different for albumin-nonselective PLGA NPs and albumin-selective SP NPs. ICG, a FDA approval diagnostic dye, is extensively cleared from the body in a short time, and the corresponding half-life is about 0.12 h. Cai et al reported that intermolecular disulfide conjugated human serum albumin (HSA)-ICG nanoparticles (HSA-ICG NPs) could slightly prolong ICG blood circulation time to some degree with t1/2 = 2.86 h, resulting from the preparation process might lead to the spatial structural changes of HSA and elite RES endocytosis in vivo [45]. In our study, the t1/2 of PLGA@BSA NPs (11.21 ± 1.70 h) was 1.94-fold prolonged compared with PLGA NPs (5.80 ± 0.92 h) and HSA-ICG NPs (t1/2 = 2.86 h), implying that preformed albumin corona could restrict the plasma proteins adsorption and lessen the complement activation, and finally extend the blood circulation time. Moreover, the t1/2 of SP@BSA NPs (11.46 ± 3.45 h) was little viable in comparison with SP NPs (11.99 ± 2.75 h), indicating that SP NPs could actively recruit endogenous albumin corona on NPs surfaces to inhibit opsonic adsorption and complement activation, which was in coincidence with the in vitro western blotting performance.
Table 2.
In vivo pharmacokinetic parameters of NPs in rats (mean ± SD, n = 5).
| Preparations | Dosage (mg/kg) | t1/2 (h) | AUC (ng/ml*h) |
|---|---|---|---|
| PLGA NPs | 4 | 5.80 ± 0.92 | 2720.28 ± 846.75 |
| PLGA@BSA NPs | 4 | 11.21 ± 1.70 | 2906.04 ± 1255.90 |
| SP NPs | 4 | 11.99 ± 2.75 | 2787.52 ± 358.06 |
| SP@BSA NPs | 4 | 11.46 ± 3.45 | 3131.34 ± 1183.57 |
Fig. 9.
Mean plasma concentration − time curves of NPs in rats after intravenous injection at a dose equivalent to 4 mg/kg ICG (mean + SD, n = 5).
4. Conclusions
Albumin-nonselective PLGA NPs and albumin-selective SP NPs were prepared by emulsion-solvent evaporation method and the resultant NPs were in spherical shape with an average diameter of 180 nm. The ICG loading efficiency of PLGA NPs and SP NPs were 78% and 85%, respectively, due to electrostatic interaction between ICG and PEI. In the presence of albumin corona, PLGA@BSA NPs exhibited higher stability, cytotoxicity, cell internalization and spheroid penetration performances in vitro, and longer blood circulation time in vivo than those of albumin-nonselective PLGA NPs. Excitingly, albumin-selective SP NPs is capable of achieving a comparable in vitro and in vivo performances with both SP@BSA NPs and PLGA@BSA NPs. Our results demonstrate that SA decorated NPs pave a versatile avenue for optimizing nanoparticle delivery without additional albumin coating.
Conflict of interest
No conflict exits in the manuscript, and the article is approved by all authors.
Acknowledgments
This work was financially supported by the National Basic Research Program of China (973 Program, No. 2015CB932100), National Natural Science Foundation of China (No. 81703451, 81573371, 81473164).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2018.07.002.
Contributor Information
Qiming Kan, Email: kanqiming1@163.com.
Jin Sun, Email: sunjin@syphu.edu.cn.
Appendix. Supplementary materials
References
- 1.Peng Q, Zhang S, Yang Q. Preformed albumin corona, a protective coating for nanoparticles based drug delivery system. Biomaterials. 2013;34(33):8521–8530. doi: 10.1016/j.biomaterials.2013.07.102. [DOI] [PubMed] [Google Scholar]
- 2.Tenzer S, Docter D, Kuharev J. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nanotechnol. 2013;8(10):772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 3.Yan Y, Gause KT, Kamphuis MM. Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines. ACS Nano. 2013;7(12):10960–10970. doi: 10.1021/nn404481f. [DOI] [PubMed] [Google Scholar]
- 4.Albanese A, Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. ACS Nano. 2014;8(6):5515–5526. doi: 10.1021/nn4061012. [DOI] [PubMed] [Google Scholar]
- 5.Saha K, Rahimi M, Yazdani M. Regulation of macrophage recognition through the interplay of nanoparticle surface functionality and protein corona. ACS Nano. 2016;10(4):4421–4430. doi: 10.1021/acsnano.6b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu SM, Gonzalez-Moragas L, Milla M. Bio-identity and fate of albumin-coated SPIONs evaluated in cells and by the C. elegans model. Acta Biomater. 2016;43:348–357. doi: 10.1016/j.actbio.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Kelly PM, Aberg C, Polo E. Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat Nanotechnol. 2015;10(5):472–479. doi: 10.1038/nnano.2015.47. [DOI] [PubMed] [Google Scholar]
- 8.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41(7):2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 9.Peng Q, Wei XQ, Yang Q. Enhanced biostability of nanoparticle-based drug delivery systems by albumin corona. Nanomedicine. 2015;10(2):205–214. doi: 10.2217/nnm.14.86. (Lond) [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Chen N, Dong C. Facile construction of near infrared fluorescence nanoprobe with amphiphilic protein-polymer bioconjugate for targeted cell imaging. ACS Appl Mater Interfaces. 2015;7(34):18997–19005. doi: 10.1021/acsami.5b05406. [DOI] [PubMed] [Google Scholar]
- 11.Treuel L, Brandholt S, Maffre P, Wiegele S, Shang L, Nienhaus GU. Impact of protein modification on the protein corona on nanoparticles and nanoparticle-cell interactions. ACS Nano. 2014;8(1):503–513. doi: 10.1021/nn405019v. [DOI] [PubMed] [Google Scholar]
- 12.Caracciolo G, Palchetti S, Colapicchioni V, Digiacomo L, Pozzi D, Capriotti AL. Stealth effect of biomolecular corona on nanoparticle uptake by immune cells. Langmuir. 2015;31(39):10764–10773. doi: 10.1021/acs.langmuir.5b02158. [DOI] [PubMed] [Google Scholar]
- 13.Yallapu MM, Chauhan N, Othman SF. Implications of protein corona on physico-chemical and biological properties of magnetic nanoparticles. Biomaterials. 2015;46:1–12. doi: 10.1016/j.biomaterials.2014.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polomska A, Gauthier MA, Leroux JC. In vitro and in vivo evaluation of PEGylated layer-by-layer polyelectrolyte-coated paclitaxel nanocrystals. Small. 2017;13(2) doi: 10.1002/smll.201602066. [DOI] [PubMed] [Google Scholar]
- 15.Park J, Sun B, Yeo Y. Albumin-coated nanocrystals for carrier-free delivery of paclitaxel. J Control Release. 2016;263:90–101. doi: 10.1016/j.jconrel.2016.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei XQ, Hao LY, Shao XR. Insight into the Interaction of graphene oxide with serum proteins and the impact of the degree of reduction and concentration. ACS Appl Mater Interfaces. 2015;7(24):13367–13374. doi: 10.1021/acsami.5b01874. [DOI] [PubMed] [Google Scholar]
- 17.Chong Y, Ge C, Yang Z. Reduced cytotoxicity of graphene nanosheets mediated by blood-protein coating. ACS Nano. 2015;9(6):5713–5724. doi: 10.1021/nn5066606. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang J, Zhang F, Yu S. Probing the binding affinity of plasma proteins adsorbed on Au nanoparticles. Nanoscale. 2017;9(14):4787–4792. doi: 10.1039/c7nr01523b. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Chen Q, Liang C. Albumin-templated biomineralizing growth of composite nanoparticles as smart nano-theranostics for enhanced radiotherapy of tumors. Nanoscale. 2017;9(39):14826–14835. doi: 10.1039/c7nr05316a. [DOI] [PubMed] [Google Scholar]
- 20.Bern M, Sand KM, Nilsen J, Sandlie I, Andersen JT. The role of albumin receptors in regulation of albumin homeostasis: Implications for drug delivery. J Control Release. 2015;211:144–162. doi: 10.1016/j.jconrel.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Chen X. Simple bioconjugate chemistry serves great clinical advances: albumin as a versatile platform for diagnosis and precision therapy. Chem Soc Rev. 2016;45(5):1432–1456. doi: 10.1039/c5cs00158g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Lu H, Khine YY, Dag A, Stenzel MH. Polyion complex micelle based on albumin-polymer conjugates: multifunctional oligonucleotide transfection vectors for anticancer chemotherapeutics. Biomacromolecules. 2014;15(11):4195–4205. doi: 10.1021/bm501205x. [DOI] [PubMed] [Google Scholar]
- 23.Jian WH, Yu TW, Chen CJ, Huang WC, Chiu HC, Chiang WH. Indocyanine green-encapsulated hybrid polymeric nanomicelles for photothermal cancer therapy. Langmuir. 2015;31(22):6202–6210. doi: 10.1021/acs.langmuir.5b00963. [DOI] [PubMed] [Google Scholar]
- 24.Sheng Z, Hu D, Zheng M. Smart human serum albumin-indocyanine green nanoparticles generated by programmed assembly for dual-modal imaging-guided cancer synergistic phototherapy. ACS Nano. 2014;8(12):12310–12322. doi: 10.1021/nn5062386. [DOI] [PubMed] [Google Scholar]
- 25.Hao Y, Wang L, Zhao Y. Targeted imaging and chemo-phototherapy of brain cancer by a multifunctional drug delivery system. Macromol Biosci. 2015;15(11):1571–1585. doi: 10.1002/mabi.201500091. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Wen T, Zhao R. Localized electric field of plasmonic nanoplatform enhanced photodynamic tumor therapy. ACS Nano. 2014;8(11):11529–11542. doi: 10.1021/nn5047647. [DOI] [PubMed] [Google Scholar]
- 27.Goodwin D, Simerska P, Chang CH. Active immunisation of mice with GnRH lipopeptide vaccine candidates: importance of T helper or multi-dimer GnRH epitope. Bioorgan Med Chem. 2014;22(17):4848–4854. doi: 10.1016/j.bmc.2014.06.052. [DOI] [PubMed] [Google Scholar]
- 28.Shi K, Liu Y, Ke L, Fang Y, Yang R, Cui F. Epsilon-poly-L-lysine guided improving pulmonary delivery of supramolecular self-assembled insulin nanospheres. Int J Biol Macromol. 2015;72:1441–1450. doi: 10.1016/j.ijbiomac.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 29.Mortimer GM, Butcher NJ, Musumeci AW, Deng ZJ, Martin DJ, Minchin RF. Cryptic epitopes of albumin determine mononuclear phagocyte system clearance of nanomaterials. ACS Nano. 2014;8(4):3357–3366. doi: 10.1021/nn405830g. [DOI] [PubMed] [Google Scholar]
- 30.Luo C, Sun J, Sun B. Facile fabrication of tumor redox-sensitive nanoassemblies of small-molecule oleate prodrug as potent chemotherapeutic nanomedicine. Small. 2014;12(46):6353–6362. doi: 10.1002/smll.201601597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleischer CC, Payne CK. Nanoparticle-cell interactions: molecular structure of the protein corona and cellular outcomes. Acc Chem Res. 2014;47(8):2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doorley GW, Payne CK. Nanoparticles act as protein carriers during cellular internalization. Chem Commun. 2012;48(24):2961–2963. doi: 10.1039/c2cc16937a. (Camb) [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Agarwal P, Zhao S, Yu J, Lu X, He X. A biomimetic hybrid nanoplatform for encapsulation and precisely controlled delivery of theranostic agents. Nat Commun. 2015;6:10081–10092. doi: 10.1038/ncomms10081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Fang S, Shi S, Deng J, Liu B, Cai L. Hybrid polypeptide micelles loading indocyanine green for tumor imaging and photothermal effect study. Biomacromolecules. 2013;14(9):3027–3033. doi: 10.1021/bm400839b. [DOI] [PubMed] [Google Scholar]
- 35.Zheng M, Zhao P, Luo Z. Robust ICG theranostic nanoparticles for folate targeted cancer imaging and highly effective photothermal therapy. ACS Appl Mater Interfaces. 2014;6(9):6709–6716. doi: 10.1021/am5004393. [DOI] [PubMed] [Google Scholar]
- 36.Ma Y, Sadoqi M, Shao J. Biodistribution of indocyanine green-loaded nanoparticles with surface modifications of PEG and folic acid. Int J Pharm. 2012;436(1–2):25–31. doi: 10.1016/j.ijpharm.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Wang M, Lei R, Zhu SF, Zhao Y. Chiral surface of nanoparticles determines the orientation of adsorbed transferrin and its interaction with receptors. ACS Nano. 2017;11(5):4606–4616. doi: 10.1021/acsnano.7b00200. [DOI] [PubMed] [Google Scholar]
- 38.Sun HP, Su JH, Meng QS. Silibinin and indocyanine green-loaded nanoparticles inhibit the growth and metastasis of mammalian breast cancer cells in vitro. Acta Pharmacol Sin. 2016;37(7):941–949. doi: 10.1038/aps.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aquino G, Sabatino R, Cantile M. Expression analysis of SPARC/osteonectin in oral squamous cell carcinoma patients: from saliva to surgical specimen. Biomed Res Int. 2013;2013 doi: 10.1155/2013/736438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoang B, Ernsting MJ, Roy A, Murakami M, Undzys E, Li SD. Docetaxel-carboxymethylcellulose nanoparticles target cells via a SPARC and albumin dependent mechanism. Biomaterials. 2015;59:66–76. doi: 10.1016/j.biomaterials.2015.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao D, Zhang H, Tao W, Wei W, Sun J. A rapid albumin-binding 5-fluorouracil prodrug with a prolonged circulation time and enhanced antitumor activity. 2017, 5(3): 502–10. [DOI] [PubMed]
- 42.He YJ, Xing L, Cui PF. Transferrin-inspired vehicles based on pH-responsive coordination bond to combat multidrug-resistant breast cancer. Biomaterials. 2017;113:266–278. doi: 10.1016/j.biomaterials.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Lin T, Zhao P, Jiang Y. Blood-brain-barrier-penetrating albumin nanoparticles for biomimetic drug delivery via albumin-binding protein pathways for antiglioma therapy. ACS Nano. 2016;10(11):9999–10012. doi: 10.1021/acsnano.6b04268. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Mao W, Lock LL. The role of micelle size in tumor accumulation, penetration, and treatment. ACS Nano. 2015;9(7):7195–7206. doi: 10.1021/acsnano.5b02017. [DOI] [PubMed] [Google Scholar]
- 45.Sheng Z, Hu D, Zheng M. Smart human serum albumin-indocyanine green nanoparticles generated by programmed assembly for dual-modal imaging-guided cancer synergistic phototherapy. ACS Nano. 2014;8(12):12310–12322. doi: 10.1021/nn5062386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.